Open Access Article
Open Access ArticleEngineering neurovascular organoids with 3D printed microfluidic chips†
Idris
Salmon
a,
Sergei
Grebenyuk
a,
Abdel Rahman
Abdel Fattah
a,
Gregorius
Rustandi
a,
Thomas
Pilkington
 b,
Catherine
Verfaillie
c and
Adrian
Ranga
b,
Catherine
Verfaillie
c and
Adrian
Ranga
 *a
*a
aLaboratory of Bioengineering and Morphogenesis, Biomechanics Section, Department of Mechanical Engineering, KU Leuven, Leuven, Belgium. E-mail: adrian.ranga@kuleuven.be
bFabLab Leuven, KU Leuven Research & Development, Belgium
cStem Cell and Developmental Biology, Department of Development and Regeneration, KU Leuven, Leuven, Belgium
First published on 25th March 2022
Abstract
The generation of tissue and organs requires close interaction with vasculature from the earliest moments of embryonic development. Tissue-specific organoids derived from pluripotent stem cells allow for the in vitro recapitulation of elements of embryonic development. However, they are not intrinsically vascularized, which poses a major challenge for their sustained growth, and for understanding the role of vasculature in fate specification and morphogenesis. Current organoid vascularization strategies do not recapitulate the temporal synchronization and spatial orientation needed to ensure in vivo-like early co-development. Here, we developed a human pluripotent stem cell (hPSC)-based approach to generate organoids which interact with vascular cells in a spatially determined manner. The spatial interaction between organoid and vasculature is enabled by the use of a custom designed 3D printed microfluidic chip which allows for a sequential and developmentally matched co-culture system. We show that on-chip hPSC-derived pericytes and endothelial cells sprout and self-assemble into organized vascular networks, and use cerebral organoids as a model system to explore interactions with this de novo generated vasculature. Upon co-development, vascular cells physically interact with the cerebral organoid and form an integrated neurovascular organoid on chip. This 3D printing-based platform is designed to be compatible with any organoid system and is an easy and highly cost-effective way to vascularize organoids. The use of this platform, readily performed in any lab, could open new avenues for understanding and manipulating the co-development of tissue-specific organoids with vasculature.
Introduction
Organoids derived from both adult and pluripotent stem cells have emerged in recent years as in vitro model systems which can recapitulate key features of physiology, with applications in drug screening and regenerative medicine.1 In particular, organoids derived from human pluripotent stem cells (hPSC) have also recapitulated key features of human embryonic development and thereby provided an unprecedented view into early human development.1,2 However, they have been limited in sized reproducibility and complexity.The presence of a vascular network has been hypothesized to be a key missing feature in these models which could account for some of these limitations. Indeed, the endothelium plays a critical role in the development of tissues and organs, functioning as a site of nutrient and waste exchange as well as a paracrine signaling center which directs differentiation, patterning and morphogenesis.3 For example, the endothelium plays an important role in the initiation of cortical neurogenesis as well as in the outgrowth of the liver bud by surrounding the embryonic hepatic endoderm, thereby delimiting the mesenchymal domain.4 In the immature brain, a relief of hypoxia mediated by endothelium invasion is shown to trigger neural stem cell differentiation.5 In addition, the expression of transcription factors such as WNT7a and WNT7b in the brain induces the acquisition of tissue-specific EC characteristics suggesting an important cross-talk between cerebral and vascular cells during nervous system development and maturation.6–9 As such, endothelial cell (EC) co-development would be a critical and necessary component for the biomimetic development of in vitro tissues such as organoids.
Early attempts at organoid vascularization focused on the transplantation of these organoids into murine hosts.10,11 Intestinal as well as cerebral organoids were shown to integrate and mature well into the host, with host vasculature seen to invade the organoid followed by the establishment of a functional perfused vascular network. While these observations underscore the importance of vasculature in the development of human organoids, the use of an in vivo host with endogenous vasculature is clearly not a practical approach for organoid vascularization, prompting a number of attempts at in vitro vascularization. The most common approach has involved the co-culture of organoids, including liver, pancreas, colon, kidney and cerebral organoids with primary ECs, with human umbilical vein endothelial cells (HUVEC) being most frequently used.12–16 However, HUVECs are already developmentally specified, in contrast to hPSC-derived ECs, which can interact and specify tissue maturation. Indeed, Carboxypeptidase M+ liver progenitor cells showed higher expression of hepatocyte marker genes such as, HNF4A and AFP, when cultured on hPSC-derived EC and hPSC-derived hepatic stellate cells, compared to co-culture on HUVECs and mesenchymal stem cells.17,18
Recent advances in organoid vascularization have also focused on the use of hPSCs engineered to overexpress the EC-specific ETV2 transcription factor. Co-aggregation of engineered and wild-type cells followed by neural differentiation with overexpression of ETV2 led to the generation of cerebral organoids with interspersed self-assembled vascular cells and enhanced functional maturation, including rudimentary blood brain barrier characteristics.19 Such co-culture models where vascular and cerebral cells are continuously grown together do not, however, mimic the characteristic spatial organization and temporal synchronization of angiogenic embryonic development, where blood vessels invade into an initially avascular neuroectodermal tissue.20,21 Microfluidic devices characterized by fluidically linked microchannels are ideally suited to provide such spatial interactions. In these devices, cells which are initially cultured separately can interact in designed spaces by migration through hydrogel substrates within the channels.22,23 However, an important limitation to the broader applicability of such devices for biological applications is that the soft lithography technologies necessary to fabricate these devices require access to dedicated clean room equipment and are time-consuming and error-prone, leading to a slow process for design iteration and optimization. High resolution 3D printing technology can overcome these limitations by offering the ability to control the spatial environment in a versatile, on-demand and easy to use manner.24,25 In particular, 3D printing technologies based on stereolithography (SLA) have seen in recent years a remarkable increases in spatial resolution and in availability of new printable resin formulations, including those which can be rendered biocompatible with post-printing processing.26,27 These technologies are now readily accessible to most labs due to significant price drops in both hardware and materials in recent years.28 Here, we explore the use of consumer-grade 3D printing technologies as a key enabling technology to generate temporally and spatially synchronized all hPSC vascularized organoid.
3D printing for microfluidic organoid vascularization
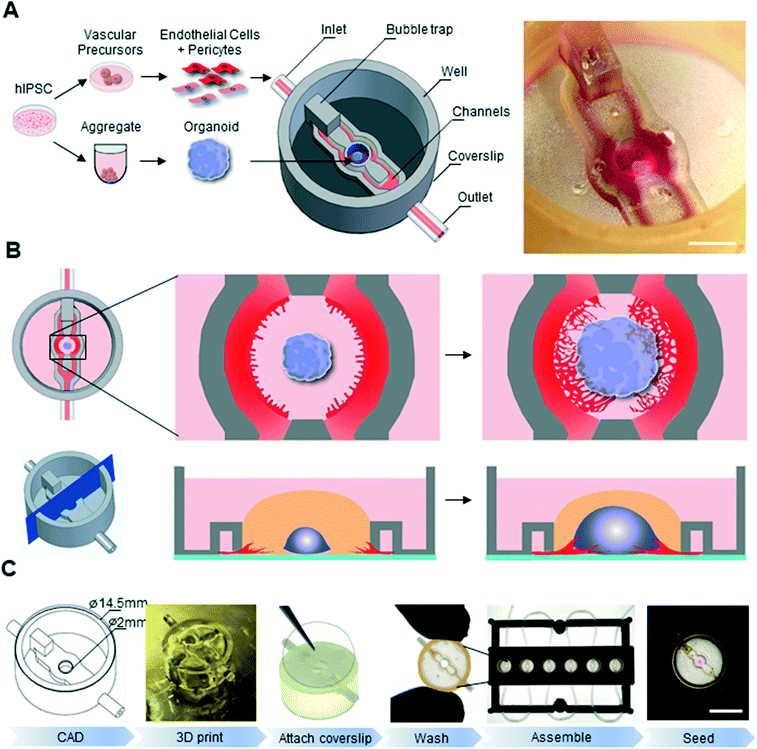
In order to tackle the challenge of both temporal and spatial synchronization between organoid and vasculature, we developed an approach based on two key aspects: the use of all-hPSC derived cell lineages and the development of customized 3D printing to engineer microfluidic chips with designed geometries. We hypothesized that by simultaneously initiating vascular and neural differentiation from PSCs, we would obtain developmentally matched lineages, which could then be grown in a spatially defined manner on a biomimetic chip (Fig. 1A).A key feature of our microfluidic chip is its “open well” design. In contrast to commonly used microfluidics where the culture chamber is closed, this design does not have any physical seal over the main culture chamber, thereby allowing for easy and direct accessibility to this compartment.16,29–31 In the context of organoid culture, this design therefore allows for direct and precise placement of an organoid in the central compartment as well as its easy removal for downstream analysis such as immunostaining. Another advantage of this design is that an individual organoid can be seeded in each chip, making it an ideal platform to study co-developmental processes at single-organoid level.32
To support vascular cell sprouting, network formation, and vascular invasion of a growing organoid on-chip, the open well microfluidic chip is designed such that microfluidic channels flanking the central organoid chamber can be seeded with vascular cells via an inlet (Fig. 1A and B). Communication of diffusible molecules and cells between the organoid chamber compartment and the microfluidic channels is ensured by a nearly complete wall separating the channels and the main compartment, leaving only a 50 μm gap between the coverslip and the 3D printed microfluidic chip (Fig. 1B bottom).
In order to print our parts, we sought an enabling printing technology and associated material which is biocompatible and allows for high-resolution printing (circa 200 μm feature size). We choose the Formlabs Form 2 printer, a consumer-grade 3D instrument based on stereolithography technology equipped with a 140 μm spot size laser which allows for printing in the micrometer range and can print a variety of biocompatible printing materials. In order to select the appropriate material for our chips, we conducted a set of experiments to quantify biocompatibility and printability of various materials. Our initial experiments indicated that hPSC culture was not compatible with the printed chips, suggesting remaining toxicity of the printed material. To render the chip biocompatible, we therefore developed an extensive washing procedure which allowed the unreacted polymers to leach out. Cell viability of cerebral organoids embedded in Matrigel and grown on various 3D printed disks after undergoing the washing procedure was assessed at day 14 of differentiation by a live/dead assay combined with flow cytometry. Cell viability was high in the control organoid-only condition (93%), as well as on disks of Dental LT (91.3%), Dental SG (91.2%), clear (90.5%) and Dental OD clear MF (88.6%) resins (ESI† Fig. S1A). Printing our microfluidic chip design with Dental LT resin resulted in closed internal microfluidic channels, crucial for EC seeding and medium perfusion, while clear resin led to gelatinous residual material (ESI† Fig. S1B Arrowheads). Only chips printed with Dental SG led to well-defined perfusable channels. This material, which had previously been reported to show no toxicity when exposed to 3T3 and hepatoma cells,24 was therefore chosen to print our microfluidic devices, allowing for an optimal combination of biocompatibility and printability (ESI† Fig. S1). To render the chip suitable for microscopy, we designed and printed a bottomless chip (Fig. 1A), which was completed in a subsequent step by the permanent attachment of glass coverslip to its bottom using a biocompatible UV-curable glue (Fig. 1C). Taking advantage of the design flexibility of 3D printing technologies, we also optimized a chip fabrication pipeline that allowed us to process tens to hundreds of chips in the same day (Fig. 1C). To parallelize the handling and image acquisition of experiments within these chips, we used inexpensive, lower resolution fused deposition modelling printing of polylactic acid (PLA) plastic to fabricate plates capable of holding 6 microfluidic devices. This multiplexed platform was instrumental in allowing on chip biological experiments in a standardized and reproducible manner.
Vascular networks on 3D printed microfluidic chip
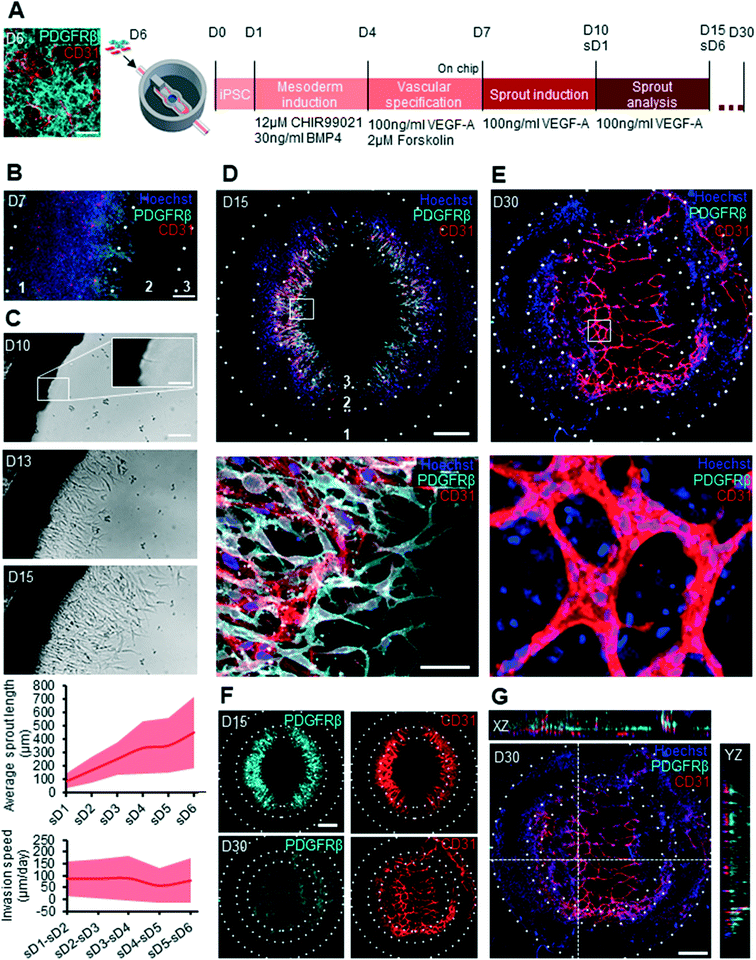
In order to produce cells for the vascular component of the chip, we aimed at generating both ECs and pericytes. Indeed, mural cell types such as pericytes play a crucial role in blood vessel stabilization in vivo and the inclusion of such cells in a microfluidic vascularization chip would therefore present an important advantage for long term vascular culture on chip.33 A recently published protocol demonstrated the directed differentiation of hPSCs towards dual EC and pericyte fates in the form of 3D blood vessel organoids.34 To rapidly obtain a large vascular cell yield for microfluidic chip seeding, we applied this protocol (medium composition and timing) to a hPSC 2D monolayer for 6 days of differentiation (Fig. 2A; ESI† Fig. S2A). In order to confirm that this 2D differentiation protocol yields both ECs and pericytes, immunohistochemistry (IHC) and gene expression analysis was performed on day 6 of differentiation, prior to cell dissociation and immediate seeding into the microfluidic chip. The presence of both pericytes, marked by PDGFRβ and ECs, marked by CD31 was confirmed at this time, thereby ensuring that the cells collected for seeding into the chip contained both vascular lineages (Fig. 2A; ESI† S2B). Furthermore, gene expression analysis of this 2D differentiated vascular cell mix showed clear upregulation of the early EC marker CD34, later EC markers CD31, CDH5 and KDR and pericyte markers such as PDGFRβ and CD73 compared to undifferentiated hPSC, while pluripotency markers NANOG and SOX2 were downregulated. This mixed vascular cell population was then used for seeding the channels of the microfluidic chip. To provide a substrate for 3D vascular invasion, we filled the central organoid chamber with Matrigel, an extracellular matrix frequently used in angiogenic sprouting assays.35 Vascular cell sprouting was triggered by the addition of 100 ng mL−1 of vascular endothelial growth factor A (VEGF-A) to the culture medium in the central compartment. Sprouting of both ECs and pericytes on chip was observed by IHC on day 7 (Fig. 2B). Vascular cells, which were initially seeded in the microfluidic channels were observed to start sprouting into the organoid culture chamber via an intermediate zone (Fig. 2B). These vascular cells were observed in the organoid culture chamber by day 10 of differentiation and progressively sprouted towards the center of the organoid culture chamber (Fig. 2C). The length of these vascular sprouts was measured over 6 days, starting from the day when they first appeared in the organoid culture chamber (sprout day 1, sD1), and was seen to increase over this observed period (Fig. 2C). This invasion speed was constant over the observed period and was calculated to be on average 80 μm day−1. To determine whether pericytes and ECs would maintain their identity over a longer differentiation period, we stained for PDGFRβ and CD31 on day 15 and day 30. Clear differences in vascular organization were evidenced between day 15 and day 30 cultures (Fig. 2D and E). At day 15 the pericytes and EC had a mixed and disorganized appearance, with pericytes and ECs in the same plane (Fig. 2F top panel). In contrast, by day 30, the vascular network was much further developed, with CD31+ cells contributing to consolidated vessel-like structures, and pericytes no longer evidenced in the same plane as the ECs and instead forming a supportive layer above the formed vascular networks (Fig. 2G; ESI† S2C).On chip sprout progression and cerebral organoid development
To demonstrate that our vascularization strategy could be applied to organoid models, we chose to incorporate the widely used cerebral organoid model into our chips.36 The generated aggregates exhibited a circular translucent band at the edges after two days of neural differentiation (ESI† Fig. S3A), suggesting successful induction of hPSCs into neural epithelial cells.36 At day 5 of neural differentiation, these neural aggregates were embedded in Matrigel and seeded into the organoid chamber of the chip for further maturation and growth. Organoid growth on chip was quantified from bright field images acquired every two days, and was seen to increase over the entire course of differentiation (ESI† Fig. S3A and S3B). Cerebral organoid maturation was verified by staining for PAX6, the earliest marker of human neural identity, and GFAP, a radial glia cell marker, on day 15, and βiii TUBULIN, a post mitotic neuron marker, on day 30 (ESI† Fig. S3B and S3C).37 In addition OLIG2+/SOX2+ motor neuron progenitors and PSD95+/SYN+ synapses were also seen in D30 cerebral organoids (ESI† Fig. S3D and S3E).38 Organoids also exhibited characteristically complex internal cytoarchitecture consisting of multiple epithelial domains. Together, these results demonstrate that 3D printed chips support the growth, differentiation and morphogenesis of cerebral organoids, as well as the development of complex vascular networks.Co-culture on 3D printed microfluidic chip
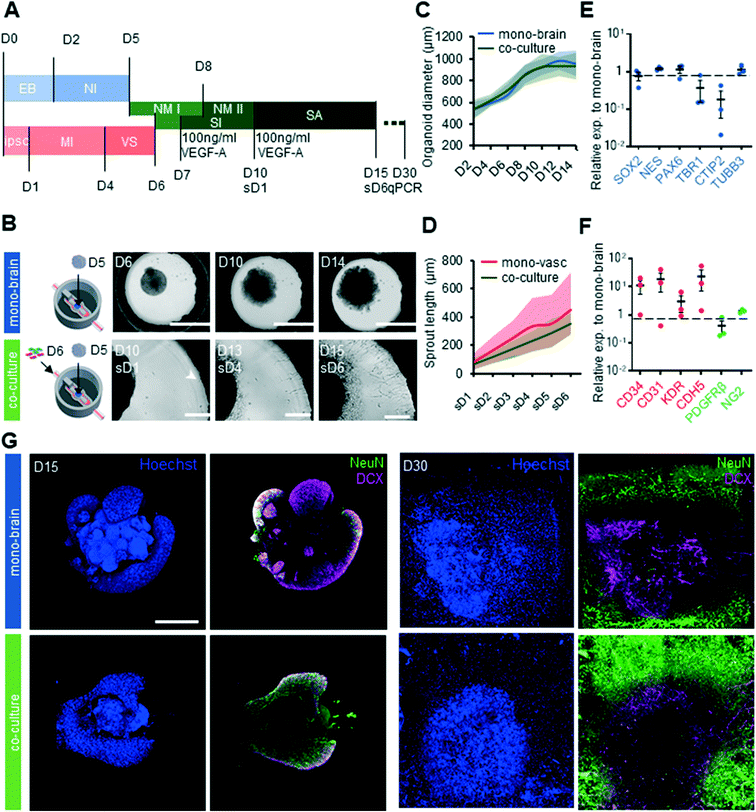
Having established that our platform was compatible with both vascular and cerebral organoid cultures, we next assessed whether these two cultures could be carried out together on chip. We therefore developed a differentiation synchronization scheme whereby both vascular differentiation as well as cerebral organoid differentiation would be initiated off chip, and would subsequently be brought together on chip at an early developmental time point.Our protocol consisted of seeding cerebral organoids on day 5 of differentiation into the organoid culture chamber, and the vascular cells on day 6 of differentiation into the microfluidic channels (Fig. 3A). In this way, co-differentiation would occur sufficiently late to allow for separate germ layer specification, but early enough that feedback such as brain-specific endothelial specification could occur. In these conditions, both cultures showed good survival, with characteristic organoid growth in the central compartment and vascular sprouting from the channels (Fig. 3B). Cerebral organoid size in co-cultures was not significantly different (p = 0.980) to that of organoids grown without vasculature, suggesting that the additional of vasculature did not impede organoid growth (Fig. 3D). Sprout length was similar between mono and co-culture systems (p = 0.367), indicating that the interaction between vascular cells and organoids did not alter sprouting characteristics (Fig. 3C). Gene expression analysis revealed that the markers of immature neurons TBR1 and deep layer neuron CTIP2 were 2.7 fold and 5.5 fold less expressed respectively in co-cultures compared to mono brain cultures (Fig. 3E). In contrast, expression levels of neural progenitor markers SOX2, NES and PAX6 in co-culture systems showed no significant changes in expression levels compared to chips harboring cerebral organoids only. Notably, EC and pericytes markers CD34, CD31, KDR, CDH5 and NG2 were highly upregulated in on chip co-culture systems (Fig. 3F). To obtain a better understanding of the maturity and the developmental dynamics of the cerebral organoids on chip we stained both mono-brain and co-culture for the immature neuron marker doublecortin (DCX) and the mature neuron marker neural nuclei (NeuN) on D15 and D30 of differentiation (Fig. 3G). In both mono and co-culture conditions at D15 the pattern of expression of both DCX and NeuN was localized in the outer layers of the organoid, concomitant with the absence of these markers in the more immature ventricular zone-like organoid core. In contrast to the mixed NeuN and DCX expression at D15, at D30 more mature NeuN+ neurons were seen on the outside of the organoid, with more immature neurons closer to the core. A strikingly higher abundance of NeuN+ cells was observed in the co-culture condition at both D15 and D30, suggesting a more rapid maturation rate due to presence of the vascular cells.
Neurovascular organoid on 3D printed microfluidic chip
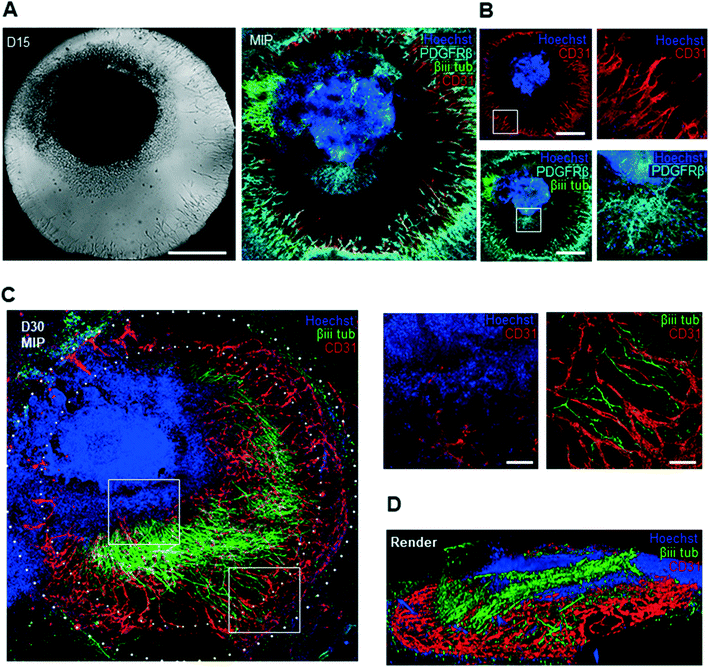
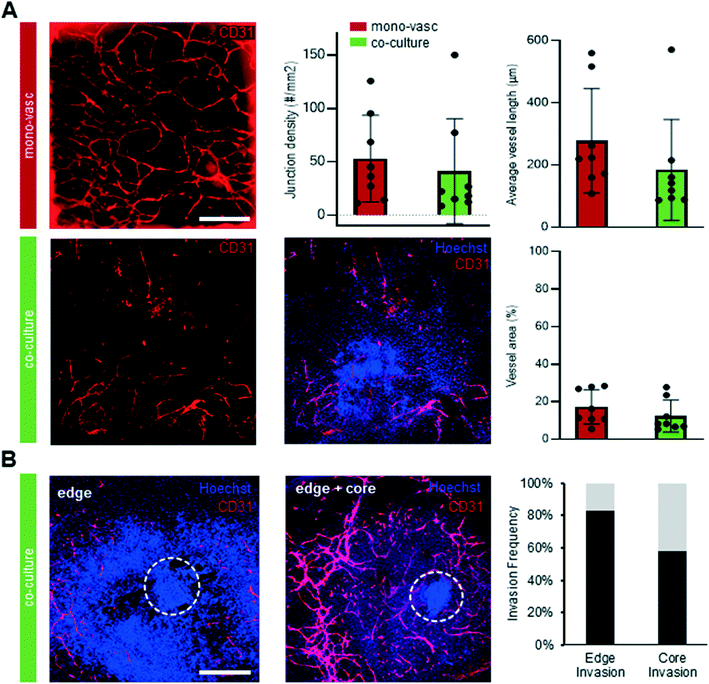
To further characterize the molecular identity and localization of both neural and vascular cells in the initial co-culture period, the neurovascular constructs were stained on day 15 of differentiation, i.e. after 9 days of on chip co-culture (Fig. 4A). βiii-TUBULIN+ post-mitotic neurons, CD31+ ECs, as well as PDGFRβ+ pericytes were all present in the co-culture on day 15, demonstrating that co-culture was permissive to the co-differentiation of these key cell types. Sprouts could be individually distinguished without overlap between each other, and were directed towards the organoid. To understand the 3D tissue organization within the chip, we analysed individual slices of a 3D confocal image stack. ECs were seen to localize near the bottom of the microfluidic chip whereas pericytes and neurons were found approximately 40 μm above the bottom of the plate (Fig. 4B). Pericytes were observed in closer contact with the organoid, in contrast to ECs, and subsequently migrated out of the organoid. A separate population of pericytes generated from a putative pool of neural crest-like cells within the organoid could be seen to migrate outward from the periphery of the organoid. In order to verify that our platform could sustain longer-term interaction between organoids and vasculature, we maintained the co-cultures for up to 30 days of differentiation. Strikingly, βiii TUBILIN+ neurites emanating from the central body of the neurovascular organoid frequently aligned with the vascular network (Fig. 4C). A 3D reconstruction of the confocal images revealed that neurites not only align with endothelial networks but also descend down from the organoid in order to come in contact with the vascular networks at the bottom of the microfluidic device (Fig. 4D). This 3D visualization additionally showed that these vascular networks formed an adjacent layer with the PDGFRβ+ pericytes (ESI† Fig. S2D), suggesting the establishment of a complex patterned architecture linking pericytes, endothelial cells and cerebral organoid. Significant phenotypic changes were observed in the latter period of differentiation, including continued organoid growth concomitant with significant expansion and consolidation of the vascular network. In order to more precisely quantify the presence and complexity of vasculature both outside and inside the cerebral organoids in both mono-vascular and co-culture conditions, we assessed vessel area, average vessel length and junctional density for vascular only and co-culture conditions, and found that vascular networks have similar features in both conditions (Fig. 5A). Additionally, endothelial cells were seen to invade the organoid edge in 83% of the analyzed co-culture organoids, and penetrated the organoid core in 42% of the organoids, as quantified by co-localization of CD31+ cells with the dense nuclear signal from the organoid core (Fig. 4C, 5B).Perfusable neurovascular organoid on 3D printed microfluidic chip
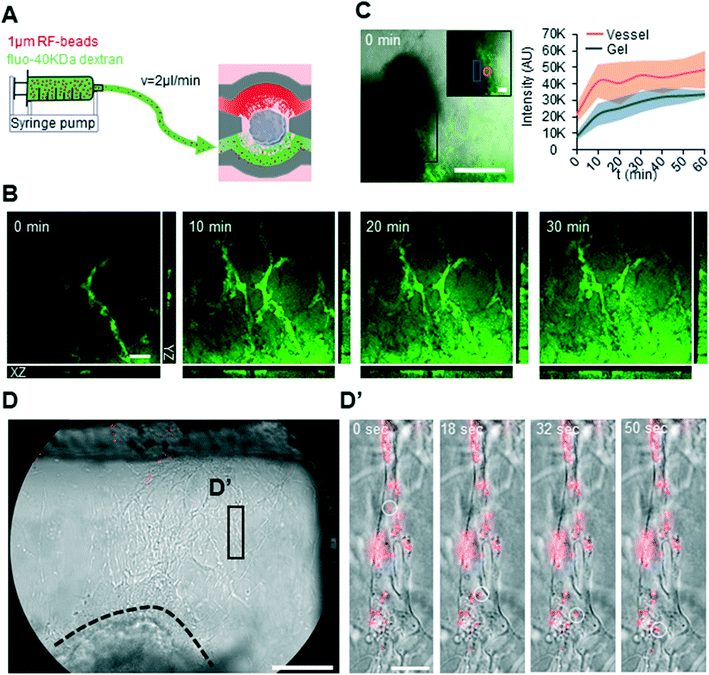
To asses both perfusion and permeability of the generated vascular networks on chip, active perfusion of the neurovascular co-culture was performed on day 20 and 25 (Fig. 6A). A strong fluorescent signal was seen within the vascular network shortly after active perfusion with fluorescein-40kDa dextran, which quickly diffused into the gel surrounding the vascular structures (Fig. 6B and C). In contrast to fluorescein-40kDa dextran, the larger 1 μm red fluorescent (RF)-beads could be tracked within single vessels and did not leak into the surrounding gel (Fig. 6D-D′) (ESI† Video S1). Taken together, these results demonstrate the functionality of the vascular networks generated within the microfluidic chip and their potential applicability for testing small, membrane-diffusible, compounds.Discussion
Here, we have demonstrated for the first time the possibility to control the physical interaction of endothelial cells, pericytes and organoids through the use of hPSCs and microfluidic technologies, in order to achieve an organoid vascularization process which is highly synchronized in time and space. The observed angiogenic sprouting and network formation in this platform are similar in size and branching architecture to those which have been reported with primary pericytes and ECs.15,39–42 The use of hPSC-derived vascular cells allows for the generation of organotypic and developmentally co-regulated vasculature, in contrast to vascular models where differentiated primary cells are used.39,40,43 We also demonstrated that cerebral organoid growth and maturation within our chip, including the presence of characteristic PAX6+, GFAP+,OLIG2+, SOX2+ and βiii-TUBULIN+ cells, as well as PSD95+/SYN+ synapses bear a close resemblance to previous studies where cerebral organoids were cultured both on a stationary PDMS micropillar plate and a PDMS microfluidic chip under flow condition.29,38,44–46 In addition, our results reveal significant difference in developmental dynamics between mono-brain and co-culture based on NEUN and DCX stains, suggesting an accelerated differentiation in co-cultures compared to mono-cultures.47We also observed that active perfusion of our vascular networks on chip show diffusion of small molecules whereas larger 1 μm-sized beads are retained in the vascular networks, in line with previous reports of perfusable vasculature on chip.39,48 In contrast to previous reports of organoid vascularization which implement vasculature with primary vascular cells or cell lines, both the vasculature and organoid in our model is exclusively derived from human PSCs, and additionally both vasculature and organoid differentiation are initiated simultaneously, suggesting that, unlike the mixed use of PSC-derived organoids and mature vascular cells, a more coordinated development is possible.
Our platform therefore demonstrates that a fully hPSC derived organoids and vasculature can be grown in 3D printed chips, thereby opening up the organoid and vascular biology fields to the possibilities offered by the design flexibility of 3D printing. Indeed, the significant price drops in recent years in both hardware and materials of such 3D printing technologies have made them readily accessible to every lab.28 In contrast to laborious conventional PDMS-based soft lithography approaches to fabricating microfluidics, the SLA-3D printing approach we present here is highly versatile and easy to use to generate microfluidic chips. We show that this approach is biocompatible, even for long-term organoid culture, and provides sufficient design flexibility to implement a device configuration amenable to co-culture and vascularization.
The generation of on-chip interactions between hPSC-derived organoid and vasculature holds great promises into the development of a complex neurovascular unit which would enable to more precisely investigate the role of ECs and pericytes in the development of human tissue, as well as for disease modelling applications with patient-specific hPSCs. Moreover, this platform has the potential to be used to model the function and perturbation of the blood brain barrier (BBB). Our standardized on-chip vascularization approach overcomes a number of limitations with other recently reported technologies.49 For example, subtractive removal of material from organoid tissue by sacrificial laser writing is a promising technique for creating perfusable channels,50 however the process of tissue ablation also has clear implications in altering tissue development. The use of a manufactured chip-like co-culture system on a re-engineered 384 well plate to study organoid- vasculature has also been recently proposed in the context of interaction between colon organoids with HUVECs and fibroblasts.15 This parallelized approach suggests that this technique could be useful for high throughput screening of drug candidates, however the use of such a standardized plate limits users flexibility in terms of the spatial control, which can be overcome using accessible 3D printing technology. Continuous long term perfusion of microvasculature on chip, which is a challenge for organoid-on-chip vascularization,51 is currently a limitation of our vascularized organoid model. Recent studies have explored various flow regimes for perfusing microvascular networks on chip over multiple days,52,53 and while similar strategies could be applied to our system, the role of flow on such a co-culture system remains to be evaluated.
The organoid vascularization approach we have shown here is designed to be generic and widely applicable, and opens a number of avenues for further research in the field of in vitro tissue vascularization. We expect that our protocol for generating ECs and pericytes from hPSC, in combination with PSC-derived organoids systems, will allow for a broader use of such developmentally synchronized co-cultures. Additionally, on-chip vascularization where the vascular and organoid components are brought together at defined times and allowed to interact starting from a defined spatial relationship could be a widely applicable strategy to generate a variety of other vascularized organoids. More broadly, the design and 3D printing of customizable microfluidic devices with consumer-grade printers and appropriately chosen and processed biocompatible materials may pave the way towards wider access and use of such technologies to generate controllable and reproducible in vitro tissues.
Materials and methods
Microfluidic device fabrication
The microfluidic device was designed in SolidEdge 2019. An .stl file was exported from SolidEdge to preform (Formlabs). The model was sliced at 50 μm and supports were attached. 3D printing was performed with Dental SG (Formlabs) material, with 8 devices printed per print. After printing, microfluidic devices were taken from the platform and placed in a 99% isopropanol (Chem-lab CL00.0901.9010) solution and sonicated for 2 minutes. Devices were rinsed two more times with fresh 99% isopropanol for 10 minutes in total. Clean microfluidic devices were dried and 5.5 μl of UV curable NOA86 (Norland products) glue was pipetted dropwise on the bottom of the microfluidic chip in order to glue a 13 mm wide round coverslips (VWR 631-0150). The glue was cured by placing the microfluidic devices on top of the LED strip at the inside of the form cure (Formlabs). Microfluidic devices were further post-cured in the form cure for 120 minutes at 60 °C to fully cure. Finally, the devices were washed for 6 days in 99% isopropanol to make the microfluidic devices fully biocompatible by placing two devices per well of a 6-well plate, with isopropanol exchange every other day.Water baths, used to avoid medium evaporation, and fit in dedicated cut-outs present on 6 well plate were also designed in Solidedge2019 and printed with clear resin (Formlabs) with a slicing distance of 50 μm. Post printing washing consisted of three short washes with 99% isopropanol and post curing for 60 minutes at 60 °C.
Finally, plates acting as holders for the microfluidic devices (6 per plate) and water baths were designed in SolidEdge2019. The design of the plate was sliced at 150 μm and 3D printed by use of a Prusa MK3S (Prusa Research) filament printer with polylactic acid (PLA) thermoplastic filament. Lids covering the plate were lasercut (Trotec Speedy100R) from 2 mm Plexiglas sheets.
hPSC cell culture
hPSC (Sigma Aldrich IPSC0028) were maintained on Matrigel-coated 6 well plates in E8 flex medium (Gibco A2858501). Upon reaching 70–80% confluency cells were split by dispase (Sigma D4693) incubation for 4 minutes at 37 °C. Colonies were scraped and split 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 onto freshly coated plates in E8 flex medium supplemented with 10 μM Y-27632 rock inhibitor (RI) (Hellobio HB2297) for 24 h. Medium was changed to E8 flex without RI and kept until splitting confluency was reached.
6 onto freshly coated plates in E8 flex medium supplemented with 10 μM Y-27632 rock inhibitor (RI) (Hellobio HB2297) for 24 h. Medium was changed to E8 flex without RI and kept until splitting confluency was reached.
Vascular mix differentiation
Vascular cells were differentiated from hPSC based on RA Wimmer et al.34 at day 0, a single cell solution of hPSC was first generated by incubation of accutase (Gibco 1110501) for 10 minutes at 37 °C. Single cells were collected, and spun down for 5 minutes at 300 RCF. Cells were counted, resuspended in E8 flex containing 10 μM RI and 250k of single cells were seeded on top of growth factor reduced (GFR) Matrigel (Becton Dickinson 354230) coated 6-well plate. The following day, when cells became confluent, medium was changed to mesoderm induction medium, a N2B27 medium supplemented with 12 μM CHIR99021 (Tocris 4423) and 30 ng mL−1 BMP-4 (R&D 5020-BP). N2B27 medium had to same composition as reported by Wimmer et al.34 on day 3 cells from a single six well were turned into a single cell solution by incubation of accutase for 5 minutes at 37 °C and spun down for 5 minutes at 300 RCF. Cells were resuspended in mesoderm induction medium and equally distributed over five six- wells coated with GFR Matrigel. The following day, the medium was changed to vascular differentiation medium composed of N2B27 medium supplemented with 100 ng mL−1 VEGF-A (Peprotech 100-20) and 2 μM Forskolin (Peprotech 6652995). On day 6 of differentiation accutase (Gibco A1110501) was pipetted for 5 minutes in the incubator to generate a single cell suspension. 6 million cells per mL were suspended in vascular differentiation medium supplemented with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 dilution of GFR Matrigel and were injected in the inlet of the microfluidic device. Afterwards the well of the microfluidic device was filled with cerebral organoid differentiation medium that contained vitamin A and 100 ng ml−1 VEGF-A to induce sprouting. This medium was changed every two days.
20 dilution of GFR Matrigel and were injected in the inlet of the microfluidic device. Afterwards the well of the microfluidic device was filled with cerebral organoid differentiation medium that contained vitamin A and 100 ng ml−1 VEGF-A to induce sprouting. This medium was changed every two days.
Cerebral organoid differentiation
Cerebral organoids were generated based on the protocol published by Lancaster et al.36 in short, medium compositions were kept the same but adjustments were made to the timing of medium delivery. On day 0, 96-well U bottom plates (Falcon 351177) were treated with anti-adherence rinsing solution (STEMCELL technologies) and subsequently rinsed twice with DMEM-F12 (Gibco 31330038). hPSC colonies were treated with accutase for 7 minutes in the incubator. Afterwards accutase was replaced by E8 Flex medium and cells were gently pipetted to obtain a single cell suspension. Cells were counted in a Bürker counting chamber and 9000 cells were seeded per well containing 10 μM RI. On day 2 of differentiation medium was changed to neural induction medium and on day 4 medium was replaced by cerebral organoid differentiation medium. Organoids were pipetted from the well and transferred to parafilm at day 5 of differentiation. Medium was aspirated and 4.5 μl of GFR Matrigel was used to lift organoids from the parafilm and to pipet them into the organoid chamber of the microfluidic device. A needle (Becton Dickinson 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600) was used to position the cerebral organoid in the centre of the chamber. Plates were turned upside down and placed in the incubator for 10 minutes to allow gelation. Afterwards 5 μl of GFR Matrigel was used to completely fill the organoid chamber with gel. Plates were put back in the incubator for 15 minutes for the gel to gelify. Afterwards 300 μl of cerebral organoid differentiation medium was pipetted into the well and water baths were filled with milliQ containing 1% vol/vol penicillin–streptomycin (Gibco 15140122). Medium was replaced at day 7 of differentiation. At day 8 of differentiation the medium was changed to cerebral organoid differentiation medium supplemented with Vitamin A. This medium was replaced every other day until day 30 of differentiation.
600) was used to position the cerebral organoid in the centre of the chamber. Plates were turned upside down and placed in the incubator for 10 minutes to allow gelation. Afterwards 5 μl of GFR Matrigel was used to completely fill the organoid chamber with gel. Plates were put back in the incubator for 15 minutes for the gel to gelify. Afterwards 300 μl of cerebral organoid differentiation medium was pipetted into the well and water baths were filled with milliQ containing 1% vol/vol penicillin–streptomycin (Gibco 15140122). Medium was replaced at day 7 of differentiation. At day 8 of differentiation the medium was changed to cerebral organoid differentiation medium supplemented with Vitamin A. This medium was replaced every other day until day 30 of differentiation.
Live dead assay
Single cell hPSC were aggregated and cerebral organoids were generated as previously described. On day 5 of differentiation neural aggregates were embedded in gel on top of 3D printed disks. These 13 mm round disks were printed with Dental SG, Dental LT, clear resin all from Formlabs and clear OD MF resin (3DResyns) on a Form 2 (Formlabs) printer and were subsequently washed for 6 days in 99% isopropanol (Chem-lab CL00.0901.9010). On day 14 of differentiation gel-embedded organoids were separated from the disks and turned into a single cell solution by incubation in TrypLE (Gibco 12605010) solution for 10 minutes at 37 °C. Afterwards, TrypLE was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in DMEM-F12 (Gibco 31330038) and every sample was spun down at 400 RCF for 5 minutes and resuspended into 200 μl of FACS buffer which was composed of 98% phosphate buffered saline (PBS) (Gibco 10010015) and 2% fetal bovine serum (Gibco 10270106). A 5 μM solution of calcein AM (Invitrogen L3224), live stain, and a 0.2 mM solution of ethidium homodimer 1 (Invitrogen L3224) were prepared in PBS and 4 μl and 8 μl of each of these solutions respectively was added to the medium in each of the samples. This staining mix was kept for 15 minutes at RT, in the dark, after which samples were spun down at 400 RCF for 5 minutes and resuspended in 200 μl of FACS buffer. 150 μl of each sample was then pipetted into a 96 flat bottom well (Falcon 351172) and 80 μl of this solution was analyzed by flow cytometry (Becton Dickinson, BD Accuri C6). Live cell percentage was calculated based on gated flow cytometer diagrams.
5 in DMEM-F12 (Gibco 31330038) and every sample was spun down at 400 RCF for 5 minutes and resuspended into 200 μl of FACS buffer which was composed of 98% phosphate buffered saline (PBS) (Gibco 10010015) and 2% fetal bovine serum (Gibco 10270106). A 5 μM solution of calcein AM (Invitrogen L3224), live stain, and a 0.2 mM solution of ethidium homodimer 1 (Invitrogen L3224) were prepared in PBS and 4 μl and 8 μl of each of these solutions respectively was added to the medium in each of the samples. This staining mix was kept for 15 minutes at RT, in the dark, after which samples were spun down at 400 RCF for 5 minutes and resuspended in 200 μl of FACS buffer. 150 μl of each sample was then pipetted into a 96 flat bottom well (Falcon 351172) and 80 μl of this solution was analyzed by flow cytometry (Becton Dickinson, BD Accuri C6). Live cell percentage was calculated based on gated flow cytometer diagrams.
Co-culture on chip
Cerebral organoids at day 5 of differentiation were seeded as previously described. The next day, vascular cells were seeded as described before. Every two days cerebral organoid differentiation medium that contained vitamin A and 100 ng ml−1 VEGF-A was changed.Sprout analysis
Starting from day 10 of differentiation sprouts could be observed in the organoid culture chamber. Brightfield images were taken from day 10 to day 15 of differentiation with an inverted microscope (Carl Zeiss MicroImaging, Zeiss Axio Observer Z1). Images were radially segmented in 18 equal segments. The longest sprout was identified and measured from the tip to the base for every segment at every time point. A pseudo-timeline was used since sprouts could also appear later than day 10 in the organoid chamber. The first day that a sprout appeared in the organoid chamber was termed sprouting day 1 (sD1).Immunohistochemistry and Angiotool analysis
Mono and co-cultures were fixed overnight at 4 °C with 4% paraformaldehyde (Sigma-Aldrich) for subsequent on chip staining. Chips were washed 3 times with PBS and inlets and outlets were sealed with parafilm to prevent leakage. Samples were blocked and permeabilized in 250 microliter of a PBS based solution that contained 5% BSA (Sigma-Aldrich), 2% Tx100 (PanREAC Applichem) and 0.05% NAN3 (Thermo scientific) for two days at RT on a shaker. Primary antibodies, PAX6 (mouse, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, DSHB), βiii TUBULIN (rabbit, 1
300, DSHB), βiii TUBULIN (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, biolegend 802001), CD31 (mouse, 1
300, biolegend 802001), CD31 (mouse, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75, Agilent M082329) and PDGFRβ (goat, 1
75, Agilent M082329) and PDGFRβ (goat, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, R&D systems AF385-SP), NeuN (mouse, 1
100, R&D systems AF385-SP), NeuN (mouse, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, Merck MAB377), DCX (rabbit, 1
300, Merck MAB377), DCX (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, cell signaling technology 4604S), PSD95 (rabbit, 1
200, cell signaling technology 4604S), PSD95 (rabbit, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 invitrogen 516900), SYN-1 (guinea pig, 1
300 invitrogen 516900), SYN-1 (guinea pig, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, synaptic systems), GFAP (Rat, 1
300, synaptic systems), GFAP (Rat, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 ThermoFisher scientific), OLIG2 (goat, 1
500 ThermoFisher scientific), OLIG2 (goat, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, Gibco A1413202) were diluted in blocking buffer and samples were placed on a shaker at RT for 2 days. Afterwards primary antibodies were washed off during 2 days with PBS containing 0.05% NAN3 and this PBS was refreshed 2 times per day. Secondary antibodies, donkey anti mouse Alexa fluor 488/555 (Invitrogen), donkey anti rabbit Alexa fluor 555 (Invitrogen), donkey anti goat Alexa fluor 647 (Invitrogen), donkey anti guinea pig 488 (Invitrogen) were diluted (all 1
300, Gibco A1413202) were diluted in blocking buffer and samples were placed on a shaker at RT for 2 days. Afterwards primary antibodies were washed off during 2 days with PBS containing 0.05% NAN3 and this PBS was refreshed 2 times per day. Secondary antibodies, donkey anti mouse Alexa fluor 488/555 (Invitrogen), donkey anti rabbit Alexa fluor 555 (Invitrogen), donkey anti goat Alexa fluor 647 (Invitrogen), donkey anti guinea pig 488 (Invitrogen) were diluted (all 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) in blocking buffer and were placed on a shaker at RT for two days. Hoechst was added (1
500) in blocking buffer and were placed on a shaker at RT for two days. Hoechst was added (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) in order to visualize nuclei. Secondary antibodies were washed of the same way as primary antibody washing. Confocal representative images were obtained with a confocal microscope (Leica microsystems, Leica SP8 Dive) in multiphoton mode. Mono-vascular and co-culture samples stained for CD31 were imaged on an inverted microscope (Invitrogen, EVOS) and images were analyzed by use of Angiotool software plugin for ImagJ for quantification of vascular network metrics.54
500) in order to visualize nuclei. Secondary antibodies were washed of the same way as primary antibody washing. Confocal representative images were obtained with a confocal microscope (Leica microsystems, Leica SP8 Dive) in multiphoton mode. Mono-vascular and co-culture samples stained for CD31 were imaged on an inverted microscope (Invitrogen, EVOS) and images were analyzed by use of Angiotool software plugin for ImagJ for quantification of vascular network metrics.54
RNA isolation and qRT PCR
RNA was isolated from both 2D, hPSC and vascular cultures, and 3D on chip cerebral organoid mono and co-cultures using TRIzol (Qiagen 79306) according to the manufacturer's instructions. 500 ng of RNA was transcribed to cDNA using superscript III (Invitrogen 11752-050), also according manufacturer's instructions. Afterwards, cDNA was combined with a primer mix containing SYBR green platinum (Invitrogen 11733-046) and analyzed in a ViiA 7 real time PCR instrument (Thermo Fisher Scientific). The primer sequences used are listed in ESI† Table S1. Fold changes were calculated compared to a reference sample (ΔΔCt method) and GAPDH was used as a house keeping gene for normalization.Microfluidic chip perfusion
Co-cultures on D20 and D25 of differentiation were connected to a syringe pump (Chemyx, Chemyx fusion 200) via tygon tubing (Saint Gobain, ACF00001) and were perfused at a rate of 2 μl min−1. The perfused solution consisted of a mix of 1 μm RF-BEADS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) (Microgenics corporation, R0100) and fluorescein-40kDa dextran (500 μg ml−1) (Sigma FD40) dissolved in PBS. Beads were visualized using an epifluorescence microscope (Nikon) and Z-stacks were made on a confocal microscope (Leica microsystems, Leica SP8 Dive) to visualize fluorescein-40 kDa dextran.
1000) (Microgenics corporation, R0100) and fluorescein-40kDa dextran (500 μg ml−1) (Sigma FD40) dissolved in PBS. Beads were visualized using an epifluorescence microscope (Nikon) and Z-stacks were made on a confocal microscope (Leica microsystems, Leica SP8 Dive) to visualize fluorescein-40 kDa dextran.
Statistical analysis
For statistical analysis, an unpaired two-tailed t-test was used with a 95% confidence interval (GraphPad Prism 8, GraphPad Software, Inc.).Author contributions
IS conducted experiments and analysis. SG,AA and GR conducted experiments. IS and TP developed the FDM 3D printed holders. IS, SG, AA, AR and CV interpreted the data and edited the manuscript. IS and AR wrote the manuscript.Conflicts of interest
No competing interests are declared.Acknowledgements
This work was supported by the FWO grants G087018N and I009718N and FWO postdoctoral fellowship 1217220 N, Interreg Biomat-on-Chip grant and Vlaams-Brabant and Flemish Government co-financing, KU Leuven grants C14/17/111 and C32/17/027, King Baudouin Foundation grant J1810950-207421 and an Allen Distinguished Inverstigator Award, a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Family Foundation.References
- J. Kim, B. K. Koo and J. A. Knoblich, Human organoids: model systems for human biology and medicine, Nat. Rev. Mol. Cell Biol., 2020, 21, 571–584 CrossRef CAS PubMed.
- S. Velasco, B. Paulsen and P. Arlotta, 3D Brain Organoids: Studying Brain Development and Disease Outside the Embryo, Annual Review of Neuroscience, Annual Reviews Inc., 2020, vol. 43, pp. 375–389 Search PubMed.
- A. Vargas-Valderrama, A. Messina, M. T. Mitjavila-Garcia and H. Guenou, The endothelium, a key actor in organ development and hPSC-derived organoid vascularization, J. Biomed. Sci., 2020, 27, 67 CrossRef PubMed.
- K. Matsumoto, H. Yoshitomi, J. Rossant and K. S. Zaret, Liver organogenesis promoted by endothelial cells prior to vascular function, Science, 2001, 294(5542), 559–563 CrossRef CAS PubMed.
- C. Lange, M. Turrero Garcia, I. Decimo, F. Bifari, G. Eelen and A. Quaegebeur, et al. Relief of hypoxia by angiogenesis promotes neural stem cell differentiation by targeting glycolysis, EMBO J., 2016, 35(9), 924–941 CrossRef CAS PubMed.
- M. F. Sabbagh, J. S. Heng, C. Luo, R. G. Castanon, J. R. Nery and A. Rattner, et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells, eLife, 2018, 7 Search PubMed , Available from: https://pubmed.ncbi.nlm.nih.gov/30188322/.
- J. M. Stenman, J. Rajagopal, T. J. Carroll, M. Ishibashi, J. McMahon and A. P. McMahon, Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature, Science, 2008, 322(5905), 1247–1250 CrossRef CAS PubMed.
- D. J. Nolan, M. Ginsberg, E. Israely, B. Palikuqi, M. G. Poulos and D. James, et al. Molecular Signatures of Tissue-Specific Microvascular Endothelial Cell Heterogeneity in Organ Maintenance and Regeneration, Dev. Cell, 2013, 26(2), 204–219 CrossRef CAS PubMed.
- R. Daneman, D. Agalliu, L. Zhou, F. Kuhnert, C. J. Kuo and B. A. Barres, Wnt/β-catenin signaling is required for CNS, but not non-CNS, angiogenesis, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(2), 641–646 CrossRef CAS PubMed.
- C. L. Watson, M. M. Mahe, J. Múnera, J. C. Howell, N. Sundaram and H. M. Poling, et al. An in vivo model of human small intestine using pluripotent stem cells, Nat. Med., 2014, 20(11), 1310–1314 CrossRef CAS PubMed.
- A. A. Mansour, J. T. Gonçalves, C. W. Bloyd, H. Li, S. Fernandes and D. Quang, et al. An in vivo model of functional and vascularized human brain organoids, Nat. Biotechnol., 2018, 36(5), 432–441 CrossRef CAS PubMed.
- T. Takebe, K. Sekine, M. Enomura, H. Koike, M. Kimura and T. Ogaeri, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant, Nature, 2013, 499(7459), 481–484 CrossRef CAS PubMed.
- T. Takebe, M. Enomura, E. Yoshizawa, M. Kimura, H. Koike and Y. Ueno, et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation, Cell Stem Cell, 2015, 16(5), 556–565 CrossRef CAS PubMed.
- Y. Shi, L. Sun, M. Wang, J. Liu, S. Zhong and R. Li, et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo, PLoS Biol., 2020, 18(5), e3000705 CrossRef CAS PubMed.
- S. Rajasekar, D. S. Y. Lin, L. Abdul, A. Liu, A. Sotra and F. Zhang, et al. IFlowPlate—A Customized 384-Well Plate for the Culture of Perfusable Vascularized Colon Organoids, Adv. Mater., 2020, 2002974 CrossRef CAS PubMed.
- K. A. Homan, N. Gupta, K. T. Kroll, D. B. Kolesky, M. Skylar-Scott and T. Miyoshi, et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro, Nat. Methods, 2019, 16(3), 255–262 CrossRef CAS PubMed.
- Y. Koui, T. Kido, T. Ito, H. Oyama, S. W. Chen and Y. Katou, et al. An In Vitro Human Liver Model by iPSC-Derived Parenchymal and Non-parenchymal Cells, Stem Cell Rep., 2017, 9(2), 490–498 CrossRef CAS PubMed.
- E. Goulart, L. C. De Caires-Junior, K. A. Telles-Silva, B. H. S. Araujo, G. S. Kobayashi and C. M. Musso, et al. Adult and iPS-derived non-parenchymal cells regulate liver organoid development through differential modulation of Wnt and TGF-β, Stem Cell Res. Ther., 2019, 10(1), 258 CrossRef PubMed.
- B. Cakir, Y. Xiang, Y. Tanaka, M. H. Kural, M. Parent and Y. J. Kang, et al. Engineering of human brain organoids with a functional vascular-like system, Nat. Methods, 2019, 16(11), 1169–1175 CrossRef CAS PubMed.
- B. Obermeier, R. Daneman and R. M. Ransohoff, Development, maintenance and disruption of the blood-brain barrier, Nat. Med., 2013, 19, 1584–1596 CrossRef CAS PubMed.
- M. Marín-Padilla, The human brain intracerebral microvascular system: development and structure, Front. Neuroanat., 2012, 13(6), 38 Search PubMed.
- S. Chung, R. Sudo, P. J. MacK, C. R. Wan, V. Vickerman and R. D. Kamm, Cell migration into scaffolds under co-culture conditions in a microfluidic platform, Lab Chip, 2009, 9(2), 269–275 RSC.
- T. Osaki, S. G. M. Uzel and R. D. Kamm, Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons, Sci. Adv., 2018, 4(10), eaat5847 CrossRef CAS PubMed.
- A. A. Yazdi, A. Popma, W. Wong, T. Nguyen, Y. Pan and J. Xu, 3D printing: an emerging tool for novel microfluidics and lab-on-a-chip applications, Microfluid. Nanofluid., 2016, 20(3), 1–18 CrossRef.
- A. I. Shallan, P. Smejkal, M. Corban, R. M. Guijt and M. C. Breadmore, Cost-effective three-dimensional printing of visibly transparent microchips within minutes, Anal. Chem., 2014, 86(6), 3124–3130 CrossRef CAS PubMed.
- N. P. MacDonald, F. Zhu, C. J. Hall, J. Reboud, P. S. Crosier and E. E. Patton, et al. Assessment of biocompatibility of 3D printed photopolymers using zebrafish embryo toxicity assays, Lab Chip, 2016, 16(2), 291–297 RSC.
- K. Piironen, M. Haapala, V. Talman, P. Järvinen and T. Sikanen, Cell adhesion and proliferation on common 3D printing materials used in stereolithography of microfluidic devices, Lab Chip, 2020, 20(13), 2372–2382 RSC.
- S. Waheed, J. M. Cabot, N. P. Macdonald, T. Lewis, R. M. Guijt and B. Paull, et al. 3D printed microfluidic devices: Enablers and barriers, Lab Chip, 2016, 16, 1993–2013 RSC.
- Y. Wang, L. Wang, Y. Guo, Y. Zhu and J. Qin, Engineering stem cell-derived 3D brain organoids in a perfusable organ-on-a-chip system, RSC Adv., 2018, 8(3), 1677–1685 RSC.
- L. Wang, T. Tao, W. Su, H. Yu, Y. Yu and J. Qin, A disease model of diabetic nephropathy in a glomerulus-on-a-chip microdevice, Lab Chip, 2017, 17(10), 1749–1760 RSC.
- M. Nikolaev, O. Mitrofanova, N. Broguiere, S. Geraldo, D. Dutta and Y. Tabata, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis, Nature, 2020, 585(7826), 574–578 CrossRef PubMed.
- K. Achberger, C. Probst, J. C. Haderspeck, S. Bolz, J. Rogal and J. Chuchuy, et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform, eLife, 2019, 8, e46188 CrossRef PubMed.
- A. Armulik, G. Genové and C. Betsholtz, Pericytes: Developmental, Physiological, and Pathological Perspectives, Problems, and Promises, Dev. Cell, 2011, 21, 193–215 CrossRef CAS PubMed.
- R. A. Wimmer, A. Leopoldi, M. Aichinger, D. Kerjaschki and J. M. Penninger, Generation of blood vessel organoids from human pluripotent stem cells, Nat. Protoc., 2019, 14(11), 3082–3100 CrossRef CAS PubMed.
- I. Arnaoutova and H. K. Kleinman, In vitro angiogenesis: Endothelial cell tube formation on gelled basement membrane extract, Nat. Protoc., 2010, 5(4), 628–635 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Generation of cerebral organoids from human pluripotent stem cells, Nat. Protoc., 2014, 9(10), 2329–2340 CrossRef CAS PubMed.
- X. Zhang, C. T. Huang, J. Chen, M. T. Pankratz, J. Xi and J. Li, et al. Pax6 is a human neuroectoderm cell fate determinant, Cell Stem Cell, 2010, 7(1), 90–100 CrossRef CAS PubMed.
- H. Kim, R. Xu, R. Padmashri, A. Dunaevsky, Y. Liu and C. F. Dreyfus, et al. Pluripotent Stem Cell-Derived Cerebral Organoids Reveal Human Oligodendrogenesis with Dorsal and Ventral Origins, Stem Cell Rep., 2019, 12(5), 890–905 CrossRef CAS PubMed.
- M. Campisi, Y. Shin, T. Osaki, C. Hajal, V. Chiono and R. D. Kamm, 3D self-organized microvascular model of the human blood-brain barrier with endothelial cells, pericytes and astrocytes, Biomaterials, 2018, 180, 117–129 CrossRef CAS PubMed.
- C. A. Bichsel, S. R. R. Hall, R. A. Schmid, O. T. Guenat and T. Geiser, Primary Human Lung Pericytes Support and Stabilize in Vitro Perfusable Microvessels, Tissue Eng., Part A, 2015, 21(15–16), 2166–2176 CrossRef CAS PubMed.
- S. Kim, H. Lee, M. Chung and N. L. Jeon, Engineering of functional, perfusable 3D microvascular networks on a chip, Lab Chip, 2013, 13(8), 1489–1500 RSC.
- J. Kim, M. Chung, S. Kim, D. H. Jo and J. H. Kim, et al. Engineering of a Biomimetic Pericyte-Covered 3D Microvascular Network, PLoS One, 2015, 10(7), e0133880 CrossRef PubMed.
- P. N. Ingram, L. E. Hind, J. A. Jiminez-Torres, A. Huttenlocher and D. J. Beebe, An Accessible Organotypic Microvessel Model Using iPSC-Derived Endothelium, Adv. Healthcare Mater., 2018, 7(2) DOI:10.1002/adhm.201700497.
- M. A. Lancaster, M. Renner, C. A. Martin, D. Wenzel, L. S. Bicknell and M. E. Hurles, et al. Cerebral organoids model human brain development and microcephaly, Nature, 2013, 501(7467), 373–379 CrossRef CAS PubMed.
- G. Quadrato, T. Nguyen, E. Z. Macosko, J. L. Sherwood, S. M. Yang and D. R. Berger, et al. Cell diversity and network dynamics in photosensitive human brain organoids, Nature, 2017, 545(7652), 48–53 CrossRef CAS PubMed.
- Y. Zhu, L. Wang, H. Yu, F. Yin, Y. Wang and H. Liu, et al. In situ generation of human brain organoids on a micropillar array, Lab Chip, 2017, 17(17), 2941–2950 RSC.
- I. Paredes, P. Himmels and C. Ruiz de Almodóvar, Neurovascular Communication during CNS Development, Dev. Cell, 2018, 45, 10–32 CrossRef CAS PubMed.
- M. Vila Cuenca, A. Cochrane, F. E. van den Hil, A. A. F. de Vries, S. A. J. Lesnik Oberstein and C. L. Mummery, et al. Engineered 3D vessel-on-chip using hiPSC-derived endothelial- and vascular smooth muscle cells, Stem Cell Rep., 2021, 16(9), 2159–2168 CrossRef CAS PubMed.
- S. Grebenyuk and A. Ranga, Engineering organoid vascularization, Frontiers in Bioengineering and Biotechnology, Frontiers Media S.A., 2019, vol. 7, p. 39 Search PubMed.
- M. A. Skylar-Scott, S. G. M. Uzel, L. L. Nam, J. H. Ahrens, R. L. Truby and S. Damaraju, et al. Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels, Sci. Adv., 2019, 5(9), eaaw2459 CrossRef CAS PubMed.
- S. Zhang, Z. Wan and R. D. Kamm, Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature, Lab Chip, 2021, 21, 473–488 RSC.
- C. Hajal, L. Ibrahim, J. C. Serrano, G. S. Offeddu and R. D. Kamm, The effects of luminal and trans-endothelial fluid flows on the extravasation and tissue invasion of tumor cells in a 3D in vitro microvascular platform, Biomaterials, 2021, 265, 120470 CrossRef CAS PubMed.
- K. Haase, F. Piatti, M. Marcano, Y. Shin, R. Visone, A. Redaelli, M. Rasponi and R. D. Kamm, Physiologic flow-conditioning limits vascular dysfunction in engineered human capillaries, Biomaterials, 2022, 280, 121248 CrossRef CAS PubMed.
- E. Zudaire, L. Gambardella, C. Kurcz and S. Vermeren, A computational tool for quantitative analysis of vascular networks, PLoS One, 2011, 6(11), e27385 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00535a |
| This journal is © The Royal Society of Chemistry 2022 |