 Open Access Article
Open Access ArticleTools for manipulation and positioning of microtissues
Emilie
Vuille-dit-Bille
 ab,
Dhananjay V.
Deshmukh
ab,
Dhananjay V.
Deshmukh
 cd,
Sinéad
Connolly
cd,
Sinéad
Connolly
 e,
Sarah
Heub
e,
Sarah
Heub
 a,
Stéphanie
Boder-Pasche
a,
Stéphanie
Boder-Pasche
 a,
Jürg
Dual
a,
Jürg
Dual
 d,
Mark W.
Tibbitt
d,
Mark W.
Tibbitt
 c and
Gilles
Weder
c and
Gilles
Weder
 *a
*a
aCentre Suisse d'Electronique et de Microtechnique SA, Neuchâtel, Switzerland. E-mail: gilles.weder@csem.ch
bMicroBioRobotic Systems Laboratory, Institute of Mechanical Engineering, EPFL, Lausanne, Switzerland
cMacromolecular Engineering Laboratory, Department of Mechanical and Process Engineering, ETH Zurich, Zurich, Switzerland
dInstitute for Mechanical Systems, Department of Mechanical and Process Engineering, ETH Zurich, Zurich, Switzerland
eLaboratory of Biosensors and Bioelectronics, Institute for Biomedical Engineering, ETH Zürich, Zurich, Switzerland
First published on 5th October 2022
Abstract
Complex three-dimensional (3D) in vitro models are emerging as a key technology to support research areas in personalised medicine, such as drug development and regenerative medicine. Tools for manipulation and positioning of microtissues play a crucial role in the microtissue life cycle from production to end-point analysis. The ability to precisely locate microtissues can improve the efficiency and reliability of processes and investigations by reducing experimental time and by providing more controlled parameters. To achieve this goal, standardisation of the techniques is of primary importance. Compared to microtissue production, the field of microtissue manipulation and positioning is still in its infancy but is gaining increasing attention in the last few years. Techniques to position microtissues have been classified into four main categories: hydrodynamic techniques, bioprinting, substrate modification, and non-contact active forces. In this paper, we provide a comprehensive review of the different tools for the manipulation and positioning of microtissues that have been reported to date. The working mechanism of each technique is described, and its merits and limitations are discussed. We conclude by evaluating the potential of the different approaches to support progress in personalised medicine.
1. Introduction
Complex three-dimensional (3D) in vitro models are gaining increasing attention as they address several limitations inherent to 2D cell culture by mimicking relevant tissue function and architecture. For example, 3D tissue models can recapitulate crucial parameters, including extensive cell–cell contacts, cell–extracellular matrix (ECM) interactions, and soluble gradients present in natural microenvironments.1,2 Owing to these characteristics, complex 3D in vitro models have the potential to reproduce biological phenomena more robustly than 2D cultures. An important example can be found in cancer research where 3D tissue models exhibit multicellular resistance to drugs, mimicking the resistance of real tumours, while 2D cultures lack this characteristic.3 In this context, microtissues, such as organoids, spheroids, and tumoroids, have emerged as promising candidates for disease modelling, drug development, and tissue engineering,4 while patient-derived microtissues hold potential in the field of personalised medicine.5 Microtissues are complex biological models that are highly sensitive to external factors and small modifications in protocols can lead to significant differences in results. These discrepancies between similar studies represent obstacles for the implementation of microtissue-based technologies. Therefore, standardisation of processes is essential for the broad use of microtissue research. To support this standardisation, platforms that generate reproducible results independently of the operator need to be developed to replace variable manual processing.To date, standardisation of microtissue processing has mainly focused on their production and maturation. Production of microtissues with controlled and uniform shape and size is critical for therapeutic efficacy and clinical reproducibility.6 Moreover, high-throughput strategies are required to produce large numbers of microtissues and to thereby support relevant drug screening assays and the biofabrication of tissue constructs. Therefore, extensive research was performed on microtissue fabrication and numerous strategies have been reported for robust batch production.7,8
Although microtissues can be produced efficiently, to unlock their full potential, effort is needed to develop effective tools for positioning microtissues. The field of microtissue manipulation is still in its infancy and current methodologies for manipulating and placing microtissues remain mainly manual, cumbersome, and time consuming. Due to the manual nature of these handling steps, experimental outcomes often depend on the skills of the operator. The scientific community is increasingly aware of this problem, motivating the need for precise and reproducible microtissue handling for the development of organ-on-chip systems, regenerative medicine, and high-throughput analysis for both preclinical testing and drug screening.9
Techniques to manipulate small objects, such as particles and cells, have been extensively developed in the past few decades. Since the potential of microtissues for personalised medicine has emerged, new techniques are sought for the manipulation of large biological entities. Tools to manipulate and position microtissues are diverse, covering a large range of applications and, have recently been driven by tissue engineering applications.9,10 The methods can be classified into four main categories: hydrodynamic techniques, bioprinting positioning, substrate modification, and non-contact active forces.
This review aims to provide a comprehensive overview of the different tools for positioning microtissues in the development of complex in vitro models. First, the importance of manipulating and positioning microtissues throughout their whole life cycle will be introduced. Then, the different techniques for manipulating and positioning microtissues will be presented. The working mechanism of each technique will be described, and its merits and limitations will be discussed. Finally, we discuss the potential of the different approaches to support progress in personalised medicine. For consistency, and since several definitions of these terms can be found in the literature,11 cell aggregates, organoids, spheroids, and tumoroids will be grouped under the general term of microtissue in this review. However, with respect to published research, the citations of any scientific articles will use the specific term used in the article.
2. Life stages of microtissues
In this section, the importance of positioning and manipulating microtissues throughout their lifecycle will be highlighted. The typical life stages of microtissues are illustrated in Fig. 1. First, microtissues are produced from cells using a range of available fabrication methods. After production, microtissues are often sorted based on different criteria and assembled into desired structures before being cultured. Analysis and testing can be performed on the microtissues prior to, during or after any processing steps.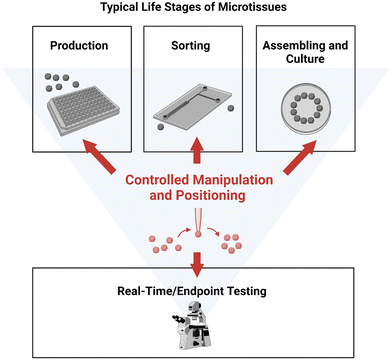 | ||
| Fig. 1 Typical life stages of microtissues. Controlled manipulation and positioning play a crucial role in the all the stages. Created with https://BioRender.com. | ||
First, numerous fabrication methods have been developed to address the needs of different microtissue types. The production methods are very diverse and range from the assembly of a few polydisperse floating microtissues in a culture flask to the batch production of monodisperse microtissues in nonadherent microwells. Interestingly, in some methods, microtissues are already positioned as they are produced in a way that facilitates subsequent handling and analysis steps. For example, batch production in microwells allows image-based microtissue assays as the microtissues are positioned in an array in one imaging plane.8 The present review concentrates on the positioning and manipulation of already formed microtissues and therefore does not discuss production methods that include positioning. For further reading on different production methods, see the review of Agrawal et al.12
Second, efficient, and automated sorting consists of the classification and the subsequent separation of microtissues into categories. Depending on the platform, the classification and the separation can be performed simultaneously or by two consecutive methods. In both cases, the separation requires the active manipulation of microtissues. Sorting based on microfluidic or acoustic forces, well-established sorting methods for cells, are examples of simultaneous classification and separation. Pick-and-place techniques can also be combined with classification methods, such as imaging with deep learning, to allow efficient sorting with individual positioning of microtissues. These advanced imaging-based methods are particularly interesting for sorting based on microtissue morphology or expression of selected fluorescent reporters.
Sorting plays an important role in regenerative medicine as it enables the identification of unhealthy autologous microtissues, and thereby reduces the risk of tumorigenesis.13 Moreover, by removing dead microtissues, sorting increases the viability of biofabricated constructs. In addition, automated sorting of microtissues upon specific morphological or physical criteria supports new research by providing more controlled parameters, and consequently leads to a deepening of our understanding of various biological mechanisms.14 Particularly, it supports drug testing assays, where chemical components are tested for a specific feature not present in every microtissue. Finally, efficient sorting also reduces the time of analysis because only the microtissue of interest would be processed.
Third, assembling is par excellence the positioning of microtissues in a pattern or in a defined location on a substrate. Assembling can be performed by various methods that either position individual microtissues or simultaneously manipulate a population of microtissues. The patterns generated can include simple arrays in 2D, complex 3D shapes and organised structures. Therefore, assembly is a crucial step for several applications, such as in regenerative medicine for the biofabrication of constructs and in drug testing for the creation of complex organ-on-chip models. The availability of standardised methods for the precise manipulation of microtissues would also strongly benefit fundamental research, which is inherently linked to personalised medicine. The assembly of microtissues obtained from different regions of a tissue with defined spatial organisation allows for the possibilities to study the communication and migration mechanisms associated with early organ development, disease morphogenesis, and tissue regeneration. Assembloids, assemblies of organoids representing different regions of an organ have recently emerged and have quickly become a leading technology in stem-cell research, particularly in the neuronal field.15–18 For such assembly approaches, sample orientation plays a key role in the fusion process where the alignment of specific features is required. In parallel, the integration of microtissues into more complexly engineered organoid-on-chip gives access to the micro-physiological environment such as fluid flow, mechanical stimulation or nutrient, and oxygen transport for the study of tissue-tissue and multi-organ interactions.19,20 Organoid-on-chip platforms rely on the precise positioning of microtissues, integration of a vasculature, and bioreactor capabilities. The importance of positioning in microtissue assembly will be further illustrated with two specific uses: tissue engineering and organ-on-chip design.
Biofabrication using microtissues as building blocks (MBB) promises to bring tissue engineering to the next level since it overcomes important limitations of using individual cells as building blocks. For example, cellular self-assembly is often slow and may take days or weeks, while microtissue self-assembly is faster and can occur within hours.21,22 Indeed, the high cell density of microtissues coupled with increased deposition of the extracellular matrix notably reduces the maturation time of a 3D construct.6 Moreover, prevascularised microtissues, which can be produced by co-culturing endothelial cells with other cell types, have the advantage of supporting the process of angiogenesis and preventing apoptosis due to oxygen deficiency.23,24 Although biofabrication using microtissues as building blocks rely on their autonomous self-organisation, accurate positioning to generate a defined pattern of microtissues with controlled spacing and organisation is essential to promote microtissue fusion.6 Moreover, the complex hierarchical architecture of in vivo tissues define their specific function and should be recreated as precisely as possible.25 Based on this observation, Eke et al. suggested the development of simulation tools that model the evolution of microtissues in culture and guide the design of the construct to achieve the desired functionality.10 In conclusion, biofabrication using MBB with precise positioning could support the production of large tissue with vascularisation, significantly reduce the time between the construct formation and assay and improve the physiological relevance of assays by better recreating in vivo tissue architecture. The possibility to build tissue constructs with complex shapes and high resolution would also open the door to in vivo replacement of injured or damaged tissues with personalised implants.
An organ-on-chip is a microfluidic-based device, which creates a dynamic environment that mimics the in vivo environment more closely than static culture in multi-well plates.26–28 Organ-on-chip devices improve drug testing assays as they enable more accurate liquid volume drug dosage and better physiological drug exposure than in static culture.29,30 To simulate the complex interactions between different organs in the body, complex microfluidic systems containing different organ models, called body-on-chips, have been developed.26 These multi-organ platforms show great potential to evaluate the efficacy of drugs and simultaneously assess their toxicity in other organs. As shown in preclinical studies, body-on-chip devices can even surpass animal models, which do not always predict human responses effectively.31 However, most of the organ-on-chip platforms rely on the in situ production of microtissues, resulting in devices that are difficult to operate and generally not suited for microtissues of different cell types.27 The positioning of externally produced microtissues would open body-on-chip platforms to all types of microtissues and thereby broaden their applications. Additionally, the positioning and immobilisation of microtissues on top of integrated sensors are essential to collect reliable data on the system.32
Finally, high-throughput analysis of microtissues remains an ongoing challenge because of the random orientation and positioning of microtissues during analysis. For example, simultaneous imaging of several microtissues requires their alignment in the same focus plane. Additionally, the rotational movements of microtissues may hinder the imaging of structural features of specific interest. Therefore, to achieve high-throughput analysis, positioning and manipulation of microtissues is needed. Additionally, simultaneous processing and imaging of microtissues reduces the variability between samples as they are all processed in the same manner and improve the reliability of the comparison between them.33 Such a property is particularly interesting for fluorescence imaging where the light exposure might change from one sample to another. A major advantage of having accurate control over the location of microtissues is the reduction of replicates, resulting in less manpower and consumables and decreasing the time of analysis. Positioning of microtissues, therefore, results in cheaper, faster, and more efficient analysis. Histology analysis, a gold-standard method for the study of the micro-anatomy of tissues, perfectly illustrates this need. Indeed, with random loading of microtissues, dozens of replicates are needed to achieve reliable results, while only 5–10 microtissues in the same focal plane could provide sufficient information for robust analysis.34
In conclusion, controlled positioning and manipulation play a key role throughout the microtissue use cycle. The ability to precisely locate microtissues can improve the efficiency of processes and investigations by reducing experimental time and by providing more controlled parameters, such as the position, orientation, and characteristic of the microtissues. Therefore, controlled positioning and manipulation of microtissues directly support the development of regenerative medicine and drug testing. The different tools that are used to position and manipulate microtissues will be presented in detail in the following sections of the review.
3. Hydrodynamic techniques
Positioning using hydrodynamic techniques involves the use of fluid flow to manoeuvre the physical position of the object of interest. This can be achieved through several means, including microfluidic chips, microfluidic droplets, pipetting-based systems, and hydrodynamic-based micromanipulators, each of which are explored in further detail below. Most microfluidic positioning techniques were first developed for inanimate microbeads or nanoparticles; however, they have since been adopted for cells and microtissues. Consequently, concerns have been raised regarding the effects that large forces present in fluidic systems can have on the viability or the functionality of living organisms.35–41 Nevertheless, studies carried out on single cells indicate that fluidic forces likely do not damage cells in suspension,42–45 implying that the same may be true for microtissues.3.1. Microfluidic chips
Microfluidic-based sorting, or inertial sorting, is carried out by injecting a suspension of particles into a channel on a microfluidic chip. The technique seeks to manipulate the physical parameters of the flow, which in turn may influence the position that suspended particles occupy or flow at in a channel. Trapping, on the other hand, relies almost completely on the longitudinal geometry of the channel, using either constricted channels to obstruct passage, or using wells or traps in the flow path to generate microscale vortices to completely halt the object of interest.46,47 Additionally, both methods can be used in conjunction with each other to direct particles into traps.In microfluidic-based sorting, a change in any of the parameters can cause a shift in the position of particles flowing in a circular channel either towards the channel centre or towards the channel walls, as shown in Fig. 2A. These parameters can relate to the suspending fluid, such as the density, viscosity or flow rate; to the channel, such as the width, length, cross-sectional geometry or the longitudinal geometry; or to the particle itself, such as its size, shape, or elastic modulus.48–51 Though this technique is primarily used for the sorting of particles or cells, it can also be used to direct larger objects of interest into a desired location within the fluid pathway. Further, sheath flow, a technique commonly used in flow cytometry, whereby two or more different fluids flow in the same direction, side-by-side, can be used with a similar effect.52
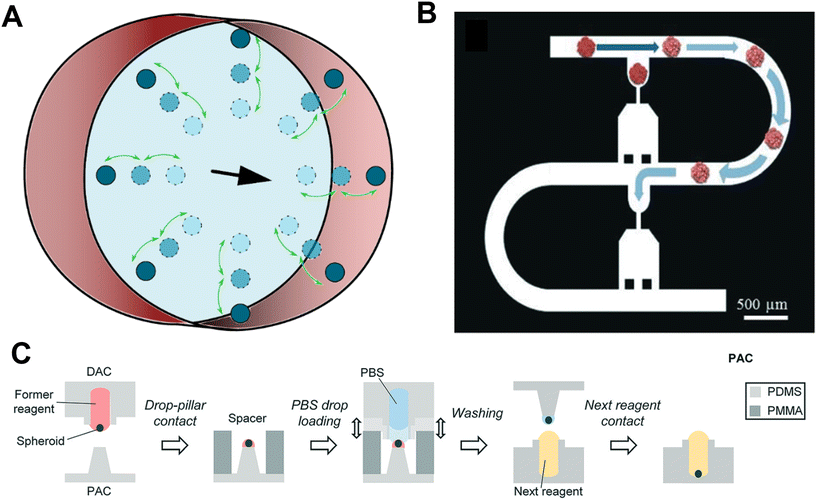 | ||
| Fig. 2 Microfluidic chips and droplets. A. Microfluidic particles flowing in a circular channel in the direction of the black arrow. In this instance the parameters of fluid flow can cause particles to move closer to the channel centre or closer to the walls. B. An example of a microfluidic device used for particle trapping. Particles are guided by hydrodynamic force, squeezed, and parked in each trap. Reproduced from ref. 28 Copyright from the Royal Society of Chemistry. C. A schematic of the DCST reagent change procedure. Reproduced from ref. 60 Copyright from the Royal Society of Chemistry. | ||
Ruppen et al. developed a microfluidic trapping device that can be used to trap microtissues (Fig. 2B).28 The device operates on the basis that the flow rate through an empty trap is relatively higher than the flow of the bypass channel, directing the particle's trajectory into the trap. Once the particle is trapped, the flow rate through the trap decreases significantly, preventing any further particles from entering. In the study, the authors were able to position 8 spheroids in a line within the same chip. In another study, larger chambers with multiple spheroid isolation units inside have also been used for microtissue isolation.53,54 A microfluidic chip that could be used to sort and entrap microtissues was also developed by Chen et al.55 The technique was expanded to differentiate between microparticles based on both size and elastic modulus. Though it has only been used with hydrogel microparticles to date, it is anticipated that it can also be used with spheroids, which are similarly sized to the particles used in this study.
A robot-integrated microfluidic chip has also been proposed as an alternative method of microfluidic positioning for microtissues.56 This involves a series of syringe pumps on either side of a microfluidic device with a visual feedback control, which can be used to position the spheroid in the desired location. Additionally, the system has the ability to measure the elasticity of a spheroid within the microfluidic chip, a parameter that can then be used for downstream sorting. This promising system has been cited as an alternative to flow cytometry for microtissues.56
Microfluidic chips have several advantages. They are biocompatible, as the channels are usually manufactured from polydimethylsiloxane (PDMS), and microtissues remain suspended in fluid, which can be cell culture medium. However, systems often have to be tailored to the specific fluid, as a change in the fluid density or viscosity can alter the microtissue trajectories. Nonetheless, this sensitivity can also be used as an advantage to sort microtissues of different sizes and mechanical properties. Microfluidic techniques enable efficient and rapid sorting and trapping. For example, the robot-integrated microfluidic chip has achieved a throughput rate of up to 92 spheroids per hour.56
These techniques also present a number of limitations. One is that the desired location of the microtissue needs to be incorporated within the microfluidic device, a constraint which may be unsuitable for certain applications, or may prohibit future access to the microtissue. Additionally, in some techniques, such as the microfluidic trapping devices outlined above, the microtissue size that can be used may be limited by the size of the microfluidic channel. Finally, microfluidic-based sorting and trapping is also quite limited spatially. Each different configuration of microtissue location requires a new microfluidic device to be manufactured which is both time-consuming and costly. Some applications, such as the robot-integrated microfluidic chip, are limited to a linear direction only, while others incorporate meandering channels, and can therefore allow for more extensive positioning over a 2D area (Fig. 2B). Positioning in three dimensions, though theoretically possible, would become extremely complex, and controlling the orientation of the microtissue is currently not possible.
3.2. Microfluidic droplets
The use of microfluidic droplets is a technique whereby particles of interest are trapped or contained within a bubble of fluid of choice. The technique has mainly been used to transport reagents for experimentation57 or to transport single cells58 in microfluidic channels. Droplet encapsulation has also been used for the development of microtissues, as it is a useful technique for precisely controlling and manipulating their local environments.59Droplet contact-based spheroid transfer (DCST) is a technique whereby a droplet array chip (a PDMS array of wells) and a pillar array chip (a corresponding PDMS array of pillars) can be used to transfer spheroid arrays between them using simple droplet contact (Fig. 2C).60 This technique allows the controlled change of reagents of individual spheroids, and therefore gives more experimental control for investigating the effects of a particular drug.60 The manipulation is, however, limited spatially as spheroid-containing microdroplets can only be transferred between pillars and wells.
Aside from DCST, the field of droplet encapsulation is very promising and could be used in a variety of different ways to manipulate microtissue position. Similar to cell-laden droplets, microtissue-laden droplets could be used in conjunction with inertial sorting techniques in microchannels, as outlined in section 3.1, to sort and separate microtissues of different physical properties.61 Further, digital microfluidics could be used to manipulate and position microtissues-laded silicone oil droplets using dielectrophoresis.62 Although, manipulating droplets containing microtissues is theoretically possible, the feasibility of the technique still needs to be demonstrated.
Droplet encapsulation can be more favourable over other microfluidic techniques, which use bulk solutions to manipulate several microtissues, because the droplet allows for the precise tailoring and control of the single microtissue environment and is therefore advantageous in drug treatment assays. Furthermore, since droplet systems do not use inertial manipulation, larger spheroids can be handled, than what is often feasible in microfluidic channels. Additionally, in opposition to microfluidic chips, the droplet encapsulation techniques are not affected by differing physical properties of the spheroids. This means that one platform can be used with a variety of different microtissue types and physical properties. Finally, by expanding the number of arrays and pillars built on to the chip, the throughput of the DCST technique can be increased. On the other hand, the droplet encapsulation technique is even more limited spatially than microfluidic channels. Currently, spheroids can only be transferred between wells and pillars of the DCST device, and so cannot be transferred to any other surfaces or devices that may be of interest to the researcher.
3.3. Pipetting-based systems
In contrast to microfluidic chips and droplets, where the positioning and manipulation is performed within a device, aspiration-based techniques use external pipettes to handle microtissues. Pipetting-based techniques utilise a hollow device with an aperture which, when brought into contact with, or into the vicinity of a microtissue, can create a negative pressure through the aperture, thereby picking up the microtissue. Depending on the size of the aperture, the microtissue can either be aspirated completely into the device or be held outside. The device can then be moved to the desired location, and with the generation of a positive pressure, the object or microtissue is released. To facilitate the positioning, microtissues are generally aspired from a solution and placed in a second solution or hydrogel at the desired location.Aspiration has been used by several different applications for manipulating microtissues. A ‘bio-gripper’, consisting of a polycarbonate cylindrical tube (9.5 mm or 4.8 mm diameter, 31.3 mm long) with a track-etched membrane fitted on the end has been used to pick up large microtissues (Fig. 3A).63,64 By attaching the opposite end of the bio-gripper to a syringe pump, a flow rate, and therefore a pressure difference, is created across the membrane, allowing it to pick microtissue structures up. Then, a reversal of the flow direction allows the microstructures to be deposited in the desired location. Spheroids of 800 μm were picked-and-placed with this device. Additionally, the bio-gripper was able to place larger, more complex toroid and honeycomb structures up to sizes of 6 mm.63,64 However, the technique seems to be limited to microtissues larger than 800 μm, a downsized version may be capable of more delicate handling of smaller sized microtissue structures.
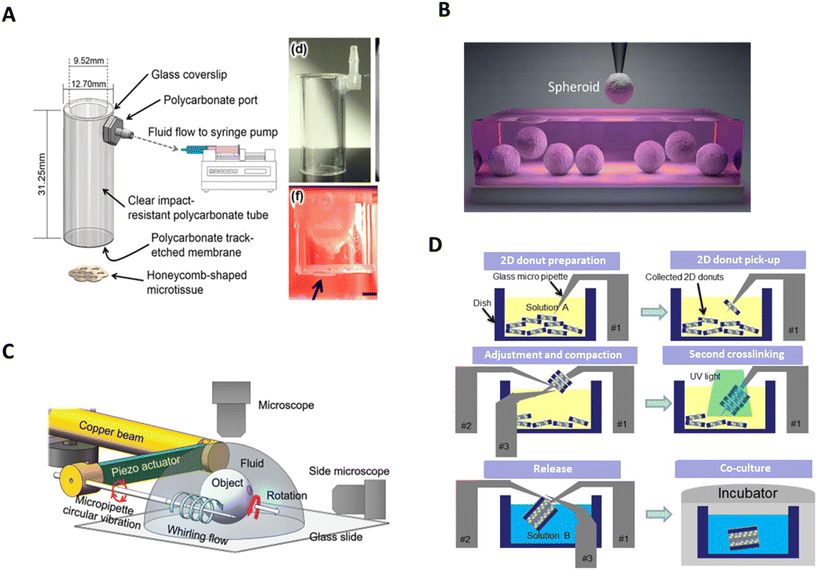 | ||
| Fig. 3 Pipetting-based systems and hydrodynamic-based micromanipulators. A. Schematic and photograph of bio-gripper. In red, side view image of a gripped honeycomb microtissue (arrow). Scale bar = 2 mm. Reproduced from ref. 64 Copyright from IOP Publishing. B. Spheroids being picked from cell media using a glass pipette in the aspiration-assisted bioprinter. Reproduced from ref. 66 Copyright from American Association for the Advancement of Science. C. Schematic illustrationof the multifunctional micromanipulation using whirling flow generated by vibration of a single piezoelectric actuator. Reproduced from ref. 86 Copyright from John Wiley and Sons. D. A schematic drawing of 3D cellular structure assembly process with rail-guided multi-microrobotic system. Reproduced from ref. 91 Copyright from SAGE Publishing. | ||
Micropipette aspiration, often used to determine the elastic modulus of single cells, has also been used to transfer spheroids between solutions.65 A micropipette aspirator with an aperture of 20 μm was used to move spheroids from a cell culture dish to a Falcon tube. The technology also has the ability to differentiate between spheroids based on size.65 This idea was further expanded to develop an aspiration-assisted bioprinter (AAB), which can extract spheroids from a bulk solution and use them to print micropatterns with a very high degree of accuracy (≈11% with respect to the spheroid size) in a gel substrate (Fig. 3B).66 Further studies using AAB have demonstrated that a variety of configurations are possible with spheroids cultivated from different cell types67–70 and the system has a throughput of approximately 40 spheroids per hour.68 However, the technology currently remains limited because a soft substrate with specific properties, typically an hydrogel, need to be used, similar to the extrusion-based in supportive medium bioprinting method (see section 5.1).
Another aspiration-based biofabrication technique, called the Kenzan method, has been successfully used to create tissue constructs with different types of microtissues.71–74 This scaffold-free biofabrication technique uses stainless steel needles, which are arranged in a geometrical design, to temporarily support a structure of impaled microtissues.75 The studies showed that the needles alter the viability of the cells in their vicinity but overall, the microtissue viability is not adversely affected.76 Though, the structural damage caused by the needle, which can be significant for small microtissues, represents a notable disadvantage compared to hydrogel substrate. Moreover, to fuse afterwards, the microtissues need to be in direct contact, restricting the microtissue size to the spacing between the needles.75
Most pipetting-based techniques manipulate only one microtissue at a time, which can be limiting for applications with large numbers of microtissues. It may be possible, however, to add simultaneously working micropipettes to reduce this problem. Fluidic forces or physical contact with the placing devices, which are inherent to their operation, may affect the viability of microtissues, however studies have shown that they either have no effect on viability80 or a very small one.66 Another limitation of some pick-and-place techniques that require the microtissue to be aspirated inside the micropipette is the additional aspiration of the adjacent liquid. As this volume will be dispensed at the same time as the microtissue, the technique might not be suited for some applications. To reduce this effect, the micropipette should be positioned in as close proximity as possible to the microtissue to minimise the aspirated liquid. Additionally, the inside of the micropipette should be carefully coated to avoid attachment of the microtissue to the wall.
Picking-and-placing also has many advantages. Cells can be manipulated in their preferred medium, or in the case of ABB, hydrogel scaffolds, both of which tend to be biocompatible environments. These methods (except the bio-gripper) can also differentiate objects of interest based on elastic moduli as well as size, allowing for a certain degree of sorting if required. Aspiration-based techniques could also be incorporated with optical sorting methods in the future, which would allow for even more refined sorting of microtissue types. Further, these systems are flexible and can generally be used for a wide range of tissue types and sizes. In the occasional event that this is not the case, changes in pipettes or probes are relatively inexpensive and straightforward procedures. The biggest advantage of aspiration-based techniques, however, is their range of motion, providing much larger manipulation areas, as well as the ability to operate in a 3D environment, unlike either microfluidic-based sorting and trapping or microfluidic droplets.
Aspiration-based techniques have the potential to be fully automated for picking-and-placing microtissues. In this case, integrated visual feedback is needed to position the micropipette over a microtissue before aspiration, as in its absence, the micropipette might go to a region without microtissue, only aspiring liquid. Although fully automated robots are used for liquid handling of cell culture83 and pick-and-place microtissue bioprinting in a specialised pre-defined environment, no automated pick-and-place technique has yet been published to transfer microtissues from standard labware such as multi-well plates. The adaptation of liquid-handling robots to microtissue transfer in standard labware is possible but will require further optimisation. The main limitation of existing liquid-handling robot resides, as stated before, in the large adjacent liquid volume during picking. For big microtissues, the generally small aperture of the tips could also be problematic and prevent aspiration.
It may be possible to adapt other methods currently used for single cell picking-and-placing to microtissue, such as FluidFM, which aspirates minimal amounts of liquid with the microtissue. FluidFM is another innovative aspiration-based technique, comprising an atomic force microscopy (AFM) cantilever with a hollow interior.84 Like micropipette aspiration, a negative pressure can be generated in the cantilever which can be used to pick up a cell, while a positive pressure deposits the cell from the tip of the probe in the desired location.85 The AFM probe has an additional advantage of providing force feedback control,77 negating the requirement to place in a hydrogel scaffold. The technique has been utilised extensively with single cells, including myoblasts, cervical cancer cells, and neurons,78–82 however, it has yet to be demonstrated with larger spheroids.
