 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
On-chip miRNA extraction platforms: recent technological advances and implications for next generation point-of-care nucleic acid tests
Loukia
Petrou
 and
Sylvain
Ladame
and
Sylvain
Ladame
 *
*
Department of Bioengineering, Imperial College London, London, W12 0BZ, UK. E-mail: s.ladame@imperial.ac.uk
First published on 5th November 2021
Abstract
Circulating microRNAs (or miRNAs) in bodily fluids, are increasingly being highlighted as promising diagnostic and predictive biomarkers for a broad range of pathologies. Although nucleic acid sensors have been developed that can detect minute concentrations of biomarkers with high sensitivity and sequence specificity, their robustness is often compromised by sample collection and processing prior to analysis. Such steps either (i) involve complex, multi-step procedures and toxic chemicals unsuitable for incorporation into portable devices or (ii) are inefficient and non-standardised therefore affecting the reliability/reproducibility of the test. The development of point-of-care nucleic acid tests based on the detection of miRNAs is therefore highly dependent on the development of an automated, on-chip, sample processing platform that would enable extraction or pre-purification of the biological specimen prior to reaching the sensing platform. In this review we categorise and critically discuss the most promising technologies that have been developed to facilitate the transition of nucleic acid tests based on miRNA detection from bench to bedside.
Introduction
miRNAs as biomarkers
Circulating microRNAs (miRNAs) are short (19–22 bp), non-coding nucleic acids that have recently emerged as promising biomarkers for a variety of diseases including cancer (for both diagnosis and origin identification) as well as heart diseases, neurological and infectious diseases.1–3The biogenesis of miRNAs starts in the nucleus where following DNA transcription, the stem-loop precursor pri-miRNAs are cleaved to form pre-miRNAs which are then transported out of the nucleus through exportins.4 In the cytoplasm, the pri-miRNAs are cleaved into miRNA duplexes with the active strand binding to Argonaute proteins and the inactive strand being degraded.5 The association with Argonaute proteins allows the miRNAs to bind to mRNA and silence its translation into proteins either through degradation or ribosome stalling.6–8 miRNAs thus play an important role in post-transcriptional gene regulation and recently have also been identified as mediators for cell-to-cell communication as certain miRNAs are preferentially sorted into exosomes.9,10
The isolation of these biomarkers from bodily fluids poses unique impediments. They circulate mainly using the two main transport mechanisms mentioned previously which protect them from degradation while in circulation. Because of this they have enhanced stability in circulation which is however compromised as soon as they are released due to their exposure to endogenous ribonucleases (RNases) which has also been previously demonstrated when injecting synthetic miRNAs or miRNAs purified from plasma back into unprocessed plasma. This led to their rapid degradation highlighting their instability when extracted from their protective transporters.11 They are still more stable than longer RNAs such as mRNAs when exposed to RNases which further highlights their utility as biomarkers.12 Promising point-of-care (POC) techniques of nucleic acid isolation have been summarised previously.13–15 Given the emerging evidence of the clinical value of miRNAs as diagnostic or prognostic biomarkers, we will focus exclusively in this review on this specific class of molecular biomarkers and recent progress made towards their isolation from clinical samples. Fig. 1 summarises the workflow of the techniques discussed in this review to illustrate the types of samples used and how these are processed. This review will focus primarily on sample processing rather than sensing, although selected technologies that successfully combined miRNA extraction and detection will also be discussed.
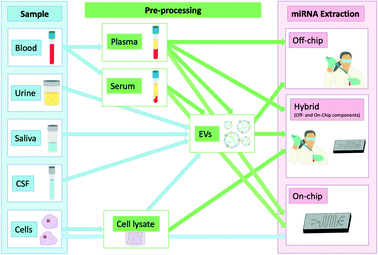 | ||
| Fig. 1 miRNAs and different locations from which they can be isolated showing any additional steps required in the extraction process. | ||
Liquid biopsies and miRNAs
Biomarkers are clinically useful in a multitude of scenarios: diagnosis, prognosis, monitoring, screening, and treatment of diseases. Liquid biopsies allow minimally or non-invasive access to important biomarkers and provide valuable alternatives to more invasive tissue biopsies.16 This involves sampling of bodily fluids (most commonly blood, saliva, urine or interstitial fluid) and can avoid complications arising from traditional tissue biopsies. Liquid biopsies can also delineate the path to diagnosis when samples are not easily accessible.17 For example, this is particularly relevant in diseases such as Alzheimer's disease or metastatic cancer lesions.18,19 Wide-spread usage of liquid biopsies in clinical settings is often limited by the sample processing required for test result attainment.Despite some clustering observed when analysing miRNAs from a variety of bodily fluids, their relative and absolute concentrations may vary with urine and saliva yielding a smaller number of miRNAs than blood or plasma.20 Weber et al. looked at the miRNA expression in 12 different bodily fluids and found that saliva had the highest numbers of unique detectable miRNAs and while some abundant miRNAs can be identified across a variety of bodily fluids, some are enriched in certain types of samples.21 This has also been shown in forensic analysis that identified certain miRNA patterns to assist in identification of the source of a bodily fluid.22 It is notable however that differences exist in the miRNA concentration between samples of similar origin such as serum and plasma from the same individual. This could be explained by the association of miRNAs with platelets that are lost in the coagulation process to obtain serum but exact causes remain largely unclear, underlining the source of error blood sample processing represents on the miRNA profile obtained.23 The complexity involved in sample processing and miRNA sensing has been highlighted previously. The transporting mechanism of miRNAs (i.e. how many are within vesicles, bound to proteins or freely circulating) for each liquid biopsy remains elusive. Arroyo et al. looked into the transport mechanism in plasma and serum and determined that the main mechanism is Argonaute2 (Ago2) proteins with only a fewer proportion of miRNAs found in vesicles.24 Turchinovich et al. looked into the transport of miRNAs in plasma and cell culture media and agreed that extracellular miRNAs are primarily transported bound to Ago2.25 This however was contradicted by the findings of Gallo et al. who determined that most miRNAs in serum and saliva are actually found within exosomes.26 These studies highlight not only the complexity of the field but also the discrepancy in the literature due at least in part to the lack of standardised protocols and technologies for sample processing, miRNA extraction as well as detection.
Herein, we discuss a variety of techniques used for miRNA extraction and their incorporation into POC devices for down-stream detection. We highlight some emerging technologies and separate them into a) those that are carried out entirely off-chip, b) those that involve some off-chip processing, and c) those that have achieved on-chip miRNA isolation and sensing in a processing-free manner (or involving minimal processing) and hold the most promise for translation into clinical settings as POC devices.
Off-chip miRNA extraction
Nucleic acid extraction
There are also obvious differences stemming from where miRNAs are isolated from which bring about differences in protocols. As described by Pritchard et al., the yield is higher from cell lysates or tissues compared to bodily fluids and the extracted miRNAs are typically of higher quality.27 Bodily fluids pose an additional challenge due to the inherent RNase activity which could degrade any extracted miRNAs if the RNases are not deactivated at the same time. Since the isolation process involves the same main steps, we will be including kits that successfully carried out miRNA isolation from cells in this review as such protocols are amendable to miRNA extraction from other sources following optimisation. Although the methodology for isolating miRNA is mostly similar to that for isolating other RNA species (e.g. mRNA or lncRNA), miRNA isolation involves specific enrichment of smaller RNAs.28 This can be achieved through gel-purification, but more recent methods harnessed by kits involve column-based techniques.
miRNA extraction kits
Recent efforts to allow faster and more efficient nucleic acid extraction have been streamlined by commercially available extraction kits. Longer RNAs or DNAs compared to miRNAs, defer in both increased length and stability and are thus generally easier to handle. This has led to the development of kits specifically targeting miRNA extraction, through incorporating additional steps to ensure adequate recovery of short RNAs. For example, the Ambion PureLink miRNA isolation kit encompasses a double column isolation method, whereby the first column is used to selectively bind larger RNA fractions while the miRNAs pass into the flow-through and are then purified by the second column. Separation by size is particularly important in samples where there is a mix of nucleic acid types. For example, serum and plasma contain mainly small RNAs and therefore do not necessitate targeted extraction of smaller RNAs.31 Some commonly used kits for a variety of samples including plasma/serum, urine, cerebrospinal fluid (CSF) and cell lysates are summarised in Table 1.| Name of kit | Phenol/chloroform extraction | Type of miRNA purification | Type of sample | Amount of sample required | Steps on protocol | Time requireda | Cost per sampleb |
|---|---|---|---|---|---|---|---|
| a Total time is an estimate given the steps and times supplied on the protocol as well as information provided by the supplier. b Cost per sample does not include the cost of additional reagents required and not supplied by the kit, or DNA spike-ins to normalise result. It is calculated as a minimum taking into account the least amount of sample/reagents required as well as using the kits that allow for the maximum samples (50 samples for most kits). In cases where more sample is required the price can be as much as 3× higher per sample than the one displayed in the table. | |||||||
| miRNeasy serum/plasma kit (QIAGEN) | Yes | Chemical and column-based | Serum/plasma | 200 μL | 15 steps | 40 min | £8.0 |
| miRNeasy serum/plasma advanced kit (QIAGEN) | No | Column-based | Serum/plasma | 200 μL | 12 steps | 25 min | £6.0 |
| Ambion® PureLink™ miRNA isolation kit (Invitrogen) | No – chloroform is optional for larger tissue amounts | Column-based | Cells | <106 cells | 24 steps | 20 min | £8.4 |
| <5 mg tissue | |||||||
| MagMAX mirVana total RNA isolation kit (Applied Biosystems) | No – chloroform is optional | Magnetic beads | Blood | 50 μL | 17 steps | 90–120 min | £4.3 |
| Tissue | 50 mg | 12 steps | |||||
| Plasma/serum | 100 μL | 17 steps | |||||
| Urine | 250 μL | 13 steps | |||||
| TaqMan™ miRNA ABC purification kit (ThermoFisher scientific) | No | Magnetic beads | Blood | 10 μL | 23 steps | 75 min | £10.8 |
| Plasma/serum | 50 μL | 22 steps | |||||
| Cells | 10–106 cells | 22 steps | |||||
| Saliva | 50 μL | 23 steps | |||||
| Urine | 50 μL | 23 steps | |||||
| mirPremier microRNA isolation kit (Sigma-Aldrich) | No | Column-based | Cells | 0.1–7 × 106 cells | 13 steps | 30 min | £5.4 |
| Total RNA purification kit (Norgen Biotek) | No | Column-based | Blood | 100 μL | 14 steps | 15 min | Not available |
| Exosome miRNA extraction kits | |||||||
| miRCURY exosome kit (QIAGEN) | No | Column-based | Plasma/serum | 500–1400 μL | 8–11 steps | 100 min | £6–£16; |
| Urine/cells/cerebrospinal fluid | 1–10 mL | 8 steps | £2.4–24 | ||||
| exoRNeasy serum–plasma Midi kit (QIAGEN) | Yes | Column-based | Plasma/serum | 0.1–1 mL | 19 steps | 60 min | £18.2 |
The use of toxic reagents such as phenol and chloroform for extraction, necessitates careful handling by trained personnel in a lab-based facility and thus limits translation into a POC device. The same applies to centrifugations which are a recurrent key step in most commercial kits and prevent direct implementation into a POC device. Recent attempts have been made to develop kits with no such chemical extraction requirements, but similar extraction capacities that are included in Table 1.
From Table 1, the average time required by kits for miRNA extraction was 52 min with the average number of steps being ∼17 and average cost per sample of ∼£9.47 (minimum based on kits for many samples). One of the kits (miRNeasy) involved phenol–chloroform extraction which has now been updated (miRNeasy Advanced) and no longer necessitates the use of such chemicals. Another extraction strategy that bypasses the need for phenol–chloroform extraction is that used by MagMAX mirVana total RNA isolation kit (Applied Biosystems) and Taqman miRNA ABC purification kit (ThermoFisher). Both kits use magnetic beads coated with oligonucleotides to selectively isolate broad panels of miRNAs. Such kits have technical limitations including their reliance on costly equipment (especially in the case of MagMAX), a rather limited capturing efficiency limited to the extraction of only currently known miRNA sequences. It is worth noting however that all kits require the use of guanidinium thiocyanate for the extraction step irrespective of sample type. This chaotropic salt is involved in the break-down of extracellular vesicles and proteins to allow miRNA release.
Previous reports have also shown that there is a lot of variability between the kits and miRNA is not always reliably extracted despite the use of spike-in controls.32,33 In the absence of a standardised protocol used by all labs, specific studies may be influenced by the miRNA isolation method used, making comparisons between studies challenging, if not impossible.34 There have also been reports that using these kits might result in partial loss of miRNAs with low GC content.35 When directly compared to each other, there does not appear to be a single most effective kit for miRNA isolation and the selection of kits generally is dictated by the type of sample and convenience.36 Finally, steps to automate extraction using the such kits have been made using a bench-top device which however would bring up the cost by thousands of pounds, making it an inaccessible option for most.
However, the main limitation of such kits is the variability and lack of reliability introduced through sample loss during the extraction process. This means that even if the kits were to be automated, unless the detection method was able to normalise the results using an internal control, the kits are limited by their lack of consistency.
Advances
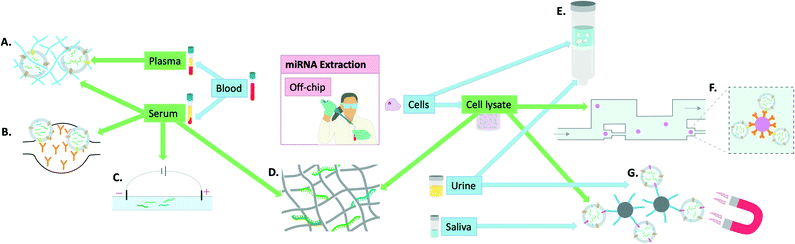 | ||
| Fig. 2 Visual summary of the off-chip miRNA extraction methodologies explored in this review and the utilised samples to achieve this: A) polymer-based exosome entrapment,45 B) PDMS functionalised with anti-CD-63 antibodies for exosome entrapment,47 C) capillary electrophoresis,42 D) titanium dioxide nanofibers for miRNA isolation,41 E) advanced glass membrane column for EV entrapment,46 F) single bead trapping array for exosome isolation (using anti-CD-63 antibodies), G) EpCAM aptamer-coated magnetic nanoparticles for exosome isolation.44 | ||
Finally, some groups have focused more on miRNA extraction specifically from exosomes and developed ways of extracting exosomes for subsequent miRNA quantification. Tayebi et al. proposed a method of exosome purification using microfluidic hydrodynamic trapping through combining immunoaffinity to confine exosomes to polystyrene microbead surfaces, with passive trapping through microfluidic channels. The sample used was cell supernatant containing exosomes and phenol–chloroform extraction was used prior to miRNA detection by RT-qPCR.43 Zhou et al. used Fe3O4@SiO2-aptamer nanoparticles to separate exosomes via magnetic separation in solution and demonstrated the utility of this method in both saliva and urine by subsequent detection on a lateral flow assay (LFA), later confirmed by RT-qPCR.44 Grunt et al. proposed a method of exosome extraction involving a mannurate–guluronate polymer to entrap and enrich exosomes from plasma, serum, urine and cell samples. For miRNA release, chaotropic salts were added and although successful the overall process remained very labour-intensive.45 Yukawa et al. took advantage of spinoidal decomposition to form a co-continuous structure to allow extracellular vesicle isolation in just 10 minutes.46 However, for subsequent miRNA extraction, commercial kits were used underlining that although a step in the right direction, this again falls short of the requisites for an on-chip miRNA extraction kit. Kanwar et al. used a chip made of PDMS that was functionalised with antibodies against CD63 (commonly over-expressed on the surface of exosomes). This allowed on-chip capturing of exosomes from serum. The exosomes were then quantified on-chip but detection of miRNAs was achieved by RT-qPCR following miRNA extraction by extraction kits.47
These recently developed techniques show a lot of promise and appear easier to translate into a POC device owing to their simplistic nature and incorporation of well-established techniques. They are, however, currently at an early stage of development and still require off-chip sample pre-processing.
Hybrid methods of miRNA extraction and detection
On-chip extraction with off-chip sample preparation
MiRNA extraction chips have been developed that still require steps such as cell lysis to be carried out off-chip before addition of the sample onto the device. This adds a step to the process which means that further optimisations are required for such devices to reach the market as a single-step POC device for miRNA extraction and sensing. As a result, they are here defined as hybrid methods (Fig. 3). An example of such a device combining both on-chip and off-chip elements is that developed by Zhong et al. This device was able to extract miRNAs, following off-chip cell lysis, to a standard identical to that of the Ambion Purelink miRNA isolation kit. This was carried out in 15 min on a PDMS microfluidic device through incorporating multiple microwells that contained buffers necessary for sample processing (i.e. RNA binding, washing and elution). The miRNAs were bound onto silica magnetic particles which were transferred between wells using a magnet. The final elution was prepared within 15 min and on-chip RT-PCR was carried out proving similar results to the extraction kit used for comparison while the process was both faster and cheaper.48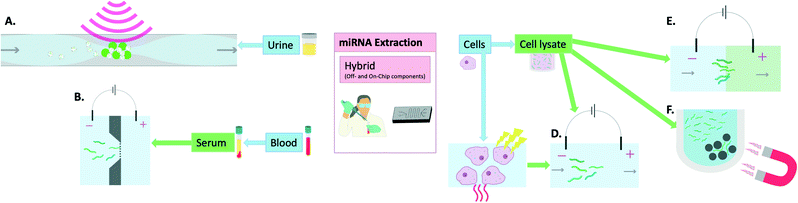 | ||
| Fig. 3 Hybrid methods (off- and on-chip components) of miRNA extraction: A) acoustic waves and seeding particles for EV entrapment,55 B) electrophoresis in buffer through nanofilter membrane,57 C) thermo-electric cell lysis,51,58 D) electrophoretic miRNA purification,58 E) isotachophoresis which involves the movement of a leading electrolyte (green) and a trailing electrolyte (blue),50 F) magnetic particles for removal of larger nucleic acids in the process of miRNA isolation.48 | ||
Slouka et al. used electrophoresis as a sample pre-treatment method following cell lysis. This was then followed by concentration of the miRNAs using a cation exchange membrane.49 Similarly, Schoch et al. used isotachophoresis with an incorporated sieving matrix to extract, isolate and preconcentrate small RNAs, including miRNAs, from cell lysates.50 The methodologies included an off-chip cell lysis procedure which was based on lysis buffers and chloroform. Dame et al. reported the use thermo-electric cell lysis followed by on-chip gel electrophoresis for miRNA separation. When compared with the mirVana kit, their method reportedly allowed 100× higher yield/efficiency when comparing the final eluates by RT-qPCR detection.51 Wei et al. also harnessed the utility of microchip electrophoresis for miRNA filtering following off-chip cell lysis by ultrasonication and centrifugation and achieved very high recoveries (94.9–108%).52 Although these electrophoretic methods seem promising, they still require manual handling at times and are thus not entirely on-chip extraction methods.
Qian et al. performed ultracentrifugation to isolate exosomes from either cell lysate or spiked into exosome-depleted serum, prior to adding the sample onto their device where they were enriched by an agarose fork-shaped microchannel through a passive capillary force. Following addition onto the device, the process of miRNA isolation including sensing via catalysed hairpin assembly took 10 minutes.53 Although quick and capable of detecting exosomal miRNAs from serum, this method in its current state requires a fair amount of sample pre-processing prior to addition onto the device and thus necessitates further optimisation.
Another approach to miRNA extraction was that developed by Deng et al. This method involves a foldable, multi-layer paper chip to extract, purify and detect miRNAs following off-chip cell lysis. Although the process took 90 min, it demonstrated great promise in its ability to achieve multiplex miRNA analysis.54
On-chip extraction with off-chip detection
On-chip miRNA extraction platforms have also been developed that required no off-chip pre-processing but were typically followed by off-chip miRNA detection (most typically via RT-qPCR). Although these extraction technologies may well be compatible with on-chip detection, this is yet to be demonstrated and they are also classified here as hybrid methods. An example of such a device is that developed by Ku et al. which incorporated acoustic trapping in an automated microfluidic platform for extracellular vesicle enrichment. Besides successfully enriching EVs, the miRNAs were subsequently extracted.55 This method of EV isolation was also used later as part of a high-throughput screening to identify miRNA biomarkers for prostate cancer.56Lee et al. used electrophoresis to extract miRNAs from serum samples through a nanofilter membrane and demonstrated comparable miRNA Cq values using RT-qPCR to a commercially available miRNA extraction kit, with the whole process of on-chip extraction lasting around 30 min.57 Behrmann et al. incorporated thermoelectric lysis onto a microfluidic device in order to extract miRNAs directly from cells which was followed by gel electrophoresis to isolate them. When compared to the mirVana isolation kit (Ambion), the chip's performance was miRNA-dependent, performing 110–220 times better for miR-16 and 8.5–14 times better for let-7a (in terms of Ct values obtained). The chip was able to extract miRNAs successfully for detection by stem-loop RT-qPCR from as little as 5 cells.58
On-chip miRNA extraction and detection
Incorporating all aspects of sample processing from miRNA extraction to amplification if necessary and finally sensing on the same platform has the potential to revolutionise the field of personalised medicine. Recently, there has been great progress on this front and unlike in the previous section where most technologies were primarily developed for extraction of cellular miRNAs, this section focuses mostly on on-chip extraction combined with on-chip detection of miRNAs from liquid biopsies, with several examples illustrated in Fig. 4. Such technologies have probably the greatest translational potential, with applications ranging from widespread public screening to longitudinal monitoring.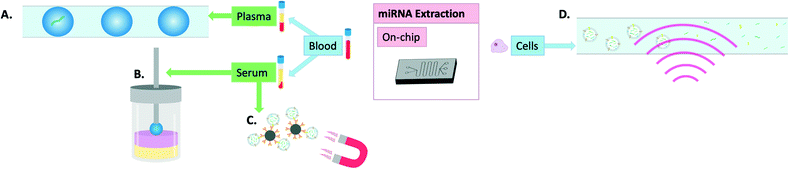 | ||
| Fig. 4 Visual summary of on-chip methods of miRNA extraction: A) miRNA encapsulated in microdroplets,60 B) magnetic single-droplet extraction where the serum sample (yellow) form hyperbranched DNA-iron oxide networks into the organic phase (pink) as they are magnetically attracted to the extraction solvent (blue) held by a magnetic rod (grey),59 C) magnetic beads coated in anti-CD-63 antibodies for exosome isolation,61,62 D) surface acoustic waves for extracellular vesicle lysis.63 | ||
Tang et al. developed a new method of single-drop microextraction whereby the analyte is transferred into a small drop of extraction solvent. This was integrated into a magnetic three-phase single-drop microextraction which enabled release and subsequent detection of miRNAs through triggering the formation of hyperbranched DNA-iron oxide networks. When applied to the detection of miRNAs from serum samples, it allowed researchers to distinguish between samples from healthy volunteers and those from patients with hepatocellular carcinoma with only 6 seconds required for extraction and 5 minutes for detection.59 Zhang et al. demonstrated the usefulness of microencapsulation in combination with a particle counter for digital analysis to detect miRNAs directly from plasma in less than 3 hours and down to 10 single copies of miRNA. The device was able to detect miRNA with a limit of detection of about 50 copies per mL of plasma and was thus able to distinguish healthy donor samples from those originating from patients with metastatic colon cancer based on detection of Let-7a levels.60
Cheng et al. and Huang et al. used anti-CD63 antibody coated magnetic beads to first bind to and then extract extracellular vesicles (EVs) from plasma.61,62 Cheng et al. incorporated the beads onto a microfluidic device and included a miRNA extraction component following EV extraction which involved EV lysis followed by capturing of the miRNAs of interest on magnetic beads coated with complementary DNA and allowed for detection down to 1 fM concentrations of miRNA using a field-effective-transistor (FET). A pneumatic-driven microfluidic device allowed automation of the aforementioned steps into an all-encompassing device for the detection of cardiovascular disease-specific miRNAs.61 Huang et al. used the same method of extraction and detection and demonstrated the utility of the device in breast cancer diagnosis.62
Ramshani et al. used an integrated microfluidic device to detect both free-floating and EV-specific miRNAs directly from plasma in 30 min. This was achieved using surface acoustic waves (SAW) to break down the EVs.63 It was previously shown that these waves, generated by electrodes, are capable of lysing both cells and exosomes to release incorporated nucleic acids.64,65 Following this lysis step, the miRNAs were concentrated using cation-exchange membranes and detected using anion-exchange membranes. To detect free-floating miRNAs, the same procedure was followed while the SAW-producing component was switched off. The device was able to generate similar results to conventional methods of extraction and detection by RT-qPCR in a fraction of the time and in a processing-free manner.63
Finally, certain sensing technologies have been developed that do not necessitate any sample pre-processing and allow detection of miRNAs directly in un-processed samples. By-passing sample pre-processing often requires sensing technologies that are both ultrasensitive (most typically with single molecule resolution) and highly specific, to detect the miRNA of interest in complex biological matrices. The absence of processing, however, may also mean that only free-circulating miRNAs can be detected, exosomal miRNAs or miRNAs involved in protein complexes being out of reach.
Cai et al. were able to detect miRNAs from the serum of prostate cancer patients using electro-optical nanopore sensing. The technology was sensitive and specific enough to distinguish patients with active disease from those in remission through multiplex miRNA profiling.66 Tavallaie et al. used gold-coated magnetic nanoparticles (Au@MNP) and electric-field-induced assembly of the DNA-Au@MNP network to detect microRNAs directly in blood in around 30 min. This allowed for detection of miRNAs from 10 aM–1 nM (in line with RT-qPCR) and was able to detect small differences in miRNA levels within blood samples obtained from mice with growing tumours.67 Kangkamano et al. used pyrrolidinyl PNA polypyrrole/silver nanofoam electrodes for electrochemical miRNA sensing. This allowed for detection of miRNA directly from plasma in the range of 0.2 fM–1 nM with a hybridisation time of 5 min,68 which is a lot faster than other similar label-free electrochemical miRNA biosensors reported.69–77 McArdle et al. used electrocatalytic platinum nanoparticles to detect miRNAs directly from human plasma and CSF on a centrifugal microfluidic device. This device was able to detect differences in miR-134 expression levels between control samples from healthy volunteers and those obtained from patients with refractory epilepsy or non-epileptic patients who experienced status epilepticus.78
Conclusions and perspectives
We have herein summarised some of the most promising methodologies and technologies for miRNA isolation developed in the last decade. All of them have demonstrated either incremental or more significant potential for simplifying complex miRNA extraction. Some appear more easily translatable into a POC device than others because of their simplicity, automation and amenity to miniaturisation. This has already led to all-in-one device prototypes capable of detecting miRNAs either directly from biological matrices or through various processing or extraction steps performed within the device itself.59–63 Such technologies could pave the way to the next generation of point-of-care nucleic acid tests with applications ranging from widespread public screening to personalised medicine. But importantly, they could also become very powerful tools to identify new miRNA biomarkers. The relative absence on the current market of diagnostic tests based on miRNAs detection from liquid biopsies can be explained by the challenges faced by researchers to identify and clinically validate miRNAs. As explained throughout this review, miRNA analysis is very strongly influenced by the way they are extracted, stored, and detected. Lifting this technical limitation should see the emergence of more clinically validated miRNAs and the demand for miRNA POC device flourish.All technologies mentioned in this review provide to some extent an alternative to the current costly, time-consuming and elaborate miRNA extraction kits (Table 2). They are also new tools made available to researchers to better understand the life cycle of miRNAs in bodily fluids. Fundamental questions such as miRNA compartmentalisation or protein complexation are critical to help develop new diagnostic or prognostic tests that are fit for purpose. For example, methodologies allow for isolation of miRNAs specifically from EVs43,46,47,53,55,59,61–63 while others are less selective toward the source of miRNAs, opening doors to both EV-specific and total miRNA biomarkers. Other technologies sensitive enough to detect miRNAs directly from bodily fluids will however be targeting a different pool of biomarkers, those miRNAs that are freely circulating. Some recurrent approaches used electrophoretic methods for miRNA isolation/purification,49–52,57 beads for extracellular vesicle isolation based on surface functionalisation43,61,62 as well as acoustic waves for either EV lysis63 or EV entrapment.55
| Liquid biopsy or cells? | Biofluid/cell lysate | Disease targeted | Biofluid processing method for miRNA isolation | Control used (e.g. commercial kit) | Efficacy compared to control | Analysis/extraction time | Amplification required? | Sensing method | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Off-chip miRNA extraction | |||||||||
| Both | Serum and cell lysate | NA | TiO2 nanofibres | Silica columns, beads and fibres | 4.2× better than the best performing control (silica fibres) | ∼20–25 min extraction | Yes | RT-qPCR | 41 |
| Liquid biopsy | Serum | Liver cancer | Protein-facilitated affinity capillary electrophoresis | NR | NA | 20 min | No | Laser-induced fluorescence | 42 |
| Cells | Cell lysate | NR – stressed or not cells | Single-bead trapping arrays | NR | NA | NR | Yes | RT-qPCR | 43 |
| Both | Lung cells, urine and saliva | Lung cancer | Fe3O4@SiO2-Aptamer nanoparticles | NR | NA | NR | Yes – duplex-specific nuclease | Fluorescence on LFAs | 44 |
| Liquid biopsy | Plasma and serum | NR | MGP-based polymer for exosome entrapment | Total exosome RNA and protein isolation kit (Invitrogen) | Higher reproducibility | NR | Yes | RT-qPCR | 45 |
| Liquid biopsy | Serum, urine and cells | NA | Advance glass membrane column (AGC) with macropores for EV isolation | Ultracentrifugation, ExoCap and ExoQuick | Higher number of distinct miRNAs detected using AGC than others | 10 min for EV extraction | Yes | RT-qPCR | 46 |
| Liquid biopsy | Serum | Pancreatic cancer | PDMS functionalized with antibodies against CD63 | NR | NA | NR | Yes | RT-qPCR/miRNA Openarray | 47 |
| Hybrid miRNA extraction and detection | |||||||||
| Liquid biopsy | Serum | Hepatocellular carcinoma | Electrophoresis through nanofilter membrane | miRNeasy serum/plasma kit (Qiagen) | Comparable Ct values | 30 min for extraction | Yes | RT-qPCR | 57 |
| Liquid biopsy | Urine | NA | Acoustic trapping for EV enrichment | Ultracentrifugation | Higher miRNA reads | NR | Yes | NGS | 55 |
| Cells | Breast cancer cells | Breast cancer | Thermo-electric lysis of cells | mirVana kit (Ambion) | Comparable | NR (<5 min) | Yes | RT-qPCR | 51 |
| Cells | Cell lysate | Breast cancer | Thermo-electric lysis of cells + on-chip gel electrophoresis | mirVana kit (Life Tech.) | 100× higher yield/efficiency | 10 min extraction | Yes | RT-qPCR | 58 |
| Cells | Cell lysate | NA | Solid phase extraction by silica magnetic particles | PureLink™ miRNA isolation kit (Invitrogen) | Identical | 15 min | Yes | On-chip RT-qPCR | 48 |
| Cells | Cell lysate | NA | Isotachophoresis after off-chip cell lysis | NR | NA | 170 s ± 10% | NA – no specific miRNAs were detected | Epifluorescent microscope | 50 |
| Cells | Cell lysate | Cancer diagnostics (miRNAs for lung cancer) | Off-chip cell lysis. miRNAs captured on paper-based device | NR | NA | 90 min (for the entire process) | Yes – HP-EXPAR | Quantum dots for optical sensing | 54 |
| Cells | Cell lysate | Bladder cancer | Off-chip cell lysis. Filtering of miRNA by microchip electrophoresis | NR | NA | NR (60 min incubation time) | No | Laser-induced fluorescence | 52 |
| Cells | Cell lysate | NR | Enrichment via agarose fork-shaped microchannel (after ultracentrifugation for exosome isolation) | NR | NA | 10 min on chip (with preparation beforehand) | No | Catalysed hairpin assembly | 53 |
| Cells | Cell lysate | Head and neck squamous cell carcinoma | Chemical lysis and electrophoresis through agarose | NR | NA | 30 min total (incl. 15 min for separation only) | No – preconcentration unit incorporated | Anion exchange membrane via surface inversion | 49 |
| On-chip miRNA extraction and detection | |||||||||
| Liquid biopsy | Serum | Hepatocellular carcinoma | Magnetic three-phase single-drop microextraction | NR – used spike in to monitor efficacy | NA | 6 s for extraction and 5 min for detection | Yes – hyperbranched DNA/Fe3O4 networks | Spectrophotometric detection | 59 |
| Both | Breast cancer cells and plasma | Cardiovascular disease | Magnetic beads coated with anti-CD63 for EV extraction | NR – calculated efficiency of extraction | NA | 5 h (240 min EV extraction, 20 min for miRNA isolation and 5 min FET-based microRNA detection) | No | Field effect transistor | 61 |
| Liquid biopsy | Plasma | Breast cancer | Magnetic beads coated with anti-CD63 for EV extraction | NR | NA | NR | No | Field effect transistor | 62 |
| Liquid biopsy | Plasma | Colon cancer | miRNAs encapsulated in microdroplets | miRNeasy serum/plasma kit (Qiagen) | 2–3 orders of magnitude lower LOD than RT-qPCR | 3 h total | Yes – isothermal exponential amplification (EXPAR) | Fluorescence | 60 |
| Liquid biopsy | Plasma and serum | Cancer diagnostics (miRNAs for pancreatic/lung cancer) | Surface acoustic waves for EV lysis | miRNeasy serum/plasma kit (Qiagen), ExoQuick® (System Biosciences Inc.) | Comparable | 30 min | No | Ion exchange membrane | 63 |
Which technology has the greatest potential is a question that cannot yet be answered with our current knowledge. But it is clear that a diversity of approaches and platforms is needed to improve our understanding of circulating cell-free miRNAs and develop new tests to detect them, at the POC or in research facilities. Whilst this review focused primarily on sample processing and miRNA extraction, how much processing and pre-purification is required in a POC nucleic acid will also be dictated by the nature of the chosen detection methodology. When ultrapure miRNA extracts are typically needed for most PCR-based tests, new on-chip miRNA sensing technologies are also being developed that are increasingly compatible with complex biological matrices and would therefore require less stringent processing prior to analysis. Processing and detection are strongly connected and only the best combinations will stand a chance to be implemented in clinical practise, providing novel routes to early diagnose as well as providing insights for better clinical management. With specific miRNA deregulation being associated with an increasingly large number of medical conditions including most cancer types, cardiovascular diseases, neurological disorders and infectious diseases,1–3 the need for new nucleic acid tests has never been more urgent.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the March of Dimes for financial support.Notes and references
- N. Rosenfeld, R. Aharonov, E. Meiri, S. Rosenwald, Y. Spector, M. Zepeniuk, H. Benjamin, N. Shabes, S. Tabak, A. Levy, D. Lebanony, Y. Goren, E. Silberschein, N. Targan, A. Ben-Ari, S. Gilad, N. Sion-Vardy, A. Tobar, M. Feinmesser, O. Kharenko, O. Nativ, D. Nass, M. Perelman, A. Yosepovich, B. Shalmon, S. Polak-Charcon, E. Fridman, A. Avniel, I. Bentwich, Z. Bentwich, D. Cohen, A. Chajut and I. Barshack, MicroRNAs accurately identify cancer tissue origin, Nat. Biotechnol., 2008, 26(4), 462–469 CrossRef CAS PubMed.
- J. Wang, J. Chen and S. Sen, MicroRNA as Biomarkers and Diagnostics, J. Cell. Physiol., 2016, 231(1), 25–30 CrossRef CAS PubMed.
- V. Kaul, K. I. Weinberg, S. D. Boyd, D. Bernstein, C. O. Esquivel, O. M. Martinez and S. M. Krams, Dynamics of Viral and Host Immune Cell MicroRNA Expression during Acute Infectious Mononucleosis, Front. Microbiol., 2018, 8, 2666 CrossRef PubMed.
- R. Yi, Y. Qin, I. G. Macara and B. R. Cullen, Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs, Genes Dev., 2003, 17(24), 3011–3016 CrossRef CAS PubMed.
- Y. Lee, C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Rådmark, S. Kim and V. N. Kim, The nuclear RNase III Drosha initiates microRNA processing, Nature, 2003, 425(6956), 415–419 CrossRef CAS PubMed.
- J. Höck and G. Meister, The Argonaute protein family, Genome Biol., 2008, 9(2), 210–210 CrossRef PubMed.
- R. W. Carthew and E. J. Sontheimer, Origins and Mechanisms of miRNAs and siRNAs, Cell, 2009, 136(4), 642–655 CrossRef CAS PubMed.
- T. P. Chendrimada, K. J. Finn, X. Ji, D. Baillat, R. I. Gregory, S. A. Liebhaber, A. E. Pasquinelli and R. Shiekhattar, MicroRNA silencing through RISC recruitment of eIF6, Nature, 2007, 447(7146), 823–828 CrossRef CAS PubMed.
- L. Xu, B. F. Yang and J. Ai, MicroRNA transport: a new way in cell communication, J. Cell. Physiol., 2013, 228(8), 1713–1719 CrossRef CAS PubMed.
- C. Villarroya-Beltri, C. Gutiérrez-Vázquez, F. Sánchez-Cabo, D. Pérez-Hernández, J. Vázquez, N. Martin-Cofreces, D. J. Martinez-Herrera, A. Pascual-Montano, M. Mittelbrunn and F. Sánchez-Madrid, Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs, Nat. Commun., 2013, 4(1), 2980 CrossRef PubMed.
- P. S. Mitchell, R. K. Parkin, E. M. Kroh, B. R. Fritz, S. K. Wyman, E. L. Pogosova-Agadjanyan, A. Peterson, J. Noteboom, K. C. Briant, A. Allen, D. W. Lin, N. Urban, C. W. Drescher, B. S. Knudsen, D. L. Stirewalt, R. Gentleman, R. L. Vessella, P. S. Nelson, D. B. Martin and M. Tewari, Circulating microRNAs as stable blood-based markers for cancer detection, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(30), 10513 CrossRef CAS PubMed.
- A. Aryani and B. Denecke, In vitro application of ribonucleases: comparison of the effects on mRNA and miRNA stability, BMC Res. Notes, 2015, 8(1), 164 CrossRef PubMed.
- C. W. Price, D. C. Leslie and J. P. Landers, Nucleic acid extraction techniques and application to the microchip, Lab Chip, 2009, 9(17), 2484–2494 RSC.
- N. Ali, R. D. C. P. Rampazzo, A. D. T. Costa and M. A. Krieger, Current Nucleic Acid Extraction Methods and Their Implications to Point-of-Care Diagnostics, BioMed Res. Int., 2017, 2017, 9306564 Search PubMed.
- S. Inamdar, R. Nitiyanandan and K. Rege, Emerging applications of exosomes in cancer therapeutics and diagnostics, Bioeng. Transl. Med., 2017, 2(1), 70–80 CrossRef CAS PubMed.
- M. Ilié and P. Hofman, Pros: Can tissue biopsy be replaced by liquid biopsy?, Transl. Lung Cancer Res., 2016, 5(4), 420 CrossRef PubMed.
- C. Alix-Panabières and K. Pantel, Liquid biopsy: from discovery to clinical application, Cancer Discovery, 2021, 11(4), 858–873 CrossRef PubMed.
- T.-R. Li, X.-N. Wang, C. Sheng, Y.-X. Li, F. Z.-T. Li, Y. Sun and Y. Han, Extracellular vesicles as an emerging tool for the early detection of Alzheimer's disease, Mech. Ageing Dev., 2019, 184, 111175 CrossRef CAS PubMed.
- G. Siravegna, S. Marsoni, S. Siena and A. Bardelli, Integrating liquid biopsies into the management of cancer, Nat. Rev. Clin. Oncol., 2017, 14(9), 531–548 CrossRef CAS PubMed.
- M. El-Mogy, B. Lam, T. A. Haj-Ahmad, S. McGowan, D. Yu, L. Nosal, N. Rghei, P. Roberts and Y. Haj-Ahmad, Diversity and signature of small RNA in different bodily fluids using next generation sequencing, BMC Genomics, 2018, 19(1), 408 CrossRef PubMed.
- J. A. Weber, D. H. Baxter, S. Zhang, D. Y. Huang, K. How Huang, M. Jen Lee, D. J. Galas and K. Wang, The MicroRNA Spectrum in 12 Body Fluids, Clin. Chem., 2010, 56(11), 1733–1741 CrossRef CAS PubMed.
- E. K. Hanson, H. Lubenow and J. Ballantyne, Identification of forensically relevant body fluids using a panel of differentially expressed microRNAs, Anal. Biochem., 2009, 387(2), 303–314 CrossRef CAS PubMed.
- K. Wang, Y. Yuan, J.-H. Cho, S. McClarty, D. Baxter and D. J. Galas, Comparing the MicroRNA spectrum between serum and plasma, PLoS One, 2012, 7, e41561 CrossRef CAS PubMed.
- J. D. Arroyo, J. R. Chevillet, E. M. Kroh, I. K. Ruf, C. C. Pritchard, D. F. Gibson, P. S. Mitchell, C. F. Bennett, E. L. Pogosova-Agadjanyan, D. L. Stirewalt, J. F. Tait and M. Tewari, Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(12), 5003 CrossRef CAS PubMed.
- A. Turchinovich, L. Weiz, A. Langheinz and B. Burwinkel, Characterization of extracellular circulating microRNA, Nucleic Acids Res., 2011, 39(16), 7223–7233 CrossRef CAS PubMed.
- A. Gallo, M. Tandon, I. Alevizos and G. G. Illei, The majority of microRNAs detectable in serum and saliva is concentrated in exosomes, PLoS One, 2012, 7(3), e30679 CrossRef CAS PubMed.
- C. C. Pritchard, H. H. Cheng and M. Tewari, MicroRNA profiling: approaches and considerations, Nat. Rev. Genet., 2012, 13(5), 358–369 CrossRef CAS PubMed.
- M. Accerbi, S. A. Schmidt, E. De Paoli, S. Park, D. H. Jeong and P. J. Green, Methods for isolation of total RNA to recover miRNAs and other small RNAs from diverse species, Methods Mol. Biol., 2010, 592, 31–50 CrossRef CAS PubMed.
- S. C. Tan and B. C. Yiap, DNA, RNA, and protein extraction: the past and the present, J. Biomed. Biotechnol., 2009, 2009, 574398 CrossRef PubMed.
- K. A. Melzak, C. S. Sherwood, R. F. B. Turner and C. A. Haynes, Driving Forces for DNA Adsorption to Silica in Perchlorate Solutions, J. Colloid Interface Sci., 1996, 181(2), 635–644 CrossRef CAS.
- P. M. Godoy, N. R. Bhakta, A. J. Barczak, H. Cakmak, S. Fisher, T. C. MacKenzie, T. Patel, R. W. Price, J. F. Smith, P. G. Woodruff and D. J. Erle, Large Differences in Small RNA Composition Between Human Biofluids, Cell Rep., 2018, 25(5), 1346–1358 CrossRef CAS PubMed.
- R. A. M. Brown, M. R. Epis, J. L. Horsham, T. D. Kabir, K. L. Richardson and P. J. Leedman, Total RNA extraction from tissues for microRNA and target gene expression analysis: not all kits are created equal, BMC Biotechnol., 2018, 18(1), 16 CrossRef PubMed.
- A. Brunet-Vega, C. Pericay, M. E. Quílez, M. J. Ramírez-Lázaro, X. Calvet and S. Lario, Variability in microRNA recovery from plasma: Comparison of five commercial kits, Anal. Biochem., 2015, 488, 28–35 CrossRef CAS PubMed.
- A. Podolska, B. Kaczkowski, T. Litman, M. Fredholm and S. Cirera, How the RNA isolation method can affect microRNA microarray results, Acta Biochim. Pol., 2011, 58(4), 535–540 CrossRef CAS PubMed.
- Y.-K. Kim, J. Yeo, B. Kim, M. Ha and V. N. Kim, Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells, Mol. Cell, 2012, 46(6), 893–895 CrossRef CAS PubMed.
- O. Bryzgunova, M. Konoshenko, I. Zaporozhchenko, A. Yakovlev and P. Laktionov, Isolation of Cell-Free miRNA from Biological Fluids: Influencing Factors and Methods, Diagnostics, 2021, 11(5), 865 CrossRef CAS PubMed.
- E. A. Lekchnov, I. A. Zaporozhchenko, E. S. Morozkin, O. E. Bryzgunova, V. V. Vlassov and P. P. Laktionov, Protocol for miRNA isolation from biofluids, Anal. Biochem., 2016, 499, 78–84 CrossRef CAS PubMed.
- A. Chanutin and R. R. Curnish, The precipitation of plasma proteins by short-chain fatty acids, Arch. Biochem. Biophys., 1960, 89(2), 218–220 CrossRef CAS PubMed.
- E. Ban, D.-K. Chae, Y. S. Yoo and E. J. Song, An improvement of miRNA extraction efficiency in human plasma, Anal. Bioanal. Chem., 2017, 409(27), 6397–6404 CrossRef CAS PubMed.
- S. Xu, S. Hossaini Nasr, D. Chen, X. Zhang, L. Sun, X. Huang and C. Qian, MiRNA Extraction from Cell-Free Biofluid Using Protein Corona Formed around Carboxyl Magnetic Nanoparticles, ACS Biomater. Sci. Eng., 2018, 4(2), 654–662 CrossRef CAS PubMed.
- L. A. Jimenez, M. A. Gionet-Gonzales, S. Sedano, J. G. Carballo, Y. Mendez and W. Zhong, Extraction of microRNAs from biological matrices with titanium dioxide nanofibers, Anal. Bioanal. Chem., 2018, 410(3), 1053–1060 CrossRef CAS PubMed.
- N. Khan, J. Cheng, J. P. Pezacki and M. V. Berezovski, Quantitative Analysis of MicroRNA in Blood Serum with Protein-Facilitated Affinity Capillary Electrophoresis, Anal. Chem., 2011, 83(16), 6196–6201 CrossRef CAS PubMed.
- M. Tayebi, Y. Zhou, P. Tripathi, R. Chandramohanadas and Y. Ai, Exosome Purification and Analysis Using a Facile Microfluidic Hydrodynamic Trapping Device, Anal. Chem., 2020, 92(15), 10733–10742 CrossRef CAS PubMed.
- P. Zhou, F. Lu, J. Wang, K. Wang, B. Liu, N. Li and B. Tang, A portable point-of-care testing system to diagnose lung cancer through the detection of exosomal miRNA in urine and saliva, Chem. Commun., 2020, 56(63), 8968–8971 RSC.
- M. Grunt, A. V. Failla, I. Stevic, T. Hillebrand and H. Schwarzenbach, A novel assay for exosomal and cell-free miRNA isolation and quantification, RNA Biol., 2020, 17(4), 425–440 CrossRef CAS PubMed.
- H. Yukawa, S. Yamazaki, K. Aoki, K. Muto, N. Kihara, K. Sato, D. Onoshima, T. Ochiya, Y. Tanaka and Y. Baba, Co-continuous structural effect of size-controlled macro-porous glass membrane on extracellular vesicle collection for the analysis of miRNA, Sci. Rep., 2021, 11(1), 8672 CrossRef CAS PubMed.
- S. S. Kanwar, C. J. Dunlay, D. M. Simeone and S. Nagrath, Microfluidic device(ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes, Lab Chip, 2014, 14(11), 1891–1900 RSC.
- R. Zhong, K. Flack and W. Zhong, Automatic extraction and processing of small RNAs on a multi-well/multi-channel(M&M) chip, Analyst, 2012, 137(23), 5546–5552 RSC.
- Z. Slouka, S. Senapati, S. Shah, R. Lawler, Z. Shi, M. S. Stack and H.-C. Chang, Integrated, DC voltage-driven nucleic acid diagnostic platform for real sample analysis: Detection of oral cancer, Talanta, 2015, 145, 35–42 CrossRef CAS PubMed.
- R. B. Schoch, M. Ronaghi and J. G. Santiago, Rapid and selective extraction, isolation, preconcentration, and quantitation of small RNAs from cell lysate using on-chip isotachophoresis, Lab Chip, 2009, 9(15), 2145–2152 RSC.
- G. Dame, J. Lampe, S. Hakenberg and G. Urban, Development of a Fast miRNA Extraction System for Tumor Analysis Based on a Simple Lab on Chip Approach, Procedia Eng., 2015, 120, 158–162 CrossRef CAS.
- K. Wei, J. Zhao, X. Luo, S. Qiu, F. He, S. Li and S. Zhao, An ultrasensitive microchip electrophoresis assay based on separation-assisted double cycling signal amplification strategy for microRNA detection in cell lysate, Analyst, 2018, 143(6), 1468–1474 RSC.
- C. Qian, Y. Xiao, J. Wang, Y. Li, S. Li, B. Wei, W. Du, X. Feng, P. Chen and B.-F. Liu, Rapid exosomes concentration and in situ detection of exosomal microRNA on agarose-based microfluidic chip, Sens. Actuators, B, 2021, 333, 129559 CrossRef CAS.
- H. Deng, X. Zhou, Q. Liu, B. Li, H. Liu, R. Huang and D. Xing, Paperfluidic chip device for small RNA extraction, amplification, and multiplexed analysis, ACS Appl. Mater. Interfaces, 2017, 9(47), 41151–41158 CrossRef CAS PubMed.
- A. Ku, N. Ravi, M. Yang, M. Evander, T. Laurell, H. Lilja and Y. Ceder, A urinary extracellular vesicle microRNA biomarker discovery pipeline; from automated extracellular vesicle enrichment by acoustic trapping to microRNA sequencing, PLoS One, 2019, 14(5), e0217507 CrossRef CAS PubMed.
- A. Ku, J. Fredsøe, K. D. Sørensen, M. Borre, M. Evander, T. Laurell, H. Lilja and Y. Ceder, High-Throughput and Automated Acoustic Trapping of Extracellular Vesicles to Identify microRNAs With Diagnostic Potential for Prostate Cancer, Front. Oncol., 2021, 11(386), 631021 CrossRef PubMed.
- K. Lee, J.-H. Kang, H.-M. Kim, J. Ahn, H. Lim, J. Lee, W.-J. Jeon, J.-H. Lee and K.-B. Kim, Direct electrophoretic microRNA preparation from clinical samples using nanofilter membrane, Nano Convergence, 2020, 7(1), 1 CrossRef CAS PubMed.
- O. Behrmann, M. Hügle, P. Bronsert, B. Herde, J. Heni, M. Schramm, F. T. Hufert, G. A. Urban and G. Dame, A lab-on-a-chip for rapid miRNA extraction, PLoS One, 2019, 14(12), e0226571 CrossRef CAS PubMed.
- S. Tang, T. Qi, Y. Yao, L. Tang, W. Chen, T. Chen, W. Shen, D. Kong, H.-W. Shi and T. Liu, Magnetic Three-Phase Single-Drop Microextraction for Rapid Amplification of the Signals of DNA and MicroRNA Analysis, Anal. Chem., 2020, 92(18), 12290–12296 CrossRef CAS PubMed.
- K. Zhang, D.-K. Kang, M. M. Ali, L. Liu, L. Labanieh, M. Lu, H. Riazifar, T. N. Nguyen, J. A. Zell and M. A. Digman, Digital quantification of miRNA directly in plasma using integrated comprehensive droplet digital detection, Lab Chip, 2015, 15(21), 4217–4226 RSC.
- H.-L. Cheng, C.-Y. Fu, W.-C. Kuo, Y.-W. Chen, Y.-S. Chen, Y.-M. Lee, K.-H. Li, C. Chen, H.-P. Ma and P.-C. Huang, Detecting miRNA biomarkers from extracellular vesicles for cardiovascular disease with a microfluidic system, Lab Chip, 2018, 18(19), 2917–2925 RSC.
- C. C. Huang, Y. H. Kuo, Y. S. Chen and G. B. Lee, An Integrated Microfluidic System for Early Diagnosis of Breast Cancer in Liquid Biopsy by Using Microrna and FET Biosensors, 2021 IEEE 34th International Conference on Micro Electro Mechanical Systems(MEMS), 2021, pp. 318–321 Search PubMed.
- Z. Ramshani, C. Zhang, K. Richards, L. Chen, G. Xu, B. L. Stiles, R. Hill, S. Senapati, D. B. Go and H.-C. Chang, Extracellular vesicle microRNA quantification from plasma using an integrated microfluidic device, Commun. Biol., 2019, 2(1), 189 CrossRef PubMed.
- J. Reboud, Y. Bourquin, R. Wilson, G. S. Pall, M. Jiwaji, A. R. Pitt, A. Graham, A. P. Waters and J. M. Cooper, Shaping acoustic fields as a toolset for microfluidic manipulations in diagnostic technologies, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(38), 15162–15167 CrossRef CAS PubMed.
- D. Taller, K. Richards, Z. Slouka, S. Senapati, R. Hill, D. B. Go and H.-C. Chang, On-chip surface acoustic wave lysis and ion-exchange nanomembrane detection of exosomal RNA for pancreatic cancer study and diagnosis, Lab Chip, 2015, 15(7), 1656–1666 RSC.
- S. Cai, T. Pataillot-Meakin, A. Shibakawa, R. Ren, C. L. Bevan, S. Ladame, A. P. Ivanov and J. B. Edel, Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum, Nat. Commun., 2021, 12(1), 3515 CrossRef CAS PubMed.
- R. Tavallaie, J. McCarroll, M. Le Grand, N. Ariotti, W. Schuhmann, E. Bakker, R. D. Tilley, D. B. Hibbert, M. Kavallaris and J. J. Gooding, Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood, Nat. Nanotechnol., 2018, 13(11), 1066–1071 CrossRef CAS PubMed.
- T. Kangkamano, A. Numnuam, W. Limbut, P. Kanatharana, T. Vilaivan and P. Thavarungkul, Pyrrolidinyl PNA polypyrrole/silver nanofoam electrode as a novel label-free electrochemical miRNA-21 biosensor, Biosens. Bioelectron., 2008, 102, 217–225 CrossRef PubMed.
- L. Liu, Y. Chang, N. Xia, P. Peng, L. Zhang, M. Jiang, J. Zhang and L. Liu, Simple, sensitive and label–free electrochemical detection of microRNAs based on the in situ formation of silver nanoparticles aggregates for signal amplification, Biosens. Bioelectron., 2017, 94, 235–242 CrossRef CAS PubMed.
- H.-A. Rafiee-Pour, M. Behpour and M. Keshavarz, A novel label-free electrochemical miRNA biosensor using methylene blue as redox indicator: application to breast cancer biomarker miRNA-21, Biosens. Bioelectron., 2016, 77, 202–207 CrossRef CAS PubMed.
- L. Liu, N. Xia, H. Liu, X. Kang, X. Liu, C. Xue and X. He, Highly sensitive and label-free electrochemical detection of microRNAs based on triple signal amplification of multifunctional gold nanoparticles, enzymes and redox-cycling reaction, Biosens. Bioelectron., 2014, 53, 399–405 CrossRef CAS PubMed.
- N. Xia, L. Zhang, G. Wang, Q. Feng and L. Liu, Label-free and sensitive strategy for microRNAs detection based on the formation of boronate ester bonds and the dual-amplification of gold nanoparticles, Biosens. Bioelectron., 2013, 47, 461–466 CrossRef CAS PubMed.
- J. Mandli, H. Mohammadi and A. Amine, Electrochemical DNA sandwich biosensor based on enzyme amplified microRNA-21 detection and gold nanoparticles, Bioelectrochemistry, 2017, 116, 17–23 CrossRef CAS PubMed.
- H.-L. Shuai, K.-J. Huang, W.-J. Zhang, X. Cao and M.-P. Jia, Sandwich-type microRNA biosensor based on magnesium oxide nanoflower and graphene oxide–gold nanoparticles hybrids coupling with enzyme signal amplification, Sens. Actuators, B, 2017, 243, 403–411 CrossRef CAS.
- N. Xia, Y. Zhang, X. Wei, Y. Huang and L. Liu, An electrochemical microRNAs biosensor with the signal amplification of alkaline phosphatase and electrochemical–chemical–chemical redox cycling, Anal. Chim. Acta, 2015, 878, 95–101 CrossRef CAS PubMed.
- Y. Zhou, Z. Zhang, Z. Xu, H. Yin and S. Ai, MicroRNA-21 detection based on molecular switching by amperometry, New J. Chem., 2012, 36(10), 1985–1991 RSC.
- H. Yin, Y. Zhou, H. Zhang, X. Meng and S. Ai, Electrochemical determination of microRNA-21 based on graphene, LNA integrated molecular beacon, AuNPs and biotin multifunctional bio bar codes and enzymatic assay system, Biosens. Bioelectron., 2012, 33(1), 247–253 CrossRef CAS PubMed.
- H. McArdle, E. M. Jimenez-Mateos, R. Raoof, E. Carthy, D. Boyle, H. ElNaggar, N. Delanty, H. Hamer, M. Dogan, T. Huchtemann, P. Körtvelyessy, F. Rosenow, R. J. Forster, D. C. Henshall and E. Spain, “TORNADO” – Theranostic One-Step RNA Detector; microfluidic disc for the direct detection of microRNA-134 in plasma and cerebrospinal fluid, Sci. Rep., 2017, 7(1), 1750 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |
