Open Access Article
Open Access ArticleInfluence of the ecological environment on the structural characteristics and bioactivities of polysaccharides from alfalfa (Medicago sativa L.)†
Chongyu
Zhang‡
a,
Eunyoung
Kim‡
b,
Jiamei
Cui
b,
Yunpeng
Wang
a,
Yunkyoung
Lee
*bc and
Guiguo
Zhang
 *a
*a
aDepartment of Nutrition and China-Korea Joint R&D center on Plant-derived polysaccharide, Shandong Agricultural University, 61 Daizong Street, Taian City 271018, China. E-mail: zhanggg@sdau.edu.cn
bDepartment of Food Science and Nutrition, and Korea-China Joint R&D center on Plant-derived polysaccharide, Jeju National University, Jeju 63243, South Korea. E-mail: lyk1230@jejunu.ac.kr
cInterdisciplinary Graduate Program in Advanced Convergence Technology & Science, Jeju National University, Jeju 63243, South Korea
First published on 24th May 2022
Abstract
Polysaccharides from alfalfa (Medicago sativa L.) (APS) exhibit a variety of bioactivities; however, little information is available on the effects of the ecological environment on the structural characteristics and bioactivities of APS. This study aimed to investigate the structural characteristics and bioactivities of two APS types isolated from alfalfa; these APSs were obtained from alfalfa cultured in normal soil (APS1) or saline–alkali soil (APS2). Results indicated that the two kinds of APS had the same monomer compositions in different molar proportions, where APS2 had greater content of arabinose and galacturonic acid than APS1. Furthermore, APS1 exhibited a greater molar mass of 1.77 × 105 g mol−1 as compared to 1.01 × 105 g mol−1 for APS2. Likewise, APS1 and APS2 had highly branched molecules with crosslinking nets composed of similar monomer residues but with different glycosidic linkages. Additionally, both APS significantly inhibited both adipogenesis and lipid accumulation in 3T3-L1 cells by downregulating mRNA expression of Ppar-γ, C/ebp-α, and Fas; APS2 had superior antiadipogenic effects as compared to APS1. Altogether, the ecological environment impacts the structural characteristics and biofunctions of APS, making them potential candidates for antiadipogenic use through functional food. These findings provide a novel perspective for the selection of phytogenic polysaccharides with specific bioactivities by considering growth environmental conditions.
1. Introduction
Plant-derived polysaccharides (PS) are a kind of macromolecular polymer synthesized in plants, which plays many vital roles in maintaining the integrity of cells and in imparting resistance against abiotic stress in plants.1,2 As a functional food/feed for humans and animals, phytogenic PS exhibit a variety of bioactivities,3,4 including antioxidative capacity,5,6 anti-inflammation,7,8 immunomodulation,9 and growth-promotion.10 Homogeneous PS are composed of a single kind of monomer by glycosidic bonding, whereas hetero-PS comprise polymers of several kinds of monosaccharides or uronic acids.11 The difference in the molecular composition of PS is believed to be a critical contributor to their diverse bioactivities.12,13 The PS isolated from brown seaweed, Laminaria japonica, dominantly consist of fucose (Fuc) and galactose (Gal) and exhibit high bile acid-binding capacity.14 Similarly, the PS from alfalfa (Medicago sativa L.) are mainly composed of glucose (Glc), galacturonic acid (GalA), and glucuronic acid (GlcA), and they perform specific activation of B cells.10 Soua et al. (2020) reported that the hydroxyl and carboxyl functionalities of uronic acid are tightly related to the antioxidant capacity of the PS that they form. This is attributable to their functioning as hydrogen donors capable of scavenging the 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical, which neutralizes oxidative stress.15 Altogether these studies indicate that the structural characteristics of phytogenic PS confer their bioactivities.13,16Most plant-derived dietary PS are resistant to degradation in the host due to the lack of relevant hydrolytic enzymes,17 therefore, these carbohydrates serve as the main energy sources for the gut microbial community and define their configuration and diversity.18–20 Patnode et al. (2019) documented that the PS from pea are primarily made up of arabinose (Ara) and promote colonization of thetaiotaomicron, whereas the PS in citrus pectin consist of GalA and selectively enhance the abundance of Vulgatus.20 Similarly, Kanwal et al. (2018) found the PS from Dictyophora indusiata are mainly composed of mannose, glucose, and galactose and are capable of significantly improving the abundance of Lactobacillaceae and Ruminococaceae in gut flora.21 Such reports assert that the different diet-derived PS with varying molecular structures can selectively shape the intestinal microbial community, and thus, is closely related to host phenotypes and disease occurrence, in general. It is, therefore, worthwhile to ascertain the molecular structure of PS to help develop precise nutrition for both animals and humans that can modulate microbial communities to enhance health-promoting effects of food.15,22
Alfalfa is a widely cultured saline-resistant, double-purpose crop that has been extensively employed as food/feed due to its rich and high-quality PS.12,23 Being a primary metabolite of plants, PS are involved in the formation of the cell wall. Their monosaccharide compositions and content significantly vary with the period of growth and the ecological environment. Our previous studies have revealed that the PS derived from alfalfa (APS) are primarily composed of GalA (57.11%), Glc (16.42%), and Ara (6.19%), and exerted antioxidant and immunomodulatory bioactivities.7,10 Contrarily, Rovkina et al. (2018) observed the major monosaccharides in APS to be Ara (43.60–46.40%) and GalA (28.20–33.40%).24 Furthermore, Ara was highlighted as an ecologically-sensitive molecule, whose synthesis in the plant was affected by ecological and environmental conditions.25,26 In another study, Ara biosynthesis was demonstrated to be a critical pathway in Arabidopsis for resistance against salt stress.27 Soil salinity is the most common abiotic stress signal that can enhance a plant's stress-tolerant capacity by triggering the biosynthesis of intracellular components.28 However, the mechanisms by which saline–alkali soils affect the composition and bioactivities of PS in alfalfa remain unclear. Therefore, it is crucial to investigate the effects of saline–alkali soils on the structural characteristics of APS for the targeted isolation and application of functional PS with required bioactivities.
We hypothesized that the saline–alkali condition of soil can influence the structural characteristics and bioactivities of APS. The present study thus aimed to investigate (1) the effects of saline–alkali soil on the structural characteristics of APS, and (2) the difference in bioactivities of two kinds of PS from alfalfa grown in either normal soil or saline–alkali soil using an in vitro model.
2. Materials and methods
2.1. Preparation of alfalfa polysaccharide
Alfalfa samples were collected from two different sites located in the temperate monsoon zone with a continental climate and diverse soil conditions. The first sampling site was located at the Chinese National Huang-Huai-Hai Regional Corn Technology Innovation Centre (N36°08′51.77′′ E116°59′49.25′′; 134 m asl); it had a brown loam soil characterized as being fine, mixed, super-active, and mesic. The average organic matter content was 18.87 mg kg−1, and the total N, Olsen P, and K in the upper 20 cm of soil in the experimental field were 3.45 g kg−1, 31.60 mg kg−1, and 78.20 mg kg−1, respectively. The second sampling site, located at Bohai Bay (N38°02′3.72′′ E118°22′5.49′′; 5 m asl), was the main saline–alkali soil area in northern China with a moderate to severe saline–alkali soil (average salt content was 0.30∼0.45%, average pH value of soil was 8.40). Organic matter content was 12.95 g kg−1, and the total N, Olsen P, and K in the upper 20 cm of soil in the experimental field were 75.95 mg kg−1, 18.42 mg kg−1, and 118.81 mg kg−1, respectively. All alfalfa samples collected at the two sites belonged to the same species (WL363, US) and were harvested at their early blooming stage (1/10 blooming).29 Fresh grass samples were immediately taken back to the lab and dried in an air-forced oven at 65 °C for further extraction and purification.10,30 The PS were extracted from alfalfa hay using hot water extraction, ethanol precipitation, and dialysis methods described in our previous report.10 Briefly, the dried alfalfa hay was chopped into 2∼3 cm long pieces and mixed with double-distilled water in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (v
30 (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) in a big glass beaker, boiled for 4 h and subsequently filtered through two layers of nylon mesh (0.2 mm mesh). The filtrate was continuously heated and kept slightly boiling to concentrate the filtrate to 1/4 volume from the initial volume. After cooling, the remaining filtrate was mixed with trichloroacetic acid (TCA) solution (5% concentrate) at a proportion of 1
v) in a big glass beaker, boiled for 4 h and subsequently filtered through two layers of nylon mesh (0.2 mm mesh). The filtrate was continuously heated and kept slightly boiling to concentrate the filtrate to 1/4 volume from the initial volume. After cooling, the remaining filtrate was mixed with trichloroacetic acid (TCA) solution (5% concentrate) at a proportion of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (v
2 (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v, filter liquid
v, filter liquid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TCA) and maintained for 2 h statically to remove protein from the liquid. After the protein was precipitated completely, the liquid fraction was centrifuged at 3000g for 10 min to collect the supernatant liquid; then, the supernatant was transferred to another container and a 4-fold volume of absolute ethyl alcohol (v
TCA) and maintained for 2 h statically to remove protein from the liquid. After the protein was precipitated completely, the liquid fraction was centrifuged at 3000g for 10 min to collect the supernatant liquid; then, the supernatant was transferred to another container and a 4-fold volume of absolute ethyl alcohol (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) was added. The mixture was kept at 4 °C for 12 h and then centrifuged at 3000g for 10 min to separate the precipitated crude PS. The fractionated crude PS was subsequently re-dissolved in d-H2O and dialyzed twice using an ultrafiltration membrane (molar mass >3500 D Mw, degree of polymerization (DP) > 10, Beijing Solarbio Science and Technology Co., Ltd, Beijing, China) against d-H2O (10 times the sample volume) at 4 °C for 48 h, changing the d-H2O every 12 h. The dialyzed liquid was collected and lyophilized to a constant weight using a vacuum dryer (Biosafer-10A, Biosafer, Nanjing, China), which was considered the crude PS from alfalfa (APS). Further purification was performed using anion-exchange column chromatography. Three grams of crude PSs was dissolved in 100 mL of distilled water and then applied to a DEAE-52 anion-exchange column (5.0 × 100.0 cm). A stepwise gradient NaCl aqueous solution (0, 0.2, 0.4 mol L−1) was used for elution at a flow rate of 0.5 mL min−1. Three fractions were collected separately and the concentration of polysaccharide in elution was detected with the phenol–sulfuric acid method.14,31 The absorbance peak of 0.4 mol L−1 was much larger than the others, therefore, the fraction eluted with 0.4 mol L−1 NaCl was further purified using gel-permeation chromatography with a Sephacryl S-500, yielding one homogeneous fraction APS.32,33 The column was washed with distilled water at a rate of 0.2 mL min−1, and eluted samples were freeze-dried.
v) was added. The mixture was kept at 4 °C for 12 h and then centrifuged at 3000g for 10 min to separate the precipitated crude PS. The fractionated crude PS was subsequently re-dissolved in d-H2O and dialyzed twice using an ultrafiltration membrane (molar mass >3500 D Mw, degree of polymerization (DP) > 10, Beijing Solarbio Science and Technology Co., Ltd, Beijing, China) against d-H2O (10 times the sample volume) at 4 °C for 48 h, changing the d-H2O every 12 h. The dialyzed liquid was collected and lyophilized to a constant weight using a vacuum dryer (Biosafer-10A, Biosafer, Nanjing, China), which was considered the crude PS from alfalfa (APS). Further purification was performed using anion-exchange column chromatography. Three grams of crude PSs was dissolved in 100 mL of distilled water and then applied to a DEAE-52 anion-exchange column (5.0 × 100.0 cm). A stepwise gradient NaCl aqueous solution (0, 0.2, 0.4 mol L−1) was used for elution at a flow rate of 0.5 mL min−1. Three fractions were collected separately and the concentration of polysaccharide in elution was detected with the phenol–sulfuric acid method.14,31 The absorbance peak of 0.4 mol L−1 was much larger than the others, therefore, the fraction eluted with 0.4 mol L−1 NaCl was further purified using gel-permeation chromatography with a Sephacryl S-500, yielding one homogeneous fraction APS.32,33 The column was washed with distilled water at a rate of 0.2 mL min−1, and eluted samples were freeze-dried.
