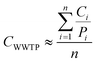Open Access Article
Open Access ArticleEmerging investigator series: meta-analyses on SARS-CoV-2 viral RNA levels in wastewater and their correlations to epidemiological indicators†
David
Mantilla-Calderon
a,
Kaiyu (Kevin)
Huang
a,
Aojie
Li
a,
Kaseba
Chibwe
a,
Xiaoqian
Yu
b,
Yinyin
Ye
 c,
Lei
Liu
d and
Fangqiong
Ling
c,
Lei
Liu
d and
Fangqiong
Ling
 *aefg
*aefg
aDepartment of Energy, Environmental and Chemical Engineering, Washington University in St. Louis, St. Louis, MO, USA. E-mail: fangqiong@wustl.edu
bCentre for Microbiology and Environmental Systems Science, Department of Microbiology and Ecosystem Science, Division of Microbial Ecology, University of Vienna, Vienna, Austria
cDepartment of Civil, Structural and Environmental Engineering, University at Buffalo, Buffalo, NY, USA
dDivision of Biostatistics, Washington University in St. Louis, St. Louis, MO, USA
eDepartment of Computer Science and Engineering, Washington University in St. Louis, St. Louis, MO, USA
fDivision of Biological and Biomedical Sciences, Washington University in St. Louis, St. Louis, MO, USA
gDivision of Computational and Data Science, Washington University in St. Louis, St. Louis, MO, USA
First published on 4th May 2022
Abstract
Background: recent applications of wastewater-based epidemiology (WBE) have demonstrated its ability to track the spread and dynamics of COVID-19 at the community level. Despite the growing body of research, quantitative synthesis of SARS-CoV-2 RNA levels in wastewater generated from studies across space and time using diverse methods has not been performed. Objective: the objective of this study is to examine the correlations between SARS-CoV-2 RNA levels in wastewater and epidemiological indicators across studies, stratified by key covariates in study methodologies. In addition, we examined the association of proportions of positive detections in wastewater samples and methodological covariates. Methods: we systematically searched the Web of Science for studies published by February 16th, 2021, performed a reproducible screening, and employed mixed-effects models to estimate the levels of SARS-CoV-2 viral RNA quantities in wastewater samples and their correlations to the case prevalence, the sampling mode (grab or composite sampling), and the wastewater fraction analyzed (i.e., solids, solid–supernatant mixtures, or supernatants/filtrates). Results: a hundred and one studies were found; twenty studies (671 biosamples and 1751 observations) were retained following a reproducible screening. The mean positivity across all studies was 0.68 (95%-CI, [0.52; 0.85]). The mean viral RNA abundance was 5244 marker copies per mL (95%-CI, [0; 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 432]). The Pearson correlation coefficients between the viral RNA levels and case prevalence were 0.28 (95%-CI, [0.01; 0.51]) for daily new cases or 0.29 (95%-CI, [−0.15; 0.73]) for cumulative cases. The fraction analyzed accounted for 12.4% of the variability in the percentage of positive detections, followed by the case prevalence (9.3% by daily new cases and 5.9% by cumulative cases) and sampling mode (0.6%). Among observations with positive detections, the fraction analyzed accounted for 56.0% of the variability in viral RNA levels, followed by the sampling mode (6.9%) and case prevalence (0.9% by daily new cases and 0.8% by cumulative cases). While the sampling mode and fraction analyzed both significantly correlated with the SARS-CoV-2 viral RNA levels, the magnitude of the increase in positive detection associated with the fraction analyzed was larger. The mixed-effects model treating studies as random effects and case prevalence as fixed effects accounted for over 90% of the variability in SARS-CoV-2 positive detections and viral RNA levels. Interpretations: positive pooled means and confidence intervals in the Pearson correlation coefficients between the SARS-CoV-2 viral RNA levels and case prevalence indicators provide quantitative evidence that reinforces the value of wastewater-based monitoring of COVID-19. Large heterogeneities among studies in proportions of positive detections, viral RNA levels, and Pearson correlation coefficients suggest a strong demand for methods to generate data accounting for cross-study heterogeneities and more detailed metadata reporting. Large variance was explained by the fraction analyzed, suggesting sample pre-processing and fractionation as a direction that needs to be prioritized in method standardization. Mixed-effects models accounting for study level variations provide a new perspective to synthesize data from multiple studies.
432]). The Pearson correlation coefficients between the viral RNA levels and case prevalence were 0.28 (95%-CI, [0.01; 0.51]) for daily new cases or 0.29 (95%-CI, [−0.15; 0.73]) for cumulative cases. The fraction analyzed accounted for 12.4% of the variability in the percentage of positive detections, followed by the case prevalence (9.3% by daily new cases and 5.9% by cumulative cases) and sampling mode (0.6%). Among observations with positive detections, the fraction analyzed accounted for 56.0% of the variability in viral RNA levels, followed by the sampling mode (6.9%) and case prevalence (0.9% by daily new cases and 0.8% by cumulative cases). While the sampling mode and fraction analyzed both significantly correlated with the SARS-CoV-2 viral RNA levels, the magnitude of the increase in positive detection associated with the fraction analyzed was larger. The mixed-effects model treating studies as random effects and case prevalence as fixed effects accounted for over 90% of the variability in SARS-CoV-2 positive detections and viral RNA levels. Interpretations: positive pooled means and confidence intervals in the Pearson correlation coefficients between the SARS-CoV-2 viral RNA levels and case prevalence indicators provide quantitative evidence that reinforces the value of wastewater-based monitoring of COVID-19. Large heterogeneities among studies in proportions of positive detections, viral RNA levels, and Pearson correlation coefficients suggest a strong demand for methods to generate data accounting for cross-study heterogeneities and more detailed metadata reporting. Large variance was explained by the fraction analyzed, suggesting sample pre-processing and fractionation as a direction that needs to be prioritized in method standardization. Mixed-effects models accounting for study level variations provide a new perspective to synthesize data from multiple studies.
Water impactRecent applications of wastewater-based epidemiology (WBE) have demonstrated its ability to track the spread and dynamics of COVID-19 at the community level. Despite the growing body of research, quantitative synthesis of SARS-CoV-2 viral RNA levels in wastewater generated from studies across space and time using diverse methods has not been performed. The meta-analysis methodology treats individual studies as members of a population of studies that all provide information on a given effect instead of drawing conclusions on exemplary studies that have shown strong positive effects. Leveraging a large sample size, meta-analysis can help move the narrative beyond statistical significance and draw attention to the magnitude, direction, and variance in effects. This study employed a meta-analysis methodology to quantitatively synthesize results among WBE studies in the first year of the COVID-19 pandemic. Positive pooled means and confidence intervals in the Pearson correlation coefficients between the SARS-CoV-2 viral RNA levels and case prevalence indicators provide quantitative evidence reinforcing the value of wastewater-based monitoring of COVID-19. Large heterogeneities among studies suggest a strong demand for experimental and computational methods to address cross-study heterogeneities. Mixed-effects models accounting for study level variations provide a new perspective to synthesize data from multiple WBE studies. |
1. Introduction
Wastewater-based virus monitoring has been shown as a promising tool for tracking disease dynamics in a large population during the ongoing COVID-19 pandemic.1 It has been reported that 39 to 65% of infected individuals may excrete viral particles through urine and feces,2–4 thus allowing wastewater-based detection. The current epidemiological approaches to estimate the COVID-19 disease prevalence rely on individualized patient testing (i.e., detection of SARS-CoV-2 in nasopharyngeal specimens). Individual testing is invasive, resource-intensive, and mostly restricted to symptomatic individuals with access to healthcare.5 Wastewater-based epidemiology (WBE) has the potential to circumvent biases caused by the varied access to individual-based testing, the presence of asymptomatic cases, and social stigma.6 Moreover, WBE has been applied in the environmental monitoring of poliovirus, effectively detecting new variants and preventing disease resurgence.7 The poliovirus experience suggests the long-term benefits of developing and refining WBE as a public health monitoring technology.As the number of WBE studies continues to grow, study-to-study variations are often encountered; thus, the growing body of data demands attention to generalizable relationships across studies. Although WBE studies focusing on the SARS-CoV-2 virus have been all conducted during the pandemic, not all tested wastewater samples provided a measurable detection when known cases were present in the associated area, thus presenting false negatives in the detection.8–20 In addition, while positive correlations between SARS-CoV-2 wastewater-based measurements and COVID-19 cases have been described,8,9,11,21–23 the strength of the correlations may vary among studies. To better describe the advantages and limitations of WBE and make evidence-based recommendations, research synthesis efforts are needed to quantify the detection rates of SARS-CoV-2 in wastewater, its RNA abundance, and their correlations to epidemiological indicators.
Meta-analysis provides an objective, quantitative, and powerful way to synthesize findings across studies.24 Instead of drawing conclusions on exemplary studies that have shown strong positive effects, meta-analyses treat individual studies as members of a population of studies that conjunctively provide information on a given effect.25 Leveraging a large sample size, a meta-analysis can help move the narrative beyond statistical significance and draw attention to the magnitude, direction, and variance in effects.24 Furthermore, the meta-analytic approach allows us to quantitatively examine the heterogeneity among study results, thus motivating the generation of new hypotheses.26 Meta-analyses systematically synthesize large quantities of data generated from multiple primary studies to reach broad generalizations. A well-conducted meta-analysis can provide a comprehensive picture of parameters of interest and their moderators that is not attainable from an individual primary study. Using statistical models to quantify the magnitude of an effect and its heterogeneity, a meta-analysis may also identify areas that require further research.
Here, we employed a meta-analytic methodology to synthesize wastewater-based SARS-CoV-2 viral RNA abundance data published by February 16th, 2021, approximately a year after the beginning of the COVID-19 pandemic. Following a PRISMA guideline,27 we synthesized and reported results from 1751 observations in 20 studies. We asked four fundamental questions: 1) what is the pooled proportion of positive detection of SARS-CoV-2 from wastewater samples; 2) what are the viral RNA levels of the SARS-CoV-2 virus in wastewater collectively and when subgrouped by key methodological variables; 3) what are the overall strengths of correlation between the positive detection or RNA levels of SARS-CoV-2 in wastewater and epidemiological indicators (daily and cumulative cases); and 4) how much of the variation in SARS-CoV-2 viral RNA abundance can be explained by COVID-19 cases alone? To account for study-level variations, mixed-effects models were employed to examine the correlation between SARS-CoV-2 viral RNA levels and positive detection.
2. Methods
Results from this systematic review and subsequent meta-analysis have been reported following the PRISMA guidelines.27 A PRISMA checklist is presented in Table S1.†2.1 Data sources
We searched the Web of Science (WoS) for publications analyzing untreated wastewater for the presence of the SARS-CoV-2 virus published by February 16th, 2021. Studies were retrieved with the search terms: TS = (SARS-CoV-2 AND (wastewater OR sewage)) from the WoS core collection. Including the keyword “COVID-19” in the search terms did not increase the number of resulting records. The following search conditions were applied: i) language was restricted to English; ii) the time span was set to “All Years”; iii) records in the Science Citation Index Expanded (SCI-Expanded) were included; and iv) the document type was set to “article”, “early access” or “letter” to retain original research (i.e., “reviews” or “editorial materials” were not selected).2.2 Study selection and eligibility criteria
Upon study retrieval from the Web of Science, duplication of records was screened by titles and authors. No duplication was found. Next, full-text records were scanned to assess for eligibility. Studies that reported nucleic-acid detections of SARS-CoV-2 from wastewater systems and associated epidemiological indicators were included. Specifically, the following inclusion criteria were applied: 1) original qPCR data in terms of SARS-CoV-2 measurements in wastewater were reported, and data were reported as quantification cycles, copy numbers per volume, genome equivalents per volume, and/or genome equivalents per weight of sample; 2) sampling locations were identified as wastewater treatment plants (WWTPs), sewage collection networks, lift stations, manholes or septic tanks; 3) SARS-CoV-2 case incidence/prevalence is reported for the associated locations during the sampling times. Rationales for each inclusion criterion are provided in Table 1. The study eligibility was assessed by David Mantilla-Calderon (DMC), Kevin Huang (KH), Aojie Li (AL), and Fangqiong Ling (FL). DMC, KH, and AL extracted and compiled the data. In case of uncertainties, these were discussed and resolved by consensus. DMC and FL curated the database.| Inclusion criteria | Rationales |
|---|---|
| C1: qPCR data were reported as quantification cycles, copy numbers per volume, genome equivalents per ml, or genome equivalents per weight | C1 provides comparable data among studies |
| C2: sampling locations for raw sewage were identified as wastewater treatment plants (WWTPs), sewage collection networks, lift stations, manholes or septic tanks | C2 allows comparisons of SARS-CoV-2 viral titers and percent positivity in wastewater within and across studies |
| C3: COVID-19 case records were reported for the associated locations during the sampling times | C3 allows comparisons of SARS-CoV-2 viral titers in wastewater within and across studies |
2.3 Data extraction
https://automeris.io/WebPlotDigitizer) was used to retrieve the information using digitized versions of figures. The qPCR/dPCR measurements themselves and the units (i.e., quantitative cycles (Ct) or viral RNA levels) were recorded. In addition, metadata about a sample was retrieved, including the study information, sample environment, and assay information. The study information included the author, title, and the year of publication. The sample environment included the following: i) the geographical location (i.e., country and city where the study was performed), ii) the sampling location within a wastewater system (i.e., samples were taken from the sewage collection systems, at the wastewater treatment plant after screenings and before sedimentation, or at the primary sedimentation tank), iii) sample processing prior to viral concentration (whether a sample was filtered, centrifuged, or left untreated), iv) the viral concentration method, v) the associated COVID case incidence or prevalence as provided in the publication, vi) the service population as provided in the publication, and vii) the date of collection of each wastewater sample. Finally, we extracted the assay information, including the choice of sampling techniques (i.e., grab or composite sampling), qPCR/dPCR gene targets, and primer sets.2.4 Data extraction and summary measures
Upon data retrieval, the SARS-CoV-2 measurements, sample environment, assay information, and COVID-19 case prevalence were recorded and converted to consistent units across studies (2.4.1–2.4.3). The proportion of positive detections was calculated (2.4.4). Annotated data are made available on https://github.com/linglab-washu/wbe-metaanalysis at the time of publication.Sampling location. Specifically, the sampling locations were annotated as i) “WWTP” if the sample was described as taken after primary screenings and before the primary sedimentation tanks, as ii) “municipal sewage network” if the sample was taken from manholes, septic tanks, or lift stations, and as iii) “in premise” if the sample was described as taken from the private sewage infrastructure of a facility such as a hospital or a dormitory.
Fractions. Wastewater and sludge samples are included in the review. Wastewater refers to samples collected from the sewage network or the WWTP, consisting predominantly of a liquid phase and to a lesser degree, a solid phase. The term sludge specifically refers to the slurry of a solid and liquid mixture collected from a primary clarifier. The liquid and solid phases of samples can be separated/fractionated by laboratory methods. Fractions resulting from sample separation processes were recorded and annotated. Specifically, a fraction was annotated as “solids” when it consisted of primary solids from a gravity thickener, or solids collected from wastewater by in-laboratory sedimentation (e.g., pellets recovered by centrifugation of the wastewater sample at >1000g). A fraction was annotated as “supernatant/filtrate” if the sample was pre-filtered at 0.22 to 0.7 μm or centrifuged at 1000–10
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g before viral concentration. A fraction was annotated as “mixture of supernatant and suspended solids” when a sample of raw unprocessed wastewater was directly used for viral concentration.
000g before viral concentration. A fraction was annotated as “mixture of supernatant and suspended solids” when a sample of raw unprocessed wastewater was directly used for viral concentration.
Viral concentration. A fraction might be subjected to a subsequent viral concentration step. Typically, viral concentration methods were applied to a mixture of supernatant and suspended solids and supernatant/filtrate fractions. Solid fractions were not typically subjected to a viral concentration step. Viral concentration methods were recorded as described in the original publication.
Gene targets. Several SARS-CoV-2 qPCR gene targets were identified during the study screening process and categorized in the “marker” variable. Targeted regions included ORF1ab, nucleoprotein (N), spike protein (S), envelope protein (E), membrane protein (M), and the RNA-dependent RNA-polymerase (RdRp) gene. The specific primer set that was used to target the marker gene was recorded under the “primer” variable using the notation Author_Marker. Two abbreviations were used in the field author, CDC, referring to the Center of Disease Control, USA (e.g., CDC_N1), and NIID, referring to the National Institute of Infectious Disease, Japan (e.g., NIID_N).
In the cases that SARS-CoV-2 measurements were performed using a multiplex qPCR/dPCR assay, the value for the variable “primer” for this specific observation was recorded by listing the primer sets employed, spaced by an underscore sign. To illustrate, if RNA levels were estimated using a duplex qPCR assay employing CDC_N1 and CDC_N2 primer sets, the value for the variable “primer” would be recorded as CDC_N1_N2. In some instances, a study may analyze multiple markers independently but report genome equivalents. A singular primer set would be recorded in the “primer” variable if it was specified in the study which primer set was used to calculate the reported genome equivalents; alternatively, the value recorded for the “primer” variable in the observation would include all the primer sets used in the study separated by a comma (e.g., CDC_N1, CDC_N2).
where CWWTP denotes the approximate cases for the WWTP, Ci is the case records for municipalityi, Pi is the population reported for municipalityi, and n is the total number of municipalities contributing wastewater to the WWTP.
Epidemiological data reported as “cumulative cases” are denoted as “cumulative cases”. Cases reported as “daily new cases”, “new cases”, “positive daily test”, “new positive daily test”, or “seven-day average cases” were denoted as “daily cases”. “Hospital admissions” and “hospitalized patients” were denoted as “hospitalized cases”. All case counts were converted to prevalence, i.e., patients per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants, to allow synthesis across studies.
000 inhabitants, to allow synthesis across studies.
2.5 Forest plot generation
Forest plots were generated using the “dmetar” package in R (ref. 28) employing a random-effects model. A random-effects meta-analysis model assumes that the observed average SARS-CoV-2 viral RNA levels can vary across studies because of real differences in the viral abundance in each system, as well as sampling variability (chance). Thus, even if all studies had an infinitely large sample size, the observed study effects would still vary because of the real differences in the sewershed's effects on viral RNA levels. Such heterogeneity in average viral RNA levels can be caused by differences in study populations (such as local COVID-19 case prevalence), the wastewater system effects on dilution or decay, the methodological differences, and other factors.29–31The weight of each study in the forest plot was calculated as
| Wi = 1/(Vi + T2) |
2.6 General linear mixed-effects model (GLMM)
General linear mixed-effects models were built to examine the epidemiological indicators (cumulative cases or daily new cases) as sources of fixed effects and studies as sources of random effects on SARS-CoV-2 measurements in wastewater. A binomial GLMM was used to model the positive detections among all observations. A linear mixed-effects model (Gaussian) was used to fit the log-transformed viral RNA levels using observations in which positive detections were made. GLMMs were built in the R package “lme4”.34 Studies were treated as sources of random effects on intercepts. Fixed-effects models were built using the same link functions to examine the significance of random effects and assess the overall fits from fixed effects. Fitting of fixed-effects models was performed using the “glm” and “lm” functions in the R stats package.35 The Akaike information criterion (AIC), Bayesian information criterion (BIC), and log-likelihoods were reported for model selection. Nakagawa's R-squared definitions were used to compute marginal and conditional R-squared values using the R package “MuMIn”.36 The studies that reported daily cases and cumulative cases were examined separately. Studies reporting solids were excluded due to a lack of replicates after being subset into studies reporting daily cases or cumulative cases (details can be found in Fig. S1†).3. Results
3.1 Systematic review
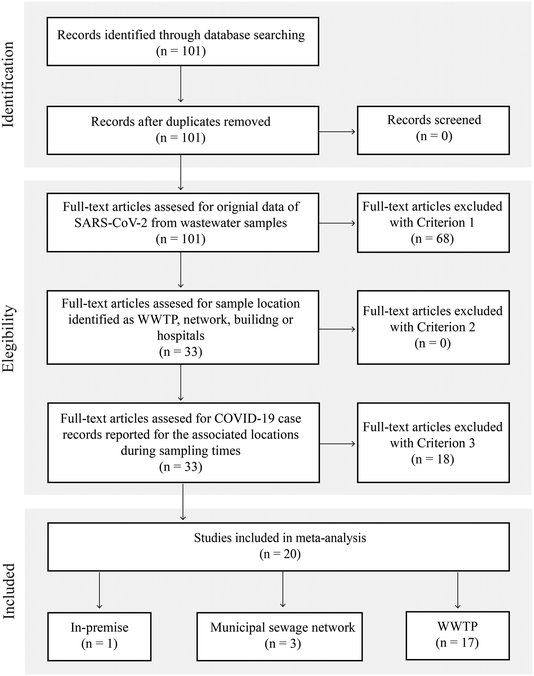
Our search identified 101 unique titles and abstracts; after screening (Fig. 1, Table S2†), 20 papers were included in this review. These studies reported SARS-CoV-2 measurements in wastewater in terms of viral RNA levels or Ct and provided epidemiological indicators. A total of 1751 observations were recorded. Fig. 1 depicts the details of the search.
Table 2 describes the basic characteristics of the included resources. Eighteen studies reported quantitative measurements for SARS-CoV-2 as gene copies per unit mass/volume,8,9,11,14–21,23,30,37–41 while two studies reported Ct values.10,13 Among the 18 quantitative studies, seventeen reported marker copies or genome equivalents per mL,8,9,14–21,23,30,37–41 and one study reported marker copies per gram of biomass.11 Epidemiological indicators were reported as daily cases in nine studies, ranging from 0.6 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants to 117 per 100
000 inhabitants to 117 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants,11,13,14,16,18,19,21,39,41 cumulative cases were reported in ten studies ranging from 1.6 per 100
000 inhabitants,11,13,14,16,18,19,21,39,41 cumulative cases were reported in ten studies ranging from 1.6 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants to 808 per 100
000 inhabitants to 808 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants,8,9,14,15,18,20,30,37,40,41 active cases were reported in four studies10,21,38,40 and hospitalized cases in two studies.13,17 Among these studies, two studies reported both daily and cumulative cases,18,41 one study reported both daily and active cases,21 and one reported both cumulative cases and hospitalized cases.13 Cumulative COVID-19 cases were the most frequently reported, followed by daily, active, and hospitalized cases. SARS-CoV-2 was detected in all studies, irrespective of case prevalence levels, albeit at varying proportions of positive detections.
000 inhabitants,8,9,14,15,18,20,30,37,40,41 active cases were reported in four studies10,21,38,40 and hospitalized cases in two studies.13,17 Among these studies, two studies reported both daily and cumulative cases,18,41 one study reported both daily and active cases,21 and one reported both cumulative cases and hospitalized cases.13 Cumulative COVID-19 cases were the most frequently reported, followed by daily, active, and hospitalized cases. SARS-CoV-2 was detected in all studies, irrespective of case prevalence levels, albeit at varying proportions of positive detections.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants unless otherwise stated. COVID-19 cases are rounded to the nearest unit. nbiosample indicates the number of unique sewage specimens analyzed, nobs indicates the number of total measurements that were made for the SARS-CoV-2 virus
000 inhabitants unless otherwise stated. COVID-19 cases are rounded to the nearest unit. nbiosample indicates the number of unique sewage specimens analyzed, nobs indicates the number of total measurements that were made for the SARS-CoV-2 virus
| Author (biosamples, observations) | Country/date of sampling | Sample collection point | Sample type | Population served | Sample fraction | Viral concentration method | Type of case (mean, min, max) |
|---|---|---|---|---|---|---|---|
| Ahmed, W. et al. (2020)12 (nbiosample = 8, nobs = 32) | Australia | Pumping station, WWTP influent | Grab and composite | 736![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 172 172 |
Supernatant and suspended solids | Electronegative membrane absorption-direct RNA extraction | Cumulative cases (50, 0, 70) |
| Feb–April, 2020 | Supernatant | Ultrafiltration (Centricon) | |||||
| Baldovin, T. et al. (2021)13a (nbiosample = 9, nobs = 18) | Italy | Municipal sewage network | Grab | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770–36 770–36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 042 042 |
Supernatant | Ultrafiltration | Cumulative cases (169, 141, 205) |
| April 23 and May 05, 2020 | Hospitalized cases (34, 30, 39) | ||||||
| D'Aoust, P. M. et al. (2021)21 (nbiosample = 22, nobs = 44) | Canada | Postgrid solids | Grab and composite | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Solids | PEG precipitation | Daily cases (117, 19, 572) |
| April–June, 2020 | Primary sludge | Alum precipitation–ultrafiltration | Active cases (19, 6, 58) | ||||
| Gonçalves, J. et al. (2021)10a (nbiosample = 15, nobs = 30) | Slovenia | Hospital sewage | Composite | N/A | Supernatant | Ultrafiltration | Cumulative casesb (2, 0, 4) |
| June, 2020 | Active casesb (2, 0, 4) | ||||||
| Gonzalez, R. et al. (2020)9 (nbiosample = 198, nobs = 594) | USA | WWTP influent | Grab and composite | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant | Hollow fiber concentrating pipet | Cumulative cases (229, 1, 2288) |
| March–May, 2020 | Adsorption–elution electronegative membrane | ||||||
| Graham, K. et al. (2020)11 (nbiosample = 89, nobs = 166) | USA | WWTP influent | Composite | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant | PEG precipitation | Daily cases (2, 1, 12) |
| March–April, 2020 | Primary settling tank | Composite | Primary solids | No concentration | |||
| March–July, 2020 | |||||||
| Haramoto, E. et al. (2020)41 (nbiosample = 5, nobs = 36) | Japan | WWTP influent | Grab | 817![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 192a 192a |
Supernatant and suspended solids | Electronegative membrane vortex–ultrafiltration | Cumulative cases (5, 0, 7) |
| March–May, 2020 | Electronegative membrane absorption-direct RNA extraction | Daily cases (1, 0, 1.0) | |||||
| Hata, A. et al. (2021)14 (nbiosample = 45, nobs = 87) | Japan | WWTP influent | Grab | 697![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant | PEG precipitation | Daily cases (8, 0, 19) |
| March–April, 2020 | Cumulative cases (15, 0, 26) | ||||||
| Kitamura, K. et al. (2021)15 (nbiosample = 32, nobs = 198) | Japan | WWTP influent, municipal sewage network | Grab | N/A | Supernatant | Adsorption–elution electronegative membrane | Cumulative casesb (122, 19, 209) |
| June–August, 2020 | PEG precipitation ultrafiltration | ||||||
| Solids | Solid precipitation–centrifugation | ||||||
| Kumar, M. et al. (2020)37 (nbiosample = 2, nobs = 6) | India | WWTP influent | Grab | N/A | Supernatant | PEG precipitation | Cumulative casesb (7793, 4912, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 674) 674) |
| May, 2020 | |||||||
| Medema, G. et al. (2020)8 (nbiosample = 25, nobs = 100) | Netherlands | WWTP influent | Composite | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant | Ultrafiltration (Centricon) | Cumulative cases (16, 0, 87) |
| Feb–March, 2020 | |||||||
| Miyani, B. et al. (2020)39 (nbiosample = 33, nobs = 33) | USA | Municipal sewage network | Grab | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant and suspended solids | Adsorption–elution electropositive column filters | Daily cases (6, 4, 8) |
| April–May, 2020 | |||||||
| Nemudryi, A. et al. (2020)16 (nbiosample = 17, nobs = 34) | USA | WWTP influent | Composite | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 831 831 |
Supernatant | Ultrafiltration | Daily cases (6, 0, 14) |
| March–June, 2020 |
| Author (biosamples, observations) | Country/date of sampling | Sample collection point | Sample type | Population served | Sample fraction | Concentration method | Type of case |
|---|---|---|---|---|---|---|---|
a Semiquantitative studies.
b Cases not normalized by 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants. 000 inhabitants.
|
|||||||
| Peccia, J. et al. (2020)23 (nbiosample = 73, nobs = 226) | USA | Primary settling tank | Grab | 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Solids | No concentration | Daily positive test (26, 3, 60) |
| March–June, 2020 | |||||||
| Randazzo, W. et al. (2020)38 (a) (nbiosample = 12, nobs = 24) | Spain | WWTP influent | Grab | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant and suspended solids | Aluminium flocculation | Active cases (80, 1, 111) |
| Feb–April, 2020 | |||||||
| Randazzo, W. et al. (2020)20 (b) (nbiosample = 42, nobs = 42) | Spain | WWTP influent | Grab | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 357 357![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 177 177 |
Supernatant and suspended solids | Aluminum hydroxide adsorption–precipitation | Cumulative cases (36, 0, 140) |
| March–April, 2020 | |||||||
| Saguti, F. et al. (2021)17 (nbiosample= 21, nobs = 21) | Sweden | WWTP influent | Composite | 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant | PS hollow fiber concentrating pipette | Newly hospitalized patients per day (9, 0, 20) |
| February–July, 2020 | Adsorption–elution electropositive cartridges–ultrafiltration | ||||||
| Sherchan, S. P. et al. (2020)18 (nbiosample = 7, nobs = 28) | USA | WWTP influent | Grab and composite | 290![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321 321 |
Supernatant and suspended solids | Adsorption–elution electronegative membrane | Cumulative cases (808, 0, 2534) |
| Jan–April, 2020 | Supernatant | Ultrafiltration | Daily cases (16, 0, 32) | ||||
| Trottier, J. et al. (2020)19 (nbiosample = 7, nobs = 14) | France | WWTP influent | Composite | 470![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Supernatant | Ultrafiltration | Daily cases (1, 0, 2) |
| May–July, 2020 | |||||||
| Westhaus, S. et al. (2021) (nbiosample = 9, nobs = 18) | Germany | WWTP influent | Composite | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 429 429![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
Supernatant | Ultrafiltration | Cumulative cases (123, 72, 220) |
| April 08, 2020 | Active cases (72, 30, 174) | ||||||
Correlations between COVID-19 cases and wastewater SARS-CoV-2 viral RNA levels were reported in six studies. This is confirmed by our analysis. We performed linear regression on each dataset. Six out of eighteen studies detected significant linear correlations between SARS-CoV-2 viral RNA levels and the respective epidemiological indicators in the study (p-value < 0.05, Table 3, Fig. S2–S4†). These six studies were conducted at WWTPs, amongst which three analyzed the solid fraction, and three analyzed the supernatant/filtrate fraction.
Daily new COVID-19 cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants 000 inhabitants |
|||
|---|---|---|---|
| Author | Linear regression | ||
| Slope | R-Squared | p-Value | |
| D'Aoust, P. M. et al. | 0.52 | 0.51 | 1.03 × 10−7 |
| Graham, K. et al. | 196.64 | 0.35 | 3.99 × 10−17 |
| Scherchan, S. P. et al. | 0.03 | 0.17 | 1.44 × 10−1 |
| Peccia, J. et al. | 1994.77 | 0.16 | 2.79 × 10−10 |
| Hata, A. et al. | 0.16 | 0.05 | 3.85 × 10−2 |
| Miyani, B. et al. | −0.20 | 0.01 | 5.10 × 10−1 |
| Haramoto, E. et al. | −4.14 | 0.00 | 7.29 × 10−1 |
| Trottier, J. et al. | 0.19 | 0.00 | 8.54 × 10−1 |
| Nemudryi, A. et al. | −0.02 | 0.00 | 7.65 × 10−1 |
Cumulative COVID-19 cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants 000 inhabitants |
|||
|---|---|---|---|
| Author | Linear regression | ||
| Slope | R-Squared | p-Value | |
| Gonzalez, R. et al. | 0.04 | 0.61 | 4.81 × 10−124 |
| Medema, G. et al. | 14.82 | 0.40 | 1.30 × 10−9 |
| Sherchan, S. P. et al. | 0.00 | 0.09 | 2.87 × 10−1 |
| Haramoto, E. et al. | −4.62 | 0.08 | 9.81 × 10−2 |
| Hata, A. et al. | 0.08 | 0.03 | 1.19 × 10−1 |
| Westhaus, S. et al. | −0.01 | 0.01 | 6.63 × 10−1 |
| Randazzo, W. et al. (b) | 0.45 | 0.01 | 5.79 × 10−1 |
Active COVID-19 cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants 000 inhabitants |
|||
|---|---|---|---|
| Author | Linear regression | ||
| Slope | R-Squared | p-Value | |
| D'Aoust, P. M. et al. | 3.85 | 0.33 | 4.70 × 10−5 |
| Randazzo, W. et al. (a) | 1.61 | 0.11 | 1.19 × 10−1 |
| Westhaus, S. et al. | −0.01 | 0.01 | 7.28 × 10−1 |
Methodological variability was present in all steps of sample collection and analysis procedures (Fig. 2). In terms of sampling locations within a wastewater system, most studies analyzed samples collected at the WWTP (16 studies).8,9,11,12,14,16–21,23,37,38,40,41 A much smaller number of studies sampled at locations in the sewage collection network (two studies)13,39 or in-premise (one study).13 Kitamura, K. et al. examined the SARS-CoV-2 virus in wastewater at both municipal sewage network locations and WWTP influent samples.15 Saguti, F. et al. monitored WWTP influent samples and upstream locations.17 Because case counts for biosamples collected at the sewage network in Saguti, F. et al. were not provided in the publication,17 these biosamples from the upstream location were not included in the meta-analysis. Among the studies sampling at WWTPs, the service population ranged from 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770 to 3.2 million individuals, and covered regions in the Americas (nstudies = 7), Asia (nstudies = 4), Europe (nstudies = 8), and Oceania (nstudies = 1).
770 to 3.2 million individuals, and covered regions in the Americas (nstudies = 7), Asia (nstudies = 4), Europe (nstudies = 8), and Oceania (nstudies = 1).
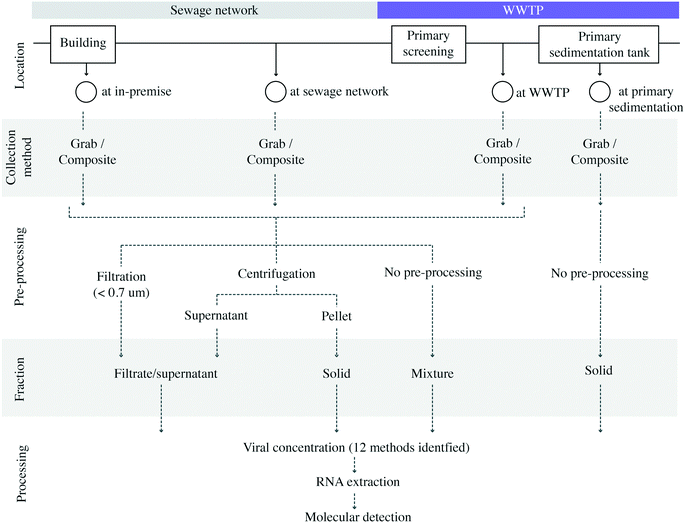 | ||
| Fig. 2 Diagram depicting reported sample collection locations, pre-processing methodologies, and their respective annotations as sampling locations and fractions in this study. | ||
Upon sample collection, studies showed great variability under sample pre-processing conditions, resulting in the enrichment of different wastewater fractions (Fig. 2). Supernatant/filtrate fractions were recovered in 12 studies using centrifugation between 1840 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g,8,9,11,14–19,30,37,40 while two studies retrieved these fractions by filtrating raw wastewater through 0.22 (ref. 13) and 0.7 μm membranes,10 respectively. Mixed supernatant and suspended solid fractions were identified in six studies where liquid wastewater samples were not subjected to any type of pre-processing. Solid fractions were retrieved in one study from influent wastewater by pellet collection after centrifugation at 1840g,15 while the remaining three studies utilizing solid fractions collected sludge samples directly from primary sedimentation tanks.11,21,23 It is important to highlight that a study may pre-process for more than one fraction (Table 2).
000g,8,9,11,14–19,30,37,40 while two studies retrieved these fractions by filtrating raw wastewater through 0.22 (ref. 13) and 0.7 μm membranes,10 respectively. Mixed supernatant and suspended solid fractions were identified in six studies where liquid wastewater samples were not subjected to any type of pre-processing. Solid fractions were retrieved in one study from influent wastewater by pellet collection after centrifugation at 1840g,15 while the remaining three studies utilizing solid fractions collected sludge samples directly from primary sedimentation tanks.11,21,23 It is important to highlight that a study may pre-process for more than one fraction (Table 2).
Once a fraction of choice was generated, a viral concentration step was usually performed prior to RNA extraction. The viral concentration protocols relied on the principles of the molecular weight cutoff achieved through ultrafiltration at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da,8,10,13,15,16,18,19,30,40 the affinity of enveloped viruses to electro-negative membranes, electro-positive membranes, or other adsorbents/flocculants such as PEG, skimmed milk, or aluminum,9,11,14,15,18,20,21,30,37–39,41 or a combination of both mechanisms sequentially.17,21,41 Some protocols did not include a concentration step and performed RNA extraction directly on the solid fraction.11,23 The methodological choices in the concentration step were highly variable, and the twelve different workflows were reported. Reviews on the viral concentration methodology and method evaluation employing surrogates can be found elsewhere.30,31,42–47
000 Da,8,10,13,15,16,18,19,30,40 the affinity of enveloped viruses to electro-negative membranes, electro-positive membranes, or other adsorbents/flocculants such as PEG, skimmed milk, or aluminum,9,11,14,15,18,20,21,30,37–39,41 or a combination of both mechanisms sequentially.17,21,41 Some protocols did not include a concentration step and performed RNA extraction directly on the solid fraction.11,23 The methodological choices in the concentration step were highly variable, and the twelve different workflows were reported. Reviews on the viral concentration methodology and method evaluation employing surrogates can be found elsewhere.30,31,42–47
Notably, the various choices in separation methods result from an underlying assumption of differential enrichment/partitioning of the viral particles within the fractions in a biosample. Therefore, we considered the fractions as subgroups in achieving pooled estimates of SARS-CoV-2 RNA levels in wastewater (3.2).
3.2 Meta-analysis on the percentage of SARS-CoV-2 positive detections from untreated wastewater
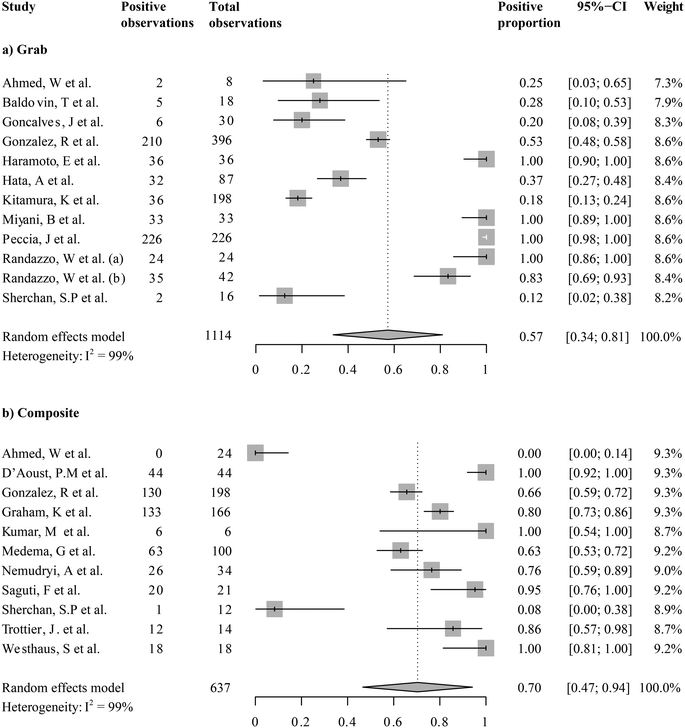
While all the current studies took place during the COVID-19 pandemic, the detections of SARS-CoV-2 from wastewater were not always positive. We first ask, what was the grand mean of positivity of detection among studies taking place in the first year of wastewater-based SARS-CoV-2 monitoring? We examined the overall positivity across 1751 observations in 20 studies, which was 0.68 (95%-CI [0.52; 0.85]). Because the sampling mode (i.e., grab or composite sampling) and fractions for analysis (i.e., supernatants/filtrates, mixed supernatant and suspended solids, and solids only) were expected to introduce variations, we examined the means of the proportion of positive detection by sampling modes (Fig. 3) or fractions analyzed (Fig. 4). Wastewater SARS-CoV-2 measurements in composite sampling mode had a detection rate of 0.70 and a 95%-CI of [0.47; 0.94], whereas those generated from the grab sampling had an average detection rate of 0.57 and a 95%-CI of [0.34; 0.81]. The SARS-CoV-2 viral detection from the composite sampling approach was significantly higher than that from the grab sample mode (one-sided t-test, pBH-adjusted = 5.63 × 10−9). When grouped by the fraction analyzed, the supernatant, mixed supernatant and suspended solids, and solid fractions exhibited positive proportions of 0.53 (95%-CI [0.32; 0.75]), 0.62 (95%-CI [0.12; 1]), and 0.82 (95%-CI [0.43; 1]), respectively. Solids and solid–supernatant mixtures had a significantly higher average positive proportion than the supernatant/filtrate fraction (pBH-adjusted < 2.00 × 10−16 and pBH-adjusted = 2.60 × 10−10 in the pairwise t-test, respectively). Solid analysis exhibited a significantly higher average positive proportion than the solid–supernatant mixtures (pBH-adjusted = 6.50 × 10−8). It should be noted that even within subgroups, high heterogeneity (I2 0.97–0.99) was revealed from the metanalysis. This could be caused by variations in COVID-19 cases or other local variables associated with a study, which will be explored in the regression analysis in section 3.4.3.3 Meta-analysis of SARS-CoV-2 RNA levels in untreated wastewater
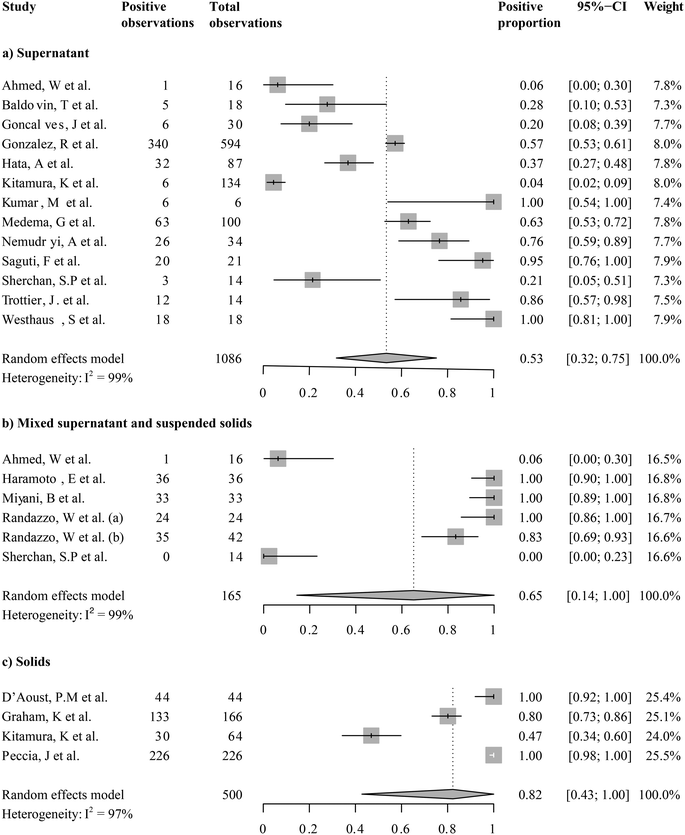
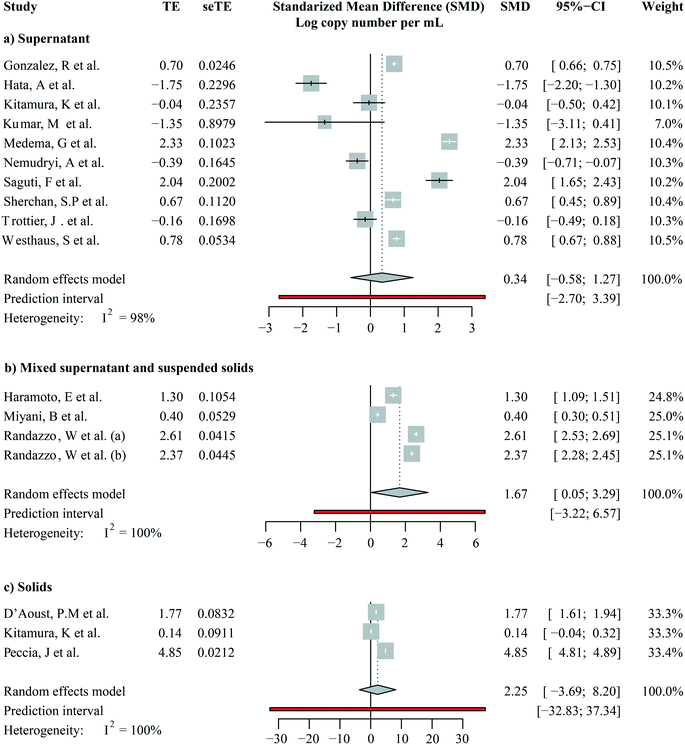
We focused on studies that reported SARS-CoV-2 RNA levels as gene copies per volume to calculate a pooled estimate of SARS-CoV-2 RNA abundance in wastewater. These are seventeen studies including a total of 1508 out of 1674 quantitative observations. Across these studies, the average SARS-CoV-2 RNA abundance was 5244 gene copies per mL (95%-CI [0; 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 432]). We then aggregated studies by the fraction analyzed, i.e., supernatants/filtrates, mixed supernatant and suspended solids, and solids. A forest plot showing the study means, weighted subgroup means, and confidence intervals is shown in Fig. 5. The average viral RNA levels in the wastewater supernatant, mixture, and solids are 50 gene copies per mL (95%-CI [0; 137], nobs = 1009), 181 gene copies per mL (95%-CI [0; 511], nobs = 165), and 30
432]). We then aggregated studies by the fraction analyzed, i.e., supernatants/filtrates, mixed supernatant and suspended solids, and solids. A forest plot showing the study means, weighted subgroup means, and confidence intervals is shown in Fig. 5. The average viral RNA levels in the wastewater supernatant, mixture, and solids are 50 gene copies per mL (95%-CI [0; 137], nobs = 1009), 181 gene copies per mL (95%-CI [0; 511], nobs = 165), and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 456 gene copies per mL (95%-CI [0; 161
456 gene copies per mL (95%-CI [0; 161![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 833], nobs = 334), respectively. The viral RNA levels from solid fractions exhibited significantly higher means than the other two groups (pBH-adjusted < 2.00 × 10−16 for both comparisons), yet the other two groups did not significantly differ (pBH-adjusted > 0.97). This finding suggests that once the viral RNA levels were beyond the detection limits, the difference between analyzed supernatants/filtrates and the mixture was not as strong.
833], nobs = 334), respectively. The viral RNA levels from solid fractions exhibited significantly higher means than the other two groups (pBH-adjusted < 2.00 × 10−16 for both comparisons), yet the other two groups did not significantly differ (pBH-adjusted > 0.97). This finding suggests that once the viral RNA levels were beyond the detection limits, the difference between analyzed supernatants/filtrates and the mixture was not as strong.
Notably, viral RNA levels varied largely even among studies investigating SARS-CoV-2 RNA levels in the same sample fractions, as shown by heterogeneity across studies (I2) higher than 95% in all subgroups (Fig. 5, S5:† cubic root transformed data). We further aggregated the observations by grab/composite sampling and focused on studies that reported WWTP observations alone (Fig. S6 and S7:† cubic root transformed data). The cross-study heterogeneity remained high (I2 >93%) even after data were aggregated in more methodologically homogeneous groups. The observed heterogeneity suggested that pandemic severity, as well as other local variables, may drive the variations in SARS-CoV-2 RNA levels among studies.
3.4 Correlation between SARS-CoV-2 viral RNA levels in wastewater and reported COVID-19 cases
The overall correlation between daily cases and SARS-CoV-2 RNA levels is 0.28 (95%-CI, [0.01; 0.51], Table S3†). The Pearson Rho between the cumulative cases and SARS-CoV-2 RNA levels was 0.29 (95%-CI, [−0.15; 0.73], Table S3†). For both kinds of epidemiological indicators, wastewater-based SARS-CoV-2 viral RNA levels exhibited an overall positive correlation. In composite samples, the Pearson Rho between the viral RNA levels and epidemiological indicators was 0.41 (95%-CI, [−0.08; 0.74]) and 0.53 (95%-CI, [−0.21; 0.88]) for daily and cumulative cases, respectively (Fig. S8†). In grab samples, the Pearson Rho between the viral RNA levels and epidemiological indicators was 0.17 (95%-CI, [−0.12; 0.43]) and 0.20 (95%-CI, [−0.33; 0.63]) for daily and cumulative cases, respectively (Fig. S8†). While composite samples showed stronger correlations, heterogeneity values remained high within subgroups of studies utilizing this sampling methodology (I2 > 81%, Fig. S8†).3.5 Covariates explaining variations in SARS-CoV-2 viral RNA levels in wastewater
We ask, how much of the large heterogeneity in the average copy numbers of SARS-CoV-2 in wastewater can be explained by the sampling mode, fraction analyzed, and COVID-19 case prevalence, respectively? To answer this question, we built univariate models focusing on each covariate, respectively. Studies reporting cumulative cases (nobs = 912, nstudies = 8) and daily cases (nobs = 500, nstudies = 8) were examined separately to ensure consistent within-group case reporting units.First, we built logistic regression models to explain the relationships between positive SARS-CoV-2 detection from sewage and each covariate considered. The models with the sampling mode and fraction analyzed as sole predictors explained 0.6% and 12.4% (Tjur's R-squared) of the total variability in SARS-CoV-2 positive detections, respectively (Table 4). The proportion of variances explained by daily and cumulative cases was 9.3% and 5.9%, respectively (Table 5).
| Univariate models | Binomial (nobs = 1508) | Gaussian (log transformation on RNA levels) (nobs = 936) | ||||
|---|---|---|---|---|---|---|
| Coefficient [95%-CI] | p-Values | Explained variance (R-squared) | Coefficient [95%-CI] | p-Values | Explained variance (R-squared) | |
| Grab_composite | 0.01 | 0.07 | ||||
| Intercept | 0.75 [0.55, 0.95] | 2.1 × 10−13*** | 2.50 [1.99, 2.98] | <2.00 × 10−16*** | ||
| Grab_composite: grab | −0.36 [−0.59, −0.12] | 2.84 × 10−3** | 2.54 [1.94, 3.14] | 3.10 × 10−16*** | ||
| Fraction | 0.12 | 0.56 | ||||
| Intercept | 0.01 [−0.11, 0.13] | 0.87 | 1.51 [1.25, 1.77] | <2.00 × 10−16*** | ||
| Fraction: solid | 2.17 [1.8, 2.56] | <2 × 10−16*** | 7.53 [7.10, 7.96] | <2.00 × 10−16*** | ||
| Fraction: solid–supernatant mixture | 1.27 [0.89, 1.67] | 1.9 × 10−10*** | 2.14 [1.56, 2.72] | 9.70 × 10−13*** | ||
| Model name | Daily case model (nobs = 500, nstudies = 8) | Cumulative case model (nobs = 912, nstudies= 8) | ||||||
|---|---|---|---|---|---|---|---|---|
| Binomial | Gaussian (log transformation on titers) | Binomial | Gaussian (log transformation on titers) | |||||
| Fixed effects | Mixed effects | Fixed effects | Mixed effects | Fixed effects | Mixed effects | Fixed effects | Mixed effects | |
| Fixed effects | b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] | b [95% CI] |
| Intercept | 0.65 [0.33, 0.96] | 3.32 [−1.35, 8.00] | 6.23 [5.63, 6.84] | 1.78 [−1.09, 4.66] | −0.03 [−0.20, 0.13] | −0.06 [−3.95, 3.83] | 1.78 [1.53, 2.04] | 1.05 [−1.25, 3.36] |
| Daily new cases | 0.06 [0.04, 0.09] | 0.11 [0.05, 0.16] | 0.01 [0.001, 0.02] | 0.01 [0.003, 0.009] | — | — | — | — |
| Cumulative cases | — | — | — | — | 0.001 [0.001, 0.002] | 0.005 [0.004, 0.007] | 0.0007 0.0001, 0.0013] | 0.002 [0.001, 0.002] |
| Random effects | ||||||||
|---|---|---|---|---|---|---|---|---|
| Study_ID (variance) | — | 23.15 | — | 17.12 | — | 29.75 | — | 10.93 |
| Adjusted R2 | 0.09 | — | 0.01 | — | 0.06 | — | 0.01 | — |
| Marginal R2 | — | 0.54 | — | 0.01 | — | 0.08 | — | 0.02 |
| Conditional R2 | — | 0.94 | — | 0.93 | — | 0.91 | — | 0.90 |
| AIC | 422 | 200 | 2579 | 1406 | 1214 | 1012 | 2381 | 1692 |
| BIC | 431 | 213 | 2591 | 1422 | 1224 | 1027 | 2394 | 1709 |
| Loglik | −209 | −97 | −1287 | −699 | −605 | −503 | −1188 | −842 |
| Random effects (p-values) | — | <2.2 × 10−16*** | — | <2.2 × 10−16*** | — | <2.2 × 10−16*** | — | <2.2 × 10−16*** |
Next, we built linear models to examine the relationships between logarithmic transformed viral RNA levels and each covariate. The variance in RNA levels explained by the sampling mode and fraction analyzed was 6.9% and a notable 56.0%, respectively, whereas the variance explained by daily and cumulative cases was 0.9% and 0.8%, respectively. In all these models, the roles of methodological variables and epidemiological indicators were significant (p < 0.05, Tables 4 and 5). The daily or cumulative cases and sampling mode explained comparable proportions of variances. Notably, the fraction analyzed explained dramatically higher variance in viral RNA levels than any other variables.
3.6 Slope coefficients in generalized linear models
Successful detection of the virus from wastewater is fundamental to WBE; our generalized linear models on positive detections can provide quantitative insights into the magnitude at which changes in each variable increase the chance of the positive detections (Table 4 binomial family models and Table 5 binomial fixed-effects model). From our models, the odds of positive detection decrease by a factor of 1.43 (95%-CI [1.81; 1.13]) when utilizing grab sampling in contrast to composite sampling. The odds of positive detection increase by a factor of 8.16 (95%-CI [6.08; 12.92]) from solid fractions in contrast to supernatants/filtrates; they increase by a factor of 3.52 (95%-CI [2.43; 5.30]) from solid–supernatant mixtures in contrast to supernatants/filtrates. With an increase in active case prevalence of 10 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants, the odds of positive detection increase by a factor of 1.06 units (95%-CI [1.04; 1.09]); with an increase in cumulative cases of 10 per 100
000 inhabitants, the odds of positive detection increase by a factor of 1.06 units (95%-CI [1.04; 1.09]); with an increase in cumulative cases of 10 per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants, they increase by a factor of 1.02 (95%-CI [1.01; 1.03]).
000 inhabitants, they increase by a factor of 1.02 (95%-CI [1.01; 1.03]).
3.7 Mixed-effects model helps account for variation by studies
While many applications of WBE rely on positive correlations between SARS-CoV-2 RNA levels in wastewater and disease prevalence, larger or comparable variability was explained by methodological covariates than the reported case prevalence in our models (Tables 4 and 5). While documenting methodological covariates in WBE studies is crucial, learning about which variables are of importance in WBE studies is an ongoing process. To address the need of building explanatory or predictive models in WBE, we considered a mixed-effects framework for modeling SARS-CoV-2 viral RNA levels from multiple studies. Here, we treat studies as a collective source of variance. We hypothesize that in addition to the role of cases as a source of fixed effects on the wastewater measurements, each study presents a source of a study-specific intercept. We tested for the significance of random effects. For both, positivity or viral RNA levels from daily or cumulative cases, the random effects from the studies were significant (p-value < 2.00 × 10−16, Table 5). Mixed-effects models also showed a lower AIC or BIC than the corresponding fixed-effects models, suggesting better fits to the data.For a mixed-effects model, we examined both the marginal R-squared, which is the proportion of variance explained by the fixed effects alone (daily or cumulative cases), and the conditional R-squared, which describes the proportion of variance explained by both the fixed and random factors (cases and the study identities respectively). Notably, mixed models exhibited conditional R-squared close to or over 0.9 for both positivity and viral RNA levels models reporting daily new cases or cumulative cases (Table 5). Thus, simultaneously considering variability across studies greatly improved our ability to explain the variation in wastewater SARS-CoV-2 measurements.
4. Discussion
Sampling modes and wastewater fractions had strong influences on the pooled means in proportions of positive detection and SARS-CoV-2 RNA levels. The sampling mode explained 0.6% in the variance of positive detections. The composite sampling mode had a higher detection rate than grab sampling, as seen from an average detection rate of 0.70 (95%-CI, [0.47; 0.94]) and 0.56 (95%-CI, [0.32; 0.79], Fig. 3), respectively. This observation is in agreement with the previous literature that showed improved SARS-CoV-2 detection in composite wastewater samples.48 George, A. et al. (2022) showed that in liquid fractions composite sampling outperformed grab sampling on smaller geographical scales, such as neighborhood, city block, and building scales. While none of the primary research studies included passive sampling, this sampling strategy is in development49 and worthy of consideration for future meta-analyses.Supernatant/filtrate, solid–supernatant mixture, and solid fractions increased by average detection rates of 0.53 (95%-CI [0.32; 0.75]), 0.62 (95%-CI [0.12; 1]), and 0.82 (95%-CI [0.43; 1]), respectively (Fig. 4). The fraction analyzed explained 12.4% of the variance in the proportions of positive detection and 56% of the variance in RNA levels. Solid fractions exhibited SARS-CoV-2 viral RNA levels that were orders of magnitude higher than supernatants/filtrates and solid–supernatant mixtures. This observation is in agreement with the previous literature showing enrichment of the SARS-CoV-2 genetic material in wastewater solids (i.e., primary settled solids).50 Given the higher proportion of SARS-CoV-2 viral RNA in solid fractions, workflows utilizing wastewater solids may be useful to track SARS-CoV-2 when infections remain at low levels in the sewershed (i.e., periods between peaks of infection, early warning detection, etc.). The large variance in viral RNA levels explained by the fraction analyzed and the large magnitudes in regression coefficients suggest that standardizing the fraction analyzed needs to be prioritized when researchers would like to design monitoring efforts across multiple labs.47 The overall detection rate and those in subgroups of any sewage fraction were below one, suggesting a need for tools to maximize the chance of SARS-CoV-2 detection from sewage samples.
In our meta-analysis, large heterogeneity was detected in all effect sizes investigated (i.e., proportions of positive detections, viral RNA levels, and Pearson Rho between RNA levels and daily or cumulative cases, Fig. 3–5, Table S3†). We hypothesize that the unexplained variations in SARS-CoV-2 RNA levels detected in wastewater can be affected by study-level factors, such as COVID-19 prevalence, lags in epidemiological data reporting,51 methodological choices,46 and differences in the wastewater collection system design. Our meta-analysis found that metadata about the collection system, such as per capita water consumption, relative contributions of domestic vs. commercial/industrial water, or sewage travel times (i.e., residence times), are currently rare. These collection system-level variables can affect the dilution of fecal materials and the genetic decay of the viral signal.31,52–54 To illustrate, domestic water consumption can vary significantly in different areas, a person in the city of Berlin generates on average 135 L of wastewater per day,55 while a person in Qatar generates on average 500 L of wastewater per day.56 Thus, the dilution of the fecal matter may vary largely in different wastewater systems. Another aspect is combined sewage in comparison to sanitary sewage. Rainfall can affect the dilution of fecal matter in a combined sewage system through stormwater run-off,57,58 while not so in a sanitary sewage system. Even among sanitary sewage, the contribution of domestic waste can vary by system, ranging from as low as 30% of the total wastewater discharge to as high as near-complete dominance.59 These design differences could lead to variations in SARS-CoV-2 RNA levels even in systems where active viral shedders were identical.
Because water usage and system design characteristics can affect SARS-CoV-2 virus measurements at the wastewater treatment plant, more detailed metadata reporting regarding the wastewater collection system is needed to better explain variations across sites. McClary-Gutierrez, J. S. et al. compiled a list of minimum reporting data for WBE applications for COVID-19,60 which can support more consistent metadata reporting across studies and facilitate the synthesis of results. Lately, it was proposed that wastewater can be viewed as an independent indicator of true prevalence, as epidemiological indicators from current reporting can be affected by under-reporting.61 Therefore, methods and tools to investigate the wastewater metagenome and derive system-level data, or bridge wastewater-based measurements to prevalence, deserve more attention.62
To address the dilution of fecal matter by various wastewater streams, normalization of SARS-CoV-2 viral RNA levels by fecal strength indicators has been performed in some studies. These propose dividing the SARS-CoV-2 viral RNA levels by the copy numbers of pepper mild mottle virus (PMMoV),21,58,63 a diet-associated RNA virus commonly found in human feces.64 Among the qualified studies included in this meta-analysis, only one study utilized normalization by PMMoV,21 thus a meta-analysis on the effect of PMMoV normalization on correlations between wastewater SARS-CoV-2 measurements and epidemiological indicators was not included in this study. The effects of normalization techniques on the performance of regression models can be a topic of future interest in meta-analysis efforts when studies employing such techniques become more abundant.
It should be noted that heterogeneity in viral RNA levels and correlations observed here may not be fully explained by recovery efficiencies of viruses from wastewater samples during viral concentration workflows. To illustrate this complexity, we discuss two studies where recovery efficiencies were reported. In one study, an average viral RNA level of 881 ± 633 marker copies per mL was detected when COVID-19 prevalence in the associated area was between 10 and 80 cumulative cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants;8 in another study, an average viral RNA level of 1.9 ± 6.0 marker copies per mL was reported within the same range of COVID-19 prevalence (10–80 cumulative cases per 100
000 inhabitants;8 in another study, an average viral RNA level of 1.9 ± 6.0 marker copies per mL was reported within the same range of COVID-19 prevalence (10–80 cumulative cases per 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 inhabitants).9 After adjusting the viral RNA levels by reported recovery efficiencies (73 and 7.7%, respectively), the adjusted copy numbers (1206 and 27 marker copies per mL, respectively) still vary by two orders of magnitude.
000 inhabitants).9 After adjusting the viral RNA levels by reported recovery efficiencies (73 and 7.7%, respectively), the adjusted copy numbers (1206 and 27 marker copies per mL, respectively) still vary by two orders of magnitude.
While the field's ability to quantify the effects of methodological variables and collection systems is an important ongoing research topic,46,47,65 mixed-effects models treating “studies” as a source of random effects can be considered useful for performing inference and prediction. Mixed-effects models handle a wide range of scenarios where observations have been sampled in a hierarchical structure rather than completely independently. In this study, treating studies as a source of random effects on intercepts profoundly improved the quality of the model, as seen in improved AIC and BIC compared to the respective fixed-effects models (Table 5). The final models reached conditional R-squared values above 0.9. The mixed-effects approach provides an alternative for researchers to leverage existing data from studies conducted elsewhere to build models useful for explaining variations in local observations.
5. Study strengths and limitations
In summary, we synthesized the available evidence on SARS-CoV-2 detection and viral RNA levels in wastewater reported during the first year of the COVID-19 pandemic. The combined detection rate across studies was 67% (95%-CI, [0.56; 0.79]). Despite the large heterogeneity in SARS-CoV-2 RNA levels among methodologically similar studies (i.e., 93% < I2< 100%), SARS-CoV-2 abundance in wastewater exhibited strong correlations with epidemiological indicators. These results reinforce that wastewater is a favorable data source to track COVID-19 dynamics in a community.Our study had several limitations. The most notable is the large amount of unexplained heterogeneity in positive detection, SARS-CoV-2 RNA levels, and Pearson correlations across studies. This is likely attributable to variability in methodological differences in SARS-CoV-2 virus measurements, wastewater-system characteristics, ways the epidemiological data were collected and reported as well as different COVID-19 incidence at the time the studies were conducted (e.g., COVID-19 waves and case fluctuations). Thus, we employed mixed-effects models to make inferences about the correlation between epidemiological indicators and viral detection/RNA levels, treating study-level variations as a source of random effects.
This systematic review and meta-analysis were performed using the Web of Science core collection focusing on the English-language literature. Other indexes such as PubMed, Medline, and Scopus are worth examining in future research. Sources such as Europe PMC which included preprint servers will increase data inclusion. As more data become available, future meta-analysis focusing on the collection of upstream sewage streams and comparisons of SARS-CoV-2 detection sensitivity between qPCR and dPCR may become possible. The present study may be used as a framework for future studies analyzing larger datasets.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the N.S.F. CAREER Award 2047470 (F. L.) and Washington University Faculty Start-up Fund (F. L.).References
- D. A. Larsen and K. R. Wigginton, Nat. Biotechnol., 2020, 38, 1151–1153 CrossRef CAS PubMed.
- Y. Chen, L. Chen, Q. Deng, G. Zhang, K. Wu, L. Ni, Y. Yang, B. Liu, W. Wang and C. Wei, J. Med. Virol., 2020, 92, 833–840 CrossRef CAS PubMed.
- F. Xiao, J. Sun, Y. Xu, F. Li, X. Huang, H. Li, J. Zhao, J. Huang and J. Zhao, Emerging Infect. Dis., 2020, 26, 1920 CrossRef CAS PubMed.
- M. Guo, W. Tao, R. A. Flavell and S. Zhu, Nat. Rev. Gastroenterol. Hepatol., 2021, 18, 269–283 CrossRef CAS PubMed.
- M. J. Binnicker, J. Clin. Microbiol., 2020, 58, e01695-20 CrossRef PubMed.
- M. Murakami, A. Hata, R. Honda and T. Watanabe, Environ. Sci. Technol., 2020, 54, 5311–5311 CrossRef CAS PubMed.
- T. Hovi, L. M. Shulman, H. Van Der Avoort, J. Deshpande, M. Roivainen and E. M. De Gourville, Epidemiol. Infect., 2012, 140, 1–13 CrossRef CAS PubMed.
- G. Medema, L. Heijnen, G. Elsinga, R. Italiaander and A. Brouwer, Environ. Sci. Technol. Lett., 2020, 7, 511–516 CrossRef CAS.
- R. Gonzalez, K. Curtis, A. Bivins, K. Bibby, M. H. Weir, K. Yetka, H. Thompson, D. Keeling, J. Mitchell and D. Gonzalez, Water Res., 2020, 186, 116296 CrossRef CAS PubMed.
- J. Gonçalves, T. Koritnik, V. Mioč, M. Trkov, M. Bolješič, N. Berginc, K. Prosenc, T. Kotar and M. Paragi, Sci. Total Environ., 2021, 755, 143226 CrossRef PubMed.
- K. E. Graham, S. K. Loeb, M. K. Wolfe, D. Catoe, N. Sinnott-Armstrong, S. Kim, K. M. Yamahara, L. M. Sassoubre, L. M. Mendoza Grijalva and L. Roldan-Hernandez, Environ. Sci. Technol., 2021,(55), 488–489 CrossRef CAS PubMed.
- W. Ahmed, N. Angel, J. Edson, K. Bibby, A. Bivins, J. W. O'Brien, P. M. Choi, M. Kitajima, S. L. Simpson and J. Li, Sci. Total Environ., 2020, 728, 138764 CrossRef CAS PubMed.
- T. Baldovin, I. Amoruso, M. Fonzo, A. Buja, V. Baldo, S. Cocchio and C. Bertoncello, Sci. Total Environ., 2021, 760, 143329 CrossRef CAS PubMed.
- A. Hata, H. Hara-Yamamura, Y. Meuchi, S. Imai and R. Honda, Sci. Total Environ., 2021, 758, 143578 CrossRef CAS PubMed.
- K. Kitamura, K. Sadamasu, M. Muramatsu and H. Yoshida, Sci. Total Environ., 2021, 763, 144587 CrossRef CAS PubMed.
- A. Nemudryi, A. Nemudraia, T. Wiegand, K. Surya, M. Buyukyoruk, C. Cicha, K. K. Vanderwood, R. Wilkinson and B. Wiedenheft, Cell Rep. Med., 2020, 1, 100098 CrossRef PubMed.
- F. Saguti, E. Magnil, L. Enache, M. P. Churqui, A. Johansson, D. Lumley, F. Davidsson, L. Dotevall, A. Mattsson and E. Trybala, Water Res., 2021, 189, 116620 CrossRef CAS PubMed.
- S. P. Sherchan, S. Shahin, L. M. Ward, S. Tandukar, T. G. Aw, B. Schmitz, W. Ahmed and M. Kitajima, Sci. Total Environ., 2020, 743, 140621 CrossRef CAS PubMed.
- J. Trottier, R. Darques, N. A. Mouheb, E. Partiot, W. Bakhache, M. S. Deffieu and R. Gaudin, One Health, 2020, 10, 100157 CrossRef PubMed.
- W. Randazzo, P. Truchado, E. Cuevas-Ferrando, P. Simón, A. Allende and G. Sánchez, Water Res., 2020, 181, 115942 CrossRef CAS PubMed.
- P. M. D'Aoust, E. Mercier, D. Montpetit, J.-J. Jia, I. Alexandrov, N. Neault, A. T. Baig, J. Mayne, X. Zhang and T. Alain, Water Res., 2021, 188, 116560 CrossRef PubMed.
- P. M. D'Aoust, T. E. Graber, E. Mercier, D. Montpetit, I. Alexandrov, N. Neault, A. T. Baig, J. Mayne, X. Zhang and T. Alain, Sci. Total Environ., 2021, 770, 145319 CrossRef PubMed.
- J. Peccia, A. Zulli, D. E. Brackney, N. D. Grubaugh, E. H. Kaplan, A. Casanovas-Massana, A. I. Ko, A. A. Malik, D. Wang and M. Wang, Nat. Biotechnol., 2020, 38, 1164–1167 CrossRef CAS PubMed.
- J. Gurevitch, J. Koricheva, S. Nakagawa and G. Stewart, Nature, 2018, 555, 175–182 CrossRef CAS PubMed.
- M. H. Murad and V. M. Montori, Jama, 2013, 309, 2217–2218 CrossRef PubMed.
- A. H. Linden and J. Hönekopp, Perspect. Psychol. Sci., 2021, 16, 358–376 CrossRef PubMed.
- M. J. Page, J. E. McKenzie, P. M. Bossuyt, I. Boutron, T. C. Hoffmann, C. D. Mulrow, L. Shamseer, J. M. Tetzlaff, E. A. Akl and S. E. Brennan, BMJ, 2021,(372), n71 CrossRef PubMed.
- M. Harrer, P. Cuijpers, T. A. Furukawa and D. D. Ebert, Doing Meta-Analysis with R: A Hands-On Guide, Chapman and Hall/CRC, 2021 Search PubMed.
- X. Bertels, P. Demeyer, S. Van den Bogaert, T. Boogaerts, A. L. van Nuijs, P. Delputte and L. Lahousse, Sci. Total Environ., 2022, 153290 CrossRef CAS PubMed.
- W. Ahmed, P. M. Bertsch, A. Bivins, K. Bibby, K. Farkas, A. Gathercole, E. Haramoto, P. Gyawali, A. Korajkic, B. R. McMinn, J. F. Mueller, S. L. Simpson, W. J. M. Smith, E. M. Symonds, K. V. Thomas, R. Verhagen and M. Kitajima, Sci. Total Environ., 2020, 739, 139960 CrossRef CAS PubMed.
- A. I. Silverman and A. B. Boehm, Environ. Sci. Technol. Lett., 2020, 7, 544–553 CrossRef CAS.
- K. Sidik and J. N. Jonkman, J. R. Stat. Soc., C: Appl. Stat., 2005, 54, 367–384 CrossRef.
- M. Borenstein, L. V. Hedges, J. P. Higgins and H. R. Rothstein, Res. Synth. Methods, 2010, 1, 97–111 CrossRef PubMed.
- D. Bates, M. Mächler, B. Bolker and S. Walker, ArXiv Prepr. ArXiv14065823, 2014.
- R Core Team, Vienna R Core Team.
- K. Barton, Httpr-Forge R-Proj. Orgprojectsmumin.
- M. Kumar, A. K. Patel, A. V. Shah, J. Raval, N. Rajpara, M. Joshi and C. G. Joshi, Sci. Total Environ., 2020, 746, 141326 CrossRef CAS PubMed.
- W. Randazzo, E. Cuevas-Ferrando, R. Sanjuán, P. Domingo-Calap and G. Sánchez, Int. J. Hyg. Environ. Health, 2020, 230, 113621 CrossRef CAS PubMed.
- B. Miyani, X. Fonoll, J. Norton, A. Mehrotra and I. Xagoraraki, J. Environ. Eng., 2020, 146, 06020004 CrossRef CAS.
- S. Westhaus, F.-A. Weber, S. Schiwy, V. Linnemann, M. Brinkmann, M. Widera, C. Greve, A. Janke, H. Hollert and T. Wintgens, Sci. Total Environ., 2021, 751, 141750 CrossRef CAS PubMed.
- E. Haramoto, B. Malla, O. Thakali and M. Kitajima, Sci. Total Environ., 2020, 737, 140405 CrossRef CAS PubMed.
- P. A. Barril, L. A. Pianciola, M. Mazzeo, M. J. Ousset, M. V. Jaureguiberry, M. Alessandrello, G. Sánchez and J. M. Oteiza, Sci. Total Environ., 2021, 756, 144105 CrossRef CAS PubMed.
- S. E. Philo, E. K. Keim, R. Swanstrom, A. Q. W. Ong, E. A. Burnor, A. L. Kossik, J. C. Harrison, B. A. Demeke, N. A. Zhou, N. K. Beck, J. H. Shirai and J. S. Meschke, Sci. Total Environ., 2021, 760, 144215 CrossRef CAS PubMed.
- G. La Rosa, L. Bonadonna, L. Lucentini, S. Kenmoe and E. Suffredini, Water Res., 2020, 179, 115899 CrossRef CAS PubMed.
- S. Bofill-Mas and M. Rusiñol, Curr. Opin. Environ. Sci. Health, 2020, 16, 7–13 CrossRef.
- B. M. Pecson, E. Darby, C. N. Haas, Y. M. Amha, M. Bartolo, R. Danielson, Y. Dearborn, G. Di Giovanni, C. Ferguson and S. Fevig, Environ. Sci.: Water Res. Technol., 2021, 7, 504–520 RSC.
- A. H. Chik, M. B. Glier, M. Servos, C. S. Mangat, X.-L. Pang, Y. Qiu, P. M. D'Aoust, J.-B. Burnet, R. Delatolla and S. Dorner, J. Environ. Sci., 2021, 107, 218–229 CrossRef CAS PubMed.
- A. D. George, D. Kaya, B. A. Layton, K. Bailey, S. Mansell, C. Kelly, K. J. Williamson and T. S. Radniecki, Environ. Sci. Technol. Lett., 2022, 9, 160–165 CrossRef CAS.
- M. Rafiee, S. Isazadeh, A. Mohseni-Bandpei, S. R. Mohebbi, M. Jahangiri-Rad, A. Eslami, H. Dabiri, K. Roostaei, M. Tanhaei and F. Amereh, Sci. Total Environ., 2021, 790, 148205 CrossRef CAS PubMed.
- S. Kim, L. C. Kennedy, M. K. Wolfe, C. S. Criddle, D. H. Duong, A. Topol, B. J. White, R. S. Kantor, K. L. Nelson and J. A. Steele, Environ. Sci.: Water Res. Technol., 2022,(8), 757–770 RSC.
- K. Bibby, A. Bivins, Z. Wu and D. North, Water Res., 2021, 117438 CrossRef CAS PubMed.
- P. Foladori, F. Cutrupi, N. Segata, S. Manara, F. Pinto, F. Malpei, L. Bruni and G. La Rosa, Sci. Total Environ., 2020, 743, 140444 CrossRef CAS PubMed.
- W. Ahmed, P. M. Bertsch, K. Bibby, E. Haramoto, J. Hewitt, F. Huygens, P. Gyawali, A. Korajkic, S. Riddell and S. P. Sherchan, Environ. Res., 2020, 191, 110092 CrossRef CAS PubMed.
- P.-Y. Hong, A. T. Rachmadi, D. Mantilla-Calderon, M. Alkahtani, Y. M. Bashawri, H. Al Qarni, K. M. O'Reilly and J. Zhou, Environ. Res., 2021, 195, 110748 CrossRef CAS PubMed.
- Berlin*, Berliner Wasserbetriebe - Water Usage at Home, https://www.bwb.de/en/2266.php, (accessed August 5, 2021).
- H. M. Baalousha and O. K. Ouda, Arabian J. Geosci., 2017, 10, 1–12 CrossRef.
- J. Tibbetts, Combined sewer systems: down, dirty, and out of date, National Institute of Environmental Health Sciences, 2005 Search PubMed.
- F. Wu, J. Zhang, A. Xiao, X. Gu, W. L. Lee, F. Armas, K. Kauffman, W. Hanage, M. Matus and N. Ghaeli, mSystems, 2020, 5, e00614-20 CrossRef PubMed.
- J. Schilling and J. Tränckner, Water, 2020, 12, 628 CrossRef.
- J. S. McClary-Gutierrez, Z. T. Aanderud, M. Al-Faliti, C. Duvallet, R. Gonzalez, J. Guzman, R. H. Holm, M. A. Jahne, R. S. Kantor and P. Katsivelis, Environ. Sci.: Water Res. Technol., 2021, 7, 1545–1551 RSC.
- S. W. Olesen, M. Imakaev and C. Duvallet, Water Res., 2021, 202, 117433 CrossRef CAS PubMed.
- L. Zhang, L. Chen, X. Yu, C. Duvallet, S. Isazadeh, C. Dai, S. Park, K. Frois-Moniz, F. Duarte, C. Ratti, E. J. Alm and F. Ling, Microbial Species Abundance Distributions Guide Human Population Size Estimation from Sewage Microbiomes, bioRxiv, 2022, DOI:10.1101/2020.12.15.390716.
- S. Feng, A. Roguet, J. S. McClary-Gutierrez, R. J. Newton, N. Kloczko, J. G. Meiman and S. L. McLellan, ACS ES&T Water, 2021, 1, 1955–1965 Search PubMed.
- M. Kitajima, H. P. Sassi and J. R. Torrey, npj Clean Water, 2018, 1, 1–9 CrossRef.
- A. Bivins, D. North, A. Ahmad, W. Ahmed, E. Alm, F. Been, P. Bhattacharya, L. Bijlsma, A. B. Boehm and J. Brown, Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19, ACS Publications, 2020 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ew00084a |
| This journal is © The Royal Society of Chemistry 2022 |