Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
An insight into an intriguing oxidative biotransformation pathway of 5-O-caffeoylquinic acid by a gut bacterium†
G.
Balaj
 a,
Z.
Tamanai-Shacoori
a,
Z.
Tamanai-Shacoori
 b,
D.
Olivier-Jimenez
b,
D.
Olivier-Jimenez
 a,
A.
Sauvager
a,
A.
Sauvager
 a,
M.
Faustin
a,
L.
Bousarghin
a,
M.
Faustin
a,
L.
Bousarghin
 b,
S.
David-Le Gall
b,
S.
David-Le Gall
 b,
S.
Guyot
b,
S.
Guyot
 c,
D.
Nebija
c,
D.
Nebija
 d,
S.
Tomasi
d,
S.
Tomasi
 *a and
M. L.
Abasq
*a and
M. L.
Abasq
 *a
*a
aUniv Rennes, CNRS, ISCR – UMR 6226, 35043 Rennes, France. E-mail: sophie.tomasi@univ-rennes1.fr
bINSERM, Univ. Rennes, INRAE, CHU Rennes, Nutrition Metabolisms and Cancer (NuMeCan), UMR-1241, Biosit, MRic/ISFR, Rennes, France
cINRAE, UR1268BIA, Team Polyphenol, Reactivity & Processing (PRP), BP35327, 35653 Le Rheu, France
dFaculty of Medicine, Department of Pharmacy, University of Prishtina, 10000 Prishtinë, Kosovo
First published on 11th May 2022
Abstract
Microbiota is known to play a pivotal role in generating bioavailable and bioactive low-molecular-weight metabolites from dietary polyphenols. 5-O-caffeoylquinic acid (5-CQA), one of the main polyphenols found in human diet, was submitted to a resting cell biotransformation study using three gut bacteria species Lactobacillus reuteri, Bacteroides fragilis and Bifidobacterium longum. These bacteria were selected according to their belonging to the main phyla found in human gut microbiota. Our study highlighted the ability of only one of the strains studied, L. reuteri, to bioconverse 5-CQA into various metabolites due to the expression of the cinnamoyl esterase enzyme as the first step. Interestingly, one known natural compound, esculetin, was described for the first time as a 5-CQA-derived metabolite after conversion by a gut bacterium, the other metabolites had already been reported. This evidence highlighted an interesting oxidative pathway occurring in vivo by intestinal microbiota leading to esculetin. This molecule was also identified after electrochemical and enzymatic oxidations of caffeic acid. The oxidation capacity of L. reuteri led to less diverse metabolites in comparison to those obtained either electrochemically and enzymatically where dimers and trimers were reported. Thus, esculetin may have interesting and benefical biological effects on gut microbiota, which should be further evaluated. Novel synbiotics could be formulated from the association of L. reuteri with 5-CQA.
1. Introduction
Polyphenols are known for their broad range of positive bioactivities in humans. These molecules have shown the ability to lower the oxidative stress involved in many diseases and positively modulate redox signalling pathways.1 Luca et al. has recently described the notion of “low bioavailability/high bioactivity paradox of polyphenols” highlighting the interest of their intestinal metabolites on human health.2Considering their superiority in terms of presence in systemic circulation compared to parent polyphenols, further studies focus nowadays on the pivotal role of microbiota in generating bioavailable low-molecular-weight metabolites.3 These gut-derived metabolites have exhibited biological activities and have shown the ability to reach the brain.4–6 Indeed, studies have highlighted them as a promising approach to prevent and attenuate many pathologies such as neurodegenerative diseases.5,6
Among dietary polyphenols, the hydroxycinnamate derivative 5-O-caffeoylquinic acid (5-CQA) according to the IUPAC name, known as chlorogenic acid, is one of the main phenolic compounds in the human daily diet, being present in various fruits and vegetables.7,8 This highly functional compound has been shown to have significant benefits for human health (i.e. cardioprotective, gastrointestinal protective, hepatoprotective, neuroprotective properties, etc.).7
The pharmacokinetic profile of 5-CQA has been already described in humans.9 After intake, one third is absorbed into the small intestine and the majority reaches the colon in an intact form. At this stage, the intestinal microbiota with cinnamoyl esterase activity biotransforms 5-CQA into caffeic acid (CA) and quinic acid, which further undergo their subsequent transformation into various microbial metabolites including m-coumaric acid and hydroxylated derivatives of phenylpropanoic, benzoic, and hippuric acids.9 These metabolites are bioavailable in the systemic circulation and reach the target organs where they can exert their biological effects.7,9 Indeed, the biotransformation of 5-CQA is dependent on the ability of bacterial species to produce this esterase as the first step. Hence, some bacteria species have already been identified as producers of this specific enzyme, such as Escherichia coli, Bifidobacterium lactis, Lactobacillus gasseri10 and L. johnsonii.11
In this work, we have investigated the biotransformation of 5-O-caffeoylquinic acid by three gut bacteria species cultivated alone: Lactobacillus reuteri (Firmicutes), Bacteroides fragilis (Bacteroidetes) and Bifidobacterium longum (Actinobacterium), belonging to the dominant bacterial phyla of the gut microbiota. The biotransformed extracts were analysed by HPLC-DAD-ESI-MS2 and the data were subjected to MZmine 2.53 and a further process creating a molecular network (MN). It is interesting to note that gut microbiota polyphenol biotransformations encompassing hydrolysis, reduction and de- or di-hydroxylation reactions are well known12,13 but there is a lack of information on possible oxidative pathways other than α- or β-oxidations. For example, one study led to the characterisation of β-oxidation of one metabolite of 5-CQA, 3-(4′-hydroxyphenyl) propanoic acid, into 4-hydroxybenzoic acid, using an in vitro experimental gastrointestinal dialysis model.14 Hence, electrochemical and enzymatic methods were adopted to generate oxidised compounds of 5-CQA and CA in order to characterise and compare their LC-MS profiles to those of the biotransformed extracts. This strategy made it possible to highlight an intriguing oxidative pathway of cinnamic acid microbiota conversion.
2. Materials and methods
2.1 Chemicals
All commercial reagents were purchased from Carlo Erba Reactifs and/or from Merck/Sigma-Aldrich (Val de Reuil, France and St Quentin Fallavier, France). 5-CQA ((1S,3R,4R,5R)-3-[(E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy-1,4,5-trihydroxycyclohexane-1-carboxylic acid in accordance with the IUPAC name and numbering) was acquired from Acros Organics (Italy) (CAS number 327-97-9) and trans-CA, esculetin, was acquired from Merck/Sigma-Aldrich. BHI (Brain Heart Infusion) broth was purchased from Bio-Rad (France). Ethyl acetate (reagent grade) and acetonitrile (≥ 99.9%, LC-MS grade) was obtained from Merck/Sigma-Aldrich. Ultrapure water was obtained using an EasyPure (Barnstead, USA) water purification system. Phosphate buffer solution (PBS) was prepared at 0.1 M by mixing 95% of 1.36% w/v KH2PO4 and 5% of 1.78% w/v Na2HPO4·2H2O and the pH was adjusted to 5.8.2.2 Bacterial strains
The bacterial strains used in our study were Lactobacillus reuteri DSM 20016, Bacteroides fragilis ATCC 25285 and Bifidobacterium longum subsp infantis DSM 20088.2.3 Growth conditions of L. reuteri, B. fragilis and B. longum
Bacteria were grown in BHI broth under anaerobic conditions using an anaerobic chamber (Macs-VA500, Don Whitley) flooded with 80% N2, 10% H2 and 10% (CO2). The kinetics of the cultures were monitored by measuring absorbance (optical density (OD) at 600 nm) and colony-forming units (CFU mL−1). The cultures of bacteria (150 mL) were established by suspending the fresh isolated bacterial strain colonies (from a 48 h culture in anaerobic conditions at 37 °C) in BHI standard medium in order to obtain an appropriate inoculum of each bacterium at 108 CFU mL−1. Then, the cultures and a flask containing only BHI standard, used as a control of the purity, were incubated in anaerobic chamber for 30 h.2.4 Biotransformation study
Resting cell biotransformation studies were carried out when the bacteria reached the stationary phase (at 30 h of growth), by adding a solution of 5-CQA (1 mM) in PBS (50 mL; pH 5.8) on the biomass (pellet) of each bacterial strain separated from the culture medium by centrifugation (3500 rpm, 4 °C, 15 min). In parallel, as controls (50 mL, pH 5.8), PBS alone or with 5-CQA, CA (1 mM) were incubated without bacteria. In addition, two more controls were prepared by either incubating bacteria in BHI or in PBS. Other biotransformation experiments were performed using the same conditions, except BHI was used instead of PBS at pH 5.8. These were incubated at 37 °C, 120 rpm under anaerobic conditions using sealed jars and anaerobic bags. After 24 h of incubation, the samples were centrifugated at 3500 rpm, at 4 °C for 15 min. The extraction process was further performed on a pellet and on a supernatant (SN) using ethyl acetate (2 × 10 mL for the pellet and 2 × 25 mL for the SN). After evaporation of the organic solvents, the resulting supernatant and pellet extracts were analysed by LC-DAD-ESI-MS2. The data of these biotransformed extracts were processed by MZmine 2.53. Then, the results were uploaded into the Python v3.7 library Matchms v0.9.2, in order to generate a molecular network (MN). The names of the identified metabolites derived from 5-CQA were given in accordance to recent recommendations.15 Nevertheless, CA was preferred to 3′,4′-dihydroxycinnamic acid for a better reading.2.5 In silico search for cinnamoyl esterase activity
The sequence of cinnamoyl esterase from L. jonhsonii was researched in the sequenced genomes of L. reuteri DSM_20016 (GCA_001436151), using Geneious Prime 2020.2, (https://www.geneious.com) and corresponded to Locus Tag named LREU_RS08790 (Fig. S1†).2.6 UV irradiation of samples
5-CQA standard at 1 mM dissolved in water (1 mL) placed in a UV-quartz cell was irradiated at 365 nm using a UV-lamp for 1 h.2.7 Electrochemical procedures
Before each measurement, a 2 mM 5-CQA or CA solution was freshly prepared in 0.1 M PBS pH 5.8 and nitrogen saturated for 5 min at room temperature. All potentials reported herein are vs. the Ag/AgCl reference electrode.Linear sweep voltammetry (LSV). The LSV (i = f(E)) were firstly recorded at a scan rate of 0.005 V s−1. UV-Vis spectra were simultaneously recorded between 200 and 600 nm.
Electrolysis. The electrolysis were performed by a chronoamperometry technique i = f(t) at a fixed potential for 1200 seconds for 5-CQA and CA. The applied electrolysis potential was fixed at +0.8 V for the 5-CQA electrolysis and +0.6 V for CA electrolysis. The UV-Vis spectra were simultaneously recorded. During electrolysis, nitrogen bubbling was maintained, first to avoid any oxidation derived from oxygen and also to mix the solution (the whole cell volume is 600 μL, whereas the optic way compartment holds approximately 200 μL of the solution). After electrolysis, the whole solution was retrieved, saturated with argon in a sealed vial and stored at −20 °C.
2.8 Enzymatic procedures by polyphenol oxidase (PPO)
The procedure was adapted from Le Bourvellec et al.17 and Cheynier et al.18 4.5 ml of 5-CQA or CA 5 mM dissolved in malate buffer 30 mM at pH 4.8 reacted with 500 μL polyphenol oxidase (PPO) malate buffer solution. The whole reaction was stirred for 5 s then filtered on PVDF 0.45 μm just before LC-DAD-ESI-MS2 analysis.2.9 LC-MS2 analysis and treatment process
3 Results and discussion
3.1 Biotransformation studies of 5-CQA
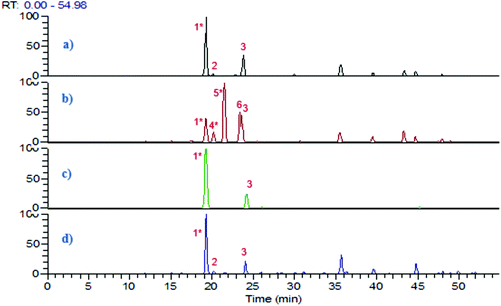
The resting cell method20 was applied for the biotransformation of 5-CQA using three gut bacteria L. reuteri, B. fragilis and B. longum. Based on the growth curve of these bacteria monitored by measuring the OD600 nm, the biotransformation reaction was performed for 24 h when the stationary phase was reached. PBS was chosen as a solvent for 5-CQA and CA since it was previously described as closer to the intracytoplasmic bacterial composition.21 The biotransformation step was performed at pH 5.8, close to the pH found in the colon. 5-CQA was dissolved in PBS at 1 mM, a concentration close to that in the colon after the intake of coffee or fruit.8 After centrifugation, the pellet and the supernatant were separately treated using ethyl acetate to obtain two extracts which were further analysed by LC-UV-ESI-MS2 in negative mode. The base peak chromatograms of the control (5-CQA incubated at 37 °C) and of the SN extracts after 5-CQA biotransformation by the three bacteria are reported in Fig. 1. While L. reuteri was able to biotransform 5-CQA, no biotransformed metabolites were detected after bioconversion by B. fragilis and B. longum (Fig. 1). To validate these results, we searched in silico for the enzyme responsible for CQA biotransformation, namely cinnamoyl esterase or feruloyl esterase (EC.3.1.1.73), which was already found in L. johnsonii22 (accession number NC_005362). A genomic comparison between the gene already described in L. jonhsonii and those present in our strain L. reuteri DSM 20016 (GCA_001436151) showed 70.7% similarity with the gene Locus Tag named LREU_RS08790 (see Fig. S1†). Nonetheless, this gene was not found in B. fragilis and B. longum, suggesting that the results from biotransformation confirmed by LC-ES-MS2 are in line with the in silico search.Research on Pfam highlighted the presence of the same domain, hydrolase_4, serine aminopeptidase, S33 in these two genes which belong to the Alpha/Beta hydrolase fold family.23 This catalytic domain is found in a very wide range of enzymes such as esterase. This cinnamoyl esterase led to the hydrolysis of 5-CQA into CA and quinic acid. CA was well detected in LC-ESI-MS2 chromatograms and quinic acid, being too polar, was either not extracted by ethyl acetate or not retained on the column (Fig. 1). The peak 3 was identified as cis-5-O-caffeoylquinic acid as suggested by Clifford et al.24 and confirmed after UV irradiation in a UV-quartz cell for 1 h, using the 5-CQA standard (see Fig. S2†).
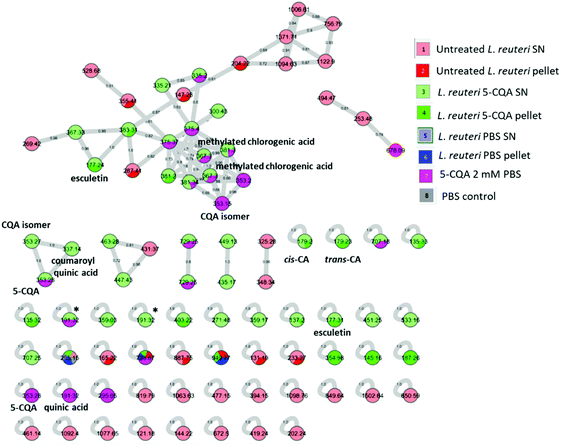
Data from the L. reuteri extract were then subjected to MZmine 2.53 and further treatment for molecular networking analysis (Fig. 2) where each tandem mass spectrum is described as a node and spectrum-to-spectrum alignments as edges defined by a cosine score (cos) (https://gnps.ucsd.edu). A high cos means high similarity between two MS/MS spectra. The created molecular network (MN) highlighted 110 nodes. One cluster encompassed the majority of connected nodes and 45 nodes corresponded to self-loops, which represent a unique feature. Specific biotransformed metabolites appeared in all the clusters but mostly as self-looped nodes.
We then annotated MN by dereplication using the GNPS library. Green nodes depicting metabolites derived from 5-CQA bioconversion by L. reuteri and showing a cos > 0.9 were displayed in Fig. 2. Among these cis-CA, trans-CA, coumaroyl quinic acid and esculetin caught our attention, due to their structure similar to 5-CQA. At this stage, we must underline that MN analysis led to the annotation of two nodes (labelled by an asterisk in Fig. 2 and found either simultaneously in the 5-CQA sample and in one biotransformed extract or in SN L. reuteri biotransformed extract alone) as quinic acid after dereplication but with a very low score (cos = 0.4) (see Table S1†). CA and coumaroyl quinic acid have already been reported after 5-CQA in vitro biotransformation by the gut microbiome.25,26 To validate some of these annotations, various analytical standards were injected as depicted in Table 1 and led to the identification of 5-CQA (ID 3), esculetin (ID 4) and trans-CA (ID 5).
| ID no. | Compound name | RT (min) | m/z | MS2 |
|---|---|---|---|---|
| a Not identified. b Identified after injection of analytical standards. c Identified by MN analysis. d Detected in MN. e Identified after comparison with data previously published. f Quinic acid comes from MS fragmentation of 5-CQA or cis-5-O-caffeoylquinic acid in the source. g Metabolites which RT could be inverted. | ||||
| 1 | 3-Hydroxybenzoic acida | 15.1 | 137.2 | 109/93 |
| 2 | Quinic acidd,f | 19.3 | 191.3 | nd |
| 3 | 5 CQAb,c | 19.3 | 353.3 | 191/179/135 |
| 4 | Esculetinb,c | 20.2 | 177.2 | 133/104 |
| 5 | trans-Caffeic acidb,c | 21.5 | 179.2 | 135 |
| 6 | Vinylcatechold,g | 21.5 | 135.3 | nd |
| 7 | 2-Phenylacetic acidd,g | 23.4 | 135.3 | nd |
| 8 | cis-Caffeic acidc | 23.5 | 179.2 | 135 |
| 9 | cis-5-O-Caffeoylquinic acidc,e | 23.6 | 353.3 | 191/179/135 |
| 10 | Quinic acidc,f | 23.6 | 191.3 | nd |
| 11 | Coumaroyl quinic acidc | 25.5 | 337.1 | 191/163 |
| 12 | 3-(3′-Hydroxyphenyl)propanoic acidd | 28.3 | 165.2 | nd |
| 13 | Methylated chlorogenic acidc,e | 35.7 | 367.3 | nd |
| 14 | Methylated chlorogenic acidc,e | 39.7 | 367.3 | nd |
Surprisingly, both cis and trans geometric isomers of CA were identified in MN. To our knowledge, it is the first report of the presence of these two CA isomers after 5-CQA biotransformation either by bacteria or human gut microbiota. Recently, Clifford expressed that only a few studies have been reported on the metabolism of cis isomers in humans.27 It should be noted that MN analysis identified cis-CA at RT: 21.5 min, highlighting a misidentification of this standard in the GNPS library. Our results concerning the attribution of ID 5 and 8 as trans-CA and cis-CA respectively, are in line with those previously published.28
The main point to highlight in our MN (Fig. 2) is the annotation of [M − H]− ions at m/z 177 as being esculetin. While Mortelé et al.26 attributed this ion as being caffeoyl quinone, we confirm the identification of esculetin using the analytical standard (Table 1). Esculetin was reported only once as an in vivo metabolite of trans-CA after oxidation through an isolated rat liver.29
Our results underline for the first time this specific mechanism by the L. reuteri DSM 20016 strain leading to the formation of a coumarin, esculetin, after ring closure of cis-caffeoyl quinone, via an interesting oxidative pathway of 5-CQA.
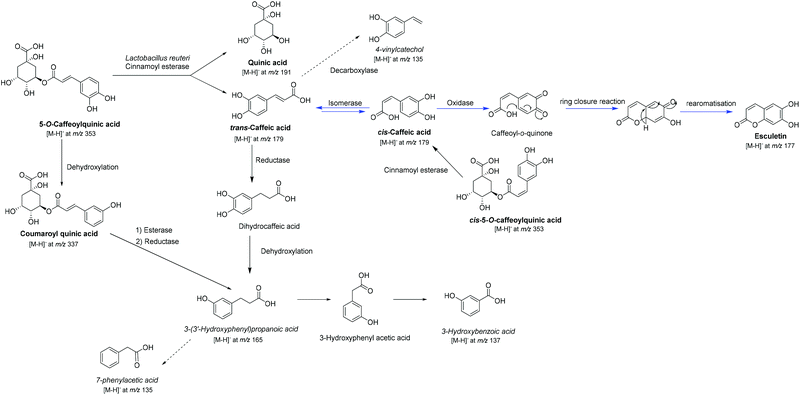
A putative 5-CQA biotransformation pathway by L. reuteri leading to esculetin is shown in Scheme 1. Due to the presence in the standard of both geometric 5-CQA isomers (ID 3 and 9), the first step, after ester cleavage, could be the geometric isomerisation of trans-CA into its cis isomer which has already been reported by photoisomerisation.30 This evidence could be supported by the detection of cis-CA (ID 8) and assume the presence of a cis–trans isomerase in L. reuteri enzymatic machinery. This kind of enzyme has already been reported in Lactobacillus31,32 and after genomic comparison an ortholog sequence in the L. reuteri studied strain (DSM 20016, GCA_001436151) was found (see Fig. S3†). The other possibility of the presence of cis-CA could come from hydrolysis of the residual cis-5-O-caffeoylquinic acid (ID 9) by cinnamoyl esterase. Similarly, with the process of oxidative cyclisation of cis-CA to esculetin after the photocatalytic isomerisation proposed previously,30 the next step corresponds to the formation of caffeoyl quinone probably by an oxidase as the biotransformation was performed under anaerobic conditions. Previous studies have reported the enzymatic conversion of cis-CA into esculetin catalysed by an oxidase, previously named phenolase33,34 for which we have found a sequence similarity with an ubiquinol oxidase in the genome of L. reuteri DSM 20016 (see Fig. S4†). The final step of ring closure can putatively occur spontaneously, followed by classical re-aromatisation to afford esculetin. It is interesting to note that Parrino et al. (2016) reported the non-formation of esculetin at pH 3.5 in comparison to the possible conversion with pH > 6.30 These data suggest that esculetin cannot be formed in the stomach. To confirm the ability of L. reuteri to produce esculetin from CA after 5-CQA biotransformation, we used BHI instead of PBS at pH 5.8 in order to prevent CA autooxidation. This phenomenon was detected after CA incubation at 37 °C in PBS at 5.8 but to a lesser degree than in biotransformed extracts (see Fig. S5†). CA autooxidation has not been checked so far in previous studies of 5-CQA and CA biotransformation. In our study, we confirm that L. reuteri was able to produce esculetin either from 5-CQA or CA in higher amounts when compared to any controls. Moreover, the absence of cis-5-O-caffeoylquinic acid 3 after 5-CQA incubation in BHI (see Fig. S5†) confirms the major formation of cis-CA by L. reuteri from isomerisation of trans-CA.
Other green colour nodes, displayed in Fig. 2, could be highlighted as tentative metabolites coming from 5-CQA biotransformation by L. reuteri by comparison to m/z data previously reported. Regarding the node at m/z 165.2, it was annotated in our MN as phenyllactic acid with a cos = 0.84 and as caffeoyl alcohol with a cos = 0.72 (see Table S1†). While Mortelé et al.26 reported a metabolite with an equivalent m/z ratio (165.0556) as 3-phenyllactic acid, other studies12,35,36 depicted this compound as 3-(3′-hydroxyphenyl)propanoic acid coming from the in vivo metabolism of 5-CQA by faecal microbiota. This compound could come either from CA after reduction into 3-(3′,4′-dihydroxyphenyl)propanoic acid then dehydroxylation12 or from coumaroyl quinic acid, one of the metabolites identified in our MN, after reduction and ester bound hydrolysis25 (Fig. 2). This could be an intermediate metabolite leading to the formation of 3-hydroxybenzoic acid, already depicted in a recent publication35 describing the metabolism study of 5-CQA in rats. The authors reported the identification of this metabolite by UHPLC-Q-TOF-MS in negative mode with an m/z at 137.023 (MS2: 136, 109, 93) which seems similar to our [M − H]− ion at m/z 137.2 with regard to their fragment ions (Table 1). Moreover, hydroxybenzoic acids have already been reported from the gut microbiota conversion of 5-CQA and CA36 and confirm this pathway. MN annotation has also led to the identification of two deprotonated molecular ions with m/z at 367.3 and at RT 35.7 and 39.7 min as already reported methylated chlorogenic acids,35 with a cos = 0.71, confirming the possible methylation of 5-CQA in our study. Nevertheless, these two ions were also detected in the 5-CQA sample dissolved in PBS suggesting that these compounds are artefacts. The formation of 5-CQA isomers during the incubation at 37 °C was previously highlighted.25 Finally, the two [M − H]− ions at m/z 135.3 present in both the pellet and SN L. reuteri biotransformed extracts, could putatively be either vinylcatechol35 coming from decarboxylation of CA or 2-phenylacetic acid.25 Nevertheless, we were not able to determine their MS/MS product ions to confirm one of these two possibilities.
3.2 Characterisation of oxidised compounds of 5-CQA and CA by electrochemical and enzymatic methods
For a better understanding of the oxidative biotransformed pathway leading to the formation of esculetin, we have performed electrochemical and enzymatic oxidation of 5-CQA and CA.Next, the controlled potential electrolysis of 5-CQA and CA were achieved in pH 5.8 0.1 M PBS in a modified UV cell at a platinum grid electrode. As a first step, UV-Visible spectra were recorded during the linear sweep voltammetry LSV at different potential values. Up to +0.75 V and +0.52 V, 5-CQA and CA remained under their native form and the wavelength of maximum absorption λmax were measured at 320 nm. Above these potentials, an absorption band was visible around 400 nm for both compounds, attesting their oxidation to their quinone form. With this knowledge, electrolysis of 5-CQA and CA were conducted at +0.8 and +0.6 V, respectively. The electrolysis was stopped after 20 minutes (corresponding to an electrolysis yield of 15%), a time for which the quinone is still present in the solution (λmax = 404 and 420 nm for 5-CQA and CA quinone respectively). All the electrolysed samples were then analysed by LC-DAD-ESI-MS2.
| Study method | Compound name | RT (min) | m/z | MS2 | λ max (nm) |
|---|---|---|---|---|---|
| Oxidation of CA by PPO | Esculetin | 20.3 | 177.2 | 133 | 343/301/291 |
| Electrochemical oxidation of CA | Esculetin | 20.2 | 177.2 | 133 | 342/301/295 |
| Biotransformation of 5-CQA by L. reuteri-(Supernatant) | Esculetin | 20.2 | 177.3 | 133/104 | 343/300 |
| Biotransformation of 5-CQA by L. reuteri (Pellet) | Esculetin | 20.2 | 177.2 | 133/104 | 344/298/252 |
Esculetin was each time identified independently of the method used underlining the stabilisation of the oxidative CA product, caffeoyl quinone, into its coumarin form, but in far lesser amounts compared to the biotransformation process. Unsurprisingly, no traces of caffeoyl quinone were found in any samples studied.
The values of λmax were in accordance to those in previous reports.39
Analysis of electrolysed and enzymatic sample of 5-CQA did not lead to the detection of esculetin, confirming that the first step of production of this coumarin is the bond ester hydrolysis which was not permitted under electrochemical and PPO conditions.
Moreover, the comparison of LC-DAD-ESI-MS2 analysis of the oxidised samples of 5-CQA and CA, obtained electrochemically and enzymatically, has demonstrated that no oxidative pathway other than the one leading to esculetin occurs by biotransformation. Indeed, for 5-CQA for example, oxidation by the electrode or by the PPO also involves dimerisation pathways. Interestingly, the LC-MS/MS profiles of the electrolysed and enzymatic solutions of 5-CQA were very similar, leading to dimers involving mechanisms described by Guyot et al.40 (see Table S2†). The oxidation of CA enzymatically also led to dimer and trimer formation.18,41
4. Conclusions
In summary, 5-CQA was biotransformed differently according to the gut bacteria strains used. Molecular network analysis led to the identification, after 5-CQA biotransformation by L. reuteri, of one oxidised derivative of cis-CA, caffeoyl quinone, which was stabilised into esculetin in its ring closure form. The inability of the two other gut bacteria studied to biotransform 5-CQA once again supports the importance of the interindividual differences of microbiota composition.42,43 The comparison with other oxidation methods such as electrochemical and enzymatic ones, confirm the formation of caffeoyl quinone followed by a ring closure into esculetin. The electrochemical and enzymatic studies have also shown that no other oxidation products of 5-CQA and CA were formed during biotransformation. These findings also highlight the interesting and specific enzymatic machinery of L. reuteri encompassing a cinnamoyl esterase, isomerase and oxidase leading to a coumarin compound. Further studies should be undertaken with the aim of identifying the potential biological effects of esculetin on the gut microbiota and will give more insights as to whether this metabolite is more active than the parent one, an evidence that we have already demonstrated with olive oil phenol compounds.16 5-CQA formulated as synbiotics44 with L. reuteri may finally be promising candidates in human health applications.Author contributions
Gentiana Balaj: conceptualisation, investigation, visualisation, formal analysis and writing – original draft preparation. Zohreh Tamanai-Shacoori: investigation, resources and writing – review & editing. Damien Ollivier: data curation, formal analysis and software. Aurélie Sauvager: investigation. M. Faustin: investigation; Latifa Bousarghin: validation and writing – review & editing. Sandrine David-Le Gall: investigation and resources. Sylvain Guyot: resources and writing – review & editing. Dashnor Nebija: funding acquisition and writing – review & editing. Sophie Tomasi: conceptualisation, funding acquisition, methodology, project administration, resources, supervision, validation, writing original draft and writing – review & editing and Marie-Laurence Abasq: conceptualiszation, funding acquisition, methodology, project administration, resources, supervision, validation, writing original draft and writing – review & editing.Abbreviations
| BHI | Brain heart infusion |
| CA | Caffeic acid |
| 5-CQA | 5-O-Caffeoylquinic acid = chlorogenic acid |
| CFU | Colony forming units |
| cos | Cosinus score |
| CV | Cyclic voltammetry |
| LSV | Linear sweep voltammetry |
| MN | Molecular network |
| OD | Optical density |
| PPO | Polyphenol oxidase |
| PBS | Phosphate buffer solution |
| RT | Retention time |
| SN | Supernatant |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Patricia Courtel, Isabelle Rouaud (COrInt), Catherine Le Lann (NuMeCan) and Helene Sotin from the P2M2 platform for their technical assistance and Solenn Ferron (COrInt) for assistance with MZmine2 treatment. We sincerely thank Campus France and the scientific committee of the Faculty of Pharmacy of Rennes 1 for a grant given to Gentiana Balaj.References
- H. Zhang and R. Tsao, Dietary polyphenols, oxidative stress and antioxidant and anti-inflammatory effects, Curr. Opin. Food Sci., 2016, 8, 33–42 CrossRef.
- S. V. Luca, I. Macovei, A. Bujor, A. Miron, K. Skalicka-Woźniak, A. C. Aprotosoaie and A. Trifan, Bioactivity of dietary polyphenols: The role of metabolites, Crit. Rev. Food Sci. Nutr., 2020, 60, 626–659 CrossRef CAS PubMed.
- G. M. Pasinetti, R. Singh, S. Westfall, F. Herman, J. Faith and L. Ho, The Role of the Gut Microbiota in the Metabolism of Polyphenols as Characterized by Gnotobiotic Mice, J. Alzheimer's Dis., 2018, 63, 409–421 CAS.
- S. L. Johnson, R. D. Kirk, N. A. DaSilva, H. Ma, N. P. Seeram and M. J. Bertin, Polyphenol Microbial Metabolites Exhibit Gut and Blood–Brain Barrier Permeability and Protect Murine Microglia against LPS-Induced Inflammation, Metabolites, 2019, 9, 78 CrossRef CAS PubMed.
- D. Carregosa, R. Carecho, I. Figueira and C. N. Santos, Low-Molecular Weight Metabolites from Polyphenols as Effectors for Attenuating Neuroinflammation, J. Agric. Food Chem., 2020, 68, 1790–1807 CrossRef CAS PubMed.
- T. R. Yamasaki, K. Ono, L. Ho and G. M. Pasinetti, Gut Microbiome-Modified Polyphenolic Compounds Inhibit α-Synuclein Seeding and Spreading in α-Synucleinopathies, Front. Neurosci., 2020, 14, 398 CrossRef PubMed.
- H. Lu, Z. Tian, Y. Cui, Z. Liu and X. Ma, Chlorogenic acid: A comprehensive review of the dietary sources, processing effects, bioavailability, beneficial properties, mechanisms of action, and future directions, Compr. Rev. Food Sci. Food Saf., 2020, 19, 3130–3158 CrossRef CAS PubMed.
- M. N. Clifford, Chlorogenic acids and other cinnamates - nature, occurrence and dietary burden, J. Sci. Food Agric., 1999, 79, 362–372 CrossRef CAS.
- M. N. Clifford, I. B. Jaganath, I. A. Ludwig and A. Crozier, Chlorogenic acids and the acyl-quinic acids: discovery, biosynthesis, bioavailability and bioactivity, Nat. Prod. Rep., 2017, 34, 1391–1421 RSC.
- D. Couteau, A. L. McCartney, G. R. Gibson, G. Williamson and C. B. Faulds, Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid: esterase - producing gut bacteria, J. Appl. Microbiol., 2001, 90, 873–881 CrossRef CAS PubMed.
- R. Bel-Rhlid, D. Thapa, K. Kraehenbuehl, C. Hansen and L. Fischer, Biotransformation of caffeoyl quinic acids from green coffee extracts by Lactobacillus johnsonii NCC 533, AMB Express, 2013, 3, 28 CrossRef PubMed.
- M. Vollmer, D. Schröter, S. Esders, S. Neugart, F. M. Farquharson, S. H. Duncan, M. Schreiner, P. Louis, R. Maul and S. Rohn, Chlorogenic acid versus amaranth's caffeoylisocitric acid – Gut microbial degradation of caffeic acid derivatives, Food Res. Int., 2017, 100, 375–384 CrossRef CAS PubMed.
- J. C. Espin, A. Gonzalez-Sarrias and F. A. Tomas-Barberan, The gut microbiota: A key factor in the therapeutic effects of (poly)phenols, Biochem. Pharmacol., 2017, 139, 82–93 CrossRef CAS PubMed.
- A. Breynaert, D. Bosscher, A. Kahnt, M. Claeys, P. Cos, L. Pieters and N. Hermans, Development and Validation of an in vitro Experimental GastroIntestinal Dialysis Model with Colon Phase to Study the Availability and Colonic Metabolisation of Polyphenolic Compounds, Planta Med., 2015, 81, 1075–1083 CrossRef CAS PubMed.
- C. D. Kay, M. N. Clifford, P. Mena, G. J. McDougall, C. Andres-Lacueva, A. Cassidy, D. Del Rio, N. Kuhnert, C. Manach, G. Pereira-Caro, A. Rodriguez-Mateos, A. Scalbert, F. Tomás-Barberán, G. Williamson, D. S. Wishart and A. Crozier, Recommendations for standardizing nomenclature for dietary (poly)phenol catabolites, Am. J. Clin. Nutr., 2020, 112, 1051–1068 CrossRef PubMed.
- M. Lambert de Malezieu, S. Ferron, A. Sauvager, P. Courtel, C. Ramassamy, S. Tomasi and M.-L. Abasq, UV-Vis Spectroelectrochemistry of Oleuropein, Tyrosol, and p-Coumaric Acid Individually and in an Equimolar Combination. Differences in LC-ESI-MS2 Profiles of Oxidation Products and Their Neuroprotective Properties, Biomolecules, 2019, 9, 802 CrossRef CAS PubMed.
- C. Le Bourvellec, J.-M. Le Quéré, P. Sanoner, J.-F. Drilleau and S. Guyot, Inhibition of Apple Polyphenol Oxidase Activity by Procyanidins and Polyphenol Oxidation Products, J. Agric. Food Chem., 2004, 52, 122–130 CrossRef CAS PubMed.
- V. Cheynier and M. Moutounet, Oxidative reactions of caffeic acid in model systems containing polyphenol oxidase, J. Agric. Food Chem., 1992, 40, 2038–2044 CrossRef CAS.
- A. Noël, A. Garnier, M. Clément, I. Rouaud, A. Sauvager, L. Bousarghin, P. Vásquez-Ocmín, A. Maciuk and S. Tomasi, Lichen-associated bacteria transform antibacterial usnic acid to products of lower antibiotic activity, Phytochemistry, 2021, 181, 112535 CrossRef PubMed.
- M. K. Julsing, D. Kuhn, A. Schmid and B. Bühler, Resting cells of recombinant E. coli show high epoxidation yields on energy source and high sensitivity to product inhibition, Biotechnol. Bioeng., 2012, 109, 1109–1119 CrossRef CAS PubMed.
- O. Mortelé, E. Iturrospe, A. Breynaert, C. Lammens, X. B. Britto, S. Malhotra-Kumar, P. Jorens, L. Pieters, A. L. N. van Nuijs and N. Hermans, Chlorogenic Acid as a Model Compound for Optimization of an In Vitro Gut Microbiome-Metabolism Model, Proceedings, 2019, 11, 31 Search PubMed.
- C. Fritsch, A. Jänsch, M. A. Ehrmann, S. Toelstede and R. F. Vogel, Characterization of Cinnamoyl Esterases from Different Lactobacilli and Bifidobacteria, Curr. Microbiol., 2017, 74, 247–256 CrossRef CAS PubMed.
- S. El-Gebali, J. Mistry, A. Bateman, S. R. Eddy, A. Luciani, S. C. Potter, M. Qureshi, L. J. Richardson, G. A. Salazar, A. Smart, E. L. L. Sonnhammer, L. Hirsh, L. Paladin, D. Piovesan, S. C. E. Tosatto and R. D. Finn, The Pfam protein families database in 2019, Nucleic Acids Res., 2019, 47, D427–D432 CrossRef CAS PubMed.
- M. N. Clifford, J. Kirkpatrick, N. Kuhnert, H. Roozendaal and P. R. Salgado, LC–MSn analysis of the cis isomers of chlorogenic acids, Food Chem., 2008, 106, 379–385 CrossRef CAS.
- F. Tomas-Barberan, R. García-Villalba, A. Quartieri, S. Raimondi, A. Amaretti, A. Leonardi and M. Rossi, In vitro transformation of chlorogenic acid by human gut microbiota, Mol. Nutr. Food Res., 2014, 58, 1122–1131 CrossRef CAS PubMed.
- O. Mortelé, E. Iturrospe, A. Breynaert, E. Verdickt, B. B. Xavier, C. Lammens, S. Malhotra-Kumar, P. G. Jorens, L. Pieters, A. L. N. van Nuijs and N. Hermans, Optimization of an in vitro gut microbiome biotransformation platform with chlorogenic acid as model compound: From fecal sample to biotransformation product identification, J. Pharm. Biomed. Anal., 2019, 175, 112768 CrossRef PubMed.
- M. N. Clifford, A. Kerimi and G. Williamson, Bioavailability and metabolism of chlorogenic acids (acyl–quinic acids) in humans, Compr. Rev. Food Sci. Food Saf., 2020, 19, 1299–1352 CrossRef CAS PubMed.
- A. Oniszczuk, LC-ESI-MS/MS Analysis and Extraction Method of Phenolic Acids from Gluten-Free Precooked Buckwheat Pasta, Food Anal. Methods, 2016, 9, 3063–3068 CrossRef.
- H. Gumbinger, U. Vahlensieck and H. Winterhoff, Metabolism of Caffeic Acid in the Isolated Perfused Rat Liver, Planta Med., 1993, 59, 491–493 CrossRef CAS PubMed.
- F. Parrino, A. Di Paola, V. Loddo, I. Pibiri, M. Bellardita and L. Palmisano, Photochemical and photocatalytic isomerization of trans -caffeic acid and cyclization of cis -caffeic acid to esculetin, Appl. Catal., B, 2016, 182, 347–355 CrossRef CAS.
- S. Kishino, J. Ogawa, K. Yokozeki and S. Shimizu, Linoleic Acid Isomerase in Lactobacillus plantarum AKU1009a Proved to Be a Multi-Component Enzyme System Requiring Oxidoreduction Cofactors, Biosci. Biotechnol. Biochem., 2011, 75, 318–322 CrossRef CAS PubMed.
- Y. Sun, Y. Tang, X. Hou, H. Wang, L. Huang, J. Wen, H. Niu, W. Zeng and Y. Bai, Novel Lactobacillus reuteri HI120 Affects Lipid Metabolism in C57BL/6 Obese Mice, Front. Vet. Sci., 2020, 7, 560241 CrossRef PubMed.
- M. Satô, Metabolism of phenolic substances by the chloroplasts—III., Phytochemistry, 1967, 6, 1363–1373 CrossRef.
- M. Sato and A. Hiraoka, Characterization of a product (Esculetin; 6,7-dihydroxycoumarin) from the phenolase-caffeic acid reaction system., Chem. Pharm. Bull., 1985, 33, 1289–1292 CrossRef CAS.
- J. Li, S.-P. Wang, Y.-Q. Wang, L. Shi, Z.-K. Zhang, F. Dong, H.-R. Li, J.-Y. Zhang and Y.-Q. Man, Comparative metabolism study on chlorogenic acid, cryptochlorogenic acid and neochlorogenic acid using UHPLC-Q-TOF MS coupled with network pharmacology, Chin. J. Nat. Med., 2021, 19, 212–224 CAS.
- M.-P. Gonthier, C. Remesy, A. Scalbert, V. Cheynier, J.-M. Souquet, K. Poutanen and A.-M. Aura, Microbial metabolism of caffeic acid and its esters chlorogenic and caftaric acids by human faecal microbiota in vitro, Biomed. Pharmacother., 2006, 60, 536–540 CrossRef CAS PubMed.
- C. Giacomelli, K. Ckless, D. Galato, F. S. Miranda and A. Spinelli, Electrochemistry of Caffeic Acid Aqueous Solutions with pH 2.0 to 8.5, J. Braz. Chem. Soc., 2002, 13, 332–338 CrossRef CAS.
- P. Hapiot, A. Neudeck, J. Pinson, H. Fulcrand, P. Neta and C. Rolando, Oxidation of caffeic acid and related hydroxycinnamic acids, J. Electroanal. Chem., 1996, 405, 169–176 CrossRef.
- M. Tattini, M. Di Ferdinando, C. Brunetti, A. Goti, S. Pollastri, C. Bellasio, C. Giordano, A. Fini and G. Agati, Esculetin and esculin (esculetin 6-O-glucoside) occur as inclusions and are differentially distributed in the vacuole of palisade cells in Fraxinus ornus leaves: A fluorescence microscopy analysis, J. Photochem. Photobiol., B, 2014, 140, 28–35 CrossRef CAS PubMed.
- S. Guyot, S. Bernillon, P. Poupard and C. M. G. C. Renard, in Recent Advances in Polyphenol Research, ed. F. Daayf and V. Lattanzio, Wiley-Blackwell, Oxford, UK, 2008, pp. 278–292 Search PubMed.
- S. Pati, I. Losito, F. Palmisano and P. G. Zambonin, Characterization of caffeic acid enzymatic oxidation by-products by liquid chromatography coupled to electrospray ionization tandem mass spectrometry, J. Chromatogr. A, 2006, 1102, 184–192 CrossRef CAS PubMed.
- L. Ho, D. Zhao, K. Ono, K. Ruan, I. Mogno, M. Tsuji, E. Carry, J. Brathwaite, S. Sims, T. Frolinger, S. Westfall, P. Mazzola, Q. Wu, K. Hao, T. E. Lloyd, J. E. Simon, J. Faith and G. M. Pasinetti, Heterogeneity in gut microbiota drive polyphenol metabolism that influences α-synuclein misfolding and toxicity, J. Nutr. Biochem., 2019, 64, 170–181 CrossRef CAS PubMed.
- T. Frolinger, S. Sims, C. Smith, J. Wang, H. Cheng, J. Faith, L. Ho, K. Hao and G. M. Pasinetti, The gut microbiota composition affects dietary polyphenols-mediated cognitive resilience in mice by modulating the bioavailability of phenolic acids, Sci. Rep., 2019, 9, 3546 CrossRef PubMed.
- K. S. Swanson, G. R. Gibson, R. Hutkins, R. A. Reimer, G. Reid, K. Verbeke, K. P. Scott, H. D. Holscher, M. B. Azad, N. M. Delzenne and M. E. Sanders, The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics, Nat. Rev. Gastroenterol. Hepatol., 2020, 17, 687–701 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1fo04304h |
| This journal is © The Royal Society of Chemistry 2022 |