The total concentration of PS in the eluent was determined by the phenol–sulfuric acid method,14,31 the protein content was determined using the Bradford method with bovine serum albumin as the reference,34 and the total phenol contents were determined using liquid chromatography electrospray ionization mass spectrometry (LC-ESI-MS).35
2.2. Determination of the monomer type in APS
All PS are formed of a series of monosaccharides or uronic acids polymerized by glycosidic linkages, which can be broken down by acid treatment. The components of PS can then be identified by ion chromatography (IC) as previously described.10 Eight monosaccharides and two uronic acids standard substrates were used in this study. These included Fuc, Ara, Gal, Glc, xylose (Xyl), mannose (Man), fructose (Fru), ribose (Ria), GalA, and GlcA.2.3. Molecular weight and polydispersity characteristics of APS
The molar mass was determined by the gel permeation chromatography-refractive index-multiangle laser light scattering (GPC-RI-MALS) method (DAWN HELEOS II, Wyatt Technology, Santa Barbara, CA, USA).10,36 The molar mass of PS can be described on the basis of the following parameters, namely, number-average of molecular weight (Mn), which is expressed as Mn = ∑(niMi)/∑ni; the weight-average of molecular weight (Mw) defined as Mw = ∑(niMi2)/∑(niMi); and the Z-average of molecular weight (Mz), calculated as Mz = ∑(niMi3)/(∑niMi2), where ni is the number of the molecules with a given molar mass, and Mi is the molar mass of the molecule under consideration.36 The polydispersity coefficient denotes the range of the molecular mass distribution and can be calculated by dividing Mw by Mn or Mz by Mn. Another molecular characteristic of PS considered in this study is the root mean square radius (RMS), which has generally been expressed as the number-average of radius (Rn), weight-average of radius (Rw), and Z-average of radius (Rz) as was done for molar mass.102.4. Determination of the glycosidic linkages in APS
Methylation analysis of PS was performed according to a previous method12,35,37 with minor modifications. In brief, 3 mg >APS powder was accurately weighed and dissolved in 1 mL d-H2O, and added into 200 μL 2-morpholine ethane sulfonic acid (0.2 M) and 200 μL carbodiimide (500 mg mL−1) solution, reacting for 2 h. The reaction mixture was subsequently added 1 mL imidazole (4 mol L−1) and divided into two equal parts, one of that then was mixed and reacted with 1 mL NaBH4 (30 mg mL−1) and the other was mixed and reacted with 1 mL NaBD4 (30 mg mL−1) for another 3 h until termination of the reaction by adding 200 μL acetic acid. The two reacted solutions were then dialyzed for 48 h followed by freeze-drying. The dried samples were individually dissolved in 500 μL anhydrous dimethyl sulfoxide (DMSO) and reacted with 50 μL DMSO/NaOH (120 mg mL−1) solution for 30 min. Ten mL of CH3I solution was added to the reaction and incubated another 10 min followed by combining with 1 mL H2O and 500 μL of dichloromethane (DCM) solution to obtain DCM phase. The dried samples were then dissolved in 100 μL trifluoroacetic acid (2 M), reacted at 121 °C for 90 min followed by evaporating to dryness at 30 °C. The sample was dissolved in 50 μL of ammonia H2O (2 M), mixed and reacted with 50 μL of 1 M NaBD4 for 2.5 h. Twenty μL of acetic acid was added to the mixture to end the reaction, followed with washing it twice with 250 μL of methanol, and dried under nitrogen. The obtained powder was mixed and reacted with 250 μL of acetic anhydride at 100 °C for 2.5 h followed by mixed and incubated with 1 mL of H2O for another 10 min. Finally, the sample was mixed with 500 μL of DCM, and centrifuged to discard aqueous phase, and the bottom DCM phase were tested on the machine. A 6890A-5975C GC-MS system (Agilent Technologies Inc. CA, USA) equipped with a BPX70 capillary column (30 mm × 0.25 mm, 0.25 μm) was used to analyze the glycosidic linkage. The gas chromatographic conditions were as follows: BPX70 chromatographic column; high purity helium gas was used as the carrier gas at a flow rate of 1.0 mL min−1; the injection volume was 1 μL, and the split ratio was 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; the initial column temperature was 140 °C, retaining this for 2 min, and procedurally increasing it to 230 °C at 3 °C min−1, with subsequent holding for 5 min; the injection temperature was 230 °C; the ion source of the mass spectrometer was set at 230 °C, and the four stage bar temperature was 150 °C. The resulting peaks of alditol acetates were identified based on their MS fragmentation patterns and the relative retention time in the GC spectrum. The molar ratios of individual linkage residues were estimated as the ratios of peak areas.
1; the initial column temperature was 140 °C, retaining this for 2 min, and procedurally increasing it to 230 °C at 3 °C min−1, with subsequent holding for 5 min; the injection temperature was 230 °C; the ion source of the mass spectrometer was set at 230 °C, and the four stage bar temperature was 150 °C. The resulting peaks of alditol acetates were identified based on their MS fragmentation patterns and the relative retention time in the GC spectrum. The molar ratios of individual linkage residues were estimated as the ratios of peak areas.
2.5. Examination of microstructures in APS
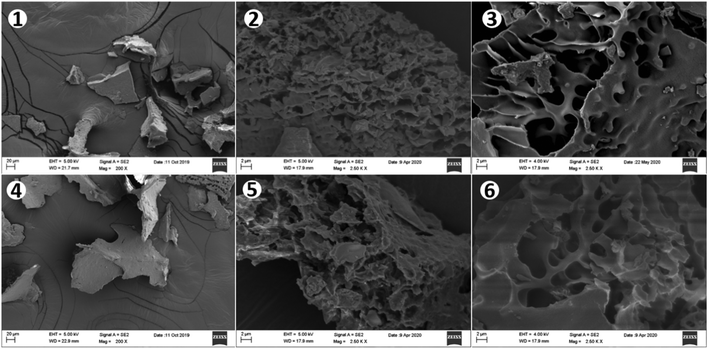
To observe microstructures in APS, purified APS samples were examined under a scanning electron microscope (SEM, SIGMA 500/VP, Zeiss Merlin Compact, Germany) and a transmission electron microscope (TEM, Tecnai G2 Spirit BIOTWIN, FEI, Hillsboro, OR, USA). In brief, APS1 and APS2 were examined by SEM model JEOL (JSM-IT100, Tokyo, Japan). Each dried PS type was mounted on a metal stub and was sputtered with gold. The images were observed at different magnifications (200× and 2500×). In the TEM investigation, a well-dispersed PS solution was first prepared using sodium dodecyl sulfate (SDS, 5 μg mL−1) to a final concentration of 5 μg mL−1. An aqueous solution of SDS was applied to dissolve APS as SDS reduces molecular aggregation. A droplet of APS solution (5 μL) was deposited on a specimen of carbon film (200 mesh, Beijing Zhongjingkeyi Technology, Beijing, China) and dried at ambient temperature and humidity. The specimen was examined using TEM at an accelerating voltage of 120 kV to observe the molecular assembly or the microstructures of APS.292.6. Spectroscopic analysis
The infrared spectrum of APS was acquired on a Fourier transform-infrared (FT-IR) spectrometer (FT-IR 650, Tianjin Gangdong Sci. & Tech. Co. Ltd, China) in the wavenumber range of 4000∼400 cm−1. The APS samples (2 mg) and KBr (200 mg) were first pressed into a pellet. One- and two-dimensional nuclear magnetic resonance (NMR) analyses of APS were carried out on an Avance Bruker III HD 600 MHz NMR spectrometer equipped with a 5 mm TCl CryoProbe at 25 °C using D2O as the solvent. The final sample concentration was 40 mg mL−1. The molecular structure of APS was determined by analyzing one- and two-dimensional NMR spectra, including the 1H-NMR, 13C-NMR, heteronuclear single quantum coherence (C,H-HSQC), single-bond proton–proton correlation spectroscopy (H,H-COSY), heteronuclear multiple bond coherence (H,C-HMBC), and nuclear Overhauser effect spectroscopy (H,H-NOESY).2.7. Antiadipogenic effects of APS
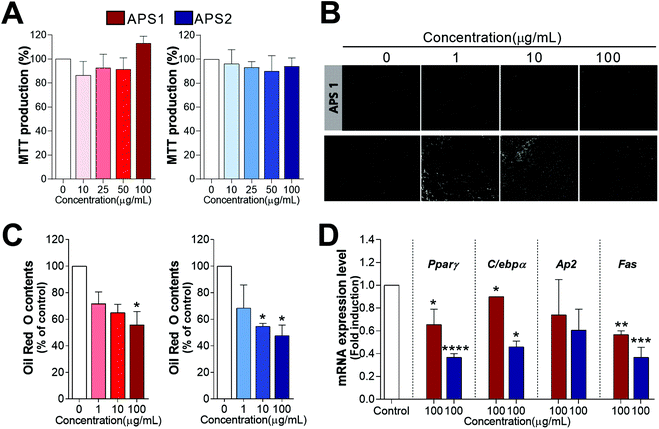
To analyze the potential antiadipogenic effects of APS1 and APS2, cells were treated with various concentrations of either APS1 or APS2 during the differentiation of 3T3-L1. On day 8 of complete adipocyte differentiation, the lipid droplet accumulation was estimated by microscopy and oil red O (ORO) staining. Briefly, the cell culture medium was removed and fixed with 10% formalin. It was finally stained with ORO. The stained cells were washed with PBS thrice. After drying the wells completely, isopropyl alcohol was added to elude ORO. Absorbance was measured by a microplate reader at 500 nm. Intracellular lipid accumulation was measured using the ORO method as described elsewhere.39
The sequences of used primers were as follows: Rplp0, 36b4: forward, 5′-GGATCTGCTGCATCTGCTTG-3′, reverse, 5′-GGCGACCTGGAAGTCCAACT-3′; Hprt: forward, 5′-TTGCTCGAGATGTCATGAAGGA-3′, reverse, 5′-AGCAGGTCAGCAAAGAACTTATAGC-3′; Pparγ: forward, 5′-GGCGATCTTGACAGGAAAGAC-3′, reverse, 5′-CCCTTGAAAAATTCGGATGG-3′; C/ebpα: forward, 5′-GGTTTTGCTCTGATTCTTGCC-3′, reverse, 5′-CGAAAAAACCCAAACATCCC-3′; Fas: forward, 5′-GGAGGTGGTGATAGCCGGTAT-3′, reverse, 5′-TGGGTAATCCATAGAGCCCAG-3′; aP2: forward, 5′-AGCATCATAACCCTAGATGGCG-3′, reverse, 5′-CATAACACATTCCACCACCAGC-3′.
2.8. Statistical analysis
The normality of observed data was verified using the Shapiro–Wilk test, where all the data were normally distributed. Results were expressed as the mean ± standard deviation (SD) of three replicates per sample. The significance of differences between means was evaluated by one-way analysis of variance (ANOVA) and Tukey's multiple comparison test using SAS version 9.0 (SAS Inst. Inc., Cary, NC, USA). Statistical significance was declared at P < 0.05.3. Results
3.1. The micromorphology and monomer compositions of APS
In this study, both APS1 and APS2 exhibited outstanding features of aggregation and precipitation, and the freeze-dried APS powder had a uniform texture and flat surface. After breaking down the outside surface, the crosslinking network or honeycombed microstructure could be observed under SEM (Fig. 1). The content of total sugar, protein, and total phenols were 95.84%, 2.36%, 1.80% in APS1, and 96.15%, 2.31%, and 1.54% in APS2, respectively (Table 1). The IC analysis showed that seven monosaccharides and two uronic acids were presented in both APS types, including Fuc, Ara, Gal, Glc, Xyl, Man, Rib, GalA, and GlcA with molar ratios of 1.43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 18.94
18.94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14.16
14.16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9.61
9.61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.18
4.18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.26
5.26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.76
0.76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 41.05
41.05![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.61 in APS1 and 0.86
4.61 in APS1 and 0.86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23.88
23.88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13.57
13.57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7.84
7.84![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.13
2.13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.60
1.60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.40
2.40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 44.99
44.99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.72 in APS2 (Fig. 2 and Table 1). Ara, GalA, and Rib had greater (P < 0.01) proportions in APS2 than in APS1. The rest of the monomer components had higher (P < 0.01) content in APS1 than in APS2. Structural analysis revealed that either type of APS contained the same monomers from the seven monosaccharides and two uronic acids mentioned above but in different molar proportions. According to the molar proportions, GlaA, Ara, Gal, and Glc were the dominant components. Their cumulative proportions accounted for 83.75% of all molar mass in APS1 and 90.27% of all molar mass in APS2. Likewise, the analysis of glycosidic residues in APS1 and APS2 revealed that the main monomer residues included GalA, Ara, Gal, and Glc; cumulatively, they formed 95.70% of all monomers in APS1 and 93.97% of all monomers in APS2.
2.72 in APS2 (Fig. 2 and Table 1). Ara, GalA, and Rib had greater (P < 0.01) proportions in APS2 than in APS1. The rest of the monomer components had higher (P < 0.01) content in APS1 than in APS2. Structural analysis revealed that either type of APS contained the same monomers from the seven monosaccharides and two uronic acids mentioned above but in different molar proportions. According to the molar proportions, GlaA, Ara, Gal, and Glc were the dominant components. Their cumulative proportions accounted for 83.75% of all molar mass in APS1 and 90.27% of all molar mass in APS2. Likewise, the analysis of glycosidic residues in APS1 and APS2 revealed that the main monomer residues included GalA, Ara, Gal, and Glc; cumulatively, they formed 95.70% of all monomers in APS1 and 93.97% of all monomers in APS2.
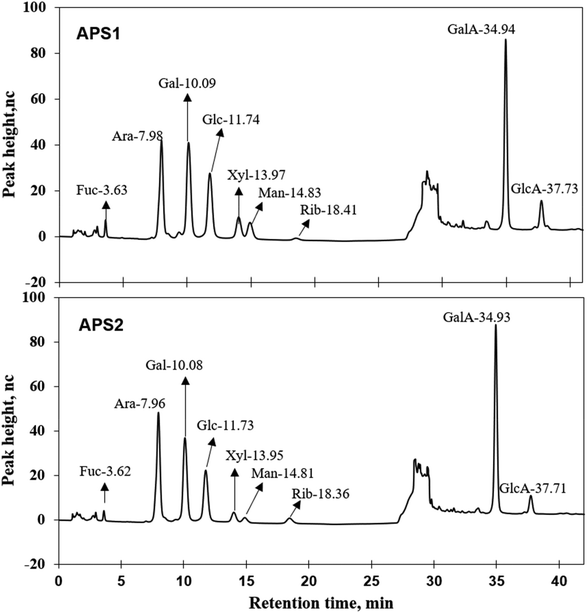 | ||
| Fig. 2 Ion chromatograms of APS1 and APS2. Each absorption peak was annotated in monosaccharide name and retention time. | ||
| Chemical component | Sample | Total sugar, % | Protein, % | Total phenols, % |
|---|---|---|---|---|
| APS1 | 95.84 | 2.36 | 1.80 | |
| APS2 | 96.15 | 2.31 | 1.54 |
| Components | RTa, min | Peak height | Peak area | Molar ratio, % | |
|---|---|---|---|---|---|
| nc | nc × min | ||||
| a RT: retention time; nc: nano coulomb (the unit of quantity of electric charge); nc × time: peak area calculated by integration. | |||||
| Fuc | APS1 | 3.63 ± 0.00 | 7.73 ± 0.23 | 1.09 ± 0.03 | 1.43 ± 0.02 |
| APS2 | 3.62 ± 0.01 | 4.20 ± 0.19 | 0.58 ± 0.03 | 0.86 ± 0.02 | |
| P-Value | 0.374 | <0.001 | <0.001 | <0.001 | |
| Ara | APS1 | 7.98 ± 0.01 | 42.20 ± 0.69 | 13.84 ± 0.26 | 18.94 ± 0.14 |
| APS2 | 7.96 ± 0.01 | 47.73 ± 1.14 | 15.55 ± 0.38 | 23.88 ± 0.36 | |
| P-Value | 0.193 | 0.014 | 0.021 | <0.001 | |
| Gal | APS1 | 10.09 ± 0.02 | 42.05 ± 0.95 | 15.73 ± 0.36 | 14.16 ± 0.19 |
| APS2 | 10.08 ± 0.01 | 36.76 ± 1.04 | 13.44 ± 0.44 | 13.57 ± 0.16 | |
| P-Value | 0.386 | 0.02 | 0.015 | 0.074 | |
| Glc | APS1 | 11.74 ± 0.03 | 28.87 ± 0.63 | 12.44 ± 0.24 | 9.61 ± 0.06 |
| APS2 | 11.73 ± 0.02 | 21.88 ± 0.75 | 9.05 ± 0.36 | 7.84 ± 0.09 | |
| P-Value | 0.682 | 0.002 | 0.001 | <0.001 | |
| Xyl | APS1 | 13.97 ± 0.04 | 8.86 ± 0.12 | 3.54 ± 0.02 | 4.18 ± 0.03 |
| APS2 | 13.95 ± 0.03 | 4.08 ± 0.16 | 1.61 ± 0.07 | 2.13 ± 0.02 | |
| P-Value | 0.751 | <0.001 | <0.001 | <0.001 | |
| Man | APS1 | 14.83 ± 0.05 | 6.61 ± 0.16 | 2.78 ± 0.06 | 5.26 ± 0.04 |
| APS2 | 14.81 ± 0.04 | 1.84 ± 0.09 | 0.76 ± 0.04 | 1.60 ± 0.04 | |
| P-Value | 0.727 | <0.001 | <0.001 | <0.001 | |
| Rib | APS1 | 18.41 ± 0.07 | 0.90 ± 0.01 | 0.53 ± 0.01 | 0.76 ± 0.00 |
| APS2 | 18.36 ± 0.05 | 2.36 ± 0.09 | 1.50 ± 0.16 | 2.40 ± 0.21 | |
| P-Value | 0.598 | <0.001 | 0.003 | 0.001 | |
| GalA | APS1 | 34.94 ± 0.02 | 82.64 ± 0.77 | 21.75 ± 0.17 | 41.05 ± 0.43 |
| APS2 | 34.93 ± 0.01 | 81.00 ± 2.69 | 21.26 ± 0.70 | 44.99 ± 0.30 | |
| P-Value | 0.536 | 0.59 | 0.53 | 0.002 | |
| GlcA | APS1 | 37.73 ± 0.02 | 12.85 ± 0.61 | 4.08 ± 0.20 | 4.61 ± 0.17 |
| APS2 | 37.71 ± 0.01 | 6.93 ± 0.63 | 2.15 ± 0.20 | 2.72 ± 0.21 | |
| P-Value | 0.618 | 0.003 | 0.002 | 0.002 | |
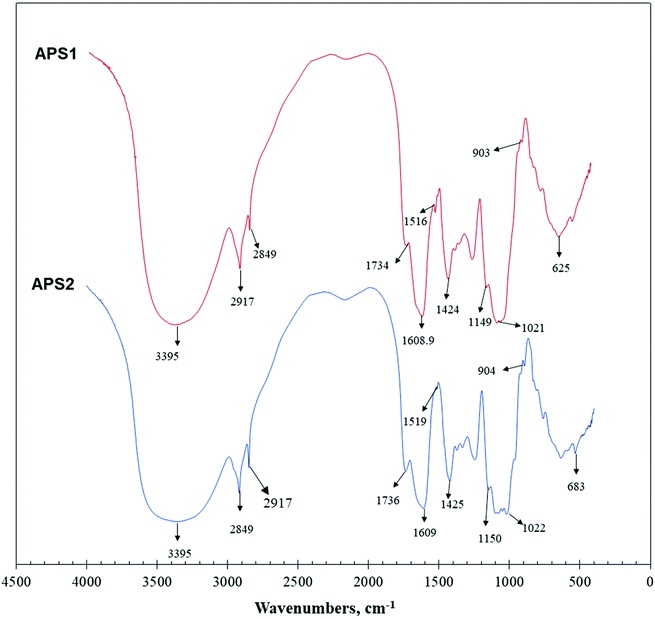
3.2. Fourier transform-infrared (FT-IR) spectra of APS
The primary maxima of absorption bands in IR spectra of APS are shown in Fig. 3. FT-IR spectroscopy is one of the important analytical techniques used to study molecular structures and conformations of macromolecules by identifying vibrations between the different atoms in molecules.APS1 and APS2 had similar transmittance spectrums with only minor variations, and the wavenumbers were mainly distributed between 400 and 4000 cm−1. As shown in Fig. 3, absorption bands at 3600∼3200 cm−1 exhibited stretching vibrations of the –OH group, while the peaks in this region showed the characteristic absorptions of PS. The absorption peak at 3395 cm−1 was attributed to the O–H stretching vibration (O–H group), which is a typical peak observed for PS. In detail, for APS1, the absorption peak at 2917 cm−1 was attributed to C–H stretching vibration (C–H group). The absorption peak at 1609 cm−1 was C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration. The absorption peak at 1516 cm−1 was identified as C
O stretching vibration. The absorption peak at 1516 cm−1 was identified as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O asymmetric stretching vibration. The absorption peaks at 1424 cm−1 and 1080 cm−1 were assigned to C–O and O–H stretching and variable angled vibrations, respectively. For APS2, the absorption peak at 2917 cm−1 was assigned to C–H stretching vibration. The absorption peak at 1609 cm−1 was attributed to C
O asymmetric stretching vibration. The absorption peaks at 1424 cm−1 and 1080 cm−1 were assigned to C–O and O–H stretching and variable angled vibrations, respectively. For APS2, the absorption peak at 2917 cm−1 was assigned to C–H stretching vibration. The absorption peak at 1609 cm−1 was attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration. The absorption peak at 1519 cm−1 was identified to occur from C
O stretching vibration. The absorption peak at 1519 cm−1 was identified to occur from C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O asymmetric stretching vibration. Additionally, the absorption peaks at 1425 cm−1 and 1080 cm−1 were attributed to C–O stretching vibration and O–H variable angle vibration, respectively.
O asymmetric stretching vibration. Additionally, the absorption peaks at 1425 cm−1 and 1080 cm−1 were attributed to C–O stretching vibration and O–H variable angle vibration, respectively.
