 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tailored point-of-care biosensors for liquid biopsy in the field of oncology
Sima
Singh
a,
Pritam Saha
Podder
b,
Matt
Russo
c,
Charles
Henry
 c and
Stefano
Cinti
c and
Stefano
Cinti
 *ad
*ad
aDepartment of Pharmacy, University of Naples Federico II, 80131 Naples, Italy. E-mail: stefano.cinti@unina.it
bDepartment of Pharmacy, Jahangirnagar University, Savar, Dhaka-1342, Bangladesh
cDepartment of Chemistry, Colorado State University, Fort Collins, CO 80523-1872, USA
dBAT Center-Interuniversity Center for Studies on Bioinspired Agro-Environmental Technology, University of Napoli Federico II, 80055 Naples, Italy
First published on 27th October 2022
Abstract
In the field of cancer detection, technologies to analyze tumors using biomarkers circulating in fluids such as blood have developed rapidly based on liquid biopsy. A proactive approach to early cancer detection can lead to more effective treatments with minimal side effects and better long-term patient survival. However, early detection of cancer is hindered by the existing limitations of conventional cancer diagnostic methods. To enable early diagnosis and regular monitoring and improve automation, the development of integrated point-of-care (POC) and biosensors is needed. This is expected to fundamentally change the diagnosis, management, and monitoring of response to treatment of cancer. POC-based techniques will provide a way to avoid complications that occur after invasive tissue biopsy, such as bleeding, infection, and pain. The aim of this study is to provide a comprehensive view of biosensors and their clinical relevance in oncology for the detection of biomarkers with liquid biopsies of proteins, miRNA, ctDNA, exosomes, and cancer cells. The preceding discussion also illustrates the changing landscape of liquid biopsy-based cancer diagnosis through nanomaterials, machine learning, artificial intelligence, wearable devices, and sensors, many of which apply POC design principles. With the advent of sensitive, selective, and timely detection of cancer, we see the field of POC technology for cancer detection and treatment undergoing a positive paradigm shift in the foreseeable future.
1. Introduction
Cancer has been recognized as one of the most significant problems in the world today because of its leading role in the overall mortality rate. According to GLOBOCAN, in 2020 there were about 19.3 million newly diagnosed cases in 185 countries, and 10 million patients died as a result of cancer.1 High cancer mortality rates are most likely due to delays in diagnosis and limited access to effective treatment. In order to minimize cancer mortality, efforts must be made to diagnose and prevent cancer early in order to monitor recurrence of tumors and respond to treatment options.2 In contrast to a reactive, symptom-based approach, this proactive paradigm requires people to perform tests to diagnose and treat cancer earlier, even before symptoms appear.3However, unfortunately, failing to detect a tumor at an earlier stage and tumor heterogeneity have led to a high mortality rate in cancer detected patients. Tumor heterogeneity accounts for differences in growth rate, invasiveness, and treatment sensitivity.4 A tissue biopsy and various imaging modalities such as color ultrasonography, computed tomography (CT), molybdenum targets, magnetic resonance imaging (MRI), and so on, are some of the approaches that can be used to evaluate tumor heterogeneity.5 However, the question arises as to why such things are not done on a regular basis. The first solutions have numerous limitations: for instance, when it comes to diagnosing cancer and determining its subtype, stage, and prognosis, a tissue biopsy is the gold standard. In many cases, especially metastatic disease such as advanced stage cancer, this is difficult to do and only a general overview of the condition is given.6 Multiple or sequential biopsies are impractical because of the difficulty of obtaining tissue samples. Due to the invasive nature of the procedure, tissue biopsy detection of cancer has several limitations. These drawbacks include patient inconvenience, associated clinical risks, sample preparation, sensitivity, precision, potential surgical problems, and financial considerations. However, biopsy of tumor tissue is associated with significant difficulties and may not accurately reflect the genome of the entire tumor mass.7 These limitations in liquid biopsies present a particular challenge when dealing with cancer cells that have developed resistance to therapy. As a result, physician are not able to identify therapeutic biomarkers at an early stage, which altered the treatment effects.8 Therefore, these techniques cannot be used for long-term clinical monitoring of cancer in resistance developed patients.
Because individual biopsies are of limited value, new approaches have been developed to monitor tumor genetics and dynamics. To this end, technological advances in cancer research and biomedical innovations have led to the development of new screening techniques using body fluid samples rather than biopsies. This technique is mostly recognized as liquid biopsy (LB).9,10 Ultrasensitive and selective detection of target molecules circulating in body fluids, such as extracellular vesicles, proteins, nucleic acids, and microRNAs (miRNA), is a strategy that is rapidly gaining acceptance as a method for cancer diagnosis. It is preferable to repeated biopsies because it has greater therapeutic efficacy, fewer side effects, and lower medical costs (Fig. 1a).11 It facilitates early cancer detection, patient selection and evaluation, prognosis, identification of tailored treatment options, and long-term prevention approaches.12,13 LB have led to the hope that spatial and temporal heterogeneity in tumor biology can be better captured by serial blood analyzes than by tissue samples from a primary tumor. This could lead to improvements in patient care and treatment outcomes.14
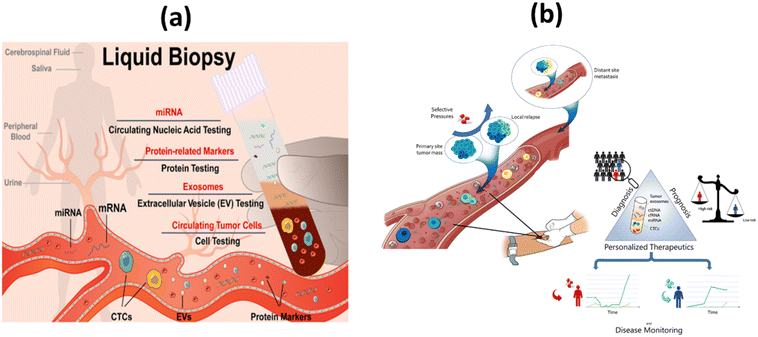 | ||
| Fig. 1 (a) Simplified representation of liquid biopsy;11 (b) the concept and clinical applications of liquid biopsy in tumor management.39 | ||
Recent study has showed that it is also prevalent in other bodily fluids, including urine, saliva, pleural effusion, and cerebrospinal fluid, as well as in the blood of cancer patients.15 Despite the technological breakthrough, the adoption of LB into clinical practice has been slow. This is mainly because the procedure to isolate and detect circulating tumor cells (also called CTCs) involves many technical complications.16 For example, certain types of CTCs can be found in human fluids, even in extremely small amounts. Several complex matrices make up the body fluids, especially the peripheral blood, which contains billions of blood cells and different proteins.17,18 Both require techniques that facilitate remarkably efficient enhancement and robust signal amplification. In order to detect fragile CTCs, a sensitive isolation and release method is required. Furthermore, the identification and analysis of CTCs with varied degrees of heterogeneity must be thorough, limiting their practical applicability.19,20 In order to overcome these problems, CTCs must be correctly, sensitively and efficiently isolated.21 To address these challenges, electrochemical sensing technology has increasingly emerged as a novel approach for CTC detection in recent years due to efficient analysis. In this regard, biosensor platforms offer attractive alternatives to existing systems. This is made possible by advanced, sensitive, fast and cost-effective biosensor technology.22,23 Over the past decade, novel architectures, configurations, and technologies have driven the development and improvement of noninvasive point-of-care (POC) technology. The combination of liquid biopsies with newly developed miniaturized platforms with engineered nano/micro materials and electrochemical/optical sensing techniques has led to the maturation of POC platforms that are both simple and cost effective.24,25
To improve the overall survival of cancer patients, we need an appropriate liquid biopsy that allows more accurate cancer diagnosis and predictive analysis. In recent decades, several papers have been published on different methods for detecting LB, such as CTCs,26,27 circulating tumor-specific nucleic acid (ctDNA and ctRNA),4,28,29 extracellular vesicles (EVs)/exosomes30,31 and proteins as cancer biomarkers using biosensor technology (Fig. 1b). All these analysts play a particularly important role in early diagnosis of tumors. Each biomarkers have strengths and limitations are listed in Table 1.32,33
| Biomarker | Strengths | Limitations |
|---|---|---|
| CTCs | • It is noninvasive in nature | • There are not enough targeted cells |
| • Suggest both diagnostic and predictive messages | • Lack of particular biomarkers | |
| • Cell morphology and molecular analysis | • Heterogeneity | |
| • Drug sensitivity and resistance cell research using in vitro culture | • Influence cell survival, phenotypic determination, and further molecular study | |
| • High cell viability and integrity | • Lack of standardization (cut-off value, detection time, etc.) | |
| • Non-restrictive to cell surface markers | • Insufficient large-scale trials | |
| • Low costs while maintaining a high output | • Notable alteration in cell size results in the omission of tiny CTTs | |
| ctDNA | • Technologically mature as compared with CTCs | • DNA fragments derived from necrotic or apoptotic cells cannot be used to represent live tumor cells |
| • Short half-life time | • Provide only genomic information | |
| • More responsive to the presence of tumors | • Low levels of gene mutation, resulting in inadequate sensitivity | |
| • Complete tumor molecular information | ||
| miRNAs | • Closely connected to cellular metabolic processes under healthy and pathological situations | • Lack of stability |
| EVs | • Excellent stability | • Challenging isolation |
| • Numerous contents | • Unknown biological qualities | |
| Proteins | • Isolation is simple | • Constrained stability |
| • Extremely sensitive | • Does not target cancer in particular | |
| • Efficient standardization |
These technologies allow researchers to control the amount of liquid and the speed of movement as well as the shape, temperature, and mechanical, electrical, and chemical effects of the substrate. These advances have led to the development of small lab-on-chip systems (LOCs). They have exceptional throughput and accuracy for the direct detection of tumor cells in body fluid samples.34
Electrochemical detection of numerous liquid biopsies for cancer has been the topic of discussion in a number of very good reviews; however, the majority of these studies mostly focused on single LB such as proteins, miRNA, ctDNA, exosomes, and cancer cells.30,35–38 A comprehensive review wrapping up the current magnificent state of the art in point-of-care (POC) and biosensor technology for all LB markers is currently awaiting publication in a single report. Consequently, the aim of this article is to provide a comprehensive summary of literature published in recent years on the development of LB biosensors and technical approaches to their use in healthcare and their management. The underlying technical obstacles and various ways to overcome them are discussed in detail. In addition, this paper reviews the performance parameters of a number of recently developed materials and technologies for LB biosensors. Identifying the shortcomings and constraints of current platforms would be beneficial to researchers. With a better knowledge of these platforms, researchers can find potential avenues for cancer monitoring biosensors for point-of-care diagnostics.
2. Emerging novel POC based approach for detection of liquid biopsy
Early detection, cancer prognosis, a treatment plan and a key to discovering the secrets of cancer growth – all this is possible with a simple blood test. The introduction of the methods of LB has revolutionized research and opened the door to countless scientific and therapeutic applications. Early and accurate diagnosis of a specific cancer is crucial to its effective treatment, where a treatment decision must be made quickly on the basis of detection. In developing and poor countries, facilities are sparsely distributed across the population. In situations with limited resources, there is a severe demand for POC automation technologies that facilitate individualized management and investigation at the point of care. In situations where few or no laboratory facilities are available, point-of-care diagnostics are used to rapidly initiate pharmacological or prognostic treatment. POC diagnostics have led to professional health care experts coming close to the patient or being performed by laypersons at home in a short period of time.40Fig. 2a and b shows the advantages of POCT in liquid biopsy-based detection of cancers.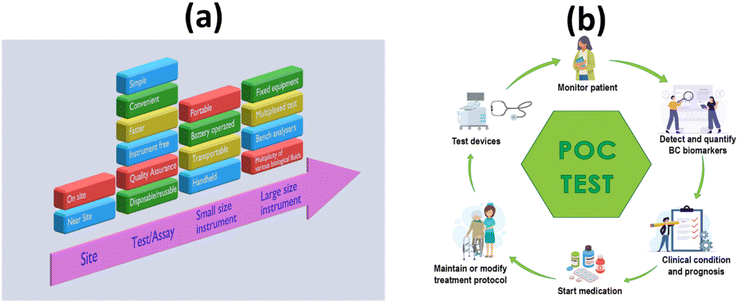 | ||
| Fig. 2 Schematic representation of the POC-based approach to the diagnostic landscape LB: (a) hierarchical properties of an appropriate POC test/device; (b) advantages of a POC test/device. | ||
Biosensors are fast, reliable, and accurate diagnostic tools used worldwide in medical and biological fields. An ideal biosensor must meet specific requirements, such as responding only to the analyte, having high sensitivity, and being able to detect small amounts of the analyte in a short period of time.41 As digitization continues to advance worldwide, combining biosensors with wireless technologies such as Bluetooth, Wi-Fi, and GPS can help provide POC-based evidence faster and at lower cost.42 In current years, widespread efforts have been made in numerous ways to develop an effective POC device for the analysis of early-stage cancer. An impressive example of the first FDA-approved POC device is the Cancer Mutation Test v2 (Roche Diagnostics Inc.). The test is based on detecting specific cancer mutations in the cfDNA. As a complementary diagnostic test, EGFR gene mutations are detected to identify patients with metastatic NSCLC eligible for Erlotinib treatment.10
In the field of traditional POC detection at the site, biosensors and nanobiosensors have been studied. Table 2 summarizes the primary sensing modalities that have been explored to date in the development of LB biosensors, the best known of which are optical and electrochemical approaches. In some contexts, overall performance has been improved through the use of additional techniques, such as the use of chips and microfluidic devices. The development of biosensors equipped with next-generation POC or biosensors is probably one of the most promising solutions to the existing difficulties and serves as the ultimate test for tailored cancer therapies at the point-of-care. Furthermore, it will be paving the way for a new home health monitoring system and providing important information for personalized treatment.
| S. no. | Target | Strategy | Response time (min) | Detection limit | Linear range | Detection medium | Ref. |
|---|---|---|---|---|---|---|---|
| Electro (bio)sensors | |||||||
| 1 | Exosomes | Immuno-magneto beads | 60 | 3 × 104 exosomes | NA | Plasma | 43 |
| 2 | CTCs | 2D molybdenum disulfide (MoS2)/FA | 30 | 0.43 cell per mL | 1 to 105 cell per mL | HeLa cells, serum from 4 patients with liver cancer, 4 persons with cervical cancer, and 8 healthy individuals | 44 |
| 3 | miRNA-21 | Gold nanoparticles (AuNPs) and the magnetic nanoparticle (Fe3O4) | 25 | 0.14 fM | 0.2 fM–1 nM | HeLa cells/MCF-7 cells | 45 |
| 4 | CTCs | 3D PdPtCuRu | — | 10 cells per mL | 10 to 2 × 104 cells per mL | A549 | 46 |
| 5 | ctDNA | CRISPR/Cas12a system and MB/Fe3O4@COF/PdAu nanocomposites | 3.3 aM | 10 aM–100 pM | The blood serum of 25 people with NSCLC | 47 | |
| Optical | |||||||
| 6 | CTCs | Antibody | 30 | 1 cell per mL | 2 to 400 cells per mL | Samples of human blood from healthy and malignant individuals | 48 |
| 7 | CTCs | Multi-functional peptide | 5 | 5 cells per mL | 5 to 100 fg mL−1 | Synthetic CTC samples | 49 |
| 8 | Exosomes | Exodisc | 60 | 1.47 × 1011 pM | — | Urine | 50 |
| 9 | miRNA-21 | MoS2 nanosheets & duplex-specific nuclease (DSN) | 150 | 426 pM | 0.5 nM to 50 nM | Human plasma | 51 |
| 10 | Exosomal protein | Utilizing aptamers, aggregation-induced emission luminogens, also known as AIEgens, and graphene oxide (GO) | 30 | 0.57 pM | 4.07 × 105 to 1.83 × 107 particles per μL | Serum | 52 |
| Microfluidics/paper based technology | |||||||
| 11 | Exosomes | Microfluidic device made of polydimethylsiloxane (PDMS) and functionalized with antibodies against CD63 | 50 | 0.5 pM | 10–15 ng | Serum | 53 |
| 12 | Exosomes | Immuno-magnetobeads | 40 | 750 exosomes per μL | — | Plasma | 54 |
| 13 | Exosomes | Nano structure immune affinity capture | 40–200 | 10 exosomes per μL | — | Plasma | 55 |
| 14 | miRNA | TiO2/CeO2 nanotube array | — | 0.6 fM | 2 fM–170 pM | Serum samples isolated from whole blood from five healthy individuals (no. 1–5) and three prostate cancer patients (no. 6–8) | 56 |
| 15 | Prostate specific antigen (PSA) protein | Immuno-magnetic detection | 180 | 34 pM | Blood | 57 | |
2.1. Proteins as biological recognition elements for biosensors in cancer
Tumor markers are usually proteins or other substances produced by cancer cells in greater amounts than by normal cells. Proteins are present in varying concentrations in samples of very different origin.58 Ideally, protein biomarkers should be detected in a minimally invasive liquid biopsy, such as a simple blood sample. Protein detection refers to the identification of various target diseases. Depending on the biosensing application, it may be useful to detect only certain proteins or a broad spectrum of proteins in a medium. For these different purposes, the key element is the biocomponent, which acts as a detection tool and determines the selectivity level of the biosensor.59 Detection of protein tumor markers in the blood is considered the gold standard compared to other methods, as it is used in most hospitals to evaluate disease progression. The fact that the expression level of biomarker proteins in biological fluids directly reflects both the physiological state of the body and the progression of disease has led to their extensive application in clinical and research settings.Human epidermal growth factor receptor 2 (HER2), also known as Neu, is a type I tyrosine kinase receptor. It is encoded by the oncogene known as ERBB2. HER2 is an oncogene that promotes proliferation, migration, and invasion in breast cancer and shares considerable homology with HER1/HER3 and HER4.60 HER2-positive tumors tend to grow more aggressively and react differently to treatment than HER2-negative tumors.61 The increasing number of functional studies provides strong evidence that these HER2-based biomarkers can be identified for early cancer detection and even used to predict clinical progression. For example, Guerrero-Esteban et al. developed a disposable electrochemical immunosensor (ECL) to detect HER2. To this end, nitrogen-rich carbon nanodots, which have two functions, have been synthesized. In Fig. 3a, the surface functional groups of covalently immobilized HER2 antibodies and co-reactants are used as carriers for the bio-recognition layer. The HER2 immunosensor shows a broad linear response with a LOD of 20.4 pg mL−1, strong selectivity, repeatability, and stability.62 Using cellulase-related sandwich assays with magnetic beads (MBs), Malecka et al. reported HER-2/neu protein in human serum. They performed the assay on modified MBs using either an antibody or an aptamer that binds HER-2/neu in a precise manner. The cellulase label degrades the nitrocellulose coating when sandwiches built on MBs are applied to a graphite electrode designed to be inexpensive and insulating. This significantly changes the electrical properties of the modified electrode. The newly developed electrochemical label-free sensor has a low detection limit (LOD), which is 1 fM in less than three hours.63
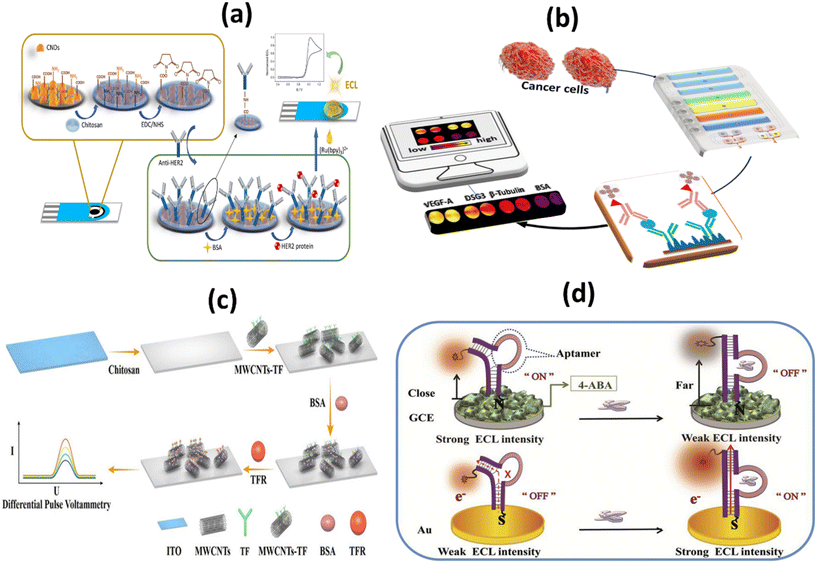 | ||
| Fig. 3 Schematic representation of: (a) HER2/BSA/anti-HER2/chit/CNDs/SPCE based immunosensor for HER2 detection;62 (b) 3D printed microfluid arrays with cell lysis on line to detect biomarkers for metastatic cancer;65 (c) ITO-Chi-MWCNTs-TF-BSA biosensor and electrochemical detection of TFR;67 (d) signal-ON and signal OFF chemiluminescence deoxyribose biosensor for the detection of NAP2 protein through DNA charge transfer mechanism.69 | ||
Desmoglein-3 (DSG-3) is overexpressed in a significant number of oral squamous cell carcinomas (OSCC). DSG-3 is a potential marker of metastasis in the lymph node for oral cancer and cervical squamous cell carcinoma (HNSCC).64 Sharafeldin and colleagues have developed the first automated 3D-printed microfluidic immunoarray. This immunoarray lyses cells with a 50 kHz cell resolver and measures the amount of released biomarker proteins associated with cells. This technique is the first microfluidic device with a detection limit below fg, on-chip lysis, and chemiluminescence detection for single cell protein quantification, it is appropriate for cancer metastasis recognition (Fig. 3b). The system achieves an LOD of 0.10 fg ml−1 of cellular resident proteins in a single cell. This was clearly supported by the increased signal amplification of the streptavidin poly-HRP labels and the ultrabright femto-luminol labels.65
Since transferrin receptors (CD71 or TFRC) are well expressed in many cancers, CD71 has become an attractive target for cancer research.66 Liu et al. developed an electronic biosensor without labels (ITO-Chi-MWCNTs-TF-BSA) with a high-performance TFR detection based on the ligand–protein interaction, as shown in Fig. 3c. The developed sensor has a LOD of 0.082 ng mL−1 and a large linear range of 1–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng mL−1. The results show linear range, sensitivity, specificity, reproducibility, and stability properties desirable for real-world applications.67
000 ng mL−1. The results show linear range, sensitivity, specificity, reproducibility, and stability properties desirable for real-world applications.67
A novel blood biomarker called neutrophil activating peptide-2 (NAP-2) has been developed to detect lung cancer in its early stages.68 To detect NAP-2, a simple and sensitive two signal-OFF/ON ECL deoxyribose sensor has been developed by Chen and colleagues (Fig. 3d). The LOD for signal-OFF ECL deoxyribose sensors was 0.008 pM and for signal-ON ECL deoxyribose sensors was 0.001 pM. Detection with this device takes only 20 minutes and ensures rapid biomarker detection. According to the dominances of the deoxyribose sensor and the ECL technique, this novel device is clearly superior to analyte-triggered large-scale structural transformations. For biomarker protein detection, deoxyribose sensors can be widely used for population-based screening activity.69
2.2. miRNAs as a target for biosensor
Micro-ribonucleic acids, or miRNAs, are known to play a key role in carcinogenesis by controlling a variety of physiological processes that occur during the cell cycle. They are particularly abundant in cancer cells. As a result, microRNAs are currently believed to play a role in both cancer genesis and prognosis.70 A schematic representation of biosensors for miRNA-based cancer detection is shown in Fig. 4a and b.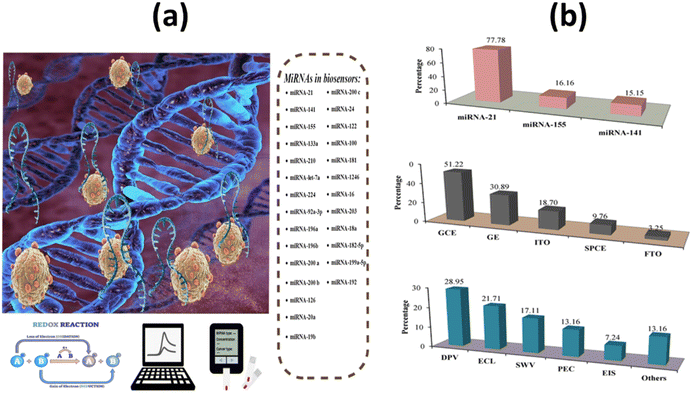 | ||
| Fig. 4 (a) Schematic design of electrochemical biosensors for miRNAs in cancer; (b) analysis and classification of the main characteristics of electrochemical biosensors for miRNA detection, including the most popular miRNAs, signal transducers, and electrochemical techniques.71 | ||
Most microRNAs are located in highly mutation-prone regions of the genome. As a direct consequence, the abnormal function of these biomolecules plays a crucial role in the progression of different cancer stages. Gene expression in cells of the immune system is mainly controlled by microRNAs (miRNAs) miRNA-21, miRNA-155, and miRNA-141. Some miRNAs can act as tumor suppressors or tumor genes, depending on their increase or decrease. miRNA levels vary in different stages of cancer, so its measurement can be a useful diagnostic tool.71 It was also discovered that miRNA-21 and miRNA-155 have a role in the pathogenic process of metastasis. However, miRNA-141 inhibits cancer cell migration, invasion, and division and typically increases the amount of this biomarker, resulting in inhibition of cancer cells.72 Researchers Li et al. used a biosensor based on a high-purity semiconducting carbon nanotube (CNT) together with polymer sorting and a field-effect transistor (FET) to find exosomal miRNA21 in breast cancer. A thiolated oligonucleotide probe was immobilized on the surface of AuNPs in the detection region. miRNA21 was subsequently detected by monitoring the current change before and after hybridization between the immobilized DNA probe and target miRNA. The method has high sensitivity (LOD: 0.87 aM) and high specificity. As shown in Fig. 5a, by distinguishing between the level of miRNA expression in cancer patients and healthy individuals, the miR-FET biosensor enables early cancer diagnosis.73 To detect miRNA-21 in the blood of breast cancer patients, Zouari and his colleagues developed an enzyme-free biosensor technology based on electrochemistry. The proposed method consists of a sandwich hybridization assay and streptavidin-modified gold nanoparticles with ferrocene coverslips to bind the DNA detection probe (Fig. 5b). The linear range was 10 fM–2 pM, while the LOD was 5 fM. The developed biosensor offers a successful combination of operational stability and selectivity, and the enzyme-free approach is less costly compared with conventional enzyme-based assays. In the AuNPs/rGO complex, the combination of the conductive and electrochemical properties of rGO and gold nanoparticles (AuNPs) improves the electron transfer rate and the amplification of the voltammetric signal.74
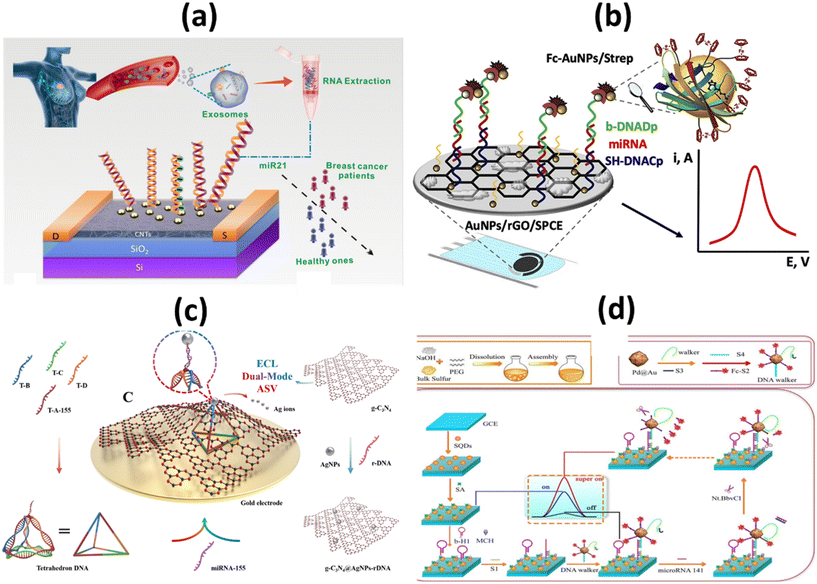 | ||
| Fig. 5 Schematic representation of (a) ultrasensitive detection of exosomal miR21 using the DNA-functionalized CNT FET biosensor.73 (b) Femtomolar direct voltammetric determination of circulating miRNAs in sera of cancer patients using an enzymeless biosensor;74 (c) ECL/AdsSV dual-mode detection capture unit, signal unit and detection principle.76 (d) Synthetic process of SQDs and fabrication process of ECL biosensor for miRNA-141 detection.80 | ||
Overexpression of miRNA-155 is primarily considered a risk factor for breast cancer. This risk factor is associated with clinicopathological markers, tumor subtypes, low survival rates, metastasis, and invasive features of breast cancer. In addition, this risk factor is thought to be associated with high tumor grade, advanced stage, and lymph node metastasis.75 As far as the specific electrochemical detection of miRNA-155 is concerned, there are few publications in the literature. Zheng et al., reported dual-mode ECL/ASV based biosensor for miRNA-155. An adsorption state was constructed using tetrahedron DNA as capture unit and g-C3N4@AgNPs-rDNA as signal unit as shown in Fig. 5c. Both ECL intensity and AdsSV stripping peak currents are linearly related to the concentration of miRNA-155, and their respective limits of quantification (LOQs) are 50 fM and 100 fM. In addition, high selectivity, repeatability, and long-term stability were achieved. In addition, to ensure the correctness and reliability of the test results, the ECL and AdsSV signals can be analyzed.76
miRNA-141 is a small, single-stranded, noncoding miRNA. It is one of the most important biomarkers for cancer as it is associated with early detection of diseases such as breast cancer, leukemia, and liver cancer.77,78 It is of utmost importance to develop a method that is not only simple but also rapid and sensitive to successfully identify miRNA-141. Wang et al. developed an ECL paper-based platform mediated by CRISPR/Cas12a (LbCpf1) on a three-dimensional (3D) DNA walker for ultrasensitive detection of miRNA-141. The AuPd NP/3D-rGO/PWE was not only highly efficient by providing a suitable environment to enhance the specific surface area. It also extends the conductivity of the electrode and increases the loading sites for biomolecules. It could be used as a signal generator to significantly improve the cathodic emission efficiency of g-C3N4NSs, as shown in Fig. 5d. The proposed multimechanical biosensor showed excellent sensitivity and specificity with LOD of 0.331 fM (S/N = 3) in the concentration range between 1 fM and 10 nM.79 Ji et al. reported an ultrasensitive SQDs-based ECL assay for the determination of miRNA-141 with double amplification of co-reaction accelerators and DNA walkers. SQDs were obtained by PEG stabilized and H2O2-assisted top-down methods. Pd@Au NPs were used as co-reaction accelerators to enhance ECL emissions from SQDs. DNA Walker significantly promoted signal amplification for built ECL sensors. The SQDs-based ECL biosensor provided a low LOD of 1.39 fM for the detection of miRNA-141.80
2.3. Circulating tumor DNA (ctDNA) as a target for biosensor
Identifying molecular biomarkers will significantly improve the accuracy of cancer diagnosis in its early stages. DNA nanomachines, which are designed and able to change DNA nanostructures, show wide potential to detect tumor biomarkers with non-invasiveness, low cost, very sensitive and very specific advantages.81 In 2016, the FDA approved a ctDNA test to detect genetic mutations of the epidermal growth factor (EGFR) receptor in patients with non-small cell lung cancer.82 Liu et al. recently developed a platform based on ctDNA to detect early lung cancer. The CRISPR/Cas12a system plays an important role in precisely identifying the target ctDNA, which is an essential target. The proposed electrochemical biosensor has a high degree of specificity, stability, and selectivity. To be more precise, the proposed biosensor has a detection limit of 3.3 aM.47With the aim of detecting the PIK3CA E545K ctDNA biomarker in the blood of breast cancer patients, Huang et al. designed an electrochemical biosensor based on the nest hybridization method to detect the biomarker more selectively and sensitively. Two dumbbell-shaped DNA units were connected by two different types of DNA probes, which triggered the nest hybridization reaction, which ultimately led to the formation of a complex DNA structure. The resulting DNA structures were captured by the target ctDNA, which eventually led to a sharp increase in the electrochemical signal. The detection limit for the biosensor is 3 pM, and under optimal conditions, the biosensor detected ctDNA in a linear range between 5 pM and 0.5 nM, which is a satisfactory analytical performance (Fig. 6a). The optimal reaction time for the hybridization chain reaction (HCR) was chosen to be 30 min. This cost-effective and easy-to-use sensing technique has also been used to detect catenae in other samples, such as spiked-in samples, pleural effusions, and serum samples from patients with malignancies. In addition, this is an appropriate and cost-effective approach because there are no heating cycles and complicated preparations. As a result of all these characteristics, the developed system can be used as a clinical platform for the detection of chronic ctDNA.83
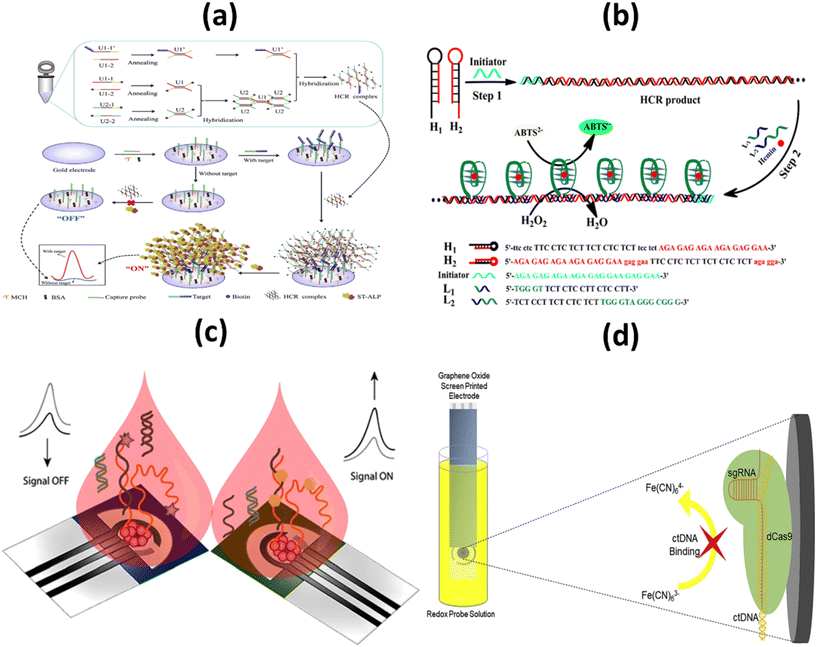 | ||
| Fig. 6 Schematic representation of: (a) the electrochemical biosensor for the detection of ctDNA based on the nest hybridization chain reaction;83 (b) colorimetric biosensor platform for selective detection of the DNA of circulating tumors;84 (c) signal ON & signal OFF platforms for electronic detection of nucleic acids based on paper;85 (d) label-free, CRISPR-powered impedimetric biosensor for the detection of mutations in ctDNA's.86 | ||
More commonly, to determine PIK3CA E542KM ctDNA from plasma of a breast cancer patient, Li and colleagues developed a selective and profound detection of ctDNA. This low-cost, user-friendly device works by forming triple-helix DNA using oligonucleotides to specifically detect dsDNA products derived from the hybridization chain reaction, creating a G-quadruplex structure that is asymmetrically cleaved. After binding with hemin, they form a complex structure known as the G-quadruplex/hemin complex, which exhibits high peroxidase-like activity. The structure catalyzes the oxidation reduction reaction between 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) and H2O2 resulting in a color change that is detected by a spectrophotometer or naked eye (Fig. 6b). The method is easy to operate with a negligible background and detection limit of 0.1 pM. The developed method was applied to detect the ctDNA from the human plasma samples with an aim to evaluate its feasibility, and it exhibited adequate accuracy as well as low relative standard deviation (below 6%) which indicates its satisfactory anti-interference performance in ctDNA detection. Moreover, the provided result ensures satisfactory accuracy and reliability in human plasma samples. Another benefit of this developed colorimetric platform is the ability to detect other biomarkers, proteins or small molecules related with the design of the capture probe's structure.84
In an attempt to improve sensitivity, Cinti et al. proposed the use of a paper-based electroanalytical strip to detect the H1047R (A3140G) missense mutation in breast cancer, using a signal ON and a signal-off method to detect single-stranded DNA (ssDNA). LOD and binding constants were in the nanomolar range which was 6 nM and 40 ± 6 nM respectively. In the approach, the signal OFF does not require any additional chemical reagents due to the integration of redox mediator in the designed platform whereas as a consequence of modifying the probe with the tag, the cost signal OFF platform is 4–5 folds higher than signal ON platform (Fig. 6c). Both the signal ON and signal OFF platform are considered promising in terms of both cost as well as miniaturization. Need of external chemical, additional washing steps and probability of electrode fouling from complex matrices are few limitations signal ON platform.85
Other tumor markers such as CRISPR associated protein 9 (Cas9) is a DNA endonuclease enzyme, which is widely utilized in various applications of genetic engineering. A mutant form of Cas9 is dCas9, through point mutations in its endonuclease domains, its endonuclease property is removed. Uygun et al. reported a CRISPR/dCas biosensor for the detection of ctDNA markers commonly assessed in the genetic evaluation of breast cancer patients (Fig. 6d). Through electrochemical impedance spectroscopy, the developed biosensor ensures rapid, selective and accurate detection of ctDNA. The limit of detection and Limit of Quantitation were calculated as 0.65 nM & 1.92 nM respectively. The CRISPRdCas9 activated impedimetric biosensor exhibited both high selectivity (as there is no increase in impedance curve with other DNA) and accuracy (greater than 96%) with ultra-rapid detection time (only 40 seconds) which has established this system as a promising tool for ctDNA detection in LB.86
2.4. Cells as a target for biosensor
It is a challenging task to develop a map linking the molecular and cellular features of living tumors with their dependencies. For the map to represent the majority of patients, pharmacological profiling data from historical long-term cell line models, innovative short-term patient models, and rapid profiling of alpha cancer cells within hours of patient sampling must be combined.87 Yang and co-workers developed a novel sandwich-type biosensor system for the label free and selective identification of MCF-7 human breast cancer cells through an electrochemical approach (Fig. 7a). The proposed biosensor was developed based on a glassy carbon electrode altered with 3-dimensional (3D) graphene as well as a hybrid of Au based nanocages (Au NCs)/amino-functionalized multiwalled carbon nanotubes (MWCNT-NH2), a potential substrate which adsorb breast cancer estrogen receptor antibodies (Ab1). This graphene-based biosensor model showed a wider linear range (up to 106 cells per mL) and a lower detection limit (80 cells per mL) as a result of the combinational advantages of DNA-labeled antibodies and a nanomaterial-based signal-amplification strategy within 60 minutes. Along with the combination of sandwich-type biomimetic interface, the developed approach can significantly amplify the detection signal with high stability which results higher capture efficiency. Therefore, for early diagnosis of cancer in clinical platform, this electrochemical cytosensing biosensor platform may be considered as a feasible and sensitive method for detecting rare cancer cells with higher specificity.88 Li et al., reported a simple, rapid and sensitive platform in a proof-of-concept study to detect melanoma-associated chondroitin sulfate proteoglycan (MCSP) on the surface of melanoma cells based on magnetic separation as well as nanozyme activity of magnetic nanoparticles (MNPs), which can be completed within 5 minutes (Fig. 7b). The nanozyme property of magnetic nanoparticles shows naked-eye identification of melanoma tumor cells down to 50 cells per mL this makes it a opportune tool for analysing cancer cells in patients. Limit of detection was calculated 13 cells per mL through analyzing the standard deviation of blank signal three times. To confirm the naked-eye observations, UV-vis spectra were used which concludes that signal intensities in patient samples were higher than in healthy control samples. The developed colorimetric based platform is more convenient and rapid analytical assessment which can be used to detect circulating tumor cells in clinical applications as well as suitable for other potential biosensing applications such as protein and nucleic acid detection in clinical analysis.89 To identify the main site of the tumor origin and to detect sensitive circulating tumor cells, Jia and colleagues proposed a strategy in combination with an in vivo capture method based on an antibody-cocktail with multicolor fluorescence imaging by using an aptamer. To identify the breast cancer (MCF-7) and hepatocellular carcinoma (SMMC 7721) CTCs, an antibody cocktail of epithelial cell adhesion molecule (EpCAM) and epidermal growth factor receptor (EGFR) was applied in vivo. The applied platform significantly improved the capture efficiency of hepatocellular CTCs from 3.17% to 26.67% whereas it slightly increased the capture efficiency from 27.00% to 29.84% in comparison with EpCAM-based detection of circulating tumor cells. On the other hand, the specific aptamer-based fluorescence probes simultaneously identified the roots of the primary tumor of breast cancer CTCs as well as hepatocellular CTCs without any signal interference (Fig. 7c). Compared with in vitro CTC detection approaches, this proposed platform has higher sensitivity as it avoids sampling error and improves the detection accuracy of circulating tumor cells for cancer patients. This biosensing platform is suitable for the early diagnosis of the cancer as well as the assessment of cancer metastasis. The efficiency of both selective identification and capture of CTC would be improved by both the single or combined application of these probes.90 Yu et al. developed a new aptamer based electrochemical cytosensor that enables rapid and highly specific detection of CML K562 cells using a specific aptamer and bio-ConA detection probe (Fig. 7d). The detection limit of the developed biosensor was calculated down to 79 cells per mL and displayed a wider linear range from 1 × 102 to 1 × 107 cells per mL. It obtained recoveries between 79.6–93.3% when it was applied to detect CML K562 cells in human blood samples, indicating the platform as an alternative approach for CML detection in clinical analysis. Because of low relative standard deviation (RSD), the developed platform ensures satisfactory reproducibility. Selectivity and sensitivity of the proof of concept-based biosensor was evaluated with a number of other cancer cells in blood samples and which results in a minimal change in the current of differential pulse voltammetry (DPV) that indicates higher specificity as well as accurate precision of the biosensor.91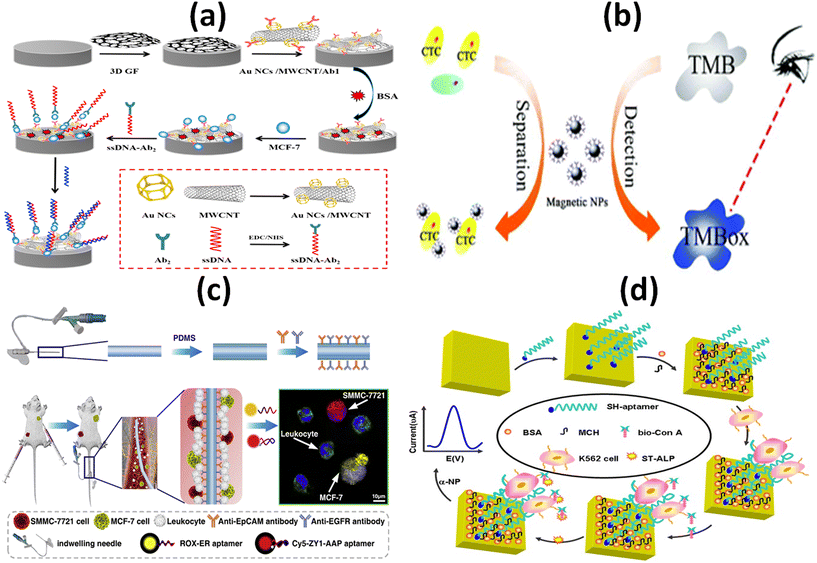 | ||
| Fig. 7 Schematic representation of: (a) electrochemical immunosensor with the DNA marker antibody for the specific detection of MCF-7 breast cancer cell;88 (b) enzyme based colorimetric biosensor for the detection of melanoma circulating tumor cell by using nanozyme based magnetic nanoparticles;89 (c) antibody cocktail-based in vivo capture and specific aptamer-based multicolor fluorescence imaging analysis of circulating tumor cells;90 (d) electrochemical aptamer cytosensor for the detection of CML K562 cell using a specific aptamer and bio-ConA detection probe.91 | ||
2.5. Exosomes and EVs as a target for biosensor
Exosomes are extracellular vesicles (EVs) produced by cells. They can be found in a variety of body fluids and play an important role in communication between cells.92 These vesicles are secreted by various cells, and their function is to trigger or inhibit various cellular and molecular processes. It is now known that exosomes released by malignant cells play a role in the recruitment and reprogramming of proximal and distal tissues, thus influencing the course of cancer at various stages of its development.93 Using bimetallic gold–silver nanodroplets, Ning et al. reported multiple, sensitive SERS detection of exosomes associated with malignancy. The presented sensor SERS is characterized by high sensitivity, excellent repeatability, good stability and simple operation, among other features. Quantitative detection of each exosome showed a clear linear relationship between the signal strength of SERS and the logarithm of the concentration of the target exosomes. A linear correlation was also found between the intensity of SERS and a LOD of 35 particles per μL.94 Suwatthanarak et al. described the collection and release of cancer-derived exosomes using a peptide–nanowire hybrid interface. They designed a peptide nanowire hybrid nanointerface to capture and release exosomes from cancer in a miniature platform. In addition, the captured exosomes have the potential to be subsequently released from the nanowires in a state that also contains neutral salt for use in future applications. Microfluidic systems for exosome-based diagnostics and treatments should benefit from this tailored surface as it enhances the effectiveness of nanowires in selective and regulated collection of exosomes from cancer.952.6. Other biological fluids
Although the use of noninvasive collection samples, particularly saliva, sputum, feces, and urine, has increased in recent years.96 Saliva is one of them that is often used to identify malignant tumors. Saliva is a noninvasive, rapidly obtained, and highly reliable sample for clinical biomarkers because it contains the same amounts of proteins, metabolites, DNA, or RNA as blood or serum.97 Exosomes from salivary tumors represent a potentially useful alternative method for detecting novel biomarkers associated with oral cancer.98 Zhao et al. conducted isolation and detection of saliva-based exosomes. The saliva was collected by a healthy donor in a salivettes according to manufacturer instructions. After proposing a new aptamer fluorescence system (Exo-AFS) for efficient detection of exosome surface proteins in lung cancer monitoring, they used an automated centrifugal microfluidic disc system with functionalized membranes (Exo-CMDS) to collect and enrich exosomes. Exo-CMDS is capable of isolating and purifying exosomes in a single step and has a highly qualified yield with an ideal exosome concentration of 5.1 × 109 particles per ml from trace blood samples (less than 300 μl) in only 8 minutes.99Urine is the second most important biological fluid after saliva because it can be collected quickly, painlessly, without special procedures or equipment, and because it can be obtained in large quantities. The best thing about using urine instead of tissue or blood is that taking urine isn't as painful as taking tissue or blood. This is especially important for patients who need samples taken more than once to see how the cancer is getting worse or how well the treatment is working.100 To this end, Wang et al. have developed a biochip based on timed light probes supported by photonic crystals (PCs) that can detect miRNA associated with bladder cancer in urine with high sensitivity. The Biochip shows excellent performance in miRNA detection in urine, with excellent recoveries using a LOD of 26.3 fM.101
3. Potential concerns in point-of-care testing technologies for cancer tests
According to the National Cancer Institute, cancer is composed of several diseases.102 Early cancer detection and screening could result to much better clinical outcomes.103 Liquid biopsy has evolved into a remarkable and minimally invasive diagnostic procedure that can reveal clinically relevant abnormalities in multiple tumors in real time. This capability has made liquid biopsy one of the most promising areas of cancer research.104 The development of POC diagnostics is a direct result of significant technological advances in the field of biosensing. These diagnostics are designed to make the testing process more advisable, user-friendly, and affordable, while allowing measurements to be taken in the field. Miniaturization is a particular strength of recent innovations such as electrochemical and fluorescence detection techniques.105,106 POC testing allows individuals to take a test that provides information quickly.103 This topic is rapidly expanding due to breakthroughs in biological knowledge and technology.The rapid growth of the POCT industry has led to a market forecast of $46.65 billion by 2021. The global point-of-care diagnostics market is expected to grow at a compound annual growth rate (CAGR) of 5.2% from $36.37 billion in 2022 to $51.94 billion in 2029.107 Tumor and cancer biomarkers are one of the many aspects in this field. This has enabled rapid diagnosis and selection in many other therapeutic areas beyond the emergency room, where it was previously unimaginable. Professionals now have more control over comprehensive patient care, as they are no longer dependent on a third party to collect a sample, forward it to a testing facility, and report the results. During both major surgery and cancer chemotherapy, recurrent, faster POC diagnostics will provide tremendous benefit to cancer patients. Typical examples of such tests include blood and urine tests. The goal of POCT is to collect the sample as quickly as possible at or near the patient's site and obtain reliable results. To develop a novel clinical test, a biomarker must be recognized and a simple and rapid detection method must be established. This biomarker then goes through several processes before it can be used clinically, including the crucial step of determining whether it can detect cancer earlier than current approaches.108
As rapid tests become more advanced in terms of speed, quality, and the capabilities of connected health and artificial intelligence, liquid biopsy would undoubtedly become an integral part of regular testing. Unfortunately, this technique has been slow to gain clinical acceptance and still needs improvement. Despite numerous improvements, the clinical usefulness of liquid biopsy for cancer diagnosis based on POCT is still debated and a major concern for clinical professionals due to its less accuracy and false-positive/negative results. Precision and accuracy of techniques remains a major issue in all markets. As CTCs, they offer the full spectrum of analysis at the DNA, RNA, and protein levels. They also make it possible to study the functions of living cells in vitro and in vivo. Similarly, extracellular vesicle analysis also provides a comprehensive result, albeit with some serious practical limitations. The ctDNA analysis is limited to drug-resistant mutations, mostly from apoptotic cells, which do not necessarily reflect the therapeutically resistant viable tumor cell clone in a cancer patient.109,110
Despite technical advances in this field, it is possible to reliably detect a small amount of ctDNA. The drawback is that many patients have low levels of CTC, especially in the early stages of the disease. These definitions are required depending on the clinical context, the type of cancer being treated, and the sensitivity of the methods to be used. For example, in NSCLC, the amount of CTCs and ctDNA is much lower than would be expected in such aggressive diseases with high metastatic potential, limiting the accuracy and reliability of POCT devices.111–113 However, recent research on ctDNA and exosomes from tumors isolated from plasma samples faces the same challenges. ctDNA assays do not provide information on many important tumor characteristics for classification and distribution and only indirectly suggest the overall tumor burden.114
The second major concern is the very low frequency of variants in liquid biopsy, which may lead to a higher likelihood of both false negative and false positive reports. This is a serious issue, as it is one of the main reasons why POCT-based liquid biopsy for biomarker detection is not widely used. Tests for conventional circulating protein biomarkers have been used in the clinical setting for many decades.10 One of the most important preanalytical goals in cfDNA analysis is to prevent the release of genomic DNA from lysing leukocytes. This DNA could potentially contribute to false-negative findings by diluting the already low proportion of tumor-specific DNA.115 The form of the cancer treated and the sensitivity of the technique used depend entirely on the clinical scenario. The key concern is that this technique is not precise for a single type of cancer and has a high false positive rate for situations that are not cancerous.116 These problems can lead to unnecessary treatments that harm patients and increase medical costs.
The detection of POCT based on liquid biopsy of ctDNA, CTCs, cfRNA, extracellular vesicles, and/or protein is a promising active field of treatment outcomes research. However, these methods need to be further developed and validated in order to be used in the clinic. Improved technology and cost-effectiveness will soon enable liquid biopsy to be a highly sensitive and specific instrument for cancer diagnosis and treatment. The difficulty is then in quality assurance, and how to ensure that the results achieved are really what a laboratory test would reveal? A better understanding of these confusing factors in the analysis of liquid biopsy will be important in order to improve our current diagnostic capabilities. Before being introduced into regular patient care, the tests must be validated. This is an important objective of the European Consortium CANCER-ID (https://www.cancer-id.eu). The quality management of liquid biopsy tests is very important to avoid the false positive and negative results.111 This improvement in sensitivity and selectivity of current POCT based devices will contribute to the development of the quality and reliability of the tests performed. It also provides assurance of error-free cancer diagnosis.
4. Challenges limiting the development point-of-care biosensors for liquid biopsy
The advent of novel diagnostic techniques and tailored medicines led to the birth of precision medicine, which in turn led to improved treatment outcomes for patients with cancer. There are a variety of factors that significantly affect the quality and precision of liquid biopsy test results. There are numerous significant barriers that must be overcome for future clinical application to promote the widespread use of POC for liquid biopsy detection in an effective and economical manner. The fact that liquid biopsy cannot be used to assess the margins, grade, size, or location of a tumor is only one of the many difficulties associated with this technique.117 The specific challenges in this field include different lever biology, technology, medicine, and regulatory. The roots of the challenges and limitations are identified in the following points:Biological challenges: under certain biological conditions, liquid biopsy exhibits low specificity and sensitivity because there is an indeterminate level of plasma biomarkers. Similarly, proteins are the most widespread biological molecules affected by disease.118 The dynamic nature and complexity of cellular proteomes from various biological sample types such as blood, serum, plasma, and tissue pose a significant analytical obstacle to the characterization and validation of candidate proteomic biomarkers in modern proteomics research.119 Considering the rarity of CTCs and the fact that an adult human should have 5 liters of total blood, it is possible that a small portion of the patient's blood (7.5 ml) is not sufficient to identify the cells. This is another example reported by Baccelli et al. Out of this, only 1.43% of individuals with advanced breast cancer had more than 500 CTCs per 7.5 ml of blood.120 It is possible that the ability to assess low levels of disease biomarkers may improve the quality of care in places where resources are limited. These results highlight functional CTC indicators that may improve the detection and treatment of metastatic breast cancer. The detection of biomarkers at low concentrations in a POC assay is a significant development: many of the most intriguing new POC prospects are based on this trend. An important future POC application is the testing of a blood sample for cancer-causing liquid biopsies. This could replace laboratory tests that take hours or days to perform.
The limited stability of POC for liquid biopsy is the next significant biological issue limiting its progress. Patients with different amounts of ctDNA have different rates of ctDNA release into the circulation due to differences in tumor size, location, and vascularity. Like, CtDNA has a relatively short half-life of about 1.5 to 2 hours in peripheral blood121 and show different levels of stability.122–124 Due to the low stability of ctDNA, processing methods for such biological samples differ significantly. Association with cell membranes, extracellular vesicles, or proteins can increase the stability of individual fragments in the circulation. This can be achieved in several ways.125
Technological challenges: several methods are currently being developed to isolate different liquid biopsy-like CTCs from abundant leukocytes and erythrocytes depending on their biological and/or physical properties. CTCs differ from normal blood cells by their size, deformability, density, or dielectric properties, all of which are used in physical separation procedures.126 Like exosomes, their main limitation is isolation and separation. Currently, there are no gold standard methods for isolation of exosomes. Exosome isolation is necessary to ensure exosome structural integrity and biological activity in order to accurately estimate their functions. The choice of isolation methods has a profound impact on identifying enriched pathways and genetic sets. Consequently, it is important to choose an appropriate isolation method to show the specific functions of the exosomes. Ultracentrifugation (UC) is now considered a “gold standard” technique for separating and isolating the exosomes of other components based on different density levels. Protein contamination can be reduced by UC. Nevertheless, throughput is low, and other particles of comparable size can be isolated.127
Another common challenge in specimen collection is contamination from activated platelet vesicles due to the physical forces involved in blood collection. To reduce shear stress and resulting platelet activation, collection areas must be standardized, and longer needles and precise blood collection can be used.128
Clinical challenges: detection of cancer based on biomarkers from liquid biopsy requires large-scale clinical trials and adequate evidence not only of the efficacy of the procedure and the reliability of the marker, but also of the clinical value of the proposed test.129 Compared to extremely low CTC counts in early stage patients, ctDNA offers the advantage of greater bioanalyte concentrations for early cancer detection. However, it is important to note that the concentration of ctDNA is also low in individuals whose cancer is at an early stage. For this reason, the development of high-sensitivity ctDNA assays is currently underway. These analyzes have revealed a background of cancer-associated mutations in normal white blood cells that can contaminate the ctDNA fraction.130 Further limits the clinical application of liquid biopsy detection.
Regulatory challenges: the life cycle of newly developed in vitro diagnostic tests begins in the discovery phase with the identification of specific biomarkers and ends when health insurers begin to reimburse hospitals and clinics for the cost of using these procedures on patients. We are now at a point where the first LB tests for cancer have proven their clinical validity after their analytical validation.131 Many liquid biopsy products are linked to companion diagnostic applications, so their approval processes are very similar to those for companion diagnostics. However, because we do not yet have a regulatory mechanism that is clearly associated with liquid biopsies, the utility of this method is limited.132
In summary, liquid biopsies are considered to be the game changer in the treatment of personalized cancer. The potential of liquid biopsies in translation cancer research is still well understood, multiple limitations, such as standardized pre- and analytical variables, need to be addressed in order to prove the role of liquid biopsy in clinical practices. Improvements in standardized and analytical variables will increase diagnostic and prognostic confidence, treatment efficiency, real-time out-of-hospital determination, and sample burden.
5. Future perspectives and concluding remarks
Liquid biopsies offer the ability to diagnose, characterize, and follow malignancies earlier than is possible with conventional methods. Due to its minimal invasiveness and excellent capacity for disease monitoring, liquid biopsy is being adopted as a beneficial technique in many domains, particularly oncology. When cancer is diagnosed at an earlier stage, successful treatment is more likely, and the patient's chances of survival increase significantly. The remarkable fact that early detection of cancer is related to survival rates has recently led to efforts to improve diagnosis and surveillance. Although existing tissue-based diagnostics are the gold standard, innovative studies are looking at liquid biopsy on a chip to identify CTCs from a blood sample. Most hematologic, clinical, and immunochemical tests are routinely performed in laboratories to detect cancer using high-throughput technologies. Detection of CTCs has evolved from commercial macro technologies to miniaturized POC-based detections that require limited reagents and allow for better sample control. Measurement of CTCs offers new opportunities for early detection of cancer and follow-up of patients based on the results. Liquid biopsy POCT diagnostics eliminate the need for a central laboratory and allow for efficient, rapid and easy automation. Timely and accurate detection of cellular and molecular biomarkers is key to effective treatment and high survival rates. This technology is also promising for early cancer detection, as it can be performed on demand to monitor dynamic changes in the molecular landscape during tumor growth or drug treatment.To promote the widespread use of POC in a powerful and cost-effective manner for detection of liquid biopsy, there are several important challenges to be addressed for further clinical implementation. Challenges in the field, current work in these areas, and where further research is needed to make early detection a reality. The low specificity and sensitivity of the various detection methods, the lack of standardization leading to experimental bias and hindering the collection of correct data, and the high financial cost are the main reasons for the limitations. The relatively low number of tumor-derived analytes found in patient biofluids is one of the major challenges for all liquid biopsy-based diagnostic approaches in early stages. Thus, it is not yet clear whether the analyte sampled at a particular time point can be used to mimic the spatial heterogeneity and temporal evolution of tumors and their microenvironment. Additionally, because of biological considerations, determining the organ e.g., absence of shedding and technical factors e.g. lack of sensitivity and/or specificity from which the analyser originates is still difficult like.15 Another significant obstacle is the standardization of processes, guidelines, and frequently insufficient technical and clinical validation for routine clinical use in the field of liquid biopsy detection.133 Standardization of analytical procedures (pre-analysis) and specific clinical trials will help to fill this gap. To achieve early detection of all cancers, many obstacles must be removed. It is important that we better understand who is most affected by cancer. In order to identify the consequences of cancer at an earlier stage, we need to better understand the origins and development of the disease.
The future of LB detection depends on automation, machine learning, and artificial intelligence. In recent years, the use of high-performance automated technology in biomedical and clinical data has increased exponentially. Precision, efficiency, and monitoring at the bedside rather than in a hospital will make significant advances in the coming decade. The use of biosensors has evolved into an autonomous diagnostic platform that enables the detection of biomarkers unique to the development, recurrence, and drug monitoring of most malignancies. Small wireless devices are transforming the way healthcare services work in a big way, ensuring that reliable detection is possible. Low-cost, wearable systems offer the potential to improve disease prevention and detection at the point of care in primary care in both high- and low-resource countries by leveraging advances in consumer electronics and wireless telecommunications.134 The field of clinical micrototal analysis has recently seen a number of notable breakthroughs, including substantial improvements in the validation, sensitivity, specificity, and integration of sample processing procedures. Liu et al. reported that machine learning is widely used in single genomic analysis. However, a single type of circulatory biomarker gives incomplete information regarding the nature of a tumor occurrence. Multi-omics detection is therefore another promising direction for early detection and treatment surveillance of cancer. The exploration ability of machine learning can allow the discovery of complex causal relationships between different molecular measurements.135 Artificial intelligence-based algorithms are able to quantify the effects of many biomarkers simultaneously and enable the detection of interactions between biomarkers that would be difficult to detect using manual approaches.15
The medical world needs to be convinced that the detection and identification of LB is valuable for medical care. The next few years will be critical for such an approach, during which scientists and engineers must present the true plausibility of this approach. This will require scaling up and demonstrating the accuracy and precision of the technology. For early detection to make a decisive advance in cancer incidence, skills beyond cancer biology are needed. These skills must include those of engineers, chemists, physicists, technology developers, behavioral scientists, and computer scientists. We, the technologists, must be open and responsive to the needs of the medical community and the industry that is bringing products to market. With significant funding and global commercial support, this method could become the next best approach to diagnosis. This will enable precise and personalized intervention strategies. The use of liquid biopsy as a diagnostic, prognostic, and predictive procedure for many cancers will become increasingly important in the future.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. C. acknowledges the MIUR Grant “Dipartimento di Eccellenza 2018-2022” to the Department of Pharmacy of University of Naples “Federico II.” This work was supported by a grant from Regione Campania-POR Campania FESR 2014/2020 “Combattere la resistanza tumorale: piattaforma integrata multidisciplinare per un approccio tecnologico innovativo alle oncoterapie-Campania Oncoterapie” (Project N. B61G18000470007). S. S. acknowledges Fondazione Umberto Veronesi postdoctoral fellowship 2022.References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef.
- M. Zhao, D. Mi, B. E. Ferdows, Y. Li, R. Wang, J. Li, D. Patel, N. Kong, S. Shi and W. Tao, Nano Today, 2022, 42, 101361 CrossRef CAS.
- R. C. Fitzgerald, A. C. Antoniou, L. Fruk and N. Rosenfeld, Nat. Med., 2022, 28, 666–677 CrossRef CAS PubMed.
- X. Li, M. Ye, W. Zhang, D. Tan, N. Jaffrezic-Renault, X. Yang and Z. Guo, Biosens. Bioelectron., 2019, 126, 596–607 CrossRef CAS PubMed.
- S. Singh, A. A. S. Gill, M. Nlooto and R. Karpoormath, Biosens. Bioelectron., 2019, 137, 213–221 CrossRef CAS PubMed.
- R. Vaidyanathan, R. H. Soon, P. Zhang, K. Jiang and C. T. Lim, Lab Chip, 2019, 19, 11–34 CAS.
- E. Heitzer, I. S. Haque, C. E. S. Roberts and M. R. Speicher, Nat. Rev. Genet., 2019, 20, 71–88 CrossRef CAS PubMed.
- E. Crowley, F. Di Nicolantonio, F. Loupakis and A. Bardelli, Nat. Rev. Clin. Oncol., 2013, 10, 472–484 CrossRef CAS PubMed.
- R. Palmirotta, D. Lovero, P. Cafforio, C. Felici, F. Mannavola, E. Pellè, D. Quaresmini, M. Tucci and F. Silvestris, Ther. Adv. Med. Oncol., 2018, 10, 1–24 Search PubMed.
- S. Bratulic, F. Gatto and J. Nielsen, Regener. Eng. Transl. Med., 2021, 7, 312–352 CrossRef.
- L. Wu, Y. Wang, X. Xu, Y. Liu, B. Lin, M. Zhang, J. Zhang, S. Wan, C. Yang and W. Tan, Chem. Rev., 2021, 121, 12035–12105 CrossRef CAS PubMed.
- J. Wang, S. Chang, G. Li and Y. Sun, Front. Med., 2017, 11, 522–527 CrossRef PubMed.
- F. Castro-Giner and N. Aceto, Genome Med., 2020, 12, 1–2 CrossRef PubMed.
- J. C. M. Wan, C. Massie, J. Garcia-Corbacho, F. Mouliere, J. D. Brenton, C. Caldas, S. Pacey, R. Baird and N. Rosenfeld, Nat. Rev. Cancer, 2017, 17, 223–238 CrossRef CAS PubMed.
- Y. Belotti and C. T. Lim, Anal. Chem., 2021, 93, 4727–4738 CrossRef CAS.
- E. S. Park, J. P. Yan, R. A. Ang, J. H. Lee, X. Deng, S. P. Duffy, K. Beja, M. Annala, P. C. Black, K. N. Chi, A. W. Wyatt and H. Ma, Lab Chip, 2018, 18, 1736–1749 RSC.
- M. Poudineh, E. H. Sargent, K. Pantel and S. O. Kelley, Nat. Biomed. Eng., 2018, 2, 72–84 CrossRef CAS PubMed.
- B. Abdulmawjood, C. Roma-Rodrigues, A. R. Fernandes and P. V. Baptista, Cancer Drug Resist., 2019, 2, 1044 Search PubMed.
- F. Tian, C. Liu, L. Lin, Q. Chen and J. Sun, TrAC, Trends Anal. Chem., 2019, 117, 128–145 CrossRef CAS.
- N. Kamyabi, R. Abbasgholizadeh, A. Maitra, A. Ardekani, S. L. Biswal and K. J. Grande-Allen, Biomed. Microdevices, 2020, 22, 1–11 CrossRef.
- S. Perakis and M. R. Speicher, BMC Med., 2017, 15, 1–2 CrossRef PubMed.
- S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Curr. Opin. Electrochem., 2018, 163, 112238 Search PubMed.
- Y. Liang, B. M. Lehrich, S. Zheng and M. Lu, J. Extracell. Vesicles, 2021, 52, 3348–3350 Search PubMed.
- S. K. Metkar and K. Girigoswami, Biocatal. Agric. Biotechnol., 2019, 17, 271–283 CrossRef.
- S. R. Adam Kratz, G. Höll, P. Schuller, P. Ertl and M. Rothbauer, Biosensors, 2019, 9, 110 CrossRef.
- K. M. Koo, N. Soda and M. J. A. Shiddiky, Curr. Opin. Electrochem., 2021, 25, 100645 CrossRef CAS.
- S. H. Hussain, C. S. Huertas, A. Mitchell, A. L. Deman and E. Laurenceau, Biosens. Bioelectron., 2022, 197, 113770 CrossRef CAS PubMed.
- C. Cai, Z. Guo, Y. Cao, W. Zhang and Y. Chen, Nanotheranostics, 2018, 2, 12–20 CrossRef PubMed.
- J. Das and S. O. Kelley, Angew. Chem., Int. Ed., 2020, 132, 2574–2584 CrossRef.
- L. Xu, N. Shoaie, F. Jahanpeyma, J. Zhao, M. Azimzadeh and K. T. Al-Jamal, Biosens. Bioelectron., 2020, 161, 112222 CrossRef CAS PubMed.
- Y. Xia, T. Chen, G. Chen, Y. Weng, L. Zeng, Y. Liao, W. Chen, J. Lan, J. Zhang and J. Chen, Talanta, 2020, 214, 120851 CrossRef CAS PubMed.
- P. Hofman, S. Heeke, C. Alix-Panabières and K. Pantel, Ann. Oncol., 2019, 30, 1448–1459 CrossRef CAS PubMed.
- M. Jiang, S. Jin, J. Han, T. Li, J. Shi, Q. Zhong, W. Li, W. Tang, Q. Huang and H. Zong, Biomark. Res., 2021, 9 Search PubMed.
- S. Campuzano, R. Barderas, M. Pedrero, P. Yáñez-Sedeño and J. M. Pingarrón, Anal. Chim. Acta, 2020, 1109, 169–190 CrossRef CAS.
- C. Liu, Y. Yang and Y. Wu, AAPS J., 2018, 20, 1–3 CrossRef CAS PubMed.
- H. Xie, K. Di, R. Huang, A. Khan, Y. Xia, H. Xu, C. Liu, T. Tan, X. Tian, H. Shen, N. He and Z. Li, Chin. Chem. Lett., 2020, 31, 1737–1745 CrossRef CAS.
- C. Lu, J. Han, X. Sun and G. Yang, Sensors, 2020, 20, 6073 CrossRef CAS PubMed.
- N. Soda, B. H. A. Rehm, P. Sonar, N. T. Nguyen and M. J. A. Shiddiky, J. Mater. Chem. B, 2019, 7, 6670–6704 RSC.
- P. Pinzani, V. D’Argenio, M. Del Re, C. Pellegrini, F. Cucchiara, F. Salvianti and S. Galbiati, Clin. Chem. Lab. Med., 2021, 59, 1181–1200 CrossRef CAS PubMed.
- C. Dincer, R. Bruch, A. Kling, P. S. Dittrich and G. A. Urban, Trends Biotechnol., 2017, 35, 728–742 CrossRef CAS.
- D. Thevenot, K. Toth, R. Durst, G. Wilson, D. Thevenot, K. Toth, R. Durst and G. Wilson, Biosens. Bioelectron., 2001, 16, 121–131 CrossRef CAS.
- R. Vashistha, A. K. Dangi, A. Kumar, D. Chhabra and P. Shukla, 3 Biotech, 2018, 8, 1–11 CrossRef.
- S. Jeong, J. Park, D. Pathania, C. M. Castro, R. Weissleder and H. Lee, ACS Nano, 2016, 10, 1802–1809 CrossRef CAS.
- Y. Chen, J. Peng, Y. Lai, B. Wu, L. Sun and J. Weng, Biosens. Bioelectron., 2019, 142, 111520 CrossRef CAS.
- X. L. Zhang, Y. Yin, S. M. Du, L. Q. Kong, Y. Q. Chai, Z. H. Li and R. Yuan, Anal. Chem., 2021, 93, 13952–13959 CrossRef CAS PubMed.
- X. Zhou, D. Bai, H. Yu, Y. Fu, L. Song, Y. Wu, K. Chen, J. Li, Y. Yang, H. Chen, Z. Wang and G. Xie, Talanta, 2023, 253, 123955 CrossRef CAS PubMed.
- F. Liu, J. Peng, Y. M. Lei, R. S. Liu, L. Jin, H. Liang, H. F. Liu, S. Y. Ma, X. H. Zhang, Y. P. Zhang, C. P. Li and H. Zhao, Sens. Actuators, B, 2022, 362, 131807 CrossRef CAS.
- Y. Yu, Y. Yang, J. Ding, S. Meng, C. Li and X. Yin, Anal. Chem., 2018, 90, 13290–13298 CrossRef CAS PubMed.
- H. Dong, D. Yao, Q. Zhou, L. Zhang and Y. Tian, Chem. Commun., 2019, 55, 1730–1733 RSC.
- H. K. Woo, V. Sunkara, J. Park, T. H. Kim, J. R. Han, C. J. Kim, H. Il Choi, Y. K. Kim and Y. K. Cho, ACS Nano, 2017, 11, 1360–1370 CrossRef CAS PubMed.
- Z. Gao, H. Yuan, Y. Mao, L. Ding, C. Y. Effah, S. He, L. He, L. E. Liu, S. Yu, Y. Wang, J. Wang, Y. Tian, F. Yu, H. Guo, L. Miao, L. Qu and Y. Wu, Analyst, 2021, 146, 1924–1931 RSC.
- B. Li, C. Liu, W. Pan, J. Shen, J. Guo, T. Luo, J. Feng, B. Situ, T. An, Y. Zhang and L. Zheng, Biosens. Bioelectron., 2020, 168, 112520 CrossRef CAS.
- S. S. Kanwar, C. J. Dunlay, D. M. Simeone and S. Nagrath, Lab Chip, 2014, 14, 1891–1900 RSC.
- Z. Zhao, Y. Yang, Y. Zeng and M. He, Lab Chip, 2016, 16, 489–496 RSC.
- P. Zhang, X. Zhou, M. He, Y. Shang, A. L. Tetlow, A. K. Godwin and Y. Zeng, Nat. Biomed. Eng., 2019, 3, 438–451 CrossRef CAS PubMed.
- L. Li, Y. Zhang, Z. Yan, M. Chen, L. Zhang, P. Zhao and J. Yu, ACS Sens., 2020, 5, 1482–1490 CrossRef CAS PubMed.
- W. Huang, C. L. Chang, N. D. Brault, O. Gur, Z. Wang, S. I. Jalal, P. S. Low, T. L. Ratliff, R. Pili and C. A. Savran, Lab Chip, 2017, 17, 415–428 RSC.
- Y. Cao, X. Zhu, J. Zhao, H. Li and G. Li, Prog. Chem., 2015, 27, 1 CAS.
- C. A. K. Borrebaeck, Nat. Rev. Cancer, 2017, 17, 199–204 CrossRef CAS PubMed.
- E. A. Perez, J. Cortés, A. M. Gonzalez-Angulo and J. M. S. Bartlett, Cancer Treat. Rev., 2014, 40, 276–284 CrossRef CAS PubMed.
- M. N. Fornier, A. D. Seidman, M. K. Schwartz, F. Ghani, R. Thiel, L. Norton and C. Hudis, Ann. Oncol., 2005, 16, 234–239 CrossRef CAS PubMed.
- T. Guerrero-Esteban, C. Gutiérrez-Sánchez, T. García-Mendiola, M. Revenga-Parra, F. Pariente and E. Lorenzo, Sens. Actuators, B, 2021, 343, 130096 CrossRef CAS.
- K. Malecka, D. Pankratov and E. E. Ferapontova, Anal. Chim. Acta, 2019, 1077, 140–149 CrossRef CAS PubMed.
- S. B. S. M. Siriwardena, T. Tsunematsu, G. Qi, N. Ishimaru and Y. Kudo, Int. J. Mol. Sci., 2018, 19, 1462 CrossRef PubMed.
- M. Sharafeldin, T. Chen, G. U. Ozkaya, D. Choudhary, A. A. Molinolo, J. S. Gutkind and J. F. Rusling, Biosens. Bioelectron., 2021, 171, 112681 CrossRef CAS PubMed.
- Y. Tan, W. Liu, Z. Zhu, L. Lang, J. Wang, M. Huang, M. Zhang and C. Yang, Anal. Bioanal. Chem., 2018, 410, 1071–1077 CrossRef CAS PubMed.
- W. Liu, S. Sun, Y. Huang, R. Wang, J. Xu, X. Liu and K. Qian, Chem. - Asian J., 2020, 15, 56–60 CrossRef CAS PubMed.
- J. Yee, M. D. Sadar, D. D. Sin, M. Kuzyk, L. Xing, J. Kondra, A. McWilliams, S. F. P. Man and S. Lam, J. Clin. Oncol., 2009, 27, 2787 CrossRef CAS PubMed.
- Y. Chen, L. Sun, X. Qiao, Y. Zhang, Y. Li and F. Ma, Anal. Chim. Acta, 2020, 1103, 67–74 CrossRef CAS PubMed.
- J. Xu, X. Wei, X. Zhang, Z. Cai, Y. Wei, W. Liu, J. Yang, Y. Li, X. Cai, T. Bai, Z. Guo, X. Qu, Q. Zhu and Y. Zhu, Sens. Actuators, B, 2021, 349, 130824 CrossRef CAS.
- M. Negahdary and L. Angnes, Coord. Chem. Rev., 2022, 464, 214565 CrossRef CAS.
- Y. Gao, B. Feng, S. Han, K. Zhang, J. Chen, C. Li, R. Wang and L. Chen, Cell. Physiol. Biochem., 2016, 38, 427–448 CrossRef CAS PubMed.
- T. Li, Y. Liang, J. Li, Y. Yu, M. M. Xiao, W. Ni, Z. Zhang and G. J. Zhang, Anal. Chem., 2021, 93, 15501–15507 CrossRef CAS PubMed.
- M. Zouari, S. Campuzano, J. M. Pingarrón and N. Raouafi, Anal. Chim. Acta, 2020, 1104, 188–198 CrossRef CAS PubMed.
- S. Mattiske, R. J. Suetani, P. M. Neilsen and D. F. Callen, Cancer Epidemiol., Biomarkers Prev., 2012, 21, 1236–1243 CrossRef CAS PubMed.
- Y. Zheng, L. Chen, X. Yin, F. Lin, Y. Xu, X. Lin and S. Weng, Microchem. J., 2021, 165, 106091 CrossRef CAS.
- T. D. Lao, Asian J. Pharm. Res. Health Care, 2018, 10, 42–49 CrossRef PubMed.
- W. Wang, G. Hong, S. Wang, W. Gao and P. Wang, Bioengineered, 2021, 12, 821–831 CrossRef CAS PubMed.
- Q. Wang, Y. Liu, J. Yan, Y. Liu, C. Gao, S. Ge and J. Yu, Anal. Chem., 2021, 93, 13373–13381 CrossRef CAS PubMed.
- K. Ji, Y. Wang, L. Mao, Y. Wang and X. Zhang, Sens. Actuators, B, 2021, 345, 130405 CrossRef CAS.
- Z. Dong, X. Xue, H. Liang, J. Guan and L. Chang, Anal. Chem., 2020, 93, 1855–1865 CrossRef PubMed.
- D. Kwapisz, Ann. Transl. Med., 2017, 5, 46 CrossRef PubMed.
- Y. F. Huang, M. L. Tao, S. H. Luo, Y. Zhang, B. Situ, X. Y. Ye, P. W. Chen, X. J. Jiang, Q. Wang and L. Zheng, Anal. Chim. Acta, 2020, 1107, 40–47 CrossRef CAS PubMed.
- R. Li, L. Zou, Y. Luo, M. Zhang and L. Ling, Sci. Rep., 2017, 7, 1–10 CrossRef PubMed.
- S. Cinti, G. Cinotti, C. Parolo, E. P. Nguyen, V. Caratelli, D. Moscone, F. Arduini and A. Merkoci, Anal. Chem., 2019, 92, 1674–1679 CrossRef PubMed.
- Z. O. Uygun, L. Yeniay and F. G
![[i with combining dot above]](https://www.rsc.org/images/entities/char_0069_0307.gif) rg
rg![[i with combining dot above]](https://www.rsc.org/images/entities/char_0069_0307.gif) n Sağın, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS PubMed.
n Sağın, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS PubMed. - Y. Y. Tseng and J. S. Boehm, Curr. Opin. Genet. Dev., 2019, 54, 33–40 CrossRef CAS PubMed.
- Y. Yang, Y. Fu, H. Su, L. Mao and M. Chen, Biosens. Bioelectron., 2018, 122, 175–182 CrossRef CAS PubMed.
- J. Li, J. Wang, Y. Wang and M. Trau, Analyst, 2017, 142, 4788–4793 RSC.
- M. Jia, Y. Mao, C. Wu, S. Wang and H. Zhang, Anal. Chim. Acta, 2019, 1082, 136–145 CrossRef CAS PubMed.
- T. Yu, H. Zhang, Z. Huang, Z. Luo, N. Huang, S. Ding and W. Feng, 2017, 29, 828-34, Electroanalysis, 2017, 29, 828–834 CrossRef CAS.
- S. Singh, N. Arshid and S. Cinti, Biosens. Bioelectron., 2022, 216, 114635 CrossRef CAS PubMed.
- S. H. Jalalian, M. Ramezani, S. A. Jalalian, K. Abnous and S. M. Taghdisi, Anal. Biochem., 2019, 571 Search PubMed.
- C. F. Ning, L. Wang, Y. F. Tian, B. C. Yin and B. C. Ye, Analyst, 2020, 145, 2795–2804 RSC.
- T. Suwatthanarak, I. A. Thiodorus, M. Tanaka, T. Shimada, D. Takeshita, T. Yasui, Y. Baba and M. Okochi, Lab Chip, 2021, 21, 597–607 RSC.
- P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Chem. Commun., 2019, 55 Search PubMed.
- S. Mishra, D. Saadat, O. Kwon, Y. Lee, W. S. Choi, J. H. Kim and W. H. Yeo, Biosens. Bioelectron., 2016, 81 Search PubMed.
- S. Principe, A. B. Y. Hui, J. Bruce, A. Sinha, F. F. Liu and T. Kislinger, Proteomics, 2013, 13 Search PubMed.
- L. Zhao, H. Wang, J. Fu, X. Wu, X. Ye Liang, X. Yu Liu, X. Wu, L. Liang Cao, Z. Yu Xu and M. Dong, Biosens. Bioelectron., 2022, 214, 114487 CrossRef CAS PubMed.
- M. Oshi, V. Murthy, H. Takahashi, M. Huyser, M. Okano, Y. Tokumaru, O. M. Rashid, R. Matsuyama, I. Endo and K. Takabe, Cancers, 2021, 13 Search PubMed.
- Y. Wang, Z. Li, Q. Lin, Y. Wei, J. Wang, Y. Li, R. Yang and Q. Yuan, ACS Sens., 2019, 4, 2124–2130 CrossRef CAS PubMed.
- https://www.cancer.gov/about-cancer/understanding/what-is-cancer .
- M. Rodrigues, I. Andrade and R. Cruz, Eur. J. Public Health, 2020, 30, ii11–ii12 Search PubMed.
- B. Trujillo, A. Wu, D. Wetterskog and G. Attard, Br. J. Cancer, 2022, 127, 1394–1402 CrossRef CAS PubMed.
- A. T. Singh, D. Lantigua, A. Meka, S. Taing, M. Pandher and G. Camci-Unal, Sensors, 2018, 18 Search PubMed.
- M. A. Khan and M. Mujahid, Sensors, 2020, 20 Search PubMed.
- https://www.fortunebusinessinsights.com/industry-reports/point-of-care-diagnostics-market-101072 .
- D. Bury, P. L. Martin-Hirsch, F. L. Martin and T. P. Dawson, Lancet, 2017, 390 Search PubMed.
- M. Heidary, M. Auer, P. Ulz, E. Heitzer, E. Petru, C. Gasch, S. Riethdorf, O. Mauermann, I. Lafer, G. Pristauz, S. Lax, K. Pantel, J. B. Geigl and M. R. Speicher, Breast Cancer Res., 2014, 16, 1–10 CrossRef.
- A. A. Davis, Q. Zhang, L. Gerratana, A. N. Shah, Y. Zhan, W. Qiang, B. S. Finkelman, L. Flaum, A. Behdad, W. J. Gradishar, L. C. Platanias and M. Cristofanilli, Breast Cancer Res., 2019, 21, 1–8 CrossRef PubMed.
- S. Mader and K. Pantel, Oncol. Res. Treat., 2017, 40 Search PubMed.
- S. Chang, J. Y. Hur, Y. La Choi, C. H. Lee and W. S. Kim, J. Pathol. Transl. Med., 2020, 54, 204–212 CrossRef PubMed.
- L. Muinelo-Romay, C. Casas-Arozamena and M. Abal, Int. J. Mol. Sci., 2018, 19 Search PubMed.
- I. Heidrich, L. Ačkar, P. Mossahebi Mohammadi and K. Pantel, Int. J. Cancer, 2021, 148 Search PubMed.
- D. Wong, S. Moturi, V. Angkachatchai, R. Mueller, G. DeSantis, D. van den Boom and M. Ehrich, Clin. Biochem., 2013, 46, 1099–1104 CrossRef CAS PubMed.
- A. Yilmaz, F. Ece, B. Bayramgürler, E. Akkaya and R. Baran, Respir. Med., 2001, 95, 666–669 CrossRef CAS PubMed.
- A. Tadimety, A. Syed, Y. Nie, C. R. Long, K. M. Kready and J. X. J. Zhang, Integr. Biol., 2017, 9 Search PubMed.
- N. L. Anderson and N. G. Anderson, Mol. Cell. Proteomics, 2002, 1 Search PubMed.
- R. Bhawal, A. L. Oberg, S. Zhang and M. Kohli, Cancers, 2020, 12 Search PubMed.
- I. Baccelli, A. Schneeweiss, S. Riethdorf, A. Stenzinger, A. Schillert, V. Vogel, C. Klein, M. Saini, T. Bäuerle, M. Wallwiener, T. Holland-Letz, T. Höfner, M. Sprick, M. Scharpff, F. Marmé, H. P. Sinn, K. Pantel, W. Weichert and A. Trumpp, Nat. Biotechnol., 2013, 31, 539–544 CrossRef CAS PubMed.
- I. Garcia-Murillas, G. Schiavon, B. Weigelt, C. Ng, S. Hrebien, R. J. Cutts, M. Cheang, P. Osin, A. Nerurkar, I. Kozarewa, J. A. Garrido, M. Dowsett, J. S. Reis-Filho, I. E. Smith and N. C. Turner, Sci. Transl. Med., 2015, 7, 302ra133 Search PubMed.
- B. K. Thakur, H. Zhang, A. Becker, I. Matei, Y. Huang, B. Costa-Silva, Y. Zheng, A. Hoshino, H. Brazier, J. Xiang, C. Williams, R. Rodriguez-Barrueco, J. M. Silva, W. Zhang, S. Hearn, O. Elemento, N. Paknejad, K. Manova-Todorova, K. Welte, J. Bromberg, H. Peinado and D. Lyden, Cell Res., 2014, 24 Search PubMed.
- Y. Jin, K. Chen, Z. Wang, Y. Wang, J. Liu, L. Lin, Y. Shao, L. Gao, H. Yin, C. Cui, Z. Tan, L. Liu, C. Zhao, G. Zhang, R. Jia, L. Du, Y. Chen, R. Liu, J. Xu, X. Hu and Y. Wang, BMC Cancer, 2016, 16, 1–9 CrossRef PubMed.
- N. Soda, K. Clack and M. J. A. Shiddiky, Sens. Diagn., 2022, 1, 343–375 RSC.
- A. R. Thierry, S. El Messaoudi, P. B. Gahan, P. Anker and M. Stroun, Cancer Metastasis Rev., 2016, 35, 347–376 CrossRef CAS PubMed.
- Q. Huang, Y. Wang, X. Chen, Y. Wang, Z. Li, S. Du, L. Wang and S. Chen, NANO, 2018, 2 Search PubMed.
- S. Li, M. Yi, B. Dong, X. Tan, S. Luo and K. Wu, Int. J. Cancer, 2021, 148, 2640–2651 CrossRef CAS PubMed.
- K. W. Witwer, E. I. Buzás, L. T. Bemis, A. Bora, C. Lässer, J. Lötvall, E. N. Nolte-’t Hoen, M. G. Piper, S. Sivaraman, J. Skog, C. Théry, M. H. Wauben and F. Hochberg, J. Extracell. Vesicles, 2013, 2, 20360 CrossRef PubMed.
- L. Gorgannezhad, M. Umer, M. N. Islam, N. T. Nguyen and M. J. A. Shiddiky, Lab Chip, 2018, 18 Search PubMed.
- J. D. Merker, G. R. Oxnard, C. Compton, M. Diehn, P. Hurley, A. J. Lazar, N. Lindeman, C. M. Lockwood, A. J. Rai, R. L. Schilsky, A. M. Tsimberidou, P. Vasalos, B. L. Billman, T. K. Oliver, S. S. Bruinooge, D. F. Hayes and N. C. Turner, J. Clin. Oncol., 2018, 36 Search PubMed.
- C. Rolfo, A. F. Cardona, M. Cristofanilli, L. Paz-Ares, J. J. Diaz Mochon, I. Duran, L. E. Raez, A. Russo, J. A. Lorente, U. Malapelle, I. Gil-Bazo, E. Jantus-Lewintre, P. Pauwels, T. Mok and M. J. Serrano, Crit. Rev. Oncol. Hematol., 2020, 151 Search PubMed.
- F. M. Goodsaid, Clin. Transl. Sci., 2019, 12 Search PubMed.
- P. Narayan, S. Ghosh, R. Philip, J. C. Barrett, R. T. McCormack, J. I. Odegaard, G. R. Oxnard, L. J. Pracht, P. M. Williams, G. J. Kelloff and J. A. Beaver, Oncologist, 2020, 25, 730–732 CrossRef PubMed.
- S. A. Boppart and R. Richards-Kortum, Sci. Transl. Med., 2014, 6 Search PubMed.
- L. Liu, X. Chen, O. O. Petinrin, W. Zhang, S. Rahaman, Z. R. Tang and K. C. Wong, Life, 2021, 1, 638 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |
