 Open Access Article
Open Access ArticleSimplifying the complex: accessible microfluidic solutions for contemporary processes within in vitro diagnostics
Nathan K.
Khosla†
 ,
Jake M.
Lesinski†
,
Jake M.
Lesinski†
 ,
Monika
Colombo
,
Léonard
Bezinge
,
Monika
Colombo
,
Léonard
Bezinge
 ,
Andrew J.
deMello
,
Andrew J.
deMello
 and
Daniel A.
Richards
and
Daniel A.
Richards
 *
*
Institute for Chemical and Bioengineering, ETH Zürich, Vladimir Prelog Weg 1, Zürich, 8093, Switzerland. E-mail: andrew.demello@chem.ethz.ch; daniel.richards@chem.ethz.ch
First published on 15th August 2022
Abstract
In vitro diagnostics (IVDs) form the cornerstone of modern medicine. They are routinely employed throughout the entire treatment pathway, from initial diagnosis through to prognosis, treatment planning, and post-treatment surveillance. Given the proven links between high quality diagnostic testing and overall health, ensuring broad access to IVDs has long been a focus of both researchers and medical professionals. Unfortunately, the current diagnostic paradigm relies heavily on centralized laboratories, complex and expensive equipment, and highly trained personnel. It is commonly assumed that this level of complexity is required to achieve the performance necessary for sensitive and specific disease diagnosis, and that making something affordable and accessible entails significant compromises in test performance. However, recent work in the field of microfluidics is challenging this notion. By exploiting the unique features of microfluidic systems, researchers have been able to create progressively simple devices that can perform increasingly complex diagnostic assays. This review details how microfluidic technologies are disrupting the status quo, and facilitating the development of simple, affordable, and accessible integrated IVDs. Importantly, we discuss the advantages and limitations of various approaches, and highlight the remaining challenges within the field.
1 Introduction
The impact of in vitro diagnostics (IVDs) on modern medicine is hard to overstate. Over the last century, IVDs have led the shift from primarily physician- and symptom-led diagnosis, a process hindered by associated biases and confounding factors,1,2 to a more empirical approach based on the identification of disease-specific biological and chemical markers. As advances in basic research have led to the discovery of biomarkers for an ever-expanding array of ailments, IVDs have in turn been developed to help clinicians detect such biomarkers and diagnose the presence and extent of a disease. The omnipresence of IVDs within modern medicine is perhaps the best indication of their impact – IVDs are routinely employed within almost every field of medicine, including infectious and parasitic diseases,3 oncology,4 cardiology,5 and endocrinology.6 Furthermore, for many diseases, IVDs are integral to the entire treatment pipeline, from initial diagnosis through to prognosis, treatment planning/monitoring, and post-treatment surveillance.7,8 IVDs are also routinely exploited in scientific research, and as such are an essential tool in the development of the next generation of therapeutics. This ubiquity has not gone unnoticed by pharmaceutical and biotech companies, and it is not uncommon to see companion IVD programs running alongside the development of novel drugs and treatments.9Whilst it is clear that IVDs have had a positive global impact, it is important to acknowledge and understand regional inconsistencies. Access to high quality IVDs is dictated by multiple social and geopolitical factors, though is broadly correlated to wealth.10–12 This leads to large discrepancies between populations. Given the well-established links between access to high quality diagnostics and overall health (both personal and public),12 addressing these discrepancies is a priority. To this end, there has been a conscious push within the research community to develop more accessible diagnostic technologies. Such a drive towards accessibility has taken many forms, including the development of completely novel assay formats, the simplification of existing assays through automation and miniaturization, integration with smartphones and other personal electronics, and improvements in prototyping and manufacturing techniques, to name a few. There have also been advancements outside of basic research, including improving user-interface and user-experience (UI/UX), enhancing communications infrastructure, streamlining governmental approval processes, and expanding transportation routes. All of these factors are important for improving diagnostic accessibility, and will be discussed throughout this review. From a scientific and engineering perspective, the vast majority of novel diagnostic assays have been developed through highly multidisciplinary and collaborative research programs. Diagnostics developers are continuously assessing novel chemical reporters,13 chemical/biochemical processes,14 and nano- and micro-scale materials for their capacity to act as biosensors for disease.15,16 These are then typically combined with emerging engineering tools to create increasingly integrated diagnostic platforms. Of these engineering tools, microfluidic technologies have arguably had the greatest impact.17 Microfluidic systems provide diagnostics developers unprecedented control over multiple aspects of their assays, such as sample filtration, analyte transport, mixing, heating, and even analysis through integrated optical and electrical sensors. Virtually every process required for an effective IVD can now be integrated within a chip-based platform and at increasingly miniaturized scales. Moreover, the vast majority of these processes can be automated and parallelized. Thus, the increasing dependence of IVDs on microfluidics is unsurprising.
Despite the impressive progress, challenges still remain. Whilst the majority of the processes required for diagnostic assays can now be performed within microfluidic devices, combining these complex operations into a fully integrated device/workflow remains a major challenge. Individual microfluidic processes can interfere with each other, a fact which is exacerbated when performing these processes in close proximity on a single device. Additionally, increasing chip complexity often leads to increased manufacturing complexity, which ultimately drives up costs and limits accessibility. Thus, despite their potential, most microfluidic-based diagnostic assays never make it to the market as fully-fledged IVDs. Indeed, the majority of the commercially viable examples perform more basic and established diagnostic assays such as lateral flow immunoassays (LFIAs) and polymerase-chain reaction (PCR). Though useful, these assays do not represent the state-of-the-art in terms of diagnostic potential.
The intention of this review is not necessarily to provide a comprehensive overview of the use of microfluidics in IVDs. Several excellent reviews have already been written on this subject; we would like to highlight recent articles from Berlanda et al. and Sachdeva et al. as being particularly informative.17,18 Rather, we aim to highlight paradigm-shifting advances in microfluidics that have the potential to disrupt the status quo by enabling increasingly complex assays to be performed on progressively simpler devices. We will first discuss how disruptive microfluidic technologies are being integrated into every aspect of IVD assays, from sample preparation through to data acquisition, and discuss the merits and limitations of the technologies. We will then highlight several integrated microfluidic systems capable of performing complex diagnostics assays in simplified formats. Finally, we will detail the remaining challenges within the field, and discuss the best routes forwards.
2 Processes
The assays which underpin IVDs are complex, multi-step processes that often require several chemical, biological, mechanical, and even electrical manipulations to work sequentially (or even simultaneously). In this section we will detail how microfluidics can be leveraged to implement these processes (Fig. 1).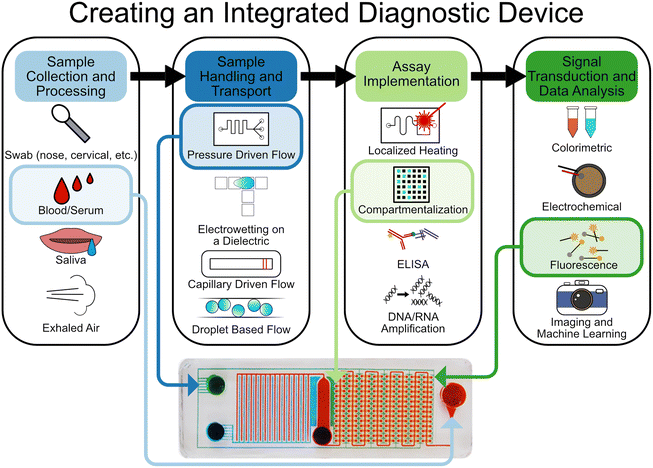 | ||
| Fig. 1 Overview and selection of the various components that contribute to forming an integrated microfluidic chip for diagnostics. Example chip: the SIMPLE chip.19 Reproduced/adapted from ref. 19 with permission from Science. | ||
2.1 Sample collection
In general, the first step in any diagnostic workflow is sample collection. It is well established that the choice of sample collection method can have a dramatic effect on the overall performance of a diagnostic assay.20,21 Despite this, traditional approaches often treat sample collection as a completely separate procedure, and give minimal thought to integration with down-stream diagnostic workflows. By enabling more efficient integration of sample collection with other diagnostic processes (such as sample processing and target detection), emerging microfluidic technologies are poised to change this modus operandi.Microfluidic approaches to sample collection can be broadly subdivided into two categories; capillary-driven and vacuum-driven, although it should be noted that researchers have recently begun to explore hydrogel-based sampling methods which rely on the swelling properties of hygroscopic materials to collect samples.22,23 Capillary-driven approaches exploit the innate properties of certain natural materials (e.g. nitrocellulose), or specifically engineered micron-scaled features and geometries (e.g. porous materials and narrow microchannels). This approach holds advantages in regard to simplicity and ease of use, as capillary action requires no external power or user input to function. Thus, it is unsurprising that capillary-driven sample collection has become the dominant strategy within the growing field of wearable sweat-based diagnostic devices. Historically, wearable sweat-based biosensors relied on naturally absorbent materials such as cotton and paper to collect sweat samples for continuous monitoring.24,25 Unfortunately, whilst fairly absorbent, these materials are not well suited for precisely transporting samples to defined locations. More recently, researchers have begun to develop wearable microfluidic devices with modern elastic materials such as silicones. These so called epidermal microfluidic systems (“Epifluidics”) were pioneered by Rogers and co-workers.26–28 Such devices are able to interface closely with human skin, and are highly efficient at sampling sweat and delivering it to a sensor. An excellent example of this was reported by Bolat et al., who fabricated a silicone-based conformal microfluidic device capable of stimulating, collecting, and analyzing sweat in an automated manner.29 The device is fully integrated and includes the supporting electronics to transmit assay results. Despite this, the technology has significant room to grow, particularly in regard to the integration of more complex fluid control for sample mixing and flow control, as well as its capacity to operate over extended periods of time.
Worthy of special mention is the growing field of aerosol collection. For diagnostic purposes, exhaled breath is the most common aerosol of interest, with target analytes typically being microbes, tumor markers and organic compounds implicated in chronic lung diseases.30,31
Historically, aerosols were collected using dedicated equipment and then analyzed offline using a variety of spectroscopic and spectrometric methods.32 More recently, researchers have begun to develop devices capable of online target detection in aerosols by embedding sensors directly into breath analyzers or wearables.33,34 These devices collect breath aerosols by wicking them via capillary action into embedded sample pads, which are integrated into microfluidic sensing assays. Research in this area has accelerated somewhat in the last two years, most likely due to the COVID-19 pandemic and the increased use of mouth coverings. Face masks provide an ideal interface for collecting aerosols, and the presence of SARS-CoV-2 virus particles in exhaled breath is well-established.35,36 Nguyen et al. recently reported a face mask-based wearable biosensor capable of detecting SARS-CoV-2 viral particles after 90 minutes of continuous mask use (Fig. 2). The device is fully integrated with sample collection, transport, lysis, and target detection being performed on the mask.37 Although the field is relatively nascent, initial results are undeniably positive.
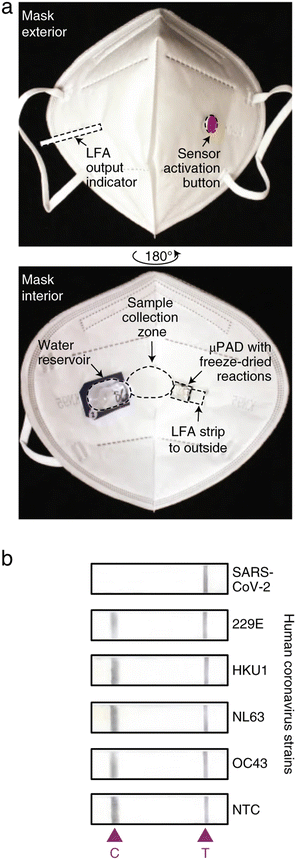 | ||
| Fig. 2 Image of lateral flow assay in a mask for aerosol sample collection. (a) Photos of a SARA-CoV-2 sensor in a mask. (b) Representative images of the LFA outputs for various human coronovirus strains. Images adapted with permission from ref. 41. Copyright © (2021) Nature Publishing Group. | ||
Whilst capillary flow is the standard approach for sweat- and breath-based sampling, where the specimen is fully aqueous and of low viscosity, the limitations of such a passive approach has hindered the efficient sampling of other specimen types (e.g. blood and saliva). In these situations, active sampling methods are required. Vacuum-driven approaches to sample collection rely on pressure differentials to drive the flow of a sample from the source to the device. This has traditionally been achieved by pre-evacuation of specific areas of the device, which are connected to the sample drawing mechanism by breakable barriers. Once the device is interfaced with the patient (for example using a microneedle), the barrier can be broken, and the sample driven in the direction of the vacuum. This approach has been iteratively improved upon over several decades. These improvements have focused on increasing blood draw volumes, introducing automation, simplifying device operation, and decreasing patient discomfort.38 An excellent example of this is the “TAP” device developed by Blicharz and coworkers, which uses an array of microneedles and a vacuum-driven draw to painlessly draw 0.1 ml of capillary blood from a patient at the push of a button.39 Although studies have shown that the blood collected by the TAP device is suitable for down-stream analysis, this step has not yet been integrated and must be performed offline. One limitation of pre-evacuated chambers is the loss of vacuum over time, rendering the device inoperable after extended periods of storage. To overcome this limitation, researchers are now exploring positive pressure generated in situ by gas-generating chemical reactions, and how this can be used to manipulate liquids within microfluidic chips.40 The next step in this process is to combine such in situ pressure generating methods into integrated devices able to sample blood directly from a patient.
2.2 Sample processing
After a patient sample has been collected, it is almost always necessary to perform some degree of sample purification and preparation to isolate the target analyte from the sample matrix. These processes are used to enrich the analyte and remove species that might inhibit downstream chemistry and hinder test performance. Historically, sample purification has normally been performed off-chip, using affinity resins on solid supports such as magnetic beads,42,43 or chromatography columns.44–46 For more complex samples, such as whole blood, a centrifugation step is often employed to separate the target from the matrix.47–50A great deal of effort has been dedicated to moving these processes to microfluidic formats. One such example is the affinity-based cell capture presented by Smejkal, et al.51 Here, microfluidic channels were coated with various recombinant protein binders that target surface receptors on cells. Cells were incubated in the channel and then washed out, with captured cells remaining bound to the surface for future analysis. Such a technique allows for cellular purification and subsequent imaging within a microfluidic device. Alternative methods of sample purification also include on-device centrifugal processes as well as novel microfluidic geometries which promote sample separation.19,52,53 Ramachandran et al. recently reported a novel electrical sample enrichment method that utilizes electric fields to perform isotachophoresis (ITP). The utility of the device was showcased by extracting and concentrating RNA from raw nasopharyngeal swabs.54 Here, the target is focused with two buffers, a high ionic mobility leading electrolyte and a low mobility trailing electrolyte. Upon application of an electric field the ions in the leading electrolyte outrun the RNA analyte ions, while the analyte moves faster than the trailing electrolyte ions. This serves to focus the analyte in an region as thin as 10 μm at the buffer interface. The RNA is subsequently utilized within a quantitative reverse transcription loop-mediated isothermal amplification (qRT-LAMP)/Cas12 CRISPR assay. Significantly, this system takes approximately 35 minutes from sample input to answer and is fully automated. However, operating the device does require the delicate tuning of electric fields, which raises questions over robustness.
Beyond simple purification, cell lysis is often used to make target analytes more available for downstream detection. Lysis is most commonly achieved chemically, by incubating the sample with specific reagents and buffers known to disrupt the cell membrane.55 This is typically done manually by a technician, and is thus susceptible to human error. A simple way of lysing cells within microfluidic devices is to directly integrate chemical lysis chambers. The sample is introduced into the lysis chamber, and mixed with lysis reagents to release the target material. Such an approach has also been used to lyse cells directly inside of droplets generated microfluidically.56,57 One limitation of chemical lysis is the requirement for liquid buffers and reagents, which increases costs and complicates device portability.
To avoid the use of liquid reagents, a range of mechanical lysis techniques have been developed. These techniques rely on breaking cell membranes through physical puncturing or mechanical homogenization through exposure to high shear forces.58–60 One recent example of note by Huang et al. uses a series of single cell channel restrictions with sharp edges to induce cell lysis.61 Cells are serially passed through eight silicon constrictions of approximately 10 μm width (placed in a significantly wider channel) with fluid flow speeds in excess of 10 m s−1, forcing membrane rupture. Through this process the authors achieve DNA yields that are statistically similar to a standard chemical lysis method. A more in-depth survey of mechanical lysing techniques can be found in the recent review article by Grigorov et al.62
2.3 Sample handling and transport
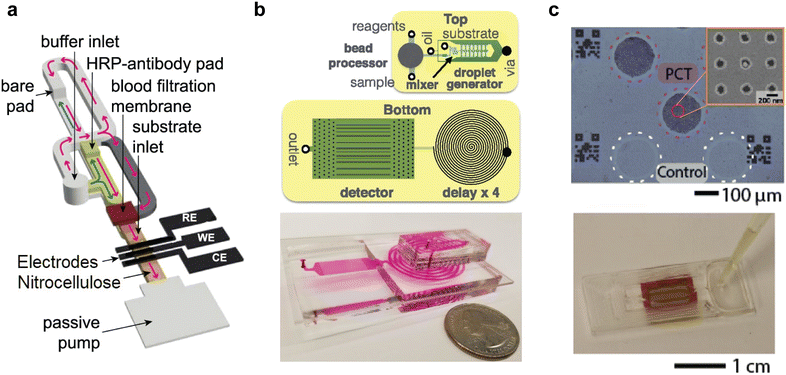
A key part of any integrated microfluidic system is the fluid handling architecture and transport system. Depending on the chemistry/biology being performed, fluid metering, particle handling, mixing and compartmentalization must be performed, and often with high precision. Multiple methods for fluid handling exist, including various pressure-driven approaches, such as segmented flows, electrowetting-on-a-dielectric (EWOD) and capillary-driven flow. Each of these locomotion methods has distinct advantages and considerations, which we will discuss here.Standard pressure-driven approaches, which employ external pumps, are cheap, ubiquitous, and robust.63,64 A good example of the use of pressure-driven flows was reported by Xu et al., who used pressure-driven flows to focus and isolate circulating tumor cells (CTCs) from whole blood prior to single cell whole exome sequencing.65 A major limitation associated with externally-driven systems is that the pumps are typically large and expensive, with the performance of the dependent assay often being heavily impacted by the performance of the pumps.66,67 Beyond these externally equipped pressure-driven systems, several techniques, such as the use of pre-pressurized chambers and hand-driven flows,68,69 have shown promise as tools for self-contained pressure-driven flows.70 Hand-driven flow systems exploit pressure gradients that result from a user applied force, typically upon a flexible membrane.71 Glynn et al. utilized finger actuation to drive fluidic transport in their point-of-care test for HIV.72 The system is both elegant and simple, utilizing a single depth channel and monolithic chip structure for ease of manufacture. Pressure is introduced to the system via manipulation of an elastomeric membrane, which can easily be done by hand. Unfortunately, whilst the device integrates all the necessary chemistry and fluidic/particle handling to perform the assay, an external bright field microscope is required for signal detection. In a similar vein, Reboud et al. utilized hand manipulation to drive a paper-based multiplexed LAMP assay for detecting malaria at the point-of-care.73 Though finger-actuated pressure-driven devices are operationally simple, they are also limited with regards to assay complexity, and questions remain regarding their robustness.
Several integrated diagnostic devices achieve fluidic locomotion via EWOD. In EWOD, droplets containing sample, reagent, buffer are spatially motivated through the application of an electric field.74 In a typical setup, the driving force behind fluid flow is electrostatic, driven by a matrix of on-chip electrodes.74 By actuating individual electrodes in a particular spatial/temporal pattern, droplets can be directed.75 EWOD platforms can easily be automated to perform complex fluidic operations.76,77 Importantly, electrodes can also be used to detect the presence or absence of fluid during an assay, effectively serving as an in-built quality control.78 However, since the movement of a droplet depends on its resistance, and therefore ionic concentration, the specific EWOD layout and applied power must be optimized for each assay/running buffer combination.79,80 Furthermore, traditional EWOD-based chips require complex electrode structures to operate, increasing manufacturing complexity, and typically rely on external power supplies for fluidic actuation.81,82 However, recent developments have begun to address these issues. Wang et al. used mechanical stimuli-controlled EWOD powered by a triboelectric nanogenerator to circumvent need for bulky external power supplies.83 Similarly, Peng et al. utilized piezo-elements and finger actuation to control and power an EWOD-based microfluidic circuit (Fig. 3).84 Both devices demonstrate progress toward more portable EWOD systems.
 | ||
| Fig. 3 Finger-actuated EWOD transport of a water droplet where actuation voltage pulses were provided by bending a series of piezoelectric elements. Reproduced/adapted from ref. 84 with permission from the Royal Society of Chemistry. | ||
Capillary-driven systems exploit specific material, channel geometries and features (such as porosity) to maximize capillary forces and direct the flow of liquids. Capillary-driven flow circumvents the need for pumps and is thus commonly employed when developing diagnostic devices for point-of-care applications. Nitrocellulose is the most commonly employed substrate for capillary-driven microfluidics, and is ubiquitous in two/three-dimensional lateral flow microfluidic systems, as well as vertical flow architectures.85–87 Unfortunately, due to the fact paper is difficult to structure into complex geometries (such as micron-sized channels and valves) paper-based analytical devices (μPADs) have historically been employed only for relatively simple assays. However, this is beginning to change. For example, Sun et al. reported a paper-based circuit that integrates cell lysis, molecular recognition, amplification and visual detection on a single device.88 Similarly, Xue et al. utilized an origami μPAD to perform cell lysis, DNA extraction, and a terminal deoxynucleotidyl transferase nick end labeling (TUNEL) assay.89 These origami μPADs integrate several steps within a compact paper-based circuit, though do require operator handling/folding and are reliant on external imaging equipment.
Beyond the use of paper, silicon and glass have been used to form micron-scale features and create more complex microfluidic circuits whilst maintaining capillary flow.90,91 For example, Hemmig et al. used silicon microfluidic circuits to create an immunoassay for the cardiac marker troponin I. This device exploits self-coalescing capillary flows to reconstitute the dried down reagents (capture antibodies, fluorescently tagged detection antibodies) and perform a bead-based immunoassay. The beads are subsequently captured in a channel and imaged.92 The operation of the device perfectly mimics that of a paper-based lateral flow immunoassay (LFIA), but the use of silicon rather than paper opens up the possibility for more extensive customization and engineering. That being said, it is worth noting that this approach relies on the use of an external camera, microscope and computer detection system. This is a disadvantage that should be addressed in future iterations of the technology.
2.4 Assay implementation
At the core of any microfluidic IVD is the assay that links the presence of a particular disease analyte to an interpretable signal. Although the fundamental chemistry/biology that governs microfluidic-based assays is often assumed to be the determining factor in assay performance, of equal importance is the supporting microfluidic architecture. In addition to facilitating automated handling of liquid samples, reagents, and buffers (see section 2.2), unique characteristics of microfludics can also be leveraged to fundamentally improve the performance of an assay. These improvements can arise from multiple factors, including more efficient heat transfer, and precise compartmentalization for ultra-sensitive detection and assay multiplexing.Efficient heat transfer in microfluidic systems is a direct result of the low-volume nature of microfluidics. Put simply, heat energy is transferred much more rapidly through small volumes as compared to larger volumes. This can be readily exploited for temperature-dependent assays (such as PCR, as discussed in section 3.2). Heating in microfluidic devices is typically achieved using external heating elements such as metal rods/blocks or laboratory hotplates and Peltiers,93 though this can cause problems when precise zonal heating or a low device footprint is desired. Another common approach is to integrate micro-Peltiers directly into devices,94,95 though this requires relative complex fabrication processes and external electrical equipment.
More elegant solutions have been developed which allow for precise zonal temperature control using features that can be integrated into the microfluidic chip. One such approach is to leverage Joule heating, which exploits the relationship between supplied potential and produced heat within conductive solids or liquids.96,97 By carefully placing a conductive material in close proximity to the zone of interest, it is possible to precisely control the local temperature. One limitation of Joule heating is the need to place additional channels or materials into the chips to facilitate the heating material. This can preclude the use of very small microfluidic chips or certain geometries. One way to circumvent this issue is through the use of electromagnetic radiation, such as infrared lasers, to induce localised heating.98 These approaches require no internal modifications to the microfluidic chip itself and are capable of rapid and precise heating.99 Several groups have reported on using laser-induced heating for droplet PCR, demonstrating the excellent localised control that can be achieved using these methods.100,101 Though electromagnetic approaches do rely on external components (such as lasers), these are themselves becoming increasingly miniaturized,102,103 and we anticipate that integration within microfluidic-based IVDs will become increasingly common.
The ability to segregate samples into micro, nano, pico, and even femtolitre volumes is frequently leveraged within microfluidic IVDs to improve performance and facilitate assay multiplexing and parallelization.104–107 Digital biosensing assays rely on compartmentalization to obtain binary ON/OFF signals – this feature grants digital assays substantially improved sensitivity and lower detection limits when compared to their analogue counterparts.108 Droplet digital PCR (ddPCR) and digital immunoassays are good examples of compartmentalization being used to significantly improve sensitivity and limit of detection (LoD), even down to the single molecule level.109,110 Compartmentalisation in microfluidic systems is commonly achieved using droplet systems, in which two immiscible phases, most commonly an aqueous solution and an oil, are mixed to form stable droplets.109 The vast majority of droplet generators rely on a specific microfluidic geometries, such as a T-junctions or narrow flow-focusing channels, to generate droplets as a function of channel width, flow rate, and fluid viscosity.111 Since the performance of these units is highly dependent on the flowrate, it is generally necessary to use bulky and expensive pumps which can precisely modulate the pressure. This massively increases the footprint of devices that rely on these droplet generators, negating many of the miniaturization benefits of microfluidics.
Realising the need for more accessible compartmentalisation methods, researchers have begun to develop droplet generators that can operate with minimal, or even no, external equipment. These systems typically rely on some form of step-emulsification, in which the dispersed phase (commonly aqueous) is flowed through rectangular channels directly into the continuous (commonly oil) phase.112 Because this process relies on interfacial tension between the two phases, rather than shear forces, to generate droplets, it is relatively invariant to flow rate.113 Thus, monodisperse droplet populations can be generated without the need for precise pumps – this is a significant boon for simple, integrated devices. Yuan et al. recently exploited step-emulsification to develop a hand-powered droplet-based LAMP assay for detecting several strains of pathogenic bacteria.114 By combining step-emulsification with a hand-powered syringe to drive flow, the group were able to do away with the majority of the equipment typically required for droplet-based assays. Though the detection aspect of the assay still required a relatively complex fluorescence microscope, this work is certainly a step in the right direction. One limitation of step emulsification compared to traditional methods is flexibility. In step emulsification, droplet diameters are determined predominately by the chip design, and cannot be altered through variation of the flow rate. This means that new devices must be redesigned and manufactured whenever a new droplet diameter is required. However, since most chips are created bespoke for their desired application, this factor is not overly limiting.
