Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hotspots of root-exuded amino acids are created within a rhizosphere-on-a-chip†
Jayde
Aufrecht
 *a,
Muneeba
Khalid
b,
Courtney L.
Walton
b,
Kylee
Tate
a,
John F.
Cahill
b and
Scott T.
Retterer
*a,
Muneeba
Khalid
b,
Courtney L.
Walton
b,
Kylee
Tate
a,
John F.
Cahill
b and
Scott T.
Retterer
 c
c
aEarth and Biological Sciences Directorate, Pacific Northwest National Laboratory, Richland, WA, USA. E-mail: jayde.aufrecht@pnnl.gov
bBiosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA
cCenter for Nanophase Materials Sciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA
First published on 28th January 2022
Abstract
The rhizosphere is a challenging ecosystem to study from a systems biology perspective due to its diverse chemical, physical, and biological characteristics. In the past decade, microfluidic platforms (e.g. plant-on-a-chip) have created an alternative way to study whole rhizosphere organisms, like plants and microorganisms, under reduced-complexity conditions. However, in reducing the complexity of the environment, it is possible to inadvertently alter organism phenotype, which biases laboratory data compared to in situ experiments. To build back some of the complexity of the rhizosphere in a fully-defined, parameterized approach we have developed a rhizosphere-on-a-chip platform that mimics the physical structure of soil. We demonstrate, through computational simulation, how this synthetic soil structure can influence the emergence of molecular “hotspots” and “hotmoments” that arise naturally from the plant's exudation of labile carbon compounds. We establish the amenability of the rhizosphere-on-a-chip for long-term culture of Brachypodium distachyon, and experimentally validate the presence of exudate hotspots within the rhizosphere-on-a-chip pore spaces using liquid microjunction surface sampling probe mass spectrometry.
Introduction
The rhizosphere, defined as the underground ecological zone influenced by the proximity of plant roots, is one of the most complex and biodiverse ecosystems on earth. It is so complex, that no two rhizospheres are expected to have the exact same physical, biological, and chemical properties.1 Within the rhizosphere, specialized microorganisms share and compete for resources, forming a dynamic trophic web with the host plant.2 Abiotic factors, such as the physical and chemical characteristics of the soil, also influence biological interactions in the rhizosphere by moderating the nutrients and space available to the organisms.3 This intricacy, compounded by spatiotemporal heterogeneity and the opacity of soil, has made rhizosphere ecology difficult to study from a systems biology perspective.In the past decade, microfluidics have enabled the study of rhizosphere organisms in reduced-complexity environments. While these setups are considerably simplified when compared with a holistic rhizosphere, microfluidics offer fully defined environmental conditions, exceptional experimental control, and high-resolution measurements in return. Notable among these microfluidic platforms are plant-on-a-chip designs which, among many applications, have enabled high-throughput phenotyping of whole plants, spatial sampling of root exudates, and organelle-scale imaging of living root cells.4–7 Distinct from axenic plant studies, microfluidic platforms have also been successfully applied to the study of plant–microbe interactions. For these studies, microfluidics are able to resolve the disparate length scales between microbes and plants, allowing the researcher to image and quantify the colonization of roots in real time and at high optical resolutions.8–11
Along with the rhizosphere organisms themselves, their interactions with the abiotic properties of the soil environment are important elements to consider when designing a synthetic habitat. Several microfluidic systems have targeted the structural aspect of the rhizosphere with designs that mimic pore space networks. These soil-on-a-chip platforms are commonly used to study microbial characteristics such as cell motility and biofilm development.12,13 Recent studies have demonstrated that biological systems exhibit different phenotypes in these structured microfluidic platforms compared to bulk media conditions.14 For example, two bacterial species which cannot co-exist at equilibrium in a spatially homogenous (i.e. well-mixed) environment, can stably coexist in a spatially heterogeneous microfluidic environment.15 These results are evidence that the retention of spatial heterogeneity is necessary to elicit the naturally occurring phenotypes of rhizosphere organisms in reduced-complexity experimental setups like microfluidic platforms.
Despite the success of microfluidics for studying individual rhizosphere components, there has yet to be a soil-mimicking microfluidic platform that is compatible with plant growth. In this article, we have designed a synthetic soil microhabitat for rhizosphere-on-a-chip studies. The structure of the microhabitat design mimics the shape statistics and grain packing of natural sand with pore spaces large enough to accommodate plant roots. We provide evidence, through computational simulation, that the structural heterogeneity of this synthetic soil microhabitat can encourage the development of sparse root exudate hotspots that exist on timescales relevant to microbial processes. We demonstrate the compatibility of this platform for growth studies with a model grass species, Brachypodium distachyon, through vegetative growth stage, and experimentally validate the presence of exuded amino acid hotspots within the pore spaces using liquid micro-junction surface sampling probe mass spectrometry (LMJ-SSP-MS).
Experimental
Device design and fabrication
The porous media design was created to replicate the shape distribution of sand particles from Kahala, Hawaii (USA) with a normal size distribution (coefficient of variance = 0.8), using a published algorithm.16 The algorithm's output bitmap image file was vectorized in LayoutEditor and scaled to a D50 of 1 mm, tiled, and cropped to an ellipse shape in AutoCAD.The design was transferred onto a chrome mask using standard photolithography techniques with a Heidelberg DWL 66 mask writer. A thin (approx. 2 μm) layer of SiO2 was grown onto a silicon wafer to enhance deep reactive ion etch selectivity. A P20 adhesion promoter was spun onto the wafer at 3000 rpm, followed by deposition of NFR-016D255cp photoresist (JSR Micro, Sunnyvale, CA) at 1000 rpm for 45 seconds. The wafer was soft-baked at 90 °C for 90 seconds, then exposed to the chrome mask for 5 seconds at 100 mJ cm−2 energy intensity. The wafer was then post-baked at 115 °C for 90 seconds and developed in CD26. Finally, the wafer was Bosch etched (Oxford RIE) to a depth of 300 μm.
Microfluidic devices were replicated from the silicon master wafer using poly-dimethylsiloxane (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 wt/wt, PDMS base to curing agent; Sylgard 184 Dow Corning) and cured for at least 2 hours at 70 °C. The inlet to each microfluidic platform was created using a 2 mm biopsy punch at approximately a 45° angle to ease the roots' growth into the synthetic soil space, similar to a published plant-on-a-chip procedure.6 Additional 1.5 mm ports were punched around the perimeter of the microfluidic device to facilitate filling. The PDMS device was then exposed to oxygen plasma and bonded to a glass slide.
1 wt/wt, PDMS base to curing agent; Sylgard 184 Dow Corning) and cured for at least 2 hours at 70 °C. The inlet to each microfluidic platform was created using a 2 mm biopsy punch at approximately a 45° angle to ease the roots' growth into the synthetic soil space, similar to a published plant-on-a-chip procedure.6 Additional 1.5 mm ports were punched around the perimeter of the microfluidic device to facilitate filling. The PDMS device was then exposed to oxygen plasma and bonded to a glass slide.
Each 2 mm inlet was fitted with a pipette tip that had been trimmed to 10 mm in length to provide support to the growing plant. Finally, the microfluidic platforms were autoclaved for sterilization and pre-filled with Murashige–Skoog basal salt media prior to use.17
Device assembly for exudate sampling
Microfluidic devices were attached to polyester track-etched (PETE) membranes (0.4 μm pore size, 12 μm thickness, 2 × 106 pores per cm2, 90 mm diameter, Sterlitech Corp.) to analyse the amino acids exuded from Brachypodium roots using LMJ-SSP-MS. To bond the PETE membrane, a published wet stamping method was used with minor modifications.5,18 A clean silicon wafer was placed on a spin coater with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PDMS to curing agent and was spun at 5000 rpm for 15 minutes to create an approximately 6 μm thick layer of PDMS film. The rhizosphere-on-a-chip PDMS device was laid, design side down, onto the wet PDMS layer, gently peeled off, then pressed against the PETE membrane, taking care to avoid wrinkling the membrane. The assembly was left in a covered Petri dish to cure at room temperature for 24 h. Once the device assembly had cured, the punched 2 mm inlet was fitted with a pipette tip. Devices were then autoclaved to sterilize and filled with 1/4× Murashige–Skoog basal salt media to prepare for seedling transfer.
1 PDMS to curing agent and was spun at 5000 rpm for 15 minutes to create an approximately 6 μm thick layer of PDMS film. The rhizosphere-on-a-chip PDMS device was laid, design side down, onto the wet PDMS layer, gently peeled off, then pressed against the PETE membrane, taking care to avoid wrinkling the membrane. The assembly was left in a covered Petri dish to cure at room temperature for 24 h. Once the device assembly had cured, the punched 2 mm inlet was fitted with a pipette tip. Devices were then autoclaved to sterilize and filled with 1/4× Murashige–Skoog basal salt media to prepare for seedling transfer.
Chemical species transport simulations
COMSOL Multiphysics® finite element modelling software was used to simulate diffusive transport of a chemical species from the boundaries of an artificial root in 2D space. Roots from a Brachypodium seedling grown within the rhizosphere-on-a-chip for two weeks were traced in AutoCAD and imported as a geometry into COMSOL. For the “open geometry” simulations, an ellipse with the same size of the rhizosphere-on-a-chip was drawn in COMSOL, and the artificial roots were subtracted from the ellipse geometry. For the “synthetic soil” geometry, the computer-aided design (CAD) file used to fabricate the rhizosphere-on-a-chip was imported into COMSOL and the artificial roots were subtracted from this design. In both simulations, the resulting fluid space geometry was given the material properties of water. The artificial root boundaries were selected and assigned a chemical species flux of 22.8 μmole m−2 s−1 with the diffusive coefficient of sucrose (5.2 × 10−6 cm2 s−1 in water at 25 °C). The simulation space was then meshed using an extra course free triangular mesh and a time-dependent, transport of diluted species study was simulated assuming diffusion as the only transport mechanism. To study the accumulation and dilution of chemical species in the rhizosphere on a chip, the root boundary flux was multiplied by a step function so that the flux was “on” for one hour then “off” for the remaining simulated 3 hours.Plant culturing
Brachypodium distachyon (L.) P. Beauv (line Bd21-3, accession W6 39233) seeds were provided by the USDA ARS plant germplasm. Seeds were de-husked and surface sterilized via immersion in 70% ethanol solution for 30 seconds followed by immersion in a 1.3% sodium hypochlorite solution for 4 minutes. The seeds were then rinsed 3 times in sterile water and transferred to a Petri dish containing full strength Murashige–Skoog basal salt mixture +0.7% phytagel. The plate was placed in a dark, 4 °C seed chamber for 48 hours to stratify the seeds and synchronize germination. After 48 hours, the seeds were transferred to a 16 h day (25/21 °C day/night) growth chamber to initiate germination. The seedlings used in the LMJ-SSP-MS experiment were germinated on a Hoagland media plate with 0.9% phytagel and grown in a 16 h day, 24 °C growth chamber.Once the seeds had germinated and before the roots were longer than 5 mm in length, the seedlings were carefully transferred to a pipette tip cut to fit into the rhizosphere-on-a-chip inlet. This transfer was done in a sterile environment (i.e. biosafety cabinet) using autoclaved utensils to prevent microbial contamination of the seedlings. For the experiments to characterize Brachypodium growth in the rhizosphere-on-a-chip, 5 seedlings were used as biological replicates (one seedling per rhizosphere-on-a-chip). Plastic containers (i.e. magenta boxes or Petri dishes) were prefilled with a mixture of water and 0.7–1% Phytogel™ to maintain humidity within the rhizosphere-on-a-chip. Once the Phytogel™ had solidified, the rhizosphere-on-a-chip was transferred to the container and oriented vertically to encourage gravitropic root growth. The container was then sealed with Parafilm and returned to the long day growth chamber. Seedlings were visually inspected every 24 hours for hydration. If needed, the containers were unsealed inside a sterile environment to expose the plants to atmospheric carbon dioxide, and sterile ¼× Murashige–Skoog basal salt media was added to the pipette tip to keep the roots submersed and hydrated.
Imaging and analysis
For optical microscopy, the rhizosphere-on-a-chips were removed from their containers and placed on a microscope stage. Optical microscopy images were acquired with an inverted Nikon TI Eclipse microscope. Root length was manually traced and measured in ImageJ.Endpoint brightfield images of the rhizosphere-on-a-chip were collected using Biotek CYTATION 5 imaging reader. The Petri dishes were placed in the holder and scanned with a 4× objective. Collected images were automatically stitched, z-projected, processed, and analysed using inbuilt Gen5™ software.
Sampling amino acids with LMJ-SSP-MS
The LMJ-SSP-MS system used here was previously described by Cahill, et al.18 The probe consists of three coaxially aligned capillaries. The outer capillary consisted of a stainless-steel tube (∼0.686 mm i.d. × ∼1.067 mm o.d. × 3.5 cm length) situated in one port of a PEEK tee (IDEX Health & Science). The second port of the tee was connected to an in-house vacuum system to provide suction of any excess solvent in the event of the solvent flow exceeding aspiration rate. The middle capillary was a stainless-steel tube (∼0.406 mm i.d. × ∼0.508 mm o.d. × 70 mm length) extended ∼1 mm outside of the outer capillary and was secured using a second PEEK tee. The second port of the tee was connected to a high performance liquid chromatography pump (model 1100, Agilent Technologies) which delivered 120 μL min−1 of 75/25/0.1 (v/v/v) acetonitrile/water/formic acid with 0.3 μM caffeine-d3 (internal standard) through the annulus region of the probe. Lastly, the third capillary was PEEK tubing (0.178 mm i.d. × 0.330 mm o.d. × 30 cm length) positioned through the middle capillary 1 mm before the end of the middle capillary. The other end was secured through the PEEK tees and connected to a heated electrospray ionization source (HESI). Caffeine-d3 was added to the solvent to normalize for any LMJ fluctuations.The HESI source utilized a stainless-steel capillary (0.178 mm i.d. × 0.356 mm o.d. × 15 cm length) to match the dimension of the connected LMJ-SSP inner capillary. The outer capillary of the LMJ-SSP was connected to electrical ground. Solvent aspiration rate was determined by the nebulizing gas (nitrogen) rate, controlled through the ion source. Solvent flow was sufficiently below the aspiration rate of the ion source to maintain the necessary liquid vortex. The HESI source was attached to a Q Exactive HF Orbitrap mass spectrometer (ThermoFisher). Mass spectrometer parameters were set as follows: sheath gas = 80, auxiliary gas = 40, capillary temp = 400 °C, source temp = 160 °C, voltage = 4.0 kV, and S-lens = 50.00 V. Mass spectra were obtained with a method consisting of three untargeted scans: m/z 100–190, m/z 190–210, m/z 250–400 and one tandem MS scan of m/z 175.12. Resolution was set to 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with an AGC target of 3e6 and a maximum injection time of 200 ms.
000 with an AGC target of 3e6 and a maximum injection time of 200 ms.
Results and discussion
Rhizosphere-on-a-chip properties
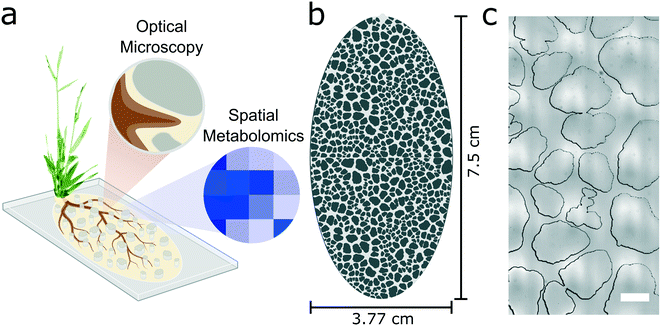
The goal of this work was to create a plant habitat that mimics structural aspects of the rhizosphere, while retaining the optical imaging and molecular sampling compatibilities of microfluidic systems (Fig. 1a). The rhizosphere-on-a-chip was designed using an approach similar to a published soil-on-a-chip platform for microbial studies.14 Briefly, a particle generator was used to create 500 unique 2-dimensional synthetic soil grains that have shape properties randomly generated from a distribution of natural sand grains from Kahala Beach, Hawaii (USA).16,19 The synthetic soil grains were oriented and packed using a constrained Voronoi tessellation to maximize anisotropy across the design.16 The output image file from the generator was converted into a CAD format and scaled so that the D50 gradation, a median measurement of soil grain size, was 1 mm, which approximates the D50 value of coarse-grained sand.19 These 500 simulated grains were tiled in a 2 × 2 layout and cropped to an ellipsoid shape (Fig. 1b). The design was fabricated using standard photolithography techniques (see Methods), etched 300 μm into a silicon wafer, and replicated in poly-dimethylsiloxane (PDMS).The resulting rhizosphere-on-a-chip has several key properties that mimic natural soils including overall anisotropy, and pore spaces that are heterogeneous in size and shape (Fig. 1c). The porosity of the microhabitat (i.e. the ratio of fluidic space to the total area including the synthetic soil) was calculated to be 0.38, which approximates that of a sandy loam.13 The major and minor axes of the synthetic soil habitat measure 75 mm and 37.7 mm, respectively, providing enough space for root growth over longer time intervals while still being a manageable size for optical microscopy. The theoretical internal volume of the pore space is 246 μL.
These soil-mimicking physical properties are expected to influence the transport and distribution of resources in the pore space, which inevitably affects the dynamics of rhizosphere organisms.20
Brachypodium growth in the rhizosphere-on-a-chip
Brachypodium distachyon (hereafter referred to by genus name only) was chosen as the target organism for the rhizosphere-on-chip for several reasons. Brachypodium is a model grass species belonging to the same family as economically important grains (e.g. wheat, oat, and rye), with a short stature and compact root system that is amenable for long-term microfluidic culture.21,22 Similarly, the generation time for Brachypodium is relatively fast with seed to reproductive stage lasting 8–12 weeks, and Brachypodium has been previously compatible with open-design microfluidic ecosystems, making it a feasible option for a rhizosphere-on-a-chip design.7We found the rhizosphere-on-a-chip design to be compatible with long-term Brachypodium growth up to one month (Fig. 2a). After one month, the roots began to grow through the outlet of the microfluidic system into the agar surrounding the rhizosphere-on-a-chip and the plant continued to develop through flowering stage. The seedlings grown within the rhizosphere-on-a-chip met several shoot developmental milestones on time, including 2–3 leaves after 10 days, and 6–7 leaves after 40 days.22 However, despite being grown under longer, warm day conditions, the plants had not yet flowered 30 days post germination as reported elsewhere.22 It is possible that the mechanical stimulation of the tight pore spaces contributed to the delay in flowering as has been described in Arabidopsis thaliana.23 The root structure of Brachypodium seedlings grown in the rhizosphere-on-a-chip also did not follow the exact developmental sequence as plants grown in soil conditions. For example, Watt et. al. reported that seedlings at the 2–3 leaf stage had one primary axile root that was 10.1–12.8 cm long, while we saw an average of two axile roots at this stage and an average total root length of 4.3 cm (n = 5, 9 days post germination).22 While there can be significant intrinsic variance in root development between seedlings, it is also possible that the mechanical stress of the tight pore spaces altered the root's structural phenotype.24,25
Because the rhizosphere-on-a-chip design consisted of various pore sizes and shapes, we anticipated that the Brachypodium roots would encounter pore spaces that were smaller than the diameter of the root. Unlike natural soil, the synthetic soil grains in the rhizosphere-on-a-chip are fixed in place and cannot be pushed aside or re-structured by the elongating root tips.26 However, these restrictions did not prevent the roots from growing through some of the smallest pore spaces in the rhizosphere-on-a-chip (Fig. 2b). In fact, roots were observed growing between synthetic soil grains which has been previously connected. To grow through these small spaces, the root cortical cells deformed to squeeze between the PDMS and glass slide while the endodermis fit within the narrow pore space (Fig. 2d). Once the root tip passed through a narrow pore, it continued to grow normally with some bulging (Fig. 2c). These cellular contortions may have an impact on root exudation rates or the ability of endophytic microorganisms to access the intercellular root space. This rhizosphere-on-a-chip system also enabled single-cell imaging of growing root hairs, which are fine-root features that are difficult to study in natural soil given their small diameter (Fig. 2e).
Once the compatibility of the rhizosphere-on-a-chip had been demonstrated with Brachypodium plants, the influence of the synthetic soil layout on the root's chemical micro-environment was examined.
Simulating exudate diffusion through the synthetic soil
Partitioned soil samples show that, although ubiquitous in the rhizosphere, microorganisms are not uniformly distributed underground.27 Instead, microbial hotspots are expected to arise, in part, due to the heterogeneous distribution of labile, or readily available, organic matter in the soil.28 A major source of this labile carbon is the plant itself through processes known as rhizodeposition, which includes the exudation of carbon compounds directly from the plant's roots.29Along with mimicking the physical structure of the soil, part of the motivation for creating a rhizosphere-on-a-chip was to re-create the chemical microenvironment of the rhizosphere, including hotspots of concentrated labile carbon. To explore the natural generation of molecular hotspots by the plant in our synthetic soil microhabitat, we simulated root exudation and the diffusion of small molecules using finite element modelling in COMSOL Multiphysics.
First, an artificial root system was drawn in AutoCAD to re-create the root architecture of a 14 day old Brachypodium seedling that was experimentally grown within the rhizosphere-on-a-chip. To understand how small molecules can accumulate in the rhizosphere-on-a-chip environment when they are exuded by the plant at a constant rate, we next assigned a constant and spatially uniform chemical species flux to the artificial root's boundaries. This flux (22.8 μmole m−2 s−1) was selected from published estimates of total belowground carbon exudation and fungal allocation from a warm-temperate forest.30 Although it is a demonstrated strategy for plants to recapture nutrients, the uptake of chemical species by the root was not modelled in this simulation due to lack consensus on molecular influx rates.31 The chemical species was assigned the same diffusion coefficient as sucrose (5.2 × 10−6 cm2 s−1 in water at 25 °C), a common root exudate.32,33 Using a time-dependent study, the accumulation and diffusion of this chemical species was simulated in two geometries: 1. An open space geometry and 2. A synthetic soil geometry generated from the same CAD file used to fabricate the rhizosphere-on-a-chip.
After an hour of simulated root exudation, the concentration profile of root exudate exhibited a markable difference between the two geometries (Fig. 3). Spatial hotspots of accumulated root exudate emerged only in the synthetic soil geometry, while the concentration profile of the open geometry remained more uniform and displayed a gradual decrease in concentration away from the roots (Fig. 3b and c and S1†). Within the synthetic soil geometry, concentrated hotspots occurred in pore spaces that approximated the diameter of the root width and, after one hour of exudation, a representative hotspot was 8× more concentrated than the same coordinate location in the open geometry due to the inhibited diffusion by the soil grains (Fig. S2†). This accumulation of root exudate within the selected hotspot followed a second order polynomial increase (R2 = 0.9996) over time.
Though little is known about how a plant's exudation rate and exudate profile change over short time-scales, such as those simulated herein, the exudate profile of a plant has been shown to change according to the plant's developmental stage and environmental conditions.34,35 Likewise, stable isotope studies on plants undergoing photosynthesis have shown that the conversion of atmospheric C to labile C compounds occurs on the order of hours.36 It is therefore feasible that root exudate profiles could switch on this same time scale. By simulating an abrupt stop in exudation, we sought to understand the temporal dynamics of chemical species switching and the persistence of molecular hotspots in the rhizosphere-on-a-chip over time.
To understand the persistence of root exudate within a molecular hotspot, the exudation flux was stopped after 1 hour and the diffusion of the chemical species throughout the pore space was simulated (Fig. 3d and e). Immediately post-exudation, the concentration of the chemical species decayed rapidly within the measured hotspot and, after 4 hours post-exudation, began to plateau at a value of 100 mM. Interestingly, this plateau value was still higher than the maximum concentration of exudate (i.e. after 1 hour of accumulation) in the same location within the open geometry. This simulated result suggests that the synthetic soil geometry layout can influence the persistence of “hot-moments” in the rhizosphere-on-a-chip.
While the simulated temporal dynamics of root exudate diffusion in the rhizosphere-on-a-chip are not on a time scale that is significant for the lifetime of a plant, these mass transport dynamics may play a significant role during the lifespan of a microorganism. For example, the average swimming speed of an E. coli cell undergoing chemotactic (i.e. chemically directed) movement is approximately 30 μm s−1, which suggests that, within 100 s of root exudation, a microbe could conceivably swim 3 mm in the direction of a plant root, although the synthetic soil grains would also act to impede chemotaxis.37,38 Though bacterial doubling times in natural soil are nearly impossible to measure, theoretical calculations estimate that fast-growing bacterium such as E. coli double in 15 hours in the wild, with experimental laboratory grown cells doubling in 20 minutes in optimal media and slightly slower in soil extract media.39,40 Given the spatial and temporal properties of the simulated hotspots and hot-moments in our rhizosphere-on-a-chip, it is reasonable that the synthetic soil design could interact with root exudation to influence microbial behaviours within the microhabitat, creating a tractable system for studying dynamic rhizosphere interactions.
These simulations suggest that the rhizosphere-on-a-chip design is capable of re-creating aspects of the chemical microenvironment found in a natural rhizosphere. Next, we sought to sample the chemical microenvironment of the rhizosphere-on-a-chip to compare experimental exudation patterns to our simulations.
Spatially sampling exuded amino acids with LMJ-SSP-MS
To experimentally validate the formation of exudate hotspots within the rhizosphere-on-a-chip, we used an established liquid micro-junction surface sampling probe mass spectrometry (LMJ-SSP-MS) method to spatially sample the chemical microenvironment.18 After germinating Brachypodium seeds overnight, three seedlings were transferred individually to three rhizosphere-on-a-chips as previously described and grown for 12 days. Two hours prior to sampling, Optima™ LC/MS grade water was added to the device inlets to wash away accumulated exudates and re-fill the pore spaces. Sampling spots of varying distance from the roots were then selected. Amino acids, which are present in most root exudates at relatively high concentrations, were targeted from the MS data.33The seedlings' roots (Fig. S3†) were traced and incorporated into the finite element analysis geometry. Because the PETE membrane was reversibly bonded to the rhizosphere-on-a-chip, the roots were occasionally able to push between the membrane and the PDMS. Where the root had grown between the synthetic soil design and the PETE membrane, individual soil grains were modified in the simulation space to account for the increased porosity in this area. The same spots from the experimental sampling were also measured in the simulation after 2 hours of constant exudation, and the concentration within each simulated spot was normalized to the spot with the highest concentration of simulated chemical species (Fig. 4a, c and e). The simulated chemical species distribution was then compared to the experimentally sampled amino acid profile (Fig. 4b, d and f).
From the experimental samples, 16 amino acids were detectable at levels >3σ of background values within the rhizosphere-on-a-chip, although not all amino acids were detected in every spot nor in every replicate. The detected amino acids overlapped with previously reported Brachypodium exudate profiles, with the exception of pyroglutamic acid, which was observed here but not previously reported.33 Note, alanine and glycine were not specifically measured in this experiment due to LMJ-SSP-MS scan time restrictions. The differences in the amino acid profile between replicate plants grown in the rhizosphere-on-a-chip may be due to intrinsic variation between seedlings or could possibly be attributed to the specific pore structure navigated by the roots, as another study has shown that exudate profiles can change based on the size of the porous substrate.25,33
Spots with the highest relative amino acid abundances were generally located adjacent to the root tips. However, not all amino acids had the same spatial distribution, with some differences between amino acid type. Basic, and hydroxylic amino acids were located proximal to the shoot or further from the roots while amidic and aliphatic amino acids were generally also located near the root tips in addition to proximal to the shoot. Interestingly, amino acid signals were generally absent from pores near the non-branched midsections of the root despite higher concentrations predicted in these spots by the simulation under the assumption of uniform exudation along the root. These results imply that amino acids are not uniformly exuded along Brachypodium roots which agrees with previous studies that have shown spatial variation in other root exudates including the amino acid tryptophan.41
The spatial variation in amino acid exudation may play an important role in recruiting beneficial microorganisms to niches in the root's microbiome. Amino acids are an important source of carbon and nitrogen for microorganisms and, some bacteria, like Bacillus subtilis, have amino acid chemoreceptors that direct cellular movement toward plant roots, resulting in spatial preferences in root colonization.8,42 Although plasma membrane localized exporters for amino acids have yet to be identified in root cells, these results and others provide support for the preferential spatial exudation of amino acids.43
While more information on exudation rates and the spatial variation in exudation would be needed to completely predict the chemical micro-environment within the rhizosphere-on-a-chip by computational simulation, the results from the LMJ-SSP-MS experiments validate that there are hotspots of root exudates created by the interaction between the roots and the synthetic soil structure.
Conclusions
We present a rhizosphere-on-a-chip platform that provides the benefits of the soil-on-a-chip structure while remaining amenable for long-term plant studies. We demonstrate this compatibility with Brachypodium distachyon, a model grass, but we expect that this platform will also be suitable for other plant species that are culturable within microfluidic systems such as Arapidopsis thaliana, Populus spp., and Oryza sativa.11,44The results of the Brachypodium growth experiments illustrate the benefit of having a tractable microhabitat for visualizing root development and phenotype plasticity within a pore space network. With its synthetic soil structure, the rhizosphere-on-a-chip will enable insight into underground processes, including emerging plant–microbe dynamics that are not possible to study in an open geometry, like surface attachment and microbial chemotaxis. Future work may aim to incorporate more complexity of the rhizosphere into this microfluidic habitat, such as solid-phase minerals, soil micro-aggregates and micro-pores, oxygen gradients, and multiple moisture regimes.
While the rhizosphere-on-a-chip in this article re-creates only the physical structure of the soil, we provide evidence that this structure can influence the plant's molecular microenvironment to create the hotspots and hot-moments that drive microbial dynamics in the rhizosphere. Pairing the rhizosphere-on-a-chip to the LMJ-SSP-MS technique will allow for non-destructive chemical sampling of the root's microenvironment over time, which could lead to the quantification of rhizosphere fluxomics in future studies.
We hope that this work will act as a foundation for rhizosphere-on-a-chip studies and that future work will continue to use the cutting-edge technologies of microfluidics to probe biological interactions while maintaining a respect for the natural environment and its ability to shape ecosystem function.
Author contributions
Conceptualization: JA and STR. Investigation: JA, MK, CLW, KT. Formal analysis and data curation: JA, MK, CLW, JFC, STR. Writing – original draft: JA. Writing – review & editing: all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A portion of the research was conducted under the Laboratory Directed Research and Development (LDRD) program at Pacific Northwest National Laboratory, a multi-program national laboratory operated by Battelle for the U.S. Department of Energy. J. Aufrecht was supported by a Linus Pauling Distinguished Postdoctoral Fellowship. A portion of this research was conducted at the Center for Nanophase Material Sciences, which is a DOE Office of Science User Facility. J. F. Cahill and C. L. Walton were funded through the U.S. Department of Energy, Office of Science, Biological and Environmental Research, Bioimaging Science Program. M. Khalid and S. Retterer received support from the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov).References
- D. L. Jones and P. Hinsinger, Plant Soil, 2008, 312, 1–6 CrossRef CAS.
- S. Shi, E. E. Nuccio, Z. J. Shi, Z. He, J. Zhou and M. K. Firestone, Ecol. Lett., 2016, 19, 926–936 CrossRef PubMed.
- M. Watt, W. K. Silk and J. B. Passioura, Ann. Bot., 2006, 97, 839–855 CrossRef PubMed.
- A. Sanati Nezhad, Lab Chip, 2014, 14, 3262–3274 RSC.
- D. E. W. Patabadige, L. J. Millet, J. A. Aufrecht, P. G. Shankles, R. F. Standaert, S. T. Retterer and M. J. Doktycz, Sci. Rep., 2019, 9, 1–10 CAS.
- J. Aufrecht, J. Ryan, D. P. Allison, A. Nebenführ, M. J. Doktycz and S. T. Retterer, J. Visualized Exp., 2017, 126, e55971 Search PubMed.
- J. Sasse, J. Kant, B. J. Cole, A. P. Klein, B. Arsova, P. Schlaepfer, J. Gao, K. Lewald, K. Zhalnina, S. Kosina, B. P. Bowen, D. Treen, J. Vogel, A. Visel, M. Watt, J. L. Dangl and T. R. Northen, New Phytol., 2019, 222, 1149–1160 CrossRef CAS PubMed.
- H. Massalha, E. Korenblum, S. Malitsky, O. H. Shapiro and A. Aharoni, Proc. Natl. Acad. Sci., 2017, 114(17), 4549–4554, DOI:10.1073/pnas.1618584114.
- J. Aufrecht, C. M. Timm, A. Bible, J. L. Morrell-Falvey, D. A. Pelletier, M. J. Doktycz and S. T. Retterer, Adv. Biosyst., 2018, 1800048, 1–12 Search PubMed.
- J. Gao, J. Sasse, K. M. Lewald, K. Zhalnina, L. T. Cornmesser, T. A. Duncombe, Y. Yoshikuni, J. P. Vogel, M. K. Firestone and T. R. Northen, J. Visualized Exp., 2018, 2018, 1–16 Search PubMed.
- M. F. Noirot-Gros, S. V. Shinde, C. Akins, J. L. Johnson, S. Zerbs, R. Wilton, K. M. Kemner, P. Noirot and G. Babnigg, Front. Plant Sci., 2020, 11, 1–11 CrossRef PubMed.
- C. E. Stanley, G. Grossmann, X. Casadevall I Solvas and A. DeMello, Lab Chip, 2015, 16, 228–241 RSC.
- J. Deng, E. P. Orner, J. F. Chau, E. M. Anderson, A. L. Kadilak, R. L. Rubinstein, G. M. Bouchillon, R. A. Goodwin, D. J. Gage and L. M. Shor, Soil Biol. Biochem., 2015, 83, 116–124 CrossRef CAS.
- J. A. Aufrecht, J. D. Fowlkes, A. N. Bible, J. Morrell-Falvey, M. J. Doktycz and S. T. Retterer, PLoS One, 2019, 14, 1–17 CrossRef PubMed.
- B. Borer, R. Tecon and D. Or, Nat. Commun., 2018, 9, 769, DOI:10.1038/s41467-018-03187-y.
- G. Mollon and J. Zhao, Granular Matter, 2012, 14, 621–638 CrossRef CAS.
- T. Murashige and F. Skoog, Physiol. Plant., 1962, 15, 473–497 CrossRef CAS.
- J. F. Cahill, M. Khalid, S. T. Retterer, C. L. Walton and V. Kertesz, J. Am. Soc. Mass Spectrom., 2020, 31, 832–839 CrossRef CAS PubMed.
- N. Das, Graduate Theses and Dissertations, University of South Florida, 2007, p. 42 Search PubMed.
- N. Nunan, H. Schmidt and X. Raynaud, Philos. Trans. R. Soc., B, 2020, 375, 20190249, DOI:10.1098/rstb.2019.0249.
- J. Brkljacic, E. Grotewold, R. Scholl, T. Mockler, D. F. Garvin, P. Vain, T. Brutnell, R. Sibout, M. Bevan, H. Budak, A. L. Caicedo, C. Gao, Y. Gu, S. P. Hazen, B. F. Holt, S. Y. Hong, M. Jordan, A. J. Manzaneda, T. Mitchell-Olds, K. Mochida, L. A. J. Mur, C. M. Park, J. Sedbrook, M. Watt, S. J. Zheng and J. P. Vogel, Plant Physiol., 2011, 157, 3–13 CrossRef CAS PubMed.
- M. Watt, K. Schneebeli, P. Dong and I. W. Wilson, Funct. Plant Biol., 2009, 36, 960–969 CrossRef PubMed.
- J. Braam, New Phytol., 2005, 165, 373–389 CrossRef PubMed.
- B. G. Forde, J. Exp. Bot., 2009, 60, 3989–4002 CrossRef CAS PubMed.
- J. Sasse, S. M. Kosina, M. de Raad, J. S. Jordan, K. Whiting, K. Zhalnina and T. R. Northen, Plant Direct, 2020, 4, 1–14 CrossRef PubMed.
- P. Hinsinger, A. G. Bengough, D. Vetterlein and I. M. Young, Plant Soil, 2009, 321, 117–152 CrossRef CAS.
- M. Watt, P. Hugenholtz, R. White and K. Vinall, Environ. Microbiol., 2006, 8, 871–884 CrossRef PubMed.
- Y. Kuzyakov and E. Blagodatskaya, Soil Biol. Biochem., 2015, 83, 184–199 CrossRef CAS.
- D. L. Jones, C. Nguyen and R. D. Finlay, Plant Soil, 2009, 321, 5–33 CrossRef CAS.
- J. E. Drake, A. Gallet-Budynek, K. S. Hofmockel, E. S. Bernhardt, S. A. Billings, R. B. Jackson, K. S. Johnsen, J. Lichter, H. R. McCarthy, M. McCormack, D. J. P. Moore, R. Oren, J. S. Pippen, S. F. Pritchard, K. K. Treseder, W. H. Schlesinger, E. H. DeLucia and A. C. Finzi, Ecol. Lett., 2011, 14, 349–357 CrossRef PubMed.
- C. R. Warren, Plant Soil, 2015, 397, 147–162 CrossRef CAS.
- K. Zhalnina, K. B. Louie, Z. Hao, N. Mansoori, U. N. da Rocha, S. Shi, H. Cho, U. Karaoz, D. Loqué, B. P. Bowen, M. K. Firestone, T. R. Northen and E. L. Brodie, Nat. Microbiol., 2018, 3, 1–11 CrossRef PubMed.
- A. Kawasaki, S. Donn, P. R. Ryan, U. Mathesius, R. Devilla, A. Jones and M. Watt, PLoS One, 2016, 11, 10, DOI:10.1371/journal.pone.0164533.
- J. M. Chaparro, D. V. Badri, M. G. Bakker, A. Sugiyama, D. K. Manter and J. M. Vivanco, PLoS One, 2013, 8, 1–10 CrossRef.
- D. V. Badri and J. M. Vivanco, Plant, Cell Environ., 2009, 32, 666–681 CrossRef CAS.
- Y. Kuzyakov and O. Gavrichkova, Glob. Chang. Biol., 2010, 16, 3386–3406 CrossRef.
- M. S. Olson, R. M. Ford, J. A. Smith and E. J. Fernandez, Environ. Sci. Technol., 2004, 38, 3864–3870 CrossRef CAS PubMed.
- R. M. Macnab and D. E. Koshland, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 2509–2512 CrossRef CAS PubMed.
- G. NandaKafle, A. A. Christie, S. Vilain and V. S. Brözel, Front. Microbiol., 2018, 9, 1–11 CrossRef PubMed.
- B. Gibson, D. J. Wilson, E. Feil and A. Eyre-Walker, Proc. R. Soc. B, 2018, 285, 20180789, DOI:10.1098/rspb.2018.0789.
- C. H. Jaegar, S. E. Lindow, W. Miller and E. Clark, Appl. Environ. Microbiol., 1999, 65, 2685–2690 CrossRef PubMed.
- R. Allard-Massicotte, L. Tessier, F. Lécuyer, V. Lakshmanan, J. F. Lucier, D. Garneau, L. Caudwell, H. Vlamakis, H. P. Bais and P. B. Beauregard, MBio, 2016, 7(6), e01664, DOI:10.1128/mBio.01664-16.
- J. Sasse, E. Martinoia and T. Northen, Trends Plant Sci., 2018, 23, 25–41 CrossRef CAS PubMed.
- N. Yanagisawa, E. Kozgunova, G. Grossmann, A. Geitmann and T. Higashiyama, Plant Cell Physiol., 2021, 00, 1–12 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00705j |
| This journal is © The Royal Society of Chemistry 2022 |