3.4. Hydrodynamic-based micromanipulators
Hydrodynamic-based micromanipulators are fully automated systems, which can control the position and, to some extent, the orientation of microtissues. To do so, these systems use one or more micropipettes to control the manipulation of microtissues.A hydrodynamic-based micromanipulator designed for microtissue guidance incorporates a manipulation system with a piezoelectric resonator.86 In this setup, a micropipette was mounted on a piezoelectric actuator, which was attached to a robotic manipulator and microtissues were placed free floating in a droplet on a glass slide beneath. The micropipette was brought into the proximity of the targeted spheroid whereby it created a whirling flow by vibrating in a circular motion.86 This caused the spheroid to become trapped in the vicinity of the microneedle without actually touching it, while gravity prevented the spheroid from rotating around the needle. From here, the needle could be moved in the x- or y-directions, carrying the spheroid with it. A schematic of the system can be seen in Fig. 3C. Changing the flow velocity gradient around the pipette, rotates the spheroid, allowing it to be visualised from multiple angles.87
The piezoelectric resonator with the micropipette has the advantage of being able to control the orientation of the spheroid of interest, however this is primarily for observation purposes, and it seems to be unable to control the orientation or the position of the spheroid accurately for placing purposes. The system can also manipulate several spheroids at once. This allows for higher throughput of spheroids, however limits manoeuvrability as, in this instance, the whirling flow causes the spheroids to accumulate at the same point, meaning spheroids are pushed into close contact and must remain side-by-side. Unlike most other systems, the spheroid does not come into physical contact with the manipulation tool at any point, which may aid in preventing physical damage. However, 3D manipulation is not yet possible with this system. Furthermore, as the technology is based on microfluidic forces, the spheroid's movement will also be influenced by its physical properties such as size and stiffness. This may be of concern in non-uniform spheroid populations.
Another hydrodynamic-based micromanipulator, the rail-guided microrobotic system, consists of a micropipette and a micromanipulator,88–90 though earlier iterations consisted of three micromanipulators.91 These are mounted on a circular rail, of approximately 100 mm diameter, surrounding the manipulation area. The micromanipulators and micropipettes can move along the rail to access the microtissues and hinged joints allow for four degrees of freedom.91 The system was originally designed to aid in the organisation of microtissues, shaped into vascular-like microchannels, by cultivating cells in toroid-shaped hydrogels. These hydrogels were placed in a petri dish at the centre of the circular rail, where they sank to the bottom of the dish. The micropipette was used to inject air at the base of the microstructure, causing it to float upwards, and to be impelled on the micromanipulator. This procedure was repeated, stacking several hydrogels on the micromanipulator, and creating a vascular microchannel. This construct was then transferred to a culture dish, and cells were allowed to proliferate and grow through the assembled structure.89 The entire process is drawn in Fig. 3D. Though the device has, so far, only been used for microtissue assembly, it may be possible to use it for their orientation and manipulation of their local position.
Another microfluidic technique using viscous finger phenomena has been developed to generate vascular-like constructs. In this technique, a less viscous fluid containing suspended cells displaces a more viscous fluid, usually a hydrogel, inside a microchannel, carving the hydrogel into a tubular structure. Recent works have shown the potential of viscous finger patterning to create mono- and multicellular and cell-laden vessels in a simple and high-throughput manner.92,93 The translation of this technique to patterning microtissues in a tubular structures is promising but requires further investigation.
The rail-guided microrobotic system is advantageous for microtissue manipulation in that it can manipulate in both 2D and 3D, however it remains to be seen how efficient or accurate the system can be. Furthermore, it is possible that physical contact between the micromanipulator and the hydrogels may inadvertently damage the cells within the microstructure. Additionally, the micromanipulators, like the piezoelectric resonator system, should allow the user to control microtissue orientation to a certain degree, a process that is difficult to achieve in most other microtissue manipulation devices. However, like for relocation, it remains to be seen how effective this approach can be in more general use.
4. Bioprinting
Bioprinting using cells as building blocks is a common biofabrication method and has been extensively studied in the past few years. This additive manufacturing method uses bioinks – a mixture of polymers and living building materials – to create viable 2D or 3D constructs. To leverage the potential of microtissues in tissue engineering, some bioprinting techniques have sought to adapt to bigger building blocks. These techniques were categorised into extrusion-based, sacrificial writing into functional tissue, droplet-based, and volumetric printing, each of which are explored in further detail below.Bioinks play a crucial role in bioprinting since they influence both the formation and the maturation of a functional tissue.94 On the mechanical aspect, a suitable bioink needs to be printable and after printing, it needs to exhibit a gelation behaviour to maintain the construct shape and support the deposition of upper layers.95–97 From a physiological aspect, the bioink should provide an appropriate microenvironment to promote cell activities and differentiation by presenting the right mechanical and chemical clues, while enabling the diffusion of nutrients and wastes. The suitable bioink properties depend on the cell type and the printing method and therefore the bioink should be optimised for each specific application. Bioink formulations are beyond the scope of this review and therefore will not be discussed further.
4.1. Extrusion-based bioprinting
Extrusion-based bioprinting techniques are characterised by the direct and continuous deposition of bioinks as filaments. The bioink is extruded from a cartridge through a nozzle by applying a pneumatic or a mechanical force. Extrusion-based techniques can be used to print bioinks in air or in a supportive medium.The dispensing of the bioink involves mechanical and thermal stresses that may have an impact on cell behaviour if they are not carefully controlled.98–101 In extrusion-based bioprinting, shear stress is inherent to the dispensing and greatly influences the viability of the cells after printing. Different printing parameters such as the nozzle diameter, the printing pressure and the viscosity of the bioink affect the shear stress level during the dispensing process.98–101 By considering several printing parameters, such as bioink composition, printing temperature and holding time, Ouyang et al. experimentally established a negative exponential relationship between cell viability and maximum shear stress level.98 To reduce adverse effects on cell viability, the properties of the bioink can be tuned. For example, shear-thinning bioinks not only facilitate the dispensing but also improve cell viability.102,103 In addition to the immediate impact on cell viability by membrane damage, shear stress influences the cell proliferation potential and viability in the long term.101 Interestingly, it was also demonstrated that cells can be printed without adverse effect if the shear stress is below a critical value. Similarly, it was established by several studies that microtissues can be printed without altering the viability of the printed spheroids or their ability to fuse and differentiate.21,22,104–111 In the context of MBB, in addition to damaging the cell membrane, printing-induced shear stress might disrupt the microtissue shape and integrity, resulting in floating single cells in the bioink.105 Consequently, the shear stress level during the dispensing process is a key parameter that should be carefully assessed to successfully dispense microtissues.
Extrusion-based bioprinting in air using MBB has been successfully used in several studies to create tissue models (Fig. 4A).22,105–110 The function and the integrity of the spheroids were not altered during the process, showing the biocompatibility of the technique. For example, Polonchuk et al. were able to measure synchronous contraction of printed cardiac spheroids upon electrical stimulation.106 Common challenges encountered during the printing of microtissues is the sedimentation of the microtissues within the hydrogel and the clogging of the nozzle. The sedimentation can be prevented by increasing the viscosity of the hydrogel, while keeping a good printability of the bioink.105 To avoid the clogging of the nozzle, monodisperse microtissues of relatively small size compared to the nozzle diameter should be used and the concentration of microtissues should be carefully optimised.
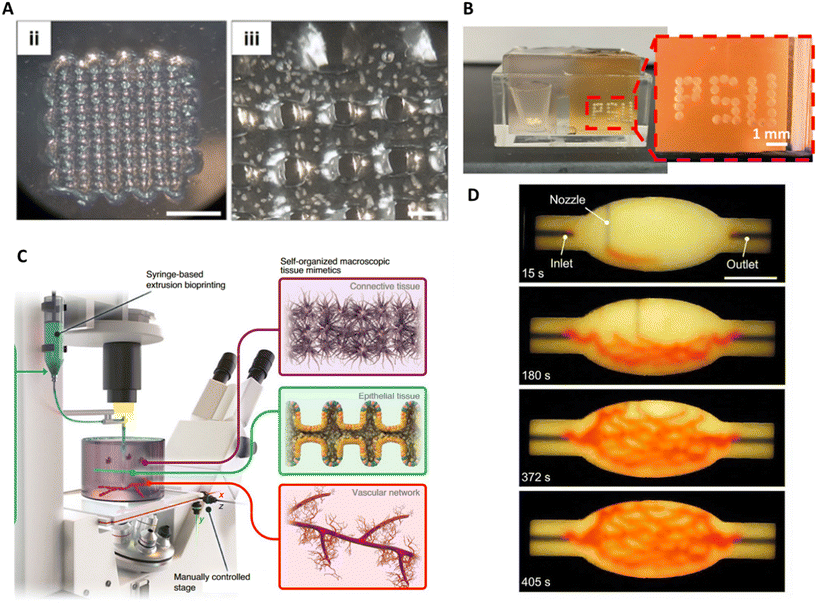 | ||
| Fig. 4 Extrusion-based bioprinting and SWFIT. A. 3D bioprinting of a spheroid-laden bioink by extrusion. Macroscopic to microscopic images of the bioprinted scaffold, scale bars = (ii) 5000 μm and (iii) 500 μm. Reproduced from ref. 22 Copyright from IOP Publishing. B. Optical photographs of 3D bioprinted spheroids using aspiration-based bioprinting. Reproduced from ref. 116 Copyright from Springer Nature. C. Illustration of the BATE concept using spontaneously self-organising building blocks to create large-scale tissues. Reproduced from ref. 117 Copyright from Springer Nature. D. An image sequence showing the embedded 3D printing of a branched, hierarchical vascular network within a compacted EB-based tissue matrix. Scale bar = 10 mm. Reproduced from ref. 118 Copyright from American Association for the Advancement of Science. | ||
In extrusion bioprinting in air, the bioink needs to show high mechanical properties after extrusion to maintain the construct shape and support the deposition of upper layers. These properties, which are essential for printing with a good resolution and accuracy, are usually achieved at the expense of the biomimicry of the matrix, restricting the tissue maturation.112 Bioprinting within a supporting medium answered this limitation by providing a platform to print mechanically weak biomimetic bioinks.112,113 To fulfil this role, the supportive medium needs to show specific properties: while exhibiting a strong shear-thinning behaviour to allow the passage of the nozzle, the supporting medium needs also to show self-healing behaviour to recover its microstructure after the printing. Most importantly, below a stress threshold, the supportive medium need to have a solid-like behaviour to support the printed materials. Particularly, the microtissues should not sediment under the action of gravity.112,114 Bioprinting in a supportive medium combined with the extrusion of bioink has been leveraged to position microtissues and create complex patterns. For example, Campos et al. created a helical structure of bioink containing small spheroids in 3 wt% elastin-like polypeptide-polyethylene glycol (ELP-RGD) inside a Pluronic bath.115 The potential of microgel support baths for the printing of microtissues was also assessed.114 In this support bath, granular microgel particles are densely packed within cell medium. The small size of the particles, between 6–7 μm diameter, allow for the printing of bioinks with fine resolution. In the study, cell aggregates were extruded in 3D arrays and matured for 8 days to produce spheroids, showing that the microgel support bath could support spheroids of 380 μm diameter. These results open the door for fast positioning of preproduced microtissues, as the authors were able to assemble 1000 spheres in a 3D array (2.5 × 2.5 × 2.5 cm3) in less than 1 hour. Additionally, theoretical calculations suggest that spheroids up to 2.5 cm diameter could be positioned without sinking, potentially making the technology suitable for a large range of microtissue sizes. A supporting bath can also be combined with other positioning techniques, such as aspiration-assisted bioprinting, to create complex patterns of microtissues (Fig. 4B).116
The ability to print a mechanically weak bioink in supporting medium was leveraged to its maximum by printing high-density organoid solutions by Brassard et al.117 Their technique, bioprinted-assisted tissue emergence (BATE), consists of an extrusion system coupled to a microscope (Fig. 4C). By having direct visual feedback on the dispensing of the bioink, the user can control and adjust the printing parameters in real time. In this study, common organoid culture matrix showing great biomimicry such as Matrigel and collagen were used as supporting medium. The printing temperature was adjusted to reach the suited properties for the matrix. This setup shows great potential to fabricate complex tissue models thanks to good positioning resolution and its low calibration requirement. Indeed, as the bioink does not contain polymers, the optimisation of the rheological properties of the bioink is minimal, while it is usually time consuming. Moreover, as microtissues are matured in common culture matrices, the adaptation of culture protocol is small. However, in the study, only very small organoids (about 50 μm diameter) were printed, and the printing of bigger organoids might require modification of the supporting matrix to avoid sedimentation while printing.
The main advantage of extrusion bioprinting using MBB is its ability to build centimetre-scale constructs with good accuracy. However, in extrusion bioprinting the microtissues cannot be manipulated individually, and their position in the hydrogel is random. Such random positioning might prevent microtissue fusion in the case of low concentration. Another limitation of extrusion-based printing is that the printing parameters should be optimised for each new bioink composition and nozzle diameter. Therefore, the extrusion of microtissue of a new size, type or in conjecture with a different polymer requires time and is not straightforward.
In conclusion, extrusion bioprinting of microtissue has been successfully established with various types of microtissues. Even though the tissue diameter was always below 400 μm, the technique should theoretically be adaptable to any size by increasing the diameter of the nozzle. Extrusion in supporting medium has been demonstrated for small microtissues and should be further investigated for organoids of bigger sizes. Particularly, the sedimentation of the microtissues during printing should be carefully studied. To further develop extrusion-based bioprinting, new strategies against clogging of the nozzle for highly concentrated microtissue bioinks need to be investigated.
4.2. Sacrificial writing into functional tissue
Skylar-Scott et al., proposed a new approach, sacrificial writing into functional tissue (SWIFT) that inverts the paradigm of extrusion in supporting medium.118 This method consists of printing a sacrificial ink into a living matrix slurry, principally composed of organoids (Fig. 4D).By combining organoids with a mixture of collagen and Matrigel, the living matrix shows a strong shear-thinning behaviour, during printing, and self-healing properties once the stress is removed. The living matrix shows analogous properties to microgels used in embedded printing approaches. After printing, the stiffening of the matrix at 37 °C enables both the external shape of the construct and the internal geometry created by the sacrificial ink to be fixed. Finally, the ink is evacuated, and the construct can be perfused. The method showed good biocompatibility, since the matrix integrity, the viability of the organoids and their microarchitecture were not adversely affected by the passage of the nozzle. The technique was leveraged to print channels in any arbitrary direction, with variable diameter depending on the printing speed. While there is theoretically no upper limit for the channel diameter, the minimum diameter is defined by the size of the spheroid. Indeed, the authors showed that when approaching the characteristic diameter of the organoids, ∼200 μm, the channel could not be printed with accuracy. The lower limit for good accuracy was a diameter of 400 μm.
The main advantage of SWIFT is the ability to create a complex vascular network (Fig. 4D) in an exceptionally dense construct (0.2 billion cells per mL) with dimensions that can exceed 40 mm × 4 mm. Additionally, the application of SWIFT extends further than vascularisation as the authors were also able to print a bioink containing organoids inside the living matrix. SWIFT has the potential to be applied to large constructs (up to 100 mL) with any type of microtissue. Therefore, it could become a standardised method to create a channel network in a biological construct. The main limitation of the technique is the dependence of the channel diameter with the microtissue size. To achieve a fine structure, the size of the microtissue should be minimised, resulting in a large number of produced microtissues for a given volume. In this case, the production of organoids may be time consuming and expensive depending on the cell source.
4.3. Droplet-based bioprinting
In contrast to extrusion-based printing where the bioink is deposited as a filament, droplet-based strategies print droplets either in a drop-on-demand or continuous mode. To print microtissues, droplets were generated by various methods: direct laser printing, micro-valve printing and acoustic printing.Laser-bioprinting was used to dispense alginate microbeads (300–400 μm diameter) containing cell aggregates.119 The cell aggregates were directly produced and grown in the alginate beads. The microbeads were first partially embedded in a gelatine layer on top of a UV-transparent substrate before the localised laser heating of the sacrificial gelatine layer ejected the microbeads onto a second substrate where they were patterned. Fig. 5A illustrates the process of laser-based bioprinting. The method is compatible with 2D patterns as well as 3D patterns by superimposing several layers of beads. The biocompatibility of the technique was ensured as it was shown that a nanosecond exposure of UV-light did not alter the viability of cells.119,120 This technique enabled the direct positioning of cell aggregates that were cultured in a 3D alginate environment without retrieving them from the alginate. However, the method is less suited for microtissues produced in other platforms as their encapsulation would require an extra step. The laser-bioprinting platform is theoretically compatible with microtissues of any size without optimisation of the protocol for each condition. This represents an advantage over extrusion-based bioprinting. In addition, the cells are not exposed to shear stress. However, major limitations are that the technique is expensive, not accessible to general users, and the preparation of the UV-transparent slide is time consuming.120,121
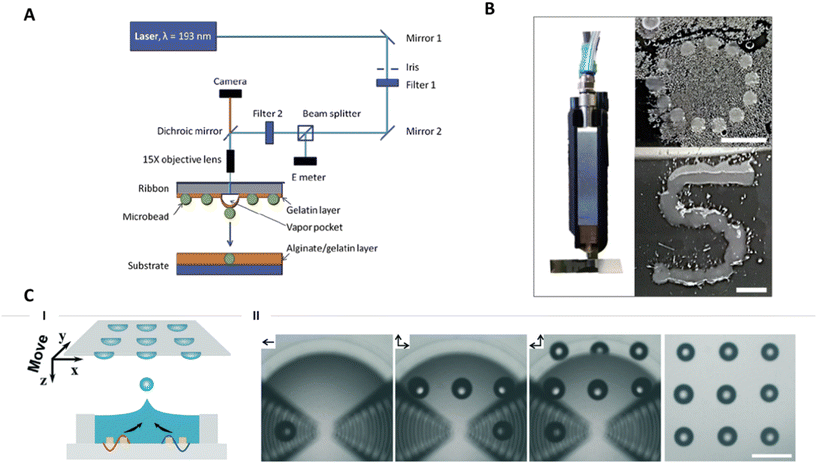 | ||
| Fig. 5 Droplet-based bioprinting. A. Schematic of laser direct-write (LDW) setup. Reproduced from ref. 119 Copyright from IOP Publishing. B. Qualitative drop-on-demand (DoD) printability tests of 3 wt% ELP-RGD printed as single drops into circular and S shapes. Scale bars = 5 mm. Reproduced from ref. 115 Copyright from Frontiers Media S.A. C. (I) schematic and (II) experimental views of the generation of the droplet pattern (“3 × 3” dots) by acoustic printing via moving the receiving substrate in the direction of the arrows. Scale bar = 500 μm. Reproduced from ref. 122 Copyright from the Royal Society of Chemistry. | ||
Drop-on-demand printing of spheroids was realised thanks to microvalves.115 Droplets containing spheroids were successfully patterned into an ‘S’ shape on a surface and printed inside an organ-on-chip (Fig. 5B). The use of a microvalve allows for a high control on the dispensed volume per droplet. This characteristic was useful to engineer organ-on-chip devices with high reproducibility. When using a 300 μm valve and applying minimum pressure, the droplet's smallest diameter was 1.4 mm. Considering the small size of the spheroids (50 μm diameter) reported in that study, several spheroids are present in the same drop. Therefore, the approach described does not provide precise positioning of individual microtissues. To improve the positioning, smaller droplets could be produced using a smaller microvalve or the size of the microtissues could be increased. In both cases, the shear stress induced on the microtissues might increase and should be thereby carefully assessed.
Finally, acoustic-based bioprinting has been developed to eject spheroids encapsulated into gelatin-methacryloyl (GelMa) droplets.122 The device generates two pulsed SAWs which interfere with each other in the middle of the fluid chamber creating an intense acoustic pressure profile (Fig. 5C). The high pressure pushes the liquid–air interface in the air and creates a droplet. In contrast to micro-valve printing, the drop size is similar to the microtissue size, and therefore only one microtissue is present per droplet. The acoustic-based printing is limited to bioink with low viscosity and leads to droplet layers with low mechanical properties. Consequently, to print 3D constructs each layer of droplets need to be first gelled by UV before being able to support a subsequent layer. This nozzle-free technique has the advantage of positioning microtissues with low cell damage because no shear stress is involved. Indeed, it was shown that the viability of the cells after printing is higher than in extrusion-based bioprinting. However, the technique is quite slow due to the individual generation of droplets and the curing time. The throughput could be improved though implementation of higher frequency and automation.
4.4. Volumetric bioprinting
Volumetric bioprinting is a light-based method, which enables building of a bioconstruct in a photocurable bioresin.123,124 In opposition to the previous bioprinting techniques, which are characterised by the mechanical deposition of the bioink, volumetric printing is based on the selective chemical crosslinking of a photoresponsive hydrogel to sculpt a 3D construct directly from the hydrogel solution. After crosslinking, the construct needs to be retrieved from the monomer solution.In volumetric bioprinting, the construct is generated by illuminating the bioresin from multiple angles with a sequence of filtered back projections. The photopolymerisation kinetic and the scattering inside of the bioresin govern the printability. This method is capable of building centimetre-scale structures in tens of seconds making it the fastest bioprinting method discussed in the present review. Additionally, overhang elements can be built with volumetric bioprinting without the help of sacrificial materials.
A recent paper has for the first time demonstrated the use of volumetric bioprinting with a microtissue-laden bioresin.123 The authors were able to print constructs with fine structure down to 50 μm in bioresin containing organoids with a diameter of 275 μm. The good resolution was achieved thanks to the addition of a refractive index matching compound, iodixanol. This compound, which was proven to be biocompatible, tunes the refractive index of the hydrogel to match the one of the microtissues, thereby reducing scattering. The optimal concentration of iodixanol needs to be optimised for each cell type as they exhibit different light scattering properties. In the study, the volumetric printed organoids showed better viability and hepatic function than extrusion printed and casted organoid controls. This difference was attributed to the minimal shear stress present in volumetric printing compared to the other techniques.
The main advantage of volumetric bioprinting is its speed and ability to print overhanging structures typical of native tissues. However, to achieve good printability, the optimisation of the light pattern and the tuning of the bioresin's refractive index is needed. The complexity of optimising the operating parameters is therefore similar to extrusion bioprinting. Finally, a disadvantage of volumetric bioprinting, is that it required a larger volume of microtissue-laden bioresin than the final volume of the construct.
5. Substrate modifications
Microtissues can be positioned through the modification of the substrate that carries them. Modifications can be classified into two main categories: structural modification of the substrate to create cavities with a specific shape and chemical modification of the surface to create cell-adhesive or cell-repellent regions.5.1. Substate structural modifications
Structuring of substrates consists of creating features with different topography levels, and it is commonly done by casting a liquid solution into a mould. This fabrication technique is readily accessible since it does not involve complex or expensive instruments.Hydrogel micro-structures have been used to compartmentalise individual microtissue into microwells, creating a tissue microarray (TMA) (Fig. 6A). The TMA enabled the positioning of multiple microtissues in a defined 2D array that increased reproducibility and throughput of histology and mass spectrometry processing.33,34,125 With this approach, micro-tissues sedimented and aligned on a plane at the bottom of the microwells. After sedimentation, the position of the microtissue was fixed by hydrogel embedding, providing stability to the TMA for further processing steps. Then, during the preparation process prior the analysis, samples were cut perpendicular to the microtissue plane in sections of a few micrometres.
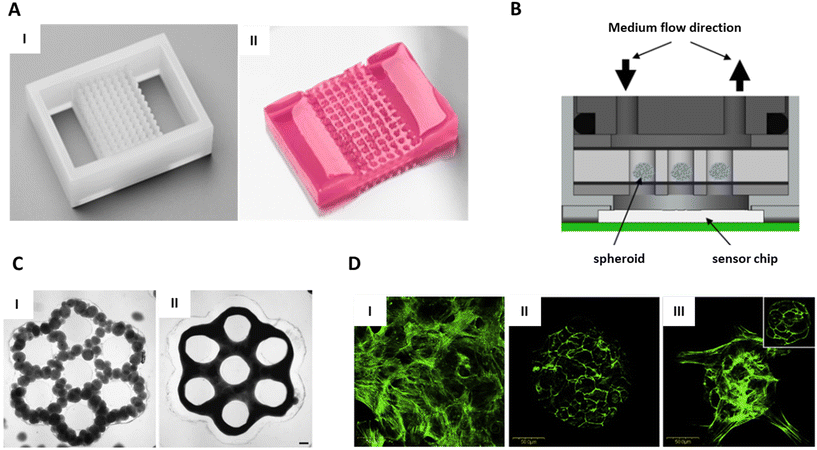 | ||
| Fig. 6 Substrate modification for positioning. A. Agarose 96-well microarray for tissue microarray. (I) Silicon negative mould. (II) Pink-dyed agarose microarray. B. Schematic design of a chip using a hydrogel microarray to immobilised spheroids on top of a sensor. Reproduced from ref. 32 Copyright from Springer Nature. C. Geometrical features in agarose to guide microtissue fusion. (I) and (III) Individual spheroids just after dispensing. II and IV) Fused tissue after 24 hours. Scale bars = 200 μm. Reproduced from ref. 126 Copyright from John Wiley and Sons. D. F-actin staining using a confocal microscopy in the bottom cell layer near the cell-substrate interface (green). (I) RGD monolayer promote cell adhesion. (II) Galactose monolayer result in spheroids formation and detachment. (III) Hybrid RGD/galactose coating result in the formation of a tethered spheroid. Scale bars = 500 μm. Reproduced from ref. 133 Copyright from Elsevier. | ||
In endpoint analysis, where thin sections are needed, precise co-planar alignment of the microtissues is crucial to maximise efficient processing. Gabriel et al. have shown that 85% of the 500 μm spheroids initially embedded in a 96-well agarose array were present within five consecutive sections demonstrating the efficiency of TMA for coplanarity alignment.33 Therefore, due to its ability to simultaneously process and analyse tens of microtissues, TMA is expected to replace manual and random microtissue embedding for micro-histology and mass spectrometry. However, with TMA, microtissues are aligned at the bottom, limiting the coplanarity of the centre of mass to monodisperse microtissue samples. In addition, manual loading of microtissues in previous works hinders the implementation of this technology for high-throughput analysis. To answer this problem, Heub et al. developed a hydrogel microarray, whose format was compatible with automated loading of microtissues by pick-and-place techniques.34 Aside from endpoint analysis, other applications of TMAs have been reported. In another study, small 9-well TMAs were placed on top of a sensor in a “spheroids-on-chip” platform (Fig. 6B). Here, spheroid immobilisation enabled continuous monitoring of pH and oxygen consumption, while a solution containing chemicals was perfused in the system.32
Patterned surfaces have also been studied as a reliable and reproducible approach for fusing multiple cell aggregates or organoids.126,127 Self-assembly is driven by the cohesive cellular activity of heterogeneous organoids. In the case of random positioning, self-assembly results in heterogenous tissues. Moreover, the time to achieve fusion is proportional to the spacing of the microtissues and is therefore variable from one sample to another. Geometrical constraints on a non-adherent substrate enable the initial contact of multiple organoids, promoting fast fusion, and guide the conformation of the assembly in the desired shape. Microtissue assembly in patterns such as concave or tubular microwells to more complex toroid or honeycomb shapes has been achieved on agarose hydrogels and elastomers (Fig. 6C).126,127 Compartmentalisation of organoids in hydrogels has also been used to study the process of fusion.126 When positioning the organoids in a line, the authors could easily quantify the kinetic and the extent of fusion by measuring the rod contraction resulting from the assembly.
The biggest advantages of substrate structuring are its ease of use, its accessibility, and its low price. But the main limitation is that each new design requires the fabrication of a new mould. Surface structuring, therefore, provides an efficient strategy for applications where a standard design can be used, such as TMA for endpoint analysis, but might become time consuming and expensive for applications requiring personalisation of designs such as biofabrication. TMA was demonstrated to have an efficient strategy to perform batch analysis of histology and mass spectrometry. However, the method is limited to monodisperse and homogeneous microtissue populations. Indeed, heterodispersed microtissues will not be aligned by their centre of mass and, since there is no control over the orientation of the microtissues, specific structural features such as buds will not be aligned. By designing microwells with a big diameter, TMA can be compatible with a large range of microtissue sizes, therefore can providing a standard method for monodisperse populations.
5.2. Surface chemical functionalisation
Chemical coatings can be used to promote or hinder cell adhesion onto a surface. The coatings are often selectively applied to a surface to create a defined pattern of cells. A standard method to pattern a coating is to use a PDMS stamp to deposit the coating on the surface.128By promoting cell adhesion, surface functionalisation has been used to pattern cells in arrays or complex geometries on various substrates such as metal, plastic, and glass.129,130 To our knowledge, this technology has not yet been applied to pattern pre-produced microtissues. However, recent studies demonstrating the possibility of generating spheroids directly tethered to a surface paved the way toward positioning through surface functionalisation.131,132 Tasmin et al. demonstrated that a galactose and Arg-Gly-Asp (RGD) hybrid coating with an optimised ratio supported the formation of hepatocyte spheroids directly tethered to a polyethylene terephthalate (PET) membrane or a polystyrene (PS) multi-well plate.131 In this system, the cells adhered strongly to RGD, while galactose was hypothesised to repel cells from the surface (Fig. 6D I and II).133 When using an optimal ratio of RGD/galactose, the RDG ligands provided sufficient binding points to anchor the bottom layer of the seeded cells to the substrate, while the bulk cells aggregated to form spheroids (Fig. 6D III). The strong anchorage of the spheroids allowed the system to be used with automated media exchange without losing spheroids.131 Similarly, Turner et al. formed tethered spheroids of different cell types on elastin-like polypeptide polyethyleneimine (ELP-PEI) coated PS well-plates.132,134 These scaffold-free approaches have several advantages for drug testing assays over multi-spheroid systems using exogenous scaffolds, such as collagen. Indeed, they prevent the aggregation of spheroids into big spheroids with a necrotic core, they decrease the spheroid loss during media exchange, and they reduce the variability in assays since they are not using collagen with significant lot-to-lot variability. Additionally, these immobilisation methods, which are compatible with materials used in standard labware, such as PS multi-well plates, support the development of standardised positioning methods for organoid culturing and organ-on-chip platforms. To push this technology toward positioning of microtissues, future work in the field should investigate if pre-formed spheroids could be immobilised on the aforementioned coatings while maintaining their integrity and specific function.
6. Non-contact active forces
Non-contact active forces, including acoustic, magnetic, dielectrophoretic, and optical forces, provide a contactless and often gentle approach to assemble and manipulate biological objects.9 Dielectrophoresis and optical traps have been used primarily to assemble multi-cellular aggregates; however, they have not yet been studied extensively for manipulation of microtissues.135,136 Therefore, in this section we focus on acoustophoresis and magnetophoresis as useful non-contact forces for the manipulation of microtissues.6.1. Magnetophoresis
Magnetic forces can be used for contactless manipulation of microtissues. The use of magnetic fields has the advantage of enabling large working distances (∼100 mm) and accuracy.137 However, only paramagnetic objects can be manipulated using magnetism. Therefore, it is necessary to consider both the manipulation techniques and the method(s) to label the microtissues (i.e., magnetise them).Magnetisation of cells is needed to enable magnetic manipulation for cell separation, enrichment, and patterning.138 One of the common approaches is to co-incubate cells with magnetic nanoparticles (MNPs), where these particles are internalised through endocytosis or phagocytosis.139 The uptake of MNPs by cells has been discussed extensively elsewhere.140 For magnetisation of spheroids, either magnetised cells can be used for spheroid formation or magnetic particles can be incorporated into spheroids.
Olsen et al. added MNPs to the cells during the formation of spheroids using the hanging-drop technique to induce magnetisation.141 They enabled fusion of magnetised smooth muscle cell spheroids by simple placement of a magnet below the culture container (Fig. 7A). This resulted in an increased cell–cell interactions and accelerated the fusion of spheroids. Since MNPs are internalised by the cells, they can interfere with intracell signalling and can cause concerns about cytotoxicity. To counter this, Bratt-Leal et al. magnetised their microtissues by pelleting embryonic stem cells (ESCs) in AggreWell 400 inserts, followed by addition of commercial magnetic polystyrene microparticles (magMPs, Ø ∼ 4 μm).142 The magMPs were trapped in the microtissues and since the magnetic particles were not internalised by the cells, this served as an attractive route for magnetisation of microtissues. They also used multiple magnets to achieve customisable patterning in microtissues (Fig. 7B). In the same work, they demonstrated controlled magnetisation of iron pillars embedded in PDMS to manipulate single spheroids (Fig. 7C). Cellular uptake of bacteriophage hydrogels has been leveraged to introduce magnetic iron oxide (embedded in the gels) to magnetise microtissues.143 They have used this technique to show fusion of astrocyte and glioblastoma microtissues. Novel approaches which do not require magnetisation of spheroids are also being explored. Instead of magnetisation of microtissues, the cell medium can be spiked with paramagnetic medium to apply magnetic forces on the cells or microtissues using negative magnetophoresis.144,145 A variety of microtissues (human stem cells (MSCs), breast adenocarcinoma cells (MDA-MB-231), and brain cancer cells) were shown to be compatible with this technique. Tocchio et al. also showed spheroid merging with single cells as well as with other spheroids (Fig. 7D).
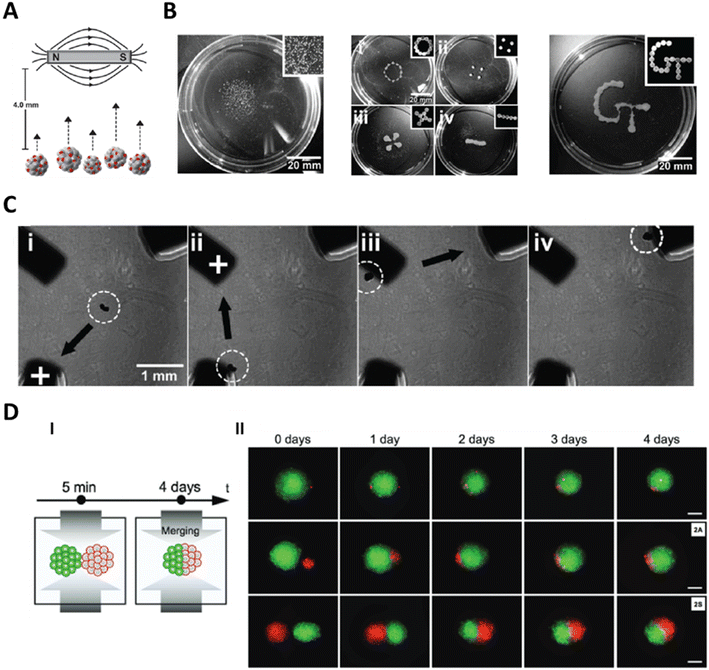 | ||
| Fig. 7 Magnetophoretic manipulation of microtissues. A. Magnetised microtissues can be manipulated by using a permanent magnet. Reproduced from ref. 142 Copyright from Oxford University Press. B. Use of multiple magnets can enable positioning of multiple microtissues and formation of custom patterns of microtissues. Reproduced from ref. 142 Copyright from Oxford University Press. C. Controlling the magnetisation of iron beams (corners of the image) enables manipulation of the microtissues towards the pillars. Reproduced from ref. 142 Copyright from Oxford University Press. D. Magnetic forces can be used to merge two microtissues together or merging a single cell with a microtissue. Scale bar = 100 μm. Reproduced from ref. 145 Copyright from John Wiley and Sons. | ||
Magnetophoresis, is a relatively rapid method of manipulation of microtissues. In magnetophoresis, the force applied on the microtissues can be tuned by changing the concentration of the magnetic component of the microtissues. This can provide a degree of control over the speed of manipulation. Another advantage is that a simple setup with a permanent magnet can be used for the manipulation of magnetic microtissue. More precise and accurate manipulation systems have also been developed for controlling micro-robots using magnetic fields and could be employed for manipulation of organoids in the future.137 One of the obvious drawbacks is that this technique requires magnetisation of microtissues, which can impact cell function. Using negative magnetophoresis offers a solution but the effect of changes to the cell medium has to be studied for each case.
6.2. Acoustophoresis
Acoustophoresis uses forces arising from acoustic fields to manipulate objects. In addition to being non-contact, acoustophoresis is also label-free and biocompatible.146Typically, acoustic standing waves are produced in a fluid where particles or cells are manipulated to the pressure nodes of these fields.147 Various forms of acoustophoretic devices have been used for spatial control of microtissues. In general, these devices can be classified in three different types based on their principle of actuation: Faraday waves, bulk acoustic waves (BAW), and surface acoustic waves (SAW).
Vibrating plate devices use Faraday waves to pattern particles or cells, much like the patterning of sand shown by Chladni in late 18th century.148 Various researchers have used similar techniques to pattern cells to encourage formation of tissues constructs.149 Faraday waves can be formed due to vertical vibration of the liquid layer (Fig. 8A). Demirci and colleagues used vibrating plates to pattern individual cells and microtissues.150–152 In their work, they patterned many spheroids (>104) using Faraday waves over a large surface area (∼4 cm2). After patterning, they characterised how individual spheroids fused together to form organoid-like structures (Fig. 8A). They patterned two different types of spheroids – composed of human umbilical vein endothelial cells (HUVECs) or NIH 3T3 murine fibroblasts – together to encourage vascularisation in patterned constructs. Further, they fabricated hepatic constructs using the acoustic assembly of liver organoids to mimic the bile canalicular network.151 While these examples demonstrate the utility of Faraday waves to organise large-scale constructs, the accessible pattern shapes are constrained by physical factors including the height of the fluid in which the patterns are created.153 Thus, Faraday waves are generally limited to producing near-surface patterns.
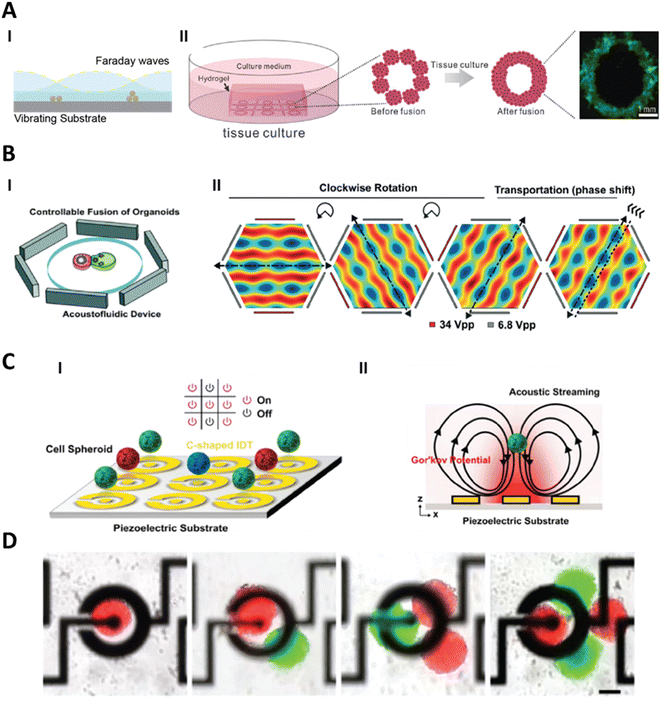 | ||
| Fig. 8 Acoustophoresis for manipulation of microtissues. A. (I) Faraday waves act through vibration of substrate plates that induces standing waves in the fluid. Hydrodynamic forces on microtissues resulting from the Faraday waves manipulate them towards the pressure nodes. (II) Such a system has been used to form a larger assembly of multiple organoids to form a tissue construct. Reproduced from ref. 151 Copyright from John Wiley and Sons. B. In bulk acoustic wave (BAW) devices, vibrations from the piezoelectric transducer are coupled with the entire structure of the acoustofluidic device. These devices are designed to produce acoustic standing waves in the fluid by the superposition of travelling waves. In this device, 3 pairs of piezoelectric transducers are used which leads to control over the translation and rotation of the microtissues. Reproduced from ref. 15 Copyright from the Royal Society of Chemistry C. Surface acoustic wave (SAW) devices use patterned electrodes on a piezoelectric substrate to form an interdigitated transducer (IDT) to produce surface waves in response to electric signals. In this device the IDTs are designed to produce acoustic radiation force and acoustic streaming. The acoustic radiation force leads to the manipulation of the microtissues towards the centre of the electrodes and the acoustic streaming leads to levitation of the microtissues. Reproduced from ref. 157 Copyright from IOP Publishing. D. This device was used not only to manipulate single organoids but also to combine multiple organoids. Reproduced from ref. 157 Copyright from IOP Publishing. | ||
In addition to the assembly of larger tissue structures, acoustic forces have been used to study the interaction between different microtissues in a controllable manner. BAW devices use a piezoelectric transducer coupled with a high-Q (high quality factor) material to produce standing acoustic waves in a fluid to enable acoustophoresis.154 High-Q materials have lower damping and are ideal for designing resonant devices. Guo and colleagues used BAW devices to form neurospheroids and then to assemble different brain organoids together.15,155 They used a hexagonal acoustofluidic device to rotate and transport organoids, allowing for controllable fusion by independently operating each of the opposing pairs of transducers (Fig. 8B). They exploited this control to fuse human forebrain organoids (hFOs) and human midbrain organoids (hMOs) to form assembloids. The rotational control over the organoids allowed them to account for the heterogeneity of the neuroepithelial buds present within hFOs and improve neurite projection when they were fused with the hMOs. While the rotation of the organoid presents unique advantages, only one organoid can be manipulated (while the other organoid is immobilised). There are however other techniques where multiple organoids can be handled simultaneously.
Like BAW devices, SAW devices have also been used for the production and handling of microtissues.156 In SAW devices, waves are produced by interdigital transducers (IDTs) on a piezoelectric substrate and the formed waves leak into the fluid as a surface wave at the solid–fluid interface. An advantage of SAW devices is that the IDTs can be patterned on the piezoelectric substrate tailored for the specific application. This was leveraged to utilise the streaming arising from surface acoustic waves to controllably manipulate, levitate, and combine organoids on a micropatterned piezoelectric substrate.157 C-shaped IDTs were used to create acoustic fields to levitate organoids at the centre of the patterned area (Fig. 8C). Many such patterned IDTs were used in tandem to enable transport and fusion of multiple organoids (Fig. 8D). Apart from the works described here, there has been an increasing interest in developing non-contact acoustic tweezers using both streaming and acoustic radiation forces arising from acoustic fields.158 While not yet demonstrated for the manipulation of microtissues, these techniques could be adapted for this aim in the future.
Overall, acoustofluidics has been used for a variety of microtissue sizes. Since the positioning of the microtissues is based on the pressure nodes formed by the acoustic fields, this technique can be used irrespective of the size variation in the microtissues. This can be advantageous as the same devices can be used for manipulating microtissues of any kind. Further, forces generated due to the acoustic fields scale with the volume of the object to be manipulated. Since microtissues are relatively large compared to cells, the speed of acoustic manipulation of microtissues is high.
While acoustophoresis has been applied broadly in the organisation and manipulation of living cells and organisms, there is a need to better characterise how the acoustic forces themselves may affect the underlying biology of microtissues during and after operation. Further, the effect of acoustics on microtissue integrity needs to be studied. Understanding what operating conditions do not disrupt microtissue structure will be necessary for broad implementation. With more interest in the study of microtissues, a better understanding of the forces involved in acoustic patterning of microtissues is also necessary. Most of the theories developed for acoustofluidics have been made assuming Rayleigh limit (i.e., wavelength of the acoustic field is much larger than the size of particles). This is a valid assumption when we consider single particles or cells but for manipulation of larger microtissues, this assumption does not hold true. In these regimes there are numerical models that can be used;159 however, a theoretical analysis is still lacking.
Both acoustophoresis and magnetophoresis offer a unique advantage and show various uses for their applications. However, both methods in their conventional forms cannot be applied selectively to individual microtissues, or separately to multiple tissues. For those purposes, manipulation using optical forces could be of interest, which have been used to manipulate single cells but not microtissues. Digitally switchable acoustic holograms do offer a unique potential for creating complex acoustic fields but are yet to be used with microtissues.160 Techniques such as electrostatic gating offer the potential for switchable magnetism in nanoparticles.161,162 Further development of such techniques and their incorporation into microtissues could lead to selective magnetic manipulation of microtissues.
7. Perspectives of manipulation and positioning tools
As previously anticipated by Ouyang et al., interest in the field of living building block assembly has increased in the last few years.9 Particularly for microtissue assembly and handling, which has intensified and the number of publications about this subject has significantly risen. The different microtissue handling techniques have enabled the formation of complex tissues, the patterning of microtissues into complex designs, as well as individual manipulation of microtissues with high precision.Despite several significant advances, the methods require additional specifications to efficiently support personalised medicine. Notably, they should be compatible with standardised workflows while allowing customisation for specific standard operating procedures (SOP). In addition to quality and safety, upscaling is also of particular importance.
While every technique has its unique advantages, we have identified the following criteria to compare them: standardisation, implementation of SOP, upscaling, complex 3D positioning, and integration in closed system (Table 1). Standardisation aims at making a process robust, by maximising compatibility, interoperability, and repeatability. Here, the analysis concentrates on two aspects. The first one is the compatibility of the labware or system with microtissues exhibiting different characteristics, such as size, shape, and stiffness. The second aspect is the potential to position the microtissues reliably. The goal is to increase the reproducibility of positioning (with a given protocol and calibrated device) independently of the operator. The customisability in SOP offered by the methods is also evaluated in two parts. First, the ease of implementing new protocols with different microtissue types or different media is considered. This criterion assesses the optimisation or calibration effort that should be made to define the new operating parameters. Second, the flexibility to generate a new positioning pattern with the same device and conditions is evaluated. Additionally, working in fully closed systems is often sought to minimise the risk of contamination. A closed environment enables working outside of a biosafety cabinet while insuring sterile conditions. In this perspective, the compatibility of the methods with closed environment is considered. The potential for upscaling is assessed based on the ability to simultaneously manipulate multiple microtissues, the potential to work with larger samples, and the ability to parallelise the technique within the same device. Finally, the potential to position and manipulate in 3D was evaluated as it is an important criterion in applications linked to in vitro model reconstruction. The complete evaluation is summarised in Table 1. The presented positioning strategies are still in the early stage of development, and consequently the analysis is based on the current state of the art, which for some methods is limited to few studies. Based on the novelty of the field, the proposed discussion aims to bring a perspective for future developments of the techniques.
| Positioning or manipulation techniques | Standardisation | New standard operating procedures implementation | Upscaling | 3D complex positioning | Integration in fully closed system | |||
|---|---|---|---|---|---|---|---|---|
| Microtissue diversity | Reliable process | Cell and/or media | Positioning pattern | |||||
| Compatibility of the labware/system with different microtissues (size and mechanical properties) | Reproducibility of the positioning with a given protocol and calibrated device but different operators | Ease to implement new protocols with different cells and/or media | Flexibility to generate a new positioning pattern with the same device and conditions | Simultaneous microtissues positioning and/or parallelisation within a device | Ability to position in 3D | Sterile conditions are insured during the positioning without a biosafety cabinet | ||
| Hydrodynamic | Microfluidic chips | − | + + | + | − − | + + | − − | Yes |
| Microfluidic droplets | + + | + + | + + | − − | + + | − − | No | |
| Pipetting-based systems | + | + + | + | + + | + | + + | No | |
| Hydrodynamic-based micromanipulators | + + | + + | − | − | − − | − | No | |
| Bioprinting | Extrusion-based printing | + | − − | − − | + + | − | + + | No |
| SWIFT | + + | − | − − | + + | − | + + | No | |
| Droplet-based printing | + + | − | − | + + | − | + | No | |
| Volumetric printing | + + | − | − − | + + | + | + + | Yes | |
| Substrate modifications | Substrate structuration | − | + + | + | − − | + + | − | Yes |
| Surface chemical functionalisation | − | + + | − | − − | + + | − − | Yes | |
| Non-contact active forces | Magnetophoresis | + + | + + | + | + | + | + + | Yes |
| Acoustophoresis | + + | + + | + + | + | + | + | Yes | |
Interestingly, the analysis shows that most of the techniques have a high to very high potential of compatibility with microtissues of different sizes and mechanical properties. Only microfluidic chips and substrate modification approaches have moderate potential but nevertheless could address diverse microtissues by adapting the design of the device. Extrusion-based bioprinting methods would also need to adapt to microtissues of different sizes by changing the nozzle sizes. Additionally, while Bhattacharjee et al. suggested that extrusion in supporting medium could be compatible with microtissues up to 2.5 cm diameter,114 the compatibility of this method with microtissues of 100–500 μm still needs to be demonstrated. Although all the methods are compatible with microtissues of different sizes, they have been, to date, only tested and validated with monodisperse microtissue populations with a small range of sizes (either small, medium or big dimensions depending on the approaches). Nonetheless, since established processes enable the production of monodisperse microtissue populations, the need for techniques working with heterogenous populations is small. Finally, reported research focused on microtissues with a round morphology and no data are available about microtissues with elongated or asymmetric morphologies. Although, standardised methods are often demonstrated with spherical microtissues populations, the morphology is inherent to the organ model and cell source. Indeed, the round morphology is not universal, and asymmetrical organoids occupy an important place in research (e.g. gut, retina, brain, cancer research).163 In conclusion, limited work about the compatibility of the methods with variable microtissues has been performed to date and their capability needs to be investigated by future experimental work.
Detailed analysis shows that, except for hydrodynamic-based approaches, the techniques often follow similar trends within a category. Therefore, Table 1 will be further evaluated by positioning family, i.e.: hydrodynamic, bioprinter, substrate modification, and non-contact active forces. Based on the features of each group, promising applications in regenerative medicine will be mentioned.
Microfluidic techniques show more variability between the different criteria than the other categories. This can be explained by the significant difference in working mechanism and system specification between the methods. The common point between these techniques is their high to very high potential to be operator-independent, thereby leading to reliable results. This property either arises from the potential of the systems to be fully automated or, in the case of microfluidic chips, from the little operating steps required. Pipetting-based systems have higher flexibility in positioning patterns and very high positional accuracy in 3D, which are comparable to bioprinters. They are however more user-friendly and operator-independent than bioprinters, as they do not require viscous bioinks and demand less optimisation steps. However, their capacity to upscal appears lower than other reported methods because every microtissue should be positioned individually. For this reason, pipetting-based systems might be advantageous to create precise patterns of a few microtissues but are still not suited to build tissue constructs in tissue engineering. The high potential of aspiration-based systems resides in pick-and-place applications either to perform transfer of microtissues between standardised labware, for drug assay or end-point analysis for instance, or by combining them with image analysis, for example, for sorting. In addition, pipettes can be readily parallelised thereby increasing the potential for upscaling for pick-and-place with standardised labware.
Bioprinters have a very high flexibility of positioning patterns, in 2D and 3D, which is inherent to additive manufacturing processes and enables the creation of complex personalised bioconstructs at a relatively low cost. However, the implementation of positioning using bioprinting is limited due to the complexity of operating the system and executing new protocols, requiring skilled operators to use the system and achieve reproducible constructs of good quality. This problem is enhanced by the lack of standard bioink formulation, which varies for each application and cell type. Additionally, current bioprinter technologies, with the exception of volumetric bioprinting, offer little potential for upscaling because they are difficult to parallelise and their operating speed is slow, thereby preventing the fabrication of large bioconstructs. Volumetric bioprinting however circumvents this speed limitation and shows higher potential of upscaling than the other bioprinting methods. Finally, bioprinters, similarly as pipetting-based systems, are in general incompatible with closed systems, and therefore should be used in combination with a safety cabinet. Overall, the positioning pattern flexibility of bioprinting is interesting for tissue engineering, where personalised bioconstructs would open the door to in vivo replacement of injured or damaged tissues. Nevertheless, positioning in non-closed systems may create safety problems and these should be carefully assessed. Therefore, bioprinters are potentially going to be deployed, but limited to highly specialised fields of use, and the availability of standardised, versatile equipment or methods is not foreseen.
The techniques related to the modification of substrates are characterised by a high potential for upscaling. Indeed, the systems could be easily parallelised and many microtissues could be positioned simultaneously. Moreover, the positioning processes do not require complex equipment, making them easily accessible, and lead to very reproducible positioning of the tissues. However, the positioning patterns are determined by the specificity of a given device, mostly confined in 2D and not flexible. To position the microtissues into a different pattern, a new device should be fabricated, which could be costly. Also, each device is limited to microtissues with a defined size range. Therefore, substrate modification is interesting for applications where standardised positioning patterns can be used. For example, it is suited for end-point analysis where a standard array of microtissues is required. Additionally, it might be more advantageous than bioprinting for tissue engineering applications where simple standard shapes are needed such as tubes for vein engineering.164
Non-contact active forces have a high to very high potential for every criterion and are therefore very promising. However, the possible cytotoxicity of magnetophoresis is not considered in Table 1 and significantly reduces its potential for regenerative medicine. For magnetophoresis, since the positioning of the microtissue is defined by the properties of the magnetic labels, the cell type and the medium do not significantly impact the positioning pattern. In the case of acoustophoresis, while the medium density and speed of sound could influence the positioning pattern, the effect is minor for water-based medium. For these reasons, new SOPs can be easily implemented with minimal optimisation of the operating parameters. Considering the positioning pattern flexibility, complex devices with several transducers would allow the generation of a wide range of 2D and 3D patterns. For example, 3D volumetric acoustophoretic displays have already been demonstrated with millimetric particles.165 However, the ability to create complex features would be more limited than that of 3D printing or aspiration-based methods. Another major advantage of acoustic and magnetic devices over other techniques is their compatibility with fully closed systems. The techniques therefore not only minimise the risk of contamination during the positioning process but can also be used with systems where the physical access to the microtissues is restricted such as organ-on-chip. Thanks to their versatility in positioning patterns and their user-friendliness, non-contact force techniques meet the needs of various research areas such as disease modelling, tissue engineering, and end-point analysis.
In addition to the engineering characteristics of the strategies previously discussed, the question of cytotoxicity and its impact on cell fate is a key factor to consider. Based on the current studies, all the different methods for positioning and manipulation do not seem to adversely affect the cell viability and cell fate. However, due to the novelty of the field, the available data are limited and disparate. There is a lack of homogeneity between the assays and parameters (time point and cell type) used to analyse the cytotoxicity and impact on cell fate. These disparities are problematic because reliable conclusions in these areas are at an early stage. Indeed, for some applications such as tissue engineering or in vitro models, the process of building constructs is inherently linked to the final goal of producing viable and functional tissues.10 One potential future solution to this challenge could be the standardisation of assays with defined parameters. While this seems feasible concerning cytotoxicity, the complexity behind the concept of cell fate makes this idea more ambitious.
8. Conclusion
The need for microtissue manipulation and positioning has emerged in the last decades and the diversity of techniques reported shows a promising future for the field. The tools to manipulate and position microtissues were used throughout all microtissue life stages, from sorting to assembling to end-point-analysis and could significantly improve the reliability and the efficiency of the research. Additionally, the range of applications of microtissues is growing and creating an increasing need for controlled positioning, thereby placing it as key player in the development of personalised medicine.Regenerative medicine and drug testing, which includes the entire process from organ and disease modelling to drug screening and preclinical trials, are major research areas in personalised medicine. Every application has specific requirements in terms of standardisation and throughput and therefore, has unique constraints for the positioning techniques. In fundamental research, the assembly of various microtissues to generate assembloids or larger multi-tissue-interaction models requires the precise positioning and orientation of the samples. Once mastered, this strategy can improve the reliability of the results and support the advances of organ mechanisms and support research linked with the evolution of the model on long term culture. In drug screening, hundreds of homogeneous microtissues are exposed to different molecules in standardised assays. High throughput is a crucial parameter, while variability in the new SOP and the flexibility of the positioning patterns is not as essential. Consequently, microfluidic chips and DCST methods have a high potential for drug screening. In preclinical trials, promising drugs are tested on a variety of microtissues to assess their performances in different conditions. Unlike drug screening, here the compatibility of a technique with a variety of microtissues and its adaptability with a new SOP is of critical importance. However, a moderate throughput is usually sufficient for preclinical trials. The implementation of pipetting-based systems and acoustophoresis methods in a preclinical trial workflow is therefore promising. In the case of regenerative medicine, small batches of microtissues derived from patients must be handled in a fully closed system to fulfil safety regulations, and to guarantee sterility. Precise assembling and flexibility in positioning patterns are also required to produce personalised tissue constructs, along with system compatibility for microtissues showing different characteristics, due to the inherent differences among donors. Acoustophoresis and volumetric bioprinting fulfil these criteria and have been shown to be good candidates for regenerative medicine.
Looking to the future, techniques for manipulation and controlled positioning of microtissues have been identified as a major bottleneck to fully exploiting and implementing microtissues in personalised medicine and will require increased attention. The analysis of currently reported tools with respect to standardisation, upscaling and integration potential enabled the most promising techniques for drug discovery and regenerative medicine to be highlighted. For the next few decades, research in this field is expected to intensify and boost the adoption of microtissues in personalised medicine.
Conflicts of interest
There are no conflicts of interest to declare.References
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef CAS PubMed.
- M. A. Kinney, T. A. Hookway, Y. Wang and T. C. McDevitt, Ann. Biomed. Eng., 2014, 42, 352–367 CrossRef PubMed.
- D. Barbone, T.-M. Yang, J. R. Morgan, G. Gaudino and V. C. Broaddus, J. Biol. Chem., 2008, 283, 13021–13030 CrossRef CAS PubMed.
- K. Schneeberger, B. Spee, P. Costa, N. Sachs, H. Clevers and J. Malda, Biofabrication, 2017, 9, 013001 CrossRef.
- E. Driehuis, K. Kretzschmar and H. Clevers, Nat. Protoc., 2020, 15, 3380–3409 CrossRef CAS PubMed.
- P. Zhuang, Y.-H. Chiang, M. S. Fernanda and M. He, Int. J. Bioprint., 2021, 7, 444 CrossRef CAS.
- M. C. Decarli, R. Amaral, D. P. dos Santos, L. B. Tofani, E. Katayama, R. A. Rezende, J. V. L. da Silva, K. Swiech, C. A. T. Suazo, C. Mota, L. Moroni and Â. M. Moraes, Biofabrication, 2021, 13, 032002 CrossRef CAS PubMed.
- N. Brandenberg, S. Hoehnel, F. Kuttler, K. Homicsko, C. Ceroni, T. Ringel, N. Gjorevski, G. Schwank, G. Coukos, G. Turcatti and M. P. Lutolf, Nat. Biomed. Eng., 2020, 4, 863–874 CrossRef CAS PubMed.
- L. Ouyang, J. P. K. Armstrong, M. Salmeron-Sanchez and M. M. Stevens, Adv. Funct. Mater., 2020, 30, 1909009 CrossRef CAS.
- G. Eke, L. Vaysse, X. Yao, M. Escudero, A. Carrière, E. Trevisiol, C. Vieu, C. Dani, L. Casteilla and L. Malaquin, Cells, 2022, 11, 1394 CrossRef CAS PubMed.
- M. C. Decarli, R. Amaral, D. P. dos Santos, L. B. Tofani, E. Katayama, R. A. Rezende, J. V. L. da Silva, K. Swiech, C. A. T. Suazo, C. Mota, L. Moroni and Â. M. Moraes, Biofabrication, 2021, 13, 032002 CrossRef CAS.
- G. Agrawal, A. Ramesh, P. Aishwarya, J. Sally and M. Ravi, Biotechnol. Prog., 2021, 37, e3126 CrossRef CAS PubMed.
- A. Bongso, C.-Y. Fong and K. Gauthaman, J. Cell. Biochem., 2008, 105, 1352–1360 CrossRef CAS PubMed.
- N. Arora, J. I. Alsous, J. W. Guggenheim, M. Mak, J. Munera, J. M. Wells, R. D. Kamm, H. H. Asada, S. Y. Shvartsman and L. G. Griffith, Development, 2017, 144, 1128–1136 CAS.
- Z. Ao, H. Cai, Z. Wu, J. Ott, H. Wang, K. Mackie and F. Guo, Lab Chip, 2021, 21, 688–699 RSC.
- C. Schmidt, Nature, 2021, 597, S22–S23 CrossRef CAS.
- A. Chen, Z. Guo, L. Fang and S. Bian, Front. Cell. Neurosci., 2020, 14, 133 CrossRef CAS PubMed.
- S. A. Sloan, J. Andersen, A. M. Paşca, F. Birey and S. P. Paşca, Nat. Protoc., 2018, 13, 2062–2085 CrossRef CAS.
- V. Carvalho, M. Bañobre-López, G. Minas, S. F. C. F. Teixeira, R. Lima and R. O. Rodrigues, Bioprinting, 2022, 27, e00224 CrossRef.
- J. M. Unagolla and A. C. Jayasuriya, Appl. Mater. Today, 2022, 29, 101582 CrossRef.
- S. Swaminathan, Q. Hamid, W. Sun and A. M. Clyne, Biofabrication, 2019, 11, 025003 CrossRef CAS.
- L. D. Moor, J. Smet, M. Plovyt, B. Bekaert, C. Vercruysse, M. Asadian, N. D. Geyter, S. V. Vlierberghe, P. Dubruel and H. Declercq, Biofabrication, 2021, 13, 045021 CrossRef.
- R. Burdis and D. J. Kelly, Acta Biomater., 2021, 126, 1–14 CrossRef CAS PubMed.
- M. Rahimnejad, N. Nasrollahi Boroujeni, S. Jahangiri, N. Rabiee, M. Rabiee, P. Makvandi, O. Akhavan and R. S. Varma, Nano-Micro Lett., 2021, 13, 182 CrossRef CAS.
- Y. Sasai, Nature, 2013, 493, 318–326 CrossRef CAS.
- J. H. Sung, Y. I. Wang, N. Narasimhan Sriram, M. Jackson, C. Long, J. J. Hickman and M. L. Shuler, Anal. Chem., 2019, 91, 330–351 CrossRef CAS.
- J.-Y. Kim, D. A. Fluri, J. M. Kelm, A. Hierlemann and O. Frey, J. Lab. Autom., 2015, 20, 274–282 CrossRef CAS.
- J. Ruppen, L. Cortes-Dericks, E. Marconi, G. Karoubi, R. A. Schmid, R. Peng, T. M. Marti and O. T. Guenat, Lab Chip, 2014, 14, 1198–1205 RSC.
- C. Ma, Y. Peng, H. Li and W. Chen, Trends Pharmacol. Sci., 2021, 42, 119–133 CrossRef CAS PubMed.
- R. Mittal, F. W. Woo, C. S. Castro, M. A. Cohen, J. Karanxha, J. Mittal, T. Chhibber and V. M. Jhaveri, J. Cell. Physiol., 2019, 234, 8352–8380 CrossRef CAS.
- R. Greek and A. Menache, Int. J. Med. Sci., 2013, 10, 206–221 CrossRef PubMed.
- F. Alexander, S. Eggert and J. Wiest, Cytotechnology, 2018, 70, 375–386 CrossRef CAS PubMed.
- J. Gabriel, D. Brennan, J. H. Elisseeff and V. Beachley, Sci. Rep., 2019, 9, 16287 CrossRef.
- S. Heub, F. Navaee, D. Migliozzi, D. Ledroit, S. Boder-Pasche, J. Goldowsky, E. Vuille-Dit-Bille, J. Hofer, C. Gaiser, V. Revol, L. Suter-Dick and G. Weder, Sci. Rep., 2022, 12, 9991 CrossRef CAS.
- C. Dong, Front. Biosci., 2005, 10, 379 CrossRef CAS PubMed.
- D. Wirtz, K. Konstantopoulos and P. C. Searson, Nat. Rev. Cancer, 2011, 11, 512–522 CrossRef CAS PubMed.
- S. P. H. Chiang, R. M. Cabrera and J. E. Segall, Am. J. Physiol., 2016, 311, C1–C14 CrossRef.
- J. G. Goetz, Science, 2018, 362, 999–1000 CrossRef CAS PubMed.
- S. Heeke, B. Mograbi, C. Alix-Panabières and P. Hofman, Cells, 2019, 8, 714 CrossRef CAS PubMed.
- M.-R. Pan, M.-F. Hou, F. Ou-Yang, C.-C. Wu, S.-J. Chang, W.-C. Hung, H.-K. Yip and C.-W. Luo, J. Clin. Med., 2019, 8, 38 CrossRef CAS PubMed.
- K. R. Bittner, J. M. Jiménez and S. R. Peyton, Adv. Healthcare Mater., 2020, 9, 1901459 CrossRef CAS.
- R. Fan, T. Emery, Y. Zhang, Y. Xia, J. Sun and J. Wan, Sci. Rep., 2016, 6, 27073 CrossRef CAS.
- U. L. Triantafillu, S. Park, N. L. Klaassen, A. D. Raddatz and Y. Kim, Int. J. Oncol., 2017, 50, 993–1001 CrossRef CAS.
- S. Connolly, K. McGourty and D. Newport, Sci. Rep., 2020, 10, 1711 CrossRef CAS PubMed.
- S. Connolly, D. Newport and K. McGourty, Sci. Rep., 2021, 11, 13997 CrossRef CAS.
- S. C. Hur, A. J. Mach and D. Di Carlo, Biomicrofluidics, 2011, 5, 022206 CrossRef.
- A. Karimi, S. Yazdi and A. M. Ardekani, Biomicrofluidics, 2013, 7, 021501 CrossRef CAS.
- H. Amini, W. Lee and D. Di Carlo, Lab Chip, 2014, 14, 2739 RSC.
- J. Zhang, S. Yan, D. Yuan, G. Alici, N.-T. Nguyen, M. Ebrahimi Warkiani and W. Li, Lab Chip, 2016, 16, 10–34 RSC.
- D. Huber, A. Oskooei, X. Casadevall i Solvas, A. deMello and G. V. Kaigala, Chem. Rev., 2018, 118, 2042–2079 CrossRef CAS PubMed.
- S. Connolly, D. Newport and K. McGourty, Biomicrofluidics, 2020, 14, 031501 CrossRef CAS PubMed.
- J. Zhou and I. Papautsky, Lab Chip, 2019, 19, 3416–3426 RSC.
- J. Zhou, J. Su, X. Fu, L. Zheng and Z. Yin, Exp. Hematol. Oncol., 2017, 6, 22 CrossRef PubMed.
- M. D. Bourn, D. V. B. Batchelor, N. Ingram, J. R. McLaughlan, P. L. Coletta, S. D. Evans and S. A. Peyman, J. Controlled Release, 2020, 326, 13–24 CrossRef CAS PubMed.
- L. Chen, J. J. Kim and P. S. Doyle, Biomicrofluidics, 2018, 12, 024102 CrossRef PubMed.
- S. Sakuma, K. Nakahara and F. Arai, IEEE Robot. Autom. Lett., 2019, 4, 2973–2980 Search PubMed.
- A. Gonzalez, D. Ciobanu, M. Sayers, N. Sirr, T. Dalton and M. Davies, Biomed. Microdevices, 2007, 9, 729–736 CrossRef CAS.
- B.-J. Jin, C. Esteva-Font and A. S. Verkman, Lab Chip, 2015, 15, 3380–3390 RSC.
- R. F.-X. Tomasi, S. Sart, T. Champetier and C. N. Baroud, Cell Rep., 2020, 31, 107670 CrossRef CAS PubMed.
- H. Kim, H. Roh, H. Kim and J.-K. Park, Lab Chip, 2021, 21, 4155–4165 RSC.
- H. Huang, Y. Yu, Y. Hu, X. He, O. Berk Usta and M. L. Yarmush, Lab Chip, 2017, 17, 1913–1932 RSC.
- J. Hong, Y. K. Kim, D.-J. Won, J. Kim and S. J. Lee, Sci. Rep., 2015, 5, 10685 CrossRef CAS PubMed.
- A. M. Blakely, K. L. Manning, A. Tripathi and J. R. Morgan, Tissue Eng., Part C, 2015, 21, 737–746 CrossRef CAS PubMed.
- B. C. Ip, F. Cui, A. Tripathi and J. R. Morgan, Biofabrication, 2016, 8, 025015 CrossRef.
- N. Arora, J. I. Alsous, J. W. Guggenheim, M. Mak, J. Munera, J. M. Wells, R. D. Kamm, H. H. Asada, S. Y. Shvartsman and L. G. Griffith, Development, 2017, 144, 1128 CAS.
- B. Ayan, D. N. Heo, Z. Zhang, M. Dey, A. Povilianskas, C. Drapaca and I. T. Ozbolat, Sci. Adv., 2020, 6, eaaw5111 CrossRef CAS PubMed.
- B. Ayan, Y. Wu, V. Karuppagounder, F. Kamal and I. T. Ozbolat, Sci. Rep., 2020, 10, 13148 CrossRef CAS PubMed.
- B. Ayan, N. Celik, Z. Zhang, K. Zhou, M. H. Kim, D. Banerjee, Y. Wu, F. Costanzo and I. T. Ozbolat, Commun. Phys., 2020, 3, 183 CrossRef CAS PubMed.
- D. N. Heo, B. Ayan, M. Dey, D. Banerjee, H. Wee, G. S. Lewis and I. T. Ozbolat, Biofabrication, 2021, 13, 015013 CrossRef CAS PubMed.
- M. H. Kim, D. Banerjee, N. Celik and I. T. Ozbolat, Biofabrication, 2022, 14, 024103 CrossRef PubMed.
- D. M. van Pel, K. Harada, D. Song, C. C. Naus and W. C. Sin, J. Cell Commun. Signaling, 2018, 12, 723–730 CrossRef PubMed.
- S. P. Grogan, E. W. Dorthé, N. E. Glembotski, F. Gaul and D. D. D'Lima, Connect. Tissue Res., 2020, 61, 229–243 CrossRef CAS PubMed.
- I. N. Aguilar, L. J. Smith, D. J. Olivos, T.-M. G. Chu, M. A. Kacena and D. R. Wagner, Bioprinting, 2019, 15, e00048 CrossRef PubMed.
- K. Arai, D. Murata, A. R. Verissimo, Y. Mukae, M. Itoh, A. Nakamura, S. Morita and K. Nakayama, PLoS One, 2018, 13, e0209162 CrossRef CAS.
- N. I. Moldovan, N. Hibino and K. Nakayama, Tissue Eng., Part B, 2017, 23, 237–244 CrossRef CAS PubMed.
- D. Murata, R. Fujimoto and K. Nakayama, Int. J. Mol. Sci., 2020, 21, 3589 CrossRef CAS PubMed.
- M. Li, L. Liu and T. Zambelli, Nano Res., 2022, 15, 773–786 CrossRef CAS.
- P. Dörig, P. Stiefel, P. Behr, E. Sarajlic, D. Bijl, M. Gabi, J. Vörös, J. A. Vorholt and T. Zambelli, Appl. Phys. Lett., 2010, 97, 023701 CrossRef.
- P. Stiefel, T. Zambelli and J. A. Vorholt, Appl. Environ. Microbiol., 2013, 79, 4895–4905 CrossRef CAS.
- O. Guillaume-Gentil, T. Zambelli and J. A. Vorholt, Lab Chip, 2014, 14, 402–414 RSC.
- E. Potthoff, D. Ossola, T. Zambelli and J. A. Vorholt, Nanoscale, 2015, 7, 4070–4079 RSC.
- V. Martinez, C. Forró, S. Weydert, M. J. Aebersold, H. Dermutz, O. Guillaume-Gentil, T. Zambelli, J. Vörös and L. Demkó, Lab Chip, 2016, 16, 1663–1674 RSC.
- H. Tegally, J. E. San, J. Giandhari and T. de Oliveira, BMC Genomics, 2020, 21, 729 CrossRef PubMed.
- A. Meister, M. Gabi, P. Behr, P. Studer, J. Vörös, P. Niedermann, J. Bitterli, J. Polesel-Maris, M. Liley, H. Heinzelmann and T. Zambelli, Nano Lett., 2009, 9, 2501–2507 CrossRef CAS PubMed.
- O. Guillaume-Gentil, T. Zambelli and J. A. Vorholt, Lab Chip, 2014, 14, 402–414 RSC.
- X. Liu, Q. Shi, Y. Lin, M. Kojima, Y. Mae, Q. Huang, T. Fukuda and T. Arai, IEEE Trans. Nanotechnol., 2018, 17, 688–691 CAS.
- X. Liu, Q. Shi, Y. Lin, M. Kojima, Y. Mae, T. Fukuda, Q. Huang and T. Arai, Small, 2019, 15, 1804421 CrossRef PubMed.
- Z. Liu, M. Takeuchi, M. Nakajima, T. Fukuda, Y. Hasegawa and Q. Huang, in 2016 International Symposium on Micro-NanoMechatronics and Human Science (MHS), IEEE, Nagoya, Japan, 2016, pp. 1–3 Search PubMed.
- J. Cui, H. Wang, Q. Shi, J. Li, Z. Zheng, T. Sun, Q. Huang and T. Fukuda, in 2018 International Conference on Manipulation, Automation and Robotics at Small Scales (MARSS), IEEE, Nagoya, Japan, 2018, pp. 1–6 Search PubMed.
- Z. Zheng, H. Wang, Q. Shi, J. Li, J. Cui, T. Sun, Q. Huang and T. Fukuda, in 2018 IEEE 8th Annual International Conference on CYBER Technology in Automation, Control, and Intelligent Systems (CYBER), IEEE, Tianjin, China, 2018, pp. 404–409 Search PubMed.
- H. Wang, Q. Shi, M. Nakajima, M. Takeuchi, T. Chen, P. Di, Q. Huang and T. Fukuda, Int. J. Adv. Robot. Syst., 2014, 11, 121 CrossRef.
- E. Delannoy, G. Tellier, J. Cholet, A. M. Leroy, A. Treizebré and F. Soncin, Biomedicines, 2022, 10, 797 CrossRef CAS PubMed.
- M.-C. Tsai, S.-Y. Wei, L. Fang and Y.-C. Chen, Adv. Healthcare Mater., 2022, 11, 2101392 CrossRef CAS PubMed.
- R. Levato, T. Jungst, R. G. Scheuring, T. Blunk, J. Groll and J. Malda, Adv. Mater., 2020, 32, 1906423 CrossRef CAS PubMed.
- M. Hospodiuk, M. Dey, D. Sosnoski and I. T. Ozbolat, Biotechnol. Adv., 2017, 35, 217–239 CrossRef CAS PubMed.
- D. Chimene, R. Kaunas and A. K. Gaharwar, Adv. Mater., 2020, 32, 1902026 CrossRef CAS.
- K. Hölzl, S. Lin, L. Tytgat, S. Van Vlierberghe, L. Gu and A. Ovsianikov, Biofabrication, 2016, 8, 032002 CrossRef PubMed.
- L. Ouyang, R. Yao, Y. Zhao and W. Sun, Biofabrication, 2016, 8, 035020 CrossRef.
- E. S. Bishop, S. Mostafa, M. Pakvasa, H. H. Luu, M. J. Lee, J. M. Wolf, G. A. Ameer, T.-C. He and R. R. Reid, Genes Dis., 2017, 4, 185–195 CrossRef CAS PubMed.
- K. Nair, M. Gandhi, S. Khalil, K. C. Yan, M. Marcolongo, K. Barbee and W. Sun, Biotechnol. J., 2009, 4, 1168–1177 CrossRef CAS.
- A. Blaeser, D. F. Duarte Campos, U. Puster, W. Richtering, M. M. Stevens and H. Fischer, Adv. Healthcare Mater., 2016, 5, 326–333 CrossRef CAS PubMed.
- B. A. Aguado, W. Mulyasasmita, J. Su, K. J. Lampe and S. C. Heilshorn, Tissue Eng., Part A, 2012, 18, 806–815 CrossRef CAS PubMed.
- A. K. Grosskopf, G. A. Roth, A. A. A. Smith, E. C. Gale, H. L. Hernandez and E. A. Appel, Bioeng. Transl. Med., 2020, 5, e10147 CAS.
- S. Swaminathan, Q. Hamid, W. Sun and A. M. Clyne, J. Visualized Exp., 2020, e61791 CAS.
- H. Horder, M. Guaza Lasheras, N. Grummel, A. Nadernezhad, J. Herbig, S. Ergün, J. Teßmar, J. Groll, B. Fabry, P. Bauer-Kreisel and T. Blunk, Cells, 2021, 10, 803 CrossRef CAS PubMed.
- L. Polonchuk, L. Surija, M. H. Lee, P. Sharma, C. L. C. Ming, F. Richter, E. Ben-Sefer, M. A. Rad, H. M. S. Sarmast, W. A. Shamery, H. A. Tran, L. Vettori, F. Haeusermann, E. C. Filipe, J. Rnjak-Kovacina, T. Cox, J. Tipper, I. Kabakova and C. Gentile, Biofabrication, 2021, 13, 045009 CrossRef CAS PubMed.
- L. De Moor, S. Fernandez, C. Vercruysse, L. Tytgat, M. Asadian, N. De Geyter, S. Van Vlierberghe, P. Dubruel and H. Declercq, Front. Bioeng. Biotechnol., 2020, 8, 484 CrossRef PubMed.
- L. Benmeridja, L. De Moor, E. De Maere, F. Vanlauwe, M. Ryx, L. Tytgat, C. Vercruysse, P. Dubruel, S. Van Vlierberghe, P. Blondeel and H. Declercq, J. Tissue Eng. Regener. Med., 2020, 14, 840–854 CrossRef CAS PubMed.
- J. Colle, P. Blondeel, A. De Bruyne, S. Bochar, L. Tytgat, C. Vercruysse, S. Van Vlierberghe, P. Dubruel and H. Declercq, J. Mater. Sci.: Mater. Med., 2020, 31, 36 CrossRef CAS PubMed.
- A. Skardal, M. Devarasetty, H.-W. Kang, I. Mead, C. Bishop, T. Shupe, S. J. Lee, J. Jackson, J. Yoo, S. Soker and A. Atala, Acta Biomater., 2015, 25, 24–34 CrossRef CAS PubMed.
- N. S. Bhise, V. Manoharan, S. Massa, A. Tamayol, M. Ghaderi, M. Miscuglio, Q. Lang, Y. S. Zhang, S. R. Shin, G. Calzone, N. Annabi, T. D. Shupe, C. E. Bishop, A. Atala, M. R. Dokmeci and A. Khademhosseini, Biofabrication, 2016, 8, 014101 CrossRef PubMed.
- A. McCormack, C. B. Highley, N. R. Leslie and F. P. W. Melchels, Trends Biotechnol., 2020, 38, 584–593 CrossRef CAS PubMed.
- W. Wu, A. DeConinck and J. A. Lewis, Adv. Mater., 2011, 23, H178–H183 CrossRef CAS PubMed.
- T. Bhattacharjee, C. J. Gil, S. L. Marshall, J. M. Urueña, C. S. O'Bryan, M. Carstens, B. Keselowsky, G. D. Palmer, S. Ghivizzani, C. P. Gibbs, W. G. Sawyer and T. E. Angelini, ACS Biomater. Sci. Eng., 2016, 2, 1787–1795 CrossRef CAS PubMed.
- D. F. D. Campos, C. D. Lindsay, J. G. Roth, B. L. LeSavage, A. J. Seymour, B. A. Krajina, R. Ribeiro, P. F. Costa, A. Blaeser and S. C. Heilshorn, Front. Bioeng. Biotechnol., 2020, 8, 374 CrossRef PubMed.
- B. Ayan, N. Celik, Z. Zhang, K. Zhou, M. H. Kim, D. Banerjee, Y. Wu, F. Costanzo and I. T. Ozbolat, Commun. Phys., 2020, 3, 183 CrossRef CAS PubMed.
- J. A. Brassard, M. Nikolaev, T. Hübscher, M. Hofer and M. P. Lutolf, Nat. Mater., 2021, 20, 22–29 CrossRef CAS PubMed.
- M. A. Skylar-Scott, S. G. M. Uzel, L. L. Nam, J. H. Ahrens, R. L. Truby, S. Damaraju and J. A. Lewis, Sci. Adv., 2019, 5, eaaw2459 CrossRef CAS PubMed.
- B. T. Vinson, T. B. Phamduy, J. Shipman, B. Riggs, A. L. Strong, S. C. Sklare, W. L. Murfee, M. E. Burow, B. A. Bunnell, Y. Huang and D. B. Chrisey, Biofabrication, 2017, 9, 025013 CrossRef PubMed.
- T. B. Phamduy, N. A. Raof, N. R. Schiele, Z. Yan, D. T. Corr, Y. Huang, Y. Xie and D. B. Chrisey, Biofabrication, 2012, 4, 025006 CrossRef PubMed.
- P. Rawal, D. M. Tripathi, S. Ramakrishna and S. Kaur, Bio-Des. Manuf., 2021, 4, 627–640 CrossRef.
- K. Chen, E. Jiang, X. Wei, Y. Xia, Z. Wu, Z. Gong, Z. Shang and S. Guo, Lab Chip, 2021, 21, 1604–1612 RSC.
- P. N. Bernal, M. Bouwmeester, J. Madrid-Wolff, M. Falandt, S. Florczak, N. G. Rodriguez, Y. Li, G. Größbacher, R.-A. Samsom, M. van Wolferen, L. J. W. van der Laan, P. Delrot, D. Loterie, J. Malda, C. Moser, B. Spee and R. Levato, Adv. Mater., 2022, 34, 2110054 CrossRef CAS PubMed.
- P. N. Bernal, P. Delrot, D. Loterie, Y. Li, J. Malda, C. Moser and R. Levato, Adv. Mater., 2019, 31, 1904209 CrossRef CAS PubMed.
- J. Johnson, J. T. Sharick, M. C. Skala and L. Li, J. Mass Spectrom., 2020, 55, e4452 CrossRef CAS PubMed.
- A. P. Rago, D. M. Dean and J. R. Morgan, Biotechnol. Bioeng., 2009, 102, 1231–1241 CrossRef CAS PubMed.
- Y. Liu, C. Dabrowska, A. Mavousian, B. Strauss, F. Meng, C. Mazzaglia, K. Ouaras, C. Macintosh, E. Terentjev, J. Lee and Y. Y. S. Huang, Adv. Sci., 2021, 8, 2003332 CrossRef PubMed.
- J. Fink, M. Théry, A. Azioune, R. Dupont, F. Chatelain, M. Bornens and M. Piel, Lab Chip, 2007, 7, 672–680 RSC.
- J. Fink, M. Théry, A. Azioune, R. Dupont, F. Chatelain, M. Bornens and M. Piel, Lab Chip, 2007, 7, 672–680 RSC.
- Y. H. Ding, M. Floren and W. Tan, Biosurf. Biotribol., 2016, 2, 121–136 CrossRef CAS PubMed.
- F. Tasnim, N. H. Singh, E. K. F. Tan, J. Xing, H. Li, S. Hissette, S. Manesh, J. Fulwood, K. Gupta, C. W. Ng, S. Xu, J. Hill and H. Yu, Sci. Rep., 2020, 10, 4768 CrossRef CAS PubMed.
- P. A. Turner, L. M. Harris, C. A. Purser, R. C. Baker and A. V. Janorkar, Biotechnol. Bioeng., 2014, 111, 174–183 CrossRef CAS PubMed.
- L. Xia, R. B. Sakban, Y. Qu, X. Hong, W. Zhang, B. Nugraha, W. H. Tong, A. Ananthanarayanan, B. Zheng, I. Y.-Y. Chau, R. Jia, M. McMillian, J. Silva, S. Dallas and H. Yu, Biomaterials, 2012, 33, 2165–2176 CrossRef CAS PubMed.
- P. A. Turner, B. Gurumurthy, J. L. Bailey, C. M. Elks and A. V. Janorkar, Process Biochem., 2017, 59, 312–320 CrossRef CAS PubMed.
- H. Tsutsui, E. Yu, S. Marquina, B. Valamehr, I. Wong, H. Wu and C. M. Ho, Ann. Biomed. Eng., 2010, 38, 3777–3788 CrossRef PubMed.
- G. R. Kirkham, E. Britchford, T. Upton, J. Ware, G. M. Gibson, Y. Devaud, M. Ehrbar, M. Padgett, S. Allen, L. D. Buttery and K. Shakesheff, Sci. Rep., 2015, 5, 1–7 Search PubMed.
- M. P. Kummer, J. J. Abbott, B. E. Kratochvil, R. Borer, A. Sengul and B. J. Nelson, IEEE Trans. Robot., 2010, 26, 1006–1017 Search PubMed.
- S. Yaman, M. Anil-Inevi, E. Ozcivici and H. C. Tekin, Front. Bioeng. Biotechnol, 2018, 19, 192 CrossRef PubMed.
- J. Kolosnjaj-Tabi, C. Wilhelm, O. Clément and F. Gazeau, J. Nanobiotechnol., 2013, 11, S7 CrossRef PubMed.
- E. Umut, in Modern Surface Engineering Treatments, ed. M. Aliofkhazraei, IntechOpen, 2013, pp. 185–208 Search PubMed.
- T. R. Olsen, B. Mattix, M. Casco, A. Herbst, C. Williams, A. Tarasidis, D. Simionescu, R. P. Visconti and F. Alexis, Acta Biomater., 2015, 13, 188–198 CrossRef CAS PubMed.
- A. M. Bratt-Leal, K. L. Kepple, R. L. Carpenedo, M. T. Cooke and T. C. McDevitt, Integr. Biol., 2011, 3, 1224–1232 CrossRef CAS PubMed.
- G. R. Souza, J. R. Molina, R. M. Raphael, M. G. Ozawa, D. J. Stark, C. S. Levin, L. F. Bronk, J. S. Ananta, J. Mandelin, M. M. Georgescu, J. A. Bankson, J. G. Gelovani, T. C. Killian, W. Arap and R. Pasqualini, Nat. Nanotechnol., 2010, 5, 291–296 CrossRef CAS PubMed.
- N. G. Durmus, H. C. Tekin, S. Guven, K. Sridhar, A. Arslan Yildiz, G. Calibasi, I. Ghiran, R. W. Davis, L. M. Steinmetz and U. Demirci, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3661–E3668 CrossRef CAS PubMed.
- A. Tocchio, N. G. Durmus, K. Sridhar, V. Mani, B. Coskun, R. El Assal and U. Demirci, Adv. Mater., 2018, 30, 1705034 CrossRef PubMed.
- M. Wiklund, Lab Chip, 2012, 12, 2018 RSC.
- H. Bruus, Lab Chip, 2012, 12, 1014–1021 RSC.
- H. J. Stöckmann, Eur. Phys. J. Spec. Top., 2007, 145, 15–23 CrossRef.
- D. Petta, V. Basoli, D. Pellicciotta, R. Tognato, J. Barcik, C. Arrigoni, E. D. Bella, A. R. Armiento, C. Candrian, R. G. Richards, M. Alini, M. Moretti, D. Eglin and T. Serra, Biofabrication, 2020, 13, 15004 CrossRef PubMed.
- P. Chen, Z. Luo, S. Güven, S. Tasoglu, A. V. Ganesan, A. Weng and U. Demirci, Adv. Mater., 2014, 26, 5936–5941 CrossRef CAS PubMed.
- P. Chen, S. Güven, O. B. Usta, M. L. Yarmush and U. Demirci, Adv. Healthcare Mater., 2015, 4, 1937–1943 CrossRef CAS PubMed.
- Y. Zhu, V. Serpooshan, S. Wu, U. Demirci, P. Chen and S. Güven, in Organoids. Methods in Molecular Biology, Humana Press Inc., 2019, vol. 1576, pp. 301–312 Search PubMed.
- D. Binks, M.-T. Westra and W. van de Water, Phys. Rev. Lett., 1997, 79, 5010–5013 CrossRef CAS.
- A. Lenshof, M. Evander, T. Laurell and J. Nilsson, Lab Chip, 2012, 12, 684 RSC.
- H. Cai, Z. Ao, L. Hu, Y. Moon, Z. Wu, H.-C. Lu, J. Kim and F. Guo, Analyst, 2020, 145, 6243–6253 RSC.
- K. Chen, M. Wu, F. Guo, P. Li, C. Y. Chan, Z. Mao, S. Li, L. Ren, R. Zhang and T. J. Huang, Lab Chip, 2016, 16, 2636–2643 RSC.
- H. Cai, Z. Wu, Z. Ao, A. Nunez, B. Chen, L. Jiang, M. Bondesson and F. Guo, Biofabrication, 2020, 12, 35025 CrossRef CAS PubMed.
- A. Ozcelik, J. Rufo, F. Guo, Y. Gu, P. Li, J. Lata and T. J. Huang, Nat. Methods, 2018, 15, 1021–1028 CrossRef CAS PubMed.
- T. Baasch, L. Leibacher and J. Dual, J. Acoust. Soc. Am., 2017, 141, 1664 CrossRef PubMed.
- Z. Ma, K. Melde, A. G. Athanassiadis, M. Schau, H. Richter, T. Qiu and P. Fischer, Nat. Commun., 2020, 11, 4537 CrossRef CAS PubMed.
- B. Huang, G. Clark, D. R. Klein, D. MacNeill, E. Navarro-Moratalla, K. L. Seyler, N. Wilson, M. A. McGuire, D. H. Cobden, D. Xiao, W. Yao, P. Jarillo-Herrero and X. Xu, Nat. Nanotechnol., 2018, 13, 544–548 CrossRef CAS PubMed.
- S. Jiang, L. Li, Z. Wang, K. F. Mak and J. Shan, Nat. Nanotechnol., 2018, 13, 549–553 CrossRef CAS PubMed.
- M. Hofer and M. P. Lutolf, Nat. Rev. Mater., 2021, 6, 402–420 CrossRef CAS PubMed.
- S. Li, D. Sengupta and S. Chien, Wiley Interdiscip. Rev. Syst. Biol. Med., 2014, 6, 61–76 CrossRef CAS PubMed.
- T. Fushimi, A. Marzo, B. W. Drinkwater and T. L. Hill, Appl. Phys. Lett., 2019, 115, 064101 CrossRef.
| This journal is © The Royal Society of Chemistry 2022 |