Assay parallelization is a process by which multiple assays are performed simultaneously. This powerful approach can be leveraged to multiplex several complementary assays on a single sample (to improve specificity or identify co-morbidities), run multiple samples against a single target (to improve throughput), or a combination of both. In any case, parallelization can dramatically increase the amount of information gleaned from samples. Unfortunately, the sheer volume of sample and reagents required for highly parallel assays can be limiting when relying on traditional plate-based assays. This is where the low-volume nature of microfluidics can be exploited to great effect. Historically, parallelization in microfluidics typically involved simply operating multiple chips in parallel i.e. scaling-out. This becomes impractical when massive parallelization (e.g. hundreds, thousands) is required.
In these instances it is more desirable to split and compartmentalize a sample within spatially separated regions of a single chip where different assays can be performed. Though this idea is not new, continuous advances in microfabrication and microfluidic control methods are facilitating the development of progressively larger and more complex arrays. An excellent example of the potential of microfluidics for generating massively multiplexed assays has been shown by Maerkl et al. The group have developed a process termed mechanically induced trapping of molecular interactions (MITOMI) to compartmentalize samples in areas patterned with distinct target-specific capture ligands.115 After incubation and capture of the targets within the samples, fluorescently-tagged antibodies are introduced to facilitate target detection. To date, the group have utilized MITOMI to develop highly-automated multiplexed arrays against multiple targets,116–118 and have shown that the method is capable of assessing up to 1024 clinical samples against 4 different biomarkers (4096 measurements) on a single chip using as little as 5 nanolitres of each sample (Fig. 4).119 One drawback of MITOMI-based devices is their complexity – their operation is made possible by a relatively complicated array of microfluidic channels, valves, and buttons. Thus, large-scale device manufacture is likely to be challenging and expensive. However, due to the incredibly low sample and reagent volumes required to achieve results, it is reasonable to assume that operational costs would be significantly decreased compared to conventional methods.
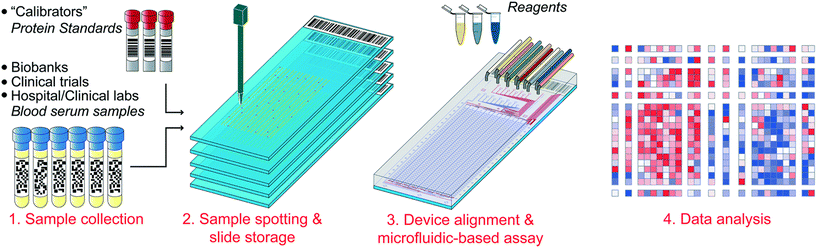 | ||
| Fig. 4 An illustration of the workflow for the parallel immunoassay. Clinical samples are spotted with a microarray robot on the same array as protein standards. The microfluidic device is aligned to the array and proper tubing is connected. The chip is then run the next day and the subsequent data is be analyzed. Reproduced/adapted from ref. 119 with permission from the Royal Society of Chemistry. | ||
2.5 Signal transduction and data acquisition
Following the generation of a signal from a chemical/biochemical assay, transduction into an analytically useful data form is generally necessary. This crucial step allows users to qualify, quantify and compare analytical signals and ultimately gain a better understanding of the presence and/or extent of disease.Given the prevalence of optical detection methods (notably, those based on colorimetric and fluorescence signals) within microfluidic-based IVDs, methods for optical transduction are by far the most developed.14,120,121 Optical detection is commonly performed using off-chip photodetectors, such as complementary metal oxide semiconductor (CMOS) cameras and photodiodes, to convert emitted or transmitted photons into an electrical signal. Since these can often be expensive and bulky, the development of fully integrated systems is somewhat challenging. Accordingly, developers have begun to take advantage of smartphone cameras and electronics, which can be more easily integrated into IVDs.122 The significant processing power, high-resolution cameras, and inbuilt connectivity make such systems ideal for developing fully integrated devices.123,124 Zhang et al. recently showed how to effectively integrate smartphone detection into a microfluidic IVD for measuring hemoglobin.125 Specifically, the researchers developed a quantum dot-based fluorescence assay for hemoglobin, and combined this with an integrated UV irradiation source and a commercial smartphone to detect the fluorescent signal. Due to their ubiquity within modern society, smartphones provide a simple and accessible method for detecting optical signals. However, their use also comes with multiple drawbacks. Smartphones are not standardized, and incorporate vastly different hardware (including cameras) and software components. This can lead to inconsistent performance between smartphone models, and can impact diagnostic accuracy.126 Equally, the power that smartphone developers have over their systems raises concerns over long-term support for smartphone-dependent IVDs, and the security of the data that is produced. Realizing this, researchers have now begun to migrate towards more open-source computing solutions, using Arduino and Raspberry Pi-based components.127 For example, Hambalek et al. developed a microfluidic device for detecting DNA methylation using a fluorescence detector connected to a Raspberry Pi computer.128 In this case, the bioassay was based around LAMP, with the fully integrated device being able to interface with a standard 96-well plate. Unfortunately, whilst the majority of device components are cheap to produce, the reliance on expensive optical filters significantly increases manufacturing costs.
The most common alternative to optical detection is electrochemical detection, which employs electrodes to transduce a change in current, potential or impedance triggered by chemical/biochemical reactions into a readable signal.120,129 When combined with microfluidic technologies, electrodes provide an attractive signal transduction pathway due to their amenability to miniaturization and ability to interface with programmable digital platforms.130 With the broad variety of microfluidic platforms (of various size, shape and material), there is no one-size-fits-all method for integrating electrodes into microfluidic substrates. Traditionally, microfabrication techniques based on photolithography have been employed for the fabrication of metal electrodes.131 Today, the trend has shifted towards low-cost and scalable alternatives, such as screen-printing. Methods for screen-printing electrodes are versatile and can be easily integrated into various materials, from polydimethylsiloxane through to nitrocellulose paper.132 That said, the performance of screen-printed electrodes remains inferior to metal electrodes,133 and since fabrication relies on a stencil, this limits its utility in rapid prototyping.134 Recently, Klunder et al. reported the use of thermoplastic electrodes in a variety of microfluidic platforms.135 These electrodes perform comparably to state-of-the-art electrochemical systems, and can be patterned using a plastic mold fabricated via laser engraving. This method has since been used to perform complex diagnostic assays, (e.g. antibody, bacteria or small molecule detection) on simple paper-based or 3D-printed microfluidic devices.136–138
Measuring electrochemical signals is often achieved using bulky and expensive potentiostats, though there are now commercial options for portable handheld devices (such as PalmSens Sensit).139 Still, their costs (>500 USD) and reliance on proprietary software remain prohibitive for many applications, particularly in resource-limited settings. With that in mind, Ainla et al. developed a universal wireless electrochemical detector (UWED), an ultra-portable potentiostat based on open-source software and consisting of low-cost and accessible electronic components.140,141 The resulting device costs less than 15 USD and is able to perform common electrochemical measurements with results comparable to that of larger benchtop potentiostats. Furthermore, the device is able to interface with computers and smartphones via Bluetooth® to wirelessly transmit data in real-time. Whilst the authors of this study acknowledge the limitations of their device in regard to operational range and accuracy, the advantages in terms of accessibility and simplicity are significant.
3 Integrated systems
Though developing highly miniaturized microfluidic solutions to individual assay steps is non-trivial, an even greater challenge is ensuring these processes reliably work in concert within a single device. In this section we will detail several devices which achieve full or partial integration of the previously detailed steps, and how these devices are leveraged to perform common diagnostic assays.3.1 Immunoassays
Protein biomarkers are one of the largest classes of diagnostic target across a broad range of diseases. Accordingly, assays that are adept at detecting proteins remain a crucial aspect of modern diagnostics. Within this class of IVDs, immunoassays are by far the most common. Immunoassays provide a wealth of information about both the presence of a disease (e.g. via antigen testing), and the bodily response to disease (e.g. via antibody testing). This makes them particularly useful for diagnosing infectious diseases, since they provide important information about an individual's current (antigen) and past (antibody) infection status. This information is absolutely essential for effective track–trace–treat pathways. Typically, an immunoassay workflow involves a capture step, followed by a labelling and signal amplification step prior to signal transduction and analysis. Each of these steps is commonly separated by a washing step or, in flow-based systems, the displacement of the carrier fluid to remove unbound molecules. Ideally, these steps are performed with no user input, so as to reduce operational variability and prevent contamination. Whist standard laboratory detection of proteins is performed using enzyme-linked immunosorbent assays (ELISA), this general method requires relatively complex lab equipment and specialized training. Over the last three decades, engineers have developed a diverse array of technologies for miniaturizing and simplifying immunoassays. The primary goal of this activity has been to develop fully integrated systems that could be operated by untrained personnel, or even the end-user themselves, i.e. at the point-of-care.145Within an abundance of techniques, paper-based LFIAs have undoubtedly emerged as the most popular PoC immunoassay format. LFIAs have become synonymous with rapid testing due to their simple format and ability to screen for the presence of antigens in a timely manner. As such, they are frequently employed as a first line of defence during infectious disease epidemics.146 This use-case is perfectly exemplified by the ubiquity of LFIA rapid testing during the COVID-19 pandemic. Despite clear advantages, the vast majority of contemporary LFIAs are still fundamentally hamstrung by the same issues as earlier iterations, i.e. low clinical sensitivities and difficulties in multiplexing.147 Recent advances in microfluidic engineering have focused on alleviating such issues without compromising the low-cost and simplicity of the format. This has required a considerable amount of creativity, but emerging tests are now able to achieve sensitivities comparable to standard lab-based tests, and even multiplexed across several disease targets.148 In paper-based immunoassays, most attention has focused on improving limits of detection, as most current solutions are unable to detect their targets at titres commonly present during the early stages of disease. This is achieved either through the use of sensitive labels (e.g. fluorescent tags)149 or through the addition of an amplification step.148,150 Integration of an amplification step, using enzymes14 or nanomaterials,151 is possible, but typically requires the introduction of additional reagents and assay steps.
Samper et al. recently showcased an integrated paper-based electrochemical device for serological testing (Fig. 5a).142 The device includes an enzymatic amplification step, effectively an in-flow ELISA, and operates through a multilayer design to achieve passive reagent delivery. The device first processes undiluted blood samples by passing them through a blood-filtration membrane. This is followed by the sequential addition of rinsing buffers and amplification reagents. In combination with electrochemical readout on a smartphone, the device enables SARS-CoV-2 immunoglobulin (Ig) G antibody quantification at a clinically relevant level using just 10 μl of whole blood. This work demonstrates that complex assays, such as ELISA, can be integrated into a low-cost paper-based portable device that can be operated with minimal user input.
 | ||
| Fig. 5 Illustrative examples of complex immunoassays performed on simple chips: (a) a capillary-driven electrochemical ELISA on a multi-layer paper device. Images adapted with permission from ref. 142. Copyright © (2021) American Chemical Society. (b) A droplet digital ELISA chip with low-cost components and high multiplexing capabilities. Images adapted with permission from ref. 143. Copyright © (2019) National Academy of Sciences. (c) A plasmonic microarray for digital immunodetection with colorimetric readout. Reproduced/adapted from ref. 144 with permission from John Wiley and Sons. | ||
Whilst paper-based LFIAs remain the most popular PoC immunoassay format, and recent advances have helped to remedy their disadvantages, multiple drawbacks remain. Paper is inflexible, and cannot be readily shaped into complex geometries. This limits paper as a substrate if more advanced microfluidic processes (such as valving to control flow) are required. Paper is also prone to fouling and blockage by more complex sample matrices (such as blood and saliva), and has an innate fluorescence that can create unwanted background noise.149 To circumvent these issues, it is evident that attention should be directed towards alternative materials that retain the accessibility of paper by mimicking its wicking properties, but allow more flexibility in terms of design and incorporation of microfluidic components.92
To perform more complex or versatile immunoassays, researchers commonly employ chip-based microfluidic systems. Of these chip-based technologies, droplet-based microfluidic systems offer particular promise for ultra-sensitive immunoassays.152,153 By using large numbers of picoliter volume droplets, signals can be quantified digitally (see section 2.4) thus allowing ultrasensitive and/or highly multiplexed detection of antigens or antibodies.154
Yelleswarapu et al. developed a low-cost PDMS-based device for droplet-based digital ELISA.143 This was achieved by combining several steps into a workflow for processing and analyzing undiluted serum samples (Fig. 5b). First, beads functionalized with antibodies were used in combination with a semi-permeable membrane to pre-concentrate and tag target proteins with an enzyme immunocomplex. Next, the sample flow was segmented at a droplet generator, the generated droplets subsequently incubated on-chip, and finally split into a highly parallelized detection zone that enables high-throughput fluorescence imaging with a smartphone (at a rate up to 1 million droplets per second). The microfluidic assay is able to detect target down to the attomolar level (a 1000-fold improvement over benchtop ELISA) at a cost of $5 per test and with an outlay of $500 for instrumentation. Furthermore, due to the segmented-flow nature of their approach, several samples can be run sequentially on a single chip.
Plasmonic microarrays offer another interesting platform for performing multiplexed or digital assays.155 These devices exploit the changes in optical properties that occur when disease targets interact with the plasmonic surface, often via a bound capture antibody. Belushkin et al. utilized a plasmonic microarray in their microfluidic chip for detecting sepsis biomarkers in serum (Fig. 5c).144 In their assay, the microarray holes are functionalized with antibodies against the target of interested, and detection is achieved in a sandwich format using antibody-coated gold nanoparticle labels. Upon capture of the target in the sandwich a shift in the plasmonic properties is detected by the integrated CMOS sensor. Importantly, the assay requires only a single user operation injection of the serum sample premixed with the gold nanoparticles. Using a portable reader with a CMOS sensor, LoDs for procalcitonin were as low as 21 pg ml−1, with assay times less than 15 min. The potential of microarray technology for highly multiplexed diagnostics is highlighted by the work of Liu and colleagues.156 The team successfully leveraged this platform for the simultaneous detection of IgG and IgM against SARS-CoV-2, as well as IgGs against SARS-CoV-1 and four other coronaviruses, in clinical samples. Additionally, the microarray format allows for the simple implementation of a control signal using anti-human IgG; a crucial but often neglected feature in low-cost PoC diagnostic devices. It should also be noted that the compatibility of similar microarrays with smartphone-based readouts has been demonstrated, opening up the potential for use at the point-of-care.157
Whilst paper-based lateral flow immunoassays represent the epitome of a simple diagnostic device, these examples highlight how new technologies can often retain the simplicity of a sample-to-answer device in a single step, yet bring added value in terms of sensitivity and multiplexing capabilities.
3.2 Nucleic acid testing
Though microfluidic molecular testing has been a key diagnostic tool since the first demonstration of PCR on a microfluidic chip in 1998,158 its importance and utility has grown given the recent COVID-19 pandemic.159,160 Key advantages of molecular tests include the ability to differentiate between different genotypes of a single virus, and the ease with which they can be adapted to virtually any molecular marker by simply designing different target-specific primers. Such tests typically rely on the enzymatic amplification of DNA, which requires cycling through multiple specific temperatures. Though highly effective, temperature cycling is energy intensive and often tricky to implement. Several isothermal amplification techniques, which employ a variety of alternative methods to initiate strand-displacement, have been developed and are becoming increasingly popular. Further analyses on the benefits and drawbacks of molecular testing (specifically for diagnostic purposes) can be found elsewhere in many excellent reviews.160–163Currently, the gold standard for clinical molecular testing is qPCR, which is often implemented at scale within semi-roboticized machines, such as the Roche cobas system.164 Such solutions allow for incredibly fast screening of hundreds to thousands of samples per day, but at the expense of accessibility and cost.165 To perform these tasks, such instruments require significant floor space and support architecture. These infrastructure requirements drive up operational costs, in addition to the cost of the machine itself. One solution to this problems is to integrate these tests onto microfluidic chips.
The Self-powered Integrated Microfluidic Low-cost Enabling (SIMPLE chip), reported by Yeh et al. in 2017, is a good example of such a chip (Fig. 6).19 The SIMPLE chip integrates many of the steps and processes outlined in section 2 within one device. The utility of the chip was showcased through the detection of methicillin-resistant Staphylococcus aureus (MRSA) within blood samples. Importantly, the chip is entirely self-contained, save for addition of the necessary reagents, which are pre-mixed with blood prior to injection. After being loaded, the sample is transported though microfluidic channels, driven by on-chip vacuum chambers and an artificial “lung” system. The vacuum voids on the chip indirectly connect to the fluid transport channels through thin, gas-permeable but liquid-impermeable membranes. During transport the sample also undergoes a hemolysis-free processing step, with red blood cells sedimenting in the main channel while plasma is “skimmed” into microwells for amplification. The SIMPLE chip generates a signal through recombinase polymerase amplification (RPA), using primers and reagents that are added to the blood prior to loading. Each microwell acts as an individual reaction chamber, allowing for 224 independent, 100 nl volume RPA amplifications. The signal is then transduced via fluorescence imaging, and analyzed. Significantly, digital readout allows quantification of target levels in a sample simply by counting “positive” and “negative” wells, with no need for a standard reference curve or calibration. Whilst the SIMPLE chip is highly integrated, a fluorescence imaging system is required. Fluorescence imaging is more complex than colorimetric or brightfield imaging, and generally requires a dedicated fluorescence reader. This increases system complexity and cost. That said, smartphone cameras are quickly becoming a viable option as inexpensive optical sensors for in-the-field or point-of-care use, as discussed in section 2.5. Though first reported 2017, the SIMPLE chip serves as an excellent reference system for fully integrated microfluidic devices. This progression towards simpler devices that produce colorimetric signals is exemplified in a study by Phillips et al., in which the authors perform a qualitative RT-LAMP-based colorimetric assay to detect HIV.166 Specifically, a small, self-contained autonomous system accepts whole blood and provides a diagnostic answer within 90 minutes. The electrical heating components are reusable, and can be powered by a smartphone, whilst the diagnostic test is done on a single-use laminated μPAD. Sample preparation is achieved using a size-exclusion membrane that captures red blood cells but allows the plasma to flow into an amplification chamber that contains the necessary dried-down reagents. The device then employs integrated heaters to perform a reverse transcriptase-LAMP (RT-LAMP) reaction over 60 minutes. A rinse buffer chamber is then opened to direct the amplified sample onto the lateral flow membrane. Fluid flow within the chip is controlled using wax valves. The valves are printed directly on the paper, and block flow until they are heated, at which point they are opened and flow resumes. Signaling is achieved by using LAMP primers tagged with a fluorophore, which when present at high enough concentrations can be seen by the naked eye, and biotin, which facilitates capture on a streptavidin test line on the lateral flow test strip. This device is an excellent example of how widely available technologies, such as a smartphone, can be integrated into a diagnostic device to simplify design and operation. Further, this work introduces all the steps of the diagnostic workflow, from sample preparation to signal readout, in a single package. This is made possible through the use of highly scalable but low-complexity wax valves, which facilitate relatively complex fluid control without the need for external pumps, implemented on a simple paper substrate. Future development of this type of device should ideally focus on the translational aspects of manufacturing, including robustness and reliability.
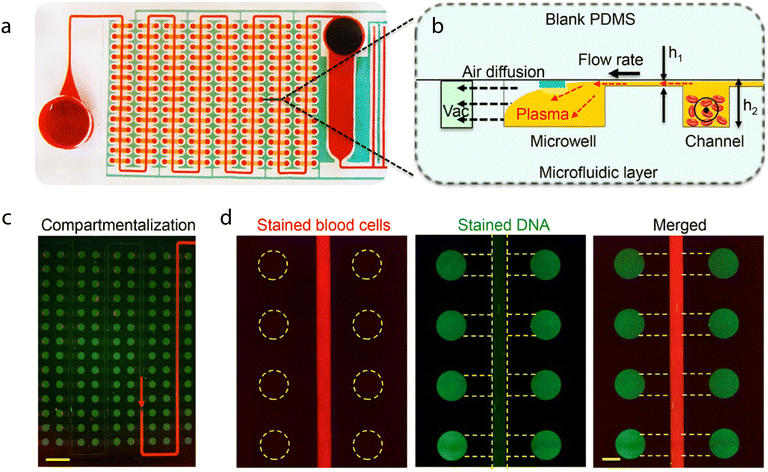 | ||
| Fig. 6 The SIMPLE Chip in operation. (a) Image of the 224 RPA reaction chambers. (b) A diagram of the vacuum-driven cell sedimentation and plasma separation channel design. (c) Illustration of compartmentalization, where the reaction wells are isolated due to an air gap after fluid flow complete. Scale bar is 2 mm (d) fluorescence image showing stained blood cells are not in the reaction wells, and that stained DNA fluorescence is obstructed by the cells. Scale bar is 500 μm. Reproduced/adapted from ref. 19 with permission from Science. | ||
Devices such as the ones presented above have been designed to address specific real-world challenges. One such challenge is the development of effective and accessible IVDs for infectious disease outbreaks, the criteria for which are described by REASSURED3 This acronym stands for ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) eal-time connectivity,
eal-time connectivity, ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ase of specimen collection,
ase of specimen collection, ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ffordable,
ffordable, ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ensitive,
ensitive, ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) pecific,
pecific, ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ser-friendly,
ser-friendly, ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) apid and robust,
apid and robust, ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) quipment-free or otherwise environmentally friendly, and
quipment-free or otherwise environmentally friendly, and ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) eliverable. Given the systemic push towards addressing these challenges, the need for diagnostic devices to be both self-contained and self-powered will become increasingly important.
eliverable. Given the systemic push towards addressing these challenges, the need for diagnostic devices to be both self-contained and self-powered will become increasingly important.
3.3 Cytology and hematology
Cytology (the study of individual cells) and hematology (the study of blood and related disorders) are fundamental tools within modern diagnostics. Sample acquisition in both cytology and hematology is dependent on biopsies, both solid and liquid, and as such an essential aspect of both cytology and hematology is the separation of target cells from complex tissue matrices. Indeed, the information that can be gleaned from these techniques is intrinsically linked to the separation methodology – this has knock-on effects for disease diagnosis and prognosis.167,168Traditionally, isolation of cells and cellular products (such as extracellular vesicles) from complex tissue samples has relied on mechanical methods such as centrifugation and filtration, or fluorescence/magnetic-activated sorting performed using complex and expensive instrumentation.169–171 Unsurprisingly, over the last two decades there has been much interest in applying microfluidic technologies to cell sorting, by employing passive, active or immunoaffinity-based approaches.172,173 Microfluidic-based cell sorters are now ubiquitous within cytology labs, and are being continuously improved to more efficiently sort, analyse and isolate micron-sized cells and even sub-micron extracellular vesicles.174
In many regards the fields of cytology and microfluidics have evolved in parallel, with many advances in microfluidics arising from challenges posed in the cytometry/cell isolation fields. Given the many benefits of microfluidics (such as automation, miniaturization, parallelization and throughput), this is unsurprising. Integrating sample manipulations within a microfluidic system minimizes experimental error and limits inter-user and inter-lab variability. Similarly, the use of single-use microfluidic chips eliminates sample carry over from previous experiments, improving both result confidence and throughput.127 Furthermore, the use of microfluidic tools facilitates the analysis of increasingly small sample volumes, which can lead to significant material and cost savings. Despite such advances, the vast majority of microfluidic-based cell sorters are still dependent on external pumps and complex optical setups this increases device footprint and upfront costs. To make this technology truly accessible and able to operate in low-resource settings whilst maintaining the advantages of larger systems, it is essential that increasingly and affordable designs be developed.
Jiang and Xiang recently reported an integrated handheld microfluidic system for high-throughput cell sorting.175 The system is comprised of three units (Fig. 7): cartridges preloaded with sample fluid and sheath fluid; a microfluidic device to stabilize the flow and separate the target cancer cells from background cells; and a 3D-printed framework encapsulating a diaphragm pump, a microfluidic chip and the electrical control circuits. The working principle of the technology relies on passive size-based separation, wherein a spiral channel exposes cells to both inertial lift forces and Dean drag forces causing larger cells to migrate to the channel wall while keeping smaller cells in flow for collection. Flow is regulated using a deformable membrane which acts as a diaphragm pump. The handheld sorter, specifically designed to replace benchtop equipment that is unavailable in resource-limited settings, is able to receive liquid biopsy samples and reliably output sorted target cells. The authors successfully applied this microfluidic technology to passively separate unlabelled malignant CTCs from clinical pleural effusions, obtaining recovery ratios of about 85% and blood cell removal ratios above 99%. Despite some clear advantages, the highlighted technology has several limitations. At the time of writing, the device has only been applied to a single cell type, so its generalizability is unproven. Given the reliance on size-based passive separation, it is safe to assume this approach would be unsuitable for separating cells of similar size, where more active sorting approaches are typically required. Whilst the device is capable of separating the cells without external equipment, down-stream analyses and data collection is still performed offline using more complex lab equipment. Solving these limitations is key for the development of fully integrated and accessible cell sorting devices. Considering the plethora of applications for single-cell analysis, the ultimate goal should be to create devices which can integrate seamlessly into downstream applications, such as cell-based imaging assays and DNA sequencing.
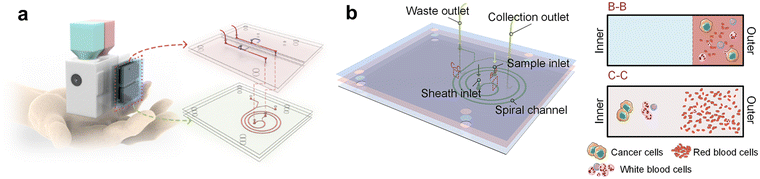 | ||
| Fig. 7 (a) Conceptual design of the integrated chip for portable cancer cell separation. (b) Microfluidic chip presenting the spiral channel, sheath and sample inlets, collection and waste outlets. In the cross-sectional views of B-B and C-C, the cell migration along the channel respectively near the inlets and outlets is illustrated. Reproduced/adapted from ref. 175 with permission from the American Chemical Society. | ||
3.4 Metabolite detection
Metabolite-based diagnostics rely on the detection of small molecules used or produced by cellular metabolism. Metabolomic biomarkers constitute arguably the most expansive and diverse subcategory of diagnostic markers; at the time of writing, the Human Metabolome Database (HMDB) lists 220![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 945 metabolites that have been found within the human body, with associations to over 625 diseases.176 In 2019, a disease-associated metabolite network (DMN) constructed using 245 diseases and 587 metabolites found 28
945 metabolites that have been found within the human body, with associations to over 625 diseases.176 In 2019, a disease-associated metabolite network (DMN) constructed using 245 diseases and 587 metabolites found 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 715 disease-metabolite associations.177 Many metabolites are associated with one or more diseases such as diabetes, cancer, cardiovascular disease, and neurological abnormalities.176 Unfortunately, the convoluted relationship between metabolites and disease makes drawing clear diagnostic lines challenging. To further complicate matters, the structural diversity of metabolites often means that a single technique cannot be exclusively employed for their detection. This is in stark contrast to genomic biomarkers for example, where techniques such as PCR (or isothermal amplification) are generalizable to a vast array of targets.
715 disease-metabolite associations.177 Many metabolites are associated with one or more diseases such as diabetes, cancer, cardiovascular disease, and neurological abnormalities.176 Unfortunately, the convoluted relationship between metabolites and disease makes drawing clear diagnostic lines challenging. To further complicate matters, the structural diversity of metabolites often means that a single technique cannot be exclusively employed for their detection. This is in stark contrast to genomic biomarkers for example, where techniques such as PCR (or isothermal amplification) are generalizable to a vast array of targets.
Current methods for analyzing metabolites rely heavily on mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. The gold standard approach is to employ tandem MS (MS/MS) coupled to liquid chromatography (LC-MS/MS) for upstream sample purification.178–180 This process is entirely dependent on complex spectrometry equipment, and requires a specialized testing lab and highly trained personnel. Though microfluidic chips have been integrated with MS metabolomic workflows, their use is mostly limited to purification prior to off-chip MS analysis.181,182 Since the technical requirements of MS currently preclude miniaturization, integrated microfluidic-based devices for sensing and analyzing metabolites have instead focused on other signaling modalities, such as optical183,184 or electrochemical.185–187
An excellent example of an integrated device for detecting metabolites is provided by Annese et al.188 The device consists of three units: a disposable chip cartridge, a reader to digitize the signal, and a graphical UI. The chip is comprised of four parallel PDMS microfluidic channels, to facilitate detection of four distinct analytes, fabricated directly on top of a CMOS sensor. The necessary reagents for each assay are dried down into separate channels, and are rehydrated upon introduction of the sample. The sample is introduced to the chip by simply applying a droplet of plasma directly to the device via an inlet. It is subsequently separated into the four fluidic channels via passive capillary pressure. Signaling within the device employs a hybrid optical-electrochemical method in which substrate-specific enzymes catalyze the oxidation of their target, producing hydrogen peroxide and inducing a color-shift in a peroxide sensitive dye. The colorimetric signal is then transduced into an electrical signal by the CMOS sensor. Finally, the signal is digitized by the attached reader and sent to a laptop running custom analysis and visualization software. The assays works directly on human plasma and produces a result in under 2 minutes. One limitation of this approach is the dependence on redox enzymes, which constrains the substrate scope. That being said, the cartridge-based approach does provide some flexibility, as the chip could be quickly modified to accommodate a different assay, provided the signal output is colorimetric. Despite these limitations, the fully integrated sample-in-answer-out device demonstrates the potential of microfluidics within this space, and is an excellent example of the direction in which the field is heading.
4 Current limitations and remaining challenges
The primary aim of this review is to convey the value that novel microfluidic technologies add to the IVD landscape. However, as with all emerging technologies, there is still room for improvement. This has been perfectly demonstrated by the ongoing COVID-19 pandemic. In the face of the rapid rise in SARS-CoV-2 infections, governments and enterprises sought diagnostic technologies that could be rapidly deployed in large numbers, often to relatively dispersed populations and at a low price point. Whilst many of the microfluidic technologies described in this review have been adapted to detect SARS-CoV-2 antigens and antibodies, the vast majority of testing is still performed using qPCR and LFIA. The reliance on these technologies, which are at this point several decades old, is telling. Clearly many of the technologies developed over the last two decades have been deemed unsuitable for mass deployment. The reasons for this are multifaceted and convoluted, ranging from technological through to logistical and geopolitical, but it is essential that they are addressed if microfluidic-based diagnostic technologies are to truly reach their full potential. In this section we provide some perspective on the challenges these new technologies face, not just for COVID but for mass adoption across many disease targets, both now and in the future.4.1 Robustness and reliability
Central to evaluating any diagnostic test is an understanding of not just its accuracy, but the conditions under which that accuracy applies. This is especially important for diagnostic assays meant for the developing world, where it may not be possible to consistently meet strict storage, handling, or usage requirements. Providing effective diagnostic technologies to these areas requires proper consideration of reagent aging, transport conditions, local storage facilities and conditions, and local clinical facilities.The difficulties faced in distributing SARS-CoV-2 diagnostics to the developing world have highlighted these issues. Notably, PCR tests contain enzymes and reagents which must be frozen to remain viable. Whereas most of North America and Europe have infrastructure to support delivery of PCR reagents (which require an uninterrupted cold chain), countries without the required infrastructure lack the ability to use these more sensitive tests. One potential solution to these problems is lyophilization, which is has the potential to help preserve reagents during long term storage.189 Similar issues also plague immunoassay deployment. Whereas cold chain storage is not often required for immunoassays, guidance from both manufacturers and regulatory bodies require test storage below 30 °C, with no guarantee of test performance after improper storage.190–192 As immunoassays are commonly employed for the diagnosis of infectious diseases that disproportionately affect populations within tropical or subtropical regions, this is a significant problem.
In addition, the reliability of diagnostic tests is critical. Highly complex instruments almost always have some form of quality control system, either integrated or in the form of standard-issue reference samples. Simplified and miniaturized microfluidic tests designed to be used by the end-user, or in resource-limited settings, must include similar self-checks if they are to be reliable. Ideally, these should be integrated within the device itself. These could include separate control channels with dried-down standards, or on-chip flow detection to ensure correct device operation and detect technical failures. An excellent example of quality control being integrated into a simple assay is the control line on a LFIA test. These control lines show no signal in the event of test failure, and limit the number of false negatives and positives. Though the system is unable to detect every type of assay failure, it goes a long way towards ensuring assay reliability, and thus trust in the end result.
Given their ability to integrate multiple complex techniques and processes into user-friendly and affordable devices, microfluidic IVDs are particularly well-suited to resource-constrained environments. However, any device which aims to have true real-world impact in these environments must address the above concerns. The impact of widely distributing an IVD with a high failure rate, or large incidences of false positive or false negatives, could be catastrophic. It is the responsibility of IVD developers to take these issues under consideration during the development stage, and plan accordingly.
4.2 Cost, scale-up, and manufacturability
In order for any technology to have significant impact, it is essential that it can be deployed at an appropriate scale and at an affordable price point for a particular disease. In the context of IVDs, scalability and affordability are determined by the complex interplay between several factors, including target disease, target populations, existing healthcare infrastructure, macro- and micro-economic factors, and trade and transport infrastructure. Gaining an in-depth understanding of these factors during, or even prior to, the development of a new diagnostic is an essential part of the process. Sadly, it is also a part that is frequently overlooked or ignored. Countless examples of highly integrated, sensitive, and specific microfluidic-based diagnostic technologies are reported in the literature every year. Though many of these devices effectively tackle an area of unmet medical need, the vast majority of these technologies remain academic curiosities due to technical or financial impracticalities which hinder their large-scale manufacture.Currently, chip-based microfluidic devices are most commonly developed using soft lithography, a technique popularized by the Whitesides lab in the 1990s.193 However, it is important to recognize that most of the core techniques still require significant infrastructure (such as cleanroom access), ultimately leading to increased costs and decreased throughput. The detrimental impact of the dependence on soft lithography is perfectly highlighted by many of the technologies discussed in this review. Take for example the SIMPLE chip (see section 3.2).19 Though an excellent example of a fully integrated sample in - answer out device for the detection of molecular biomarkers, the manufacturing process requires multiple steps which are not amenable to large-scale manufacture. These include an overnight curing step, reagent patterning, and vacuum sealing in total the process takes over 6 days. Given the single-use nature of the device, it is unlikely that supply would be able to meet demand in the case of a large-scale infectious disease outbreak. The same can be said of the droplet-based digital ELISA platform developed by Yelleswarapu et al.143 This mobile platform is able to achieve an impressive limit of detection at a relatively affordable price point, but manufacture of the device requires the careful alignment of multiple PDMS and glass components. This relatively complex manufacturing process is hard to replicate on a large-scale. These examples highlight the importance of moving towards manufacturing technologies that do not require stringent cleanroom conditions and specialized equipment, particularly if large-scale manufacturing is required. The research community has taken note of this, and multiple technologies aimed towards solving these issues are under development. Amongst these, lamination, injection moulding, hot embossing, 3D-printing, and nanoimprint lithography all show promise, though they are not without their own limitations. Each technology is limited in terms of the materials which can be manipulated, and many are incapable of achieving sub- or even low- micron resolution.194,195 Put simply, as it stands there is not one single technology which can satisfy the criteria for each and every microfluidic diagnostic platform. If the eventual aim is to apply a particular technology within the real-world, researchers must carefully consider the limitations of these scalable technologies and design their chips and assays accordingly.
4.3 Regulations and data integrity
As we drive towards the goal of broadly accessible diagnostics, it is essential that we consider relevant regulatory issues, particularly in regard to data integrity. Such restrictions are relatively easy to address in centralized laboratories, where testing is done on-site by healthcare professionals and can be easily assessed by regulatory bodies. However, these issues become more complex as tests move to non-centralized locations (such as doctor's offices, medical centers, airports, and concert/sporting venues). Governments may struggle to formulate regulations that support mass adoption and testing while also maintaining strict control over test and result integrity. This dichotomy is apparent in the US (and many other) governments' responses to the COVID-19 pandemic. The technology exists for people to test for SARS-CoV-2 at home, but activities such as travel almost universally require tests with certified results, administered in some way by a healthcare professional.196,197Fortunately, these issues are already being addressed. Guidelines, such as the REASSURED criteria for infectious disease diagnostics,3 have been published to help IVD developers. Additionally, regulatory bodies have thorough and extensive criteria governing the approval of diagnostic devices. In general, these criteria consider issues such as device quality control, clinical specificity and sensitivity, as well as setting the testing requirements and performance targets on the path to device approval. Full information on the regulatory frameworks can be found at the respective governing body, such as the US Food and Drug Administration IVD documentation or the European Commission's directive on IVD devices.198,199 Finally, multiple non-government organizations exist to guide researchers through the IVD development process, including FIND and PATH.200,201
However, the regulations surrounding diagnostic devices often do not end with approval of the device itself. This is perfectly exemplified by the aforementioned SARS-CoV-2 tests, which must be administered by a professional even if the test format is approved for self-use. These policy decisions have been backed by data, which show that self-administration can reduce sensitivity by up to 10 percentage points.20 Data like this suggests there may well be a gap between what researchers consider to be user-friendly, and what is actually usable by the general public. Consulting with UI/UX specialists during device development and performing thorough usability tests is key to solving these issues. There is also the issue of trust with self-reported tests, particularly when there is an incentive for dishonesty (e.g. granting the ability to travel or to attend public events). Solving these issues is non-trivial, and finding an appropriate solution will undoubtedly require significant cooperation between IVD developers and policy makers.
Lastly, the drive to widespread adoption of medical tests raises significant security and privacy concerns. Whereas today the majority of an individual's health information can be found in well-secured central repositories, such as their hospital's electronic health record system, it is easy to imagine a future where diagnostics devices are offered by many private companies each with their own approach to data storage. In the US and EU, the laws governing health data storage are complex and highly specific, with separate responsibilities for healthcare providers, data warehouses, and others who interact with personal data.202 As companies blur the line between diagnostic manufacture and these more regulated roles, they will need to proceed with full awareness of their different responsibilities to both the law and their patients.
For diagnostic tests to be effective, they need to be trusted by patients and doctors, and conform to the relevant regulations. Further, governmental agencies need scientific input to craft appropriate checks and processes to efficiently ensure public safety. For these reasons it is important that scientists and policy makers work together when it comes to regulating diagnostic devices.
5 Conclusion and outlook
The field of in vitro diagnostics is an excellent exemplar of what can be achieved when powerful engineering techniques are applied to medical problems. By leveraging the benefits of microfluidics, namely miniaturization, automation, integration, and parallelization, researchers have been able to create increasingly simple and more accessible IVDs. The devices detailed in this review are testament to the potential of microfluidics-based IVDs, and should serve as an inspiration for what can be achieved when the creativity and ingenuity of researchers is applied to an important task. However, despite substantial progress, significant work towards the goal of fully integrated sample-in-answer-out diagnostic devices remains. The majority of IVD devices reported in the literature every year are able to perform certain aspects of a diagnostic test well (e.g. fluid transport, assay operation, and signal transduction/analysis), but require crucial steps (such as sample preparation) to be performed off-device using traditional laboratory equipment. This ultimately compromises the utility and potential of the devices. Moving forwards, researchers should focus not just on the development of novel microfluidic techniques, but also on how these techniques can be integrated with existing technologies. Similarly, a focus on robustness and reliability is often lacking in novel microfluidic IVDs. Regardless of the sensitivity, LoD, and specificity of an assay within a device, if the supporting architecture is not able to operate reliably then the device is not fit for purpose. Finally, as IVDs are simplified and made more accessible, it is essential that developers address important questions regarding the use and storage of personal data. Though many of the issues presented herein are non-trivial and unlikely to be overcome with engineering alone, the challenges they present form a clear path forward for researchers to ultimately achieve the goal of truly accessible IVDs.Abbreviations
| CMOS | Complementary metal oxide semiconductor |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CTC | Circulating tumor cell |
| DMN | Disease-associated metabolite network |
| ELISA | Enzyme-linked immunosorbent assay |
| EWOD | Electrowetting-on-dielectric |
| Ig | Immunoglobulin |
| ITP | Isotachophoresis |
| IVD | In vitro diagnostic |
| LAMP | Loop-mediated isothermal amplification |
| LC | Liquid chromatography |
| LE | Leading electrolyte |
| LFIA | Lateral flow immunoassay |
| LoD | Limit of detection |
| MDA | Multiple displacement amplification |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance |
| PAD | Paper-based analytical devices |
| PCR | Polymerase-chain reaction |
| PDMS | Polydimethylsiloxane |
| PoC | Point-of-care |
| REASSURED | ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) eal-time connectivity, eal-time connectivity, ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) ase of specimen collection, ase of specimen collection, ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ffordable, ffordable, ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) ensitive, ensitive, ![[S with combining low line]](https://www.rsc.org/images/entities/char_0053_0332.gif) pecific, pecific, ![[U with combining low line]](https://www.rsc.org/images/entities/char_0055_0332.gif) ser-friendly, ser-friendly, ![[R with combining low line]](https://www.rsc.org/images/entities/char_0052_0332.gif) apid and robust, apid and robust, ![[E with combining low line]](https://www.rsc.org/images/entities/char_0045_0332.gif) quipment-free or otherwise environmentally friendly, and quipment-free or otherwise environmentally friendly, and ![[D with combining low line]](https://www.rsc.org/images/entities/char_0044_0332.gif) eliverable eliverable |
| RPA | Recombinase polymerase amplification |
| TE | Trailing electrolyte |
| UI/UX | User-interface and user-experience |
| UWED | Universal wireless electrochemical detector |
Author contributions
Writing – original draft: N. K. Khosla, J. M. Lesinski, M. Colombo, L. Bezinge, D. A. Richards; writing – review & editing: A. J. deMello, D. A. Richards; supervision: D. A. Richards, M. Colombo, A. J. deMello.Conflicts of interest
There are no conflicts to declare.Acknowledgements
D. A. R. acknowledges funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement 840232. M. C. acknowledges funding from Foundation Botnar (Basel, Switzerland).Notes and references
- E. O'Sullivan and S. Schofield, J. R. Coll. Physicians Edinb., 2018, 48, 225–232 CrossRef PubMed.
- G. Saposnik, D. Redelmeier, C. C. Ruff and P. N. Tobler, BMC Med. Inf. Decis. Making, 2016, 16, 138 CrossRef PubMed.
- K. J. Land, D. I. Boeras, X. S. Chen, A. R. Ramsay and R. W. Peeling, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS PubMed.
- D. Crosby, S. Bhatia, K. M. Brindle, L. M. Coussens, C. Dive, M. Emberton, S. Esener, R. C. Fitzgerald, S. S. Gambhir, P. Kuhn, T. R. Rebbeck and S. Balasubramanian, Science, 2022, 375, eaay9040 CrossRef CAS PubMed.
- K. R. King, L. P. Grazette, D. N. Paltoo, J. T. McDevitt, S. K. Sia, P. M. Barrett, F. S. Apple, P. A. Gurbel, R. Weissleder, H. Leeds, E. J. Iturriaga, A. K. Rao, B. Adhikari, P. Desvigne-Nickens, Z. S. Galis and P. Libby, JACC Basic Transl. Sci., 2016, 1, 73–86 CrossRef PubMed.
- J. Ehrenkranz, Endocrinol. Metab. Clin. North Am., 2017, 46, 615–630 CrossRef PubMed.
- U.-P. Rohr, C. Binder, T. Dieterle, F. Giusti, C. G. M. Messina, E. Toerien, H. Moch and H. H. Schäfer, PLoS One, 2016, 11, e0149856 CrossRef PubMed.
- A. Jensen-Doss, E. M. B. Haimes, A. M. Smith, A. R. Lyon, C. C. Lewis, C. F. Stanick and K. M. Hawley, Adm. Policy Ment. Health, 2018, 45, 48–61 CrossRef PubMed.
- L. P. Orellana García, F. Ehmann, P. A. Hines, A. Ritzhaupt and A. Brand, Front. Med., 2021, 8, 753187 CrossRef PubMed.
- K. A. Fleming, S. Horton, M. L. Wilson, R. Atun, K. DeStigter, J. Flanigan, S. Sayed, P. Adam, B. Aguilar, S. Andronikou, C. Boehme, W. Cherniak, A. N. Cheung, B. Dahn, L. Donoso-Bach, T. Douglas, P. Garcia, S. Hussain, H. S. Iyer, M. Kohli, A. B. Labrique, L.-M. Looi, J. G. Meara, J. Nkengasong, M. Pai, K.-L. Pool, K. Ramaiya, L. Schroeder, D. Shah, R. Sullivan, B.-S. Tan and K. Walia, Lancet, 2021, 398, 1997–2050 CrossRef.
- P. Ondoa, L. Oskam, M. M. Loembe and I. N. Okeke, Lancet, 2021, 398, 1947–1949 CrossRef.
- W. A. Fischer and D. A. Wohl, Lancet Infect. Dis., 2022, 22(6), 754–756 CrossRef PubMed.
- L. C. Lopes, A. Santos and P. R. Bueno, Sensors and Actuators Reports, 2022, 4, 100087 CrossRef.
- A. Suea-Ngam, L. Bezinge, B. Mateescu, P. D. Howes, A. J. deMello and D. A. Richards, ACS Sens., 2020, 5, 2701–2723 CrossRef CAS PubMed.
- J. Gomez-Marquez and K. Hamad-Schifferli, Nat. Nanotechnol., 2021, 16, 484–486 CrossRef CAS PubMed.
- P. D. Howes, R. Chandrawati and M. M. Stevens, Science, 2014, 346, 1247390 CrossRef PubMed.
- S. F. Berlanda, M. Breitfeld, C. L. Dietsche and P. S. Dittrich, Anal. Chem., 2021, 93, 311–331 CrossRef CAS PubMed.
- S. Sachdeva, R. W. Davis and A. K. Saha, Front. Bioeng. Biotechnol., 2021, 8, 602659 CrossRef PubMed.
- E.-C. Yeh, C.-C. Fu, L. Hu, R. Thakur, J. Feng and L. P. Lee, Sci. Adv., 2017, 3, e1501645 CrossRef PubMed.
- S. Wuerstle, C. D. Spinner, F. Voit, D. Hoffmann, S. Hering, S. Weidlich, J. Schneider, A. Zink, M. Treiber, R. Iakoubov, R. M. Schmid, U. Protzer and J. Erber, Infection, 2021, 49, 927–934 CrossRef CAS PubMed.
- M.-L. W. Kinshella, P. Tilley, G. N. Al-Rawahi, J. A. Srigley, I. Kayda, M. Canes, M. McLennan, J. N. Bone, M. Dittrick, V. J. Gadkar, L. M. Hoang and D. M. Goldfarb, Diagn. Microbiol. Infect. Dis., 2022, 102, 115566 CrossRef CAS PubMed.
- B. Ying and X. Liu, iScience, 2021, 24, 103174 CrossRef PubMed.
- I. Y. Jung, J. S. Kim, B. R. Choi, K. Lee and H. Lee, Adv. Healthcare Mater., 2017, 6, 1601475 CrossRef PubMed.
- W. Ji, J. Zhu, W. Wu, N. Wang, J. Wang, J. Wu, Q. Wu, X. Wang, C. Yu, G. Wei, L. Li and F. Huo, Research, 2021, 2021, 1–19 CrossRef PubMed.
- N. Promphet, J. P. Hinestroza, P. Rattanawaleedirojn, N. Soatthiyanon, K. Siralertmukul, P. Potiyaraj and N. Rodthongkum, Sens. Actuators, B, 2020, 321, 128549 CrossRef CAS.
- J. T. Reeder, Y. Xue, D. Franklin, Y. Deng, J. Choi, O. Prado, R. Kim, C. Liu, J. Hanson, J. Ciraldo, A. J. Bandodkar, S. Krishnan, A. Johnson, E. Patnaude, R. Avila, Y. Huang and J. A. Rogers, Nat. Commun., 2019, 10, 5513 CrossRef CAS PubMed.
- J. Choi, Y. Xue, W. Xia, T. R. Ray, J. T. Reeder, A. J. Bandodkar, D. Kang, S. Xu, Y. Huang and J. A. Rogers, Lab Chip, 2017, 17, 2572–2580 RSC.
- J. Choi, R. Ghaffari, L. B. Baker and J. A. Rogers, Sci. Adv., 2018, 4, eaar3921 CrossRef PubMed.
- G. Bolat, E. De la Paz and N. F. Azeredo, et al. , Anal. Bioanal. Chem., 2022, 414, 5411–5421 CrossRef CAS PubMed.
- K. P. Fennelly, C. Acuna-Villaorduna, E. Jones-Lopez, W. G. Lindsley and D. K. Milton, Chest, 2020, 157, 540–546 CrossRef CAS PubMed.
- J. Xi, W. Zhao, J. E. Yuan, J. Kim, X. Si and X. Xu, PLoS One, 2015, 10, e0139511 CrossRef PubMed.
- O. Gould, N. Ratcliffe, E. Król and B. de Lacy Costello, J. Breath Res., 2020, 14, 041001 CrossRef CAS PubMed.
- D. Maier, E. Laubender, A. Basavanna, S. Schumann, F. Güder, G. A. Urban and C. Dincer, ACS Sens., 2019, 4, 2945–2951 CrossRef CAS PubMed.
- P.-I. Gouma, L. Wang, S. Simon and M. Stanacevic, Sensors, 2017, 17, 199 CrossRef PubMed.
- I. B. Shlomo, H. Frankenthal, A. Laor and A. K. Greenhut, EClinicalMedicine, 2022, 45, 101308 CrossRef PubMed.
- M. Malik, A.-C. Kunze, T. Bahmer, S. Herget-Rosenthal and T. Kunze, Int. J. Infect. Dis., 2021, 110, 105–110 CrossRef CAS PubMed.
- P. Q. Nguyen, L. R. Soenksen, N. M. Donghia, N. M. Angenent-Mari, H. de Puig, A. Huang, R. Lee, S. Slomovic, T. Galbersanini, G. Lansberry, H. M. Sallum, E. M. Zhao, J. B. Niemi and J. J. Collins, Nat. Biotechnol., 2021, 39, 1366–1374 CrossRef CAS PubMed.
- G.-S. Liu, Y. Kong, Y. Wang, Y. Luo, X. Fan, X. Xie, B.-R. Yang and M. X. Wu, Biomaterials, 2020, 232, 119740 CrossRef CAS PubMed.
- T. M. Blicharz, P. Gong, B. M. Bunner, L. L. Chu, K. M. Leonard, J. A. Wakefield, R. E. Williams, M. Dadgar, C. A. Tagliabue, R. El Khaja, S. L. Marlin, R. Haghgooie, S. P. Davis, D. E. Chickering and H. Bernstein, Nat. Biomed. Eng., 2018, 2, 151–157 CrossRef CAS PubMed.
- J. Yang, X. Liu, Y. Pan, J. Yang, B. He, Y. Fu and Y. Song, Sens. Actuators, B, 2019, 291, 192–199 CrossRef CAS.
- P. Q. Nguyen, L. R. Soenksen, N. M. Donghia, N. M. Angenent-Mari, H. de Puig, A. Huang, R. Lee, S. Slomovic, T. Galbersanini, G. Lansberry, H. M. Sallum, E. M. Zhao, J. B. Niemi and J. J. Collins, Nat. Biotechnol., 2021, 39, 1366–1374 CrossRef CAS PubMed.
- C. Stemmer, M. Beau-Faller, E. Pencreac'h, E. Guerin, A. Schneider, D. Jaqmin, E. Quoix, M.-P. Gaub and P. Oudet, Clin. Chem., 2003, 49, 1953–1955 CrossRef CAS PubMed.
- K. Rudi, M. Kroken, O. J. Dahlberg, A. Deggerdal, K. S. Jakobsen and F. Larsen, BioTechniques, 1997, 22, 506–511 CrossRef CAS PubMed.
- N. Shi, X. Bu, M. Zhang, B. Wang, X. Xu, X. Shi, D. Hussain, X. Xu and D. Chen, Molecules, 2022, 27, 2702 CrossRef CAS PubMed.
- A. Vernerová, L. K. Krčmová, O. Heneberk, V. Radochová and F. Švec, J. Pharm. Biomed. Anal., 2022, 212, 114644 CrossRef PubMed.
- F. Oesch, H. Wagner, K. Platt and M. Arand, J. Chromatogr. B: Biomed. Sci. Appl., 1992, 582, 232–235 CrossRef CAS.
- M. S. Bhamla, B. Benson, C. Chai, G. Katsikis, A. Johri and M. Prakash, Nat. Biomed. Eng., 2017, 1, 0009 CrossRef CAS.
- J. T. Papu, M. J. Rust and A. W. Browne, Point Care, 2014, 13, 48–53 CrossRef.
- S. S. Rao, J. S. Ramesh, R. Huliyappa, Z. War, B. S. A. Kumar, A. S. Tauheed and R. DrSouza, 2019 11th International Conference on Communication Systems & Networks (COMSNETS), Bengaluru, India, 2019, pp. 864–869 Search PubMed.
- H. Yuan, T.-T. Tsai, H.-P. Wang, Y.-S. Chien, C.-A. Chen, C.-C. Chu, C.-T. Ho, P.-H. Chu and C.-F. Chen, Chem. Eng. J., 2021, 409, 128131 CrossRef CAS.
- J. Smejkal, P. Malý, M. Kuchař, N. Panova, A. Semerádtová, P. Aubrecht, M. Štofik and J. Malý, Biosens. Bioelectron., 2021, 172, 112784 CrossRef CAS PubMed.
- I. Banerjee, S. G. Aralaguppe, N. Lapins, W. Zhang, A. Kazemzadeh, A. Sönnerborg, U. Neogi and A. Russom, Lab Chip, 2019, 19, 1657–1664 RSC.
- M. Vázquez, D. Brabazon, F. Shang, J. O. Omamogho, J. D. Glennon and B. Paull, TrAC, Trends Anal. Chem., 2011, 30, 1575–1586 CrossRef.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- M. Shehadul Islam, A. Aryasomayajula and P. Selvaganapathy, Micromachines, 2017, 8, 83 CrossRef.
- Q. Ruan, et al. , Sci. Adv., 2020, 6, eabd6454 CrossRef CAS PubMed.
- S. C. Kim, I. C. Clark, P. Shahi and A. R. Abate, Anal. Chem., 2018, 90, 1273–1279 CrossRef CAS PubMed.
- J. Kim, J. W. Hong, D. P. Kim, J. H. Shin and I. Park, Lab Chip, 2012, 12, 2914 RSC.
- J. Siegrist, R. Gorkin, M. Bastien, G. Stewart, R. Peytavi, H. Kido, M. Bergeron and M. Madou, Lab Chip, 2010, 10, 363–371 RSC.
- H. Kido, M. Micic, D. Smith, J. Zoval, J. Norton and M. Madou, Colloids Surf., B, 2007, 58, 44–51 CrossRef CAS PubMed.
- X. Huang, X. Xing, C. N. Ng and L. Yobas, Micromachines, 2019, 10, 488 CrossRef PubMed.
- E. Grigorov, B. Kirov, M. B. Marinov and V. Galabov, Micromachines, 2021, 12, 498 CrossRef PubMed.
- C. Mercer, A. Jones, J. F. Rusling and D. Leech, Electroanalysis, 2019, 31, 208–211 CrossRef CAS PubMed.
- P. Hardinge, D. K. Baxani, T. McCloy, J. A. H. Murray and O. K. Castell, Sci. Rep., 2020, 10, 21886 CrossRef CAS PubMed.
- M. Xu, H. Zhao, J. Chen, W. Liu, E. Li, Q. Wang and L. Zhang, Cytometry, Part A, 2020, 97, 46–53 CrossRef CAS PubMed.
- S. A. M. Shaegh, Z. Wang, S. H. Ng, R. Wu, H. T. Nguyen, L. C. Z. Chan, A. G. G. Toh and Z. Wang, Microfluid. Nanofluid., 2015, 19, 557–564 CrossRef CAS.
- M. M. Gong and D. Sinton, Chem. Rev., 2017, 117, 8447–8480 CrossRef CAS PubMed.
- M. Ochoa, R. Rahimi, J. Zhou, H. Jiang, C. K. Yoon, D. Maddipatla, B. B. Narakathu, V. Jain, M. M. Oscai, T. J. Morken, R. H. Oliveira, G. L. Campana, O. W. Cummings, M. A. Zieger, R. Sood, M. Z. Atashbar and B. Ziaie, Microsyst. Nanoeng., 2020, 6, 1–16 CrossRef PubMed.
- T. Kokalj, Y. Park, M. Vencelj, M. Jenko and L. P. Lee, Lab Chip, 2014, 14, 4329–4333 RSC.
- M. Boyd-Moss, S. Baratchi, M. D. Venere and K. Khoshmanesh, Lab Chip, 2016, 16, 3177–3192 RSC.
- K. Iwai, R. D. Sochol and L. Lin, 2011 IEEE 24th International Conference on Micro Electro Mechanical Systems, 2011, pp. 1131–1134 Search PubMed.
- M. T. Glynn, D. J. Kinahan and J. Ducrée, Lab Chip, 2014, 14, 2844–2851 RSC.
- J. Reboud, G. Xu, A. Garrett, M. Adriko, Z. Yang, E. M. Tukahebwa, C. Rowell and J. M. Cooper, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4834–4842 CrossRef CAS PubMed.
- M. Abdelgawad, S. L. S. Freire, H. Yang and A. R. Wheeler, Lab Chip, 2008, 8, 672–677 RSC.
- H. Moon, A. R. Wheeler, R. L. Garrell, J. A. Loo and C.-J. C. Kim, Lab Chip, 2006, 6, 1213–1219 RSC.
- S. Petralia, D. Motta and S. Conoci, Biotechnol. Bioeng., 2019, 116, 2087–2094 CrossRef CAS PubMed.
- H. Norian, R. M. Field, I. Kymissis and K. L. Shepard, Lab Chip, 2014, 14, 4076–4084 RSC.
- H. Ding, S. Sadeghi, G. J. Shah, S. Chen, P. Yuin Keng, C.-J. Kim and R. M. van Dam, Lab Chip, 2012, 12, 3331–3340 RSC.
- Y.-J. Li and B. P. Cahill, Langmuir, 2017, 33, 13139–13147 CrossRef CAS PubMed.
- D. Jiang, Z. Fan, H. Wang, M. Xu, G. Chen, Y. Song and Z. L. Wang, ACS Nano, 2020, 14, 15394–15402 CrossRef PubMed.
- C. Peng, Z. Zhang, C.-J. Kim and Y. S. Ju, Lab Chip, 2014, 14, 1117–1122 RSC.
- A. Rival, D. Jary, C. Delattre, Y. Fouillet, G. Castellan, A. Bellemin-Comte and X. Gidrol, Lab Chip, 2014, 14, 3739–3749 RSC.
- C. Wang, X. Li, Y. Qiu, L. Wang, C. Li, G. Liu, G. Liu, Q. Zheng, X. Chen, H. Tian, C. Wang and J. Shao, Nano Energy, 2022, 98, 107310 CrossRef CAS.
- C. Peng, Z. Zhang, C.-J. C. Kim and Y. S. Ju, Lab Chip, 2014, 14, 1117–1122 RSC.
- K. M. Schilling, D. Jauregui and A. W. Martinez, Lab Chip, 2013, 13, 628–631 RSC.
- A. W. Martinez, S. T. Phillips, Z. Nie, C.-M. Cheng, E. Carrilho, B. J. Wiley and G. M. Whitesides, Lab Chip, 2010, 10, 2499–2504 RSC.
- J. Park and J.-K. Park, Sens. Actuators, B, 2017, 246, 1049–1055 CrossRef CAS.
- Y. Sun, Y. Chang, Q. Zhang and M. Liu, Micromachines, 2019, 10, 531 CrossRef PubMed.
- W. Xue, D. Zhao, Q. Zhang, Y. Chang and M. Liu, Chem. Commun., 2021, 57, 11465–11468 RSC.
- M. Rocca, Y. Temiz, M. L. Salva, S. Castonguay, T. Gervais, C. M. Niemeyer and E. Delamarche, Lab Chip, 2021, 21, 3573–3582 RSC.
- O. Gökçe, S. Castonguay, Y. Temiz, T. Gervais and E. Delamarche, Nature, 2019, 574, 228–232 CrossRef PubMed.
- E. Hemmig, Y. Temiz, O. Gökçe, R. D. Lovchik and E. Delamarche, Anal. Chem., 2020, 92, 940–946 CrossRef CAS PubMed.
- L. Chen, J. West, P.-A. Auroux, A. Manz and P. J. R. Day, Anal. Chem., 2007, 79, 9185–9190 CrossRef CAS PubMed.
- G. Maltezos, A. Gomez, J. Zhong, F. A. Gomez and A. Scherer, Appl. Phys. Lett., 2008, 93, 243901 CrossRef.
- G. Maltezos, M. Johnston, K. Taganov, C. Srichantaratsamee, J. Gorman, D. Baltimore, W. Chantratita and A. Scherer, Appl. Phys. Lett., 2010, 97, 264101 CrossRef PubMed.
- D. Vigolo, R. Rusconi, R. Piazza and H. A. Stone, Lab Chip, 2010, 10, 795 RSC.
- A. J. de Mello, M. Habgood, N. L. Lancaster, T. Welton and R. C. R. Wootton, Lab Chip, 2004, 4, 417 RSC.
- M.-S. Hung, C.-C. Ho and C.-P. Chen, J. Biomed. Opt., 2016, 21, 087003 CrossRef PubMed.
- V. Miralles, A. Huerre, F. Malloggi and M.-C. Jullien, Diagnostics, 2013, 3, 33–67 CrossRef PubMed.
- Z. Wang, X. Liang, H. Su, S. Li and Y. Chen, Ind. Eng. Chem. Res., 2021, 60, 14341–14353 CrossRef CAS.
- H. Kim, S. Vishniakou and G. W. Faris, Lab Chip, 2009, 9, 1230 RSC.
- W. Wang, C. Zhou, T. Zhang, J. Chen, S. Liu and X. Fan, Lab Chip, 2015, 15, 3862–3869 RSC.
- Z. Feng and L. Bai, Micromachines, 2018, 9, 122 CrossRef PubMed.
- C. Dincer, R. Bruch, A. Kling, P. S. Dittrich and G. A. Urban, Trends Biotechnol., 2017, 35, 728–742 CrossRef CAS PubMed.
- K. R. Mitchell, J. E. Esene and A. T. Woolley, Anal. Bioanal. Chem., 2022, 414, 167–180 CrossRef CAS PubMed.
- X. Wang, X.-Z. Hong, Y.-W. Li, Y. Li, J. Wang, P. Chen and B.-F. Liu, Mil. Med. Res., 2022, 9, 11 Search PubMed.
- L. Rosenfeld, T. Lin, R. Derda and S. K. Y. Tang, Microfluid. Nanofluid., 2014, 16, 921–939 CrossRef.
- J. Mok, M. N. Mindrinos, R. W. Davis and M. Javanmard, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 2110–2115 CrossRef CAS PubMed.
- S.-Y. Teh, R. Lin, L.-H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198 RSC.
- L. Cohen, N. Cui, Y. Cai, P. M. Garden, X. Li, D. A. Weitz and D. R. Walt, ACS Nano, 2020, 14, 9491–9501 CrossRef CAS PubMed.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed.
- L. Liu, N. Xiang, Z. Ni, X. Huang, J. Zheng, Y. Wang and X. Zhang, BioTechniques, 2020, 68, 114–116 CrossRef CAS PubMed.
- Z. Shi, X. Lai, C. Sun, X. Zhang, L. Zhang, Z. Pu, R. Wang, H. Yu and D. Li, Chem. Commun., 2020, 56, 9056–9066 RSC.
- H. Yuan, J. Tian, Y. Chao, Y.-S. Chien, R.-H. Luo, J.-Y. Guo, S. Li, Y.-J. Chou, H. C. Shum and C.-F. Chen, ACS Sens., 2021, 6, 2868–2874 CrossRef CAS PubMed.
- S. J. Maerkl and S. R. Quake, Science, 2007, 315, 233–237 CrossRef CAS PubMed.
- F. Volpetti, J. Garcia-Cordero and S. J. Maerkl, PLoS One, 2015, 10, e0117744 CrossRef PubMed.
- Z. Swank, G. Michielin, H. M. Yip, P. Cohen, D. O. Andrey, N. Vuilleumier, L. Kaiser, I. Eckerle, B. Meyer and S. J. Maerkl, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2025289118 CrossRef CAS PubMed.
- J. L. Garcia-Cordero, C. Nembrini, A. Stano, J. A. Hubbell and S. J. Maerkl, Integr. Biol., 2013, 5, 650–658 CrossRef CAS PubMed.
- J. L. Garcia-Cordero and S. J. Maerkl, Lab Chip, 2014, 14, 2642–2650 RSC.
- K. R. Mitchell, J. E. Esene and A. T. Woolley, Anal. Bioanal. Chem., 2022, 414, 167–180 CrossRef CAS PubMed.
- C. Dincer, R. Bruch, E. Costa-Rama, M. T. Fernández-Abedul, A. Merkoçi, A. Manz, G. A. Urban and F. Güder, Adv. Mater., 2019, 31, 1806739 CrossRef PubMed.
- M. Pohanka, Rev. Anal. Chem., 2020, 39, 20–30 CrossRef CAS.
- A. P. Nsabimana, B. Uzabakiriho, D. M. Kagabo, J. Nduwayo, Q. Fu, A. Eng, J. Hughes and S. K. Sia, JMIR Public Health Surveill., 2018, 4, e11203 CrossRef PubMed.
- V. Turbé, C. Herbst, T. Mngomezulu, S. Meshkinfamfard, N. Dlamini, T. Mhlongo, T. Smit, V. Cherepanova, K. Shimada, J. Budd, N. Arsenov, S. Gray, D. Pillay, K. Herbst, M. Shahmanesh and R. A. McKendry, Nat. Med., 2021, 27, 1165–1170 CrossRef PubMed.
- L. Zhang, Z. Tian, H. Bachman, P. Zhang and T. J. Huang, ACS Nano, 2020, 14, 3159–3169 CrossRef CAS PubMed.
- B. Hunt, A. J. Ruiz and B. W. Pogue, J. Biomed. Opt., 2021, 26, 040902 Search PubMed.
- M. Vázquez, L. Anfossi, H. Ben-Yoav, L. Diéguez, T. Karopka, B. D. Ventura, S. Abalde-Cela, A. Minopoli, F. D. Nardo, V. Kumar Shukla, A. Teixeira, A. Tvarijonaviciute and L. Franco-Martínez, Lab Chip, 2021, 21, 4330–4351 RSC.
- J. A. Hambalek, J. E. Kong, C. Brown, H. E. Munoz, T. Horn, M. Bogumil, E. Quick, A. Ozcan and D. Di Carlo, ACS Sens., 2021, 6, 3242–3252 CrossRef CAS PubMed.
- B. Gil Rosa, O. E. Akingbade, X. Guo, L. Gonzalez-Macia, M. A. Crone, L. P. Cameron, P. Freemont, K.-L. Choy, F. Güder, E. Yeatman, D. J. Sharp and B. Li, Biosens. Bioelectron., 2022, 203, 114050 CrossRef CAS PubMed.
- A. Fernández-la-Villa, D. F. Pozo-Ayuso and M. Castaño-Álvarez, Curr. Opin. Electrochem., 2019, 15, 175–185 CrossRef.
- H. A. Abdulbari and E. A. M. Basheer, ChemBioEng Rev., 2017, 4, 92–105 CrossRef.
- M. A. Mohd Asri, A. N. Nordin and N. Ramli, Biomicrofluidics, 2021, 15, 061502 CrossRef CAS PubMed.
