Open Access Article
Open Access ArticleCsAKT1 is a key gene for the CeO2 nanoparticle's improved cucumber salt tolerance: a validation from CRISPR-Cas9 lines†
Yuquan
Peng
a,
Linlin
Chen
b,
Lan
Zhu
b,
Lvjun
Cui
a,
Li
Yang
a,
Honghong
Wu
 *bcd and
Zhilong
Bie
*a
*bcd and
Zhilong
Bie
*a
aKey Laboratory of Horticultural Plant Biology, Ministry of Education/College of Horticulture and Forestry Sciences, Huazhong Agricultural University, Wuhan, China 430070. E-mail: biezl@mail.hzau.edu.cn
bMOA Key Laboratory of Crop Ecophysiology and Farming System in the Middle Reaches of the Yangtze River, College of Plant Science & Technology, Huazhong Agricultural University, Wuhan, China 430070. E-mail: honghong.wu@mail.hzau.edu.cn
cHubei Hongshan Laboratory, Wuhan, China 430070
dCollege of Agronomy and Biotechnology, China Agricultural University, Beijing 100083, China
First published on 24th October 2022
Abstract
Salinity is one of the main factors limiting the crop growth and yield. The application of nanomaterials such as cerium oxide nanoparticles (nanoceria) improves salt tolerance in many plant species. The known mechanisms of the nano-improved crop salt tolerance mostly occur at the physiological and hormonal levels but not at the gene level. The lack of proper methods to precisely investigate the key genes involved in the nano-improved crop salt tolerance is one of the main reasons for this. We found that both the leaf and root applications of PNC (polyacrylic acid-coated nanoceria) improved the cucumber salt tolerance, showing higher shoot and root dry weight and better photosynthetic performance than the control plants. Under salinity stress, PNC-treated cucumber plants showed a significant lower malondialdehyde (MDA) content of leaf (46.7% and 13.3% for leaf and root application of PNC, respectively) and root (31.4% and 19.9% for leaf and root application of PNC, respectively) and reactive oxygen species (ROS) levels such as the H2O2 content of leaf (46.6% and 22.0% for leaf and root application of PNC, respectively) and root (50.0% and 29.9% for leaf and root application of PNC, respectively). Further experiments showed that under the salinity stress, compared with control plants, PNC-treated cucumber plants had significantly higher leaf (216.1% and 59.5% for leaf and root application of PNC, respectively) and root (148.2% and 71.8% for leaf and root application of PNC, respectively) K+ content. The RNA seq showed that under the salinity stress, modulation on AKT1 is more responsible for the PNC improved K+ maintenance in cucumber. Further, the CsAKT1 depletion experiment (CRISPR-Cas9 lines) confirmed that under salinity stress, CsAKT1 plays an important role in the potassium uptake in cucumber plants treated with PNC. Our results showed that foliar PNC delivery enabled stronger cucumber salt tolerance than the root application associated with a better maintained K+/Na+ ratio. CRISPR-Cas9 lines further confirmed that CsAKT1 is a key gene involved in the PNC-improved cucumber salt tolerance. Our work not only adds more knowledge to the mechanisms underlying nano-improved plant salt tolerance but also suggests a CRISPR-Cas9 approach to study the key genes involved in it.
Environmental significanceSalinity is a major environmental stress limiting the crop growth and yield. The use of nanomaterials to improve the plant salt tolerance is an emerging approach to facilitate the sustainable development of agriculture. The cerium oxide nanoparticle (nanoceria)-improved plant salt tolerance was widely reported in many species, such as rice, rapeseed, cotton, and Arabidopsis. The omics technique helped to identify the up- or down-regulated genes in nanoceria treated plants under salinity. However, our knowledge about the key genes (identified and then validated) involved in nanoceria-improved plant salt tolerance is insufficient. Herein, we created CRISPR/Cas9 lines and validated that CsAKT1 is a key gene responsible for nanoceria-improved cucumber salt tolerance. Our work not only showed that CsAKT1 increased the leaf K+ content and thus helped to maintain the K+/Na+ ratio in nanoceria treated plants than control under salinity but also suggests that CRISPR-Cas9 is an efficient approach to study key genes involved in the nanomaterial-improved plant salt tolerance. |
1. Introduction
The global population is expected to rise to 9.3 billion by 2050, which is a big challenge for the food supply.1 Due to climate change, the abuse of fertilizers and pesticides and irrigation, land salinization has intensified. More than 1.0 × 107 km2 of land around the world is affected by salt, resulting in abundant crop yield.2–4 Therefore, improving plant salt tolerance is important for securing the food supply. To address food security issues in the background of increased soil salinization, nano-improved salt stress tolerance for plants could be an approach.5,6Nano-improved salt tolerance has been reported in many crop species, including rice,7 wheat,8 barley,9 potatoes,10,11 rapeseed,12–14 cotton,15,16 and cucumber.17–20 For example, cerium oxide nanoparticles (nanoceria) are potent ROS (reactive oxygen species) scavengers. The formed oxygen deficit due to the co-existence of Ce3+ and Ce4+ on the surface of nanoceria can scavenge H2O2 (hydrogen peroxide), O2˙− (superoxide anion), and OH˙ (hydroxyl radical) as non-radical counterparts.21–23 The known mechanisms behind nanoceria-improved plant salt tolerance are mainly associated with maintaining ROS homeostasis to reduce oxidative damage, an improvement in K+ retention and Na+ exclusion to maintain the K+/Na+ ratio, an increase in gas signaling molecules such as nitric oxide, and modulation at the plant hormone level and α-amylase activities.7,12,13,16,24,25 However, most of the above-mentioned studies are conducted at physiological, metabolic, and hormonal levels. Our knowledge about the insightful understanding of nano-improved plant salt tolerance at the transcription level is insufficient. The events that happened in nano-improved plant salt tolerance at the transcriptional level might be much earlier than those at physiological, metabolic, and hormonal levels. The RNA seq and qPCR (quantitative real-time PCR) results can point out the modulation of nanomaterials on gene expression levels in plants under salinity stress.7,13,15,16,20,26,27 While, our knowledge about the validated key genes which are responsible for nanoceria-improved plant salt tolerance is still limited.
Under salinity stress, over-accumulated Na+ not only inhibits K+ absorption in plants but also competes with K+ for the binding sites of enzymes, which specifically requires K+ as a co-factor.28,29 Therefore, the ability to maintain a high K+/Na+ ratio in the cytoplasm under salt stress is a common index for plant salt tolerance.16,30 K+ is an essential nutrient for the plant growth. It involves many physiological processes such as photosynthesis, protein synthesis, osmotic regulation, and the control of cell membrane polarization.31 Thus, maintaining K+ homeostasis is important for plant salt tolerance. To maintain K+ homeostasis in the cell cytosol, mainly two routes are involved in increasing the K+ influx from the growth medium to cytosol and reducing the K+ efflux from the cytosol to the apoplast. The known genes for controlling K+ influx are AKT (K+ transporter) and HAK (high-affinity K+ transporter) families.32–34GORK (guard cell outwardly rectifying K+ channel) and NSCC (non-selective cation channels) channels are known to play an important role in the process of K+ efflux under salinity stress.32–34 Further, our knowledge about how nanoparticles affect these genes and which gene might be the most responsive for nano-improved plant salt tolerance is still insufficient.
In this study, poly(acrylic) acid-coated nanoceria (PNC) were synthesized and used. Its role in maintaining K+/Na+ homeostasis to improve cucumber salt tolerance was investigated. Furthermore, unlike the RNA-seq data obtained at 0 h salt stress in our previous study,20 in this study, we performed RNA-seq to investigate the changes in the expressions level of K+ transport-related genes in nanoceria-treated cucumber plants with 7 days' salinity stress. CRISPR/Cas9 lines were created to verify the role of the K+ transport-related gene with the most significant change of the expression level in nanoceria-improved cucumber salt tolerance. Overall, we found that CsAKT1 is a key gene responsible for nanoceria-improved cucumber salt tolerance.
2. Materials and methods
2.1. Plant materials
This study used the salt-sensitive cucumber (Cucumis sativus L.) variety “Jin Chun No. 4” from Tianjin Kerun Agricultural Science and Technology Co., LTD. Cucumber seeds were sterilized according to earlier investigations by soaking in clean water at 25 °C for three hours followed by at 55 °C for 15 minutes.35 Then, seeds were added to Petri dishes (10 cm in diameter) that had three layers of filter papers that had been moistened with water. For seed germination, Petri dishes containing seeds were placed in an incubator set at 28 °C. The seeds were sown in the sponge and grown in containers with adequate aeration in 1/2 Hoagland nutrient solution. The seedlings were then transferred to a hydroponic cup with the following dimensions: mouth diameter: 90 mm, a bottom diameter: 57 mm, height: 135 mm, and 400 mL 1/2 modified Hoagland nutritional solution. The mixture contains 1 mM MgSO4·7H2O, 4 mM CaCl2, 10 mM KNO3, 0.5 mM Ca(H2PO4)2·H2O, and 74.93 mg L−1 coolaber trace element solids (DZPM0059-500G) (pH 6.8 adjusted with 1 mM KOH). The growing environment was configured to have a temperature range of 28 ± 1 °C during the day and 18 ± 1 °C at night, a light intensity of 250 μmol m−2 s−1, a day/night cycle of 14/10 hours, and a humidity range of 70–75%. Every experiment employed three replicates, each with two plants unless otherwise stated.2.2. Synthesis and characterization of PNC
Following the procedure outlined by Wu et al.,21 poly (acrylic) acid-coated nanoceria (PNC) was synthesized and characterised. In a 50 mL conical tube, 1.08 g cerium(III) nitrate (Sigma Aldrich, 99%) and 4.5 g poly (acrylic) acid (1800 MW) were quickly dissolved in 2.5 mL and 5.0 mL of deionized water, respectively. A vortex mixer was used to completely combine the solutions at 2000 rpm for 15 minutes. 15 mL of ammonium hydroxide solution (30%, Sigma Aldrich) was added dropwise to the combined solution in a 50 mL beaker. For 24 hours at room temperature, the mixture was stirred at 500 rpm. To eliminate any big agglomerates and other debris, the mixture was centrifuged at 4000 rpm for 1 h. To purify the supernatant, 10 K Amicon cells (MWCO 10 K, Millipore Inc.) were spun at 4500 rpm for 45 minutes, six times. A UV-vis spectrophotometer (UV-1800, AOE) was used for measurements and using Beer–Lambert's law the concentration of PNC was determined by measuring its peak absorbance at 271 nm. A Brookhaven Zeta-sizer (NanoBrook 173 Plus) instrument was used to measure zeta potential and the hydrodynamic diameter of 0.1 mM PNC suspended in dd H2O. Then, for later processing, the acquired PNC product was kept in a freezer at 4 °C. 20 μL PNC was mounted on a copper grid with holes covered in carbon for TEM (transmission electron microscope) imaging, then an FEI Talos microscope was set to 300 kV to capture images of the samples. The samples were then kept at 4 °C for future use.2.3. Synthesis and confocal imaging of DiI-PNC
DiI-PNC was synthesized according to our previous publication,20 and the final product was stored at 4 °C for further use. Confocal imaging of DiI-PNC was performed 3 hours after DiI-PNC application to cucumber leaves and roots. The confocal imaging was conducted by following the procedures, which are described in our previous publication.202.4. Incubating cucumber with foliar-delivered or root-applied nanoceria and the salt stress treatment
PNC was administered to cucumber seedlings either foliarly or through the roots (at the period of the growth of the second true leaf). 0.1 mM PNC was utilized, which was determined from a PNC concentration screening experiment.20 The true leaves of the cucumber were sprayed with 1 mL of 0.1 mM PNC (a total of 0.1 mol PNC in the solution) for foliar delivery.20 A pipette with clipped tips was used to spray the solution. To aid in the incubation of PNC in leaves, silwet L-77 (0.05% v/w) was utilized. Kimwipes were employed to prevent any solution from dripping onto untreated areas of the plant. For the root application, the seedling roots were placed for 30 minutes in a 50 mL centrifuge tube with 25 mL of 4 μM PNC (containing a total of 0.1 mol PNC in the suspension). As a control, 1 mL of water was equally sprayed on the second true leaves. A 50 mL centrifuge tube with 25 mL of 4 μM PNC (a total of 0.1 mol PNC in the solution) was used for the root treatment, and 1 mL of water was uniformly sprayed on the second true leaves as a control. For applying salt stress, 75 mM NaCl was added to the culture fluid after 3 hours of adaptation under room light.2.5. Assessing phenotypic and photosynthetic performance of cucumber plants with PNC under salt stress
For measuring, the dry weight of the shoots and roots, the first true leaf's SPAD levels, maximal photochemical efficiency (Fv/Fm), net photosynthetic rate (Pn), stomatal conductance (Gs), and intercellular CO2 concentration (Ci) of salt-stressed (75 mM NaCl, 7 days) cucumber were carried out according to the protocols described in previous publications.20,362.6. Determination of cerium, K+, and Na+ content
After NaCl treatment for 7 days, cerium, K+, and Na+ content samples of the leaves, roots, and whole plant were measured as described in our previous study.20 Briefly, 0.1 g of the samples were digested with sulfuric acid. Then, the volume of the collected digestion solution was set to 10 mL by using ultrapure water. ICP-MS (Inductively Coupled Plasma-Mass Spectrometry) (Wuhan Qijunchicheng Technology Co., LTD.) technique was used to measure the cerium content. K+ and Na+ contents were measured using an atomic absorption spectrophotometer (6300C, Shimadzu).372.7. Determination of ROS content, MDA, and REC
After 7 days of salt stress (75 mM NaCl), H2O2, O2˙−, and MDA (malondialdehyde) content and REC (relative electrical conductivity) in leaf and root samples were measured as described in our previous study.20,39–42 Briefly, H2O2 content was determined using a hydrogen peroxide kit from the Nanjing Jiancheng Institute of Biological Engineering.38 The hydroxylamine hydrochloride oxidation technique was used to measure the concentration of O2˙−.39 The content of O2˙− was then determined using the standard curve for O2˙− standard after measuring the solution's absorbance value at 530 nm. DAB (diaminobenzidine) and NBT (nitrotetrazolium blue chloride) stainings were performed according to the protocols reported in the previous publication.40 The MDA content was measured following the method described in aprevious publication.41 The amount of MDA in samples was calculated using the formula: C (nmol g−1) = (6.45 (A532 − A600) − 0.56 × A450) × 5/0.1. The relative electrical conductivity was determined by following the previous study.2.8. Transcriptome assay
After 7 days of salt stress (75 mM NaCl), the leaves and roots were sampled. The samples were frozen in liquid nitrogen and pulverized into powder before being transferred to a business that specializes in transcriptome sequencing (Megi Bio, Shanghai). Differential expression analysis was performed using the cucumber (Chinese Long) genome V3 (https://cucurbitgenomics.org/). The statistical table of the genome alignment results is shown in Table S2.† The software DESeq2 was used to compute and identify differential genes.43 TPM (Transcription Per Million) was utilized to determine gene expression, and the criteria for differential gene screening were set as Log 2 (foldchange value) > 1, p-value 0.05.44 The RNA seq data was uploaded to NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA874600), and the accession number is PRJNA874600.2.9. Quantitative real-time PCR analysis
After 7 days of salt stress (75 mM NaCl), the leaves and roots were sampled for real-time fluorescent quantitative PCR (qRT-PCR). RNA was extracted using the TransZol kit from TransGen in Beijing, and mRNA was reverse transcribed using Hiscript Reverse Transcriptase from Vazyme in China. Reference genes were applied after reverse transcription to assess the reverse transcription's quality. For the next experiment, cDNA and primers that could amplify the reference gene bands were used. ABI6500 (ABI, USA), 10 μL system with 1 μL diluted cDNA, 5 μL 2× TransStartTM Top Green qRT-PCR SuperMix (TransGen, Beijing), and 1 μL forward and reverse primers each were used to perform qRT-PCR. It took 40 cycles of 94 °C pre-denaturation for 30 s, 94 °C denaturation for 5 s, 56 °C annealing for 30 s, and 72 °C extensions for 10 s to complete the PCR. The expression analysis was calculated using 2−ΔΔct. Primers were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). For the absolute quantification of CsGORK, CsAKT1, and CsHAK5;3, we prepared their respective standard substances. Using cucumber cDNA as a template, full-length amplification of CsGORK, CsAKT1, and CsHAK5;3 was performed using high-fidelity Taq enzyme (I-5™ 2× High-fidelity Master Mix, Engine, China). The PCR amplification procedure was as follows: 95 °C for 3 min, 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min 40 s, and 40 cycles. The obtained PCR reaction products were detected using 1% agarose gel electrophoresis. After confirming the correct band size, the gel was cut and the target band was recovered using the AxyPrepTM DNA gel recovery kit (AxyGEN, USA). Then, the recycled glue samples were sent to the company for sequencing. After the sequencing was correct, the samples were diluted to 100 ng μL−1 standard. Then, the standard was diluted to 10−1, 10−2, 10−3, 10−4, 10−5 of the previous concentration with dd H2O. These samples were used for qRT-PCR quantification and standard curves were made (Fig. S1a–c†). The absolute quantification of CsGORK, CsAKT1, and CsHAK5;3 was calculated using the standard curve of their standard. The used primers are shown in Table S1.†2.10. CsAKT1 sequence analysis and subcellular localization
Using TMHMM (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) for analysis of the amino acid sequence of the transmembrane area of CsAKT1 (CsaV3_1G029650). DNAMAN 9.0 was used for sequence alignment of CsAKT1. NCBI (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used to predict CsAKT1 protein structure. Subcellular localization methods refer to previous studies.45,46 1305.4 vectors were used for subcellular localization. Full-length amplification primers were designed based on the Bgl II restriction site sequence and CsAKT1 CDS sequence on the vector. The recombinant plasmid of CsAKT1 was transformed into Escherichia coli 5α. The method of recombination and transformation was performed according to the operation manual of the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing). Escherichia coli was cultured overnight, and several plaques were randomly selected for colony PCR. The positive plaques identified were sent to Optimus for sequencing. Enough recombinant plasmids were obtained by expanding the culture and extracting plasmids from the correctly sequenced E. coli. The recombinant plasmid and control plasmid (Empty vector) were transformed into Agrobacterium GV3101 by the freeze–thaw method. The transient expression test was carried out using Agrobacterium tumefaciens containing recombinant plasmid, control plasmid, plasmid PM-RK CD3-1007 with cell membrane marker mCherry, and auxiliary plasmid P19. The injected tobacco was placed in an incubator for 48–72 h, followed by observations using a confocal laser microscope (Leica TCS-SP8, Germany).2.11. CsAKT1 yeast heterologous expression
According to our previous method,47 the yeast heterologous expression vector was pYES2-NTA. The CDS (coding sequence) sequence of CsAKT1 was amplified and inserted into the Hind III restriction site of the pYES2-NTA vector by homologous recombination using cucumber cDNA as a template. The salt-sensitive yeast strain G19 with recombinant pYES2-NTA–CsAKT1 and empty pYES2-NTA (experimental control) were cultured in the SD-ura liquid medium overnight to OD600 = 0.7, and then the yeasts were diluted to 10−1, 10−2, 10−3, 10−4 of previous concentration with ddH2O. 5 μL of the solution was pipetted and placed dropwise on the plate containing 0, 100 mM NaCl AP solid medium, after dripping, the plate was placed on an ultra-clean workbench and air was blown. After the plate was blown dry, it was placed upside down in a 30 °C incubator to grow for 4 days and the growth of yeast was observed. The recombinant pYES2-NTA–CsAKT1 and empty pYES2-NTA vectors were transferred into K+ uptake-deficient yeast strain WΔ6, cultured in the SD-ura liquid medium overnight to OD600 = 0.7, and then diluted with dd H2O to 101, 102, 103, 104 times, 5 μL liquid was drawn and placed dropwise on a plate of the AP solid medium containing 0, 100 mM KCl. After dripping, the plate was placed on the ultra-clean workbench to blow. After the plate was blown dry, it was placed upside down in a 30 °C incubator to grow for 4 days, and the growth of yeast was observed. A test buffer was prepared as follows, 10 mM MES, 2% galactose, and Ca(OH)2 to adjust the pH to 6.0. 200 μL of yeast WΔ6 was then added with CsAKT1 to 50 mL of the AP liquid medium containing 50 mM KCl, which was cultured overnight at 28 °C, 220 r min−1, to OD600 = 1.0, then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 r min−1 for 1 min; the supernatant was discarded and 50 mL of dd H2O was added to resuspend the yeast, followed by centrifugation at 10
000 r min−1 for 1 min; the supernatant was discarded and 50 mL of dd H2O was added to resuspend the yeast, followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 r min−1 for 1 min and discarding the supernatant, and then adding 50 mL of the AP liquid medium without K+ and incubation for 4 h for K+ starvation treatment. After the starvation treatment was completed, the solution was centrifuged at 10
000 r min−1 for 1 min and discarding the supernatant, and then adding 50 mL of the AP liquid medium without K+ and incubation for 4 h for K+ starvation treatment. After the starvation treatment was completed, the solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 r min−1 for 1 min, the supernatant was discarded, 50 mL of the test buffer was added, and the yeast was resuspended. Three treatments were set up, i.e., inoculating the yeast with the liquid AP medium containing 200 μM NaCl, 200 μM KCl, and 200 μM NaCl + 200 μM KCl. Then, the liquids were placed on a 30 °C shaker, 220 r min−1, and 4 mL of bacterial solution was drawn every 5 min, centrifuged at 12
000 r min−1 for 1 min, the supernatant was discarded, 50 mL of the test buffer was added, and the yeast was resuspended. Three treatments were set up, i.e., inoculating the yeast with the liquid AP medium containing 200 μM NaCl, 200 μM KCl, and 200 μM NaCl + 200 μM KCl. Then, the liquids were placed on a 30 °C shaker, 220 r min−1, and 4 mL of bacterial solution was drawn every 5 min, centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 r min−1 for the 30 s, and the supernatant was collected for the determination of Na+ and K+ contents.
000 r min−1 for the 30 s, and the supernatant was collected for the determination of Na+ and K+ contents.
2.12. Creation of CRISPR/Cas9-induced CsAKT1 deletion plants
CsAKT1 (CsaV3_1G029650) genome sequence was downloaded and constructed from the Cucurbit Genomics Database (https://cucurbitgenomics.org/), and then submitted for sequencing to the Geneious software to obtain a collection of sgRNA (small guide RNA) sequences. The target was preferentially selected in the first external display sgRNA sequences with a low off-target rate. According to the sequence design, sgRNA primers were obtained and sent to a primer synthesis company (Tsingke, Beijing) for synthesis. After the synthetic primers were obtained, annealing and ligation of the vector were performed. After the reaction was completed, the ligation products were transformed into Escherichia coli 5α. The pKSE402 empty plasmids and pKSE402-containing CsAKT1 sgRNA were used to transform Agrobacterium K599 (Weidi, Shanghai) referring to the K599 instruction manual. Then, the cucumber was infected. After the infection was successful, the non-GFP roots were removed every week, and the GFP roots were kept. The GFP roots were sampled, DNA was extracted, and PCR amplification was performed. The PCR products were subjected to Hi-TOM sequencing to detect gene editing efficiency.37 The transformed cucumber seedlings were obtained, cultivated to 2 leaves, and subjected to 75 mM NaCl treatment. The morphological and physiological indices of cucumber were measured at 75 mM NaCl treatment for 7 d.2.13. Data analysis and image processing
The statistical analysis and sketching of the images were performed using Rstudio 4.03 software. For a significant comparison, the Duncan-type new complex range method was employed. The images were typeset using Photoshop 6.0 software.3. Results
3.1. PNC characterization and its improvement on the performance of cucumber under salinity stress
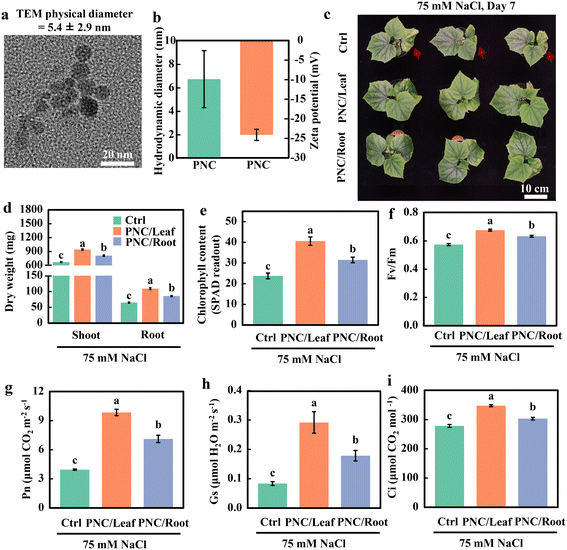
In this study, we synthesized poly (acrylic) acid-coated nanoceria (PNC) with a size below 10 nm. Characterization data showed that the TEM size, hydrodynamic diameter (measured in ddH2O), and zeta potential (measured in ddH2O) of PNC were 5.4 ± 2.9 nm (Fig. 1a), 6.6 ± 2.4 nm (Fig. 1b), and −24.0 ± 1.4 mV (Fig. 1b), respectively. Confocal imaging results showed that compared with no signals in the control group, clear DiI signals were detected in the leaves and roots of cucumber plants treated with DiI-PNC (Fig. S2a and b†).Our results showed that compared with control plants under salinity (75 mM NaCl, 7 days), foliar spray or root application of PNC improved cucumber salt tolerance, showing better phenotypic performance (Fig. 1c), increased shoot (40.5 and 20.7% for leaf and root application of PNC, respectively) and root (67.8 and 31.8% for leaf and root application of PNC, respectively) dry weight (Fig. 1d), and higher chlorophyll content (70.8 and 32.5% for leaf and root application of PNC, respectively) (Fig. 1e). Also, cucumber plants with PNC application showed higher total areas of leaf (85.3% and 39.8% for leaf and root application of PNC, respectively) (Fig. S3a†), bigger total lengths of root (41.3% and 13.0% for leaf and root application of PNC, respectively) (Fig. S3b†) and total surface areas of root (33.4% and 15.3% for leaf and root application of PNC, respectively) (Fig. S3c†), and increased root volume (83.3% and 43.0% for leaf and root application of PNC, respectively) (Fig. S3d†) than control plants under salinity stress. Gas exchange experiments showed that compared with control plants under salinity (75 mM NaCl, 7 days), foliar spray or root application of PNC improved the photosynthetic performance of cucumber under salt stress, showing the increase of Fv/Fm (17.8% and 10.3% for leaf and root application of PNC, respectively) (Fig. 1f and S4a†), Pn (149.3% and 80.3% for leaf and root application of PNC, respectively) (Fig. 1g), Gs (248.6% and 113.1% for leaf and root application of PNC, respectively) (Fig. 1h), Ci (24.8% and 8.9% for leaf and root application of PNC, respectively) (Fig. 1i) and Tr (89.5% and 51.6% for leaf and root application of PNC, respectively) (Fig. S4b†). Together, these results suggest that both leaf and root application of PNC could improve the growth and photosynthetic performance of cucumber under salt stress. More importantly, PNC leaf application showed better improvement in cucumber salt tolerance compared to PNC root application. ICP-MS results showed that after 7 days of salinity stress (75 mM NaCl), cerium content in the whole cucumber (20.3 vs. 6.3 μg g−1, 3.2 folds), leaf (23.4 vs. 4.0 μg g−1, 5.8 folds) and root (8.4 vs. 13.4 μg g−1, 62.3%) are significantly higher in the foliar-delivered PNC than the root applied one (Fig. S5a and b†), further confirming that foliar application of PNC is more optimal than the root application, at least for improving cucumber salt tolerance.
3.2. PNC application decreased ROS content in cucumber under salinity stress
Compared with control plants under salinity stress (75 mM NaCl, 7 days), PNC-treated cucumber plants showed a decrease in H2O2 content in leaf (46.6% and 22.0% for leaf and root application of PNC, respectively) and root (50.0% and 29.9% for leaf and root application of PNC, respectively) (Fig. S6a†), and significantly lower O2˙− content in leaf (58.7% and 26.4% for leaf and root application of PNC, respectively) and root (53.2% and 33.0% for leaf and root application of PNC, respectively) (Fig. S6b†). Also, significantly lower MDA content in leaf (46.7% and 13.3% for leaf and root application of PNC, respectively) and root (31.4% and 19.9% for leaf and root application of PNC, respectively) (Fig. S6c†) was found in cucumber plants treated with PNC than the control under salinity stress. This is also observed in REC (relative electric conductivity), showing that the PNC-treated cucumber plants have significantly lower REC in leaf (50.6% and 32.8% for leaf and root application of PNC, respectively) and root (33.6% and 13.6% for leaf and root application of PNC, respectively) than the control plants under salinity stress (Fig. S6d†). DAB and NBT staining experiments further confirmed that cucumber plants with leaf or root application of PNC have decreased H2O2 and O2˙− content compared to control plants under salinity stress (Fig. S6e and f†).3.3. PNC-induced CsAKT1 – mediated absorption of K+ in cucumber under salinity stress
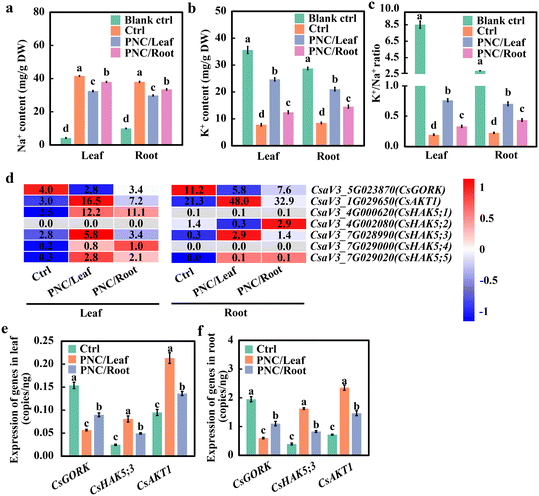
No doubt, compared with a blank control (0 mM NaCl, 7 days), cucumber plants showed increased Na+ content in leaf (8.9 folds) and root (2.8 folds), decreased K+ content of leaf (78.1%) and root (70.5%), and decreased K+/Na+ ratio content of leaf (97.8%) and root (92.3%) after salinity stress (75 mM NaCl, 7 days) (Fig. 2a–c). Compared with salt control plants (75 mM NaCl, 7 days), the PNC-treated cucumber showed a decrease in Na+ content in leaf (21.8% and 8.7% after leaf and root application of PNC, respectively) and root (21.5% and 11.8% after leaf and root application of PNC, respectively) (Fig. 2a). While, the PNC-treated cucumber showed an increase in K+ content in leaf (216.1% and 59.5% for leaf and root application of PNC, respectively) and root (148.2% and 71.8% for leaf and root application of PNC, respectively) compared to control plants under the salinity stress (Fig. 2b). In terms of the K+/Na+ ratio, under salinity stress, the PNC-treated cucumber plants showed increased K+/Na+ ratio in leaf (304.5% and 74.7% for leaf and root application of PNC, respectively) and root (216.1% and 95.0% for leaf and root application of PNC, respectively) compared with the control plants (Fig. 2c). Overall, these results suggest that to maintain K+/Na+ ratios under salinity stress, PNC application enabled K+ retention, contributing to more than its improved Na+ extrusion in cucumber plants.RNA seq and qRT-PCR experiments were further conducted to investigate the changes in the expression level of K+ transporters in cucumber plants with PNC application under salinity stress. Before RNAseq and qRT-PCR, we tested the quality of RNA and found that the integrity of RNA was good without obviously observed degradation (Fig. S7a and b†). Then, PCA analysis (Fig. S8a and b†) was performed on leaf and root transcriptomes of salt-stressed (75 mM NaCl, 7 days) cucumber plants treated with PNC. It showed that the samples with three replicates were all located in 95% confidence ellipses, indicating good repeatability of the samples. qRT-PCR analysis (Fig. S8c†) and correlation analysis (Fig. S8d†) showed that the results of qRT-PCR and RNAseq (Fig. S9†) were consistent. Also, the correlation coefficient R2 was 0.93 (Fig. S8c†), indicating that the transcriptome results were accurate and reliable. Further data analysis showed that the number of up-regulated genes in leaves and roots was significantly higher than down-regulated genes, regardless of salt-stressed cucumber plants with PNC foliar spray or root application (Fig. S10a and b†). GO enrichment analysis (P < 0.05) showed that DEGs in leaves and roots of cucumber with either foliar sprayed or root-applied PNC could be enriched into groups such as cation binding and transporter activity (Fig. S9c–f†).
The phylogenetic tree analysis of cucumber and Arabidopsis Thaliana revealed that cucumber GORK and AKT1 had 1 member and HAK5 had 5 members (Fig. S11†). The heat map analysis showed that compared with H2O control, potassium ion channel protein genes, such as GORK, HAK5, and AKT1 were mostly up-regulated in leaves and roots of cucumber with either foliar delivered or root applied PNC (Fig. 2d). Furthermore, compared with the root application, foliar sprayed PNC downregulated CsGORK, upregulated CsHAK5;3 and CsAKT1. qRT-PCR results showed that PNC did not induce changes in the gene expression levels of CsGORK, CsHAK5;3, and CsAKT1 in cucumber under non-saline conditions (Fig. S12†). While compared with the control plants under salinity (75 mM NaCl, 7 days), PNC-treated cucumber plants showed a decreased expression level of CsGORK (36.8% decrease in leaf and 45.8% decrease in the root of plants with PNC leaf spray over its root application), increased expression level of CsHAK5;3 (36.2% increase in leaf and 60.8% increase in the root of plants with PNC leaf spray over its root application), and increased expression level of CsAKT1 (63.8% increase in leaf and 96.4% increase in the root of plants with PNC leaf spray over its root application) (Fig. 2e and f). Together, these results not only further confirm that foliar-delivered PNC is more optimal than the root application in improving cucumber salt tolerance, but also suggest that CsAKT1 is more responsible for PNC improved cucumber salt tolerance.
3.4. Expression analysis and functional analysis of CsAKT1 in yeasts
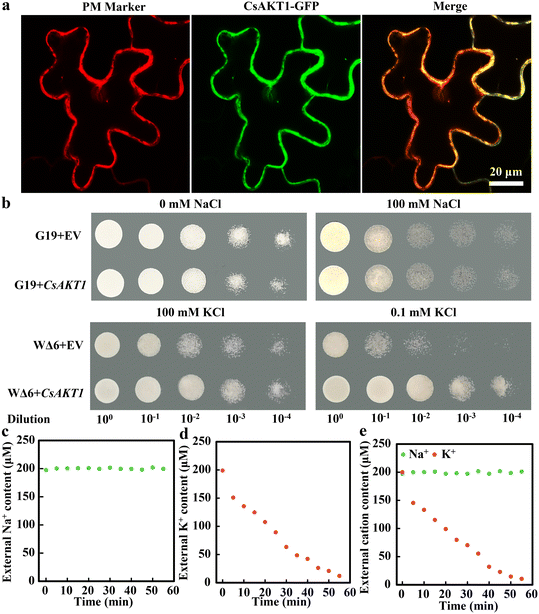
TMHMM (prediction of transmembrane helices in proteins) was used to predict the transmembrane structure of CsAKT1. Our results showed that CsAKT1 contained five transmembrane domains with a long C-terminal (Fig. S13a and b†), which is consistent with the case in Arabidopsis. Further analysis showed that CNBD (cyclic nucleotide-binding domain) of CsAKT1 was located between 399–485 amino acid sites, anchored repeat domain was located between 532–688 amino acid sites, and KHA (rich in hydrophobic and acidic amino acids) domain is located between the 804 and 866 amino acid sites (Fig. S13c†). Subcellular localization experiments showed that CsAKT1 was localized to the plasma membrane (Fig. 3a). Yeast G19 (salt-sensitive strain) and WΔ6 (K+ deficient strain) strains were used to further verify whether CsAKT1 could transport K+. Our results showed that under 100 mM of NaCl treatment, no significant difference in the growth status was found between G19 transformed with CsAKT1 and the G19 empty (Fig. 3b), indicating that CsAKT1 could not transport Na+. Under 100 mM and 0.1 mM KCl treatment, WΔ6 yeast transformed with CsAKT1 grew significantly better compared to WΔ6 empty (Fig. 3c), indicating that CsAKT1 could transport K+. Furthermore, the ion depletion experiment showed that compared with no change in Na+ content in 200 μM NaCl buffer, K+ content decreased significantly in CsAKT1 transformed WΔ6 yeast inoculated into the liquid AP medium supplemented with either 200 μM KCl or 200 μM NaCl + 200 μM KCl (Fig. 3c–e). This further confirmed the role of CsAKT1 in K+ transport.3.5. Knockout CsAKT1 impaired PNC enabled improvement of salt tolerance on cucumber
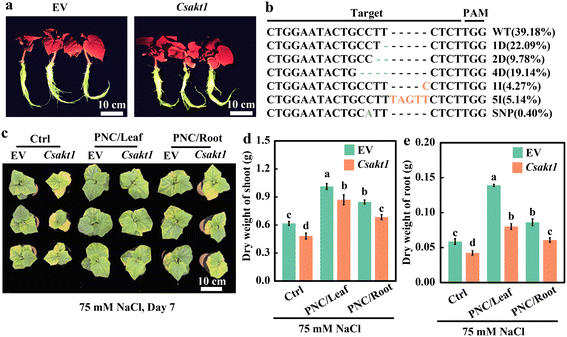
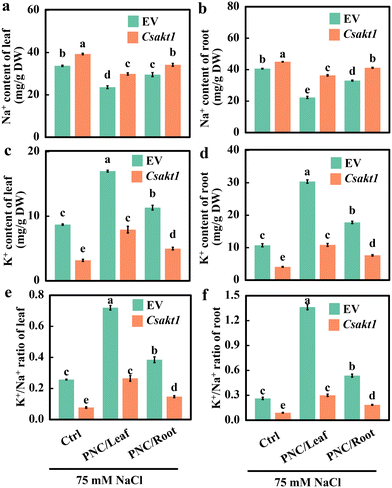
By using the root transformation system, CsAKT1 was knocked out in the cucumber rootstock. Hi-TOM sequencing showed that the editing efficiency was 60.82% (Fig. 4a and b). Protein translation analysis showed that knock-out lines could induce mismatch and early termination of protein translation (Fig. S14a†). Together, it suggests that Cas9 successfully edited CsAKT1 of cucumber with high editing efficiency. Compared with EV (Empty vector) plants under salinity (75 mM NaCl, 7 days), CsAKT1 knockout plants showed worse phenotypic performance (Fig. 4c), significantly lower dry weight of shoot (21.9%, 14.1%, and 19.2% for control, leaf and root application of PNC, respectively, Fig. 4d) and root (27.8%, 42.4%, and 29.5% for control, leaf and root application of PNC, respectively, Fig. 4e), and significantly smaller total area of the leaf (31.7%, 19.6%, and 22.5% for control, leaf and root application of PNC, respectively) (Fig. S15a†). It also showed a significantly decreased total length of root (22.8%, 22.2%, and 24.9% for control, leaf, and root application of PNC, respectively, Fig. S15b†), the total surface area of root (27.2%, 27.0%, and 22.7% for control, leaf and root application of PNC, respectively, Fig. S15c†), and root volume (35.8%, 32.1%, and 28.1% for control, leaf and root application of PNC, respectively, Fig. S15d†) in CsAKT1 knockout plants than the EV plants under salinity.K+ and Na+ content data showed that compared with EV plants under salinity (75 mM NaCl, 7 days), CsAKT1 knockout cucumber plants showed significantly increased Na+ content regardless of the treatment with the control (33.7 ± 0.3 vs. 39.3 ± 0.3 mg g−1 in leaf and 40.6 ± 0.2 vs. 45.0 ± 0.1 mg g−1 in root), or foliar PNC delivery (23.6 ± 0.7 vs. 29.9 ± 0.5 mg g−1 in leaf and 22.3 ± 0.7 vs. 36.4 ± 0.5 mg g−1 in root), or root PNC application (29.6 ± 1.0 vs. 34.2 ± 0.7 mg g−1 in leaf and 33.0 ± 0.3 vs. 41.2 ± 0.3 mg g−1 in root) (Fig. 5a and b). While, under salinity stress, significant lower K+ content was found in CsAKT1 knockout cucumber plants than the EV plants with control (8.7 ± 0.1 vs. 3.2 ± 0.2 mg g−1 in leaf and 10.7 ± 0.5 vs. 4.1 ± 0.1 mg g−1 in root), foliar PNC application (17.0 ± 0.1 vs. 7.9 ± 0.5 mg g−1 in leaf and 30.4 ± 0.5 vs. 10.9 ± 0.4 mg g−1 in root), or root application of PNC (11.3 ± 0.4 vs. 5.0 ± 0.2 mg g−1 in leaf and 17.8 ± 0.4 vs. 7.6 ± 0.2 mg g−1 in root) (Fig. 5c and d). Together, not surprisingly, compared with EV plants under salinity (75 mM NaCl, 7 days), CsAKT1 knockout cucumber plants showed decreased K+/Na+ ratios in leaf (69.9%, 63.2%, and 61.7% for control, leaf and root application of PNC, respectively, Fig. 5e) and root (65.7%, 78.0%, and 65.6% for control, leaf and root application of PNC, respectively, Fig. 5f).
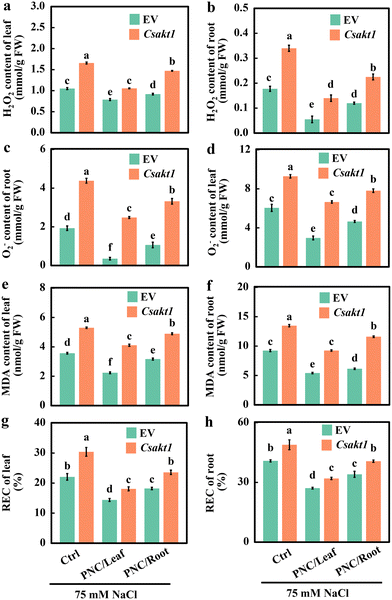
Similarly, we found that CsAKT1 knockout cucumber plants showed significantly higher H2O2 content in leaf (57.9%, 34.0%, and 60.5% for control, leaf and root application of PNC, respectively, Fig. 6a) and root (91.5.9%, 154.5%, and 87.5% for control, leaf and root application of PNC, respectively, Fig. 6b), the O2˙− content in leaf (53.5%, 124.7%, and 67.9% for control, leaf and root application of PNC, respectively) (Fig. 6c) and root (127.1%, 603.5%, and 212.5% for control, leaf and root application of PNC, respectively, Fig. 6d), and MDA content in leaf (48.7%, 83.9%, and 54.3% for control, leaf and root application of PNC, respectively, Fig. 6e) and root (45.9%, 70.8%, and 88.9% for control, leaf and root application of PNC, respectively, Fig. 6f) than the EV plants under salinity (75 mM NaCl, 7 days). This is the same case in the REC results, showing that compared with the EV plants under salinity, CsAKT1 knockout cucumber plants have significantly increased REC in leaf (37.8%, 25.7%, and 29.5% for control, leaf and root application of PNC, respectively, Fig. 6g) and root (19.8%, 17.9%, and 19.3% for control, leaf and root application of PNC, respectively, Fig. 6h).
4. Discussion
4.1. Leaf but not root nanoceria application resulted in higher improvement in cucumber salt tolerance is associated with a better maintained K+/Na+ ratio
As mentioned previously, the K+/Na+ ratio is a hallmark of plant salt tolerance.49,50 Similarly, in nano-improved plant salt tolerance, a better maintained K+/Na+ ratio was observed in the nanomaterials treated group than in the control. Cotton plants treated with nanoceria showed a significantly higher K+/Na+ ratio than the control under salt stress.16 Rapeseed primed with nanoceria also showed a significantly higher shoot and root K+/Na+ ratio than the control under salt stress.12,51,52 This is also found in this work. Cucumber plants with either foliar or root applied nanoceria showed a significantly higher K+/Na+ ratio than the control under salt stress (Fig. 2b). Together, it suggests that maintaining the K+/Na+ ratio could be a commonly employed mechanism in nano-improved plant salt tolerance.Many studies have shown that both leaf and root application can improve the salt tolerance of plants such as Arabidopsis,24 cotton,16 and rapeseed.13,25,53 In this study, we found that foliar application of nanoceria resulted in a better improvement of cucumber salt tolerance compared to the root application (Fig. 1c). In our previous study, we found that PNC-induced early stimulation of the antioxidant system (before the onset of salt stress) could be a reason for better salt resistance in cucumber plants with foliar but not root application of PNC.20 Here, we found that after 7 days of salt treatment, cucumber with foliar PNC application showed a significantly higher K+/Na+ ratio than the one with root PNC application (Fig. 2b). It suggests that after salt stress, the ability to maintain the K+/Na+ ratio could be a reason for the better salt resistance in cucumber plants with foliar PNC application than the root application. The variations of the K+/Na+ ratio were also shown in different organs of nanoparticles treated plants under salt stress. For example, under salt stress, the second true leaf showed a significantly higher K+/Na+ ratio than the first true leaf in cotton plants with foliar nanoceria application.16 Overall, it suggests that the variation of ability to maintain the K+/Na+ ratio could be a reason behind the better-improved salt resistance in cucumber plants with foliar PNC application than the root application.
4.2. CsAKT1 could be a target of nano-improved plant salt tolerance
AKT1, as a class of inwardly rectifying K+ channels, is widely existing in plants and is important for maintaining K+ homeostasis.54,55 It is a typical member of the Shaker family and plays an important role in plant response to salinity stress. Both the N-terminus and C-terminus of Shaker channel proteins are located in the cytoplasm. The N-terminus is a very short domain composed of about 60 amino acids. The C-terminal, at the end of the sixth transmembrane segment, contains a C-junction consisting of about 80 amino acid residues, a cyclic nucleotide-binding domain (CNBD), and an anchor protein domain (Ankyrin-related domain; ANKY) and a KHA domain rich in hydrophobic acid residues.56 Members of the Shaker family have six transmembrane chains (S1–S6), and the four transmembrane chains S1–S4 are closely related to the ability of potassium ion channels to sense and respond to changes in the membrane potential.57,58 There is a positively charged arginine or lysine repeat (Arg/Lys–Xaa–Xaa–Arg/Lys) at every two amino acid residues in the S4 transmembrane chain. The presence of this structure enables S4 to move across the membrane, resulting in a conformational change of the membrane channel, which controls the opening and closing of the channel.59 There is a pore domain between S5 and S6, which consists of a polypeptide fragment embedded in the cell membrane to form a channel. Generally, a single potassium ion channel is a homotetramer structure, consisting of four subunits symmetrically, and can only allow one potassium ion to pass through.60,61 Shaker-type potassium ion channels can form heterotetramer structures, which is also an important characteristic of the Shaker channel. This structure allows plants to regulate the intracellular potassium transport activity in each organ or tissue relatively independently.62 In different species, the AKT1 has the function of the potassium ion transport, e.g. tomato,63 rice,64 maize,65 soybean,54 and Arabidopsis.66 This further confirms that AKT1 plays an important role in the absorption of K+, maintenance of K+ content in plants, and enhancement of plant salt tolerance.Previous studies have shown that GORK, AKT1, and HAK5 are important potassium ion transporters in response to salt stress. Among them, GORK can mediate potassium efflux,67 while AKT1 and HAK5 can mediate active absorption of potassium.66,68,69 In this study, we found that both leaf and root application of PNC could induce downregulation of GORK, which is different from no significant change in CsGORK gene expression level in PNC-treated Arabidopsis under salinity stress.24 Similar results were shown in the PNC-treated rapeseed under salt stress, showing no significant change in the relative expression level of BnaGORK between PNC-treated rapeseed and control plants under salt stress, either in the root or in the leaf.70 While, PNC treatment significantly upregulated BnaHAK5 in the shoots but not in the root, compared with control plants under salt stress.70 A similar case was shown in this study. Both leaf and root application of PNC could induce up-regulation of CsAKT1 and CsHAK5, while the absolute expression of CsAKT1 was significantly higher than that of CsHAK5, indicating that CsAKT1 is the most responsive gene for nanoceria-improved cucumber salt tolerance. Moreover, to date, our knowledge about how nanoparticles induce gene expression in plants under stress is still insufficient. The direct or indirect (via ROS or Ca2+ signals) modulation effect of nanoparticles on transcription factors might be a reason. This is worthy to be further investigated in future studies.
Domain analysis of CsAKT1 showed that it has CNBD, anchored repeat domain, and KHA domain, which are consistent with the basic characteristics of Shaker-type channel protein.62 Unlike the common six transmembrane chains in Shaker-type channel protein,71,72 the predicted structure analysis showed that CsAKT1 contains only five transmembrane structures, which may be due to the shorter third transmembrane region. Consistent with the function of AKT1 reported in other species,63,65,66 here, we found that AKT1 is localized in the plasma membrane of cucumber, and executes the transportation of potassium ions. With the creation of CRISPR/Cas9 lines, we confirmed that CsAKT1 is a key gene to nanoceria-improved cucumber salt tolerance. Our work suggests that AKT1 could be a target for nano-improved plant salt tolerance.
4.3. Genome editing is a powerful tool for studying the key genes involved in nano-improved plant stress tolerance
Genome editing is a powerful technique for functional gene analysis. The Cas9-mediated gene editing system is one of the important tools.48,73,74 Applications of Cas9-mediated gene editing in crops include 1) improving the yield and quality of crops,75 2) improving the disease resistance76 and insect resistance77 of crops, 3) improving herbicide resistance of crops,78 4) improving the abiotic stress resistance of crops,79 such as drought resistance and salt tolerance.80 To our surprise, such techniques have not been properly utilized in investigating and verifying the key genes in nano-improved plant salt tolerance.Previously, we developed a highly efficient root transformation system for Cucurbitaceae that can be used to verify the function of cucumber genes.37 In this study, in order to verify whether CsAKT1 is a key gene for nanoceria-improved cucumber salt tolerance, we used Cas9 gene editing technology to obtain cucumber seedlings with root deletion of CsAKT1. Our results showed that root deletion of CsAKT1 significantly reduced the enhancement of nanoceria-improved cucumber salt tolerance (no matter with leaf or root application mode). To the best of our knowledge, this is the first study using the CRISPR/Cas9 technique to verify the key genes in nano-improved plant salt tolerance. This shows that besides making mutants, genome editing such as CRISPR/Cas9 technique could be another powerful tool in verifying the key genes in nano-improved plant stress tolerance. This new approach should be encouraged to be adopted in future studies.
5. Conclusions
This work adds new knowledge about the mechanisms behind nanoceria-improved cucumber salt tolerance. Our results showed that the leaf application of PNC was more effective than the root application of PNC in improving cucumber salt tolerance, which is associated with its better ability to maintain ROS homeostasis and K+/Na+ ratio. More interestingly, we found that CsAKT1 is a key gene for PNC improved salt tolerance in cucumber. Our results not only add more knowledge about the mechanisms behind plant-nanoceria interaction but also provide an approach to investigating the key genes related to nano-improved plant stress tolerance.Author contributions
Conceptualization, H. W., and B. Z.; writing-original draft preparation, H. W., Y. P., and B. Z.; conducting experiments, Y. P., L. C., L. Z., and L. C.; data analysis, H. W., B. Z., Y. P., and L. Y.; and all authors have read and agreed to the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFD1000800), National Natural Science Foundation of China (32072653, 32071971, 31901464), the Hubei Agricultural Science and Technology Innovation Center Program (2021-620-000-001-032), joint project SZYJY2021008 from Huazhong Agricultural University and Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, project 2662020ZKPY001 supported by the Fundamental Research Funds for the Central Universities, and the Natural Science Foundation of Hubei Province (2019CFA017). We thank Mr. Jianbo Cao and Mrs. Limin He for their help in TEM imaging at the Public Laboratory of Electron Microscopy, Huazhong Agricultural University.References
- J. Bailey-Serres, J. E. Parker, E. A. Ainsworth, G. E. D. Oldroyd and J. I. Schroeder, Genetic strategies for improving crop yields, Nature, 2019, 575, 109–118 CrossRef CAS PubMed.
- A. Litalien and B. Zeeb, Curing the earth: A review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation, Sci. Total Environ., 2020, 698, 134235 CrossRef CAS PubMed.
- M. Liu, T. Pan, S. I. Allakhverdiev, M. Yu and S. Shabala, Crop halophytism: an environmentally sustainable solution for global food security, Trends Plant Sci., 2020, 25, 630–634 CrossRef CAS PubMed.
- A. Singh, Soil salinization management for sustainable development: A review, J. Environ. Manage., 2021, 277, 111383 CrossRef CAS PubMed.
- L. J. Zhao, L. Lu, A. D. Wang, H. L. Zhang, M. Huang, H. H. Wu, B. S. Xing, Z. Y. Wang and R. Ji, Nano-biotechnology in agriculture: use of nanomaterials to promote plant growth and stress tolerance, J. Agric. Food Chem., 2020, 68, 1935–1947 CrossRef CAS PubMed.
- H. H. Wu and Z. H. Li, Recent advances in nano-enabled agriculture for improving plant performance, Crop J., 2022, 10, 1–12 CrossRef.
- H. Zhou, H. H. Wu, F. Zhang, Y. Su, W. X. Guan, Y. J. Xie, J. P. Giraldo and W. B. Shen, Molecular basis of cerium oxide nanoparticle enhancement of rice salt tolerance and yield, Environ. Sci.: Nano, 2021, 8(11), 3294–3311 RSC.
- I. Wahid, S. Kumari, R. Ahmad, S. J. Hussain, S. Alamri, M. H. Siddiqui and M. I. R. Khan, Silver nanoparticle regulates salt tolerance in wheat through changes in ABA concentration, ion homeostasis, and defense systems, Biomolecules, 2020, 10(11), 1506 CrossRef CAS PubMed.
- A. Karami and A. Sepehri, Nano titanium dioxide and nitric oxide alleviate salt induced changes in seedling growth, physiological and photosynthesis attributes of barley, Zemdirbyste-Agriculture, 2018, 105, 123–132 CrossRef.
- S. M. H. Gowayed, H. S. M. Al-Zahrani and E. M. R. Metwali, Improving the salinity tolerance in potato (solanum tuberosum) by exogenous application of silicon dioxide nanoparticles, Int. J. Agric. Biol., 2017, 19, 183–192 CrossRef.
- A. W. M. Mahmoud, E. A. Abdeldaym, S. M. Abdelaziz, M. B. I. El-Sawy and S. A. Mottaleb, Synergetic effects of zinc, boron, silicon, and zeolite nanoparticles on confer tolerance in potato plants subjected to salinity, Agronomy, 2019, 10(1), 19 CrossRef.
- M. N. Khan, Y. H. Li, Z. Khan, L. L. Chen, J. H. Liu, J. Hu, H. H. Wu and Z. H. Li, Nanoceria seed priming enhanced salt tolerance in rapeseed through modulating ROS homeostasis and alpha-amylase activities, J. Nanobiotechnol., 2021, 19, 276 CrossRef CAS PubMed.
- Y. H. Li, J. H. Liu, C. C. Fu, M. N. Khan, J. Hu, F. M. Zhao, H. H. Wu and Z. H. Li, CeO2 nanoparticles modulate Cu–Zn superoxide dismutase and lipoxygenase-IV isozyme activities to alleviate membrane oxidative damage to improve rapeseed salt tolerance, Environ. Sci.: Nano, 2022, 9, 1116–1132 RSC.
- M. N. Khan, Y. H. Li, C. C. Fu, J. Hu, L. L. Chen, J. Yan, Z. Khan, H. H. Wu and Z. H. Li, CeO2 nanoparticles seed priming increases salicylic acid level and ROS scavenging ability to improve rapeseed salt tolerance, Glob. Chall., 2022, 2200025, DOI:10.1002/gch2.202200025.
- J. An, P. G. Hu, F. J. Li, H. H. Wu, Y. Shen, J. C. White, X. L. Tian, Z. H. Li and J. P. Giraldo, Emerging investigator series: molecular mechanisms of plant salinity stress tolerance improvement by seed priming with cerium oxide nanoparticles, Environ. Sci.: Nano, 2020, 7, 2214–2228 RSC.
- J. H. Liu, G. J. Li, L. L. Chen, J. J. Gu, H. H. Wu and Z. H. Li, Cerium oxide nanoparticles improve cotton salt tolerance by enabling better ability to maintain cytosolic K+/Na+ ratio, J. Nanobiotechnol., 2021, 19, 153 CrossRef CAS PubMed.
- A. Alsaeedi, H. El-Ramady, T. Alshaal, M. El-Garawany, N. Elhawat and A. Al-Otaibi, Silica nanoparticles boost growth and productivity of cucumber under water deficit and salinity stresses by balancing nutrients uptake, Plant Physiol. Biochem., 2019, 139, 1–10 CrossRef CAS PubMed.
- L. Lu, M. Huang, Y. X. Huang, P. F. X. Corvini, R. Ji and L. J. Zhao, Mn3O4 nanozymes boost endogenous antioxidant metabolites in cucumber (Cucumis sativus) plant and enhance resistance to salinity stress, Environ. Sci.: Nano, 2020, 7, 1692–1703 RSC.
- T. A. Shalaby, E. Abd-Alkarim, F. El-Aidy, E. S. Hamed, M. Sharaf-Eldin, N. Taha, H. El-Ramady, Y. Bayoumi and R. A. R. Dos, Nano-selenium, silicon and H2O2 boost growth and productivity of cucumber under combined salinity and heat stress, Ecotoxicol. Environ. Saf., 2021, 212, 111962 CrossRef CAS PubMed.
- L. L. Chen, Y. Q. Peng, L. Zhu, Y. Huang, Z. L. Bie and H. H. Wu, CeO2 nanoparticles improved cucumber salt tolerance is associated with its induced early stimulation on antioxidant system, Chemosphere, 2022, 134474 CrossRef CAS PubMed.
- H. H. Wu, N. Tito and J. P. Giraldo, Anionic cerium oxide nanoparticles protect plant photosynthesis from abiotic stress by scavenging reactive oxygen species, ACS Nano, 2017, 11, 11283–11297 CrossRef CAS PubMed.
- G. M. Newkirk, H. H. Wu, I. Santana and J. P. Giraldo, Catalytic scavenging of plant reactive oxygen species in vivo by anionic cerium oxide nanoparticles, J. Visualized Exp., 2018, 138, e58373 Search PubMed.
- Y. Wu and H. T. Ta, Different approaches to synthesising cerium oxide nanoparticles and their corresponding physical characteristics, and ROS scavenging and anti-inflammatory capabilities, J. Mater. Chem. B, 2021, 9, 7291–7301 RSC.
- H. H. Wu, L. Shabala, S. Shabala and J. P. Giraldo, Hydroxyl radical scavenging by cerium oxide nanoparticles improves Arabidopsis salinity tolerance by enhancing leaf mesophyll potassium retention, Environ. Sci.: Nano, 2018, 5, 1567–1583 RSC.
- L. Rossi, W. L. Zhang and X. M. Ma, Cerium oxide nanoparticles alter the salt stress tolerance of Brassica napus L. by modifying the formation of root apoplastic barriers, Environ. Pollut., 2017, 229, 132–138 CrossRef CAS PubMed.
- Z. M. Almutairi, Effect of nano-silicon application on the expression of salt tolerance genes in germinating tomato ('Solanum lycopersicum' L.) seedlings under salt stress, Plant Omics, 2016, 9(1), 106–114 CAS.
- H. Hernández-Hernández, A. Juárez-Maldonado, A. Benavides-Mendoza, H. Ortega-Ortiz, G. Cadenas-Pliego, D. Sánchez-Aspeytia and S. González-Morales, Chitosan-PVA and copper nanoparticles improve growth and overexpress the SOD and JA genes in tomato plants under salt stress, Agron., 2018, 8(9), 175 CrossRef.
- C. Zhao, H. Zhang, C. Song, J. K. Zhu and S. Shabala, Mechanisms of plant responses and adaptation to soil salinity, Innovation, 2020, 1, 100017 Search PubMed.
- H. Zhang, J. Zhu, Z. Gong and J. K. Zhu, Abiotic stress responses in plants, Nat. Rev. Genet., 2022, 23, 104–119 CrossRef CAS PubMed.
- F. Y. Yan, H. M. Wei, W. W. Li, Z. H. Liu, S. Tang, L. Chen, C. Q. Ding, Y. Jiang, Y. F. Ding and G. H. Li, Melatonin improves K+ and Na+ homeostasis in rice under salt stress by mediated nitric oxide, Ecotoxicol. Environ. Saf., 2020, 206, 111358 CrossRef CAS PubMed.
- J. Cui and G. Tcherkez, Potassium dependency of enzymes in plant primary metabolism, Plant Physiol. Biochem., 2021, 166, 522–530 CrossRef CAS PubMed.
- H. H. Wu, X. C. Zhang, J. P. Giraldo and S. Shabala, It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress, Plant Soil, 2018, 431, 1–17 CrossRef CAS.
- D. Lhamo, C. Wang, Q. Gao and S. Luan, Recent advances in genome-wide analyses of plant potassium transporter families, Curr. Genomics, 2021, 22(3), 164–180 CrossRef CAS PubMed.
- T. Y. Yang, H. M. Feng, S. Zhang, H. J. Xiao, Q. D. Hu, G. Chen, W. Xuan, N. Moran, A. Murphy, L. Yu and G. H. Xu, The potassium transporter OsHAK5 alters rice architecture via atp-dependent transmembrane auxin fluxes, Plant Commun., 2020, 1, 100052 CrossRef PubMed.
- Y. Huang, H. S. Cao, L. Yang, C. Chen, L. Shabala, M. Xiong, M. L. Niu, J. Liu, Z. Zheng, L. J. Zhou, Z. W. Peng, Z. L. Bie and S. Shabala, Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae, J. Exp. Bot., 2019, 70, 5879–5893 CrossRef CAS PubMed.
- M. L. Niu, Y. Huang, S. T. Sun, J. Y. Sun, H. S. Cao, S. Shabala and Z. L. Bie, Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure, J. Exp. Bot., 2018, 69, 3465–3476 CrossRef CAS PubMed.
- S. Y. Geng, H. Sohail, H. S. Cao, J. Y. Sun, Z. Chen, L. J. Zhou, W. B. Wang, R. W. Ye, L. Yang and Z. L. Bie, An efficient root transformation system for CRISPRCas9-based analyses of shoot–root communication in cucurbit crops, Hortic. Res., 2022, uhab082, DOI:10.1093/hr/uhab082.
- Y. J. Li, Z. H. Tang, Z. Y. Pan, R. G. Wang, X. Wang, P. Zhao, M. Liu, Y. X. Zhu, C. Liu, W. C. Wang, Q. Liang, J. Gao, Y. C. Yu, Z. Y. Li, B. F. Lei and J. Sun, Calcium-mobilizing properties of salvia miltiorrhiza-derived carbon dots confer enhanced environmental adaptability in plants, ACS Nano, 2022, 16, 4357–4370 CrossRef CAS PubMed.
- J. Yang, Y. Cao and N. D. Zhang, Spectrophotometric method for superoxide anion radical detection in a visible light (400-780 nm) system, Spectrochim. Acta, Part A, 2020, 239, 118556 CrossRef CAS PubMed.
- D. Kumar, M. A. Yusuf, P. Singh, M. Sardar and N. B. Sarin, Modulation of antioxidant machinery in alpha-tocopherol-enriched transgenic brassica juncea plants tolerant to abiotic stress conditions, Protoplasma, 2013, 250, 1079–1089 CrossRef CAS PubMed.
- S. P. Kashyap, N. Kumari, P. Mishra, M. D. Prasad, M. Aamir, B. Singh and H. C. Prasanna, Transcriptional regulation-mediating ROS homeostasis and physio-biochemical changes in wild tomato (Solanum chilense) and cultivated tomato (Solanum lycopersicum) under high salinity, Saudi J. Biol. Sci., 2020, 27, 1999–2009 CrossRef CAS PubMed.
- W. Zhou and M. Leul, Uniconazole-induced alleviation of freezing injury in relation to changes in hormonal balance, enzyme activities and lipid peroxidation in winter rape, Plant Growth Regul., 1998, 26(1), 41–47 CrossRef CAS.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- L. Zhou, J. H. Chen, Z. Z. Li, X. X. Li, X. D. Hu, Y. Huang, X. K. Zhao, C. Z. Liang, Y. Wang, L. Sun, M. Shi, X. H. Xu, F. Shen, M. S. Chen, Z. J. Han, Z. Y. Peng, Q. Zhai, J. Chen, Z. F. Zhang, R. L. Yang, J. X. Ye, Z. C. Guan, H. M. Yang, Y. T. Gui, J. Wang, Z. M. Cai and X. Q. Zhang, Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma, PLoS One, 2010, 5, e15224 CrossRef CAS PubMed.
- B. K. Nelson, X. Cai and A. Nebenführ, A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants, Plant J., 2007, 51, 1126–1136 CrossRef CAS PubMed.
- Y. V. Sheludko, Y. R. Sindarovska, I. M. Gerasymenko, M. A. Bannikova and N. V. Kuchuk, Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression, Biotechnol. Bioeng., 2007, 96, 608–614 CrossRef CAS PubMed.
- J. Y. Sun, H. S. Cao, J. T. Cheng, X. M. He, H. Sohail, M. L. Niu, Y. Huang and Z. L. Bie, Pumpkin CmHKT1;1 controls shoot Na+ accumulation via limiting Na+ transport from rootstock to scion in grafted cucumber, Int. J. Mol. Sci., 2018, 19, 2648 CrossRef PubMed.
- Z. Y. Lv, R. Jiang, J. F. Chen and W. S. Chen, Nanoparticle-mediated gene transformation strategies for plant genetic engineering, Plant J., 2020, 104, 880–891 CrossRef CAS PubMed.
- Z. Chen, I. Newman, M. Zhou, N. Mendham, G. Zhang and S. Shabala, Screening plants for salt tolerance by measuring K+ flux: a case study for barley, Plant, Cell Environ., 2005, 28, 1230–1246 CrossRef CAS.
- T. J. Sun, N. Ma, C. Q. Wang, H. F. Fan, M. X. Wang, J. Zhang, J. F. Cao and D. M. Wang, A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean, Front. Plant Sci., 2021, 12, 638340 CrossRef PubMed.
- M. M. Rady, E. M. Desoky, S. M. Ahmed, A. Majrashi, E. F. Ali, S. Arnaout and E. Selem, Foliar nourishment with nano-selenium dioxide promotes physiology, biochemistry, antioxidant defenses, and salt tolerance in phaseolus vulgaris, Plants, 2021, 10(6), 1189 CrossRef CAS PubMed.
- M. B. Hassanpouraghdam, L. V. Mehrabani, M. R. Rahvar, L. Khoshmaram and A. Soltanbeigi, Mollifying salt depression on Anethum graveolens L. by the foliar prescription of Nano-Zn, KNO3, Methanol, and Graphene Oxide, J. Soil Sci. Plant Nutr., 2022, 1–13 Search PubMed.
- L. Rossi, W. L. Zhang, L. Lombardini and X. M. Ma, The impact of cerium oxide nanoparticles on the salt stress responses of Brassica napus L, Environ. Pollut., 2016, 219, 28–36 CrossRef CAS PubMed.
- C. Feng, C. M. He, Y. F. Wang, H. H. Xu, K. H. Xu, Y. Zhao, B. W. Yao, Y. H. Zhang, Y. Zhao, C. K. F. Idrice, J. Luo, D. Q. Sun, H. T. Gao, F. W. Wang, X. W. Li, W. C. Liu, Y. Y. Dong, N. Wang, Y. G. Zhou and H. Y. Li, Genome-wide identification of soybean Shaker K+ channel gene family and functional characterization of GmAKT1 in transgenic Arabidopsis thaliana under salt and drought stress, J. Plant Physiol., 2021, 266, 153529 CrossRef CAS PubMed.
- X. S. Wang, J. L. Zhao, Q. W. Fang, X. C. Chang, M. Y. Sun, W. B. Li and Y. G. Li, GmAKT1 is involved in K+ uptake and Na+/K+ homeostasis in Arabidopsis and soybean plants, Plant Sci., 2021, 304, 110736 CrossRef CAS PubMed.
- F. Yang, T. Wang and L. Liu, Pollen germination is impaired by disruption of a Shaker K+ channel OsAKT1.2 in rice, J. Plant Physiol., 2020, 248, 153140 CrossRef CAS PubMed.
- M. Nieves-Cordones, A. Chavanieu, L. Jeanguenin, C. Alcon, W. Szponarski, S. Estaran, I. Cherel, S. Zimmermann, H. Sentenac and I. Gaillard, Distinct amino acids in the C-linker domain of the Arabidopsis K+ channel KAT2 determine its subcellular localization and activity at the plasma membrane, Plant Physiol., 2014, 164, 1415–1429 CrossRef CAS PubMed.
- M. Nieves-Cordones and I. Gaillard, Involvement of the S4-S5 linker and the C-linker domain regions to voltage-gating in plant Shaker channels: comparison with animal HCN and Kv channels, Plant Signaling Behav., 2014, 9(10), e972892 CrossRef PubMed.
- B. Benito, R. Haro, A. Amtmann, T. A. Cuin and I. Dreyer, The twins K+ and Na+ in plants, J. Plant Physiol., 2014, 171, 723–731 CrossRef CAS PubMed.
- G. Pilot, F. Gaymard, K. Mouline, I. Chérel and H. Sentenac, Regulated expression of Arabidopsis Shaker K+ channel genes involved in K+ uptake and distribution in the plant, Plant Mol. Biol., 2003, 51(5), 773–787 CrossRef CAS PubMed.
- H. Kashtoh and K. H. Baek, Structural and functional insights into the role of guard cell ion channels in abiotic stress-induced stomatal closure, Plants, 2021, 10(12), 2774 CrossRef CAS PubMed.
- F. Gambale and N. Uozumi, Properties of shaker-type potassium channels in higher plants, J. Membr. Biol., 2006, 210, 1–19 CrossRef CAS PubMed.
- H. Gao, W. J. Yang, C. X. Li, X. G. Zhou, D. M. Gao, U. Khashi, M. Rahman, N. H. Li and F. Z. Wu, Gene expression and K+ uptake of two tomato cultivars in response to sub-optimal temperature, Plants, 2020, 9, 65 CrossRef CAS PubMed.
- I. Ahmad, A. Mian and F. J. Maathuis, Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance, J. Exp. Bot., 2016, 67, 2689–2698 CrossRef CAS PubMed.
- W. Han, Y. Ji, W. Wu, J. K. Cheng, H. Q. Feng and Y. Wang, ZMK1 is involved in K+ uptake and regulated by protein kinase ZmCIPK23 in zea mays, Front. Plant Sci., 2021, 12, 517742 CrossRef PubMed.
- M. Nieves-Cordones, A. Lara, R. Rodenas, J. Amo, R. M. Rivero, V. Martinez and F. Rubio, Modulation of K+ translocation by AKT1 and AtHAK5 in Arabidopsis plants, Plant, Cell Environ., 2019, 42, 2357–2371 CrossRef CAS PubMed.
- M. Jayakannan, J. F. B. O. Bose, O. F. R. Z. Babourina, Z. F. S. S. Rengel and S. Shabala, Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel, J. Exp. Bot., 2013, 64(8), 2255–2268 CrossRef CAS PubMed.
- T. Y. Yang, S. Zhang, Y. B. Hu, F. C. Wu, Q. D. Hu, G. Chen, J. Cai, T. Wu, N. Moran, L. Yu and G. H. Xu, The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels, Plant Physiol., 2014, 166, 945–959 CrossRef PubMed.
- A. Lara, R. Rodenas, Z. Andres, V. Martinez, F. J. Quintero, M. Nieves-Cordones, M. A. Botella and F. Rubio, Arabidopsis K+ transporter HAK5-mediated high-affinity root K+ uptake is regulated by protein kinases CIPK1 and CIPK9, J. Exp. Bot., 2020, 71, 5053–5060 CrossRef CAS PubMed.
- Y. Li, J. Hu, J. Qi, F. Zhao, J. Liu, L. Chen, L. Chen, J. Gu, H. Wu and Z. Li, Improvement of leaf K+ retention is a shared mechanism behind CeO2 and Mn3O4 nanoparticles improved rapeseed salt tolerance, Stress Biol., 2022 Search PubMed (provisionally accepted).
- G. D. Chen, Q. Chen, K. J. Qi, Z. H. Xie, H. Yin, P. Wang, R. Z. Wang, Z. Huang, S. L. Zhang, L. Wang and J. Y. Wu, Identification of Shaker K+ channel family members in Rosaceae and a functional exploration of PbrKAT1, Planta, 2019, 250, 1911–1925 CrossRef CAS PubMed.
- Y. Yang, J. L. Han, Y. Zhang, S. Z. Lin, M. X. Liang, L. Z. Zhao and Z. Z. Song, Genome-wide identification and characterization of the Shaker-type K+ channel genes in Prunus persica (L.) batsch, Int. J. Genomics, 2022, 2022, 5053838 Search PubMed.
- A. Razzaq, F. Saleem, M. Kanwal, G. Mustafa, S. Yousaf, A. H. M. Imran, M. K. Hameed, M. S. Khan and F. A. Joyia, Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox, Int. J. Mol. Sci., 2019, 20(16), 4045 CrossRef CAS PubMed.
- X. Ma, X. Zhang, H. Liu and Z. Li, Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9, Nat. Plants, 2020, 6, 773–779 CrossRef CAS PubMed.
- T. Wang, C. Zhang, H. Zhang and H. Zhu, CRISPR/Cas9-mediated gene editing revolutionizes the improvement of horticulture food crops, J. Agric. Food Chem., 2021, 69, 13260–13269 CrossRef CAS PubMed.
- V. M. G. Borrelli, V. Brambilla, P. Rogowsky, A. Marocco and A. Lanubile, The enhancement of plant disease resistance using CRISPR/Cas9 technology, Front. Plant Sci., 2018, 9, 1245 CrossRef PubMed.
- S. Tyagi, K. Kesiraju, M. Saakre, M. Rathinam, V. Raman, D. Pattanayak and R. Sreevathsa, Genome editing for resistance to insect pests: an emerging tool for crop improvement, ACS Omega, 2020, 5, 20674–20683 CrossRef CAS PubMed.
- H. R. Dong, Y. Huang and K. J. Wang, The development of herbicide resistance crop plants using CRISPR/Cas9-mediated gene editing, Genes, 2021, 12, 912 CrossRef CAS PubMed.
- M. U. Farooq, M. F. Bashir, M. U. S. Khan, B. Iqbal and Q. Ali, Role of crispr to improve abiotic stress tolerance in crop plants, Biol. Clin. Sci. Res. J., 2021, 2021, 1 Search PubMed.
- V. V. Santosh Kumar, R. K. Verma, S. K. Yadav, P. Yadav, A. Watts, M. V. Rao and V. Chinnusamy, CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in indica mega rice cultivar MTU1010, Physiol. Mol. Biol. Plants, 2020, 26, 1099–1110 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2en00688j |
| This journal is © The Royal Society of Chemistry 2022 |