 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
You will know by its tail: a method for quantification of heterogeneity of bacterial populations using single-cell MIC profiling†
Natalia
Pacocha‡
a,
Marta
Zapotoczna‡
b,
Karol
Makuch
 *a,
Jakub
Bogusławski
cde and
Piotr
Garstecki
*a,
Jakub
Bogusławski
cde and
Piotr
Garstecki
 *a
*a
aInstitute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: kmakuch@ichf.edu.pl; pgarstecki@ichf.edu.pl
bDepartment of Molecular Microbiology, Institute of Microbiology, Faculty of Biology, Biological and Chemical Research Centre, University of Warsaw, Żwirki i Wigury 101, 02-089 Warsaw, Poland
cInternational Centre for Translational Eye Research, Institute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland
dDepartment of Physical Chemistry of Biological Systems, Institute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland
eLaser & Fiber Electronics Group, Faculty of Electronics, Wrocław University of Science and Technology, Wybrzeze Wyspianskiego 27, 50-370 Wroclaw, Poland
First published on 5th October 2022
Abstract
Severe non-healing infections are often caused by multiple pathogens or by genetic variants of the same pathogen exhibiting different levels of antibiotic resistance. For example, polymicrobial diabetic foot infections double the risk of amputation compared to monomicrobial infections. Although these infections lead to increased morbidity and mortality, standard antimicrobial susceptibility methods are designed for homogenous samples and are impaired in quantifying heteroresistance. Here, we propose a droplet-based label-free method for quantifying the antibiotic response of the entire population at the single-cell level. We used Pseudomonas aeruginosa and Staphylococcus aureus samples to confirm that the shape of the profile informs about the coexistence of diverse bacterial subpopulations, their sizes, and antibiotic heteroresistance. These profiles could therefore indicate the outcome of antibiotic treatment in terms of the size of remaining subpopulations. Moreover, we studied phenotypic variants of a S. aureus strain to confirm that the profile can be used to identify tolerant subpopulations, such as small colony variants, associated with increased risks for the development of persisting infections. Therefore, the profile is a versatile instrument for quantifying the size of each bacterial subpopulation within a specimen as well as their individual and joined heteroresistance.
Introduction
Heterogeneity of an infecting bacterial population is an important consideration for the identification of an effective antibiotic treatment.1,2 Some of the most common infectious diseases are caused by more than one co-colonizing bacterial pathogen, including soft tissue infections, peritonitis, cystic fibrosis, urinary tract infections, and endocarditis.3 An estimated 10% of antibiotic-resistant subpopulations is undetectable by current diagnostic tests leading to treatment failure.4 Moreover, polymicrobial infections doubled the number of amputations in diabetic foot infections,5 while polymicrobial bloodstream infections were shown to increase the mortality rates from 24 to 47%.6Standard methods for antibiotic susceptibility testing fail to inform on the level of distribution of antibiotic resistance of the entire bacterial population of co-colonizing species. An example of co-infecting pathogens is Staphylococcus aureus and Pseudomonas aeruginosa abundant and prevailing in the airways and lungs of cystic fibrosis patients.7,8 Inadequate antibiotic regimens are likely to interfere with the population's complexity in an uncontrollable manner and may promote survival or emergence of resistant subpopulations or remove the non-pathogenic susceptible microflora.9
Moreover, in response to environmental stress bacteria can transiently modify their transcriptional profiles or acquire mutations. This type of stress-induced phenotypic heterogeneity has been known since the middle of the 20th century.10 These mutations often lead to the reduction in metabolic activity or changes to the cell membrane potential that result in an altered phenotypes, such as a longer lag time, increased tolerance to antibiotics. Stress-induced phenotypic variants are also often associated with chronic infections.11–15 Nicoloff et al. reported that in ca. 27.4% of 766 bacteria-antibiotic combinations with 28 antibiotics, the bacterial strains were heteroresistant.16 Heterogeneity of a bacterial strain in response to antibiotics has been shown to depend on changing the number of copies of antibiotic resistance genes. Gene copy number oscillation of plasmid- and chromosomally-encoded antibiotic resistance genes was shown responsible for changing the levels of resistance of bacterial cells within a bacterial population.17,18
Previous works have demonstrated the advantages of droplet microfluidics for the cultivation of bacteria and the determination of antibiotic breakpoints at the single-cell level.19–23 Encapsulation of single bacterial cells with various antibiotic concentrations was used to determine the single-cell minimum inhibitory concentration (scMIC).24 Lyu et al.25 quantified bacterial heteroresistance by measuring the fraction of bacteria with MIC higher than antibiotic concentration c. In this paper, we denote the same fraction by FR(c) and refer to it as a ‘scMIC profile’. The scMIC profile is a function starting from 1 when all bacterial cells proliferate and monotonically decrease, reaching the value 0 when the antibiotic concentration is sufficiently high to inhibit the growth of all bacteria. The concentration corresponding to complete inhibition of the whole population determines ‘scMIC’, as defined by Artemova et al.24 The setup proposed by Lyu et al. for quantification of heteroresistance consisted of single bacterial cells encapsulated with antibiotic into ca. 75 pL droplets, which were then incubated under conditions optimal for bacterial growth following their probing with alamarBlue for detection of viability-dependent fluorescence signal in each droplet.25 Lyu's approach was a high-resolution method enabling detection of a resistant subpopulation as low as 10−6 of the entire population, yet the method required labelling, which limited its use. Similarly, a high-resolution droplet-based method for the determination of the scMIC and the distribution of scMIC was used by Scheler et al.,26 who also went beyond a single cell level where the amount – not the concentration – of the antibiotic influences the bacteria inhibition. Heteroresistance profiles at a single cell level discussed by Scheler et al. referred to the probability distribution of MIC, p(MIC). The MIC distribution can be obtained from scMIC profile26 by taking the derivative, p(c) = −dFR(c)/dc. Bacterial labeling was a factor that limited its experimental use to a single bacterial strain.
Recently, we reported that scattering can be used as a label-free detection of bacterial growth in nanoliter droplets and for the determination of scMIC.27 Here, we extended the experimental setup by the implementation of an additional label-free detector of natural light emission which is specific to some bacterial species. We utilized the double detection device to investigate the scMIC profiles of phenotypically complex bacterial populations, including polymicrobial samples, and confirmed that the scMIC profiling efficiently informs antibiotic susceptibility and other parameters of population complexity, including the size of phenotypically diverse subpopulations. We hope that our model paves the way for a better characterization of antibiotic susceptibility in complex bacterial samples.
Results
Single-cell MIC profiles of binary bacterial populations
To investigate scMIC profiles of polymicrobial samples we designed and built a microfluidic system for label-free detection of bacterial growth in nanoliter droplets. The optical setup shown in Fig. S1† was used for the simultaneous detection of scattered light as well as the native fluorescence within the 510–550 nm range. Bacterial strains differ in fluorescence excitation and emission spectra, and this feature can be used to distinguish them. The sensitivity of detection of autofluorescence was validated using droplets containing diverse bacterial species. Fig. S2† represents signal intensity readouts (peaks) for emulsions containing various bacterial isolates (1–5). Each sample contains both, empty- and bacteria filled droplets. Peaks of increased height (increased intensity) represent droplets containing bacteria producing native fluorescence (1–4) while equal height peaks in sample 5 indicate that the bacterium is not producing detectable levels of native fluorescence. Table S1† lists all the bacterial species tested and their detectable levels of native fluorescence. Native fluorescence was detected in all tested bacteria apart from isolates of Staphylococcus spp. Previously it has been shown that S. aureus produces autofluorescence in red spectral range28 (>600 nm, outside the detection bandwidth of our system) due to the presence of porphyrins.29 At the same time, e.g., Pseudomonas aeruginosa emits a yellowish-green autofluorescence28 (within the detection bandwidth) from pyoverdine.30We sought to use this label-free detection consisting of two selective detectors to determine the antibiotic susceptibility profiles of samples consisting of pre-mixed suspensions of S. aureus and P. aeruginosa. Both opportunistic pathogens are frequently co-isolated from infections of catheters, endotracheal tubes, skin, eyes, and the respiratory tract, including the airways of people with cystic fibrosis. A range of antibiotic concentrations was added to the pre-mixed suspensions prior to experiment. Emulsification of single bacterial cells from the sample into 1 nL droplets achieving ca. 10% of single inoculated droplets containing at least one cell capable of proliferation. In further discussion, we refer to these droplets as “non-empty”. Approx. 30 thousand droplets were produced from each sample. Readouts of scattered light and autofluorescence intensities were taken for each droplet after incubation for 18 hours at 37 °C [Fig. 1a]. The number of droplets with multiplying bacteria was scored based on their high intensity of either signal and the fractions of positive droplets, f+, were calculated for each antibiotic concentration (see ESI† for more details). We define scMIC by the concentration that inhibits all bacteria (here determined at the point where f+ ≤ 0.1%). The scMIC for the samples made of a pre-mixed population was determined at 1.5 μg mL−1 for ciprofloxacin and 4 μg mL−1 for tobramycin [Fig. 1c and d], which corresponded to the breakpoint concentrations of the less susceptible strain, as verified by comparing to the breakpoints values determined for single strains using the standard microdilution method [Fig. 1b]. It is worth mentioning that Artemova et al.24 determined scMIC in a similar way, although by plating cells on agar and counting colony growth. This could lead to different scMIC values due to the limited amount of antibiotic in droplets.26 The application of an autofluorescence detector allowed for discrimination between bacterial subpopulations and for quantitative estimation of the proportion of either strain across antibiotic concentrations in a studied sample. Droplets containing S. aureus were detected by scattering whilst P. aeruginosa containing could be detected by both detectors. At the region of low antibiotic concentrations (up to 0.125 μg mL−1 for ciprofloxacin, or 0.5 μg mL−1 for tobramycin) the f+ values from scattered light corresponded to the growth of the entire mixed bacterial population [Fig. 1c and d]. The scMIC profiles determined by scattering and autofluorescence for ciprofloxacin were superimposed above the S. aureus breakpoint c of ca. 0.5 μg mL−1, thus represented subpopulation of recoverable P. aeruginosa [Fig. 1c]. The scMIC profile for tobramycin above the 1 μg mL−1 was determined only from scattering as no autofluorescence could be detected above this concentration, suggesting the remaining recoverable cells were all S. aureus. We estimated the proportions of both species in the samples based on the fraction of positive droplets where bacteria were not treated with antibiotic. To obtain the sizes of P. aeruginosa populations we designated the ratios of the droplets recognized by high native fluorescence intensity to the positive droplets detected by the high signal of scattered light. We determined P. aeruginosa population at 50% ± 5% and 9% ± 1% for samples treated with ciprofloxacin and tobramycin, respectively. The remaining portions of the samples correspond to the sizes of S. aureus population.
