Open Access Article
Open Access ArticleResazurin-based high-throughput screening method for the discovery of dietary phytochemicals to target microbial transformation of L-carnitine into trimethylamine, a gut metabolite associated with cardiovascular disease†
Carolina
Simó
 a,
Tiziana
Fornari
b,
Mónica R.
García-Risco
b,
Ainize
Peña-Cearra
cd,
Leticia
Abecia
a,
Tiziana
Fornari
b,
Mónica R.
García-Risco
b,
Ainize
Peña-Cearra
cd,
Leticia
Abecia
 cd,
Juan
Anguita
cd,
Juan
Anguita
 c,
Héctor
Rodríguez
c and
Virginia
García-Cañas
c,
Héctor
Rodríguez
c and
Virginia
García-Cañas
 *a
*a
aMolecular Nutrition and Metabolism, Institute of Food Science Research (CIAL), Spanish National Research Council (CSIC), Madrid, 28049, Spain. E-mail: virginia.garcia@csic.es; Tel: +34-910017900
bInstitute of Food Science Research (CIAL), Autonomous University of Madrid, Madrid, 28049, Spain
cCIC bioGUNE. Bizkaia Science and Technology Park, bld 801 A, 48160, Derio, Bizkaia, Spain
dImmunology, Microbiology and Parasitology Department, Medicine and Nursing Faculty, University of the Basque Country (UPV), 48940, Leioa, Spain
First published on 12th April 2022
Abstract
Nowadays, there is great interest in the discovery of food compounds that might inhibit gut microbial TMA production from its methylamine precursors. In this work, an innovative novel screening strategy capable of rapidly determining the differences in the metabolic response of Klebsiella pneumoniae, a bacteria producing TMA under aerobic conditions, to a library of extracts obtained from food and natural sources was developed. The proposed high-throughput screening (HTS) method combines resazurin reduction assay in 384-well plates and Gaussian Processes as a machine learning tool for data processing, allowing for a fast, cheap and highly standardized evaluation of any interfering effect of a given compound or extract on the microbial metabolism sustained by L-carnitine utilization. As a proof-of-concept of this strategy, a pilot screening of 39 extracts and 6 pure compounds was performed to search for potential candidates that could inhibit in vitro TMA formation from L-carnitine. Among all the extracts tested, three of them were selected as candidates to interfere with TMA formation. Subsequent in vitro assays confirmed the potential of oregano and red thyme hexane extracts (at 1 mg mL−1) to inhibit TMA formation in bacterial lysates. In such in vitro assay, the red thyme extract exerted comparable effects on TMA reduction (∼40%) as 7.5 mM meldonium (∼50% TMA decrease), a reported L-carnitine analogue. Our results show that metabolic activity could be used as a proxy of the capacity to produce TMA under controlled culture conditions using L-carnitine to sustain metabolism.
1. Introduction
The metabolic function of the gut microbial community plays a crucial role in health and disease. In recent years, there has been an increasing interest in the metabolite trimethylamine N-oxide (TMAO). This metabolite is present in some foods, particularly in seafood, but it can also be generated through a meta-organismal stepwise process that involves (i) the microbial production of trimethylamine (TMA) in the gut from dietary precursors and (ii) its subsequent oxidation to TMAO by flavin-containing monooxygenases (FMOs) in the liver.1 Circulating levels of TMAO have been associated with increased risk of CVD through several mechanisms.2–4 Choline, L-carnitine, betaine, and other TMA-containing compounds are the major dietary precursors of microbial TMA, therefore, foods such as red meat, eggs, dairy products, and saltwater fish, are potential precursors of TMAO in humans. Considering the multifactorial nature of TMAO production, various strategies may be followed to interfere with different biochemical and physiological steps that constitute the TMA/TMAO pathway in order to reduce circulating TMAO levels and prevent or reduce the risk of TMAO-related diseases. Limiting the dietary consumption of food enriched in dietary trimethylamines, inhibition of hepatic FMO activity and modulation of gut microbiota composition and/or its TMA production capacity are some of the strategies proposed to reduce circulating TMAO levels.5Different gut microorganisms have diverse abilities to generate TMA from dietary precursors as they harbour genes related to the synthesis of enzymes implicated in TMA production. In the last few years, the structure and function of some of those enzymes have been investigated providing valuable information about these potential targets for the modulation of TMA-related microbial function.6–13 In this regard, various research efforts have been directed to discover new candidate molecules that could act as inhibitors of the gut microbial TMA biosynthesis from choline as the precursor, aiming at reducing circulating TMAO levels and its linked deleterious effects on health.11,14–16 On the other side, TMA production from L-carnitine has been less explored. With regard to this precursor, two main pathways have been described. A predominant anaerobic pathway governed by two sets of bacterial proteins encoded by the Cai operon and the Bbu gene cluster has recently been elucidated.13 The other pathway is constituted by carnitine monooxygenase (CntA) and its associated reductase (CntB), which are known to produce TMA and malic semialdehyde from L-carnitine under aerobic conditions.17 Meldonium, the anti-ischemic drug and structural analogue of L-carnitine and γ-butyrobetaine (GBB), has been reported to interfere with TMA formation through this pathway.18 The structure of CntAB from Acinetobacter baumannii has been recently elucidated as well as its substrate specificity and inhibition, setting the molecular basis for the future structure-guided discovery of inhibitors.8,9 Indeed, results from a random screening of drug libraries based on an enzymatic assay with purified recombinant CntA revealed three inhibitor candidates.9 However, only one compound was able to significantly inhibit TMA production in living TMA-producing bacterial cells in the presence of L-carnitine. Therefore, there is a need to assess the inhibitory potential of inhibitor candidates in living bacteria, in which the metabolic pathway under study is often more complex and subjected to the influence of more factors than in an in vitro enzymatic reaction.
The ability of specific foods, food constituents and phytochemicals to lower the levels of circulating TMAO has been mostly demonstrated in animal models, as it has been recently reviewed.5 However, only in a few cases, the association between changes in circulating TMAO levels and the gut microbiota has been explained, and the mechanisms underlying the effects of the dietary elements as the object of the study remain to be elucidated. Besides the mentioned scarcity of mechanistic studies, the lack of methods to screen for natural compounds that inhibit microbial TMA production is another factor that has precluded the development of novel functional food ingredients to target microbial TMA generation. In most published reports, detection of TMA in culture media is the preferred approach to measure the ability of a microorganism to produce TMA under different experimental conditions.19,20 However, this procedure is not straightforward and costly mass spectrometry instrumentation is required for its unequivocal detection and accurate quantitation. Using this approach, Bresciani et al. observed inhibition of choline and L-carnitine degradation when blonde orange juice was tested, and attributed that inhibition mainly to the sugar content.19 As plausible hypotheses to explain their findings, authors suggested that the presence of sugars naturally contained in juices might push the microbial enzymatic activity towards more metabolically favourable pathways rather than metabolizing choline and L-carnitine to produce TMA; or alternatively, sugars might be converted into short chain fatty acids, reducing pH and inhibiting choline and L-carnitine bioconversion.
In spite of these research efforts, there is a clear need to discover new compounds with the ability to interfere with gut microbial TMA generation. Thus, observations such as (i) the obvious involvement of diet on TMA formation and circulating TMAO levels, (ii) the recent findings regarding the potential mitigating effects of certain foods on the generation of these biologically relevant metabolites, and (iii) the scarce investigation on dietary compounds that might inhibit microbial TMA production from L-carnitine, motivated us to explore new methods for screening new food sources to attenuate gut microbial TMA generation. Therefore, this work is aimed at developing an innovative screening strategy capable of rapidly capturing differences in the metabolic response of Klebsiella pneumoniae, a microorganism producing TMA under aerobic conditions, to a library of pure natural compounds and plant extracts. We developed a high-throughput screening (HTS) method that combines resazurin reduction assay in 384-well plates and machine learning tools for data processing, allowing for a fast, cheap and highly standardized evaluation of any interfering effect of a given extract or compound on the microbial metabolism sustained by L-carnitine utilization. As a proof-of-concept of this strategy, a pilot screening of 39 natural extracts and 6 pure compounds, tested at different concentrations, was performed to search for potential candidates that could inhibit in vitro TMA formation from L-carnitine.
2. Materials and methods
2.1 Bacterial strain and growth media
K. pneumoniae from human fecal sample was kindly donated by Laboratorio Clínico Central (San Sebastián de los Reyes, Spain). This strain harbours functional CntAB and was able to produce a significant amount of TMA when cultivated in the presence of L-carnitine.21 The bacterial isolate was maintained at −80 °C in 10% w/v glycerol in a brain–heart infusion (BHI) media. K. pneumoniae cells were grown aerobically at 37 °C in a defined chemical medium (DM) based on the medium described previously7 with some modifications, having the following composition: 1 g L−1 NH4Cl, 0.5 g L−1 NaCl, 3 g L−1 KH2PO4, 17.1 g L−1 Na2HPO4·12H2O, 0.6 mg L−1 Na2MoO4·2H2O, 0.22 g L−1 CaCl2·6H2O, 50 μM FeCl3, 0.24 g L−1 MgSO4, 0.02 mg L−1 biotin, 0.02 mg L−1 folic acid, 0.1 mg L−1 pyridoxine hydrochloride, 0.05 mg L−1 thiamine hydrochloride, 0.05 mg L−1 riboflavin, 0.05 mg L−1 nicotinic acid, 0.05 mg L−1 pantothenic acid, 0.001 mg L−1 vitamin B12, 0.05 mg L−1 4-aminobenzoic acid, and 0.05 mg L−1 lipoic acid. L-Carnitine or glucose was added to the DM as the sole carbon and energy source.2.2. Growth curves of bacteria and counting of viable bacteria
Bacterial growth was estimated by means of turbidity measurements in transparent microtiter plates. Changes in turbidity of cell suspensions were determined by measuring the optical density at 600 nm wavelength (OD600) using a microplate reader SynergyTM (BioTek Instruments Inc., USA). Bacterial viability was determined as colony forming units (CFU) per mL by counting the number of viable bacteria by the formation of visible colonies CFU on Brain Heart Infusion agar plates incubated at 37 °C for 24 h, following the single plate-serial dilution spotting approach.222.3. Preparation of the standardized bacterial inoculum
A frozen stock of K. pneumoniae was pre-grown in 5 mL BHI at 37 °C with continuous shaking for 4–6 h. To induce L-carnitine-to-TMA metabolism, the culture was then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 with DM containing 20 mM L-carnitine, which was then incubated at 37 °C with continuous shaking for 12–14 h. A subculture was prepared by inoculating 100 μL in 10 mL DM with 20 mM L-carnitine. This subculture was incubated at 37 °C for 8 h, and the pellet was collected by centrifugation at 4500g for 10 min and suspended in 5 mL DM without the carbon source. Optical density at 570 nm (OD570) was measured using a MultiscanTM FC microplate reader (Thermo Scientific, USA) for calculating bacterial density in suspension. Thus, OD570 value was adjusted with DM to the equivalent of 2.33 × 108 colony forming units per mL (CFU mL−1), which was determined from the calibration curve. The resulting standardized microbial suspension was used as starting bacterial inoculum for the resazurin-based HTS assay.
100 with DM containing 20 mM L-carnitine, which was then incubated at 37 °C with continuous shaking for 12–14 h. A subculture was prepared by inoculating 100 μL in 10 mL DM with 20 mM L-carnitine. This subculture was incubated at 37 °C for 8 h, and the pellet was collected by centrifugation at 4500g for 10 min and suspended in 5 mL DM without the carbon source. Optical density at 570 nm (OD570) was measured using a MultiscanTM FC microplate reader (Thermo Scientific, USA) for calculating bacterial density in suspension. Thus, OD570 value was adjusted with DM to the equivalent of 2.33 × 108 colony forming units per mL (CFU mL−1), which was determined from the calibration curve. The resulting standardized microbial suspension was used as starting bacterial inoculum for the resazurin-based HTS assay.
2.4. Plant extract preparation and chemical characterization
A series of bench-scale extraction tests were conducted from selected edible plants and fruits. Different solvents such as water, ethanol and hexane were used for ultrasound-assisted extraction (UAE) for the extraction of plant compounds based on polarity. The plant samples in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 plant–solvent ratio were subjected to ultrasound (Branson Digital Sonifier 550 model, Danbury, USA) frequency of 20 kHz, 550 W and 60% of sonication output amplitude pretreatment for 20 min and 50 °C or 30 °C.
10 plant–solvent ratio were subjected to ultrasound (Branson Digital Sonifier 550 model, Danbury, USA) frequency of 20 kHz, 550 W and 60% of sonication output amplitude pretreatment for 20 min and 50 °C or 30 °C.
Supercritical fluid extraction (SFE) was carried out using a pilot-plant extractor (SF2000 Thar Technology, Pittsburgh, USA), with independent control of temperature and pressure. Different sequential extractions were accomplished, with pure CO2 and CO2 using 15% ethanol as the cosolvent, at 40 °C, 80 g min−1 CO2 flow rate, and pressures in the range 10–30 MPa and time in the range 40–120 min. Following the extraction process, the extracts were evaporated under reduced pressure using a rotary evaporator and finally the concentrated extracts were stored at −4 °C. Stock extract solutions (25, 100 or 200 mg mL−1) were prepared by dissolving the extracted material into a homogeneous solution using sterile dimethyl sulfoxide (DMSO) or water. Additional experimental conditions for the extraction procedures are listed in the ESI, Table S1.†
Extracts selected as candidates from the screening assay were characterized by gas chromatography-mass spectrometry (GC-MS) in a GC 7890A system (Agilent Technologies, USA) with a mass spectrometer detector 5975C triple-axis. An HP-5MS capillary column (30 m × 0.25 mm i.d. and 0.25 μm phase thickness) was used. The chromatographic method started with an initial temperature of 40 °C, then increased to 150 °C, at 3 °C min−1 and was held at 150 °C for 10 min. The method finished with a 3 min post-run at 300 °C. The injection volume was 1 μl in the splitless mode. Helium (99.99%) was employed as the carrier gas. The temperatures were 260 °C for the injector, 230 °C for the mass spectrometer ion source, 280 °C for the interface and 150 °C for the quadrupole. The mass spectrometer was operated under the electron impact mode (70 eV) and it was used in total ion current (TIC) mode and scanned the mass range from 40 to 500 m/z. GC-Ms chromatograms and identified compounds in selected extracts are shown in the ESI, Fig. S1 and Table S2,† respectively.
2.5. Resazurin HTS assay
The resazurin metabolization assays were performed using black, polystyrene, clear bottom 384-well plates (Nunclon1 surface, Nunc, Roskilde, Denmark). To avoid water condensation on the microplate lids during the measurements, the lids were coated with 0.05% Triton X-100 in 20% ethanol.23 Bacterial inoculum, resazurin concentration and analysis time were initially optimized. During this optimization stage, the growth of K. pneumoniae in DM with 20 mM L-carnitine was quantified by measuring the OD570. For each resazurin concentration tested, the bacterial culture was diluted to an optical density equivalent to 4 × 107 CFU mL−1 in MD containing a given concentration of resazurin (ranging from 0.25 to 5 μg mL−1). These suspensions were used as stocks to obtain various two-fold dilution series of bacteria ranging from 4 × 107 to 0.25 × 106 CFU mL−1 in MD containing the same concentration of resazurin as the respective stock. The assay was performed by adding 35 μl of the bacterial suspensions into the wells containing 35 μl of DM with 40 mM L-carnitine for a 384-well assay plate in ten different replicates. The DM medium with 20 mM L-carnitine and different resazurin concentrations were used as blanks to correct for any background interfering signal. The 384-well microplate was placed in a microplate reader Cytation™ (BioTek Instruments Inc., USA) and incubated at 37 °C with orbital shaking. The fluorescence (RFU) of microbial-generated resorufin was recorded at λex = 520 nm/λem = 590 nm (bottom-read) immediately after adding standardized inoculum (or DM for blanks) with resazurin to all wells, and then again in 15 min periods for 12 h.For each assay in 384-well-plates under optimal selected conditions, testing wells were loaded in five technical replicates per condition as follows: 35 μL of natural extracts or pure components prepared in DM with the selected carbon source (5 mM glucose or 40 mM L-carnitine) was added per well. Two-fold serial dilutions of each compound or extract were performed before the addition of the inoculum. Then, 35 μL of K. pneumoniae suspension (2 × 107 CFU mL−1) in MD containing 5 μg mL−1 resazurin was added to each well. Each plate had controls with bacterial inoculum containing the vehicle (DMSO or water) and blanks containing the culture media with the testing bioactives, but without bacterial inoculum. The 384-well microplate was placed in a microplate reader Cytation™ and incubated at 37 °C with orbital shaking. The fluorescence (RFU) of microbial-generated resorufin was recorded as mentioned above.
2.6. Microbial TMA determination
TMA was determined in the growth medium by capillary electrophoresis with the UV detection (CE-UV) method as described previously.21 Briefly, the bacterial culture or bacterial lysate was mixed with derivatizing agent 80 mm 2,4′-dibromoacetophenone (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, v/v) in a 2 ml tube, and incubated at 70 °C for 60 min. The derivatized sample was evaporated using a nitrogen stream or vacuum concentrated with SpeedVac to dryness. Then, water (same volume as the derivatizing agent) was added and vortex-shaken for 15 min. The sample was subjected to centrifugation at 15
3, v/v) in a 2 ml tube, and incubated at 70 °C for 60 min. The derivatized sample was evaporated using a nitrogen stream or vacuum concentrated with SpeedVac to dryness. Then, water (same volume as the derivatizing agent) was added and vortex-shaken for 15 min. The sample was subjected to centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 15 °C, and the supernatant was directly analyzed by CE-UV. Analyses were carried out on a P/ACE 5010 capillary electrophoresis system (Beckman Coulter Instruments, CA, USA) with UV detection at 254 nm.
000g for 10 min at 15 °C, and the supernatant was directly analyzed by CE-UV. Analyses were carried out on a P/ACE 5010 capillary electrophoresis system (Beckman Coulter Instruments, CA, USA) with UV detection at 254 nm.
2.7. L-Carnitine oxygenase/reductase activity in the K. pneumoniae lysate
A frozen stock of K. pneumoniae was grown in 5 mL BHI broth and the culture was then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 with 10 mL of DM supplemented with 20 mM L-carnitine and incubated overnight at 37 °C with continuous shaking (150 rpm). The culture was then diluted 1
100 with 10 mL of DM supplemented with 20 mM L-carnitine and incubated overnight at 37 °C with continuous shaking (150 rpm). The culture was then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 with 1 L of DM supplemented with 20 mM L-carnitine, which was grown at 37 °C overnight. Cells were harvested by centrifugation (9000g, 20 min) and resuspended in lysis buffer (20 mM Tris, 50 mM NaCl, pH 8) supplemented with a protease inhibitor cocktail (Sigma-Aldrich). The cells were lysed by agitation with glass beads with a high-speed homogenizer FastPrep-24™ 5G (MP Biomedicals, CA, USA). Three cycles of agitation (40 s) were interspersed with cooling on ice (1 min). The lysate was cleared by centrifugation and the protein content of the supernatant was determined using a commercially available Qubit Assay kit and the readings were obtained with a Qubit Fluorometer 3.0 (Thermo Fisher Scientific, MA, USA). The assay was performed on 96-well plates. The total volume of the reaction was 200 μL. A volume of 100 μL of the pure compound, extract or meldonium was added to all test wells, the lysate solution (50 μL per well) was added to all test wells, prior to the start of the reaction with the addition of the substrate L-carnitine and NADH as the cofactor (50 μL per well). The plate was incubated at 37 °C for 20 h. After incubation, 100 μL of the reaction mixture was subjected to TMA determination by CE-UV.
100 with 1 L of DM supplemented with 20 mM L-carnitine, which was grown at 37 °C overnight. Cells were harvested by centrifugation (9000g, 20 min) and resuspended in lysis buffer (20 mM Tris, 50 mM NaCl, pH 8) supplemented with a protease inhibitor cocktail (Sigma-Aldrich). The cells were lysed by agitation with glass beads with a high-speed homogenizer FastPrep-24™ 5G (MP Biomedicals, CA, USA). Three cycles of agitation (40 s) were interspersed with cooling on ice (1 min). The lysate was cleared by centrifugation and the protein content of the supernatant was determined using a commercially available Qubit Assay kit and the readings were obtained with a Qubit Fluorometer 3.0 (Thermo Fisher Scientific, MA, USA). The assay was performed on 96-well plates. The total volume of the reaction was 200 μL. A volume of 100 μL of the pure compound, extract or meldonium was added to all test wells, the lysate solution (50 μL per well) was added to all test wells, prior to the start of the reaction with the addition of the substrate L-carnitine and NADH as the cofactor (50 μL per well). The plate was incubated at 37 °C for 20 h. After incubation, 100 μL of the reaction mixture was subjected to TMA determination by CE-UV.
2.8. Data analysis
All the fluorescence data points were subtracted with their respective blank well signals at each time point. Then, background corrected fluorescence signals were fitted and used to compute the time derivative curves accounting for estimated resazurin reduction rate as a function of time using the publicly available Fitderiv software.24 Fitderiv also estimated some characteristics of the curves, including (a) the maximum resazurin reduction signal (MaxRS), (b) the time to reach the maximum resazurin reduction rate (Time-MaxRate), (c) the maximum resazurin reduction rate (MaxRate) as the maximum value from the estimated resazurin metabolization as a function of time, and (d) the lag time (Lag-Time) as the intercept of the line parallel to the time axis that passes through the initial fluorescence signal and the tangent to the curve from the point on the curve with the maximum reduction rate.24 During assay optimization, these parameters were evaluated by curve fitting of data obtained with each resazurin concentration and density inoculum, and by calculating the reduction rates as a function of time. Graphs were plotted using Matplotlib25 and Seaborn26 libraries in Python, and correlation and regression analyses were performed with statmodels27 and Pingouin libraries.28 Fluorescence curves are represented as the mean of blank-subtracted fluorescence data (n = 5) and the 95% confidence interval. In vitro data are expressed as the mean and standard deviation (SD). Statistical comparisons were performed using the Student's t-test. A probability (p) value of <0.05 was considered statistically significant.3. Results
3.1. Bacterial growth and TMA production in the microplate culture with L-carnitine as the sole carbon source
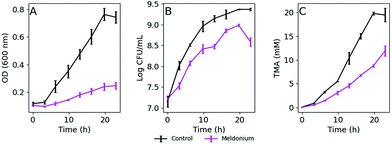
K. pneumoniae was grown in a defined medium (DM) with 20 mM L-carnitine (DM + C) as the sole carbon source as described in the Materials and Methods section. Bacterial growth measuring the OD600, viable cells reported as CFU mL−1, and TMA produced over 24 h in DM + C are summarized in Fig. 1A–C. As is shown in Fig. 1B (black line), the bacterial growth under these limiting nutrient conditions was exponential, but several hours were necessary to double the number of bacterial cells. As expected, TMA formation in culture media followed a similar trend (Fig. 1C), whereas the presence of meldonium, a drug that has been reported to decrease intestinal microbiota-dependent production of TMA from L-carnitine,18 decreased TMA formation as well as bacterial growth (purple lines in Fig. 1). Correlation analysis revealed good correlation between both, CFU mL−1 and OD600 measurements, and TMA concentration in culture media as indicated by Spearman's correlation coefficients (r = 0.96 and 0.94, respectively, n = 16, p-value <0.05).In response to the need for a fast, cheap and simple method that could enable the screening of compounds that may influence TMA metabolism in K. pneumoniae, we envisaged the adaptation of the routinely used resazurin reduction assay for bacterial viability. In this case, the adaptation of the resazurin reduction assay would be exploited to track potential changes in the microbial metabolism under strict growing conditions that lead to in vitro TMA production in the presence of various extracts. Ideally, this method would continuously monitor the metabolic activity of bacterial cells sustained by L-carnitine as the carbon source, avoiding additional sampling steps during the assay.
3.2. Optimization of the resazurin HTS assay
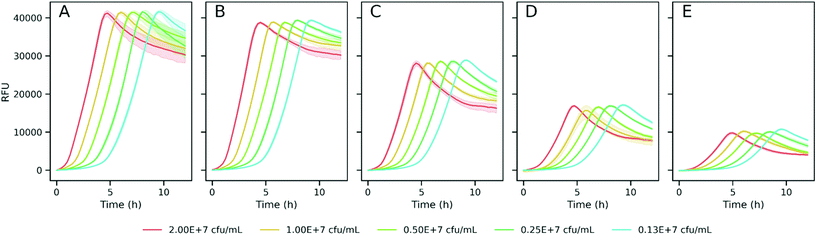
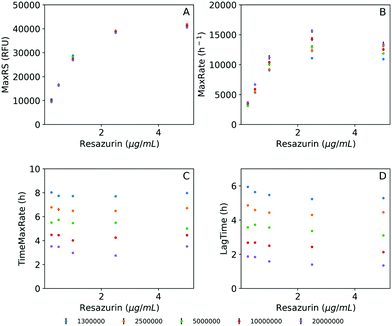
To explore the suitability of resazurin reduction to our goal, we first optimized the method to establish adequate assay conditions that can provide reproducible, sensitive and accurate data for bacterial metabolic activity. Thus, the reduction of resazurin by K. pneumoniae was monitored by the acquisition of fluorescence signals (λex = 520 nm/λem = 590 nm) in 384-well plates to investigate the influence of resazurin concentration, initial inoculum density, and incubation time on the fluorescence signal over the time. Fig. 2 shows the background subtracted fluorescence curves accounting for the resazurin transformation to resorufin in bacterial suspensions. As expected, the three variables, analysis time, resazurin concentration and inoculum density had a significant effect on the signal. Thus, in order to analyze in detail the effect of these variables on the resazurin reduction curves, Fitderiv software, based on non-parametric Gaussian Process (GP) analysis, was used to fit the time-series data from 384-well plate micro-cultures, and to infer the corresponding time derivatives.24 Using this approach, it was possible to model resazurin reduction by K. pneumoniae and determine several parameters. MaxRS and LagTime were estimated from the model fits, whereas, the MaxRate and TimeMaxRate were obtained from the time derivative curve as indicators of the maximum reduction rate and the time point at which that maximum is reached, respectively.Examples of the obtained fluorescence curves, model fits and time derivatives, from which these parameters were estimated, are shown in the ESI, Fig. S2.† In addition, Fig. 3 illustrates the four parameters derived from these data plotted as a function of resazurin concentration. Fig. 3A suggests that the maximum fluorescence signal greatly increased with resazurin concentration reaching a plateau at concentrations above 2.5 μg mL−1, with poor dependence on the assayed inoculum density interval. Following a similar trend, the estimated maximal rate values at which resazurin is reduced, increased with concentration until reaching a maximum at 2.5 μg mL−1 (Fig. 3B). Furthermore, this parameter (MaxRate) showed a linear dependence on the logarithm of the inoculum density (R2 = 0.99) in the assays performed with 2.5 μg mL−1 resazurin (ESI Fig. S3†). On the other side, the time-associated parameters (TimeMaxRate and LagTime) exhibited less dependence on resazurin concentration, but a strong inverse dependence on bacterial inoculum density (Fig. 3C and D). This observation can also be confirmed by the best-fit semi-logarithmic curves of these data providing a high (>0.99) coefficient of determination with most of the resazurin concentrations assayed (ESI Fig. S3†). According to these results and in order to set the optimum resazurin concentration that allows a good correlation of the inferred parameters with the initial bacterial load, a concentration of 2.5 μg mL−1 resazurin was selected for further experiments. In addition, a density of 107 CFU mL−1 of bacteria was also set as a trade-off between the time of analysis and expenditure of time and material resources to obtain sufficient biomass for the assay.
Under the selected experimental conditions, we investigated the strength of the relationship between fluorescence measurements and TMA formation. To achieve this, a 384-well plate was prepared to incubate standardized K. pneumoniae inoculum (equivalent to 107 CFU mL−1) in DM + C containing 2.5 μg mL−1 resazurin, and 7.5 mM meldonium or vehicle. Incubation was performed in a microplate reader at 37 °C with continuous orbital shaking. Every 15 minutes, fluorescence was recorded at λex = 520 nm/λem = 590 nm and every 30 minutes culture aliquots were withdrawn from the wells for TMA analysis. Then, the TMA peak areas obtained from CE-UV analyses and the fluorescence signals measured at the respective time points were subjected to correlation analysis. Spearman's rho indicated a strong correlation (r = 0.97, n = 65 and p-value <0.01) between TMA peak areas and all those RFU values ranging from 1200 to 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 that were acquired at time points before reaching MaxRS (ESI Fig. S4†).
000 that were acquired at time points before reaching MaxRS (ESI Fig. S4†).
3.3. Performance of the method to track changes in K. pneumoniae metabolism
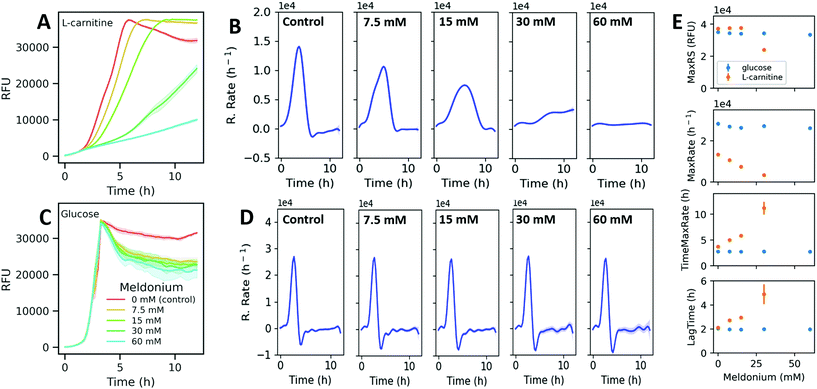
Next, the change in the model parameters obtained from assays under different culturing conditions was evaluated. First, a set of assays were carried out in DM + C under selected optimal conditions with doubling dilutions of meldonium (60 mM–7.5 mM). Preliminary assays showed that this range of meldonium concentrations inhibited TMA generation by K. pneumoniae in MD + C in a dose-dependent manner (ESI Fig. S5†). The presence of meldonium in the growing media induced concentration-dependent changes in the fluorescence curves (Fig. 4A). Resazurin reduction rates were computed and the parameters derived from the curves were also determined. The time derivatives estimated from the model fits obtained with meldonium displayed obviously different reduction rate profiles compared to the reduction rate curve obtained without meldonium (Fig. 4B). Then, to test the selectivity of the observed meldonium effects on L-carnitine sustained metabolism, a set of assays were performed under the same experimental conditions but replacing L-carnitine with glucose as the sole carbon source. The corresponding fluorescence and reduction rate curves obtained with glucose and different meldonium concentrations did not show significant differences compared to those obtained in the controls (Fig. 4C and D). A comparison of the parameters estimated from processing these data revealed that both TimeMaxRate and LagTime increased with meldonium concentration in DM + C, whereas the values for these parameters remained unaltered when DM was supplemented with glucose (DM + G, Fig. 4E). Similarly, MaxRS and MaxRate parameters did not show remarkable differences under various experimental conditions assayed in DM + G. By contrast, with DM + C, MaxRate followed a negative linear relationship with meldonium concentration, as deduced by the simple linear regression fitting of the data (y = −331x + 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 072, R2 = 0.989). The MaxRS parameter remained unaffected for lower meldonium concentrations (7.5 and 15 mM) indicating that despite the decreased reduction rates observed at higher meldonium dilutions, resazurin was totally reduced within the incubation period (12 h). In contrast, higher meldonium concentrations provide extremely low reduction rates (MaxRates < 3500 RFUs h−1) and prevented the fluorescence signals to reach the maximal resazurin signal even after the whole incubation period (12 h, Fig. 4A). This observation also seemed to have a negative effect on the dispersion of estimated time-related parameters (Fig. 4E). As indicated by the MaxRate values, bacteria incubated in DM + G were able to reduce resazurin two times faster than in the presence of L-carnitine (28
072, R2 = 0.989). The MaxRS parameter remained unaffected for lower meldonium concentrations (7.5 and 15 mM) indicating that despite the decreased reduction rates observed at higher meldonium dilutions, resazurin was totally reduced within the incubation period (12 h). In contrast, higher meldonium concentrations provide extremely low reduction rates (MaxRates < 3500 RFUs h−1) and prevented the fluorescence signals to reach the maximal resazurin signal even after the whole incubation period (12 h, Fig. 4A). This observation also seemed to have a negative effect on the dispersion of estimated time-related parameters (Fig. 4E). As indicated by the MaxRate values, bacteria incubated in DM + G were able to reduce resazurin two times faster than in the presence of L-carnitine (28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327 and 13
327 and 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 352 RFU h−1, respectively). It is also interesting to note that under both culture conditions, after the fluorescence signal reached the maximum, the resazurin reduction rate rapidly decreased reaching even negative values under the incubation conditions with glucose (Fig. 4D). Such negative rates in glucose-supplemented media are presumably due to a secondary reduction of resorufin to the colorless and non-fluorescent degradation product hydroresorufin.
352 RFU h−1, respectively). It is also interesting to note that under both culture conditions, after the fluorescence signal reached the maximum, the resazurin reduction rate rapidly decreased reaching even negative values under the incubation conditions with glucose (Fig. 4D). Such negative rates in glucose-supplemented media are presumably due to a secondary reduction of resorufin to the colorless and non-fluorescent degradation product hydroresorufin.
3.4. Screening of natural extracts with potential to interfere with microbial TMA production from L-carnitine
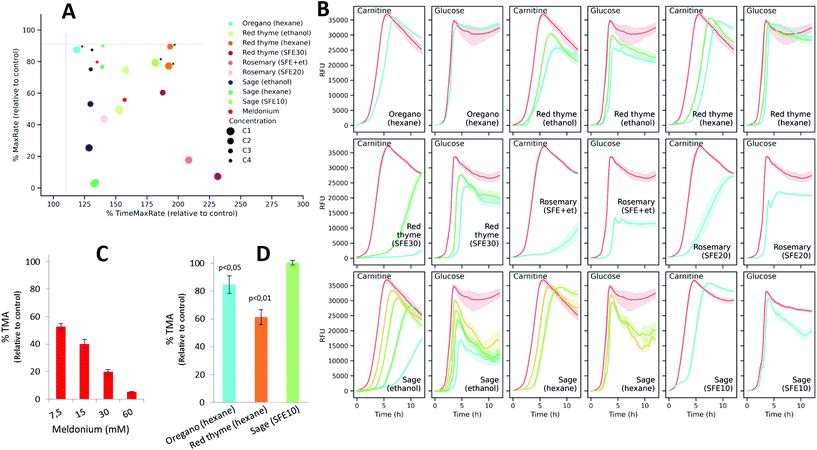
Next, the applicability of the screening method was evaluated with 39 extracts obtained from several food and natural sources including spirulina algae, spinach, olive leaf, juniper berry, garlic, red onion, red thyme, oregano, sage and rosemary, using different extraction technologies and conditions. Extracts were prepared at four different concentration levels, which depended on their solubility, and were tested by the resazurin reduction assay developed in this work. The information about the extraction conditions and the assayed concentrations are summarized in ESI Table S1.† Fitderiv software was used to fit time-series fluorescence data obtained from the incubations, and to compute time derivatives as well as the related curve parameters (ESI Table S3†). First, we examined the effect of the extracts on the maximum fluorescence signal achieved relative to the control during the assay. Fig. 5A displays the percentages of MaxRS obtained from each given extract and concentration, relative to those values obtained from controls. Data points are colored according to the type of extraction method used. As a general observation, extracts obtained by ultrasound-assisted extraction (UAE) with ethanol as the solvent (black dots in Fig. 5A) caused the highest decrease in MaxRS values compared to the extracts obtained with water (blue dots) or hexane (pink dots), regardless of the natural material extracted. Similarly, increasing pressure during the sequential supercritical fluid extraction (SFE) process or adding ethanol as cosolvent induced higher reductions in MaxRS compared to the control. Fig. 5B displays a 3D scatter plot that represents the percentage of TimeMaxRate, MaxRate and MaxRS obtained with a given compound relative to the control values. This graph revealed some trends of the extracts according to the different extraction conditions. Various water and ethanolic extracts decreased MaxRS values by more than 5% relative to controls. This reduction was specifically more evident in extracts obtained from red onion, spinach, juniper berry, red thyme and garlic (5–25% reduction in MaxRS values relative to control). However, a closer examination of data revealed that this effect was accompanied by an early acceleration of resazurin reduction, characterized by MaxRate increments higher than 25% and TimeMaxRate decreases down to 60% to the respective parameter values obtained from controls (vertical-axis in Fig. 5B). In these cases, the inspection of reduction rates curves (time derivatives) revealed that after rapidly reaching the maximal value, the resazurin reduction rates exhibited a fast decrease down to negative values, a similar phenomenon described above in glucose-supplemented medium, suggesting that those conditions were favorable for the fast conversion of resorufin into hydroresorufin. These findings allow us to hypothesize that sequential reductions of resazurin and resorufin could be boosted by the presence of compounds favoring these reactions or other carbon sources in the extracts, a notion that seems compatible with water or ethanol extraction. However, this hypothesis should be further investigated.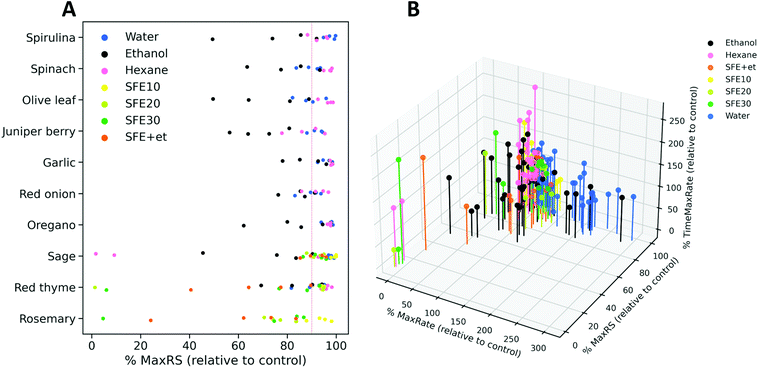 | ||
| Fig. 5 (A) Percentage of MaxRS values obtained with the extracts relative to the respective value of control assays. Dots represent an extract at one concentration and are grouped based on the starting source material. The color code refers to the different extraction conditions used as indicated by the inner legend (water, ethanol and hexane are the solvents used in UAE, SFE followed by a number referring to the pressure used for extraction (ESI Table S1†), and SFE + et refers to the use of ethanol as a cosolvent during SFE extraction). (B) 3D scatter plot of the estimations obtained for three parameters derived from model fits (MaxRS) and reduction rate as a function of time (MaxRate and TimeMaxRate). Estimated values obtained from the assays with extracts were expressed as percentages relative to those obtained from control conditions. | ||
As the main goal of the proposed screening is to find natural extracts able to induce changes in resazurin reduction as an indirect measurement of interference with L-carnitine metabolization and TMA production, the extracts showing high MaxRate values (>90% of those obtained in controls) and low TimeMaxRate values (<110%) were excluded as potential inhibitors. Using these criteria, thirty extracts were filtered out as candidates (Fig. 6A). Also, in order to exclude those conditions that could exert effects independent of L-carnitine metabolization, the remaining nine extracts (at different selected concentrations; depicted in Fig. 6A) were also assayed on DM + G. Fig. 6B shows the fluorescence curves obtained with the extracts in L-carnitine and glucose-supplemented media. Among the remaining nine extracts, only three of them, including oregano (hexane, C1), red thyme (hexane, C1 and C2), and sage (SFE10, C1), fulfilled the aforementioned criteria in DM + C in addition to not inducing evident changes in the signals (MaxRS > 90% of that of the control) when glucose is used to fuel bacterial metabolism.
Next, to validate these results, meldonium and the three extracts selected as candidates were assayed in vitro for their ability to inhibit enzymatic TMA production using a K. pneumoniae cell lysate. As expected, meldonium was also effective at inhibiting TMA generation by K. pneumoniae cell lysates in a dose-dependent manner. It was observed that even the lowest meldonium concentration (7.5 mM) reduced TMA production to 52.8% relative to the non-treated control (Fig. 6C). The incubation of the cell lysate with any of the two hexane extracts (obtained from oregano and red thyme) at the highest concentrations, significantly (p-value <0.05) reduced in vitro TMA generation to 84.6% and 61.3% relative to the control, respectively, whereas the extract obtained from sage using SFE did not induce a significant effect (Fig. 6D).
To exclude that these observations were not due to toxic effects of the extracts, K. pneumoniae was grown in nutrient broth media in the presence of oregano (hexane, C1) and red thyme (hexane, C1). When compared to the non-treated controls, K. pneumoniae exhibited similar growth profiles when treated with oregano and red thyme extracts (ESI Fig. S6†), thus discarding a possible toxic effect.
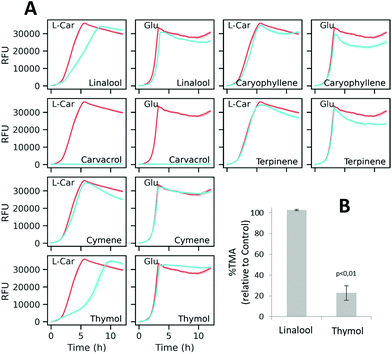
According to the GC-MS characterization, the major compounds tentatively identified in the red thyme extract were o-cymene, γ-terpinene, β-linalool, thymol, carvacrol and caryophyllene (ESI Fig. S1 and Table S2†). These compounds, with unknown influence on the microbial metabolization of L-carnitine, were assayed by the optimized resazurin reduction method. Fig. 7A shows the fluorescence curves obtained by incubating K. pneumoniae with each compound in either DM + C or DM + G. As deduced from the resazurin reduction curves, caryophyllene, terpinene, and cymene did not induce obvious changes in the fluorescence curves. On the other side, carvacrol had a strong impact on the curves, exhibiting a flat profile with both carbon sources which is indicative of strong suppression of metabolic activity (and possibly toxic activity). The assayed concentrations of thymol and linalool showed a differential effect on the curves obtained with L-carnitine compared to those obtained with glucose. As shown in Fig. 7B, only thymol significantly inhibited TMA formation in K. pneumoniae clarified lysates, suggesting a possible involvement of this compound in the observed activity of the extract.
4. Discussion
The search for new food ingredients able to interfere with gut microbial TMA generation is gaining more interest in recent years.5 The discovery of novel food ingredients with the ability to specifically target gut microbial TMA production would greatly benefit from the availability of suitable screening strategies. Also, there is a need for assessing the inhibitory potential of inhibitor candidates on living bacteria, in which the metabolic pathway under study is often more complex and subjected to the influence of more factors than in an in vitro enzymatic reaction. In this work, we present a novel high-throughput screening method that allows the identification of food and natural extracts that specifically interfere with the metabolic activity of a microorganism under in vitro growing conditions optimized to produce TMA from L-carnitine.It has been previously reported that some K. pneumoniae strains can rapidly uptake and degrade high concentrations of L-carnitine to TMA.23 Carbon source utilization assays using well-plate readers have shown to be extremely useful in providing information about the rate of carbon source consumption, which is directly linked to the metabolic activity. In the present work, we show that metabolic activity could be used as a proxy of the capacity to produce TMA under controlled culture conditions using L-carnitine to sustain metabolism. In many reports, the metabolic activity is measured using bacterial growth as a proxy. Optical density measurement of bacterial cultures is an accepted method, but is often prone to aggregation problems, and non-homogeneous bacterial suspensions are known to result in a low signal-to-noise ratio. Moreover, when plant extracts are added to microbial culture media, precipitation may occur contributing to the variation of the OD600 values measured, complicating the interpretation of the results. Although there is also the possibility of combining optical detection along with other alternative testing approaches (i.e., plating bacteria) to assay bacterial growth, it is not compatible with an HTS format and does not allow for the required time resolution. In our hands, the growth of a TMA producing K. pneumoniae was slow in DM containing L-carnitine as a sole carbon source. Zhu et al. have previously demonstrated that the TMA producer A. baumannii, which harbors cntA/B genes, can also grow in defined medium with L-carnitine as the sole carbon source, whereas mutants lacking either cntA or cntB can no longer grow on carnitine as a sole carbon and energy source.7 In the present work, although OD600 measurements allowed the possibility of investigating the kinetics of K. pneumoniae growth and TMA generation in the presence of meldonium, bacterial growth was slow and growth curves did not show the typical sigmoidal shape, and therefore, the use of parametric primary models to fit the data was discarded.
As an alternative to optical density, a great variety of methods using fluorescent dyes may be implemented with well-plate readers for the indirect monitoring of bacterial growth and metabolism.29 Among them, resazurin allows the detection of microbial growth in extremely small volumes of solution in microtiter plates. The resazurin molecule (oxidized form, blue, nonfluorescent), also commercially known as alamarBlue™ and PrestoBlue™, is reduced to resorufin (pink, fluorescent) in the medium as a result of cellular activity derived from cell growth. Despite its widespread use, some questions regarding the enzymes involved in resazurin reduction and the cellular location where it takes place still remain open. In mammalian cells, its reduction has been linked to mitochondrial reductases as well as diverse diaphorases located in the cytoplasm.30 Diaphorases are also found in bacteria which makes them potential candidates for resazurin reduction; however, it has been demonstrated that other reductases can reduce resazurin in vitro. With regard to cellular location, Chen et al. demonstrated that resazurin reduction to resorufin occurs intracellularly, whereas resorufin reduction to dihydroresorufin can also occur extracellularly in anaerobic cultures of Enterobacter faecalis.31
Despite its fast and sensitive properties to track bacterial metabolism, the more frequent applications of resazurin are aimed at assessing bacterial viability and growth with end-point measurements.32–35 However, it is well recognized that metabolic activity is not always related to growth, and therefore, it is essential to distinguish between amounts (values measured at defined time points) and activities (i.e., rates). Following this rationale, we aimed at developing a novel resazurin reduction method to track the metabolism of living bacteria under aerobic TMA producing conditions. Tracking the dynamics of resazurin reduction, with time resolution and without any additional steps allowed us to indirectly identify extracts affecting the metabolism of living K. pneumoniae cells sustained by L-carnitine utilization that could be selected as candidates for further assessment as inhibitors of TMA production. Our results show that resazurin reduction assay is well suited for tracking the metabolic activity of slow growing bacterial cells; however, it requires optimization of some of the parameters affecting fluorescence signal as well as taking into account several precautions.
As it has been reported previously, the reduction of fluorescent resorufin into a further reduced non-fluorescent dihydroresorufin may lead to aberrant results in which metabolically active bacterial cells produce a weak signal, whereas dying cells, which could not sustain further reduction, yield a high fluorescence signal.36 To model resazurin reduction by K. pneumoniae, we optimized the resazurin concentration and initial bacterial load, variables that have been reported to affect resorufin reduction into dihydroresorufin. Also, it has been reported that species with thiol functional groups may cause the reduction of resazurin to resorufin in the absence of cells.37 Therefore, to avoid such interferences and deviations, it is highly recommended to implement appropriate controls and blanks that ensure that the assays are truly representative of the interaction between the analyte and cells and not the analyte and assay reagents. Our results were in line with previously reported data, showing that fluorescence signals were highly dependent on both bacterial density and resazurin concentration.34,38 Furthermore, whereas bacterial density mostly impacted the time parameters, resazurin concentration almost exclusively affected the maximum signal and maximum reduction rate. Under the selected optimal conditions, a good correlation was obtained between the fluorescence signals and TMA concentration within the time interval that preceded the time at which the maximal fluorescence signal was reached. After that point, the loss of correlation is indicative of resazurin exhaustion and/or dihydroresorufin generation. In spite of the optimization, negative reduction rate values were observed in the assays with certain extracts. However, due to the efficient methodology to process data, such phenomenon indicative of resorufin reduction could be objectively detected (reduction rates <0).
With regard to data analysis, different methods have been described to extract meaningful data from the resazurin-reduction curves that help in estimating microbial density or viability. For instance, Mariscal et al. used regression analysis to calculate the time needed to reach 50% of the maximum fluorescence signal in biofilms.39 This approach showed the different resazurin reduction kinetics of various microorganisms suggesting the need to characterize the critical parameters for each assay. A similar approach based on regression analysis was followed by Travnickova et al. to estimate the number of viable bacteria on electrospun nanofiber filtration membranes.35 In that case, the time needed to reach a fixed fluorescence signal was preferred for the generation of standard curves against the log of bacterial plate counts. Then, time to reach the established fluorescent value derived from resazurin reduction curve was used for the calculation of the bacterial concentration in test samples from the respective calibration curve. In another report, the kinetics of resazurin reduction was modeled to detect differences among various toxicants under anaerobic conditions.38 In that case, the authors demonstrated in their system that the pseudo-first-order rate constant for the reduction of resazurin to resorufin was a good parameter to measure toxicity in fresh anaerobic sludges. In the present work, we applied a non-parametric approach developed by Swain et al.24 to achieve our goal of modelling microbial metabolism under various conditions. This approach uses GPs to infer the first time derivatives from time-series data. GPs are powerful statistical machine learning models that can efficiently capture complex nonlinear process dynamics.40 This tool has recently attracted much attention in the field of computational data modelling and it has been successfully applied to model diverse biological processes.24,41–45 It does not require knowledge of the underlying process and can capture many temporal trends in the data. The advantage of this strategy over other existing non-parametric methods is that it systematically combines data from replicate experiments and predicts errors both in the estimations of derivatives and in any summary statistics. In addition, GP can be trained on small data sets, which in addition to the other methodological advantages motivated us to apply this tool to model our experimental data. The algorithm used all experimental replicates to infer the resazurin reduction rate as a function of time and the associated estimated errors. Model parameters MaxRate and TimeMaxRate were obtained from the time derivative curve as indicators of the maximum reduction rate and the time point at which that maximum is reached, respectively. MaxRS and LagTime were estimated from the model fits, the former being especially useful to discard those conditions that, for the purpose of this screening method, might have unintended effects on the system. This was exemplified by the observed drops in MaxRS values induced by those extracts that had strong effects on reduction rates, either by initially accelerating the resazurin reduction or by almost suppressing it.
In our system, reduction of MaxRate and increase of TimeMaxRate (and LagTime) are theoretically indicative of deleterious effects on resazurin reduction curves, and in turn, on the bacterial metabolic activity. We found meldonium suitable to evaluate the behavior of the model parameters upon interference of metabolism sustained by L-carnitine utilization. Meldonium was first reported to inhibit TMA production without affecting L-carnitine uptake into K. pneumoniae cells under microaerobic conditions and bacterial growth.18 More recently, it has been described as an oxidizable substrate of CntA with a reported lower affinity than L-carnitine (Km values of 152 nM and 117 nM, respectively).9 Our results showed that K. pneumoniae growth and TMA production decreased in the presence of L-carnitine and meldonium compared to growth obtained in presence of L-carnitine alone. A possible explanation for these partially discrepant results on bacterial growth is the difference in the nutritive conditions, which in our assay are very limiting (DM + L-carnitine) compared to those used in the previous report.18 Meldonium was able to decrease MaxRates and increase TimeMaxRates and LagTimes in a concentration-dependent fashion in the resazurin assay, whereas MaxRS values were only affected at high meldonium concentrations probably due to the fact that at very low reduction rates, the time to reach MaxRS is longer than 12 h. Interestingly, meldonium did not induce evident effects when L-carnitine was substituted by glucose corroborating its selective interference with L-carnitine metabolization (and TMA production). This effect on TMA formation was further confirmed by TMA quantification by CE-UV in both K. pneumoniae culture and lysates.
To explore the applicability of this approach, we used the method to identify the differential effects 39 extracts (at four concentration levels, respectively) on the dynamics of resazurin reduction. Most of the extracts obtained with water and some extracts with ethanol exhibited an unwanted accelerating effect on resazurin reduction, which in turn was associated with the occurrence of negative reduction rate values. On the other hand, a group of nine extracts, in at least one assayed concentration, showed substantial MaxRate reduction when compared to controls. Complementary assays in DM + G served to discard those conditions that also exerted similar effects in the presence of glucose as those observed with L-carnitine. This allowed us to narrow down the number of extracts that only inhibit microbial metabolism sustained by L-carnitine utilization, and to exclude as much as possible other unrelated inhibitory effects. Thus, extracts from both oregano (1 mg mL−1) and red thyme (0.5 and 1 mg mL−1) obtained with hexane, and SFE sage extract (1 mg mL−1) obtained with CO2 under 10 MPa pressure, fulfilled the selected criteria and were considered as candidates with the potential to interfere with L-carnitine metabolization and TMA production. Subsequent assays confirmed the potential of both oregano and red thyme hexane extracts (at 1 mg mL−1) to interfere with TMA formation in bacterial lysates. In this in vitro assay, the red thyme extract exerted comparable effects on TMA reduction (∼40%) as 7.5 mM meldonium (∼50% TMA decrease), whereas the oregano extract showed a milder effect (∼15% TMA reduction compared to control TMA levels). In addition, both extracts neither affect the growth nor were bactericidal at the assayed concentrations, and therefore, the observed effects on resazurin reduction and TMA production were not due to toxic effects. To further investigate the activity of the red thyme extract constituents, a set of six pure compounds tentatively identified in the extract were also assayed. Interestingly, thymol and linalool interfered with the bacterial metabolic activity with L-carnitine but not with glucose as the sole carbon source. Lysate assays further confirmed that thymol inhibited TMA formation, which allows us to hypothesize that this compound, which is abundant in the red thyme extract and is also present in the one obtained from oregano, could contribute to the observed activity of both extracts. Among the many activities attributed to thymol, its antimicrobial and antifungal properties are the more frequently reported.46 It has been suggested that the antimicrobial effect of this phenolic compound can result in part from a perturbation in the lipid fraction of microorganism plasma membrane, but this effect seems to be dependent on lipid composition and net surface charge of microbial membranes47 which might explain the divergent MIC values reported for different bacterial strains. Besides, thymol has also been involved in bacterial metabolism and showed to be a potent inhibitor of L-lactate production in some ruminal microorganisms.48 Our findings lead us to speculate that thymol might also interfere with L-carnitine metabolization in the TMA producer K. pneumoniae, providing new perspectives about the biological activity of this bioactive compound. Carvacrol is also capable of expanding and partitioning the lipids of the bacterial cell membrane. Despite this common mechanism often attributed to thymol and carvacrol, their interaction modes with the bacterial surface seem to be different.49 Compared to the effect exerted by thymol, our observations suggest that carvacrol completely abolished the metabolic response of K. pneumoniae regardless of the type of carbon source in the medium, suggesting a strong bactericidal/bacteriostatic effect. Carvacrol and thymol are structural isomers, but the hydroxyl group in carvacrol is more exposed than the hydroxyl group in thymol, which makes the former less hydrophobic than the latter, a feature that likely affects the membrane permeability. Our results suggest that the position of the hydroxyl group in their molecular structure plays a crucial role in the effect of these compounds on K. pneumoniae metabolism, which is in line with previous reports that observe differential effects of this compounds in in vitro assays.50
Finally, we should note that although the CntA gene is present in certain Gram-negative bacteria belonging to the Proteobacteria phylum,51 the results obtained in the present screening cannot be directly extrapolated to other CntA/B containing bacteria. Our preliminary results with Serratia marcescens, another Proteobacteria harbouring functional CntA/B (data not shown), indicate that other species can be used to implement this screening strategy; however, the optimization of some of the parameters affecting fluorescence signals and time of analysis is required. It is essential to mention that the proposed method presents obvious limitations inherent in any screening method using a redox dye and living cells. For instance, (i) the rate of resazurin reduction might depend not only on the cell metabolic status, but also on cell permeability; and (ii) the effect of the metabolic function on resazurin reduction might be multifactorial, with various metabolic reactions and cofactors involved, and therefore, the observation of the wanted effects (MaxRate reduction and/or TimeMaxRate increase) with the candidates cannot be directly associated with CntAB inhibition. Indeed, to draw more relevant biological conclusions about the bioactivity of the candidate extracts on the metabolic function of gut microbiota, their evaluation under other experimental frameworks would be mandatory.
Author contributions
C.S. and V.G.-C. were involved in conceptualization, performed experiments, processed data, administered the project funding and wrote the manuscript. T.F., M.R.G., A.P., L.A., J.A., and H.R. performed experiments and revised the manuscript. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank J. M. Prados for the computer support. This work is jointly supported by research grant AGL2017-89055-R (Ministerio de Ciencia e Innovación, MICINN). We acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).References
- Z. Wang, E. Klipfell, B. J. Bennett, R. Koeth, B. S. Levison, B. Dugar, A. E. Feldstein, E. B. Britt, X. Fu, Y. M. Chung, Y. Wu, P. Schauer, J. D. Smith, H. Allayee, W. H. Tang, J. A. DiDonato, A. J. Lusis and S. L. Hazen, Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease, Nature, 2011, 472, 57–63 CrossRef CAS PubMed.
- R. A. Koeth, Z. Wang, B. S. Levison, J. A. Buffa, E. Org, B. T. Sheehy, E. B. Britt, X. Fu, Y. Wu, L. Li, J. D. Smith, J. A. DiDonato, J. Chen, H. Li, G. D. Wu, J. D. Lewis, M. Warrier, J. M. Brown, R. M. Krauss, W. H. W. Tang, F. D. Bushman, A. J. Lusis and S. L. Hazen, Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis, Nat. Med., 2013, 19, 576–585 CrossRef CAS PubMed.
- W. Zhu, J. C. Gregory, E. Org, J. A. Buffa, N. Gupta, Z. Wang, L. Li, X. Fu, Y. Wu, M. Mehrabian, R. B. Sartor, T. M. McIntyre, R. L. Silverstein, W. H. W. Tang, J. A. DiDonato, J. M. Brown, A. J. Lusis and S. L. Hazen, Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk, Cell, 2016, 165, 111–124 CrossRef CAS PubMed.
- C. Roncal, E. Martínez-Aguilar, J. Orbe, S. Ravassa, A. Fernandez-Montero, G. Saenz-Pipaon, A. Ugarte, A. Estella-Hermoso de Mendoza, J. A. Rodriguez, S. Fernández-Alonso, L. Fernández-Alonso, J. Oyarzabal and J. A. Paramo, Trimethylamine-N-oxide (TMAO) predicts cardiovascular mortality in peripheral artery disease, Sci. Rep., 2019, 30, 15580 CrossRef PubMed.
- C. Simó and V. García-Cañas, Dietary bioactive ingredients to modulate the gut microbiota-derived metabolite TMAO. New opportunities for functional food development, Food Funct., 2020, 11, 6745–6776 RSC.
- S. Cracium and E. P. Balskus, Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 21307–21312 CrossRef PubMed.
- Y. Zhu, E. Jameson, M. Crosatti, H. Schäfer, K. Rajakumar, T. D. H. Bugg and Y. Chen, Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 4268–4273 CrossRef CAS PubMed.
- M. Massmig, E. Reijerse, J. Krausze, C. Laurich, W. Lubitz, D. Jahn and J. Moser, Carnitine metabolism in the human gut: Characterization of the two-component carnitine monooxygenase CntAB from Acinetobacter baumannii, J. Chem. Biol., 2020, 295, 13065–13078 CrossRef CAS PubMed.
- M. Quareshy, M. Shanmugam, E. Townsed, E. Jameson, T. D. H. Bugg, A. D. Cameron and Y. Chen, Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer, J. Biol. Chem., 2021, 296, 100038 CrossRef CAS PubMed.
- G. Falony, S. Vieira-Silva and J. Raes, Microbiology meets big data: The case of gut microbiota-derived trimethylamine, Annu. Rev. Microbiol., 2015, 69, 305–321 CrossRef CAS PubMed.
- Z. Wang, A. B. Roberts, J. A. Buffa, B. S. Levison, W. Zhu, E. Org, X. Gu, Y. Huang, M. Zamanian-Daryoush, M. K. Culley, A. J. DiDonato, X. Fu, J. E. Hazen, D. Krajcik, J. A. DiDonato, A. J. Lusis and S. L. Hazen, Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis, Cell, 2015, 163, 1585–1595 CrossRef CAS PubMed.
- R. A. Koeth, B. S. Levison, M. K. Culley, J. A. Buffa, Z. Wang, J. C. Gregory, E. Org, Y. Wu, L. Li, J. D. Smith, W. H. W. Tang, J. A. Didonato, A. J. Lusis and S. L. Hazen, γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO, Cell Metab., 2014, 20, 799–812 CrossRef CAS PubMed.
- L. J. Rajakovich, B. Fu, M. Bollenbach and E. P. Balskus, Elucidation of an anaerobic pathway for metabolism of L-carnitine-derived γ-butyrobetaine to trimethylamine in human gut bacteria, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2101498118 CrossRef CAS PubMed.
- A. B. Roberts, X. Gu, J. A. Buffa, A. G. Hurd, Z. Wang, W. Zhu, N. Gupta, S. M. Skye, D. B. Cody, B. S. Levison, W. T. Barrington, M. W. Russell, J. M. Reed, A. Duzan, J. M. Lang, X. Fu, L. Li, A. J. Myers, S. Rachakonda, J. A. DiDonato, J. M. Brown, V. Gogonea, A. J. Lusis, J. C. Garcia-Garcia and S. L. Hazen, Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential, Nat. Med., 2018, 24, 1407–1417 CrossRef CAS PubMed.
- M. Orman, S. Bodea, M. A. Funk, A. Martínez-del Campo, M. Bollenbach, C. L. Drennan and E. P. Balskus, Structure-guided identification of a small molecule that inhibits anaerobic choline metabolism by human gut bacteria, J. Am. Chem. Soc., 2019, 141, 33–37 CrossRef CAS PubMed.
- C. Su, X. Li, Y. Yang, Y. Du, X. Zhang, L. Wang and B. Hong, Metformin alleviates choline diet-induced TMAO elevation in C57BL/6J mice by influencing gut-microbiota composition and functionality, Nutr. Diabetes, 2021, 11, 27 CrossRef CAS PubMed.
- G. Kalnins, E. Sevostjanovs, D. Hartmane, S. Grinberga and K. Tars, CntA oxygenase substrate profile comparison and oxygen dependecy of TMA production in Providencia rettgeri, J. Basic Microbiol., 2018, 58, 52–59 CrossRef CAS PubMed.
- J. Kuka, E. Liepinsh, M. Makrecka-Kuka, J. Liepins, H. Cirule, D. Gustina, E. Loza, O. Zharkova-Malkova, S. Grinberga, O. Pugovics and M. Dambrova, Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting L-carnitine microbial degradation, Life Sci., 2014, 117, 84–92 CrossRef CAS PubMed.
- L. Bresciani, M. Dall'asta, C. Favari, L. Calani, D. Del Rio and F. Brighenti, An in vitro exploratory study of dietary strategies based on polyphenol-rich beverages, fruit juices and oils to control trimethylamine production in the colon, Food Funct., 2018, 9, 6470–6483 RSC.
- L. Iglesias-Carres, L. A. Essenmacher, K. C. Racine and A. P. Neilson, Development of a high-throughput method to study the inhibitory effect of phytochemicals on trimethylamine formation, Nutrients, 2021, 13, 1466 CrossRef CAS PubMed.
- V. García-Cañas, E. Aznar and C. Simó, Screening gut microbial trimethylamine production by fast and cost-effective capillary electrophoresis, Anal. Bioanal. Chem., 2019, 411, 2697–2705 CrossRef PubMed.
- P. Thomas, A. C. Sekhar, R. Upreti, M. M. Mujawar and S. S. Pasha, Optimization of single plate-serial dilution spotting (SP-SDS) with sample anchoring as an assured method for bacterial and yeast CFU enumeration and single colony isolation from diverse samples, Biotechnol. Rep., 2015, 8, 45–55 CrossRef PubMed.
- J. D. Brewster, A simple micro-growth assay for enumerating bacteria, J. Microbiol. Methods, 2003, 53, 77–86 CrossRef PubMed.
- P. Swain, K. Stevenson, A. Leary, L. F. Montano-Gutierrez, I. B. N. Clark, J. Vogel and T. Pilizota, Inferring time derivatives including cell growth rates using Gaussian processes, Nat. Commun., 2016, 7, 13766 CrossRef CAS PubMed.
- J. D. Hunter, Matplotlib: A 2D Graphics Environment, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- M. L. Waskom, Seaborn: statistical data visualization, J. Open Source Softw., 2021, 6, 3021 CrossRef.
- S. Seabold and J. Perktold, Statsmodels: Econometric and statistical modeling with python, In 9th Python in Science Conference. 2010 Search PubMed.
- R. Vallat, Pingouin: statistics in Python, J. Open Source Softw., 2018, 3, 1026 CrossRef.
- O. Braissant, M. Astasov-Frauenhoffer, T. Waltimo and G. Bonkat, A review of methods to determine viability, vitality, and metabolic rates in microbiology, Front. Microbiol., 2020, 11, 547458 CrossRef PubMed.
- S. N. Rampersad, Multiple Applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays, Sensors, 2012, 12, 12347–12360 CrossRef CAS PubMed.
- J. L. Chen, T. W. J. Steel and D. C. Stuckey, Metabolic reduction of resazurin, location within the cell for cytotoxicity assays, Biotechnol. Bioeng., 2017, 115, 351–358 CrossRef PubMed.
- H. J. Kim and S. Jang, Optimization of a resazurin-based microplate assay for large-scale compound screenings against Klebsiella pneumoniae, Biotechnology, 2018, 8, 3 Search PubMed.
- O. Tyc, L. Tomás-Menor, P. Garbeva, E. Barrajón-Catalán and V. Micol, Validation of the AlamarBlue assay as a fast screening method to determine the antimicrobial activity of botanical extracts, PLoS One, 2016, 11, e0169090 CrossRef PubMed.
- M. Vukomanovic and E. Torrents, High time resolution and high signal-to-noise monitoring of the bacterial growth kinetics in the presence of plasmonic nanoparticles, J. Nanobiotechnol., 2019, 17, 21 CrossRef PubMed.
- E. Travnickova, P. Mikula, J. Oprsal, M. Bohacova, L. Kubac, D. Kimmer, J. Soukupova and M. Bittner, Resazurin assay for assessment of antimicrobial properties of electrospun nanofiber filtration membranes, AMB Express, 2019, 9, 183 CrossRef PubMed.
- J. O′brien, I. Wilson, T. Orton and F. Pognan, Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mamalian cell cytotoxicity, Eur. J. Biochem., 2000, 267, 5421–5426 CrossRef PubMed.
- B. H. Neufeld, J. B. Tapia, A. Lutzke and M. M. Reynolds, Small molecule interferences in resazurin and MTT-based metabolic assays in the absence of cells, Anal. Chem., 2018, 90, 6867–6876 CrossRef CAS PubMed.
- J. L. Chen, T. W. J. Steel and D. C. Stuckey, Modeling and application of a rapid fluorescence-based assay for biotoxicity in anaerobic digestion, Environ. Sci. Technol., 2015, 49, 13463–13471 CrossRef CAS PubMed.
- A. Mariscal, R. M. Lopez-Gigosos, M. Carnero-Varo and J. Fernandez-Crehuet, Fluorescent assay based on resazurin for detection of activity of disinfectants against bacterial biofilm, Appl. Microbiol. Biotechnol., 2009, 82, 773–783 CrossRef CAS PubMed.
- C. E. Rasmussen and C. K. I. Williams, Gaussian Process for Machine Learning, The MIT Press, USA, 2006 Search PubMed.
- A. J. H. Fedorec, T. Ozdemir, A. Doshi, Y. K. Ho, L. Rosa, J. Rutter, O. Velazquez, V. B. Pinheiro, T. Danino and C. P. Barnes, Two new plasmid post-segregational killing mechanisms for the implementation of synthetic gene networks in Escherichia coli, iScience, 2019, 14, 323–334 CrossRef CAS PubMed.
- J. González, M. Salvador, O. Ozkaya, M. Spick, K. Reid, C. Costa, M. J. Bailey, C. Avignone Rossa, R. Kümmerli and J. I. Jiménez, Loss of pyoverdine secondary receptor in Pseudomonas aeruginosa results in a fitter strain suitable for population invasion, ISME J., 2021, 15, 1330–1343 CrossRef PubMed.
- F. Pietsch, G. Heidrich, N. Nordholt and F. Schreiber, Prevalent synergy and antagonism among antibiotics and biocides in Pseudomonas aeruginosa, Front. Microbiol., 2021, 11, 615618 CrossRef PubMed.
- D. Cai, W. Liu, L. Ji and S. Dawei, Bayesian optimization assisted meal bolus decision based on Gaussian processes learning and risk-sensitive control, Control Eng. Pract., 2021, 114, 104881 CrossRef.
- A. A. Baiz, H. Ahmadi, F. Shariatmadari and M. A. Karimi Torshizi, A Gaussian process regression model to predict energy contents of corn for poultry, Poult. Sci., 2021, 99, 5838–5843 CrossRef PubMed.
- A. Marchese, K. E. Orhan, M. Daglia, R. Barbieri, A. Di Lorenzo, S. F. Nabavi, O. Gortzi, M. Izadi and S. M. Nabavi, Antibacteiral and antifungal activities of thymol: a brief review of the literature, Food Chem., 2016, 210, 402–414 CrossRef CAS PubMed.
- D. Trombetta, F. Castelli, M. G. Sarpietro, V. Venuti, M. Cristani, C. Daniele, A. Saija, G. Mazzanti and G. Bisignano, Mechanisms of antibacterial action of three monoterpenes, Antimicrob. Agents Chemother., 2005, 49, 2474–2478 CrossRef CAS PubMed.
- J. D. Evans and S. A. Martin, Effects of thymol on ruminal microorganisms, Curr. Microbiol., 2000, 41, 336–340 CrossRef CAS PubMed.
- O. Y. Althunibat, H. Qaralleh, S. Y. A. Al-dalin, M. Abboud, K. Khleifat, I. S. Majali, H. D. Aldi'in, W. A. Rayyan and A. Jaafraa, Effect of Thymol and Carvacrol, the Major Components of Thymus capitatus on the Growth of Pseudomonas aeruginosa, J. Pure Appl Microbiol., 2016, 10, 367–374 CAS.
- M. Jukic, O. Politeo, M. Maksimovic, M. Milos and M. Milos, In Vitro Acetylcholinesterase Inhibitory Properties of Thymol, Carvacrol and their Derivatives Thymoquinone and Thymohydroquinone, Phytother. Res., 2017, 21, 259–261 CrossRef PubMed.
- S. Rath, B. Heidrich, D. H. Pieper and M. Vital, Uncovering the trimethylamine-producing bacteria of the human gut microbiota, Microbiome, 2017, 5, 54 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2fo00103a |
| This journal is © The Royal Society of Chemistry 2022 |