- E. P. Randviir, Electrochim. Acta, 2018, 286, 179–186 CrossRef CAS.
- A. García-Miranda Ferrari, S. J. Rowley-Neale and C. E. Banks, Talanta Open, 2021, 3, 100032 CrossRef.
- K. J. Klunder, Z. Nilsson, J. B. Sambur and C. S. Henry, J. Am. Chem. Soc., 2017, 139, 12623–12631 CrossRef CAS PubMed.
- L. A. Pradela-Filho, E. Noviana, D. A. G. Araújo, R. M. Takeuchi, A. L. Santos and C. S. Henry, ACS Sens., 2020, 5, 274–281 CrossRef CAS PubMed.
- V. Katseli, M. Angelopoulou and C. Kokkinos, Adv. Funct. Mater., 2021, 31, 2102459 CrossRef CAS.
- E. Noviana, K. J. Klunder, R. B. Channon and C. S. Henry, Anal. Chem., 2019, 91, 2431–2438 CrossRef CAS PubMed.
- PalmSens, PalmSens – Compact Electrochemical Interfaces, https://www.future-science.com/doi/abs/10.2144/97223rr01, Accessed 1 July 2022 Search PubMed.
- A. Ainla, M. P. S. Mousavi, M.-N. Tsaloglou, J. Redston, J. G. Bell, M. T. Fernández-Abedul and G. M. Whitesides, Anal. Chem., 2018, 90, 6240–6246 CrossRef CAS PubMed.
- A. Nemiroski, D. C. Christodouleas, J. W. Hennek, A. A. Kumar, E. J. Maxwell, M. T. Fernández-Abedul and G. M. Whitesides, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 11984–11989 CrossRef CAS PubMed.
- I. C. Samper, A. Sánchez-Cano, W. Khamcharoen, I. Jang, W. Siangproh, E. Baldrich, B. J. Geiss, D. S. Dandy and C. S. Henry, ACS Sens., 2021, 6, 4067–4075 CrossRef CAS PubMed.
- V. Yelleswarapu, J. R. Buser, M. Haber, J. Baron, E. Inapuri and D. Issadore, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4489–4495 CrossRef CAS PubMed.
- A. Belushkin, F. Yesilkoy, J. J. González-López, J. C. Ruiz-Rodríguez, R. Ferrer, A. Fàbrega and H. Altug, Small, 2020, 16, 1906108 CrossRef CAS PubMed.
- F. Li, M. You, S. Li, J. Hu, C. Liu, Y. Gong, H. Yang and F. Xu, Biotechnol. Adv., 2020, 39, 107442 CrossRef CAS PubMed.
- B. G. Andryukov and B. G. Andryukov, AIMS Microbiol., 2020, 6, 280–304 CAS.
- B. Adamson, R. Sikka, A. L. Wyllie and P. Premsrirut, medRxiv, 2022, preprint, DOI:10.1101/2022.01.04.22268770, https://www.medrxiv.org/content/10.1101/2022.01.04.22268770v1.full.pdf+html.
- Y. Liu, L. Zhan, Z. Qin, J. Sackrison and J. C. Bischof, ACS Nano, 2021, 15, 3593–3611 CrossRef CAS PubMed.
- B. S. Miller, L. Bezinge, H. D. Gliddon, D. Huang, G. Dold, E. R. Gray, J. Heaney, P. J. Dobson, E. Nastouli, J. J. L. Morton and R. A. McKendry, Nature, 2020, 587, 588–593 CrossRef CAS PubMed.
- C. N. Loynachan, M. R. Thomas, E. R. Gray, D. A. Richards, J. Kim, B. S. Miller, J. C. Brookes, S. Agarwal, V. Chudasama, R. A. McKendry and M. M. Stevens, ACS Nano, 2018, 12, 279–288 CrossRef CAS PubMed.
- L. Bezinge, A. Suea-Ngam, A. J. deMello and C.-J. Shih, Mol. Syst. Des. Eng., 2020, 5, 49–66 RSC.
- Y. Ding, P. D. Howes and A. J. deMello, Anal. Chem., 2020, 92, 132–149 CrossRef CAS PubMed.
- L. Cohen, N. Cui, Y. Cai, P. M. Garden, X. Li, D. A. Weitz and D. R. Walt, ACS Nano, 2020, 14, 9491–9501 CrossRef CAS PubMed.
- J. Poorbaugh, T. Samanta, S. W. Bright, S. E. Sissons, C.-Y. Chang, P. Oberoi, A. J. MacDonald, A. P. Martin, K. L. Cox and R. J. Benschop, J. Immunol. Methods, 2019, 466, 9–16 CrossRef CAS PubMed.
- K. Yang, X. Yao, B. Liu and B. Ren, Adv. Mater., 2021, 33, 2007988 CrossRef CAS PubMed.
- T. Liu, J. Hsiung, S. Zhao, J. Kost, D. Sreedhar, C. V. Hanson, K. Olson, D. Keare, S. T. Chang, K. P. Bliden, P. A. Gurbel, U. S. Tantry, J. Roche, C. Press, J. Boggs, J. P. Rodriguez-Soto, J. G. Montoya, M. Tang and H. Dai, Nat. Biomed. Eng., 2020, 4, 1188–1196 CrossRef CAS PubMed.
- X. Wang, T.-W. Chang, G. Lin, M. R. Gartia and G. L. Liu, Anal. Chem., 2017, 89, 611–615 CrossRef CAS PubMed.
- M. U. Kopp, A. J. de Mello and A. Manz, Science, 1998, 280, 1046–1048 CrossRef CAS PubMed.
- W. H. Organization, Recommendations for National SARS-CoV-2 Testing Strategies and Diagnostic Capacities, 2021, https://apps.who.int/iris/bitstream/handle/10665/342002/WHO-2019-nCoV-lab-testing-2021.1-eng.pdf Search PubMed.
- C. Y. Yu, K. G. Chan, C. Y. Yean and G. Y. Ang, Diagnostics, 2021, 11, 53 CrossRef CAS PubMed.
- B. Schweitzer and S. Kingsmore, Curr. Opin. Biotechnol., 2001, 12, 21–27 CrossRef CAS PubMed.
- P. Craw and W. Balachandran, Lab Chip, 2012, 12, 2469–2486 RSC.
- A. Afzal, J. Adv. Res., 2020, 26, 149–159 CrossRef CAS PubMed.
- F. Hoffmann-La Roche Ltd, Cobas® 6000 Analyzer Series, https://diagnostics.roche.com/global/en/products/systems/cobas_-6000-analyzer-series.html, Accessed 1 July 2022 Search PubMed.
- G. C. Dash, U. K. Rout, R. R. Nanda, D. Parai, H. R. Choudhary, S. Kanungo, S. K. Palo, J. S. Kshatri, J. Turuk, B. K. Mishra, S. Pati and D. Bhattacharya, J. Clin. Lab. Anal., 2021, 35, e23835 CrossRef CAS PubMed.
- E. A. Phillips, T. J. Moehling, K. F. K. Ejendal, O. S. Hoilett, K. M. Byers, L. A. Basing, L. A. Jankowski, J. B. Bennett, L.-K. Lin, L. A. Stanciu and J. C. Linnes, Lab Chip, 2019, 19, 3375–3386 RSC.
- W. Li, Y. Zhou, Y. Deng and B. L. Khoo, Cancers, 2022, 14, 818 CrossRef CAS PubMed.
- P. Sandbhor Gaikwad and R. Banerjee, Analyst, 2018, 143, 1326–1348 RSC.
- C. Gardiner, D. D. Vizio, S. Sahoo, C. Théry, K. W. Witwer, M. Wauben and A. F. Hill, J. Extracell. Vesicles, 2016, 5(1), 32945 CrossRef PubMed.
- M. Kersaudy-Kerhoas and E. Sollier, Lab Chip, 2013, 13, 3323–3346 RSC.
- N. Norouzi, H. C. Bhakta and W. H. Grover, PLoS One, 2017, 12, e0180520 CrossRef PubMed.
- G. Li, W. Tang and F. Yang, Biotechnol. J., 2020, 15, 1900225 CrossRef CAS PubMed.
- Y. Meng, M. Asghari, M. K. Aslan, A. Yilmaz, B. Mateescu, S. Stavrakis and A. J. deMello, Chem. Eng. J., 2021, 404, 126110 CrossRef CAS.
- M. Asghari, X. Cao, B. Mateescu, D. van Leeuwen, M. K. Aslan, S. Stavrakis and A. J. deMello, ACS Nano, 2020, 14, 422–433 CrossRef CAS PubMed.
- F. Jiang and N. Xiang, Anal. Chem., 2022, 94, 1859–1866 CrossRef CAS PubMed.
- K. Mi, Y. Jiang, J. Chen, D. Lv, Z. Qian, H. Sun and D. Shang, Front. Bioeng. Biotechnol., 2020, 8, 1–10 CrossRef PubMed.
- Y. Wang, L. Juan, J. Peng, T. Zang and Y. Wang, BMC Bioinf., 2019, 20, 574 CrossRef PubMed.
- Z. Yi and Z.-J. Zhu, Computational Methods and Data Analysis for Metabolomics, Springer US, New York, NY, 2020, pp. 139–148 Search PubMed.
- S. Heiles, Anal. Bioanal. Chem., 2021, 413, 5927–5948 CrossRef CAS PubMed.
- S. C. Nanita and L. G. Kaldon, Anal. Bioanal. Chem., 2016, 408, 23–33 CrossRef CAS PubMed.
- J. R. Kraly, R. E. Holcomb, Q. Guan and C. S. Henry, Anal. Chim. Acta, 2009, 653, 23–35 CrossRef CAS PubMed.
- L. Zhang, T. Xu, J. Zhang, S. C. C. Wong, M. Ritchie, H. W. Hou and Y. Wang, Anal. Chem., 2021, 93, 10462–10468 CrossRef CAS PubMed.
- T. D. Oblak, J. A. Meyer and D. M. Spence, Analyst, 2009, 134, 188–193 RSC.
- T. Songjaroen, T. Maturos, A. Sappat, A. Tuantranont and W. Laiwattanapaisal, Anal. Chim. Acta, 2009, 647, 78–83 CrossRef CAS PubMed.
- C.-J. Huang, J.-L. Lin, P.-H. Chen, M.-J. Syu and G.-B. Lee, Electrophoresis, 2011, 32, 931–938 CrossRef CAS PubMed.
- C.-Y. Liu, J. Rick, T.-C. Chou, H.-H. Lee and G.-B. Lee, Biomed. Microdevices, 2008, 11, 201 CrossRef PubMed.
- Z. Liao, J. Wang, P. Zhang, Y. Zhang, Y. Miao, S. Gao, Y. Deng and L. Geng, Biosens. Bioelectron., 2018, 121, 272–280 CrossRef CAS PubMed.
- V. F. Annese, S. B. Patil, C. Hu, C. Giagkoulovits, M. A. Al-Rawhani, J. Grant, M. Macleod, D. J. Clayton, L. M. Heaney, R. Daly, C. Accarino, Y. D. Shah, B. C. Cheah, J. Beeley, T. R. J. Evans, R. Jones, M. P. Barrett and D. R. S. Cumming, Microsyst. Nanoeng., 2021, 7, 21 CrossRef CAS PubMed.
- M. J. Hammerling, K. F. Warfel and M. C. Jewett, Biotechnol. J., 2021, 16, 2000572 CrossRef CAS PubMed.
- N. H. Service, Advanced Service – Community Pharmacy COVID-19 Lateral Flow Device Distribution Service, National Health Service advanced service specification, 2021 Search PubMed.
- I. ACON Laboratories, COVID-19 Antigen Home Test Package Insert for Healthcare Providers, ACON Laboratories, Inc., Technical Report IN195000 Rev. 42022/02 Search PubMed.
- I. Abbot Diagnostics Scarborough, BinaxNOW COVID-19 Ag Card (PN 195-000) – Instructions For Use, Abbot Diagnostics Scarborough, Inc., Technical Report IN195000 Rev. 42022/02 Search PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- B. K. Gale, A. R. Jafek, C. J. Lambert, B. L. Goenner, H. Moghimifam, U. C. Nze and S. K. Kamarapu, Inventions, 2018, 3, 60 CrossRef.
- V. Faustino, S. O. Catarino, R. Lima and G. Minas, J. Biomech., 2016, 49, 2280–2292 CrossRef PubMed.
- Center for Disease Control, Self-Testing At Home or Anywhere, 2022, https://www.cdc.gov/coronavirus/2019-ncov/testing/self-testing.html, Accessed 1 July 2022 Search PubMed.
- Center for Disease Control, Travel, CDC, https://www.cdc.gov/coronavirus/2019-ncov/travelers/index.html, Accessed 1 July 2022 Search PubMed.
- Center for Devices and Radiological Health, Overview of IVD Regulation, Mon, 10/18/2021 – 10:09, https://www.fda.gov/medical-devices/ivd-regulatory-assistance/overview-ivd-regulation, Accessed 1 July 2022 Search PubMed.
- E. Union, Regulation (EU) 2017/746 of the European Parliament and of the Council of 5 April 2017 on in Vitro Diagnostic Medical Devices and Repealing Directive 98/79/EC and Commission Decision 2010/227/EU (Text with EEA Relevance), https://eur-lex.europa.eu/eli/reg/2017/746/oj, Accessed 1 July 2022 Search PubMed.
- FIND, Home – FIND, https://www.finddx.org/, Accessed 1 July 2022 Search PubMed.
- PATH, PATH, https://www.path.org/, Accessed 1 July 2022 Search PubMed.
- S. Tovino, Seton Hall Law Rev., 2017, 47, 973–994 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |