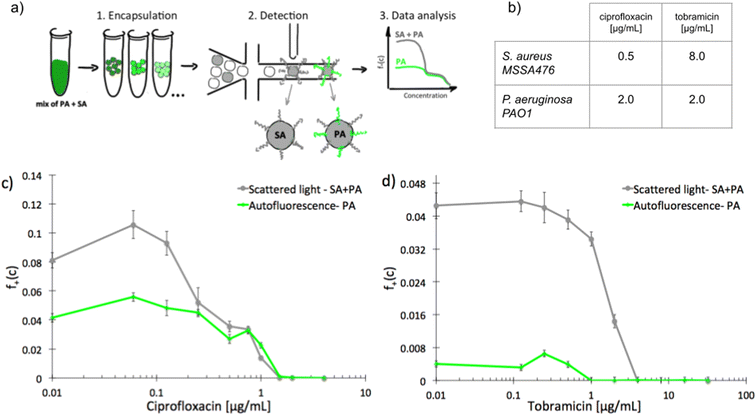 | ||
| Fig. 1 Experimental determination of scMIC profiles of polymicrobial samples of pre-mixed S. aureus MSSA47 and P. aeruginosa PAO1; a) schematic overview of the workflow for label-free determination of scMIC profile, b) MIC values determined by standard microdilution method for ciprofloxacin and tobramycin against studied strains, c and d) scMIC profiles of pre-mixed samples determined as a fraction of positive droplets, a function of antibiotic concentration measured based on scattered light (grey line) and autofluorescence (green line) intensity. The breakpoints values for both premixed samples are 1.5 and 4 μg ml−1 for ciprofloxacin and tobramycin, respectively. Statistical treatment of error analysis is described in detail in ESI.† | ||
These results demonstrate the effectiveness of our method in the determination of the scMIC profile of a sample consisting of two bacterial subpopulations. Moreover, we observed that scMIC profiles obtained using scattering alone are sufficient to inform on the population complexity (existence of more than one subpopulation of bacteria with different level of antibiotic tolerance). Yet, in mixed populations of a large disproportion in subpopulation sizes (one strain over the other), autofluorescence is essential to discriminate.
scMIC profiles of normal cells and small colony variants
Next, we used scattering-based detection to determine the scMIC profiles of phenotypically heterogeneous samples of a clonal strain. We selected normal morphology colonies (normal colony phenotype, NCP) of strain S. aureus SH1000 grown under optimal conditions, as well as colonies representing the small colony variant (SCV) phenotype, which we triggered with gentamicin [Fig. 2a]. Isolated SCV colonies were characterized by phenotypic changes, including aminoglycoside tolerance, small colony size, longer lag time, and reduced pigmentation [Fig. 2d, S3†]. Both populations, consisting of cells harvested from either NCP or SCV colonies, were separately subjected to scMIC testing [Fig. 2b] and the respective scMIC profiles were determined (see Materials and methods for more details). The NCP population required 2 μg mL−1 for the inhibition of the entire population while the scMIC of the SCV population was determined at 32 μg mL−1 [Fig. 2c]. Both scMIC values determined for either NCP or SCV were consistent with the breakpoint concentrations measured for the respective populations using the standard microdilution method [Table S2†].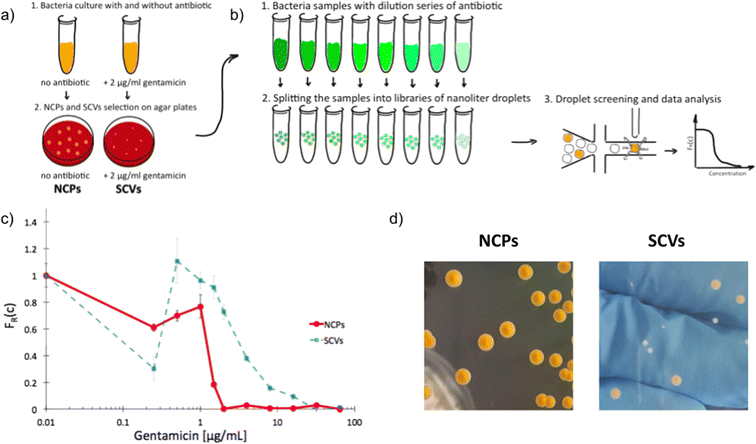 | ||
| Fig. 2 Experimental determination of single-cell MIC profile for Staphylococcus aureus of normal colony phenotype (NCPs) and small colony variants (SCVs); a) schematic overview of the experimental workflow of SCVs triggering and b) determination of scMIC including sample analysis towards phenotypic heterogeneity, c) comparison of single-cell MIC profiles determined for NCPs (red) and SCVs (blue), d) photographs of representative NCP and SCV colonies on tryptic soy agar, respectively. Statistical treatment of error analysis is described in detail in ESI.† | ||
Distinctive scMIC profiles, consisting of two regions, were observed for NCPs and SCVs [Fig. 2c]. The region of low antibiotic concentrations (up to gentamycin of ca. 1 μg mL−1) with a slowly decaying fraction of recovering bacteria FR(c) was similar for both, NCP and SCV. The second region which we referred to as a transition region started at gentamicin of 1 μg mL−1 and was different in both populations. For NCPs the transition region ended at 2 μg mL−1 and had a sharp decline, while for SCVs it reached 32 μg mL−1 and had a much milder slope. Therefore, the transition region for SCVs was about 16 times broader than for NCPs. In this sense our SCVs population was 16 times more resistant than NCPs, suggesting it consists of bacterial cells of larger distribution of antibiotic susceptibility. In the low antibiotic region with a high proportion of positive droplets the FR(c) values fluctuations were observed. They could be related to the decreased stability of non-empty droplets (droplets containing bacteria) likely to be caused by the cell aggregation, which is characteristic of the strain S. aureus SH1000.
‘Tails’ in the scMIC profiles
As we characterized the scMIC profiles of either NCPs or gentamicin-triggered SCVs, we wanted to investigate if they could determine their respective contribution within pre-mixed populations of a known proportion. The homogenous suspensions of NCPs and SCVs were combined into heterogeneous mixtures consisting of either 12% SCVs or 50% of SCVs and the remaining portion of NCPs, respectively [Fig. 3a]. As expected, the scMIC profiles of mixtures were combinations of profiles of either population. The profile of 12% SCV declined sharply in the transition region around 1 μg mL−1 to the FR(c) value of 0.08 and continued on with a minimal reduction in the shape of a tail between 1 and 8 μg mL−1. The height of the tail at 0.08 directly corresponded to the size of SCV subpopulation and it amounts to 8% [Fig. 3b]. Whereas the scMIC distribution for the mixture of 50% SCV was broader, yet equally sharp starting at 2 μg mL−1 down to FR(c) ca. 0.37 and continuing as a tail ending at 16 μg mL−1 [Fig. 3b]. Based on the height of the tail we estimated the SCV cells at 37% of the entire population. The difference between 50% and 37% follows from the precision of our method that we discuss in “Method precision” below.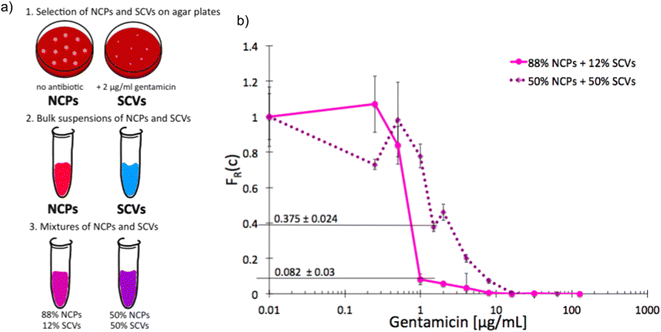 | ||
| Fig. 3 Single-cell MIC measurement of mixtures of phenotypic variants of S. aureus SH1000; a) schematic workflow of sample preparation, b) single-cell MIC profile of phenotypically heterogeneous mixtures containing respectively 12% (solid pink) and 50% (dotted purple) small colony variants (SCV) mixed with normal phenotype cells (NCPs). Statistical treatment of error analysis is described in detail in ESI.† | ||
These results indicate that the tail represents the size of SCVs subpopulation, whether it is low height corresponding to the low SCV content or high with a larger size subpopulation of tolerant bacteria. Consequently, the shape of scMIC profile is an indicator of coexistence of subpopulations of different distributions of antibiotic susceptibility and their respective sizes.
“Tail” can inform phenotypic heterogeneity of a biofilm
The recalcitrance of S. aureus biofilms is commonly associated with the emergence of slow-growing subpopulations, such as the SCVs. We sought to investigate whether our label-free method is suitable for the detection of antibiotic tolerant subpopulations in a biofilm and to determine an adequate antibiotic dose for inhibition of this phenotypically heterogeneous population. We started by determining the unique scMIC profiles of NCPs and SCVs isolated from dispersed in vitro biofilm [Fig. 4a]. The scMIC of NCPs and SCVs were estimated at 1.5 and 32 μg mL−1, respectively, which corresponded to the scMIC profiles of either population isolated from suspension cultures [Fig. 2]. Moreover, the distinctive shapes of transition regions for either NCPs or SCVs were confirmed as a sharp decline and a milder slope, respectively [Fig. 4b]. Subsequently, we used scMIC profiles to estimate the size of SCV contribution in biofilms formed in vitro. The absence of a transition region (absence of a tail) in the scMIC profile of biofilm formed without gentamicin indicated it represents mostly a homogenous population. On the other hand, the biofilm formed in the presence of gentamicin had a transition tail with a base at 0.068 indicating the SCV content at 6.8% ± 1.9%, demonstrating the role of sub-MIC concentrations of gentamicin in triggering heteroresistance within a biofilm culture. [Fig. 4c and d].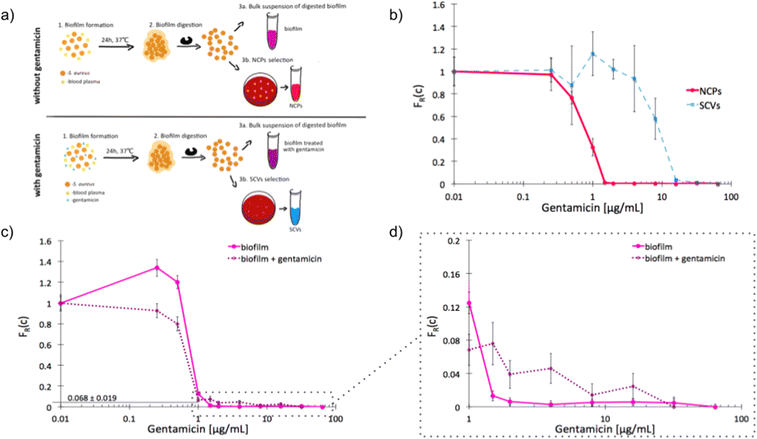 | ||
| Fig. 4 Single-cell MIC profiles of S. aureus dispersed from the biofilm. a) Schematic workflow of biofilms formation together with NCPs and SCVs selection in the absence (top) and presence (bottom) of gentamicin; b) heterogeneity profiles for NCPs (solid red) and SCVs (dotted blue) isolated from a biofilm cultured for 24 hours in the presence of gentamicin; c) scMIC profiles of biofilms cultured in the presence (dotted purple) and absence (solid pink) of gentamicin; d) the tails of scMIC profiles shown in c). Statistical treatment of error analysis is described in detail in ESI.† | ||
Method precision
Measurements of scMIC profile require counting of droplets and determination of the number of positives. In an ideal experiment, we expect that the error of the fraction of resistant bacteria, σFR(c), decreases as the square root of the number of non-empty droplets,26Nf+(0), as follows,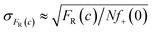 . Therefore, one thousand non-empty droplets typically used in our experiments would give FR(c) fluctuations below 3%. The scMIC profile would be smooth – up to 3% fluctuation – monotonically decreasing curve. However, more significant fluctuations were observed in our experiments. For example, in Fig. 2c the fluctuations were sometimes larger than the error bars that we determined with the method described in ESI.† Therefore, the error bars shown in the paper do not account for the variation between repeated experiments. The error bars account for the uncertainty in the calculation of the scMIC distribution from a single experiment, yet do not account for the variability between repeated experiments. This second factor introduces additional noise stemming from e.g. coalescence of droplets observed at late stages of culturing of S. aureus in droplets. For this reason, we estimated the accuracy of our method also by performing three repetitions of the same experiment. We cultured S. aureus biofilm for 72 hours in the presence of gentamicin. The biofilm was dispersed and subjected to scMIC experiments in three technical replicates measured one by one. Fig. S4† shows the heterogeneity profiles of all repetitions. Droplets without antibiotics or containing a low concentration of gentamicin were affected by more significant standard deviations. It is related to decreasing droplet stability with an increasing number of bacteria cells which we observed to be particularly visible for droplets containing S. aureus SH1000 cells as they tend to aggregate. These data show the highest deviation from the mean, 38%, in the transition region for concentration c = 0.5 μg ml−1. The remaining concentrations give the deviation of FR smaller than 7%. Our numerical analysis revealed the following SCVs concentrations in repetitions no 1–3: 17.9 ± 1.4%, 31.3 ± 2.9%, 29.8 ± 3.0%, respectively. The average of obtained results is 26% and the standard deviation is 7.3%. The relative error is 7.3/26 = 28%.
. Therefore, one thousand non-empty droplets typically used in our experiments would give FR(c) fluctuations below 3%. The scMIC profile would be smooth – up to 3% fluctuation – monotonically decreasing curve. However, more significant fluctuations were observed in our experiments. For example, in Fig. 2c the fluctuations were sometimes larger than the error bars that we determined with the method described in ESI.† Therefore, the error bars shown in the paper do not account for the variation between repeated experiments. The error bars account for the uncertainty in the calculation of the scMIC distribution from a single experiment, yet do not account for the variability between repeated experiments. This second factor introduces additional noise stemming from e.g. coalescence of droplets observed at late stages of culturing of S. aureus in droplets. For this reason, we estimated the accuracy of our method also by performing three repetitions of the same experiment. We cultured S. aureus biofilm for 72 hours in the presence of gentamicin. The biofilm was dispersed and subjected to scMIC experiments in three technical replicates measured one by one. Fig. S4† shows the heterogeneity profiles of all repetitions. Droplets without antibiotics or containing a low concentration of gentamicin were affected by more significant standard deviations. It is related to decreasing droplet stability with an increasing number of bacteria cells which we observed to be particularly visible for droplets containing S. aureus SH1000 cells as they tend to aggregate. These data show the highest deviation from the mean, 38%, in the transition region for concentration c = 0.5 μg ml−1. The remaining concentrations give the deviation of FR smaller than 7%. Our numerical analysis revealed the following SCVs concentrations in repetitions no 1–3: 17.9 ± 1.4%, 31.3 ± 2.9%, 29.8 ± 3.0%, respectively. The average of obtained results is 26% and the standard deviation is 7.3%. The relative error is 7.3/26 = 28%.
It is worth mentioning that performing repetitions is not the only way to estimate the method's reproducibility. As we expect, all scMIC curves should be monotonically decaying functions. Therefore, nonmonotonicity around each point (i.e. how much the fraction of resistant bacteria is bigger than the measurement for the nearest lower concentration) also indicates the magnitude of the error. From this perspective, performing the repeated experiment in Fig. S4† was unnecessary. But it tests whether the method replicability can be inferred from the nonmonotonicity: notice that the value of nonmonotonicity of each of the scMIC curves from Fig. S4† is of the order of the differences between the different curves. That shows how the method reproducibility can be estimated from every single scMIC curve.
As we discussed above, the error could, in principle, be estimated from the nonmonotonicity of a scMIC curve. Such an estimation requires many measurements for concentrations around a given scMIC value. Because we also deal with situations with a relatively small number of measured concentrations in the region of interest (e.g. only two measured point around scMIC = 0.5 in Fig. 3b, dotted curve), we use the experiment with the highest number of measurements, Fig. S4,† to estimate the relative error due to coalescence, which is 28% as discussed above.
Assuming a similar precision of the method (e.g. the relative error 28%) for about twice larger content (50% of SCV's which corresponds to Fig. 3b) this precision explains the discrepancy between the prepared sample of 50% of SCV's and the measurement based on a height of the tail (37%) in the discussion of Fig. 3b.
We justify the usage of the above relative error in case of different samples as follows. We assume that the error due to the coalescence of droplets is proportional to the fraction of nonempty droplets. As a result, the relative error of scMIC does not depend on scMIC value. Assuming also that the coalescence is on a similar level for different samples, we used the experiment with best statistics (three scMIC curves in Fig. S4†) to estimate the relative error due to coalescence in Fig. 3b.
Discussion
Even in a clonal population, bacteria may differ from each other in their response to an antibiotic. On the one hand, a single bacterium in a droplet with an antibiotic can proliferate and colonize the droplet. On the other hand, the same antibiotic amount may inhibit the growth of another cell of the same bacterium. After incubation, the former situation leads to a droplet with plenty of bacteria (positive). In the latter case, the droplet contains a negligible number of bacteria (negative). FR(c) denotes the fraction of single-cell bacteria that “recover” from the treatment. In other words, FR(c) describes the inhibition of bacteria population at a single cell level which we refer to as the scMIC profile.Here we proposed a dual label-free detection, which to the best of our knowledge, has never been reported before for quantification of bacterial growth in droplets. We found that the shape of the scMIC profile of a bacterial sample, estimated based on joint detection using scattering and autofluorescence, can inform about the coexistence of diverse subpopulations representing different autofluorescence (genus), sizes of respective subpopulations, the level of their respective antibiotic tolerance, and heteroresistance. These parameters are of key importance for the characterization of the response of complex bacterial populations to antibiotics. Although bacterial samples we investigated were derived from in vitro cultures, to achieve a controlled method validation, they were carefully selected to represent challenges in terms of diagnosis and treatment. We showed that the scMIC profile determined using our method was effective in sample discrimination between strains of P. aeruginosa and S. aureus in terms of their subpopulation sizes and antibiotic susceptibilities. Quantifying the size of subpopulations of co-infectious pathogens and their combined susceptibility to antibiotics, which may differ from individual ones, could be a breakthrough in infection control and make better use of existing antibiotic resources.
Similarly, we investigated scMIC profiles of small colony variants known to arise in vivo and promote chronic infection due to their altered phenotypes. Their transient nature presents a challenge for their identification and quantification, both in the clinics and research environment. Profiles of a population of SCVs triggered with gentamicin during exponential growth in vitro demonstrated a large level of heterogeneity. This was reflected by the breath of the scMIC profile (16 times more resistant response) of this population and suggests that the response of exponentially growing S. aureus to gentamicin which leads to the formation of SCVs may be in itself heterogeneous as it leads to different gene expression patterns resulting in antibiotic heteroresistance.
We found that populations containing SCVs could be identified by tails in their scMIC profiles. The shape of the scMIC profile tail, which we define as the region of scMIC profile starting at the bottom of a sharp decline of the fraction of recovering bacteria informed on the presence of SCVs. The tail height directly corresponded to the SCVs quantity thus confirming that the method was effective in the estimation of SCV subpopulations as small as 6.8%. To our knowledge, this is the first method that allows for quantitative estimation of this type of phenotypic variants which are of clinical relevance and associated with persistent infections.
Finally, the scMIC profile was informative in terms of the composition of a biofilm known for its heterogeneous nature, informing the size of either subpopulation within the biofilm culture. Application of our method could, therefore, inform on the absolute concentration of an antibiotic necessary to inhibit the growth of complex bacterial populations and provide an early stage indicator of the risk of heteroresistant pathogens associated with increased risks of complications.
Our approach in the current form is a research tool, which can be used for a better characterisation of antibiotic response of heterogeneous populations. However, we believe that it can be improved for routine clinical use after automation of droplet generation and detection, improving the stability of droplets, and introduction of simplified (e.g. machine learning based) protocols for the analysis of the scMIC profiles.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains of Staphylococcus aureus SH1000, S. aureus MSSA476 and Pseudomonas aeruginosa PAO1 were cultured in tryptic soy broth (Biocorp, Poland) for determination of scMIC. Escherichia coli ATCC25922, Staphylococcus intermedius ATCC29663, Staphylococcus aureus Newman, Staphylococcus epidermidis ATCC12228, Pseudomonas aeruginosa ATCC27853, Klebsiella pneumoniae ATCC13883 and Acinetobacter baumanii ATCC19606 were cultured in lysogeny broth (Carl Roth, Germany) while Salmonella arizonae ATCC13314, Shigella sonnei ATCC29930 and Listeria monocytogenes ATCC19115 were cultured in brain heart infusion (Biocorp, Poland) for autofluorescence measurement.Triggering small colony variants
Small colony variants (SCVs) were triggered and isolated as described previously.31 Briefly, tryptic soy broth containing gentamicin at 2 μg mL−1 was inoculated with a single colony of S. aureus SH1000 allowing culture growth at 37 °C for 18 h. SCVs were subsequently isolated by plating serial dilutions of the culture on tryptic soy agar plates not permitting the growth of normal cells but selecting for the spontaneous persistent variants of SCVs. The phenotypic switch was confirmed as smaller colony size, lack of pigmentation, and deficiency in beta-hemolysis. The SCVs colonies were harvested with a loop, suspended in 30% (v/v) glycerol solution, or directly subjected to experimental procedures described.In vitro biofilm formation
Biofilms were grown in microtiter plates (Nunclon™ Delta Surface, Thermo Scientific Nunc) either uncoated or pre-coated with human plasma. For plasma pre-coated wells a total of 100 μL of 20% (v/v) blood plasma in 50 mM carbonate buffer, pH 9.6, was incubated in the wells for 2 h at 37 °C prior inoculation with bacteria. Overnight cultures of S. aureus were diluted 1:200 in tryptic soy broth and inoculated at 100 μL in microtiter wells. Biofilms were formed for either 24 h or 72 h at 37 °C. After washing off unattached cells, biofilm structures were dispersed with proteinase K, added to the wells at 100 μL for 30 min at 37 °C. Digested biofilms were subjected to thorough pipetting following viable counts to determine their densities on either TSA or TSA containing 2 μg mL−1 of gentamicin. Gram-staining was performed to confirm that the aggregates were separated into single cells.Reagents
Chip fabrication: PDMS precursor and initiator (Dow Corning, USA) and tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (United Chemical Technologies, USA), Novec 1720 (3 M, USA). Generation and reading of droplets: Novec HFE 7500 fluorocarbon oil (3 M, USA) and triblock PFPE–PEG–PFPE surfactant (synthesized based on procedure in US/2010/0022414 A1). Antibiotics: gentamicin sulfate solution (TOKU-E, Japan and Sigma-Aldrich, USA), ciprofloxacin (TOKU-E, Japan), tobramycin sulfate (TOKU-E, Japan). Human plasma was purchased from the Regional Blood Donation Centre in Warsaw, Poland.Microfluidic chip fabrication
Microfluidic chips for droplet formation and detection were prepared in a four-step procedure. First, a polycarbonate (PC) mold is prepared by milling the microfluidic channels in 0.5 cm PC plate using a CNC machine (MSG4025, Ergwind, Poland). In the second step, polydimethylsiloxane (PDMS) is poured onto the PC chip, polymerized at 75 °C for 1 h, and the surface is activated by Laboratory Corona Treater (BD 20 AC, Electro-Technic Products, USA) and silanized in the vapors of tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (United Chemical Technologies, USA) for 30 min under 10 mbar pressure. In the third step, the silanized mold is filled with liquid PDMS, polymerized at 75 °C for 1 h, and then the chips are bonded to 1 mm thick glass slides using oxygen plasma. In the last step, the microfluidic channels are hydrophobically modified by introducing Novec 1720 (3 M, USA), evaporation at room temperature, and baking the chips at 75 °C for 1 h.scMIC determination
The following assumptions were made to determine the scMIC profile, FR(c). For a given antibiotic concentration c, the same suspension of bacteria was encapsulated in N(c) droplets. They contained Nne(c) non-empty droplets (typically containing a single bacterium, the issue which we discuss below). After the incubation with an antibiotic of concentration c, they gave N+(c) positives. Thus, the fraction of recovering bacteria is given by FR(c) = N+(c)/Nne(c). The number of droplets N and the number of positives N+ could be determined from the detector and they determine the fraction of positive droplets, f+(c) = N+(c)/N(c). But FR(c) required the number of non-empty droplets, Nne(c), as well. To determine the fraction of bacteria that recovered, FR(c), we used the experiment without antibiotic. Here, the number of non-empty droplets was equal to the number of positives, Nne(0) = N+(0), therefore, the fraction of non-empty droplets among all droplets is, f+(0) = N+(0)/N(0). Using the same bacteria suspension in the experiment with concentration c, we calculated the number of non-empty droplets by Nne(c) = f+(0)N(c). We then obtain, FR(c) = f+(c)/f+(0). It is worth mentioning, that this relation between FR and f+ does not take into account droplets with two, three, and more bacteria. However, as we explain in Supporting Information, in our experiments, most non-empty droplets contain a single bacterium, about 5% of them contain two, and 0.15% three bacteria. We also estimate, that neglecting droplets with two and more bacteria by using formula FR(c) = f+(c)/f+(0), may change the value of FR(c) by 10%.The initial bacterial samples were diluted to the densities of 0.5–2 × 105 CFU mL−1 providing a high probability of single-cell encapsulation (5–20% of positive droplets). Antibiotics were added to the sample at a range of concentrations the following emulsification into 1 nL droplets. The droplets were collected in 0.5 mL test tubes separately for each antibiotic concentration and incubated overnight at 37 °C. We believe that separation of the droplets according to the antibiotic concentration reduces the diffusion of the droplet content to the continuous phase and most importantly minimizes the transfer of antibiotics between droplets. After incubation, the droplets were scanned and the intensity of scattered or native fluorescence light was measured.
It was not always possible to determine from the detector signal (Fig. S2†) whether the signal corresponds to a positive or to a negative droplet. It requires introducing a threshold that leads to a systematic error and an alternative approach to determine the scMIC profile. A detailed description of the determination of the scMIC profile with an explanation of all errors is shown in Fig. 1–4 (including the error related to the threshold) is in ESI.†
Scattered light intensity detection
Bacterial growth in droplets was measured based on scattered light intensity following the procedure described by Pacocha, Bogusławski et al. 2020. Positive and negative droplets were characterized by high and low intensity of scattered light, respectively. The number of positive compartments and a total number of droplets was used for the calculation of recovering bacteria fraction, FR(c), and scMIC profile determination.Autofluorescence intensity detection
Autofluorescence signal was detected in positive droplets by illumination with a 473 nm laser beam delivered by a fiber, collimated with a lens (focal length 19 mm), and focused by a 20× microscope objective. Native fluorescence light is collected by the objective, reflected on a dichroic mirror (a cut-off wavelength of 490 nm), passes a bandpass filter (central wavelength of 530 and 43 nm of bandwidth), pinhole, and targets a photomultiplier (Hamamatsu H5783-20). In this configuration, similar to a confocal microscope, only fluorescence originating from the focal volume will reach the photomultiplier. Thus, background autofluorescence (e.g., of PDMS) will be rejected. The optical setup shown in Fig. S1† was used for simultaneous detection of scattered and autofluorescence light intensity.Negative droplets are visible due to the autofluorescence of culturing medium, while the presence of a large number of bacteria cells causes a pronounced increase in the fluorescence intensity. The intrinsic fluorescence of bacteria is linked with fluorescent amino acids (tryptophan, tyrosine, and phenylalanine) and coenzymes such as nicotinamide adenine dinucleotide hydride (NADH).32 It's important to emphasize that the detection of bacteria from S. aureus genus is possible by optical setup modification. The excitation and spectrum of this bacteria lies in the UV spectral range (centered at approx. 280 nm and 330 nm, respectively).32,33 Thus, the detection of autofluorescence of S. aureus would require a different light source, and changing bandpass filter placed in front of the photomultiplier.
Author contributions
N. P. performed experiments, M. Z. designed the project, K. M. analyzed the data, N. P. and J. B. developed the autofluorescence-based detection of bacteria growth in the droplets, N. P., M. Z., K. M., J. B., P. G. wrote the manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
N. P. was supported by the National Science Centre funding based on decision numbers DEC-2014/12/W/NZ6/00454 (Symphony) and Maestro 10 2018/30/A/ST4/00036. M. Z. was funded by the National Science Centre, Poland grant no. 2018/31/D/NZ6/02648 (SONATA14), K. M. acknowledges NCN Maestro 10 2018/30/A/ST4/00036 and the support from the Foundation for Polish Science within the Team-Net POIR.04.04.00-0016ED/18-00 program, P. G. was supported by the Foundation for Polish Science (FNP) within TEAM TECH POIR.04.04.00-00-2159/16-00 and by The National Science Centre (NCN) within Maestro 10 2018/30/A/ST4/00036, J. B. was funded by the International Research Agendas program of the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund.Notes and references
- A. Leligdowicz and M. A. Matthay, Heterogeneity in sepsis: new biological evidence with clinical applications, J. Crit. Care, 2019, 231(23), 1–8 Search PubMed.
- C. Pereira, J. Larsson, K. Hjort, J. Elf and D. I. Andersson, The highly dynamic nature of bacterial heteroresistance impairs its clinical detection, Commun. Biol., 2021, 41(4), 1–12 Search PubMed.
- K. A. Brogden, J. M. Guthmiller and C. E. Taylor, Human polymicrobial infections, Lancet, 2005, 365, 253–255 CrossRef.
- K. M. Kuper, D. M. Boles, J. F. Mohr and A. Wanger, Antimicrobial susceptibility testing: a primer for clinicians, Pharmacotherapy, 2009, 29, 1326–1343 CrossRef PubMed.
- S. A. S. Hitam, S. A. Hassan and N. Maning, The Significant Association between Polymicrobial Diabetic Foot Infection and Its Severity and Outcomes, Malays. J. Med. Sci., 2019, 26, 107 Search PubMed.
- F. E. McKenzie, Case mortality in polymicrobial bloodstream infections, J. Clin. Epidemiol., 2006, 59, 760–761 CrossRef CAS PubMed.
- K. Bisht, J. Baishya and C. A. Wakeman, Pseudomonas aeruginosa polymicrobial interactions during lung infection, Curr. Opin. Microbiol., 2020, 53, 1–8 CrossRef CAS PubMed.
- A. J. Fischer, et al. Sustained Coinfections with Staphylococcus aureus and Pseudomonas aeruginosa in Cystic Fibrosis, Am. J. Respir. Crit. Care Med., 2021, 203, 328–338 CrossRef CAS PubMed.
- M. K. Wieneke, et al. Association of Diverse Staphylococcus aureus Populations with Pseudomonas aeruginosa Coinfection and Inflammation in Cystic Fibrosis Airway Infection, mSphere, 2021, 6, 1–22 CrossRef PubMed.
- J. Bigger, Treatment of Staphylococcal Infections with Penicillin by Intermittent Sterilisation, Lancet, 1944, 244, 497–500 CrossRef.
- B. C. Kahl, Small colony variants (SCVs) of Staphylococcus aureus--a bacterial survival strategy, Infect., Genet. Evol., 2014, 21, 515–522 CrossRef CAS PubMed.
- C. Vulin, N. Leimer, M. Huemer, M. Ackermann and A. S. Zinkernagel, Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters, Nat. Commun., 2018, 9, 1–8 CrossRef CAS PubMed.
- G. Loss, et al. Staphylococcus aureus Small Colony Variants (SCVs): News From a Chronic Prosthetic Joint Infection, Front. Cell. Infect. Microbiol., 2019, 0, 363 CrossRef CAS PubMed.
- L. Tuchscherr, B. Löffler and R. A. Proctor, Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs), Front. Microbiol., 2020, 0, 1028 CrossRef PubMed.
- F. Peyrusson, et al. Intracellular Staphylococcus aureus persisters upon antibiotic exposure, Nat. Commun., 2020, 111(11), 1–14 Search PubMed.
- H. Nicoloff, K. Hjort, B. R. Levin and D. I. Andersson, The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification, Nat. Microbiol., 2019, 4, 1–13 CrossRef PubMed.
- H. Nicoloff, K. Hjort, B. R. Levin and D. I. Andersson, The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification, Nat. Microbiol., 2019, 4, 504–514 CrossRef CAS PubMed.
- D. Belikova, A. Jochim, J. Power, M. T. G. Holden and S. Heilbronner, “Gene accordions” cause genotypic and phenotypic heterogeneity in clonal populations of Staphylococcus aureus, Nat. Commun., 2020, 111(11), 1–15 Search PubMed.
- J. Q. Boedicker, L. Li, T. R. Kline and R. F. Ismagilov, Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics, Lab Chip, 2008, 8, 1265–1272 RSC.
- L. Baraban, et al. Millifluidic droplet analyser for microbiology, Lab Chip, 2011, 11, 4057–4062 RSC.
- X. Liu, et al. High-throughput screening of antibiotic-resistant bacteria in picodroplets, Lab Chip, 2016, 16, 1636–1643 RSC.
- T. S. Kaminski, O. Scheler and P. Garstecki, Droplet microfluidics for microbiology: Techniques, applications and challenges, Lab Chip, 2016, 16, 2168–2187 RSC.
- A. M. Kaushik, et al. Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform, Biosens. Bioelectron., 2017, 97, 260–266 CrossRef CAS PubMed.
- T. Artemova, Y. Gerardin, C. Dudley, N. M. Vega and J. Gore, Isolated cell behavior drives the evolution of antibiotic resistance, Mol. Syst. Biol., 2015, 11, 822 CrossRef.
- F. Lyu, et al. Phenotyping antibiotic resistance with single-cell resolution for the detection of heteroresistance, Sens. Actuators, B, 2018, 270, 396–404 CrossRef CAS.
- O. Scheler, et al. Droplet-based digital antibiotic susceptibility screen reveals single-cell clonal heteroresistance in an isogenic bacterial population, Sci. Rep., 2020, 10, 3282 CrossRef CAS PubMed.
- N. Pacocha, et al. High-Throughput Monitoring of Bacterial Cell Density in Nanoliter Droplets: Label-Free Detection of Unmodified Gram-Positive and Gram-Negative Bacteria, Anal. Chem., 2020, 93, 843–850 CrossRef.
- Y. C. Wu, et al. Autofluorescence imaging device for real-time detection and tracking of pathogenic bacteria in a mouse skin wound model: preclinical feasibility studies, J. Biomed. Opt., 2014, 19, 085002 CrossRef.
- W. K. Philipp-Dormston and M. Doss, Comparison of Porphyrin and Heme Biosynthesisin Various Heterotrophic Bacteria, Enzyme, 1973, 16, 57–64 CrossRef CAS PubMed.
- C. D. Cox and P. Adams, Siderophore activity of pyoverdin for Pseudomonas aeruginosa, Infect. Immun., 1985, 48, 130–138 CrossRef CAS PubMed.
- A. M. Edwards, Phenotype switching is a natural consequence of Staphylococcus aureus replication, J. Bacteriol., 2012, 194, 5404–5412 CrossRef CAS PubMed.
- P. M. Molyneux, S. Kilvington, M. J. Wakefield, J. I. Prydal and N. P. Bannister, Autofluorescence Signatures of Seven Pathogens: Preliminary in Vitro Investigations of a Potential Diagnostic for Acanthamoeba Keratitis, Cornea, 2015, 34, 1588–1592 CrossRef PubMed.
- H. K. Peguda, et al. The Autofluorescence Patterns of Acanthamoeba castellanii, Pseudomonas aeruginosa and Staphylococcus aureus: Effects of Antibiotics and Tetracaine, J. Pathog., 2021, 10, 894 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Scheme of optical setup, exemplary autofluorescence signals, growth curves of normal colony and small colony variant phenotype, precision of scMIC curve determination, list of bacteria strains tested towards native fluorescence intensity, breakpoint concentrations of NCPs and SCVs, details regarding determination of scMIC profile. See DOI: https://doi.org/10.1039/d2lc00234e |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |
