Direct carbon capture for the production of high-performance biodegradable plastics by cyanobacterial cell factories†
Chunlin
Tan
,
Fei
Tao
* and
Ping
Xu
 *
*
The State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic and Developmental Sciences and School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China. E-mail: taofei@sjtu.edu.cn; pingxu@sjtu.edu.cn; Fax: +86-21-34206723; Tel: +86-21-34204066
First published on 5th April 2022
Abstract
Plastic pollution caused by non-biodegradable plastics is one of the most widely discussed and notable challenges of the 21st century. Developing biodegradable plastics, such as polylactic acid (PLA), is broadly accepted as the ultimate way of alleviating this problem. However, current PLA production technology relies heavily on food-producible materials such as corn, which leads to competition for resources between material production and food supply. Carbon dioxide whose excessive emission aggravates global warming is an abundant underexplored carbon resource. Herein, we developed a cyanobacterial cell factory for the de novo biosynthesis of PLA directly from CO2, using combinational strategies of metabolic engineering and high-density cultivation (HDC). The heterologous pathway for PLA production, which involves engineered D-lactic dehydrogenase, propionate CoA-transferase, and polyhydroxyalkanoate synthase, was introduced into Synechococcus elongatus PCC7942. Subsequently, various metabolic engineering strategies, including promoter optimization, acetyl–CoA self-circulation, and carbon-flux redirection, were systematically applied, resulting in an approximately 19-fold increase of PLA to 15.0 mg g−1 dry cell weight (DCW) compared to the control. Finally, the PLA titer of 108.0 mg L−1 was obtained after using the HDC strategy, approximately 270 times higher than that obtained from the initially constructed strain. Surprisingly, the molecular weight (Mw, 62.5 kDa; Mn, 32.8 kDa) of our cyanobacterial PLA is among the highest reported levels, which is superior for commercialization. This study sheds light on the prospects of autotrophic plastic production from CO2 using cyanobacterial cell factories.
Introduction
Nowadays, the most widely used plastics produced from fossil fuels are non-degradable, causing substantial environmental damage, called plastic pollution.1–3 A total of 6.3 billion tons of plastic waste was generated from 1950 to 2015, of which 9% was recycled, 12% was incinerated, and the remaining 79% was either stored in landfills or directly released into the environment.4 Plastic pollution has become one of the most pressing environmental issues, as the rapidly increasing production of disposable plastic products has overwhelmed the world's ability to deal with them. Therefore, there is an urgent need to develop biodegradable plastics such as polylactic acid (PLA), polyhydroxybutyrate (PHB), and polyε-caprolactone (PCL).5 PLA is a non-natural polyester and is considered to be one of the most promising “green plastics” because of its several outstanding properties, such as biocompatibility, biodegradability, and high mechanical strength.6–8 PLA has been widely used in disposable plastic products. Moreover, PLA can also be extensively utilized in chemical, medical, pharmaceutical, and 3D printing industries.1 It is now increasingly recognized that PLA polyesters will play a key role in solving the problem of plastic pollution.9So far, PLA has mainly been produced via a chemical process consisting of ring-opening polymerization of lactide (a ring dimer of lactic acid) or direct polymerization of lactic acid.8,10 Unfortunately, the lactide and lactic acid production methods require sugar-based feedstocks, leading to competition for resources between PLA production and food supplies.11,12 Therefore, it is essential to use non-edible materials as feedstocks for PLA production. Excessive CO2 emissions aggravate global warming and climate change due to industrial activities and the continuous use of fossil fuels.13 Meanwhile, CO2 is also one of the most abundant natural carbon resources and has recently been recognized as the ideal feedstock for third-generation biorefineries.5 Therefore, it would be wise to produce PLA directly from CO2.
Cyanobacteria have emerged as an essential microbial chassis since they can produce value-added chemicals utilizing only sunlight and CO2.14,15 These value-added chemicals include lactic acid,11,16,17 1,3-propanediol,18 and 2-phenylethanol.19 However, the production concentrations of such low-molecular-weight chemicals by cyanobacteria are usually low, which is the most significant barrier to the commercialization of cyanobacterial cell factories. For example, D-lactic acid reached the highest titer of 226.6 g L−1 using heterotrophic B. licheniformis MW3,20 while the relatively lower titer of 1.31 g L−1 was obtained using an autotrophic cyanobacterium.11 It is also time-consuming, laborious, and costly to harvest and purify these water-soluble low-molecular-weight chemicals at such low concentrations, thus restricting commercialization. It is well known that the cell density of cyanobacteria is relatively lower than that of other heterotrophic bacteria such as E. coli.21 For example, in the case of S. elongates PCC7942, the cell density at OD730 nm was approximately 2.3 within ten days when cultured in the BG11 medium.11,22 The low cell density is considered to be the main reason behind the low productivity of cyanobacteria.
Notably, microbial cells make up only a tiny proportion of the culture broth volume, and the cellular inclusion granules can easily be harvested together with the cells.23 Moreover, the harvesting technique of flocculation can provide a low-energy, cost-effective, and environmentally friendly option to harvest cyanobacterial cells.24 Therefore, enabling product accumulation inside cyanobacterial cells would be an effective way to bypass the barrier of low titer. The pathway for the biosynthesis of PLA in vivo has been established in various hosts such as E. coli, Yarrowia lipolytica, and Saccharomyces cerevisiae,3,8,25–29 and involves propionyl–CoA transferase (PCT) converting lactic acid into lactyl–CoA, and polyhydroxyalkanoate (PHA) synthase polymerizing D-lactyl–CoA to produce PLA.27 The PLA homopolymer and its copolymer poly-3HB-co-LA were produced in the engineered E. coli.26,28,30 Subsequently, Lajus et al. engineered the yeast Yarrowia lipolytica for poly (D-lactic acid) (PDLA) production, yielding 26 mg g−1-DCW of PDLA.8 The yeast Saccharomyces cerevisiae was then engineered to produce the PLA and P(LA-co-3HB) copolymer, yielding up to 0.73% PDLA of the cell dry weight.29 Overall, it would be both promising and feasible to produce PLA in cyanobacteria.
It is broadly accepted that the insufficient availability of light and/or CO2 is responsible for the low growth rate and cell density of cyanobacteria.31 With the development of LED technology, controllable and strong light can be provided in photoreactors.32 Meanwhile, concentrated CO2 is also readily available from the waste gas of power plants and steel plants, which can be used as the carbon source for cyanobacteria.33 Therefore, it is worth using both customizable light and concentrated CO2 to achieve high-density cultivation (HDC),34 which would be crucial for improving cyanobacterial productivity.35
In this study, we developed a cyanobacterial cell factory for PLA production directly from CO2, using a combination of metabolic engineering and HDC. Multi-omics analysis was carried out to explore the factors affecting the PLA production. By optimizing expression levels of the key enzymes (the PCT and PHA synthases), and simultaneously knocking down four genes for redirecting carbon flux to the PLA biosynthesis using small RNAs (sRNAs), the production of PLA was significantly increased. Then, the production of PLA was further improved using the HDC strategy. Flocculation was also used to simplify the product harvest. In total, the production of PLA was increased by 270-fold, compared to the initially constructed strain, yielding 108.0 mg L−1 PLA (corresponding to 23.0 mg g−1 DCW).
Results
Engineering of cyanobacterial cell factories for the production of PLA
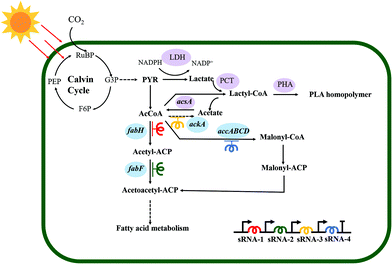
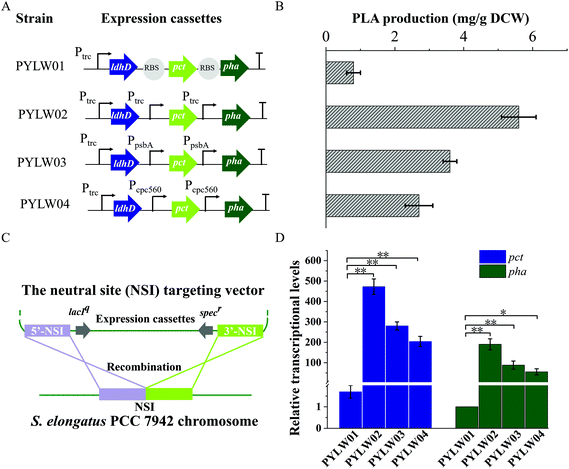
Herein, an autotrophic microbial cell factory was first constructed for the de novo biosynthesis of PLA from CO2, shown in Fig. 1. The cassette of biosynthesis for PLA consists of D-lactate dehydrogenase (LDH, encoded by ldhD), propionate CoA-transferase (PCT, encoded by pct), and PHA synthase (PHA, encoded by pha). Cyanobacteria produce NADPH as the primary carrier for reducing equivalents.11 Then, LDH was engineered to switch the coenzyme specificity from NADH to NADPH, thus making it more efficient for producing D-lactate (this work was done previously11). Subsequently, the engineered ldhD, the evolved pct and pha, which came from Lactobacillus bulgaricus,11Clostridium propionicum, and Pseudomonas sp.26 respectively, were cloned into the plasmid pAM-MCS12 under the control of inducible promoter Ptrc. The resulting plasmid pAM-PYLW01 was used for recombination at the neutral site I (NSI) of the S. elongatus chromosomes (Fig. 2A and C). Correct chromosome integration was confirmed by colony PCR (Fig. S1†) and DNA sequencing.Characterization of PLA synthesized by engineered S. elongatus
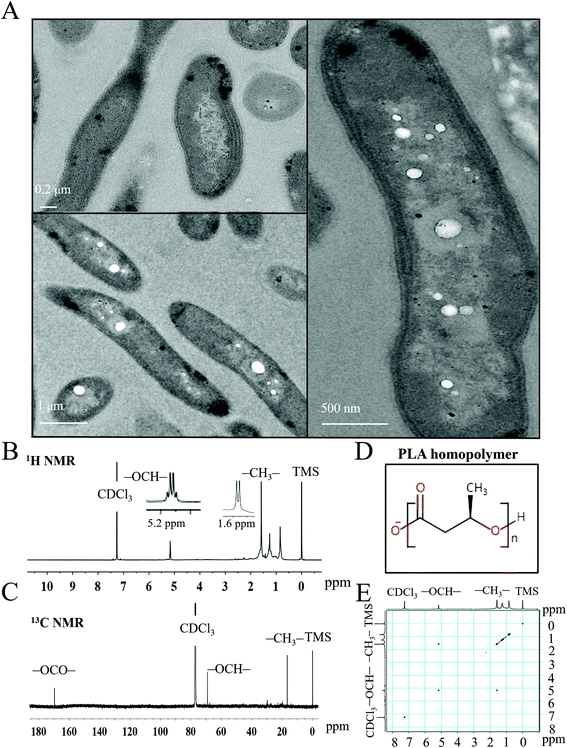
To characterize the PLA granules synthesized by engineered S. elongatus, we conducted the transmission electron microscopy analysis using the PYLW01 strain. The PLA polymers accumulated as inclusion granules in engineered S. elongatus cells (bottom left and right panels of Fig. 4A) were significantly different from the wild type (top left panel of Fig. 4A). This result is consistent with a previous study.28 1D (1H and 13C) and 2D (1H–1H) COSY NMR spectroscopy was performed to test the composition and monomer sequence of the inclusion granules. As shown in Fig. 4B, the 600 MHz 1H spectrum displays the specific quadruplet of PLA homopolymers; the oxymethine proton (–OCH–) and methyl protons (–CH3) of PLA are assigned at 5.2 and 1.6 ppm in the 600 MHz 1H spectrum, respectively. Moreover, the 1H–1H COSY spectrum shows that proton signals at 5.2 ppm (–OCH–) and 1.6 ppm (–CH3) are coupled to each other, while other peaks are not associated with a different one (Fig. 4C and E). In the 13C NMR spectrum, carbonyl carbon (–OCO–), oxymethylene carbon (–OCH–), and methyl carbon (–CH3) signals typically occur in the region of δ 170, δ 69, and δ 17, respectively, representing the configurational structure of PLA. This result shows that the product is the PLA polymer (Fig. 4D), which is consistent with a previous study.28 The molecular weight of PLA was determined by both the GPC and MALDI-TOF mass spectroscopy (Fig. S2 and S3†).36 From the GPC spectrum analysis, the average molecular weight (Mw) is 62.5 kDa, and the polydispersity of 1.9 (Fig. S2†). The result is consistent with the MALDI-TOF mass spectrum (Fig. S3†).Promoter optimization for the PLA biosynthesis pathway
The recombinant S. elongatus PYLW01 harboring the metabolic engineering pathway for PLA biosynthesis could produce PLA at very low efficiency, yielding 0.8 mg g−1 DCW PLA (Fig. 2B). Based on the previous report,11,37 together with our study, the rate-limitation steps of PLA biosynthesis are believed to be the generation of lactyl–CoA and the polymerization step. Thus, the expression of PCT and PHA synthases was optimized under the control of Ptrc, PpsbA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 38 and Pcpc560, compared with the three genes under the control of one promoter. Quantitative reverse transcription PCR (RT-qPCR) was then performed to determine the transcriptional levels of the genes of pct and pha among the four constructed strains. As shown in Fig. 2D, the two genes were all successfully transcribed under the different promoters. It was found that the transcriptional level of the two rate-limiting genes was the highest under the control of three independent promoters of Ptrc out of the four constructs. Furthermore, the four different recombinants were cultivated in the BG11 medium to induce the heterologous enzymes with constant light exposure to produce PLA. Subsequently, the highest level of production was achieved under the control of three independent promoters of Ptrc. The yield of the resulting strain PYLW02 increased approximately 7-fold to 5.6 mg g−1 DCW PLA (Fig. 2B). Thus, it appears that the Ptrc promoter is most effective for producing PLA, judging from both the expression levels and the yield data (Fig. 2B and D).
38 and Pcpc560, compared with the three genes under the control of one promoter. Quantitative reverse transcription PCR (RT-qPCR) was then performed to determine the transcriptional levels of the genes of pct and pha among the four constructed strains. As shown in Fig. 2D, the two genes were all successfully transcribed under the different promoters. It was found that the transcriptional level of the two rate-limiting genes was the highest under the control of three independent promoters of Ptrc out of the four constructs. Furthermore, the four different recombinants were cultivated in the BG11 medium to induce the heterologous enzymes with constant light exposure to produce PLA. Subsequently, the highest level of production was achieved under the control of three independent promoters of Ptrc. The yield of the resulting strain PYLW02 increased approximately 7-fold to 5.6 mg g−1 DCW PLA (Fig. 2B). Thus, it appears that the Ptrc promoter is most effective for producing PLA, judging from both the expression levels and the yield data (Fig. 2B and D).
Designing an acetyl–CoA self-sufficient system to increase the CoA pool
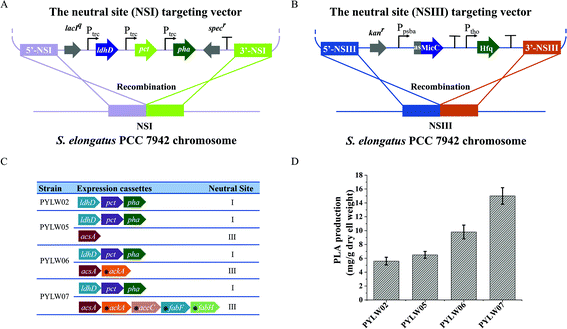
Based on the in silico genome-scale metabolic flux analysis done in the previous study,26 the surplus acetyl–CoA pool was expected to increase the PLA production, since it might be one of the bottlenecks for PLA synthesis. To further enlarge the acetyl–CoA pool and enhance the PLA production, we constructed an acetyl–CoA self-sufficient system in the strain PYLW02. Firstly, the native promoter of acsA encoding the acetyl–CoA synthetase was replaced with the stronger Ptrc promoter. Then, the Ptrc-acsA expression cassette was integrated into the neutral site III, resulting in the strain PYLW05 (Fig. 3A and C). The sRNA regulatory tool has been successfully applied to the cyanobacteria to knock down four genes (with up to 96% blockage), while hardly imposing any metabolic burden on host cells.39,40 Considering that the gene ackA encoding acetate kinase can convert acetyl–CoA to acetate, we knocked down ackA using the sRNA tool, generating the strain of PYLW06 (Fig. 3C). The PLA production was slightly increased after the overexpression of acetyl–CoA synthetase and was significantly improved upon knocking down the gene ackA; the PLA yield of the strain PYLW06 increased approximately 12-fold to 9.8 mg g−1 DCW, compared with the strain PYLW01 (Fig. 3D). These results indicate that the biosynthesis efficiency of PLA can be enhanced by increasing the amount of acetyl–CoA pool.Redirection of carbon flux to the PLA biosynthetic pathway
As the fatty acid biosynthesis competes for the carbon flux with the biosynthesis of bioproducts,41,42 it is necessary to regulate the expression of crucial fatty acid pathway enzymes, so that more carbon flux can be directed to the PLA biosynthesis. In this study, sRNA was also used to limit the amount of acetyl–CoA channeling to fatty acid synthesis and increase PLA production further (Fig. 3B). In cyanobacteria, FabH converting acetyl–CoA to acetyl–ACP, FabF converting acetyl–ACP to acetoacetyl–ACP, and ACC (acetyl–CoA carboxylase, encoding by accABCD genes) converting acetyl–CoA to malonyl–CoA have been shown to be essential for fatty acid biosynthesis.43,44 Previously, the downregulation of the critical genes fabD, fabB, fabF, and fabH successfully decreased fatty acid production, thus increasing malonyl–CoA levels up to 5-fold using PTRNAs.41 In this study, accC, fabF, and fabH were downregulated to redirect the carbon flux to the PLA biosynthesis pathway using sRNA, as shown in Fig. 1 and 3C. The resulting strain PYLW07 was cultivated in the BG11 medium for the PLA production after being induced by IPTG and theophylline, resulting in an approximately 19-fold increase to 15.0 mg g−1 DCW compared to the strain PYLW01 (Fig. 3D).The HDC strategy for increasing PLA production
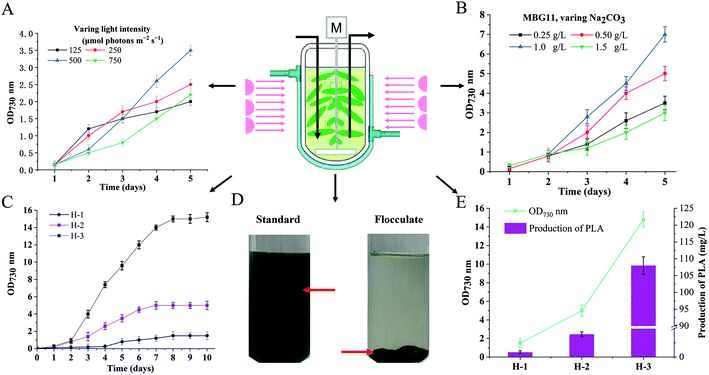
The highest yield-producing strain PYLW07 was used for the HDC experiments. The cultivation strategy of shaking flasks has been the traditional method for cultivating cyanobacteria. However, it suffers from insufficient supply of CO2, inability to adjust the intensity of light, and inconvenient supplementation of fresh medium, thus resulting in low photosynthetic efficiency.35,45,46 This study developed the HDC strategy to improve PLA production by integrating CO2 supply enhancement, nutrient supply optimization, and illumination strategy improvement, using a specially designed photobioreactor (Fig. 5).Different light intensities were determined to investigate their effects on the growth of cyanobacteria in the photobioreactor. As shown in Fig. 5A, the highest cell density at OD730 nm was 3.5, obtained from the light intensity of 500 μmol photons per m2 per s. A longer lag phase was observed when the strain was cultivated with 600 μmol photons per m2 per s. Considering that a high intensity of light can cause a more extended growth delay period when the bacteria are in the early stages of growth, we adjusted the light intensity from 125 μmol photons per m2 per s initially, to 500 μmol photons per m2 per s (Fig. S10†) after two days with the ventilation of CO2/air (5%, v/v, 1 vvm) gas mixture. The growth rate of engineered S. elongatus in the photoreactor was higher than that in the flask-shake system, reaching OD730 = 5.0 (Fig. 5C) and accumulating 40 mg L−1 PLA, which was almost 100-fold greater than that of the strain PYLW01 cultivated under shaking flasks (Fig. 5E).
It has been previously reported that modifying the medium composition can improve biomass production using the engineered cyanobacteria, especially by modifying the levels of sulfur, nitrate, and phosphate, which were identified as the primary limiting nutrients for high-density growth using the BG11 medium.47 Based on these reports,31,48,49 the BG11 medium was optimized with regard to nitrate, phosphate, sulfate, and carbonate levels, resulting in the MBG11 medium, which is more nutritious than the 5 × BG11,49 followed by Na2CO3 (0.2 g L−1), MgSO4·7H2O (5 g L−1), K2HPO4 (1 mM), NaNO3 (7.4 g L−1), and a 5-fold increase in trace minerals, compared to the BG11 medium. The cell density was improved from OD730 nm of 1.7 to OD730 nm of 3.0 upon using the MBG11 medium (Fig. S5†). A previous study showed that NaHCO3 in the BG11 medium had a very positive impact on cell growth.49 To investigate whether the high concentration of Na2CO3 could further improve the cell density of cyanobacteria, different concentrations of Na2CO3 were added to the modified BG11 (MBG11) medium. Notably, the highest cell density was achieved when the strain was cultivated in the MBG11 medium containing 1.0 g L−1 Na2CO3 (Fig. 5B). The highest cell density of the engineered PYLW07 strain was observed at the OD730 nm of 15.0 within 10 days under the optimal culture conditions, yielding 108.0 mg L−1 of PLA (Fig. 5E).
Biomass harvesting using polyampholyte flocculants
As shown in Fig. 5D, the PLA-producing strain PYLW07 was successfully harvested using polyacrylamide flocculant and coagulant aids. Previous studies have shown that the culture's pH considerably affects flocculation efficiency when the polyampholyte flocculants are used. The optimal flocculation effect was observed when the pH of the culture was adjusted from 11.0 to 8.0.50 Interestingly, the culture pH for PLA production was exactly 8.0. Therefore, the optimal flocculation effect was observed without adjusting the pH. Surprisingly, the flocculation efficiency of the PYLW07 strain was greater than about 99% after gravitational sedimentation for 5 min (Fig. 5D). It should be noted that the higher flocculation efficiency may lead to better PLA product recovery, since PLA is in inclusion granules inside the S. elongatus PCC7942 cells. As a result, harvesting by flocculation holds significant promise as a replacement for the traditional method of centrifugation, which is energy-intensive and costly for large-scale applications.LC-MS based metabolomic analysis
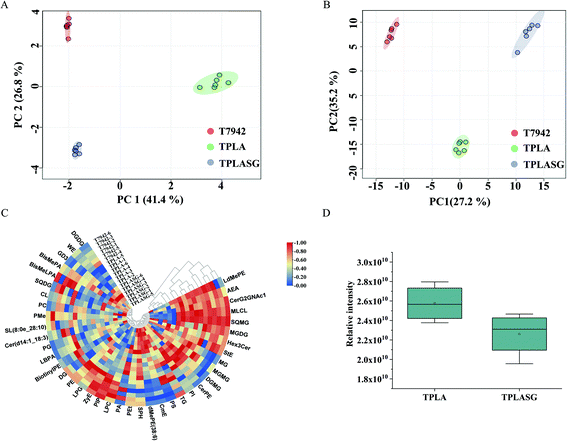
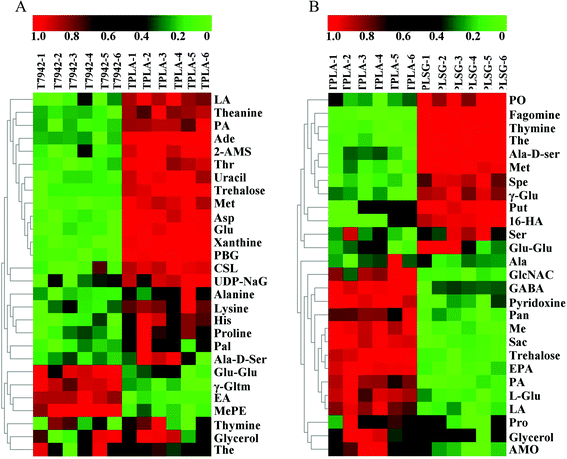
In order to further explore the factors influencing PLA production, untargeted metabolomics was conducted to analyze the differences among the T7942 (wild type), the TPLA (harboring PLA pathway), and the TPLASG (integrating the promoter optimization and redirection of carbon flux to PLA biosynthesis into the TPLA strain), as shown in Fig. S6, S8 and S9.† Principal component analysis (PCA) was carried out to analyze the overall distribution of the metabolites among the three strains. As shown in Fig. 6A and B, the six biological replicates of each sample clustered well, indicating that the analysis possesses good repeatability. Compared to T7942, the high accumulation of lactic acid was observed in TPLA (Fig. 7A). This intracellular abundance of lactic acid indicates that it was excessive for PLA production. This result is consistent with the finding that PCT and PHA synthases are the rate-limiting enzymes for PLA biosynthesis.37 The metabolomic analysis also revealed that the accumulation of alanine, aspartate, glutamine, histidine, lysine, methionine, proline, and threonine increased after introducing the PLA biosynthesis pathway (Fig. 7A). It is reported that enhanced CO2 fixation could lead to increased demand for ATP and NADPH, thus relieving the phototoxicity of cyanobacteria.19 Hence, it is reasonable to suggest that the incorporation of the PLA synthesis pathway made the cyanobacterial metabolism more vigorous, thereby increasing the production of a large number of amino acids.Compared with the TPLA, the lactic acid accumulation decreased in TPLASG while the PLA production increased (Fig. 7B). It shows that the systematic metabolic engineering operation in TPLASG is effective. On the contrary, the intracellular abundances of amino acids decreased in the TPLASG strain, such as alanine, glutamine, methionine, proline, and serine L-2-aminoadipate. Previous studies have shown that amino acid synthesis and introduced product synthesis compete for carbon flux and energy.51,52 Thus, the further enhancement of PLA biosynthesis may decrease amino acid biosynthesis after its carbon depriving effect surpasses the metabolic promoting effect. Based on this, the reduced production of amino acids may be caused by the higher production of PLA in the TPLASG strain.
Notably, the heatmaps showed significant differences in the lipid content between the TPLA and TPLASG strains (Fig. 6C and Fig. S7†). The total amount of lipid in the TPLASG strain was less than that in the TPLA strain, suggesting that the carbon flux of the fatty acid pathway decreased as the sRNA tool was used on the TPLASG strain (Fig. 6D). This result is direct proof that the sRNA tool works well for knocking down genes in cyanobacteria. Some other significant differences have also been observed. Compared with the TPLA strain, the intracellular amount of trehalose decreased by around 80% in the TPLASG strain (Fig. 7B). Trehalose can be stored as a carbohydrate reserve and energy source in the cyanobacteria.53,54 Hence, it is reasonable to suggest that the energy utilization to synthesize PLA may lead to a decreased amount of trehalose in cyanobacteria. As shown in Fig. 7B, the amount of putrescine in TPLASG was increased by 53-fold compared to the TPLA group. It has been reported that putrescine has essential effects on growth, development, and resistance to adverse environmental factors.55 Therefore, the increment of putrescine amount may be a positive cellular response for coping with the metabolic stress caused by the high accumulation of PLA.
Discussion
Disposable plastic products are ubiquitous worldwide today. Unfortunately, most used plastics produced from fossil resources are non-degradable, causing severe environmental issues. Therefore, degradable plastics will inevitably take the place of regular plastics with time, as the concerns regarding the depletion of fossil resources and environmental pollution grow.56 Meanwhile, atmospheric CO2 levels have increased sharply in the past 50 years to approximately 414 ppm, after staying stable at around 200–280 ppm for around 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 years.5,57 It may significantly contribute to global warming, rising sea levels, and even the extinction of ∼24% of plant and animal species.2 It is a worldwide consensus that effective strategies for carbon capture need to be developed to meet the severe climate-change mitigation challenge and adhere to the Paris Agreement of 2015.58,59 In this study, a cyanobacterial cell factory was successfully designed and constructed to produce the biodegradable plastic poly (D-lactic acid) (PDLA, Fig. S11†) directly from CO2, resulting in 108.0 mg L−1 PDLA (corresponding to 23.0 mg g−1 DCW). This study provides a feasible alternative solution to address plastic pollution and excessive CO2 emissions simultaneously.
000 years.5,57 It may significantly contribute to global warming, rising sea levels, and even the extinction of ∼24% of plant and animal species.2 It is a worldwide consensus that effective strategies for carbon capture need to be developed to meet the severe climate-change mitigation challenge and adhere to the Paris Agreement of 2015.58,59 In this study, a cyanobacterial cell factory was successfully designed and constructed to produce the biodegradable plastic poly (D-lactic acid) (PDLA, Fig. S11†) directly from CO2, resulting in 108.0 mg L−1 PDLA (corresponding to 23.0 mg g−1 DCW). This study provides a feasible alternative solution to address plastic pollution and excessive CO2 emissions simultaneously.
Furthermore, PDLA is a vital feedstock to improve the thermal resistance, mechanical performance, and hydrolysis resistance of PLA–based materials.60 Poly (L-lactic acid) (PLLA) and PDLA can form stereocomplex (SC) crystals with a high melting point that can be used to synthesize the high-performance PLA.61 In addition, no metal catalyst is involved in our biosynthesis of PDLA, which makes it suitable for use in the pharmaceutical and medical industries.62,63 Surprisingly, the highest reported average molecular weights for in vivo PDLA production (Mw, 62.5 kDa, Mn, 32.8 kDa, and dispersity of 1.9) were obtained in this study (Fig. S2 and S3†). In accordance with previous studies, the highest melting peak was obtained when the number-average molecular weights (Mn) of PDLA ranged from 23 to 50 kDa. The Mn of the PDLA produced by cyanobacteria was in the optimal range, which can facilitate its utilization in high-performance PLA production and endow extra commercial value on the corresponding products.64
Currently, the production of PLA is mainly carried out by the polymerization of lactic acid, which is primarily produced by the microbial fermentation of sugars. Therefore, the large-scale production of PLA in the traditional way will unavoidably compete with global food supplies.65 Compared with the sugar-based feedstock (called first-generation feedstock) and food-related biomass (called second-generation feedstock), CO2 (called the third-generation feedstock) is increasingly desirable for bioproduction processes.5 Therefore, the utilization of CO2 for the production of PLA appears to be more attractive.
Autotrophic cyanobacteria are a promising platform for the production of chemicals directly from CO2.66 However, the production concentration and productivity have been relatively low in cyanobacteria, compared with heterotrophic biological chassis.19 It makes the product harvest highly difficult, especially when the produced chemicals are secreted into the culture medium, thus greatly restricting the commercial process. Compared to low-molecular-weight chemicals such as ethanol, succinate, glycerol, and 2,3-butanediol,22 polymer granules synthesized in vivo are obviously easier to harvest. Meanwhile, polyelectrolyte flocculants can be used to harvest cyanobacteria more conveniently and cost-effectively.50 Therefore, it is reasonable to suggest that polymer production in cyanobacteria is more promising for the industrial process than low-molecular-weight chemicals. In this study, the PLA production was increased from 0.4 mg L−1 to 108.0 mg L−1 using an engineered cyanobacterial cell factory, involving combinational strategies of metabolic engineering and HDC (Fig. 5). The yield is highly comparable to that of the heterotrophic yeast Y. lipolytica, and even higher than that of the yeast S. cerevisiae8,29 using glucose as the feedstock. These results strongly suggest that it is promising to produce PLA using such cyanobacterial cell factories.
Moreover, the molecular weights of PLA produced by our cyanobacteria are higher than that produced by heterotrophic strains such as E. coli and yeast. Many aspects affect the molecular weight, including the types and expression levels of active PHA synthase, esterase, and lipase, and the chain transfer reactions during polymerization.28,67,68 Previous studies have shown that cyanobacteria are natural PHB polyester producers.69,70 Thus, the natural intracellular environment may be more suitable for the polymerization of PLA. For example, the high abundance of ATP in cyanobacteria may facilitate the biosynthesis of products in numerous enzymatic steps,42 such as the polymerization reaction steps. Moreover, the cell sizes of the cyanobacteria are generally larger than those of other bacteria, usually 3 to 10 μm in diameter, with the largest reaching up to 60 μm. These larger cell sizes may also accommodate larger PLA granules, since they are present in the form of inclusion bodies in tiny intracellular spaces.71 Therefore, it is reasonable to conclude that the cyanobacterial cell factory is a promising platform for the efficient production of polymers such as PLA-like polyesters.
Several strategies were combined to improve PLA production, including promoter optimization, redirection of carbon flux using sRNA tools, and the HDC strategy. The code-optimization and promoter-optimization strategies increased the PLA polymer content by 7-fold compared to the control (Fig. 2B). The PLA production was further increased by approximately 19-fold compared to the initially constructed strain, using the acetyl–CoA self-sufficient module and the carbon flux redirection strategy (Fig. 2B and Fig. 3D). The metabolomic analysis shows that the overall lipid content declined after inhibiting the essential fatty acid synthesis genes, indicating that the sRNA tool functions well in the cells (Fig. 6D). Our method offers effective combinational strategies for increasing the PLA production. Notably, the PLA production increased by 270-fold compared to the control (the PYLW01 strain cultivated in shake flasks) in a self-designed photobioreactor (Fig. 5E). The engineered S. elongatus strain PYLW07 could grow to an OD730 of 15 after 10-day cultivation (Fig. 5C) using the HDC strategy, which is comparable with the heterotrophic yeast cultivation under the optimized conditions. The HDC strategy served as an “amplifier” that enlarged the positive effects of the metabolic engineering strategy. Among the several factors affecting the growth of the cyanobacterium in the HDC strategy, artificial light with the enhancement of the red spectral region was outstanding (Fig. S10†). The cyanobacteria mainly absorb light in the wavelength range of ∼300 nm to ∼750 nm.72 Besides, the relatively higher concentration of Na2CO3 (1 g L−1) could increase the cyanobacterial cell density and maintain stable pH in the culture medium while bubbling 5% CO2 (Fig. 5B). These indicate that integrating metabolic engineering and HDC strategies are a powerful approach for improving the productivity of cyanobacterial cell factory, facilitating their commercialization.
Based on the previous studies,73,74 together with our analysis, the PHA synthase plays a crucial role in affecting the polymers’ production and molecular weights. Therefore, the engineering of PHA synthase could contribute to the improvement of PLA production. In addition, copolymerization has been well known to be an effective strategy for improving the properties of polymers. For example, PHB has the disadvantage of brittleness,75 while the P(3HB-co-LA) copolymer is composed of 3-hydroxybutyrate (3HB) and lactic acid (LA), showing better flexibility, as well as other favorable physical and mechanical properties.28,76 The stereocomplex of PLLA and PDLA could significantly enhance the heat resistance and mechanical properties of PLA.77 Poly (lactic acid-co-glycolic acid) (PLGA) is one of the most effective biodegradable polymeric nanoparticles that could be used as a cancer drug delivery system.78 To achieve copolymerization, PHA synthase also needs to be engineered to polymerize various substrates. Therefore, the engineering of PHA synthase is worth exploring and could further improve polymer production and accelerate commercialization.
Chloroform was selected for the product analysis on a laboratory scale since it was the most reported method and may endorse the comparability of our results to the literature.8,26–28 However, since chloroform is indeed a non-green solvent,79 the extraction of PLA by several green solvents was also attempted. We identified four green solvents for effective PLA extraction (Table S6†), suggesting their further use instead of the non-green solvent chloroform. Moreover, it was also reported that polyhydroxyalkanoate (PHA) could be obtained by the inorganic aqueous phase extraction.79 It is possible to extract PLA in such a green method on an industrial scale since PLA has similar solubility with PHA. On the other hand, the E+-factor was reported to be an important measure for the environmental impact of (bio)chemical reactions.80 As shown in Table S5,† the E+-factor of directly using solar light is a negative number; the E+-factors of using clean energy (nuclear, wind, and solar power) are also very low; even the E+-factors of burning fossil fuels are in the relatively low range according to the literature.80 In summary, direct carbon capture for biodegradable plastics production may have a high potential of being green and sustainable, especially when sunlight is used.
Materials and methods
Chemicals and reagents
Unless otherwise specified, all chemicals were purchased from Sigma-Aldrich (St Louis, MO) or Macklin (GmbH, Germany). The restriction enzymes, T4 DNA ligase (New England Biolabs Inc, USA), 2 × Phanta Max Master Mix (Vazyme Biotech Co., Ltd), 2 × CloneExpress Mix (Vazyme Biotech Co., Ltd) were used for cloning and plasmid construction. The polyacrylamide flocculant and polyelectrolyte flocculant were purchased from the Jieyuan Chemical Co., Ltd (Cangzhou, China). Genomic DNA was extracted using the Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA) and other reagents of high quality were obtained from general suppliers in Shanghai.Strains and cultural conditions
The characteristics of the strain used in this study are listed in Table S1.† The BG11 medium was used as a fundamental medium for S. elongatus PCC7942.18 For the high-density cultivation strategy,34 the engineered S. elongatus PCC7942 was grown in the modified BG11 medium (MBG11 medium) at 30 °C using a photobioreactor with an initial illumination intensity of 125 μmol photons per m2 per s, which was then increased to 500 μmol photons per m2 per s after two days. For cultivation with bubbling air containing 5% CO2, gases were bubbled through 5 μm micropores at a flow rate of 50 mL min−1. For the PLA production, the engineered S. elongatus cells in the exponential phase were diluted to 0.05 (OD730) in 500 mL of BG-11 medium containing 20 mg L−1 spectinomycin in 800 mL flask and continuously provided with CO2-enriched air (5%, v/v) using a mixed ventilator. The cultures were then induced with 1 mM IPTG11 and 2 mM theophylline81 after growing to an OD730 of 0.4–0.6. The BG11 medium was further modified for the high-density cultivation by adding the following chemicals (L−1): Na2CO3 1 g, MgSO4·7H2O 5 g, NaNO3 5 g, KH2PO4 (1 M) 2 mL, and five-fold of trace minerals compared with the original BG11 medium. The trace minerals of the intracellular extract and the remaining medium after fermentation in BG11 medium and modified BG11 medium were measured using inductively coupled plasma optical emission spectrometry (ICP-OES, Optima 8300, PerkinElmer, Germany, Table S4†).Process for flocculation
The polyacrylamide flocculant and assistant flocculant were dissolved in water at room temperature in the ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 (w/w) and 5
300 (w/w) and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (w/w), respectively. Then, the dissolved flocculant was subject to the fermentation broth obtained from the high-density cultivation at the ratio of 1.5% (v/v).50 The mixed culture was stirred for 2 min. Next, the dissolved assistant flocculant was added to the solution and vortexed for 30 s. The culture was then subjected to gravitational sedimentation for a while. The flocculation efficiency was characterized by the optical density measurements before and after settling at a wavelength of OD730 nm.50,82
100 (w/w), respectively. Then, the dissolved flocculant was subject to the fermentation broth obtained from the high-density cultivation at the ratio of 1.5% (v/v).50 The mixed culture was stirred for 2 min. Next, the dissolved assistant flocculant was added to the solution and vortexed for 30 s. The culture was then subjected to gravitational sedimentation for a while. The flocculation efficiency was characterized by the optical density measurements before and after settling at a wavelength of OD730 nm.50,82
DNA manipulation
All DNA manipulation and general molecular biology techniques were performed according to standard protocols.83 The plasmids and primers used in this study are listed in Tables S2 and S3† and briefly described. The pEASY-Blunt cloning vector (Transgen, China) was used for subcloning genes. The shuttle vector pAM-MCS12 was used for inserting the gene cassettes into the neutral site I (NS I) of the S. elongatus PCC7942 chromosome.42 The Hfq-MicC expressing plasmid, inserting the small RNA tools into the neutral site III (NS III) of the S. elongatus PCC7942 genome, was a kind gift from Dr Tao Sun.81 The ldhD gene encoding D-lactate dehydrogenase was engineered to switch the coenzyme specificity from NADH to NADPH, which has been done before,11 thus promoting NADPH utilization in cyanobacteria for D-lactate production. The genes encoding propionyl–CoA transferase (PCT) from Clostridium propionicum and the PHA synthase from Pseudomonas sp. were synthesized by GenScript (Nanjing, China) with codon optimization based on the codon bias of Synechococcus elongatus.26 The gene acsA encoding acetyl–CoA synthetase was from the native genome of the S. elongatus PCC7942. Plasmid pAM-PYLW01 was constructed by inserting ldhD, pct, and pha into the pAM-MCS12 plasmid18 between the EcoRI and XhoI sites under the control of the IPTG-inducible promoter Ptrc. Thereafter, the plasmid pAM-PYLW02, pAM-PYLW03, pAM-PYLW04 were generated by adding the independent Ptrc promoter, PpsbA promoter and Pcpc560 promoter to target pct and pha genes integrated at NSI, respectively. In addition, the strain of PYLW05 was generated by insertion of the acsA into the NSIII site using the pBA3031 M plasmid. Moreover, knockdown of the ackA (acetate kinase), accC (acetyl–CoA carboxylase), fabF (ketosynthase), fabH (β-ketoacyl-ACP synthase III) genes was performed using the inducible Hfq-MicC system integrated at NSIII,81 generating the strain of PYLW06 and PYLW07.Strain construction and transformation
The strains used in this study are listed in Table S1.† All the operations of chromosomes were carried out by homologous recombination of the plasmid DNA into the S. elongatus PCC7942 chromosomes at the neutral site I or neutral site III.84 Strain PYLW01 (TPLA), PYLW02, PYLW03, and PYLW04 were constructed by recombination of plasmids pAM-PYLW01, pAM-PYLW02, pAM-PYLW03 and pAM-PYLW04 into the chromosomes of S. elongatus PCC7942, respectively. In addition, the strain PYLW05 was then constructed by recombination of plasmid DNA of pBA-acsA into the strain PYLW02 chromosomes at neutral site III. Moreover, the expression cassettes of sRNA were introduced into the chromosomes at neutral site III using the Hfq-MicC tool. The PpsbA promoter was used for expressing all sRNAs and the chaperone hfq was under the control of a theophylline-responsive riboswitch. The genes of ackA, accC, fabF, and fabH were downregulated by sRNA tools. Schematics of the PYLW06 and PYLW07 (TPLASG) strains are listed in Fig. 3C. S. elongatus PCC7942 was transformed using previously described protocols.85 All the transformants were cultivated in the BG11 medium with 20 mg L−1 spectinomycin for successful selection recombination. The right candidate transformants were further analyzed by PCR using the primers listed in Table S3† to verify that the targeted genes were correctly integrated into the NSI or NSIII of S. elongatus PCC7942.Extraction and analysis of PLA
The PLA-produced culture was centrifuged at 8000 rpm for 15 min, and then the cells were washed twice with distilled water and freeze-dried overnight at −80 °C. PLA was extracted using the Soxhlet apparatus with chloroform.27 About 2.0 g of lyophilized cells were filled with 200 volumes of the chloroform refluxed in the Soxhlet apparatus for 24 h, and the extracted materials were collected. The chloroform–polymer mixed solution was filtered. The polymer was washed twice in the ten volumes of hexane and collected by centrifugation.86GC-MS determined the concentration of the polymers accumulated in the cells (Fig. S4†). The cultures were centrifuged at 8000 rpm for 15 min, and then the cells were washed twice with distilled water and freeze-dried overnight at −80 °C. About 60 mg of samples were dissolved in acidified methanol, i.e., containing 15% (v/v) H2SO4 and chloroform in a hydrothermal synthesis reactor at 120 °C for 4 h. The resulting methyl esters of constituent lactate were subsequently used for the GC-QQQMS analysis.28,87 The GC-MS analysis was conducted by Thermo & TSQ8000 system equipped with TraceGOLD TG-WAXMS (30 m × 0.25 mm × 0.25 μm). The GC oven temperature was initially at 50 °C for 5 min and ramped to 230 °C at 7.5 °C min−1. Then the oven temperature was increased to 260 °C with a gradient of 10 °C min−1 and held for 5 min.
The composition of the polymer was determined by NMR on a Bruker AVANCE III HD 600 MHz (Bruker, Rheinstetten, Germany) using tetramethylsilane (TMS) as an internal standard. The polymer was diluted in CDCl3 and the NMR spectra were recorded at 298 k. The 1H NMR spectrum was acquired using an excitation flip angle of 30° at the delay time of 10 s. In the 13C NMR experiment, the 30° pulse width was 3.5 μs at the delay time of 2.5 s. The 2D COSY experiment was performed with 1.3 s delay time, 16 scans and processing data size of 2 k × 1 k.
The molecular weights of the polymer were determined by gel permeation chromatography (GPC) at 35 °C using a Waters 1525 system equipped with Waters 2414 detectors and Agilent PLgel Sum MIXED-C column. The elution solvent used was chloroform at a flow rate of 1 mL min−1. Samples were resuspended in chloroform and filtered through a 0.4 μm filter. The MALDI-TOF mass spectrum was carried out using the DCTB as a matrix, CF3CO2Na as a cationization agent and chloroform as a solvent in a MALDI-7090 (SHIMADZU, Japan) mass spectrometer.36
Transmission electron microscopy analysis
The wild type and engineered S. elongatus PCC7942 cells were fixed with glutaraldehyde and then post-fixed with potassium permanganate for 1 h. Then, the cells were dehydrated in graded ethanol and propylene oxide, respectively. The Leica EM UC7 system conducted the ultrathin sections after being stained with lead citrate and uranyl acetate.69 The sample was then examined using biological transmission electron microscopy (Tecnai G2 spirit Biotwin system) at 80 kHz.RT-qPCR analysis of synthetic pathway genes
The engineered S. elongatus PCC7942 strains were grown in BG11 medium at 30 °C with an illumination intensity of 100 mol photons per m2 per s when the OD730 reached 0.4–0.6, 1 mM IPTG was added to the culture to induce the targeted genes. The total RNA was then extracted using an RNAprep Pure Cell/Bacteria Kit (Tiangen Biotech, China). The extracted total RNA was then treated with DNase I to remove the genomic DNA and used as a template for cDNA synthesis with random primers and SuperScriptTMIII Reverse Transcriptase (Invitrogen, China). The quantitative PCR was conducted using the SuperReal PreMix SYBR Green kit (Tiangen Biotech, China) and the CFX96 Real-Time system (Bio-Rad, Hercules, CA, USA). Beacon Designer was used to design the primers for qPCR and the rnpB gene was used as a control.Extraction and measurement of the metabolome
Based on the previous studies,88,89 the extraction of the metabolites was carried out for the cyanobacteria cells with minor modifications. Briefly, the cells were quenched and extracted rapidly with 1 mL of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 MeOH/H2O (containing 0.1% formic acid, −80 °C) and then frozen in liquid nitrogen. Then, the supernatants of the samples were collected for LC-MS analysis after being frozen-thawed three times to extract the metabolites.
20 MeOH/H2O (containing 0.1% formic acid, −80 °C) and then frozen in liquid nitrogen. Then, the supernatants of the samples were collected for LC-MS analysis after being frozen-thawed three times to extract the metabolites.
The three kinds of samples, the wild types of strains T7942, TPLA, and TPLASG, were subjected to the Thermo UPLC Q-Extractive (QE) coupled with an ESI ion source for metabolomics analyses according to the previous study.90 The chromatographic separation was achieved with the RP Zorbax Eclipse XBD-C18 column. Mobile phase A is aqueous of 0.1% formic acid in diluted water, and mobile phase B is the organic phase of 0.1% formic acid in pure acetonitrile. The LC gradient elution program was as follows: t = 0.0 min, 99% A; t = 5.0 min, 99% A; t = 5.5 min, 70% A; t = 9 min, 100% B; t = 11 min, 100% B; and t = 12.1 min, 99% A. The mass parameters of the C18-ESIMS are as follows: full mass spectrometry scanning range 80–1000 m/z, the capillary voltage of 4500 V, the gas flow rate was 1.6 bar, the velocity of dry gas at 220 °C was 6.0 L min−1. According to the previous studies,90 the network tool of MetaboAnalyst91 was used to conduct the metabolome data analysis. And the OmicsBean Multiomics was used to conduct the pathway enrichment analysis.92 Then, the lipid identification was based on LipidSearch software.
Conclusion
In this study, we developed a light-driven autotrophic cyanobacterial cell factory to produce bio-degradable plastic PLA directly from CO2. A combination of systematic metabolic engineering and high-density cultivation strategies significantly increased the PLA yield by approximately 270 times, compared to the initially constructed strain. Meanwhile, the polyelectrolyte flocculant was introduced to harvest the cells efficiently, easily, and quickly. The calculation of the energy parameter (E+-factor) inspires us that future efforts are to directly use solar light and CO2 for biodegradable material production. The PLA production by cyanobacteria was highly comparable to that of heterotrophic microorganisms. Moreover, the molecular weight of PLA produced by cyanobacteria in this study is even higher than that produced by heterotrophic organisms. Considering its availability and practicality for future engineering for producing high-performance copolymers, our approach may possess excellent potential for commercialized production. Overall, our study provides a feasible alternative solution to address plastic pollution and excessive CO2 emissions simultaneously.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the grants of National Key R&D Program of China (2018YFA0903600) and the NSFC (31870088 and 32170105). We thank Dr Tao Sun for his kind gift of sRNA plasmid.References
- T. L. de Albuquerque, J. E. Marques Junior, L. P. de Queiroz, A. D. S. Ricardo and M. V. P. Rocha, Int. J. Biol. Macromol., 2021, 186, 933–951 CrossRef CAS PubMed.
- T. P. Hughes, A. H. Baird, D. R. Bellwood, M. Card, S. R. Connolly, C. Folke, R. Grosberg, O. Hoegh-Guldberg, J. B. C. Jackson, J. Kleypas, J. M. Lough, P. Marshall, M. Nystrom, S. R. Palumbi, J. M. Pandolfi, B. Rosen and J. Roughgarden, Science, 2003, 301, 929–933 CrossRef CAS PubMed.
- T. H. Yang, Y. K. Jung, H. O. Kang, T. W. Kim, S. J. Park and S. Y. Lee, Appl. Microbiol. Biotechnol., 2011, 90, 603–614 CrossRef CAS PubMed.
- C. J. Rhodes, Sci. Prog., 2018, 101, 207–260 CrossRef PubMed.
- Z. H. Liu, K. Wang, Y. Chen, T. W. Tan and J. Nielsen, Nat. Catal., 2020, 3, 274–288 CrossRef CAS.
- M. S. Singhvi, S. S. Zinjarde and D. V. Gokhale, J. Appl. Microbiol., 2019, 127, 1612–1626 CrossRef CAS PubMed.
- G. Li, M. H. Zhao, F. Xu, B. Yang, X. Y. Li, X. X. Meng, L. S. Teng, F. Y. Sun and Y. X. Li, Molecules, 2020, 25, 5023–5041 CrossRef CAS PubMed.
- S. Lajus, S. Dusseaux, J. Verbeke, C. Rigouin, Z. P. Guo, M. Fatarova, F. Bellvert, V. Borsenberger, M. Bressy, J. M. Nicaud, A. Marty and F. Bordes, Front. Bioeng. Biotechnol., 2020, 8, 954–969 CrossRef PubMed.
- A. Z. Naser, I. Deiab and B. M. Darras, RSC Adv., 2021, 11, 17151–17196 RSC.
- R. Mehta, V. Kumar, H. Bhunia and S. N. Upadhyay, J. Macromol. Sci., Polym. Rev., 2005, C45, 325–349 CrossRef CAS.
- C. Li, F. Tao, J. Ni, Y. Wang, F. Yao and P. Xu, Sci. Rep., 2015, 5, 9777–9788 CrossRef CAS PubMed.
- K. Okano, Q. Zhang, S. Shinkawa, S. Yoshida, T. Tanaka, H. Fukuda and A. Kondo, Appl. Environ. Microbiol., 2009, 75, 462–467 CrossRef CAS PubMed.
- A. Jaiswal, V. Babu, B. Baishya and P. Jaiswal, J. Environ. Pathol., Toxicol. Oncol., 2020, 39, 317–334 CrossRef.
- J. T. Broddrick, D. G. Welkie, D. Jallet, S. S. Golden, G. Peers and B. O. Palsson, Metab. Eng., 2019, 52, 42–56 CrossRef CAS PubMed.
- X. Y. Gao, T. Sun, G. S. Pei, L. Chen and W. W. Zhang, Appl. Microbiol. Biotechnol., 2016, 100, 3401–3413 CrossRef CAS PubMed.
- Y. Hirokawa, R. Goto, Y. Umetani and T. Hanai, J. Biosci. Bioeng., 2017, 124, 54–61 CrossRef CAS PubMed.
- R. Hidese, M. Matsuda, T. Osanai, T. Hasunuma and A. Kondo, ACS Synth. Biol., 2020, 9, 260–268 CrossRef CAS PubMed.
- Y. Wang, F. Tao, J. Ni, C. Li and P. Xu, Green Chem., 2015, 17, 3100–3110 RSC.
- J. Ni, H. Y. Liu, F. Tao, Y. T. Wu and P. Xu, Angew. Chem., Int. Ed., 2018, 57, 15990–15994 CrossRef CAS PubMed.
- C. Li, F. Tao and P. Xu, ChemBioChem, 2016, 17, 1491–1494 CrossRef CAS PubMed.
- L. Bahr, A. Wustenberg and R. Ehwald, J. Appl. Phycol., 2016, 28, 783–793 CrossRef.
- H. Y. Liu, J. Ni, P. Xu and F. Tao, ACS Synth. Biol., 2018, 7, 2436–2446 CrossRef CAS PubMed.
- B. Maestro and J. M. Sanz, Microb. Biotechnol., 2017, 10, 1323–1337 CrossRef CAS PubMed.
- L. Van Haver and S. Nayar, Algal Res., 2017, 24, 167–180 CrossRef.
- S. J. Park, S. Y. Lee, T. W. Kim, Y. K. Jung and T. H. Yang, Biotechnol. J., 2012, 7, 199–212 CrossRef CAS PubMed.
- T. H. Yang, T. W. Kim, H. O. Kang, S. H. Lee, E. J. Lee, S. C. Lim, S. O. Oh, A. J. Song, S. J. Park and S. Y. Lee, Biotechnol. Bioeng., 2010, 105, 150–160 CrossRef CAS PubMed.
- Y. K. Jung and S. Y. Lee, J. Biotechnol., 2011, 151, 94–101 CrossRef CAS PubMed.
- Y. K. Jung, T. Y. Kim, S. J. Park and S. Y. Lee, Biotechnol. Bioeng., 2010, 105, 161–171 CrossRef CAS PubMed.
- A. Ylinen, H. Maaheimo, A. A. Hakala, M. Penttila, L. Salusjarvi and M. Toivari, J. Ind. Microbiol. Biotechnol., 2021, 48, 28–40 CrossRef PubMed.
- S. Taguchi, M. Yamadaa, K. Matsumoto, K. Tajima, Y. Satoh, M. Munekata, K. Ohno, K. Kohda, T. Shimamura, H. Kambe and S. Obata, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17323–17327 CrossRef CAS PubMed.
- R. L. Clark, L. L. McGinley, H. M. Purdy, T. C. Korosh, J. L. Reed, T. W. Root and B. F. Pfleger, Metab. Eng., 2018, 47, 230–242 CrossRef CAS PubMed.
- L. Fathurrahman, A. H. Hajar, D. W. Sakinah, Z. Nurhazwani and J. Ahmad, Pak. J. Biol. Sci., 2013, 16, 1517–1523 CrossRef CAS PubMed.
- J. H. Duarte, E. G. D. Morais, E. M. Radmann and J. A. V. Costa, Bioresour. Technol., 2017, 234, 472–475 CrossRef CAS PubMed.
- H. Takano, H. Takeyama, N. Nakamura, K. Sode, J. G. Burgess, E. Manabe, M. Hirano and T. Matsunaga, Appl. Biochem. Biotechnol., 1992, 34, 449–458 CrossRef.
- D. Dienst, J. Wichmann, O. Mantovani, J. S. Rodrigues and P. Lindberg, Sci. Rep., 2020, 10, 5932–5948 CrossRef CAS PubMed.
- C. J. Chen, Z. Y. Bai, Y. Q. Cui, Y. Cong, X. B. Pan and J. C. Wu, Macromolecules, 2018, 51, 6800–6809 CrossRef CAS.
- K. Matsumoto, K. Tobitani, S. Aoki, Y. Song, T. Ooi and S. Taguchi, AMB Express, 2014, 4, 83–88 CrossRef PubMed.
- U. Nair, C. Thomas and S. S. Golden, J. Bacteriol., 2001, 183, 1740–1747 CrossRef CAS PubMed.
- S. M. Gaida, M. A. A. Hinai, D. C. Indurthi, S. A. Nicolaou and E. T. Papoutsakis, Nucleic Acids Res., 2013, 41, 8726–8737 CrossRef CAS PubMed.
- T. Sun, S. Li, X. Song, J. Diao, L. Chen and W. Zhang, Biotechnol. Adv., 2018, 36, 1293–1307 CrossRef CAS PubMed.
- Y. Yang, Y. Lin, L. Li, R. J. Linhardt and Y. Yan, Metab. Eng., 2015, 29, 217–226 CrossRef CAS PubMed.
- J. Ni, F. Tao, Y. Wang, F. Yao and P. Xu, Green Chem., 2016, 18, 3537–3548 RSC.
- J. B. Parsons and C. Rock, Prog. Lipid Res., 2013, 52, 249–276 CrossRef CAS PubMed.
- J. Kuo and C. Khosla, Metab. Eng., 2014, 22, 53–59 CrossRef CAS PubMed.
- P. C. Lin and H. B. Pakrasi, Planta, 2019, 249, 145–154 CrossRef CAS PubMed.
- K. Vavitsas, M. Fabris and C. E. Vickers, Genes, 2018, 9, 11–30 CrossRef PubMed.
- H. U. Kim, P. Charusanti, S. Y. Lee and T. Weber, Nat. Prod. Rep., 2016, 33, 933–941 RSC.
- P. V. Alphen, H. A. Najafabadi, F. B. D. Santos and K. J. Hellingwerf, Biotechnol. J., 2018, 13, 1700764–1700772 CrossRef PubMed.
- A. Wlodarczyk, T. T. Selao, B. Norling and P. J. Nixon, Commun. Biol., 2020, 3, 1–14 CrossRef PubMed.
- C. Niemi and F. G. Gentili, Physiol. Plant, 2021, 173, 536–542 CrossRef CAS PubMed.
- S. Sato and S. Yanagisawa, Plant Cell Physiol., 2014, 55, 306–319 CrossRef CAS PubMed.
- S. K. Masakapalli, N. J. Kruger and R. G. Ratcliffe, Plant J., 2013, 74, 569–582 CrossRef CAS PubMed.
- S. Wu, L. He, R. Shen, X. Zhang and Q. Wang, J. Microbiol. Biotechnol., 2011, 21, 830–837 CrossRef CAS PubMed.
- Y. Qiao, W. H. Wang and X. F. Lu, Metab. Eng., 2020, 62, 161–171 CrossRef CAS PubMed.
- W. Raksajit, P. Maenpaa and A. Incharoensakdi, J. Biochem. Mol. Biol., 2006, 39, 394–399 CAS.
- S. Y. Choi, M. N. Rhie, H. T. Kim, J. C. Joo, I. J. Cho, J. Son, S. Y. Jo, Y. J. Sohn, K. A. Baritugo, J. Pyo, Y. Lee, S. Y. Lee and S. J. Park, Metab. Eng., 2020, 58, 47–81 CrossRef CAS PubMed.
- J. R. Petit, J. Jouzel, D. Raynaud, N. I. Barkov, J. M. Barnola, I. Basile, M. Bender, J. Chappellaz, M. Davis, G. Delaygue, M. Delmotte, V. M. Kotlyakov, M. Legrand, V. Y. Lipenkov, C. Lorius, L. Pepin, C. Ritz, E. Saltzman and M. Stievenard, Nature, 1999, 399, 429–436 CrossRef CAS.
- I. Pikaar, J. de Vrieze, K. Rabaey, M. Herrero, P. Smith and W. Verstraete, Sci. Total Environ., 2018, 644, 1525–1530 CrossRef CAS PubMed.
- N. Mac Dowell, P. S. Fennell, N. Shah and G. C. Maitland, Nat. Clim. Change, 2017, 7, 243–249 CrossRef CAS.
- X. Han, K. M. Huang, H. Z. Tang, J. Ni, J. Q. Liu, P. Xu and F. Tao, Biotechnol. J., 2019, 14, 1800656–1800667 CrossRef PubMed.
- T. X. Lv, C. B. Zhou, J. Q. Li, S. Y. Huang, H. Y. Wen, Y. F. Meng and S. C. Jiang, J. Appl. Polym. Sci., 2018, 135, 45663–45670 CrossRef.
- J. Jozwicka, K. G. Jagiela, A. Gutowska, K. T. Schmidt and M. Cieplinski, Fibres Text. East. Eur., 2012, 20, 135–141 CAS.
- R. Ata, A. Aladdin, N. Othman, R. A. Malek, O. Leng, R. Aziz and H. El Enshasy, J. Chem. Pharm. Res., 2015, 2015, 51–63 Search PubMed.
- J. Shao, S. Xiang, X. C. Bian, J. R. Sun, G. Li and X. S. Chen, Ind. Eng. Chem. Res., 2015, 54, 2246–2253 CrossRef CAS.
- A. M. Varman, Y. Yu, L. You and Y. J. J. Tang, Microb. Cell Fact., 2013, 12, 117–125 CrossRef PubMed.
- P. Farrokh, M. Sheikhpour, A. Kasaeian, H. Asadi and R. Bavandi, Biotechnol. Prog., 2019, 35, 2835–2851 CrossRef PubMed.
- L. Jurasek and R. H. Marchessault, Appl. Microbiol. Biotechnol., 2004, 64, 611–617 CrossRef CAS PubMed.
- B. H. A. Rehm, Biochem. J., 2003, 376, 15–33 CrossRef CAS PubMed.
- M. Koch, J. Bruckmoser, J. Scholl, W. Hauf, B. Rieger and K. Forchhammer, Microb. Cell Fact., 2020, 19, 1–12 CrossRef PubMed.
- M. Lackner, D. Kamravamanesh, M. Krampl, R. Itzinger, C. Paulik, I. Chodak and C. Herwig, Int. J. Biobased Plast., 2019, 1, 48–59 CrossRef.
- G. Q. Chen and X. R. Jiang, Curr. Opin. Biotechnol., 2018, 53, 20–25 CrossRef CAS PubMed.
- M. Y. Ho, N. T. Soulier, D. P. Canniffe, G. Z. Shen and D. A. Bryant, Curr. Opin. Plant Biol., 2017, 37, 24–33 CrossRef CAS PubMed.
- M. F. Chek, A. Hiroe, T. Hakoshima, K. Sudesh and S. Taguchi, Appl. Microbiol. Biotechnol., 2019, 103, 1131–1141 CrossRef CAS PubMed.
- H. B. Zou, M. X. Shi, T. T. Zhang, L. Li, L. Z. Li and M. Xian, Appl. Microbiol. Biotechnol., 2017, 101, 7417–7426 CrossRef CAS PubMed.
- L. Lv, Y. L. Ren, J. C. Chen, Q. Wu and G. Q. Chen, Metab. Eng., 2015, 29, 160–168 CrossRef CAS PubMed.
- J. W. Ye, D. K. Hu, X. M. Che, X. R. Jiang, T. Li, J. C. Chen, H. Q. M. Zhang and G. Q. Chen, Metab. Eng., 2018, 47, 143–152 CrossRef CAS PubMed.
- H. W. Pan, J. J. Kong, Y. J. Chen, H. L. Zhang and L. S. Dong, Int. J. Biol. Macromol., 2019, 122, 848–856 CrossRef CAS PubMed.
- F. S. T. Mirakabad, K. N. Koshki, A. Akbarzadeh, M. R. Yamchi, M. Milani, N. Zarghami, V. Zeighamian, A. Rahimzadeh, S. Alimohammadi, Y. Hanifehpour and S. W. Joo, Asian Pac. J. Cancer Prev., 2014, 15, 517–535 CrossRef PubMed.
- M. A. Hassan, S. D. Ensan, P. T. Voon, L. Y. Phang and Y. Shirai, US Pat, 2011/0160427A1, 2011 Search PubMed.
- F. Tieves, F. Tonin, E. Fernández-Fueyo, J. M. Robbins, B. Bommarius, A. S. Bommarius, M. Alcalde and F. Hollmann, Tetrahedron, 2019, 75, 1311–1314 CrossRef CAS.
- T. Sun, S. Li, X. Song, G. Pei, J. Diao, J. Cui, M. Shi, L. Chen and W. Zhang, Biotechnol. Biofuels, 2018, 11, 26–42 CrossRef PubMed.
- K. L. Morrissey, M. I. Keirn, Y. Inaba, A. J. Denham, G. J. Henry, B. W. Vogler, M. C. Posewitz and M. P. Stoykovich, Algal Res., 2015, 11, 304–312 CrossRef.
- J. Sambrook, E. F. Fritsch and T. Maniatis, Molecular cloning: a laboratory manual, Cold Spring Harbor Laboratory, NY, 1989, vol. 2 Search PubMed.
- S. A. Bustos and S. S. Golden, J. Bacteriol., 1991, 173, 7525–7533 CrossRef CAS PubMed.
- S. S. Golden, J. Brusslan and R. Haselkorn, Methods Enzymol., 1987, 153, 215–231 CAS.
- C. T. Nomura, K. Taguchi, S. Taguchi and Y. Doi, Appl. Environ. Microbiol., 2004, 70, 999–1007 CrossRef CAS PubMed.
- G. Braunegg, B. Sonnleitner and R. M. Lafferty, Eur. J. Appl. Microbiol. Biotechnol., 1978, 6, 29–37 CrossRef CAS.
- J. Cui, T. Sun, S. Li, Y. Xie, X. Song, F. Wang, L. Chen and W. Zhang, Front. Bioeng. Biotechnol., 2020, 8, 500–511 CrossRef PubMed.
- Y. Wang, M. Shi, X. Niu, X. Zhang, L. Gao, L. Chen, J. Wang and W. Zhang, Microb. Cell Fact., 2014, 13, 151–163 CrossRef PubMed.
- X. Meng, X. Zhao, X. Ding, Y. Li, G. Cao, Z. Chu, X. Su, Y. Liu, X. Chen, J. Guo, Z. Cai and X. Ding, J. Agric. Food Chem., 2020, 68, 6776–6787 CrossRef CAS PubMed.
- Z. Pang, G. Zhou, J. Chong and J. Xia, Metabolites, 2021, 11, 44–58 CrossRef CAS PubMed.
- X. Bao, X. Guo, M. Yin, M. Tariq, Y. Lai, S. Kanwal, J. Zhou, N. Li, Y. Lv, C. Pulido-Quetglas, X. Wang, L. Ji, M. J. Khan, X. Zhu, Z. Luo, C. Shao, D. H. Lim, X. Liu, N. Li, W. Wang, M. He, Y. L. Liu, C. Ward, T. Wang, G. Zhang, D. Wang, J. Yang, Y. Chen, C. Zhang, R. Jauch, Y. G. Yang, Y. Wang, B. Qin, M. L. Anko, A. P. Hutchins, H. Sun, H. Wang, X. D. Fu, B. Zhang and M. A. Esteban, Nat. Methods, 2018, 15, 213–220 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1gc04188f |
| This journal is © The Royal Society of Chemistry 2022 |