 Open Access Article
Open Access Article4-Hydroxydibenzyl: a novel metabolite from the human gut microbiota after consuming resveratrol
C. E.
Iglesias-Aguirre
 a,
F.
Vallejo
a,
D.
Beltrán
a,
J.
Berná
a,
F.
Vallejo
a,
D.
Beltrán
a,
J.
Berná
 b,
J.
Puigcerver
b,
M.
Alajarín
b,
J.
Puigcerver
b,
M.
Alajarín
 b,
M. V.
Selma
b,
M. V.
Selma
 a and
J. C.
Espín
a and
J. C.
Espín
 *a
*a
aLaboratory of Food & Health, Research Group on Quality, Safety and Bioactivity of Plant Foods, CEBAS-CSIC, P. O. Box 164, 30100 Campus de Espinardo, Murcia, Spain. E-mail: jcespin@cebas.csic.es
bDepartment of Organic Chemistry, Faculty of Chemistry, University of Murcia, 30100 Murcia, Spain
First published on 23rd June 2022
Abstract
Resveratrol (RSV) was known to be metabolised by the gut microbiota to dihydroresveratrol, lunularin (LUNU), and (or) 3,4′-dihydroxy-trans-stilbene (DHST). We describe here for the first time that LUNU can be further dehydroxylated, but only at the 3-position, to yield 4-hydroxydibenzyl, a novel metabolite found in human urine after RSV intake in 41 out of 59 healthy participants. In contrast, DHST was not further dehydroxylated, and thus, 4-hydroxy-trans-stilbene was not detected as a gut microbial metabolite of RSV. Faecal in vitro incubations confirmed the in vivo results.
Introduction
The stilbene trans-resveratrol (3,5,4′-trihydroxy-trans-stilbene, resveratrol, RSV) has been reported to exert health benefits in different clinical studies.1–5 However, the metabolism of RSV by the human gut microbiota has been scarcely approached so far. An in vitro pioneering study by Jung et al.6 screened 43 animal- or human-associated bacterial strains for RSV metabolism. Among the strains metabolizing RSV, Eggerthella lenta ATCC 43055 and Bacteroides uniformis ATCC 8492 converted RSV efficiently into dihydroresveratrol (DHRSV).A further in vitro and in vivo study conducted by Bode et al.7 showed that RSV's main gut microbial metabolites were DHRSV, 3,4′-dihydroxydibenzyl (also known as lunularin, LUNU), and 3,4′-dihydroxy-trans-stilbene (DHST). After consuming a single RSV dose, these metabolites were found in hydrolysed urine samples from 12 healthy volunteers. The bacteria Slackia equolifaciens and Adlercreutzia equolifaciens converted RSV into DHRSV, although the microbial groups involved in the biotransformation of RSV to other metabolites remain mostly unknown.7 Recently, a bacterial strain isolated from human faeces, capable of reducing RSV to DHRSV has been described and proposed as Adlercreutzia rubneri sp. nov. with the type and only strain ResAG-91T.8
Recently, Jarosova et al.9 described the metabolism in vitro of RSV, among other stilbenes, using faecal samples from 5 individuals. In contrast to Bode et al.,7 DHRSV was the only metabolite detected, but not LUNU or DHST.
LUNU and DHST are produced by a single dehydroxylation at the 3- or 5-position (it is the same theoretical position as the resorcinol nucleus is symmetrical) in DHRSV and RSV, respectively.7 Thus, we hypothesized that a similar further dehydroxylation could convert LUNU into 4-hydroxydibenzyl (4HDB) and DHST into 4-hydroxy-trans-stilbene (4HST). Therefore, we aimed to identify the presence of both monohydroxylated metabolites, 4HDB and 4HST, in urine and faeces as possible novel RSV-derived metabolites from the human gut microbiota.
Materials and methods
Materials
trans-Resveratrol (3,5,4′-trihydroxy-trans-stilbene, resveratrol, RSV, 99% purity), 6,7-dihydroxycoumarin (98%), β-glucuronidase (≥100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units per mL), and sulfatase (>10
000 units per mL), and sulfatase (>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 units per g solid) from Helix pomatia were purchased from Sigma-Aldrich (St. Louis, MO, USA). 4-Hydroxy-trans-stilbene (4HST, 98%) was obtained from ThermoFisher Sci. (Madrid, Spain). Organic solvents were purchased from Merck (Darmstadt, Germany) and Milli-Q ultrapure water from Millipore Corp. (Bedford, MA, USA).
000 units per g solid) from Helix pomatia were purchased from Sigma-Aldrich (St. Louis, MO, USA). 4-Hydroxy-trans-stilbene (4HST, 98%) was obtained from ThermoFisher Sci. (Madrid, Spain). Organic solvents were purchased from Merck (Darmstadt, Germany) and Milli-Q ultrapure water from Millipore Corp. (Bedford, MA, USA).
Synthesis and spectroscopy data of resveratrol-derived metabolites
1H NMR spectra were recorded at 25 °C on Avance 300 and 400 MHz instruments (Bruker, Karlsruhe, Germany). 1H NMR chemical shifts are reported relative to tetramethylsilane (Me4Si) and were referenced via residual proton resonances of the corresponding deuterated solvent. Abbreviations of coupling patterns are as follows: br, broad; s, singlet; d, doublet; t, triplet; q, quadruplet; m, multiplet. Coupling constants (J) are expressed in Hz.Dihydroresveratrol (>97%; DHRSV) was synthesized as previously described,10 and showed identical spectroscopic data as those reported therein: 1H NMR (400 MHz, DMSO-d6) δ 9.11 (s, 1H), 9.02 (s, 2H), 6.99 (d, J = 8.4 Hz, 2H), 6.65 (d, J = 8.5 Hz, 2H), 6.06 (d, J = 2.1 Hz, 2H), 6.02 (t, J = 2.1 Hz, 1H), 2.64 (m, 4H).
4-Hydroxydibenzyl (or 4-phenethylphenol, 4HDB, >97%) was synthesized according to Camaioni et al.,11 showing the following spectroscopic data: 1H NMR (400 MHz, DMSO-d6) δ 9.12 (s, 1H), 7.30–7.12 (m, 5H), 6.99 (d, J = 8.5 Hz, 2H), 6.64 (d, J = 8.4 Hz, 2H), 2.84–2.71 (m, 4H).
3-Hydroxydibenzyl (or 3-phenethylphenol, 3HDB, >97%) was synthesized as described elsewhere,12 and showed identical spectroscopic data as those reported therein: 1H NMR (300 MHz, CDCl3) δ 7.34–7.25 (m, 2H), 7.24–7.12 (m, 4H), 6.77 (dd, J = 7.6, 1.0 Hz, 1H), 6.71–6.65 (m, 2H), 4.71 (s, 1H), 2.96–2.83 (m, 4H).
3,4′-Dihydroxydibenzyl (or 3-(4-hydroxyphenethyl)phenol, lunularin, LUNU, >97%) was synthesized following the procedure described by Ali et al.,13 and showed identical spectroscopic data as those reported therein: 1H NMR (300 MHz, CDCl3) δ 7.15 (td, J = 7.6, 0.7 Hz, 1H), 7.07–7.01 (m, 2H), 6.78–6.72 (m, 3H), 6.69–6.63 (m, 2H), 4.66 (s, 1H), 4.61 (s, 1H), 2.83 (br s, 4H).
3,4′-Dihydroxy-trans-stilbene (or 3-(4-hydroxystyryl)phenol, DHST, >97%) was synthesized as previously described,14 showing identical spectroscopic data as those reported therein: 1H NMR (400 MHz, DMSO-d6) δ 9.56 (s, 1H), 9.36 (s, 1H), 7.41 (d, J = 8.5 Hz, 2H), 7.13 (t, J = 7.8 Hz, 1H), 7.03 (d, J = 16.3 Hz, 1H), 6.98–6.88 (m, 3H), 6.76 (d, J = 8.5 Hz, 2H), 6.63 (d, J = 6.8 Hz, 1H) ppm.
Subjects and study design
This dietary intervention followed the ethical guidelines outlined in the Helsinki Declaration of 1975 and its amendments. This study belongs to an ongoing larger trial (MetaboGut), approved (reference PI-042) by IMDEA-Food (Madrid, Spain) and the Spanish National Research Council's Bioethics Committee (Madrid). The present study was not designed to evaluate specific health effects on the volunteers but to increase the current knowledge about the metabolism of RSV by the human gut microbiota.Volunteers with no diagnosed chronic disease were recruited. Table 1 shows the main characteristics of the participants. Exclusion criteria were as follows: pregnancy/lactation, recent use of antibiotics (within 1-month prior to the study), history of smoking (recent past or present), diagnosed chronic illness, previous gastrointestinal surgery, or taking medication (within 1-month prior to the study). The study was explained to the participants who provided written informed consent.
Participants consumed one daily hard gelatin capsule in the evening for 7 days. The capsules contained 150 mg RSV from Polygonum cuspidatum and were manufactured by Laboratorios Admira S.L. (Alcantarilla, Murcia, Spain) following the European Union's Good Manufacturing Practices requirements. The RSV content of the extract was certified by the company (98.7% purity by HPLC) and was free of anthraquinones, metals, solvents, and pathogens.
Faecal cultures
Baseline stool samples were provided by 9 volunteers randomly chosen from the group (7 males and 2 females normoweight subjects). Preparation of faecal suspensions and subsequent culturing experiments were conducted with minor modifications as previously described.15Aliquots were prepared with 10 g of stool samples in filter bags and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (w/v) with L-cysteine hydrochloride supplemented (0.05%) Nutrient Broth using a stomacher for homogenization. Filtered faecal suspensions (50 μL) were inoculated into 5 mL of Wilkins-Chalgren anaerobe medium (WAM, Oxoid) added with 0.05% L-cysteine and containing a 30 μM solution of each standard in DMSO (0.6% DMSO in the final culture medium). The compounds (RSV, DHRSV, DHST, LUNU, 4HDB, and 4HST) were individually added to the broth and incubated under anoxic conditions in an anaerobic chamber (Concept 400, Baker Ruskin Technologies Ltd, Bridgend, South Wales, UK) with an atmosphere consisting of N2/H2/CO2 (85
10 (w/v) with L-cysteine hydrochloride supplemented (0.05%) Nutrient Broth using a stomacher for homogenization. Filtered faecal suspensions (50 μL) were inoculated into 5 mL of Wilkins-Chalgren anaerobe medium (WAM, Oxoid) added with 0.05% L-cysteine and containing a 30 μM solution of each standard in DMSO (0.6% DMSO in the final culture medium). The compounds (RSV, DHRSV, DHST, LUNU, 4HDB, and 4HST) were individually added to the broth and incubated under anoxic conditions in an anaerobic chamber (Concept 400, Baker Ruskin Technologies Ltd, Bridgend, South Wales, UK) with an atmosphere consisting of N2/H2/CO2 (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) at 37 °C. Incubations of faecal cultures without added compounds, and incubations of the compounds without fecal inocula, were used as controls. Three replicates were carried out using each faecal suspension and compound. Samples were collected after 7 days of incubation and processed for UPLC-QTOF-MS and(or) GC-MS analyses.
10) at 37 °C. Incubations of faecal cultures without added compounds, and incubations of the compounds without fecal inocula, were used as controls. Three replicates were carried out using each faecal suspension and compound. Samples were collected after 7 days of incubation and processed for UPLC-QTOF-MS and(or) GC-MS analyses.
Sampling procedure and processing
Volunteers provided urine and faeces at baseline and after 7 days. Samples were frozen at −80 °C until further analysis. The urine samples were centrifuged, filtered through a 0.22 μm PVDF filter, and diluted with acidified water (0.1% formic acid) before analysis by UPLC-QTOF-MS. The urinary excretion of creatinine was measured to allow standardisation of diuresis, as reported elsewhere.16For the complete deconjugation of phase-II conjugated metabolites, urine samples were treated overnight with glucuronidase and sulfatase from Helix pomatia, as previously described.16 Hydrolysed urine samples were analysed by UPLC-QTOF-MS and GC-MS.
Stool samples (1 g) were processed as previously described.17 Faecal samples (5 mL) from fermentation experiments were extracted with 5 mL of ethyl acetate plus 1.5% formic acid and processed, as described elsewhere.15 Both faecal and in vitro fermented samples were analysed by LC-MS and GC-MS.
In the case of GC-MS analyses, urine, faeces, and faecal fermentation samples were extracted as described above and analyzed with and without derivatisation. The evaporated residues were dissolved in 30 μL pyridine and converted to trimethylsilyl derivatives by adding 30 μL of N,O-bis-(trimethylsilyl) trifluoroacetamide (BSTFA) containing 1% trimethylchlorosilane (TMCS) and incubated at 100 °C for 15 min. The sample residues after speed vacuum evaporation were dissolved in acetone and injected into the GC-MS equipment for the analyses without derivatization.
Analysis of resveratrol and derived metabolites
The first methodology was optimised for non-silylated metabolites, 3HDB and 4HDB, and the MS parameters were the following: SIM acquisition type with both 198 and 107 m/z ions selected, segment retention times fixed between 21 and 24 min with high-resolution mode, dwell time of 25 ms and cycle time of 13.96 Hz, and finally, a calculated EMV of 1700.
The second methodology was used for silylated metabolites, i.e., LUNU, DHP, RSV, DHRSV, DHST, and 4HST, and the MS parameters were the following: SCAN acquisition type with a threshold of 1000, scan mass range from 50 to 800 Da at 2.0 scan per s, a scan speed of 1.562 u/s, a cycle time of 502.60 ms and finally, a calculated EMV of 1360.
Compounds were identified by direct comparison with the available standards and confirmed by their spectral properties, molecular mass, and fragmentation pattern. The quantification of RSV and derived metabolites and their conjugates were determined by interpolation in the calibration curves obtained with their corresponding available standards in the urine and faecal matrices.
Results
Resveratrol metabolites in urine and faeces
The UPLC-ESI-QTOF-MS analysis of urine samples allowed the tentative identification of 25 RSV-derived metabolites, mostly phase-II derived from both human and microbial origin (results not shown). Thus, the hydrolysis of urine samples was carried out to identify the parent (unconjugated) RSV and its derived gut microbial metabolites using the available standards. As a result, RSV, cis-RSV, DHRSV, LUNU, and DHST were identified in urine. Fig. 1 shows representative extracted ion chromatograms of these metabolites, although high inter-individual variability was observed since RSV, cis-RSV, and DHRSV were observed in all the samples, while LUNU was detected in 41 and DHST only in 14 of the 59 volunteers. The same metabolites were also detected in faeces (results not shown). However, no monohydroxylated metabolites were found in urine or faeces despite different modifications of the UPLC-ESI-QTOF protocol, or UPLC-QQQ analyses were tested (results not shown).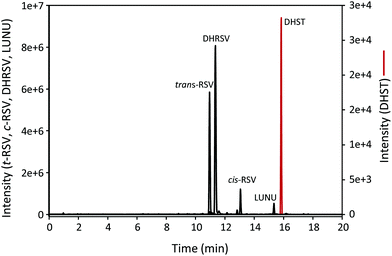 | ||
| Fig. 1 UPLC-ESI-QTOF-MS extracted ion chromatograms of hydrolysed urine samples after RSV intake for 7 days. | ||
Next, we searched for the possible monohydroxylated metabolites using GC-MS. Table 2 shows the compounds detected using GC-MS in silylated and non-silylated samples. Silylation allowed for determining the same metabolites detected by UPLC-QTOF-MS (Table 2 and Fig. 2). The available standard of the monohydroxylated stilbene 4HST was detected by GC-MS, but its ion mass was not found in any urine or faeces sample, which also excluded the presence of the isomer 3-hydroxy-trans-stilbene (3HST). Since detecting the monohydroxylated dibenzyls 3HDB and 4HDB remained elusive, we next analyzed the hydrolysed urine samples by GC-MS with no silylation. Finally, both available 3HDB and 4HDB standards were detected in non-silylated samples (Table 2 and Fig. 3A). The metabolite 4HDB, with an ion mass of 198 and the main fragment of 107, was found in the urine of 23 individuals after consuming RSV but not in their control urine samples (Fig. 3B). The comparison of the peak detected in the participants’ urine with the available standard allowed us the identification of 4HDB as a novel metabolite produced by the human gut microbiota after consuming RSV (Table 2 and Fig. 3). In contrast, the isomer 3HDB was not detected in any sample.
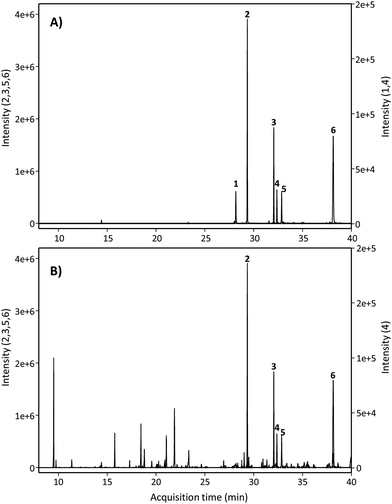 | ||
| Fig. 2 GC-MS extracted ion chromatograms after silylation of (A) standards and (B) hydrolysed urine samples. Peak numbers are listed in Table 2 (silylated samples). | ||
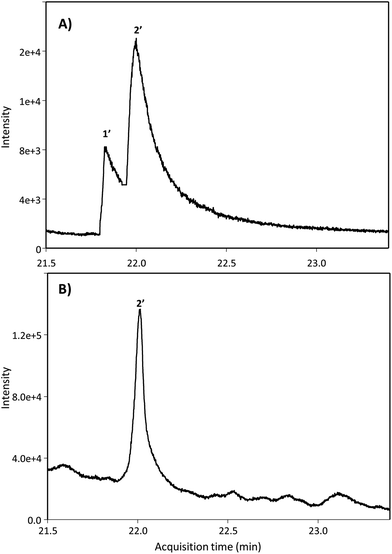 | ||
| Fig. 3 GC-MS extracted ion chromatograms of non-silylated samples of (A) dibenzyl standards and (B) hydrolysed urine. Peak numbers are listed in Table 2 (non-silylated samples). | ||
| No. | Compound | RT (min) | Mass | Target ion | LOD; LOQ (nM) | Occurrence | Urine (μg mg−1 creatinine)b | Faeces (μg g−1)b |
|---|---|---|---|---|---|---|---|---|
| U, urine; F, faeces; FC, faecal culture; ND, not detected; D, detected but not quantified; 4HST, 4-hydroxy-trans-stilbene; LUNU, lunularin; DHRSV, dihydroresveratrol; DHST, 3,4′-dihydroxy-trans-stilbene; RSV (trans-resveratrol); 3HDB, 3-hydroxydibenzyl; 4HDB, 4-hydroxydibenzyl.a Hydrolysed samples.b Mean ± SD.c Tentatively quantified as RSV.d Mass without silylation. | ||||||||
| Silylated | ||||||||
| 1 | 4HST | 28.1 | 268 | 268 | 200; 500 | ND | — | — |
| 2 | LUNU | 29.3 | 358 | 179 | 50; 100 | U, F, FC | 85.7 ± 129.5 | 30.1 ± 29.3 |
| 3 | cis-RSV | 32.0 | 444 | 444 | — | U | 268.2 ± 150.3c | — |
| 4 | DHRSV | 32.4 | 446 | 179 | 100; 250 | U, F, FC | 909.2 ± 649.2 | 33.6 ± 50.2 |
| 5 | DHST | 32.9 | 356 | 356 | 200; 500 | U, F | 10.0 ± 4.4 | D |
| 6 | RSV | 38.2 | 444 | 444 | 100; 250 | U, F, FC | 464.8 ± 266.3 | 1.5 ± 3.0 |
| Non-silylated | ||||||||
| 1′ | 3HDB | 21.8 | 198d | 107 | 200; 500 | ND | — | — |
| 2′ | 4HDB | 22.1 | 198d | 107 | 100; 250 | U, FC | 12.5 ± 36.6 | — |
Faecal cultures to confirm the production of 4HDB and (or) 4HST
Next, we separately incubated RSV, DHRSV, LUNU, DHST, and 4HST in faecal cultures from 9 participants randomly chosen to detect the corresponding derived metabolites. After testing different incubation times, we chose 7 days since longer incubation times did not modify the results (results not shown). Again, interindividual variability was observed, as in the case of the urine and faeces analysed. After 7 days of incubation, RSV yielded DHRSV in all the samples (something readily observed at shorter incubation times). LUNU and 4HDB were also detected, but only in faecal cultures from 6 of 9 individuals (steps a–d, Fig. 4), thus confirming the in vivo results. 4HST was not detected in any sample, also confirming the lack of 4HST derivatives in vivo after RSV intake.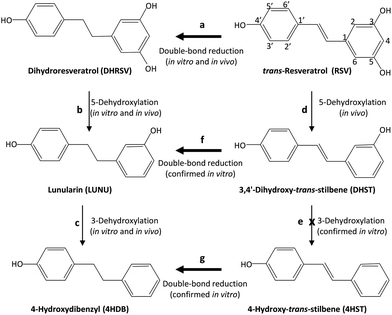 | ||
| Fig. 4 In vivo and in vitro production of 4HDB, but not 4HST, by the gut microbiota from RSV. Thicker arrows indicate favoured reactions. | ||
The incubation of DHRSV produced the metabolites LUNU and 4HDB (steps b and c, Fig. 4), but only in some volunteers. However, 3HDB was not detected in any sample. Next, the incubation of LUNU confirmed the lack of 3HDB production, and only 4HDB was detected in some samples.
Finally, the incubation of DHST did not produce 4HST but LUNU (step f, Fig. 4), something observed in all the samples analysed, and 4HDB in 7 samples (step c, Fig. 4). No further degradation of 4HDB was observed in any sample (results not shown). Similarly, the incubation of 4HST only produced 4HDB in all the individuals (step g, Fig. 4). Overall, and considering a high interindividual variability in RSV metabolism, faecal incubations confirmed the results obtained in vivo after RSV intake.
Discussion
Since Jang et al.18 reported the RSV's anticancer activity, RSV has been described as a compound that could increase lifespan and prevent or reduce many pathological processes or diseases in preclinical and clinical studies.19–22The human gut microbiota catabolises RSV to produce DHRSV and, to a lower extent, LUNU, and DHST, showing interindividual variability.7 In this regard, as recently suggested, different human gut microbiota compositions might affect the outcome of trials with RSV since the biological activity of the microbial metabolites might differ from that of RSV,23,24 which might explain the lack of consensus on RSV as a health-promoting polyphenol.25
In the present study and, in agreement with Bode et al.,7 we detected the gut microbial metabolites DHRSV, LUNU, and DHST after RSV intake and confirmed the interindividual variability of RSV metabolism in a group of 59 volunteers.
However, confirming this study's hypothesis, we describe here for the first time that LUNU can be further dehydroxylated at the 3-position to yield 4HDB as a novel metabolite produced by the human gut microbiota from RSV. We have adopted the IUPAC nomenclature to name this metabolite, i.e., 4-hydroxydibenzyl instead of 4-hydroxybibenzyl since the dibenzyl core is 1,2-diphenylethane, being correct names 4-(2-phenylethyl)phenol, 4-hydroxydibenzyl, 4-phenethylphenol, p-phenylethylphenol. In this regard, we have used 4-hydroxydibenzyl to highlight the position of the –OH group. Notably, 4HDB was a minor metabolite found only in those participants that produced LUNU. In contrast, LUNU was not dehydroxylated at the 4-position to yield 3-hydroxydibenzyl (3HDB), which was not detected in vivo or in vitro. Therefore, these results suggest that the human gut microbiota lacks the capacity to dehydroxylate stilbenes and dibenzyls at the 4-position. The specificity of dehydroxylases is also relevant in the metabolism of bile acids26 and the sequential dehydroxylation pathway of urolithins and different phenolics.27,28 Although this requires further research, we hypothesize the different capability to dehydroxylate DHRSV and LUNU is a hallmark of inter-individual variability of RSV metabolism. Whether this metabolism, including the production of 4HDB, impacts health upon RSV intake deserves further investigation.
The double bond enzymatic reduction of the 4-styrylphenol core seemed to be the most favoured step since DHRSV was detected in the urine and faeces of all individuals after RSV intake, which was confirmed in vitro after incubation of the stilbenes RSV, DHST, and 4HST to produce DHRSV, LUNU, and 4HDB, respectively. 4HST reduction only occurred in the presence of faecal inocula in the in vitro studies. Therefore, the most abundant substrates for dehydroxylases were dibenzyls (DHRSV and LUNU), which might explain the sporadic detection of DHST (14 individuals out of 59). Similarly, 4HST was not detected in any in vivo or in vitro samples because DHST was reduced to LUNU before producing 4HST, which, if any were produced, would be immediately reduced to 4HDB, preventing 4HST accumulation. Therefore, assuming 4HST could be potentially an RSV-metabolite produced by the human gut microbiota, other more favoured reactions prevented its detection, i.e., double bond reduction of the 4-styrylphenol core, even after 7 days of static in vitro incubation. This finding agrees with the metabolic profile of RSV-derived metabolites in non-hydrolysed samples previously reported, including the tentative identification of different LUNU and DHST glucuronide and sulfate conjugates, but not 4HST conjugates.16,29
4HDB was not found in faeces, probably due to (i) its rapid absorption and phase-II conjugation to be excreted in the urine, (ii) its low content (below LOD), and (or) (iii) possible interferences between stool matrix and GC-MS analysis without silylation. In contrast, 4HDB was detected in hydrolysed urine samples. However, we could not detect the corresponding 4HDB conjugates because of the lack of signal in UPLC-QTOF-MS and UPLC-QQQ. Besides, as expected, 4HDB conjugates were not detected by GC-MS either because they lack the volatility required for analysis by GC-MS.30 Although challenging, we acknowledge that 4HDB detection should be improved in further studies.
Conclusions
The compound 4-hydroxydibenzyl (4HDB) is described here for the first time as a novel metabolite produced by the human gut microbiota after consuming RSV and specifically produced after dehydroxylation of lunularin (LUNU) at the 3-position. GC-MS was crucial for detecting this metabolite, which had remained undetected despite many investigations on RSV metabolism.Our results suggest that the human gut microbiota could produce, at least theoretically in some volunteers, the compound 4-hydroxy-trans-stilbene (4HST). However, this metabolite was not detected in any sample because reducing the double bond of the 4-styrylphenol core of its potential precursors DHST and RSV was preferential, thus favouring the 3-dehydroxylation of the dibenzyls DHRSV and LUNU resulting from that reduction.
Further research on the biological activity and safety issues of 4HDB is warranted. In addition, our ongoing research addresses the observed high variability in RSV metabolism by the human gut microbiota, emphasizing why not all individuals can dehydroxylate stilbenes or dibenzyls.
Author contributions
Conceptualization: J. C. E.; investigation: C. E. I. A., F. V., D. B.; methodology: F. V., J. B., J. P., M. A.; visualization: C. E. I. A., F. V., M. V. S.; validation: F. V., C. E. I. A., J. C. E.; resources: J. C. E, M. V. S.; supervision: J. C. E.; writing original draft: J. C. E.; writing – review & editing: all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research has been supported by Project PID2019-103914RB-I00 from the Ministry of Science and Innovation (MICINN, Spain). C. E. I. A. and J. P. are the holders of predoctoral grants FPU18/03961 and FPU19/05419, respectively, from MICINN.References
- J. Tomé-Carneiro, M. Gonzálvez, M. Larrosa, M. J. Yáñez-Gascón, F. J. García-Almagro, J. A. Ruiz-Ros, F. A. Tomás-Barberán, M. T. García-Conesa and J. C. Espín, Grape resveratrol increases serum adiponectin and downregulates inflammatory genes in peripheral blood mononuclear cells: a triple-blind, placebo-controlled, one-year clinical trial in patients with stable coronary artery disease, Cardiovasc. Drugs Ther., 2013, 27, 37–48 CrossRef PubMed.
- C. Moussa, M. Hebron, X. Huang, J. Ahn, R. A. Rissman, P. S. Aisen and R. S. Turner, Resveratrol regulates neuro-inflammation and induces adaptive immunity in Alzheimer's disease, J. Neuroinflammation, 2017, 14, 1 CrossRef PubMed.
- A. Hoseini, G. Namazi, A. Farrokhian, Ž. Reiner, E. Aghadavod, F. Bahmani and Z. Asemi, The effects of resveratrol on metabolic status in patients with type 2 diabetes mellitus and coronary heart disease, Food Funct., 2019, 10, 6042–6051 RSC.
- R. H. Wong, J. J. Thaung Zaw, C. J. Xian and P. R. Howe, Regular Supplementation With Resveratrol Improves Bone Mineral Density in Postmenopausal Women: A Randomized, Placebo-Controlled Trial, J. Bone Miner. Res., 2020, 35, 2121–2131 CrossRef CAS PubMed.
- J. J. Thaung Zaw, P. R. Howe and R. H. Wong, Long-term effects of resveratrol on cognition, cerebrovascular function and cardio-metabolic markers in postmenopausal women: A 24-month randomised, double-blind, placebo-controlled, crossover study, Clin. Nutr., 2021, 40, 820–829 CrossRef CAS PubMed.
- C. M. Jung, T. M. Heinze, L. K. Schnackenberg, L. B. Mullis, S. A. Elkins, C. A. Elkins, R. S. Steele and J. B. Sutherland, Interaction of dietary resveratrol with animal-associated bacteria, FEMS Microbiol. Lett., 2009, 297, 266–273 CrossRef CAS PubMed.
- L. M. Bode, D. Bunzel, M. Huch, G.-S. Cho, D. Ruhland, M. Bunzel, A. Bub, C. M. Franz and S. E. Kulling, In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota, Am. J. Clin. Nutr., 2013, 97, 295–309 CrossRef CAS PubMed.
- D. A. Stoll, N. Danylec, S. T. Soukup, B. Hetzer, S. E. Kulling and M. Huch, Adlercreutzia rubneri sp. nov., a resveratrol-metabolizing bacterium isolated from human faeces and emended description of the genus Adlercreutzia, Int. J. Syst. Evol. Microbiol., 2021, 71, 004987 CAS.
- V. Jarosova, O. Vesely, P. Marsik, J. Jaimes, K. Smejkal, P. Kloucek and J. Havlik, Metabolism of Stilbenoids by Human Faecal Microbiota, Molecules, 2019, 24, 1155 CrossRef PubMed.
- V. Cardile, L. Lombardo, C. Spatafora and C. Tringali, Chemo-enzymatic synthesis and cell-growth inhibition activity of resveratrol analogues, Bioorg. Chem., 2005, 33, 22–33 CrossRef CAS PubMed.
- D. M. Camaioni and J. A. Franz, Carbon-hydrogen vs. carbon-carbon bond cleavage of 1,2-diarylethane radical cations in acetonitrile-water, J. Org. Chem., 1984, 49, 1607–1613 CrossRef CAS.
- A. Wilhelm, P. Kendrekar, A. E. M. Noreljaleel, E. T. Abay, S. L. Bonnet, L. Wiesner, C. de Kock, K. J. Swart and J. H. van der Westhuizen, Synthesis and in vitro antiplasmodial activity of aminoalkylated chalcones and analogues, J. Nat. Prod., 2015, 78, 1848–1858 CrossRef CAS PubMed.
- M. A. Ali, K. Kondo and Y. Tsuda, Synthesis and Nematocidal Activity of Hydroxystilbenes, Chem. Pharm. Bull., 1992, 40, 1130–1136 CrossRef CAS.
- R. Martí-Centelles, E. Falomir, J. Murga, M. Carda and J. A. Marco, Inhibitory effect of cytotoxic stilbenes related to resveratrol on the expression of the VEGF, hTERT and c-Myc genes, Eur. J. Med. Chem., 2015, 103, 488–496 CrossRef PubMed.
- D. Beltrán, M. D. Frutos-Lisón, J. C. Espín and R. García-Villalba, Re-examining the role of the gut microbiota in the conversion of the lipid-lowering statin monacolin K (lovastatin) into its active β-hydroxy acid metabolite, Food Funct., 2019, 10, 1787–1791 RSC.
- M. Á. Ávila-Gálvez, R. García-Villalba, F. Martínez-Díaz, B. Ocaña-Castillo, T. Monedero-Saiz, A. Torrecillas-Sánchez, B. Abellán, A. González-Sarrías and J. C. Espín, Metabolic Profiling of Dietary Polyphenols and Methylxanthines in Normal and Malignant Mammary Tissues from Breast Cancer Patients, Mol. Nutr. Food Res., 2019, 63, 1801239 CrossRef PubMed.
- R. García-Villalba, J. C. Espín and F. A. Tomás-Barberán, Chromatographic and spectroscopic characterization of urolithins for their determination in biological samples after the intake of foods containing ellagitannins and ellagic acid, J. Chromatogr. A, 2016, 1428, 162–175 CrossRef PubMed.
- M. Jang, L. Cai, G. O. Udeani, K. V. Slowing, C. F. Thomas, C. W. W. Beecher, H. H. S. Fong, N. R. Farnsworth, A. D. Kinghorn, R. G. Mehta, R. C. Moon and J. M. Pezzuto, Cancer chemopreventive activity of resveratrol, a natural product derived from grapes, Science, 1997, 275, 218–220 CrossRef CAS PubMed.
- J. Tomé-Carneiro, M. Larrosa, A. González-Sarrías, F. Tomás-Barberán, M. García-Conesa and J. C. Espín, Resveratrol and clinical trials: the crossroad from in vitro, studies to human evidence, Curr. Pharm. Des., 2013, 19, 6064–6093 CrossRef PubMed.
- F. Fogacci, G. Tocci, V. Presta, A. Fratter, C. Borghi and A. F. G. Cicero, Effect of resveratrol on blood pressure: A systematic review and meta-analysis of randomized, controlled, clinical trials, Crit. Rev. Food Sci. Nutr., 2019, 59, 1605–1618 CrossRef CAS PubMed.
- F. M. Delpino and L. M. Figueiredo, Resveratrol supplementation and type 2 diabetes: a systematic review and meta-analysis, Crit. Rev. Food Sci. Nutr., 2021, 1–16 Search PubMed.
- D. Delmas, C. Cornebise, F. Courtaut, J. Xiao and V. Aires, New Highlights of Resveratrol: A Review of Properties against Ocular Diseases, Int. J. Mol. Sci., 2021, 22, 1295 CrossRef CAS PubMed.
- K. Pallauf, D. Chin, I. Günther, M. Birringer, K. Lüersen, G. Schultheiß, S. Vieten, J. Krauß, F. Bracher, N. Danylec, S. T. Soukup, S. E. Kulling and G. Rimbach, Resveratrol, lunularin and dihydroresveratrol do not act as caloric restriction mimetics when administered intraperitoneally in mice, Sci. Rep., 2019, 9, 4445 CrossRef PubMed.
- I. Günther, G. Rimbach, C. I. Mack, C. H. Weinert, N. Danylec, K. Lüersen, M. Birringer, F. Bracher, S. T. Soukup, S. E. Kulling and K. Pallauf, The Putative Caloric Restriction Mimetic Resveratrol has Moderate Impact on Insulin Sensitivity, Body Composition, and the Metabolome in Mice, Mol. Nutr. Food Res., 2020, 64, 1901116 CrossRef PubMed.
- P. C.-T. Tang, Y.-F. Ng, S. Ho, M. Gyda and S.-W. Chan, Resveratrol and cardiovascular health–promising therapeutic or hopeless illusion?, Pharmacol. Res., 2014, 90, 88–115 CrossRef CAS PubMed.
- M. Funabashi, T. L. Grove, M. Wang, Y. Varma, M. E. McFadden, L. C. Brown, C. Guo, S. Higginbottom, S. C. Almo and M. A. Fischbach, A metabolic pathway for bile acid dehydroxylation by the gut microbiome, Nature, 2020, 582, 566–570 CrossRef CAS PubMed.
- M. V. Selma, J. C. Espín and F. A. Tomás-Barberán, Interaction between phenolics and gut microbiota: role in human health, J. Agric. Food Chem., 2009, 57, 6485–6501 CrossRef CAS PubMed.
- R. García-Villalba, D. Beltrán, M. D. Frutos, M. V. Selma, J. C. Espín and F. A. Tomás-Barberán, Metabolism of different dietary phenolic compounds by the urolithin-producing human-gut bacteria Gordonibacter urolithinfaciens and Ellagibacter isourolithinifaciens, Food Funct., 2020, 11, 7012–7022 RSC.
- M. Á. Ávila-Gálvez, A. González-Sarrías, F. Martínez-Díaz, B. Abellán, A. J. Martínez-Torrano, A. J. Fernández-López, J. A. Giménez-Bastida and J. C. Espín, Disposition of Dietary Polyphenols in Breast Cancer Patients’ Tumors, and Their Associated Anticancer Activity: The Particular Case of Curcumin, Mol. Nutr. Food Res., 2021, 65, 2100163 CrossRef PubMed.
- J. L. Ordóñez, G. Pereira-Caro, I. Ludwig, J. M. Muñoz-Redondo, M. J. Ruiz-Moreno, A. Crozier and J. M. Moreno-Rojas, A critical evaluation of the use of gas chromatography- and high performance liquid chromatography-mass spectrometry techniques for the analysis of microbial metabolites in human urine after consumption of orange juice, J. Chromatogr. A, 2018, 1575, 100–112 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |
