 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tailor-made enzyme consortium segregating sclerenchyma fibre bundles from willow bark†
Dou
Jinze
 *a,
Wang
Jincheng
*a,
Wang
Jincheng
 b,
Zhao
Jian
b,
Zhao
Jian
 *b and
Vuorinen
Tapani
*b and
Vuorinen
Tapani
 a
a
aDepartment of Bioproducts and Biosystems, Aalto University, Espoo, Finland. E-mail: jinze.dou@aalto.fi; Tel: +358 413115001
bState Key Laboratory of Microbial Technology, Shandong University, Qingdao, China. E-mail: zhaojian@sdu.edu.cn; Tel: +86 13573158538
First published on 23rd February 2022
Abstract
We report for the first time that pectin-degrading enzymes could be tailored for wood bark based on the chemical features of pectin. Besides wood, stems of trees contain 10–20% bark that remains one of the largest underutilized biomasses on the planet. Unique extractive compounds, suberin, pectin, sclerenchyma fibres, etc. form a major part of the bark that is today mainly combusted for energy production. In certain trees, such as willow, lignified sclerenchyma fibres organize in continuous, thin bundles or bast fibres which are surrounded by the non-lignified ground tissue. Random screening of lignocellulose-degrading enzymes is the mainstream, suitable for uniform and simple substrates like cellulose, but the pectin chemistry is more complex. The structure of pectin was first elucidated after which pectin-degrading enzymes were tailored. Surprisingly, the applied pectinases alone were able to fully liberate the fibre bundles from the bark under mild conditions. When the pectinases were used together with hemicellulases, fibre bundles with an abnormally low surface lignin content of 10% were obtained. Overall, the novel findings of this study give promise for commercial valorisation of the underappreciated bark biomass in the future without the need to build huge plants with their expensive chemical recovery systems. Most importantly, the “tailor-made enzyme consortium based on the structural features of the substrate” concept may be a revolutionary breakthrough in precisely designing biochemical degradation strategies particularly for the recalcitrant macromolecule component (such as pectin) of lignocellulosic biomass.
Introduction
Lignocellulosic biomass represents perhaps the only sustainable carbon source that can be valorised to produce energy, chemicals, and functional materials in large scale.1 Advanced microbial technologies have been applied mainly to produce high-value products from the main lignocellulose components, namely cellulose, hemicelluloses and lignin, in the last decades.2–4 However, wood bark has merely been used as an energy supply in the forest industry and no microbial treatments tailored for the bark have been reported, possibly because of its unique recalcitrant macromolecules such as suberin and condensed polyphenols. Additionally, chemical pulping of the bark requires harsh conditions as its guaiacyl-type lignin is less reactive than the syringyl-type of lignin of hardwood xylem.5,6 Importantly, significant amounts of another recalcitrant macromolecule, pectin, has been reported to be present in the bark of spruce,7 willow8 and Scots pine.9Calcium ions present in the plant cell wall increase its rigidity through forming ionic bridges between the pectin molecules.10 Efficient non-enzymatic deconstruction of pectin can be realized by chemical means. Application of aqueous NaHCO311 or NaOH12 at elevated temperatures leads to depolymerization of methyl esterified galacturonan chains through a β-elimination reaction. Specifically, the β-elimination is known to break down interunit linkages in methyl esterified pectin. The solubilization of pectin fragments in the compound middle lamella leads to intercellular weakening and cell softening.13 However, these changes typically require the use of alkaline conditions and energy and lead to partial degradation of the sclerenchyma fibre bundles that are present in the bark of several species, such as willow.14 Breakthroughs in the isolation of intact sclerenchyma fibre bundles from the bark by selective biotechnical means could make it a more attractive source of functional fibre bundles. For example, willow bark fibre bundles are spinnable into yarns and able to provide excellent protection from ultraviolet radiation and against Gram-positive pathogens like Staphylococcus aureus.11
Pectin plays an important role in the initial lignification of the plant cell wall,15 bonds covalently to lignin via ester linkages and bridges lignin with hemicelluloses.16 The covalent bonds between pectin, lignin and hemicelluloses5 may limit the reactivity of the matrix towards pectinases that could potentially be applied for selective solubilization of pectin. Pectin is abundantly available in reaction wood17 and present mainly in the primary wall and middle lamella of plant cells. In non-lignified cells, the thin middle lamella layer is mainly composed of pectin, which binds the adjacent neighbouring cells together. Pectin is synthesized during the first stage of primary cell wall growth and contributes to the firmness and mechanical strength of both the cell wall alone and intercellular adhesion of the cells. Moreover, pectin together with hemicelluloses (and lignin) forms a matrix around cellulose fibrils in the primary cell wall contributing to its ductility. Pectin may also play a role in the defence mechanisms of plants against pathogens.18 Therefore, understanding the pectin chemistry is an indispensable step in designing customized pectinase treatments for the isolation of fibre bundles from the bark.
The presence of 1,4-linked α-D-galacturonic acid (GalA) units is a dominant feature of the complex pectin macromolecule. The prevailing structural representation of pectin includes subsegments of homogalacturonan (HG), xylogalacturonan, rhamnogalacturonan I (RG-I) and rhamnogalacturonan II (RG-II). HG, the most abundant domain, is a linear polymer chain composed solely of GalA units. The backbone of RG-I, the second most abundant domain of pectin, comprises alternating 1,4-linked GalA and 1,2-linked α-L-rhamnopyranose units.17,18 Due to the specific catalytic activity of the enzyme, the pectinases need to be tailored in degrading pectin according to the structural features of the substrate although the ‘random screening’ of the microorganisms is nowadays a conventional strategy.
Such a ‘random screening’ strategy (Table 1) is suitable for the cell wall components like cellulose as cellulose is uniform and simple;19 however the chemical structure of the macromolecular pectin varies a lot between species, even differs at different positions of the same plant (or wood). These components can also be acetylated (or methylated) to different degrees.18 Thus, the microbial consortium should be tailored based on the special features of the targeted pectin substrate, for example the pectin-degrading enzymes suitable for catalysing the depolymerization of the pectin with various degrees of methylation can be tailored (Table 2) according to the methylation degree of the substrate. There has been no such tailored pectinase microbial strategy reported for any lignocellulosic biomass, including grass fibres like ramie in Table 1.
| Methodology | Mechanism (or design elements) | Microorganism selection | Temp. (°C); time (h) | Substrate | Ref. | |
|---|---|---|---|---|---|---|
| Conventional strategy | Chemical degumming | Cleavage of the Ca2+/pectin crossbridges by alkali | — | 120; 5 | Ramie | 2 and 20 |
| — | 100; 1 | Wood bark | 11 and 12 | |||
| Microbial degumming | Microbial growth, metabolism, enzymatic degradation | Random screening | 30; 16 | Ramie | 2 and 21 | |
| Enzymatic degumming | Pectinase- and hemicellulosic-degrading enzymes | Random screening | 50; 4 | Ramie | 2 and 22 | |
| Our strategy | Enzymatic degumming (microbial consortium design based on pectin features of wood bark) | Average degree of methylation (ca. 50.7); unevenly distributed methylation degree | Tailored PelA + PelC/PelB | 50; 4 | Wood bark | This study |
| High proportion of RG-I domains and their arabinan side chains | Tailored arabinanase | |||||
| Main hemicelluloses (xylan and glucomannan) | Tailored xylanase; mannanase |
| Enzyme | Gene | EC number | Cazy family | Function |
|---|---|---|---|---|
| Pectate lyase A | pelA | 4.2.2.2 | PL 1 | Catalyzes the depolymerization of non-esterified polygalacturonate (DM = 0%) |
| Pectate lyase C | pelC | 4.2.2.2 | PL 3 | Catalyzes the depolymerization of both polygalacturonate and pectin with low degree of methylation (optimal for DM = 22%), with an endo mode of action28 |
| Pectin lyase | pelB | 4.2.2.10 | PL 1 | Catalyzes the depolymerization of pectin with DM = 85% (optimal), with an endo mode of action; cannot degrade polygalacturonate29 |
| endo-1,4-β-Mannosidase | gmuG | 3.2.1.78 | GH 26 | Catalyzes the endo hydrolysis of mannan, galactomannan and glucomannan |
| Rhamnogalacturonan endolyase | rhgW | 4.2.2.23 | PL 11 | Catalyzes the degradation of the RG-I backbone by a β-elimination mechanism |
| endo-1,5-α-L-Arabinosidase | abnA | 3.2.1.99 | GH 43 | Catalyzes the internal cleavage of linear 1,5-α-L-arabinan and of branched sugar beet arabinan |
| endo-1,4-β-Xylanase | xynA | 3.2.1.8 | GH 11 | Catalyzes the endo hydrolysis of xylan |
Microbial pectinase conversion has been mostly applied for degumming of grass fibres like ramie22 (Table 1) or textile fibres from flax23 and juices (grape, citrus, and potato) for the food industry.18 It is worth noting that the chemical composition of the ramie or flax is completely different in comparison with the wood bark (Table S1†). Ramie or flax biomass contains mostly holocellulose (85–87 wt%) and pectin (2 wt%); there is a negligible amount (ca. 0.5 wt%) of lignin24 present in these grass-type biomasses. However, Klason-lignin represents 17–26 wt% of the wood bark.6 More specifically, willow tree's bark, similar to spruce bark, contains a complex mixture of Klason lignin (24–31 wt%),5 extractives (10–15 wt%), suberin (5 wt%),25 starch (1.3 wt%), pectin (3 wt%) and other traditional components (i.e. holocellulose represents less than 50 wt%), which means that the covalent linkages between pectin, lignin and hemicelluloses contribute more complex to the recalcitrance of the matrix towards non-tailored pectin-degrading enzymes which could potentially be applied for selective solubilization of pectin from wood (or willow) bark than the grass-type biomass. Moreover, the bioavailability of the tree bark is million-fold more abundant than grass fibres.7–9 No paper on enzymatic segregation of fibre bundles from wood bark has been published.
This study aims at tailored microbial valorisation of willow bark through separating its sclerenchyma fibre bundles. Furthermore, the thick-walled individual sclerenchyma fibres from willow bark are relatively longer and provide much higher stiffness and strength than fibres from its wooden parts at the same sheet density.26 We hypothesized that pectin is mostly present in the parenchyma tissue of willow bark26 and that the presence of covalent bonds between pectin, lignin and/or hemicelluloses5,16 restricts the reactivity of pectinases. Hemicellulases were speculated to possibly improve the accessibility of the pectinases in the bark without degrading its sclerenchyma fibre bundles, which could happen in hot alkaline treatments. Several spectroscopic and chromatographic techniques were applied to gain insight into the types of enzymes involved in the depolymerization and to elucidate the chemical composition of pectin and the obtained fibre bundles. Surprisingly, all fibre bundles present in willow bark were fully liberated and most of the pectin was solubilized when the tailored pectinase action precisely targeted the HG and RG-I domains.
Experimental
Materials and chemicals
One-year old willow hybrid “Klara” stems were harvested from the field of Carbons Finland Oy located at Kouvola, Finland, on May 18, 2017. The exterior bark was manually stripped off in the field and cut into 15 cm pieces for storage at −20 °C before further use. Citric acid, hydrogen chloride, sodium hydroxide, 95% ethanol, dichloromethane, acetone and sodium bicarbonate were from Sinopharm Chemical Reagent Co., Ltd. Deuterium oxide, acetic acid-d4, α-amylase, amyloglucosidase, and tetramethylsilane were supplied by Macklin China. Methanol-d4 was purchased from Tenglong Weibo Technology China. Glycine, D-galacturonic acid, D-glucose, D-xylose, D-galactose, D-mannose, L-rhamnose, L-arabinose, konjac glucomannan, xylan, apple pectin, and citrus peel pectin were supplied by Sigma Aldrich China.Growth of microorganisms (enzyme activity and organisms)
Bacterial strains, plasmids, primers, and enzyme functions used in this study are listed in Tables S2–S4† and Table 2. Pre-culturing of the Bacillus subtilis 7-3-3 cells (isolated from the soil in Shandong University and preserved in the China Center for Type Culture Collection No. M200038) was aerobically conducted on Luria–Bertani agar plates at 37 °C for 12 h. The components of Luria–Bertani media (pH 7.2) were tryptone (10 g L−1); yeast extract (5 g L−1); and NaCl (10 g L−1).22 The media were further introduced into 5 mL of seed medium inside 15 mL screw-capped glass tubes. The components of the seed medium (pH 6.8) were tryptone (5 g L−1); yeast extract (5 g L−1); NaCl (5 g L−1); and K2HPO4 (10 g L−1); D-glucose (10 g L−1). The culture medium was supplemented with 5 μg mL−1 chloramphenicol when necessary (Sangon, China) and further cultivated by shaking in an incubator (200 rpm) at 37 °C for 12 h to produce crude enzymes. The production medium I was prepared using22 bran (54 g L−1); corn flour (42 g L−1); (NH4)2SO4 (3 g L−1); MgSO4·7H2O (2 g L−1); K2HPO4 (1 g L−1); and Na2CO3 (1 g L−1). The fermentation was initiated by inoculating 3% v/v seed media into 500 mL glass bottles containing production medium I (Table S3†). The production medium I fermentation was carried out by shaking the solution in an incubator (200 rpm) at 34 °C for 72 h. The production medium II (i.e. 2× Super-rich) contained tryptone (30 g L−1), yeast extract (50 g L−1), and K2HPO4 (6 g L−1) (Table S3†). The production medium II fermentation was carried out in an incubator (200 rpm) at 37 °C for 24 h. After the fermentation, the cell-free culture supernatant was centrifuged and used as crude enzymes for the experiment.Assay of enzyme activities
Mannase and xylanase activities were measured using konjac glucomannan and xylan, respectively, as the substrate (30 min at 50 °C). Filter paper activity was acquired for 60 min at 50 °C using Whatman filter paper (1 cm × 6 cm, 50 mg) as the substrate according to the dinitrosalicylic acid method.31 Pectate lyase activity was determined using polygalacturonic acid or apple pectin as the substrate.32 For all the enzyme activities, one unit of enzyme activity was defined as the amount of enzyme that liberated 1 μmol min−1 reducing sugars under the applied conditions. The enzyme activity was measured in glycine–NaOH buffer (pH 9.6). These water-soluble standard substrates were used to evaluate the expression level of enzymes.Bark treatments
Understanding the pectin structure is essential in designing customized enzymes for liberating the sclerenchyma fibre bundles from willow bark. For analytical separation of pectin, the willow bark (WB) was ground (1 mm mesh size) before its extraction successively with water, dichloromethane, and acetone for a complete elimination of extractives (Fig. 1a). The extract-free WB was treated with aqueous citric acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, w/v, pH 2) at 90 °C for 60 min. The slurry was first filtered through a filter membrane (pore size: 15–20 μm) and further centrifuged at 8000 rpm for obtaining a clear supernatant. The citric acid treated willow bark (CAW) was kept for further analyses. Ethanol was added to the supernatant (final ethanol concentration: 75 v/v%) and the mixture was kept under cold conditions (+4 °C) for precipitation of pectin. Centrifugation (8000 rpm) and further lyophilization were carried out to obtain freeze dried crude citric acid pectin (CAP). The crude pectin was further purified from smaller carbohydrate molecules in dialysis bags (Biosharp, cutoff 14 kDa, 96 h). Finally, the collected pectin precipitates were centrifuged and lyophilized to obtain dialyzed citric acid pectin (DCAP).
30, w/v, pH 2) at 90 °C for 60 min. The slurry was first filtered through a filter membrane (pore size: 15–20 μm) and further centrifuged at 8000 rpm for obtaining a clear supernatant. The citric acid treated willow bark (CAW) was kept for further analyses. Ethanol was added to the supernatant (final ethanol concentration: 75 v/v%) and the mixture was kept under cold conditions (+4 °C) for precipitation of pectin. Centrifugation (8000 rpm) and further lyophilization were carried out to obtain freeze dried crude citric acid pectin (CAP). The crude pectin was further purified from smaller carbohydrate molecules in dialysis bags (Biosharp, cutoff 14 kDa, 96 h). Finally, the collected pectin precipitates were centrifuged and lyophilized to obtain dialyzed citric acid pectin (DCAP).
 | ||
| Fig. 1 Experimental flow for (a) pectin isolation and (b) sclerenchyma fibre bundle separation from the willow bark. Fractions marked with dashed lines were not further investigated in this study. For abbreviations, see Table S5.† | ||
For the fibre bundle separation, the willow inner bark (WIB) was cut to 15 cm long pieces before treating with hot water (80 °C, liquid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) solid = 25
solid = 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 30 min. The hot water extract contains biologically active components that can be purified by chromatography33 and have potential use in pharmaceutics.34 Part of the hot water extracted WIB was treated with 3 wt% NaHCO3 (liquid
1) for 30 min. The hot water extract contains biologically active components that can be purified by chromatography33 and have potential use in pharmaceutics.34 Part of the hot water extracted WIB was treated with 3 wt% NaHCO3 (liquid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) solid = 15
solid = 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100 °C, 60 min) to increase the accessibility of the bark to enzymes. The solid residue (NAWIB) was preserved for enzymatic treatments while pectin (NA-P) was precipitated from the filtrate with 75 v/v% ethanol (final concentration). Enzyme treatments of WIB and NAWIB were carried out by mixing (150 rpm) 1 g of the substrate and the enzyme mixture in 50 mL of 0.2 M glycine–NaOH buffer solution (pH 9.6) in an Erlenmeyer flask at 50 °C for 4 h (the detailed components are summarized in Table S5†). The enzymatic treatments were conducted in triplicate to ensure reproducibility of the results. The slurry was filtered to separate the liquid and solid residue (fibre bundles). Pectin was isolated from the filtrate by precipitation with 75 v/v% ethanol (final concentration) overnight at 4 °C followed by centrifugation at 8000 rpm and lyophilization. The solid residue (fibre bundles) was washed with distilled water and ethanol and the gravimetric yield was determined.
1, 100 °C, 60 min) to increase the accessibility of the bark to enzymes. The solid residue (NAWIB) was preserved for enzymatic treatments while pectin (NA-P) was precipitated from the filtrate with 75 v/v% ethanol (final concentration). Enzyme treatments of WIB and NAWIB were carried out by mixing (150 rpm) 1 g of the substrate and the enzyme mixture in 50 mL of 0.2 M glycine–NaOH buffer solution (pH 9.6) in an Erlenmeyer flask at 50 °C for 4 h (the detailed components are summarized in Table S5†). The enzymatic treatments were conducted in triplicate to ensure reproducibility of the results. The slurry was filtered to separate the liquid and solid residue (fibre bundles). Pectin was isolated from the filtrate by precipitation with 75 v/v% ethanol (final concentration) overnight at 4 °C followed by centrifugation at 8000 rpm and lyophilization. The solid residue (fibre bundles) was washed with distilled water and ethanol and the gravimetric yield was determined.
Analytical methods
To determine their carbohydrate composition, the solid pectin and fibre bundle samples were hydrolyzed according to NREL/TP-510-42618.35 High-performance liquid chromatography (Shimadzu LC-20AT) was adopted to quantify the monosaccharides (galacturonic acid, glucose, xylose, galactose, mannose, rhamnose, and arabinose) in the hydrolysates of the pectin and fibre bundle samples using Biorad Aminex HPX-87P (mobile phase: 5 mM H2SO4) and Biorad Aminex HPX-87H (mobile phase ultrapure water) columns, respectively. A constant eluent flow rate of 0.2 mL min−1 was applied at room temperature. The determination of galacturonic acid content by acid hydrolysis could ultimately cause a degradation of galacturonic acid;18,36 therefore the recovery factor of galacturonic acid (59.2 ± 0.007%) was calculated and used for further calculations.Proton (1H) nuclear magnetic resonance (NMR) spectroscopy was applied to determine the degree of methylation (DM) and degree of acetylation (DA) of the pectin samples.37 They were first treated for 2 h with 0.8 mL of 0.4 M NaOH in D2O, containing 0.2 mg mL−1 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TSP-d4) as an internal standard for chemical shift (δC/δH, 0/0 ppm) calibration and quantification. After saponification, the slurry was centrifuged (Eppendorf Centrifuge 5424 R, Germany) and the supernatant was transferred to an NMR tube. 1H NMR spectra were acquired with a Bruker AVANCE III 600 MHz instrument (Bruker Biospin, USA) equipped with a cryoprobe. The following parameters were used for 1H NMR: a relaxation delay of 5 s, a spectral width of 19 ppm, 16 transients, and 64K data points.
Two-dimensional heteronuclear single quantum coherence (HSQC) spectra of the pectin samples were acquired at 22 °C with spectral widths of 19.8 ppm and 240 ppm for 1H and 13C NMR, respectively. A relaxation delay of 1.5 s, d24 delay of 0 ms and 1K data points were applied for HSQC (hsqcetgpsisp.2 pulse sequence from the Bruker Library).
Fourier transform infrared spectroscopy (FT-IR) with attenuated total reflection (ATR) (PerkinElmer, UK) was used to scan the IR absorption spectra of the pectin and fibre bundle samples within a range of 4000–500 cm−1 with an acquisition time of 30 s.
X-ray photoelectron spectroscopy (XPS) with a KRATOS AXIS SUPRA instrument was used for determining the atomic surface composition of the fibre bundles. Pure cellulose filter paper (Whatman) was used as a reference. After the measurements, atomic concentrations were calculated using CasaXPS software and an energy shift correction was done relatively to the C–C peak (284.8 eV). The XPS data processing was interpreted using Shirley background and Voigt function as a convolution of 70% Gaussian and 30% Lorentzian which is common for carbon-based samples. Table S6† contains the peak positions and full width at half maximum (FWHM) of the fit done with constraints on peak positions and FWHM, which is in full agreement with the literature.38
For the quantification of starch, the samples were treated with α-amylase and amyloglucosidase according to NREL/TP-510-42624 and the formed glucose was quantified with the dinitrosalicylic acid method.31
FEI Quanta250 FEG operating at 5.0 kV was chosen to acquire the scanning electron microscopy (SEM) images of the fibre bundles. The specimens were sputter coated with gold to increase their electrical conductivity and to avoid further charging.
A high performance size-exclusion chromatography (HP-SEC) system (L2130, HITACHI, Japan) with multiangle laser light scattering (MALLS) detection was utilized to determine the molecular weight distributions of the pectin samples. The samples were solubilized in 0.1 M NaCl and filtered through a 0.22 μm membrane prior to injection.36Mn (number-averaged molecular weight) and Mw (weight-averaged molecular weight) were measured and further calculated using the ASTRA 5.3.4 software (Wyatt Technology).
The samples prepared for the antibacterial tests for the growth inhibition of Staphylococcus aureus ATCC 25923 included cotton yarn (control), PWI-F (Fig. 1), and WB fibre bundles produced with NaHCO3 (20 wt% on WB)11 according to GB/T 20944.3-2008. Briefly, the autoclaved fabric samples (0.75 g) were inoculated with 5 mL of bacterial suspension (3–4 × 105 CFU mL−1) and 70 mL phosphate-buffered saline in a 250 mL Erlenmeyer flask under shaking (i.e. 150 rpm) at 25 °C for 18 hours. Samples (1 mL) of the suspensions withdrawn at 0 h and 18 h were serially diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and further plated using the agar pour plate method. The agar plates were incubated at 37 °C for 24 h and the colonies were counted from plates with 30–300 colonies. The antibacterial ratio Y was calculated using the formula: Y = (Wt − Qt)/Wt × 100%, where Wt and Qt (CFU mL−1) refer to the number of bacteria counted after 18 h incubation on the cotton control and test samples, respectively. The antibacterial effect was considered effective when Y ≥ 70%.
10) and further plated using the agar pour plate method. The agar plates were incubated at 37 °C for 24 h and the colonies were counted from plates with 30–300 colonies. The antibacterial ratio Y was calculated using the formula: Y = (Wt − Qt)/Wt × 100%, where Wt and Qt (CFU mL−1) refer to the number of bacteria counted after 18 h incubation on the cotton control and test samples, respectively. The antibacterial effect was considered effective when Y ≥ 70%.
Fibre bundle samples were characterized for the phase purity and crystal structure by X-ray diffraction (XRD; PANalytical X'Pert PRO MPD Alpha-1; Cu Kα1 radiation).39 The relative crystallinity index was computed using an amorphous fitting method40 applied to the scattering angle range of 13–50°. The crystalline part of the fibre bundle is modelled by using 20 Gaussian functions in the positions defined by the cellulose I beta crystal reflections (these gaussian peaks can be seen in Fig. S1† with grey dashed lines). The Segal crystallinity index41 was also calculated.
Results and discussion
Structural characteristics of pectin
Aqueous solutions of citric acid, HCl and NaOH were used to extract the pectic polysaccharides from the willow bark. As expected, the highest pectin yield (2.8%, Table S7†) was obtained with citric acid due to its ability to bind calcium ions.42 Extraction using the weak citric acid may bring a major advantage of achieving high extraction efficiency and preserving its native chemical structure compared to other strong mineral acids.18 Yet, this amount was only a fraction of the original amount of pectin in WB. The citric acid treatment solubilized approximately half of the pectin (Table 3) of which only a part was recovered by ethanol precipitation.| WB | WDAW | CAW | NAWIB | WIB | PN-F | PHN-F | PWI-F | PHWI-F | |
|---|---|---|---|---|---|---|---|---|---|
| FB yield (% on WB) | — | — | — | — | — | 19.6 (1.6) | 17.6 (1.7) | 19.4 (2.2) | 19.2 (0.4) |
| Carbohydrate composition (mg g−1 WB) | |||||||||
| Galacturonic acid | 115 (4) | 113 (6) | 65 (4) | 37 (4) | 48 (8) | 5 (1) | 5 (1) | 7 (0) | 7 (1) |
| Glucose | 302 (11) | 265 (7) | 217 (8) | 196 (18) | 165 (25) | 94 (0) | 86 (0) | 92 (0) | 88 (2) |
| Xylose | 29 (2) | 35 (2) | 29 (1) | 31 (3) | 25 (5) | 17 (1) | 13 (4) | 11 (0) | 15 (0) |
| Galactose | 23 (2) | 19 (2) | 16 (1) | 10 (1) | 10 (1) | 2.5 (0) | 1.9 (0) | 2.3 (0) | 2.7 (0) |
| Mannose | 34 (0) | 26 (2) | 18 (1) | 11 (1) | 12 (1) | 5 (1) | 9 (6) | 11 (0) | 7 (2) |
| Rhamnose | 12 (2) | 11 (1) | 8 (0) | 5 (0) | 6 (1) | 1 (0) | 2 (0) | 2 (0) | 2 (1) |
| Arabinose | 41 (2) | 39 (2) | 17 (1) | 17 (2) | 14 (2) | 2 (0) | 2 (1) | 3 (0) | 3 (1) |
| Sum | 557 | 507 | 369 | 308 | 278 | 127 | 118 | 128 | 123 |
Although purification of the isolated citric acid pectin (CA-P) by dialysis had hardly any effect on the relative monosaccharide composition, the treatment led to double degree of esterification in the sample (DCA-P) (Table 4). The molar ratio of galacturonic acid and rhamnose indicated that both HG and RG-I domains were present in significant amounts. The contents of galactose and arabinose were comparable and the high (Gal + Ara)/Rha ratio of 6.2 suggested that the RG-I domains are highly branched in the willow bark compared to the citric acid pectin from grape peel (ratio 4.1).36 No xylose (xylan) was present, but the glucose content of the sample was high. Although the RG-I domain assisted solubilization of cellulose fibrils has been presented in the literature,43 the glucose in CA-P and DCA-P seemed to originate from starch, the content of which was relatively high (1.3 wt%) in the willow bark. A similar occurrence of glucose was earlier reported for hawthorn berry pectin.44 Protein residues remain to be the dominant unknown compound in ethanol-precipitated pectin samples (Table 4) as proteins are prone to alcohol precipitation,45 and the cell wall of the willow bark has been particularly reported to be rich in protein.5 In comparison with the citric acid, the aqueous NaHCO3 treatment removed the pectin of NAWIB more completely (Table 3). However, the yield of the pectin (NA-P) precipitated by ethanol was lower, obviously due to the much lower molecular weight of NA-P compared to that of CA-P (Table 4). The alkaline extraction conditions resulted in increased content of RG-I in the recovered pectin. Moreover, significantly less glucose was present in the pectin (NA-P) extracted from WIB with aqueous NaHCO3 (Table 4).
| CA-P | DCA-P | NA-P | PN-P | PHN-P | PWI-P | PHWI-P | |
|---|---|---|---|---|---|---|---|
| Pectin yield (% WB) | 2.8 (0.2) | 1.5 (0.6) | 2.5 (0.1) | 2.1 (0.5) | 2.4 (0.3) | 2.6 (0.4) | 2.9 (0.6) |
| M w (kDa) | 263 (1) | 264 (2) | 16 (0.3) | 51 (0.5) | — | 42 (1) | — |
| M w/Mn | 5.3 (0.1) | 3.4 (0.03) | 4.3 (0.6) | 4.3 (0.2) | — | 3.6 (0.3) | — |
| Monosaccharides (mg g−1) | |||||||
| Galacturonic acid (GalA) | 455 (13.2) | 333 (29.3) | 483 (36.2) | 261 (—) | 212 (—) | 224 (—) | 255 (—) |
| Glucose | 224 (6.4) | 164 (3.6) | 40 (2.8) | 79 (—) | 87 (—) | 58 (—) | 50 (—) |
| Xylose | 0 | 0 | 0 | 41 (—) | 15 (—) | 16 (—) | 25 (—) |
| Galactose (Gal) | 122 (2.9) | 93 (11.7) | 85 (5.7) | 85 (—) | 72 (—) | 70 (—) | 60 (—) |
| Mannose | 0 | 0 | 0 | 33 (—) | 32 (—) | 13 (—) | 27 (—) |
| Rhamnose (Rha) | 37 (2.8) | 27 (2.7) | 23 (2.6) | 56 (—) | 41 (—) | 36 (—) | 41 (—) |
| Arabinose (Ara) | 111 (5.7) | 79 (8.9) | 50 (3.5) | 119 (—) | 84 (—) | 69 (—) | 73 (—) |
| Overall | 951 | 696 | 681 | 673 | 543 | 485 | 529 |
| Molar composition | |||||||
| Rha/GalA | 0.10 (0.005) | 0.10 (0.001) | 0.06 (0.002) | 0.25 (—) | 0.23 (—) | 0.19 (—) | 0.19 (—) |
| (Gal + Ara)/Rha | 6.2 (0.23) | 6.28 (0.12) | 5.76 (0.3) | 3.74 (—) | 3.90 (—) | 3.87 (—) | 3.27 (—) |
| HG (%) | 53.0 (0.6) | 53.0 (0.7) | 64.3 (0.1) | 34.2 (—) | 36.7 (—) | 42.2 (—) | 44.8 (—) |
| RG-I (%) | 47.0 (0.6) | 47.0 (0.7) | 29.6 (0.2) | 65.8 (—) | 63.3 (—) | 57.8 (—) | 55.2 (—) |
| DM (%) | 50.7 (8.0) | 93.2 (2.5) | 47.0 | 68.3 (7.1) | 82.5 (4.1) | 79.8 (1.1) | 65.4 (8.3) |
| DA (%) | 14.2 (1.9) | 27.4 (1.4) | 13.6 | 29.5 (3.2) | 36.2 (1.2) | 35.7 (2.6) | 32.8 (5.8) |
The solution-state 2D HSQC NMR spectra (Fig. 2 and Table S8†) of DCA-P revealed information on typical interunit linkages of pectin, consisting of galacturonic acid, arabinose, rhamnose and galactose. All spectra were assigned according to the literature.46–51 Three strong and well resolved signals (Fig. 2a) at δC/δH of 55.9/3.82, 23.6/2.16 and 19.6/1.26 ppm indicated the presence of methyl groups of 1,4-α-D-GalpA(OMe), 1,4-α-D-GalpA(OAc) and rhamnose, respectively. Non-anomeric methine signals of galacturonic acid (1,4-α-D-GalpA) were present at 69.3/3.8 ppm, 69.7/3.9 ppm, 74.2/4.73 and 83.2/4.27 ppm (Fig. 2b). Specific non-anomeric and anomeric (Fig. 2c) methine signals revealed the presence of terminal and 1,3-, 1,5-, 1,2,5-, 1,3,5- and 1,2,3,5-linked arabinofuranosyl groups while only terminal and 1,4-linked galactopyranosyl groups were detected. Strong starch (1,4-α-D-Glcp)48 signals were also present which may suggest that starch was covalently linked with pectin.52 The anomeric (poly)galacturonic acid signal obviously overlapped with that of starch because the pectin was not neutralized after isolation with citric acid unlike in many other studies.46 The detailed assignment of the spectral information of Fig. 2b and c is summarized in Table S8.† The HSQC spectra of other pectin samples (PN-P, PWI-P, PHWI-P and CA-P) are presented in Fig. S2.†
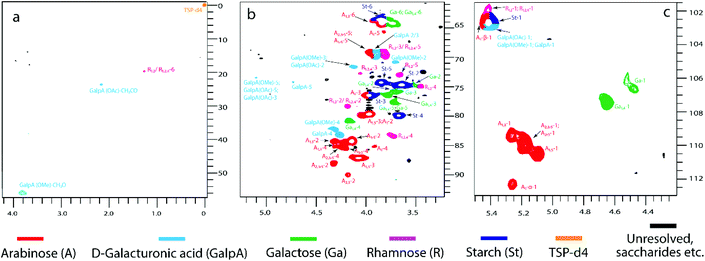 | ||
| Fig. 2 (a) Methyl, (b) non-anomeric methylene and methine and (c) anomeric methine group regions of 2D heteronuclear single quantum coherence (HSQC) NMR spectrum of dialyzed citric acid extracted pectin (DCA-P) of the willow bark (see Fig. 1). For abbreviations, see Table 5. | ||
Approximate linkage type quantification of the main neutral sugars of pectin (Table 5) was based on volume integration of the corresponding HSQC contours. The high ratio of 1,2,4- and 1,2-linked rhamnopyranosyl units indicated that RG-I domains were originally highly branched (CA-P and DCA-P). Galactopyranosyl groups were mostly 1,4-linked with relatively few terminal groups, corresponding to linear galactan side chains. The absence of branched and 1,3-linked galactopyranosyl groups showed that arabinogalactans were not present in the willow bark pectin.27 The arabinofuranosyl groups were mostly 1,5-linked although branches at O-2 and O-3 were also present. The relatively low share of terminal arabinofuranosyl residues supported the less branched arabinan side-chain structure.
| Linkages | Abbreviation | DCA-P | CA-P | NA-P | PN-P | PWI-P | PHWI-P | |
|---|---|---|---|---|---|---|---|---|
| a Integration of C1/H1 and C2/H2. b Integration of C4/H4. c Integration of C1/H1 and C4/H4. | ||||||||
| Arabinose interunit linkagesa (%) | ||||||||
| →5)-α-L-Araf-(1→ | A1.5 | 90 (3) | 87 (6) | 90 (0) | 89 (9) | 87 (6) | 93 (1) | |
| →3)-α-L-Araf-(1→ | A1.3 | 4 (1) | 2 (1) | 4 (2) | 2 (1) | 2 (1) | 1 (0) | |
| →2,3,5)-α-L-Araf-(1→ | A2.3.5 | 3 (1) | 5 (2) | 3 (1) | 5 (4) | 5 (2) | 2 (0) | |
| →2,5)-α-L-Araf-(1→ | A2.5 | 1 (0) | 2 (1) | 2 (1) | 3 (2) | 4 (2) | 2 (1) | |
| α-L-Araf-(1→ | A1-α | 2 (1) | 5 (4) | 1 (0) | 2 (1) | 2 (1) | 2 (1) | |
| Rhamnose interunit linkagesb (%) | ||||||||
| →2)-α-L-Rhap-(1→ | R1.2 | 25 (9) | 7 (1) | 90 (5) | 96 (2) | 99 (1) | 99 (1) | |
| →2,4)-α-L-Rhap-(1→ | R1.2.4 | 75 (9) | 93 (1) | 10 (5) | 4 (2) | 1 (1) | 1 (1) | |
| Galactose interunit linkagesc (%) | ||||||||
| β-D-Galp-(1→ | Ga | 14 (2) | 22 (11) | 89 (9) | 4 (3) | 4 (0) | 6 (1) | |
| →4)-β-D-Galp-(1→ | Ga1.4 | 86 (2) | 78 (11) | 11 (9) | 96 (3) | 96 (0) | 94 (1) | |
Enzyme consortium design based on the pectin structure
Pectinases show specific catalytic activity in degrading pectins depending on their structural features (Table 2), such as the degree of methylation of HG domains. The high proportion of RG-I domains and their arabinan and galactan side chains are characteristic of willow bark pectin, and therefore arabinanase and arabinogalactanase were speculated to promote pectin degradation. However, arabinogalactanase was not used here because of its low optimal pH (4–6).27A preliminary study was carried out to evaluate the effectiveness of individual natural and engineered pectinases and their combinations in the treatment of WIB. The gravimetric yields of the treatments and chemical characteristics of the obtained pectin and the solid residue (fibre bundles) were compared after each pectinase and blank treatment (Table S9†). A high yield of ethanol precipitated pectin and a low yield of fibre bundles were considered markers of an effective treatment. Pectate lyase B (PelB), which is selective towards highly methylated pectin, was less effective than pectin lyase A (PelA) and pectin lyase C (PelC), and the combination of PelA and PelC activities (PelAPelC) resulted in most comprehensive removal of pectins, similar to the synergetic action of PelA and PelC in degumming ramie.22 PelB and PelAPelC were used together (Table 1) for achieving the synergistic action for depolymerizing pectin having an unevenly distributed methylation degree.46 Arabinanase (AbnA) had a small catalytic effect over a blank treatment and also when used with PelA. Rhamnogalacturonan endolyase (RhgW) showed an almost negligible effect of depolymerizing the backbone of the RG-I domain based on a blank treatment using RhgW alone and also when used together with PelA, which may indicate that the RhgW cannot even reach the substrate (i.e. RG-I backbone) as the RG-I domains of willow bark pectin are highly branched.
Furthermore, possible covalent linkages (complexes) between pectin (or the side chains of RG-I domains), lignin and hemicelluloses5 might contribute to the recalcitrance of the matrix. Because xylan and glucomannan were the main hemicelluloses of the willow bark (Table 3), xylanase (XynA) and mannanase (GmuG) were used to potentially assist in the depolymerization of the covalent complexes15 and increase the accessibility of the cell wall. Thus, based on the initial screening, a mixture of PelAPelC, PelB and AbnA was selected for further pectinase treatments and XynA and GmuG were added into the enzyme mixture (Table 1) in combined pectinase and hemicellulase treatments (Table S5†).
Enzymatic degumming of the willow bark
The yield and characteristics of the ethanol precipitated pectin from each pectinase (PN-P and PWI-P) and combined pectinase and hemicellulase treatment (PHN-P and PHWI-P) are summarized in Table 4. The characteristic sugars (galacturonic acid, arabinose, rhamnose, and galactose) of pectin were present in all samples which proved the success of the enzyme mixtures used. A pretreatment with NaHCO3 resulted in a 2.5 wt% pectin (NA-P) yield, and a similar amount of pectin was recovered after subsequent treatments with pectinases (PN-P) or pectinases and hemicellulases (PHN-P), corresponding to an overall pectin yield of 4.6–4.9 wt%. On the other hand, the approach without alkaline pretreatment (PWI-P and PHWI-P) yielded 2.6–2.9 wt% pectin, similar to CA-P (2.8 wt%). The simultaneous use of hemicellulases with pectinases resulted in a slightly higher yield of pectin without affecting much its hemicellulose sugar (xylose and mannose) content. In contrast, xylose and mannose were not present in the pectin released by NaHCO3.The enzyme treatments did not really decrease the degree of methylation or acetylation of the pectin (Table 4), which was confirmed qualitatively through the respective methyl signals in the 2D HSQC NMR spectra (Fig. S2†). In most cases, the mildly alkaline pretreatment slightly decreased the DM and DA of the pectin because of the competing hydrolysis and β-elimination reactions.13 Although it yielded extensively degraded pectin, the pretreatment did not affect the molecular weight of the enzymatically released pectin. While NaHCO3 increased the share of HG in the recovered pectin, the pectinase treatments led to enrichment of RG-I domains in the solubilized polymers. Some changes were also observed in the linkage patterns of the neutral sugars (Table 5). Most remarkably, branched rhamnopyranosyl units were scarce after the pectinase treatments and the number of terminal galactopyranosyl units was reduced.
While the FTIR spectra of the purified pectin DCA-P and commercial citrus pectin were very similar, the pectin samples PN-P and PWI-P had strong additional bands at ca. 1600, 1500, 1410 and 1330 cm−1 (Fig. 3). These signals indicated the coprecipitation of glycine (buffer)53,54 which is sparingly soluble in ethanol. A 13C/1H correlation peak at 44.5/3.5 ppm in the HSQC spectra of PN-P, PWI-P and PHWI-P (Fig. S2†) confirmed the presence of glycine. The same signal was absent in the HSQC spectra of CA-P (Fig. S2†) and DCA-P (Fig. 2) which were obtained without using the glycine buffer. The absorption band at 1735 cm−1 confirmed the presence of ester groups in DCA-P, PN-P and PWI-P.36,42,55 In CA-P the absorption maximum was shifted to 1725 cm−1, which could be explained by some coprecipitation of citric acid/citrate. A 13C/1H correlation peak at 47/2.8 ppm in the HSQC spectrum of CA-P (Fig. S2†) confirmed the presence of citric acid/citrate53,54 in the impure citric acid extracted pectin. The intensity of the ester band at 1735 cm−1 was relatively weak for PN-P and PWI-P due to their contamination by the glycine buffer and unidentified carbohydrates that gave additional signals in the NMR spectra (Fig. S2†).
 | ||
| Fig. 3 FTIR spectra of commercial citrus pectin and ethanol-precipitated pectin samples obtained after treatments of the willow bark with citric acid (CA-P), citric acid followed by dialysis (DCA-P), pectinases (PWI-P) and NaHCO3 followed by pectinases (PN-P). For further clarification of the abbreviations see Table S5† and Fig. 1. | ||
Generally, the fibre bundle yield after the enzymatic treatments (Table 3) was close to the previously reported yield in NaHCO311 and NaOH12 treatments. Almost complete absence of galacturonic acid (GalA) in the fibre bundles demonstrated the efficiency in their separation with the pectinases. Similarly, the content of neutral pectic sugars (arabinose, rhamnose and galactose) was significantly reduced. For comparison, almost half of GalA remained in the solid residue after treating the bark with citric acid. The additional effect of hemicellulases on the yield and composition of the fibre bundles was minor and within experimental error. The effect of the NaHCO3 pretreatment was also small, and surprisingly, the pectinase consortium alone was sufficient to separate the fibre bundles without any prior chemical treatment. Interestingly, the crystallinity of the fibre bundles after the entire enzymatic treatments (PWI-F) was increased compared to the fibre bundles under the treatment of aqueous NaHCO3 followed by pectinases (i.e. PN-F) shown in Table 6 and Fig. S3,† which confirms that the Ca2+/pectin crossbridge may play the role of preventing the coalescence of cellulose fibre bundles in the cell wall.56,57 Overall, the increase in the crystallinity index could possibly be attributed to an increase of semicrystalline cellulose in the fibre bundles due to the quantitative removal of pectin shown in Table 4; the effect of the crystallinity index on the macroscopic properties and applicability of the enzymatically treated fibre bundles is unknown and this is out of scope of this present study. Fig. 4 and Fig. S4–S7† show SEM images of the fibre bundles after the enzymatic treatments. The surfaces of the bundles appeared to be smoother and cleaner than the surfaces of the chemically separated fibre bundles.11
 | ||
| Fig. 4 SEM images of fibre bundles isolated from the willow bark after treatment with (a) aqueous NaHCO3 followed by pectinases (PN-F), (b) aqueous NaHCO3 followed by pectinases and hemicellulases (PHN-F), (c) pectinases (PWI-F) and (d) pectinases and hemicellulases (PHWI-F). For further clarification of the abbreviations, see Table S5† and Fig. 1. | ||
| Sample | Relative CrI (SD) | Segal CI |
|---|---|---|
| PWI-F | 0.28 (0.03) | 0.58 |
| PN-F | 0.25 (0.03) | 0.48 |
High-resolution X-ray photoelectron spectroscopy (XPS) of acetone-extracted fibre bundles provided data on the atomic surface (top 10 nm) composition of the obtained fibre bundles (Fig. 5 and Table S10†). The share of the binding energy component C–C (284.8 eV) was used as a quantitative marker of the surface lignin content.58 The surface lignin content of the fibre bundles was estimated to be ca. 20 and 10%, respectively, after the treatment with pectinases alone (PN-F and PWI-F) and pectinases and hemicellulases together (PHN-F and PHWI-F). For comparison, a treatment with aqueous NaHCO3 yielded fibre bundles with ca. 40% surface lignin content although condensed polyphenolic tannins could possibly contribute to the surface ‘lignin’ content.11 Anyway, the enzymatic approach not only facilitated the isolation of fibre bundles but also purification of their surfaces from lignin or its complexes with pectin and hemicelluloses. On the other hand, the cleaner pectinase treated fibre bundles (PWI-F) did not provide as excellent antimicrobial protection against Staphylococcus aureus ATCC 29213 as the NaHCO3 (20 wt% dosage) treated fibre bundles did (the antibacterial efficiencies were 85.0 and 96.5%, respectively) (Fig. S8†). In addition, the pectinase (and hemicellulases) treated fibre bundles lacked the characteristic reddish hue of the NaHCO3 treated fibre bundles (Table S11 and Fig. S9†).11
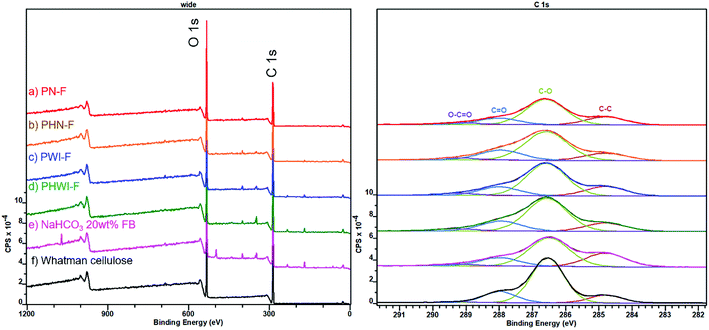 | ||
| Fig. 5 XPS spectra of fibre bundles isolated from the willow bark after treatment with (a) aqueous NaHCO3 followed by pectinases (PN-F), (b) aqueous NaHCO3 followed by pectinases and hemicellulases (PHN-F), (c) pectinases (PWI-F), (d) pectinases and hemicellulases (PHWI-F) and (e) aqueous NaHCO3 (20 wt% dosage).11 The spectrum of pure cellulose (f) is included as a reference. For further clarification of the abbreviations see Table S5† and Fig. 1. | ||
Overall, the molecular weight changes (Table 4, Table S12 and Fig. S10†) induced by the enzyme treatment strongly indicate the effectiveness of the designed consortium in depolymerizing the pectin macromolecules into molecular fragments. The XPS, brightness, and antibacterial activity results suggest that the functionalities of the fibre bundles can be maximally protected through the entire biochemical approach.
Conclusions
Although this study provided completely new biological information on full valorisation of pure fibre bundles from the willow bark, it is the success in elucidating the structural features of the willow bark pectin that opens new interesting scenarios for designing tailor-made biochemical degradation strategies for lignocellulosic biomass. The knowledge on the pectin structure was, and continues to be, important in designing tailored enzymes and biotechnical processes for the solubilization of the willow bark pectin. Surprisingly, the customized pectinases were not only able to fully separate the fibre bundles but also modify their surfaces without any chemical treatment. The bark fibre bundles with their unique antibacterial properties could potentially find use in specific applications. In comparison with the commercially most important wood species, the fast-growing willow is still a small crop that does not fit into the scale of chemical pulp mills. However, its unique extractive profile, excellent fibres, etc. make the willow bark an attractive fraction in the manufacture of special products. The debarked willow wood could be used like any wood for different products that would fit into the concept of a mini-mill that would not require expensive chemical recovery processes.With the aid of hemicellulases the surface lignin content of the sclerenchyma fibres reached an abnormally low level of 10%. Possibly the enzymes were able to degrade the native lignin-carbohydrate complexes on the fibre surfaces, which remains an interesting question to be clarified and applied in other connections. Further effort is required to gain insights into the chemical features of hemicellulose from willow bark and then the tailored hemicellulases will be employed maximally together with pectinases so that the screening efforts are reduced and the accuracy of effectiveness for the biocatalysts is improved, which is more advantageous. The response surface methodology (RSM) will then be systematically conducted to evaluate and identify the best enzyme consortium compositions of pectinases and hemicellulases in segregating sclerenchyma fibre bundles from the willow bark. Because of the specific catalytic activity of the enzyme, there has been no such “tailored pectinase strategy” reported for any lignocellulosic biomass according to the chemical features of their substrate (i.e. pectin), including grass fibres like ramie or flax. Most likely the knowledge and concept presented in this paper could be replicated and further applied for the bark of other pectin-rich wood species.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors would like to thank Yingjie Guo and Haiyan Sui from Shandong University for their skilful assistance in HPLC and NMR analyses, respectively. Appreciation is also extended to Leena-sisko Johansson and Zborowski Charlotte from Aalto University for the XPS interpretation. This work made use of the NMR and XPS premises from Shandong University. This work also made use of the RawMatTERS Finland infrastructure (RAMI) facilities based at Aalto University. The authors would like to give a special acknowledgement for the support from Svedström Kirsi from the Department of Physics, University of Helsinki for the interpretation of crystallinity analysis results. This study was partly supported by the National Key Research and Development Program of China (2019YFC1905902). This work was a part of the Academy of Finland's Flagship Programme under Projects No. 318890 and No. 318891 (Competence Center for Materials Bioeconomy, FinnCERES).References
- F. H. Isikgor and C. R. Becer, Polym. Chem., 2015, 6, 4497–4559, 10.1039/C5PY00263J.
- M. Shahid, F. Mohammad, G. Chen, R. C. Tang and T. Xing, Green Chem., 2016, 18, 2256–2281, 10.1039/C6GC00201C.
- I. Kataeva, M. B. Foston, S. J. Yang, S. Pattathil, A. K. Biswal, F. L. Poole II, M. Basen, A. M. Rhaesa, T. P. Thomas, P. Azadi, V. Olman, T. D. Saffold, K. E. Mohler, D. L. Lewis, C. Doeppke, Y. Zeng, T. J. Tschaplinski, W. S. York, M. Davis, D. Mohnen, Y. Xu, A. J. Ragauskas, S. Y. Ding, R. M. Kelly, M. G. Hahn and M. W. W. Adams, Energy Environ. Sci., 2013, 6, 2186–2195, 10.1039/C3EE40932E.
- C. Xu, R. A. D. Arancon, J. Labidi and R. Luque, Chem. Soc. Rev., 2014, 43, 7485–7500, 10.1039/C4CS00235K.
- J. Dou, H. Kim, Y. Li, D. Padmakshan, F. Yue, J. Ralph and T. Vuorinen, J. Agric. Food Chem., 2018, 66, 7294–7300, DOI:10.1021/acs.jafc.8b02014.
- D. M. Neiva, J. Rencoret, G. Marques, A. Gutiérrez, J. Gominho, H. Pereira and J. C. del Río, ChemSusChem, 2020, 13, 4537–4547, DOI:10.1002/cssc.202000431.
- M. L. Normand, B. Rietzler, F. Vilaplana and M. Ek, Polymers, 2021, 13, 1106, DOI:10.3390/polym13071106.
- R. Toman, Š. Karácsonyi and M. Kubačková, Carbohydr. Res., 1975, 43, 111–116, DOI:10.1016/S0008-6215(00)83977-4.
- L. Valentín, B. Kluczek-Turpeinen, S. Willför, J. Hemming, A. Hatakka, K. Steffen and M. Tuomela, Bioresour. Technol., 2010, 101, 2203–2209, DOI:10.1016/j.biortech.2009.11.052.
- K. Thor, Front. Plant Sci., 2019, 10, 440, DOI:10.3389/fpls.2019.00440.
- J. Dou, M. Rissanen, P. Ilina, H. Mäkkylä, P. Tammela, S. Haslinger and T. Vuorinen, Ind. Crops Prod., 2021, 164, 113387, DOI:10.1016/j.indcrop.2021.113387.
- J. Dou, J. Paltakari, L. S. Johansson and T. Vuorinen, ACS Sustainable Chem. Eng., 2019, 7, 2964–2970, DOI:10.1021/acssuschemeng.8b04001.
- A. Ng and K. W. Waldron, J. Sci. Food Agric., 1997, 73, 503–512 CrossRef CAS.
- C. J. Knill and J. F. Kennedy, Carbohydr. Polym., 2003, 51, 281–300, DOI:10.1016/S0144-8617(02)00183-2.
- D. Lairez, B. Cathala, B. Monties, F. Bedos-Belval, H. Duran and L. Gorrichon, Biomacromolecules, 2005, 6, 763–774, DOI:10.1021/bm049390y.
- X. Kang, A. Kirui, M. C. D. Widanage, F. Mentink-Vigier, D. J. Cosgrove and T. Wang, Nat. Commun., 2019, 10, 347, DOI:10.1038/s41467-018-08252-0.
- E. Donev, M. L. Gandla, L. J. Jönsson and E. J. Mellerowicz, Front. Plant Sci., 2018, 9, 1537, DOI:10.3389/fpls.2018.01537.
- F. Dranca and M. Oroian, Food Res. Int., 2018, 113, 327–350, DOI:10.1016/j.foodres.2018.06.065.
- Y. H. Percival Zhang, M. E. Himmel and J. R. Mielenz, Biotechnol. Adv., 2006, 24, 452–481, DOI:10.1016/j.biotechadv.2006.03.003.
- P. Fan, F. He, Y. Yang, M. Ao, J. Ouyang, Y. Liu and L. Yu, Biochem. Eng. J., 2015, 97, 50–58, DOI:10.1016/j.bej.2014.12.010.
- T. Shu, Y. Bai, Y. Wang, H. Wang, P. Li, M. Xiang, T. Yu, H. Xu and L. Yu, J. Cleaner Prod., 2020, 276, 124217, DOI:10.1016/j.jclepro.2020.124217.
- M. Zou, X. Li, J. Zhao and Y. Qu, PLoS One, 2013, 8, e79357, DOI:10.1371/journal.pone.0079357.
- S. Alix, L. Lebrun, S. Marais, E. Philippe, A. Bourmaud, C. Baley and C. Morvan, Carbohydr. Polym., 2012, 87, 177–185, DOI:10.1016/j.carbpol.2011.07.035.
- R. Thakur, C. R. Sarkar and R. Sarmah, Indian J. Fibre Text. Res., 1999, 24, 276–278 CAS.
- J. Dou, D. V. Evtuguin and T. Vuorinen, J. Agric. Food Chem., 2021, 69, 10848–10855, DOI:10.1021/acs.jafc.1c04112.
- J. Dou, L. Galvis, U. Holopainen-Mantila, M. Reza, T. Tamminen and T. Vuorinen, ACS Sustainable Chem. Eng., 2016, 4, 3871–3876, DOI:10.1021/acssuschemeng.6b00641.
- A. G. J. Voragen, G. J. Coenen, R. P. Verhoef and H. A. Schols, Struct. Chem., 2009, 20, 263, DOI:10.1007/s11224-009-9442-z.
- M. Soriano, P. Diaz and F. I. J. Pastor, Microbiology, 2006, 152, 617–625, DOI:10.1099/mic.0.28562-0.
- M. Soriano, A. Blanco, P. Díaz and F. I. J. Pastor, Microbiology, 2000, 146, 89–95, DOI:10.1099/00221287-146-1-89.
- M. Zou, X. Li, W. Shi, F. Guo, J. Zhao and Y. Qu, Process Biochem., 2013, 48, 1143–1150, DOI:10.1016/j.procbio.2013.05.023.
- M. Kapoor, Q. K. Beg, B. Bhushan, K. S. Dadhich and G. S. Hoondal, Process Biochem., 2000, 36, 467–473, DOI:10.1016/S0032-9592(00)00238-7.
- Z. Wang, Y. Wang, D. Zhang, J. Li, Z. Hua, G. Du and J. Chen, Bioresour. Technol., 2010, 101, 1318–1323, DOI:10.1016/j.biortech.2009.09.025.
- J. Dou, J. Heinonen, T. Vuorinen, C. Xu and T. Sainio, Sep. Purif. Technol., 2021, 261, 118247, DOI:10.1016/j.seppur.2020.118247.
- K. K. Kesari, A. Dhasmana, S. Shandilya, N. Prabhakar, A. Shaukat, J. Dou, J. M. Rosenholm, T. Vuorinen and J. Ruokolainen, Antioxidants, 2020, 9, 552, DOI:10.3390/antiox9060552.
- J. B. Sluiter, R. O. Ruiz, C. J. Scarlata, A. D. Sluiter and D. W. Templeton, J. Agric. Food Chem., 2010, 58, 9043–9053, DOI:10.1021/jf1008023.
- J. Cui, W. Ren, C. Zhao, W. Gao, G. Tian, Y. Bao, Y. Lian and J. Zheng, Carbohydr. Polym., 2020, 229, 115524, DOI:10.1016/j.carbpol.2019.115524.
- J. Müller-Maatsch, A. Caligiani, T. Tedeschi, K. Elst and S. Sforza, J. Agric. Food Chem., 2014, 62, 9081–9087, DOI:10.1021/jf502679s.
- T. R. Gengenbach, G. H. Major, M. R. Linford and C. D. Easton, J. Vac. Sci. Technol., A, 2021, 39, 013204, DOI:10.1116/6.0000682.
- H. S. Kousar, D. Srivastava, M. Karppinen and G. C. Tewari, J. Phys.: Condens. Matter, 2019, 31, 405704, DOI:10.1088/1361-648X/ab27b0.
- P. Ahvenainen, I. Kontro and K. Svedström, Cellulose, 2016, 23, 1073–1086, DOI:10.1007/s10570-016-0881-6.
- S. Nam, A. D. French, B. D. Condon and M. Concha, Carbohydr. Polym., 2016, 135, 1–9, DOI:10.1016/j.carbpol.2015.08.035.
- J. Yang, T. Mu and M. Ma, Food Chem., 2018, 244, 197–205, DOI:10.1016/j.foodchem.2017.10.059.
- A. W. Zykwinska, M. J. Ralet, C. D. Garnier and J. J. Thibault, Plant Physiol., 2005, 139, 397–407, DOI:10.1104/pp.105.065912.
- L. Roman, M. Guo, A. Terekhov, M. Grossutti, N. P. Vidal, B. L. Reuhs and M. M. Martinez, Food Hydrocolloids, 2021, 113, 106476, DOI:10.1016/j.foodhyd.2020.106476.
- H. Yoshikawa, A. Hirano, T. Arakawa and K. Shiraki, Int. J. Biol. Macromol., 2012, 50, 865–871, DOI:10.1016/j.ijbiomac.2011.11.005.
- E. N. Makarova and E. G. Shakhmatov, Carbohydr. Polym., 2020, 246, 116544, DOI:10.1016/j.carbpol.2020.116544.
- E. G. Shakhmatov, P. V. Toukach, E. A. Michailowa and E. N. Makarova, Carbohydr. Polym., 2014, 113, 515–524, DOI:10.1016/j.carbpol.2014.07.037.
- H. N. Cheng and T. G. Neiss, Polym. Rev., 2012, 52, 81–114, DOI:10.1080/15583724.2012.668154.
- F. Dourado, S. M. Cardoso, A. M. S. Silva, F. M. Gama and M. A. Coimbra, Carbohydr. Polym., 2006, 66, 27–33, DOI:10.1016/j.carbpol.2006.02.020.
- G. L. Sassaki, M. Guerrini, R. V. Serrato, A. P. S. Filho, J. Carlotto, F. Simas-Tosin, T. R. Cipriani, M. Iacomini, G. Torri and P. A. J. Gorin, Carbohydr. Polym., 2014, 104, 34–41, DOI:10.1016/j.carbpol.2013.12.046.
- H. Y. Y. Yao, J. Q. Wang, J. Y. Yin, S. P. Nie and M. Y. Xie, Food Res. Int., 2021, 143, 110290, DOI:10.1016/j.foodres.2021.110290.
- D. Gawkowska, J. Cybulska and A. Zdunek, Polymers, 2018, 10, 762, DOI:10.3390/polym10070762.
- E. L. Ulrich, H. Akutsu, J. F. Doreleijers, Y. Harano, Y. E. Ioannidis, J. Lin, M. Livny, S. Mading, D. Maziuk, Z. Miller, E. Nakatani, C. F. Schulte, D. E. Tolmie, R. K. Wenger, H. Yao and J. L. Markley, Nucleic Acids Res., 2008, 36, 402–408, DOI:10.1093/nar/gkm957.
- T. Öman, M. B. Tessem, T. F. Bathen, H. Bertilsson, A. Angelsen, M. Hedenström and T. Andreassen, BMC Bioinf., 2014, 15, 413, DOI:10.1186/s12859-014-0413-z.
- L. Monfregola, M. Leone, V. Vittoria, P. Amodeo and S. D. Luca, Polym. Chem., 2011, 2, 800–804, 10.1039/C0PY00341G.
- J. D. Crowe, P. Hao, S. Pattathil, H. Pan, S. Ding, D. B. Hodge and J. K. Jensen, Front. Plant Sci., 2021, 12, 737690, DOI:10.3389/fpls.2021.737690.
- J. Berglund, D. Mikkelsen, B. M. Flanagan, S. Dhital, S. Gaunitz, G. Henriksson, M. E. Lindström, G. E. Yakubov, M. J. Gidley and F. Vilaplana, Nat. Commun., 2020, 11, 4692, DOI:10.1038/s41467-020-18390-z.
- L. S. Johansson and J. M. Campbell, Surf. Interface Anal., 2004, 36, 1018–1022, DOI:10.1002/sia.1827.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2gc00188h |
| This journal is © The Royal Society of Chemistry 2022 |