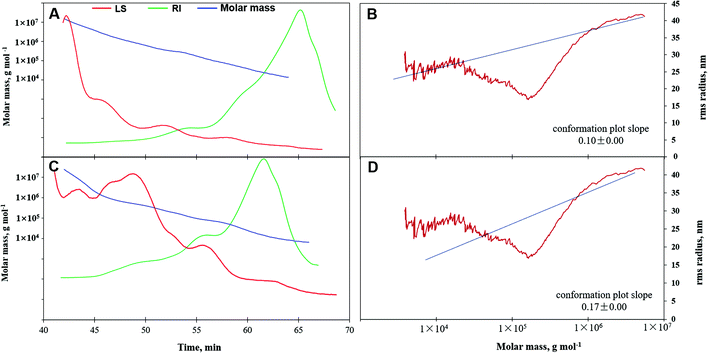
3.3. Molecular weight, polydispersity, and conformation of APS
Molar mass, variation tendency, and conformation characteristics from GPC-RI-MALS data are shown in Table 2 and Fig. 4. The average molecular weight denoted as Mn, Mw, or Mz of APS1 were (3.15 ± 0.05) × 104, (1.77 ± 0.01) × 105, and (3.10 ± 0.01) × 106 g mol−1, which were greater than those for APS2, which has Mn, Mw, and Mz of (1.41 ± 0.05) × 104, (1.01 ± 0.01) × 105, and (2.48 ± 0.01) × 106 g mol−1, respectively. This demonstrated that APS1 had greater molar mass as compared to APS2. However, the polydispersity coefficient of APS2 (7.18) was greater than that of APS1 (5.61) (P < 0.05), which suggested a wider variation range of molar mass for the former and smaller molecules in APS2 as compared to APS1.| Item | APS1 | APS2 | P-Values | |
|---|---|---|---|---|
| Molar mass (g mol−1) | M n | (3.15 ± 0.05) × 104 | (1.41 ± 0.05) × 104 | <0.001 |
| M w | (1.77 ± 0.01) × 105 | (1.01 ± 0.01) × 105 | <0.001 | |
| M z | (3.10 ± 0.01) × 106 | (2.48 ± 0.01) × 106 | <0.001 | |
| Polydispersity | M w/Mn | 5.61 ± 0.05 | 7.18 ± 0.05 | <0.001 |
| M z /Mn | 98.53 ± 6.50 | 175.75 ± 7.84 | <0.001 | |
| Root mean square radius, RMS (nm) | R n | 42.80 ± 0.09 | 30.60 ± 0.02 | <0.001 |
| R w | 39.30 ± 0.07 | 26.20 ± 0.02 | <0.001 | |
| R z | 40.80 ± 0.02 | 26.20 ± 0.04 | <0.001 | |
This was further confirmed by the molar mass trend curve (Fig. 4A & C), where APS2 had a gradually decreasing multi-angle laser scattering (red line) curve, which had a rapidly ascending refractive index (RI, green line) as compared to APS1, indicating a wider range of molecular weights in APS2. In addition, the RMS values displayed by Rn, Rw, and Rz were 42.80, 39.30, and 40.80 for APS1 and 30.60, 26.20, and 26.20 for APS2, respectively. When the RMS value of a polymer is more than 10 (upon considering log (Mw) as the horizontal coordinate and log (RMS) as the vertical coordinate to fit the conformation plot), the slope would indicate the molecular configuration.40 As shown in Fig. 4, the slope was 0.10 in the APS1 conformation plot and 0.17 in the APS2 conformation plot, indicating that both APS1 and APS2 exhibited complex crosslinking with highly branched PS macromolecules.
3.4. Glycosidic linkages between monosaccharides of APS
The findings of GC-MS analysis suggested that APS1 was mainly composed of nine different glycosidic residues, including three nonreducing terminals of t-Ara, t-Glc, and t-GalA with the molar ratios of 7.79%, 4.18%, and 9.92%, as well as other intrachain residues, involving 1,2-Rha; 1,5-Ara; 1,4-GlaA; 1,4-Glc; 1,3,4-GlaA, 1,3,6-Gla, in molar ratios of 4.30%, 7.28%, 36.46%, 18.12%, 4.23%, and 7.71%, respectively (Table 3). Similarly, APS2 consisted of seven monomer residues, including two non-reducing terminals of t-Ara and t-GalA, in molar ratios of 9.49% and 12.57%. Other residues, 1,2-Rha; 1,5-Ara; 1,4-GalA; 1,4-Glc; and 1,3,6-Gal, were present in molar ratios of 6.03%, 11.42%, 39.52%, 15.00%, and 5.97%, respectively, and were identified to be intrachain residues. The cumulative proportions of major residues, including GalA, Ara, Gal, and Glc, were 95.70% in APS1 and 93.97% in APS2.| Serial number | RT, min | nc | nc × min | Glycosidic linkagesa | Mol, % |
|---|---|---|---|---|---|
| a The glycosidic residues marked with an asterisk (*) means the corresponding component was a uronic acid in the PS molecules. | |||||
| APS1 | |||||
| 1 | 6.61 | 4 261 690.58 | 14 784 412.41 | t-Ara(f) | 7.79 |
| 2 | 9.57 | 2 000 582.51 | 9 391 561.58 | 1,2-Rha(p) | 4.30 |
| 3 | 9.74 | 1 720 293.14 | 9 185 892.06 | t-Glc(p) | 4.18 |
| 4 | 10.79 | 5 491 190.89 | 21 775 359.38 | *t-Gal(p)A | 9.92 |
| 5 | 11.45 | 3 716 643.52 | 15 194 805.55 | 1,5-Ara(f) | 7.28 |
| 6 | 14.72 | 18 530 303.05 | 86 978 621.68 | *1,4-Gal(p)A | 36.46 |
| 7 | 15.02 | 10 019 103.24 | 43 216 516.88 | 1,4-Glc(p) | 18.12 |
| 8 | 16.74 | 1 923 428.48 | 10 888 505.28 | *1,3,4-Gal(p)A | 4.23 |
| 9 | 19.96 | 4 652 262.25 | 19 853 895.59 | 1,3,6-Gal(p) | 7.71 |
| APS2 | |||||
| 1 | 6.59 | 5 041 644.96 | 18 048 302.66 | t-Ara(f) | 9.49 |
| 2 | 9.56 | 3 275 072.03 | 13 192 902.45 | 1,2-Rha(p) | 6.03 |
| 3 | 10.77 | 6 937 374.61 | 27 839 272.69 | *t-Gal(p)A | 12.57 |
| 4 | 11.44 | 6 069 245.46 | 23 902 732.31 | 1,5-Ara(f) | 11.42 |
| 5 | 14.72 | 19 947 942.16 | 95 103 104.48 | *1,4-Gal(p)A | 39.52 |
| 6 | 15.00 | 8 445 426.07 | 35 900 958.33 | 1,4-Glc(p) | 15.00 |
| 7 | 19.94 | 3 195 852.97 | 15 417 443.64 | 1,3,6-Gal(p) | 5.97 |
In addition, the Ara and GalA residues ratio was 20.91% and 52.09% in APS2, which were significantly higher than those in APS1 of 15.08% and 50.61%. This finding further verified that Ara and GalA were ecologically-sensitive components, and their synthesis was strengthened when the alfalfa was subjected to the saline–alkali stress. The main branching points were at 1,3,4-GlcA and 1,3,6-Gla in APS1 molecule, as well at 1,3,6-Gla in APS2 molecule. The ratio between the terminal units and the branching points was 1.83 in APS1 and 3.69 in APS2, suggesting that the number of the terminal unit was more than that of the branching points in the two kinds of PS molecules. In addition, the degree of branching (DB) was 33.83% in APS1 and 28.03% in APS2, which was the proportion of the number of terminal and branch residues accounting for the total amount of saccharide residues.29
3.5. Nuclear magnetic resonance (NMR) spectrum of APS
Linkages between glycosyl residues in APS1 and APS2 were further investigated by NMR analyses, including 1H NMR, 13C NMR, and HSQC spectra (Fig. 5) with the assistance COSY, HMBC, and NOESY (not shown). Based on the analysis of APS1, the 1H NMR spectrum signals were mainly distributed across 1.5∼6.3 ppm; the chemical shift (δ) from 3.5 to 4.5 ppm was due to protons of the glycosidic ring, and the δ of 4.09, 4.40, 4.81, 5.26, 5.83 were assigned to terminal protons, while other peaks (chemical shift) ranged in δ 1.5∼2.0, 2.5∼3.0, and 4.3∼5.5 ppm. The 13C NMR (500 MHz, D2O) spectrum revealed that the carbon signals were primarily distributed at chemical shift δ of 50 to 110 ppm. The primary anomeric carbons were at δ of 101.30, 102.50, 103.84, 104.48, 104.50, 104.90, 104.69, 105.30, 110.62 ppm, ranging from 100 to 110 ppm. On the other hand, the main signals peaks of including the 84.61, 81.96, 81.02, 80.59, 79.55, 79.01, 78.71, 77.54, 76.70, 73.31, 71.20, 67.69, 65.80, and 62.02 were distributed across δ of 60 to 105 ppm. In addition, regarding the correlations of C/H in the HMBC spectrum (Fig. 5C), eighteen cross signals with the 1H being from 4.5 to 7.0 ppm and 13C in the range of 90 to 120 ppm further demonstrated the presence of A–J residues in APS1 molecule that is represented in Fig. 5 and summarized in Table 4.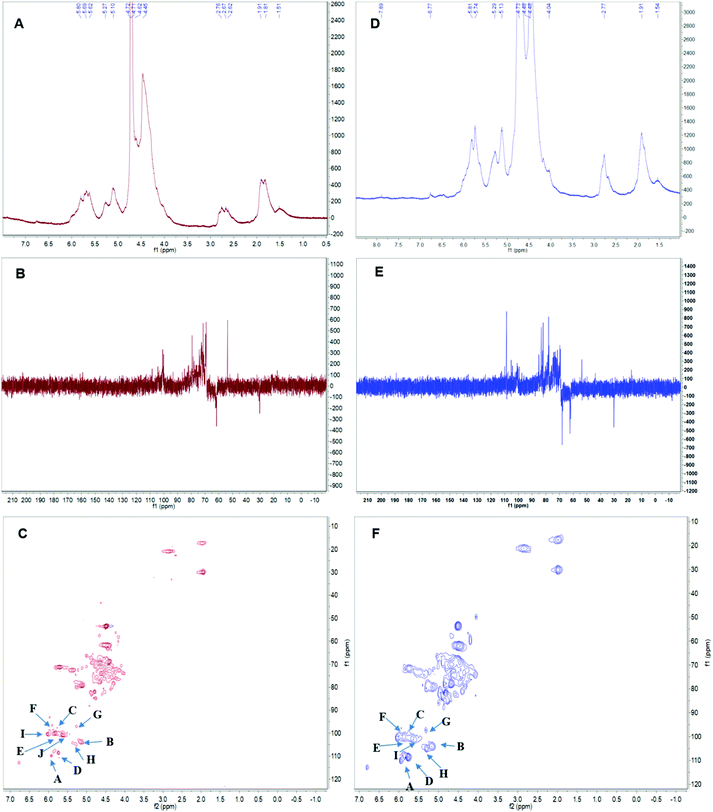 | ||
| Fig. 5 The nuclear magnetic resonance (NMR) spectrum of APS in D2O solution at 25 °C. Panels A & D show the 1H spectra of APS1 and APS2. Panels B & E depict the 13C spectra of APS1 and APS2. Panels C & F depict the heteronuclear multiple bond coherence (HMBC) spectrum of APS1 and APS2. A, B, C … J in Panels C & F depict the chemical shift of carbon1 (C1) and hydrogen1 (H1) in residue A. The order of residues, A, B, C … J, are consistent with Table 4. | ||
| Number | Glycosyl residues | Chemical shift (ppm), δ | ||||||
|---|---|---|---|---|---|---|---|---|
| H1/C1 | H2/C2 | H3/C3 | H4/C4 | H5/C5 | H6/C6 | H6b | ||
| APS1 | ||||||||
| A | α-L-Araf-(1→ | 5.17 | 4.13 | 3.87 | 4.06 | 3.76 | 3.64 | |
| 110.62 | 82.62 | 77.97 | 85.22 | 62.64 | ||||
| B | β-D-Glcp-(1→ | 4.30 | 3.28 | 3.46 | 3.21 | 3.6 0 | 3.54 | 3.45 |
| 104.48 | 74.66 | 76.62 | 83.4 | 77.36 | 64.00 | |||
| C | β-D-Galp-(1→ | 4.67 | 3.70 | 3.90 | 3.75 | 3.63 | 3.74 | 3.62 |
| 102.50 | 71.50 | 70.05 | 71.71 | 76.40 | 62.45 | |||
| D | →5)-α-D-Araf-(1→ | 5.03 | 4.42 | 4.56 | 4.02 | 4.23 | 4.12 | |
| 108.26 | 81.40 | 84.50 | 82.50 | 67.50 | ||||
| E | →3)-α-D-Galp-(1→ | 4.63 | 3.70 | 3.80 | 3.86 | 3.57 | 3.63 | 3.74 |
| 105.30 | 71.42 | 83.10 | 69.96 | 74.14 | 62.45 | |||
| F | →4)-α-D-GalpA-(1→ | 4.66 | 3.96 | 3.75 | 3.73 | 3.49 | 3.85 | 3.64 |
| 101.30 | 71.78 | 72.34 | 77.69 | 75.88 | 62.14 | |||
| G | →4)-β-D-Glcp-(1→ | 4.31 | 3.30 | 3.63 | 3.68 | 3.47 | 3.95 | 3.79 |
| 103.84 | 74.17 | 71.58 | 77.76 | 76.69 | 61.67 | |||
| H | →6)-β-D-Galp-(1→ | 4.37 | 3.44 | 3.58 | 3.86 | 3.88 | 3.95 | 3.83 |
| 104.90 | 72.16 | 73.93 | 74.96 | 69.87 | 70.50 | |||
| I | →3,4)-α-D-GalpA-(1→ | 5.13 | 3.23 | 4.78 | 4.65 | 4.32 | 4.45 | 4.32 |
| 104.58 | 77.25 | 76.63 | 62.21 | 75.42 | 70.15 | |||
| J | →3,6)-β-D-Galp-(1→ | 4.45 | 3.57 | 3.63 | 4.16 | 3.84 | 3.96 | 3.86 |
| 104.69 | 71.31 | 81.50 | 69.82 | 74.81 | 70.76 | |||
| APS2 | ||||||||
| A | α-L-Araf-(1→ | 5.17 | 4.13 | 3.87 | 4.06 | 3.76 | 3.64 | |
| 110.62 | 82.62 | 77.97 | 85.22 | 62.64 | ||||
| B | β-D-Glcp-(1→ | 4.30 | 3.28 | 3.46 | 3.21 | 3.60 | 3.54 | 3.45 |
| 104.48 | 74.66 | 76.62 | 83.40 | 77.36 | 64.00 | |||
| C | β-D-Galp-(1→ | 4.67 | 3.70 | 3.90 | 3.75 | 3.63 | 3.74 | 3.62 |
| 102.5 | 71.50 | 70.05 | 71.71 | 76.40 | 62.45 | |||
| D | →5)-α-D-Araf-(1→ | 5.03 | 4.42 | 4.56 | 4.02 | 4.23 | 4.12 | |
| 108.26 | 81.40 | 84.50 | 82.50 | 67.50 | ||||
| E | →3)-α-D-Galp-(1→ | 4.63 | 3.70 | 3.80 | 3.86 | 3.57 | 3.63 | 3.74 |
| 105.30 | 71.42 | 83.10 | 69.96 | 74.14 | 62.45 | |||
| F | →4)-α-D-GalpA-(1→ | 4.66 | 3.96 | 3.75 | 3.73 | 3.49 | 3.85 | 3.64 |
| 101.30 | 71.78 | 72.34 | 77.69 | 75.88 | 62.14 | |||
| G | →4)-β-D-Glcp-(1→ | 4.31 | 3.30 | 3.63 | 3.68 | 3.47 | 3.95 | 3.79 |
| 103.84 | 74.17 | 71.58 | 77.76 | 76.69 | 61.67 | |||
| H | →6)-β-D-Galp-(1→ | 4.37 | 3.44 | 3.58 | 3.86 | 3.88 | 3.95 | 3.83 |
| 104.90 | 72.16 | 73.93 | 74.96 | 69.87 | 70.50 | |||
| I | →3,6)-β-D-Galp-(1→ | 4.45 | 3.57 | 3.63 | 4.16 | 3.84 | 3.96 | 3.86 |
| 104.69 | 71.31 | 81.50 | 69.82 | 74.81 | 70.76 | |||
In the HSQC spectrum, according to the one-dimensional and two-dimensional NMR spectra, the glycosidic bond signals of APS1 were assigned as follows: there was a correlative signal peak between the anomeric hydrogen of glycosidic bond →4)-α-D-GalpA-(1→ and the C4, suggesting there was the →4)-α-D-GalpA-(1 → 4)-α-D-GalpA-(1→ intrachain linkage. There was correlative signal peak between the anomeric hydrogen of glycosidic bond, β-D-Galp-(1→ and C3 of →3)-α-D-Galp-(1→, indicating that there was β-D-Galp-(1 → 3)-α-D-Galp-(1→ intrachain linkage. The anomeric hydrogen of glycosidic bond →3)-α-D-Galp-(1→ correlated to the C3 of -4)-α-D-GalpA-(1→, indicating the intrachain structure of →3)-α-D-Galp-(1–4)-α-D-GalpA-(1→ existing. There was correlative signal peak between the anomeric hydrogen of glycosidic bond →4)-α-D-GalpA-(1→ and C3 of →3,6)-β-D-Galp-(1→, suggesting that there was →4)-α-D-GalpA-(1,3,6)-β-D-Galp-(1→ intrachain structure. The signal peaks of anomeric hydrogen of glycosidic bond →4)-α-D-GalpA→ correlated to that of C6 in the →3,6)-β-D-Galp-(1→, indicating →4)-α-D-GalpA-(1–6,3)-β-D-Galp-(1→ to exist. There was the correlative signal peak between the anomeric hydrogen of glycosidic bond →4)-α-D-GalpA-(1- and C4 of →3,4)-α-D-GalpA-(1→, indicating →4)-α-D-GalpA-(1,4,3)-α-D-GalpA-(1→ existing. The anomeric hydrogen of glycosidic bond →4)-α-D-GalpA-(1→ correlate to the C3 of →3,4)-α-D-GalpA-(1→, indicating →4)-α-D-GalpA-(1,3,4)-α-D-GalpA- (1→ existing.
In APS2 analysis, the 1H NMR spectrum signals are mainly distributed in 1.5∼6.3 ppm, the chemical shift between δ 3.2 to 4.5 ppm were glycosidic ring protons, and the δ of 4.09, 4.45, 4.65, 4.95, 5.42, 5.64, and 5.78 were assigned to terminal protons, while other signals peaks (chemical shift) were distributed across δ of 1.5∼2.0, 2.5∼3.0, and 4.3∼5.5 ppm. An analysis of the 13C NMR spectrum revealed that the carbon signals were primarily distributed across δ of 50 to 100 ppm. The anomeric carbons were at δ of 101.30, 102.50, 103.84, 104.48, 104.90, 104.69, 105.30, and 110.62 ppm. The signals of anomeric carbons were mainly found at δ of 100 to 110 ppm. Additionally, the main signals peaks of chemical shift distributed in the range of 60 to 105 ppm, including δ of 84.61, 81.96, 81.02, 80.59, 79.55, 79.01, 78.71, 77.54, 76.70, 73.31, 71.20, 67.69, 65.80, and 62.02 ppm. In addition, regarding the correlations of C/H in the HSQC spectrum (Fig. 5F), nine cross signals with the 1H being from 4.6 to 6.5 ppm and 13C in the range of 90 to 115 ppm further demonstrated the presence of A–I residues as shown in Fig. 5 and summarized in Table 4. There was a correlative signal peak between the anomeric hydrogen of glycosidic bond, →4)-α-D-GalpA-(1→ and its C4, indicating the existence of→4)-α-D-Galp-(1 → 4)-α-D-GalpA-(1→ in the APS2 molecular chain. There was a correlative signal peak between the anomeric hydrogen of glycosidic bond β-D-Galp-(1→ and C3 of -3)-α-D-Galp-(1→, indicating there is the β-D-Galp-(1 → 3)-α-D-Galp-(1→ in APS2 chain. The signal peak of anomeric hydrogen in →3)-α-D-Galp-(1→ correlated with C3 in -4)-α-D-GalpA-(1→, indicating there are →3)-α-D-Galp-(1–4)-α-D-GalpA-(1→ in APS2 molecule. There was correlative signal peak between the anomeric hydrogen of glycosidic bond in →4)-α-D-GalpA-(1→ to that of C3 in -3,6)-β-D-Galp-(1→, indicating there are →4)-α-D-GalpA-(1,3,6)-β-D-Galp-(1→ in the APS2 molecule. There was correlative signal peak between the anomeric hydrogen of the glycosidic bond in →4)-α-D-GalpA-(1→ and C6 in -3,6)-β-D-Galp-(1-, indicating there is →4)-α-D-GalpA-(1–6,3)-β-D-Galp-(1→ intrachain structure. Therefore, based on the results of glycosidic linkage and NMR spectra analysis, the possible repeating structural unit of APS1 and APS2 could be shown in Table 4.
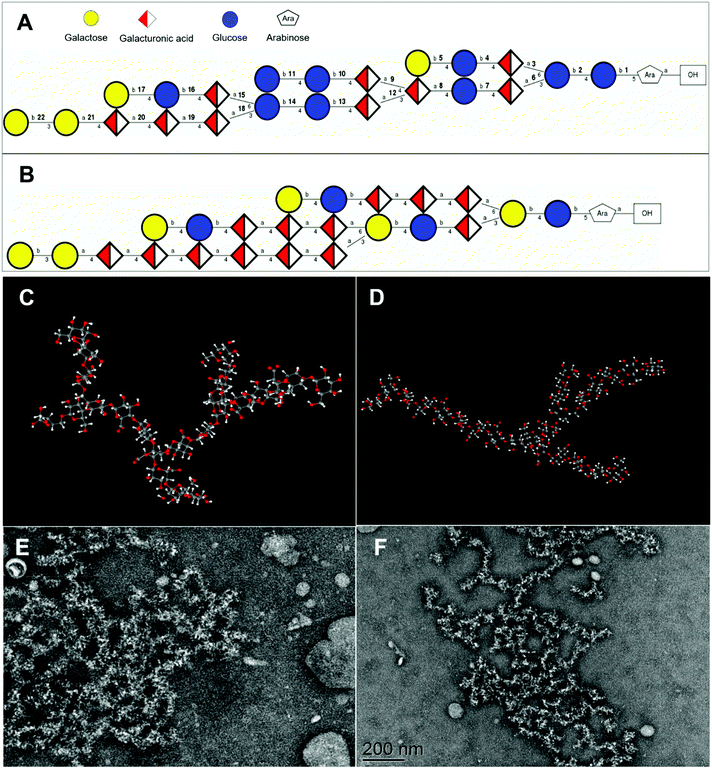
3.6. Three-dimensional structure and spatial conformation of APS
The potential three-dimensional molecular structures of APS1 and APS2 were predicted by combining all the structural parameters, such as monosaccharide composition, molar mass, glycosidic type, sequences, and linking position of the branched chain. The repeating unit of APS1 was composed of Gal, GalA, Glc, and Ara in molar proportions of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9
9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9
9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 6A) and molar mass of 4236 D, while the repeating units of APS2 consisted of Gal, GalA, Glc, and Ara in molar proportions of 6
1 (Fig. 6A) and molar mass of 4236 D, while the repeating units of APS2 consisted of Gal, GalA, Glc, and Ara in molar proportions of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 6B) and a molar mass of 4472 D. At the same time, considering the molecular weight of APS1 and APS2, it can be deduced that the APS1 molecule contained 41 repeating units and APS2 molecule contained 22 repeating units, suggesting both APS1 and APS2 to be long-chain macromolecules. Additionally, the three-dimensional spatial conformation showed the repeating units of both APS1 and APS2 to be densely interconnected and highly branched (Fig. 6C & D, ESI Video† of APS1 & APS2 3-D structure). The overall shape profile and spatial conformation of APS1 and APS2 were inspected by TEM and are presented in Fig. 6E & F. The unfolded partial molecules of APS1 exhibited a crosslinking net or grid pattern with branched structures, whereas APS2 appeared to be a random coil in SDS solution. This finding reconciles well with the molecular configuration plot inferring that a highly branched structure exists in APS1 and APS2 molecules.
1 (Fig. 6B) and a molar mass of 4472 D. At the same time, considering the molecular weight of APS1 and APS2, it can be deduced that the APS1 molecule contained 41 repeating units and APS2 molecule contained 22 repeating units, suggesting both APS1 and APS2 to be long-chain macromolecules. Additionally, the three-dimensional spatial conformation showed the repeating units of both APS1 and APS2 to be densely interconnected and highly branched (Fig. 6C & D, ESI Video† of APS1 & APS2 3-D structure). The overall shape profile and spatial conformation of APS1 and APS2 were inspected by TEM and are presented in Fig. 6E & F. The unfolded partial molecules of APS1 exhibited a crosslinking net or grid pattern with branched structures, whereas APS2 appeared to be a random coil in SDS solution. This finding reconciles well with the molecular configuration plot inferring that a highly branched structure exists in APS1 and APS2 molecules.
The structural characteristics and spatial conformation confer bioactivities of PS.41 However, due to their complex compositions, highly branched structure, high molecular weight, viscosity, and aggregating properties in solution, it is a difficult task to characterize the spatial conformation of PS molecules.42 In this study, we made a preliminary exploration and comparison of the spatial structure of APS1 and APS2 from the different ecological environments. However, the relationship between this unique molecular conformation and its biological activities must be studied further.
3.7. Cellular toxicity and antiadipogenic effects of APS1 and APS2
The potential cellular toxicity of APS in 3T3-L1 cells indicated 100 μg mL−1 of APS1 or APS2 to have no significant toxic effect as shown in Fig. 7A. Both APS1 and APS2 did not alter MTT production as compared to the control at concentrations from 0 to 100 μg mL−1. Subsequently, to analyze potential antiadipogenic effects of APS, the preadipocyte 3T3-L1 cells were cultured with a differentiation medium in the presence of either APS1 or APS2. Lipid droplet accumulation in fully differentiated adipocytes was stained with ORO and observed microscopically (Fig. 7B & C). Both APS1 and APS2 treatment significantly inhibited adipogenesis and lipid accumulation in 3T3-L1 cells with superior effects by APS2 as compared to APS1. Namely, the lipid droplet accumulation in 3T3-L1 adipocytes treated with 100 μg mL−1 APS1 and APS2 decreased to 34.21 ± 7.18% and 44.37 ± 5.60%, respectively, as compared to the untreated cells.Next, we evaluated the transcriptional regulation of adipocyte marker genes upon APS treatment in 3T3-L1 cells. It is well known that the process of adipocyte differentiation is associated not only with intracellular lipid accumulation but also with the regulation of various transcription factors. There are several transcription factors, such as C/ebpα, C/ebpβ, and Pparγ, documented to be sequentially and cooperatively expressed during the differentiation. Especially, Pparγ and C/ebpα are considered master regulators that are highly expressed during differentiation.43 In addition, they have been proposed to be expression regulators of various proteins, such as adipocyte fatty acid-binding protein (Ap2) and fatty acid synthase (Fas), in adipocytes. As shown in Fig. 7D, APS2 effectively decreased the mRNA expression level of Pparγ (P<0.0001), C/ebpα (P < 0.05), and Fas (P < 0.001) as compared with the non-treated group (control), whereas APS1 affected the mRNA expression levels of Pparγ (P < 0.05), C/ebpα (P < 0.05), and Fas (P < 0.01).
4. Discussion
Polysaccharides are polymers comprising one or several types of monosaccharides or uronic acids. They commonly occur in the cell wall of plants.44 Interestingly, PS and their monomer compositions can act as bio-functional components, which continuously change with environmental conditions to help the plant cope with increasing environmental stress.45 Alfalfa is an extensively cultured double-purpose crop with excellent nutrient qualities and rich PS content. PS isolated from alfalfa have been reported to have multiple bioactivities, including immunomodulation,10 anti-inflammation,29 antioxidation, anticancer,46 and growth-promotion.30 Our previous studies have also demonstrated the immunomodulating effects of APS by specific activation of B cells and its involvement in attenuating oxidative stress in mouse embryonic fibroblast (MEFs) cells by modulating the MAPK/p38 and NF-κB pathways.7In this study, two kinds of water-soluble PS were isolated from alfalfa grown in two different soils (denoted as APS1) and saline–alkali soil (denoted as APS2). As a result, APS1 and APS2 exhibited similar physical features with good aggregation and precipitation. The SEM inspection revealed that both APS1 and APS2 had a similar flat surface and honeycomb or porous microstructure. This was consistent with the work of Soua et al. (2020),15 who reported the PS extracted from Ephedra alata stems appeared to be high-porosity particles. However, Cheng and Feng (2013) reported a flat and smooth structure of PS extracted from Epimedium acuminatum.47 The different findings regarding the PS microstructure can be explained by the difference in sources and the isolation methods.24 The porous microstructures of APS1 and APS2 have been speculated to be the basis for some of their specific properties, including water-holding or oil holding capacities, adsorption capacity, and gel-forming tendency.48–50 The IC analysis indicated the two kinds of APS to contain the same monomers, including seven monosaccharides and two uronic acids, arranged in different molar fractions. According to the molar proportions, GlaA, Ara, Gal, and Glc were the dominant components that formed 83.75% of the total mass in APS1 and 90.27% in APS2. This is similar to our previous study on APS isolated from alfalfa harvested in the USA10 that indicated APS to be composed of eight monomer types, including Fuc, Ara, Gal, Glc, Xyl, Man, GlaA, and GlcA in molar ratios of 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8.0
8.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.7
4.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21.3
21.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.2
3.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.0
1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 74.2
74.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14.9. Similarly, Rovkina et al. (2018)24 reported two water-soluble PS, which were fractioned from alfalfa, to have the same chemical components (Fuc, Ara, Gal, Glc, GalA, and GlcA) arranged in a molar ratio of 3.0
14.9. Similarly, Rovkina et al. (2018)24 reported two water-soluble PS, which were fractioned from alfalfa, to have the same chemical components (Fuc, Ara, Gal, Glc, GalA, and GlcA) arranged in a molar ratio of 3.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43.6
43.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.1
6.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11.6
11.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.6
2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33.4 and 3.6
33.4 and 3.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 46.4
46.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7.0
7.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13.8
13.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.1
1.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28.2. Overall, these findings from our study and others suggest that PS from alfalfa cultured in different environmental conditions contain similar components; bulk of the PS contains GlaA, Ara, Gal, and Glc making up more than 83% of the molar mass. Thus, these four monomers may be the species-specific intrachain monosaccharide components of APS.
28.2. Overall, these findings from our study and others suggest that PS from alfalfa cultured in different environmental conditions contain similar components; bulk of the PS contains GlaA, Ara, Gal, and Glc making up more than 83% of the molar mass. Thus, these four monomers may be the species-specific intrachain monosaccharide components of APS.
Another interesting result was that APS2 had a notably increased Ara and GlaA ratio as compared to APS1. A recent study revealed that Arabidopsis has greater Ara biosynthesis capacity under salt stress, and the concentration of Ara in the cell wall is enhanced when Arabidopsis is subjected to high salinity conditions.27 Furthermore, when alfalfa is exposed in Cd-rich soil, the abundance of xylogalacturonan cross-linked with xylose and galacturonic acid substantially increases in the cell wall preventing Cd from entering and binding to the cell wall.25 Similarly, saline–alkali soil may have induced higher Ara and GalA biosynthesis in alfalfa, which can explain the higher content of Ara and GalA in APS2 as it was harvested from alfalfa grown in saline–alkali soil as compared to APS1. Similarly, the analysis of glycosidic residues in APS1 and APS2 revealed that the main monomer residues in APS included GalA (50.61%, 52.09%%), Ara (15.08%, 20.91%), Gal (7.71%, 5.97%), and Glc (22.30%, 15.00%), with their cumulative proportions being 95.70% in APS1 and 93.97% in APS2. Amongst these four monosaccharide residues, increased proportions of Ara and GalA residues were observed in APS2, which corresponded well to the component and spatial conformation analysis. The repeating units and molecular structure obtained from NMR and other structural analyses similarly indicated the increased proportions of Ara and GalA. Therefore, Ara and GalA might be two ecologically sensitive molecules that can be effectively induced to be synthesized under the saline–alkali conditions of the soil in alfalfa.
The average molar mass and polydispersity of PS are pivotal molecular characteristics that influence their dynamic properties and bioactivities.47 In this study, we demonstrated that APS1 has higher molar mass and RMS values as compared to APS2, whereas polydispersity appeared to have a wider range in APS2 as compared to in APS1, i.e., APS1 was a bigger molecule as compared to APS2. The abundance of smaller molecules in APS2 as compared to in APS1 reflects the stress resistance of alfalfa to its growing environment.51,52 The greater polydispersity coefficient of APS2 indicated smaller molecules to exist in this PS homolog that might have a close relationship with stress resistance in alfalfa.13,52
In the current study, both APS1 and APS2 exhibited no cellular toxicity to 3T3-L1 cells, when applied in the range of 0 to 100 μg mL−1. While both APS1 and APS2 exhibited antiadipogenic effects in 3T3-L1 cells, APS2 had a superior antiadipogenic effect as compared to APS1. This finding is corroborated well by the regulation of the lipid accumulation-related genes. For example, APS1 significantly reduced the expression Fas, while APS2 decreased the expression of Pparγ, C/ebpα, and Fas in 3T3-L1 cells. These data were also in agreement with the ORO staining results from APS1 or APS2 during adipogenesis. C/ebpβ and C/ebpδ are expressed in the early stages of adipocyte differentiation and regulate Pparγ and C/ebpα expression.53Srebp-1c is a transcription factor that regulates the biosynthesis of fatty acids, cholesterol, and triglycerides. Furthermore, Srebp-1c is activated during the induction of Ppar-γ and C/ebpα gene expression in 3T3-L1 adipocyte differentiation.54,55 As shown in Fig. 7D, APS2 effectively decreases mRNA expression levels of Pparγ, C/ebpα, and Fas as compared to the non-treated group (control). APS1 and APS2 also attenuate the mRNA expression levels of Fas among adipocyte-specific genes regulated by C/ebpa or Pparγ during adipocyte differentiation. These results suggest that APS2 prevents adipocyte differentiation and lipid accumulation partially via inhibition of adipogenesis-related marker genes, which is more effective than APS1.53 Likewise, some previous studies have documented the capacity of plant-derived PS to inhibit pre-adipocyte proliferation and reduce the accumulation of lipids in cells or tissues.56 In a parallel study, Suraiya et al. (2019) documented the polysaccharide of Saccharina japonica at the dose of 100 to 500 μg mL−1 suppressed the adipogenesis and decreased the expression of relative genes Pparγ and C/ebpα.57 Dietary supplement with 2% brown algae (Padina tetrastromatica) polysaccharides decreased the differentiation of adipocytes and down-regulated the expression of p-Akt, p38 and Pparγ in visceral adipose tissue of diet-induced obese C57BL6 mice.58 A previous study reported that crude fucoidans composed of fucose, glucuronic acid, galactose, xylose, and sulfate exerted antiadipogenic activities, and thus, inhibited the 3T3-L1 cell differentiation.59 Flaxseed PS fermented by fecal bacteria also exhibited anti-adipogenic effects with a decrease in lipid accumulation in 3T3-L1 cells by downregulation of mRNA and protein expression of Pparγ, C/ebpα, and C/ebpβ in differentiating adipocytes.60 Therefore, plant-derived PS may be potential candidates for the use as antiadipogenic agents in functional foods that target obesity.53
Lastly, it is worthwhile to note that there are not enough data for us to draw a conclusion for a bioavailability of APS1 and APS2. Others have reported that the molecular size and polarity of PS are the crucial elements to affect their absorption rate and/or bioavailability.60 Based on our findings, we can predict that the relatively large molecular mass (APS1 = 1.77 × 105 g mol−1, APS2 = 1.01 × 105 g mol−1) and the strong acidic property by containing high proportion of GalA and GlcA of APS1 and APS2 play an important role in regulating the biological availability in digestible tract of animals.61 However, further studies including a pharmacokinetic assay and direct/indirect interaction with receptor(s) are necessary to understand the bioavailability of APS.
Our findings support the hypothesis that saline–alkali soil affects monomer components, structural characteristics, and bioactivities of APS. The results also provide novel insights into the possibility of targeted isolation of phytogenic PS of selected compositions capable of performing specific bioactivities under different ecological environments.
5. Conclusions
In summary, nine monomer types of Fuc, Ara, Gal, Glc, Xyl, Man, Rib, GlaA, and GlcA were identified in APS1 and APS2 molecules in different proportions. GalA, Ara, Gal, and Glc were predominant constituents of both APS, while APS2 had a greater content of Ara and GalA than did APS1. Additionally, APS1 exhibited a greater molar mass of 1.77 × 105 g mol −1 (Mw) than APS2, which has a molar mass of 1.01 × 105 g mol −1. Furthermore, APS2 had a wider range of molar mass distribution than APS1, allowing for greater polydispersity (98.54 vs. 175.75). Both APS1 and APS2 significantly inhibited adipogenesis and lipid accumulation in 3T3-L1 cells by downregulating mRNA expression levels of Pparγ, C/ebpα, and Fas. Furthermore, APS2 exhibited stronger antiadipogenic effects as compared to APS1. Altogether, the ecological environment influences the structural characteristics and biofunctions of APS. Therefore, APS may be a potential candidate for use as an agent or functional food against obesity. These findings also provide a scientific perspective on targeted isolation of phytogenic PS with specific bioactivities achieved by putting them under specific growth and environmental conditions.Author contributions
Chongyu Zhang: investigation, formal analysis; Eunyoung Kim: investigation, formal analysis; Jiamei Cui: investigation, formal analysis; Yunpeng Wang: resources, investigation; Yunkyoung Lee: conceptualization, supervision, funding acquisition, writing (review and editing); Guiguo Zhang: conceptualization, supervision, funding acquisition, writing (original draft).Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the National Key R&D program of China-Korea cooperative project (2019YFE0107700, NRF-2019K1A3A1A20081146), the National Research Foundation Grant of Korea (NRF-2017R1D1A3B03031665, NRF-2020R1A2C2004144, and 2021R1A6A3A13046827), the Forage Industrial Innovation Team Project (SDAIT-23-05), Shandong Province Technology Innovation Guidance Plan (2019YFE0107700), and the Excellent Seed Project (2019LZGC012) of China.Notes and references
- H. Le Gall, F. Philippe, J. M. Domon, F. Gillet, J. Pelloux and C. Rayon, Cell wall metabolism in response to abiotic stress, Plants, 2015, 4, 112–166 CrossRef PubMed.
- Y. Ge, S. Ahmed, W. Yao, L. You, J. Zheng and K. Hileuskaya, Regulation effects of indigestible dietary polysaccharides on intestinal microflora: An overview, J. Food Biochem., 2021, 45, e13564 CrossRef CAS PubMed.
- J. Roewe, G. Stavrides, M. Strueve, A. Sharma, F. Marini, A. Mann, S. A. Smith, Z. Kaya, B. Strobl, M. Mueller, C. Reinhardt, J. H. Morrissey and M. Bosmann, Bacterial polyphosphates interfere with the innate host defense to infection, Nat. Commun., 2020, 11, 4035 CrossRef CAS PubMed.
- Z. Li, C. Zhang, B. Li, S. Zhang, F. G. Haj, G. Zhang and Y. Lee, The modulatory effects of alfalfa polysaccharide on intestinal microbiota and systemic health of Salmonella serotype (ser.) Enteritidis-challenged broilers, Sci. Rep., 2021, 11, 10910 CrossRef CAS PubMed.
- S. Wang, X. Dong and J. Tong, Optimization of enzyme-assisted extraction of polysaccharides from alfalfa and its antioxidant activity, Int. J. Biol. Macromol., 2013, 62, 387–396 CrossRef CAS PubMed.
- P. Shao, X. Chen and P. Sun, Improvement of antioxidant and moisture-preserving activities of Sargassum horneri polysaccharide enzymatic hydrolyzates, Int. J. Biol. Macromol., 2015, 74, 420–427 CrossRef CAS PubMed.
- L. Wang, Y. Xie, W. Yang, Z. Yang, S. Jiang, C. Zhang and G. Zhang, Alfalfa polysaccharide prevents H2O2-induced oxidative damage in MEFs by activating MAPK/Nrf2 signaling pathways and suppressing NF-kappaB signaling pathways, Sci. Rep., 2019, 9, 1782 CrossRef PubMed.
- W. Chaisuwan, Y. Phimolsiripol, T. Chaiyaso, C. Techapun, N. Leksawasdi, K. Jantanasakulwong, P. Rachtanapun, S. Wangtueai, S. R. Sommano, S. You, J. M. Regenstein, F. J. Barba and P. Seesuriyachan, The Antiviral Activity of Bacterial, Fungal, and Algal Polysaccharides as Bioactive Ingredients: Potential Uses for Enhancing Immune Systems and Preventing Viruses, Front. Nutr., 2021, 8, 772033 CrossRef PubMed.
- W. Liu, Y. Liu, R. Zhu, J. Yu, W. Lu, C. Pan, W. Yao and X. Gao, Structure characterization, chemical and enzymatic degradation, and chain conformation of an acidic polysaccharide from Lycium barbarum L, Carbohydr. Polym., 2016, 147, 114–124 CrossRef CAS PubMed.
- C. Zhang, Z. Li, C. Y. Zhang, M. Li, Y. Lee and G. G. Zhang, Extract Methods, Molecular Characteristics, and Bioactivities of Polysaccharide from Alfalfa (Medicago sativa L.), Nutrients, 2019, 11, 1181 CrossRef CAS PubMed.
- R. Yi, L. Deng, J. Mu, C. Li, F. Tan and X. Zhao, The Impact of Antarctic Ice Microalgae Polysaccharides on D-Galactose-Induced Oxidative Damage in Mice, Front. Nutr., 2021, 8, 651088 CrossRef PubMed.
- P. Gao, J. Bian, S. Xu, C. Liu, Y. Sun, G. Zhang, D. Li and X. Liu, Structural features, selenization modification, antioxidant and anti-tumor effects of polysaccharides from alfalfa roots, Int. J. Biol. Macromol., 2020, 149, 207–214 CrossRef CAS PubMed.
- S. S. Ferreira, C. P. Passos, P. Madureira, M. Vilanova and M. A. Coimbra, Structure-function relationships of immunostimulatory polysaccharides: A review, Carbohydr. Polym., 2015, 132, 378–396 CrossRef CAS PubMed.
- J. Gao, L. Lin, B. Sun and M. Zhao, Comparison Study on Polysaccharide Fractions from Laminaria japonica: Structural Characterization and Bile Acid Binding Capacity, J. Agric. Food Chem., 2017, 65, 9790–9798 CrossRef CAS PubMed.
- L. Soua, M. Koubaa, F. J. Barba, J. Fakhfakh, H. K. Ghamgui and S. E. Chaabouni, Water-Soluble Polysaccharides from Ephedra alata Stems: Structural Characterization, Functional Properties, and Antioxidant Activity, Molecules, 2020, 25, 2210 CrossRef CAS PubMed.
- B. Li, F. Lu, X. Wei and R. Zhao, Fucoidan: structure and bioactivity, Molecules, 2008, 13, 1671–1695 CrossRef CAS PubMed.
- Z. Zhang, H. Liu, B. Yu, H. Tao, J. Li, Z. Wu, G. Liu, C. Yuan, L. Guo and B. Cui, Lycium barbarum polysaccharide attenuates myocardial injury in high-fat diet-fed mice through manipulating the gut microbiome and fecal metabolome, Food Res. Int., 2020, 138, 109778 CrossRef CAS PubMed.
- M. S. Desai, A. M. Seekatz, N. M. Koropatkin, N. Kamada, C. A. Hickey, M. Wolter, N. A. Pudlo, S. Kitamoto, N. Terrapon, A. Muller, V. B. Young, B. Henrissat, P. Wilmes, T. S. Stappenbeck, G. Nunez and E. C. Martens, A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility, Cell, 2016, 167, 1339–1353 CrossRef CAS PubMed.
- E. D. Sonnenburg, H. Zheng, P. Joglekar, S. K. Higginbottom, S. J. Firbank, D. N. Bolam and J. L. Sonnenburg, Specificity of Polysaccharide Use in Intestinal Bacteroides Species Determines Diet-Induced Microbiota Alterations, Cell, 2010, 141, 1241–1252 CrossRef CAS PubMed.
- M. L. Patnode, Z. W. Beller, N. D. Han, J. Cheng, S. L. Peters, N. Terrapon, B. Henrissat, S. Le Gall, L. Saulnier, D. K. Hayashi, A. Meynier, S. Vinoy, R. J. Giannone, R. L. Hettich and J. I. Gordon, Interspecies Competition Impacts Targeted Manipulation of Human Gut Bacteria by Fiber-Derived Glycans, Cell, 2019, 179, 59–73 CrossRef CAS PubMed.
- S. Kanwal, T. P. Joseph, L. Owusu, R. Xiaomeng, L. Meiqi and X. Yi, A Polysaccharide Isolated from Dictyophora indusiata Promotes Recovery from Antibiotic-Driven Intestinal Dysbiosis and Improves Gut Epithelial Barrier Function in a Mouse Model, Nutrients, 2018, 10, 1003 CrossRef PubMed.
- F. Zhao, Q. Liu, J. Cao, Y. Xu, Z. Pei, H. Fan, Y. Yuan, X. Shen and C. Li, A sea cucumber (Holothuria leucospilota) polysaccharide improves the gut microbiome to alleviate the symptoms of type 2 diabetes mellitus in Goto-Kakizaki rats, Food Chem. Toxicol., 2020, 135, 110886 CrossRef CAS PubMed.
- J. Zhang, B. Yin, Y. Xie, J. Li, Z. Yang and G. Zhang, Legume-Cereal Intercropping Improves Forage Yield, Quality and Degradability, PLoS One, 2015, 10 Search PubMed , e0144813.
- K. I. Rovkina, S. V. Krivoshchekov, A. M. Guryev, M. S. Yusubov and M. V. Belousov, Water-Soluble Polysaccharides of Alfalfa (Medicago sativa (Fabaceae)) of Flora of Krasnoyarsk Krai, Russ. J. Bioorg. Chem., 2018, 44, 854–859 CrossRef CAS.
- A. Gutsch, K. Sergeant, E. Keunen, E. Prinsen, G. Guerriero, J. Renaut, J.-F. Hausman and A. Cuypers, Does long-term cadmium exposure influence the composition of pectic polysaccharides in the cell wall of Medicago sativa stems?, BMC Plant Biol., 2019, 19, 271 CrossRef PubMed.
- O. Douchiche, A. Driouich and C. Morvan, Spatial regulation of cell-wall structure in response to heavy metal stress: cadmium-induced alteration of the methyl-esterification pattern of homogalacturonans, Ann. Bot., 2010, 105, 481–491 CrossRef CAS PubMed.
- C. Zhao, O. Zayed, F. Zeng, C. Liu, L. Zhang, P. Zhu, C.-C. Hsu, Y. E. Tuncil, W. A. Tao, N. C. Carpita and J.-K. Zhu, Arabinose biosynthesis is critical for salt stress tolerance in Arabidopsis, New Phytol., 2019, 224, 274–290 CrossRef CAS PubMed.
- M. Krzeslowska, The cell wall in plant cell response to trace metals: polysaccharide remodeling and its role in defense strategy, Acta Physiol. Plant., 2011, 33, 35–51 CrossRef CAS.
- L. Chen, J. Liu, Y. Zhang, B. Dai, Y. An and L. L. Yu, Structuralthermal, and anti-inflammatory properties of a novel pectic polysaccharide from alfalfa (Medicago sativa L.) stem, J. Agric. Food Chem., 2015, 63, 3219–3228 CrossRef CAS PubMed.
- C. Y. Zhang, L. P. Gan, M. Y. Du, Q. H. Shang, Y. H. Xie and G. G. Zhang, Effects of dietary supplementation of alfalfa polysaccharides on growth performance, small intestinal enzyme activities, morphology, and large intestinal selected microbiota of piglets, Livest. Sci., 2019, 223, 47–52 CrossRef.
- X. Li, J. Wang, H. Zhang and Q. Zhang, Renoprotective effect of low-molecular-weight sulfated polysaccharide from the seaweed Laminaria japonica on glycerol-induced acute kidney injury in rats, Int. J. Biol. Macromol., 2017, 95, 132–137 CrossRef CAS PubMed.
- X. Q. Zha, L. Xue, H. L. Zhang, M.-N. Asghar, L. H. Pan, J. Liu and J. P. Luo, Molecular mechanism of a new Laminaria japonica polysaccharide on the suppression of macrophage foam cell formation via regulating cellular lipid metabolism and suppressing cellular inflammation, Mol. Nutr. Food Res., 2015, 59, 2008–2021 CrossRef CAS PubMed.
- X. Y. Li, Q. M. Li, Q. Fang, X. Q. Zha, L. H. Pan and J. P. Luo, Laminaria japonica Polysaccharide Inhibits Vascular Calcification via Preventing Osteoblastic Differentiation of Vascular Smooth Muscle Cells, J. Agric. Food Chem., 2018, 66, 1821–1827 CrossRef CAS PubMed.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- J. Cui, Y. Wang, E. Kim, C. Zhang, G. Zhang and Y. Lee, Structural Characteristics and Immunomodulatory Effects of a Long-Chain Polysaccharide From Laminaria japonica, Front. Nutr., 2022, 9, 762596 Search PubMed.
- J. T. Oberlerchner, T. Rosenau and A. Potthast, Overview of Methods for the Direct Molar Mass Determination of Cellulose, Molecules, 2015, 20, 10313–10341 CrossRef CAS PubMed.
- L. Chen, J. Liu, Y. Zhang, B. Dai, Y. An and L. L. Yu, Structural, thermal, and anti-inflammatory properties of a novel pectic polysaccharide from alfalfa (Medicago sativa L.) stem, J. Agric. Food Chem., 2015, 63, 3219–3228 CrossRef CAS PubMed.
- E. Kim, J. Cui, I. Kang, G. Zhang and Y. Lee, Potential Antidiabetic Effects of Seaweed Extracts by Upregulating Glucose Utilization and Alleviating Inflammation in C2C12 Myotubes, Int. J. Environ. Res. Public Health, 2021, 18, 1367 CrossRef CAS PubMed.
- S. H. Seo, S.-M. Jo, T. T. M. Truong, G. Zhang, D.-S. Kim, M. Lee, Y. Lee and I. Kang, Peanut sprout rich in p-coumaric acid ameliorates obesity and lipopolysaccharide-induced inflammation and the inhibition of browning in adipocytes via mitochondrial activation, Food Funct., 2021, 5361–5374 RSC.
- D. Lee and S. K. R. Williams, Thermal field-flow fractionation and multiangle light scattering of polyvinyl acetate with broad polydispersity and ultrahigh molecular weight microgel components, J. Chromatogr., 2010, 1217, 1667–1673 CrossRef CAS PubMed.
- L. Yang and L.-M. Zhang, Chemical structural and chain conformational characterization of some bioactive polysaccharides isolated from natural sources, Carbohydr. Polym., 2009, 76, 349–361 CrossRef CAS.
- R. Zhong, X. Wan, D. Wang, C. Zhao, D. Liu, L. Gao, M. Wang, C. Wu, S. M. Nabavid, M. Daglia, E. Capanoglu, J. Xiao and H. Cao, Polysaccharides from Marine Enteromorpha: Structure and function, Trends Food Sci. Technol., 2020, 99, 11–20 CrossRef CAS.
- J. E. Lee, H. Schmidt, B. Lai and K. Ge, Transcriptional and Epigenomic Regulation of Adipogenesis, Mol. Cell. Biol., 2019, 39, e00601–18 CAS.
- D. A. Samac, L. Litterer, G. Temple, H. J. Jung and D. A. Somers, Expression of UDP-glucose dehydrogenase reduces cell-wall polysaccharide concentration and increases xylose content in alfalfa stems, Appl. Biochem. Biotechnol., 2004, 113–116, 1167–1182 CrossRef CAS PubMed.
- K. H. Caffall and D. Mohnen, The structure, function, and biosynthesis of plant cell wall pectic polysaccharides, Carbohydr. Res., 2009, 344, 1879–1900 CrossRef CAS PubMed.
- S. Wang, X. Dong and J. Tong, Optimization of enzyme-assisted extraction of polysaccharides from alfalfa and its antioxidant activity, Int. J. Biol. Macromol., 2013, 62, 387–396 CrossRef CAS PubMed.
- H. Cheng, S. Feng, X. Jia, Q. Li, Y. Zhou and C. Ding, Structural characterization and antioxidant activities of polysaccharides extracted from Epimedium acuminatum, Carbohydr. Polym., 2013, 92, 63–68 CrossRef CAS PubMed.
- D. Sui and B. Wang, Transcriptome Analysis Reveals Complex Defensive Mechanisms in Salt-Tolerant and Salt-Sensitive Shrub Willow Genotypes under Salinity Stress, Int. J. Genomics, 2020, 2020, 6870157 Search PubMed.
- S. de Jong and F. van de Velde, Charge density of polysaccharide controls microstructure and large deformation properties of mixed gels, Food Hydrocolloids, 2007, 21, 1172–1187 CrossRef CAS.
- W. Wang, M. Shen, L. Jiang, Q. Song, S. Liu and J. Xie, Influence of Mesona blumes polysaccharide on the gel properties and microstructure of acid-induced soy protein isolate gels, Food Chem., 2020, 313, 126125 CrossRef PubMed.
- R. S. Aquino, C. Grativol and P. A. S. Mourão, Rising from the Sea: Correlations between Sulfated Polysaccharides and Salinity in Plants, PLoS One, 2011, 6, e18862 CrossRef CAS PubMed.
- G. S. Lorda, R. C. Castano, A. B. Pordomingo, M. D. Pastor and A. P. Balatti, Growth and polysaccharide formation in Sinorhizobium meliloti strains in an air-lift-type fermentor. Effect on nodulation velocity in alfalfa plants, Rev. Argent. Microbiol., 2003, 35, 57–61 CAS.
- B. R. Sharma, H. J. Kim, M. S. Kim, C. M. Park and D. Y. Rhyu, Caulerpa okamurae extract inhibits adipogenesis in 3T3-L1 adipocytes and prevents high-fat diet–induced obesity in C57BL/6 mice, Nutr. Res., 2017, 47, 44–52 CrossRef CAS PubMed.
- A. R. Ryu, Y. W. Kim and M. Y. Lee, Chlorin e6-mediated photodynamic therapy modulates adipocyte differentiation and lipogenesis in 3T3-L1 cells, Photodiagn. Photodyn. Ther., 2020, 31, 101917 CrossRef CAS PubMed.
- D. J. Steger, G. R. Grant, M. Schupp, T. Tomaru, M. I. Lefterova, J. Schug, E. Manduchi, C. J. Stoeckert Jr. and M. A. Lazar, Propagation of adipogenic signals through an epigenomic transition state, Genes Dev., 2010, 24, 1035–1044 CrossRef CAS PubMed.
- J. P. Li, Y. Yuan, W. Y. Zhang, Z. Jiang, T. J. Hu, Y. T. Feng and M. X. Liu, Effect of Radix isatidis polysaccharide on alleviating insulin resistance in type 2 diabetes mellitus cells and rats, J. Pharm. Pharmacol., 2019, 71, 220–229 CrossRef CAS PubMed.
- S. Suraiya, Y. B. Choi, H. D. Park, W. J. Jang, H.-H. Lee and I.-S. Kong, Saccharina japonica fermented by Monascus spp. inhibit adipogenic differentiation and gene expression analyzed by real-time PCR (Q-PCR) in 3T3-L1 cell, J. Funct. Foods, 2019, 55, 371–380 CrossRef CAS.
- P. P. Sharma and V. Baskaran, Polysaccharide (laminaran and fucoidan), fucoxanthin and lipids as functional components from brown algae (Padina tetrastromatica) modulates adipogenesis and thermogenesis in diet-induced obesity in C57BL6 mice, Algal Res., 2021, 54, 102187 CrossRef.
- R. M. Oliveira, R. B. G. Camara, J. F. S. Monte, R. L. S. Viana, K. R. T. Melo, M. F. Queiroz, L. G. A. Filgueira, L. M. Oyama and H. A. O. Rocha, Commercial Fucoidans from Fucus vesiculosus Can Be Grouped into Antiadipogenic and Adipogenic Agents, Mar. Drugs, 2018, 16, 193 CrossRef PubMed.
- X. Lin, W. Xu, L. Liu, S. Ou and X. Peng, In vitro fermentation of flaxseed polysaccharide by fecal bacteria inhibits energy intake and adipogenesis at physiological concentration, Food Res. Int., 2021, 139, 109920 CrossRef CAS PubMed.
- L. Zhe, L. N. Wang, X. Lin, S. Lan and F. Yi, Drug delivery for bioactive polysaccharides to improve their drug-like properties and curative efficacy, Drug delivery, 2017, 24, 70–80 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo00371f |
| ‡ These authors contributed equally to this study and share the first authorship. |
| This journal is © The Royal Society of Chemistry 2022 |