Open Access Article
Open Access ArticleDoes size matter? A proteomics-informed comparison of the effects of polystyrene beads of different sizes on macrophages†
Véronique
Collin-Faure
a,
Bastien
Dalzon
a,
Julie
Devcic
a,
Hélène
Diemer
bc,
Sarah
Cianférani
bc and
Thierry
Rabilloud
 *a
*a
aChemistry and Biology of Metals, CEA, IRIG-LCBM, Univ. Grenoble Alpes, CNRS UMR5249, F-38054 Grenoble, France. E-mail: thierry.rabilloud@cnrs.fr
bLaboratoire de Spectrométrie de Masse BioOrganique (LSMBO), IPHC UMR 7178, Université de Strasbourg, CNRS, 67087 Strasbourg, France
cInfrastructure Nationale de Protéomique ProFI - FR2048, 67087 Strasbourg, France
First published on 5th July 2022
Abstract
Plastics are one of the most preoccupying emerging pollutants. Macroplastics released into the environment degrade into microplastics and nanoplastics. Because of their small size, these micro and nano plastic particles can enter the food chain and, in addition to their ecotoxicological effects, contaminate humans with still unknown biological effects. Plastics being particulate pollutants, they are handled in the human body by scavenger cells such as macrophages, which are important players in the immune system. In order to get a better appraisal of the effects of plastic particles on macrophages, we have studied the effects of unmodified polystyrene particles of 0.1, 1 and 10 micrometers of diameter, by a combination of proteomics and targeted validation experiments. Proteomics showed important adaptive changes of the proteome in response to exposure to plastics, with more than one third of the detected proteins showing a significance change in their abundance in response to cell exposure to at least one plastic bead size. These changes affected for example mitochondrial, lysosomal or cytoskeletal proteins. Although an increase in the mitochondrial transmembrane potential was detected in response to 10 micrometer beads, no alteration in cell viability was observed. Similarly, no lysosomal dysfunction and no alteration in the phagocytic capability of the cells was observed in response to exposure to plastics. When the inflammatory response was examined, no increase in the secretion of tumor necrosis factor or interleukin 6 was observed. Oppositely, the secretion of these cytokines in response to lipopolysaccharide was observed after exposure to plastics, which suggested a decreased ability of macrophages to respond to bacterial infection. In conclusion, these results provide a better understanding of the responses of macrophages to exposure to polystyrene particles of different sizes.
Environmental significanceThe presence of micro and nano plastics in our environment is a developing ecological concern, as they can be easily ingested by organisms ranging from invertebrates to humans, and impact them. Whatever the exposure route, the particles that will cross the biological barriers will be taken up by innate immune cells such as macrophages, with a potential impact on their properties. We analyzed the responses of macrophages exposed to polystyrene beads of different sizes (0.1 to 10 μm) by proteomics and targeted validation experiments. Proteomics showed extensive adaptive changes, although no inflammatory response was observed. However, polystyrene-exposed cells showed a slightly decreased ability to respond to a bacterial stimulus, suggesting slightly impaired immune functions of macrophages after exposure to plastics. |
1. Introduction
Because of their attractive technical properties (e.g. resistance to water and to mechanical stress), plastics are widely used in a plethora of products, including single use products such as food packaging. The overall worldwide production of plastics is hundreds of millions of metric tons per year.1 Unfortunately, a fair proportion of these plastics ends up in the environment, with estimates above 500 Mt per year.2 While the accumulation of plastics in seas has been long documented and is still under active study,3–6 they are also widely present in freshwaters, including rivers.7–9 Their accumulation in soils is less documented, but results obtained on marine sediments10,11 do not bring optimism, especially for environments such as landfills or floodplains.12,13 Upon environmental degradation, the plastic wastes degrade into smaller particles, first microplastics (in the millimeter range) and then nanoplastics (micrometer and sub-micrometer range). These very small size plastic particles can enter the food chain,14 and microplastics have indeed been observed in human feces.15 Furthermore, airborne microplastics have been found in human lungs,16 and translocation of plastics in intestinal models has been documented,17,18 leading to the detection of microplastics in human blood19 and placenta.20 This contamination poses in turn the problem of the effects of these micro and nanoplastics on human health.1,21,22 In order to gain further insights into these potential effects, mechanistic studies are desirable, and are more easily conducted in vitro. The epithelial barriers (e.g. intestine) are obvious targets and have thus been studied.17,18 As these studies have documented a translocation across the intestinal barrier, then the immune system becomes an obvious target too, and has indeed been studied.17,23 The interest to study the effects of micro and nanoplastics is indeed reinforced by their detection in lungs,16 where alveolar macrophages are a target cell type of choice.The type of plastic to be tested is also an important parameter. While microplastics collected from the environment would be an ideal target, their fine composition (including their contaminants24–26) is likely to change from one sample to another. Thus, a model system is needed. Although a model system will provide data for a single chemical composition only, and cannot take into account synergistic/cocktail effects as observed for endocrine disruptors,27 it can provide data related to the effects of size, surface chemistry, etc., provided that these parameters can be controlled. However, this plastic model should be taken from the most dominant plastics found in waste, whose composition is known.4,6 Taking these parameters into account, polystyrene appears as the most attractive choice. It can be produced by emulsion polymerization in a variety of sizes ranging from a few tens of nanometers to a few tens of micrometers, can be labelled by a variety of tracers, including metals,28 and its surface chemistry can be controlled.29 Furthermore, polystyrene is known to degrade into nanoplastics30 and seems to be more prevalent in nanoplastics than in microplastics.4 Thus polystyrene has been widely used as a plastic model to investigate several parameters, such as the effects of surface charge,29,31,32 or size.33
Indeed, various effects of polystyrene nanoparticles have been described on macrophages. However, most papers reporting effects have used either very high doses, above 100 μg ml−1 (ref. 33) or surface chemistry with high charge densities (above 20 μeq g−1) of positive or negative groups.29,31,32,34,35 Only a few papers have reported effects with uncharged polystyrene at moderate concentrations (e.g. in ref. 36). Conversely, several papers have reported an absence of clear effects of polystyrene nanobeads on macrophages.17,23,37
In order to get deeper insights into the effects of polystyrene particles on macrophages, we chose to apply a comparative proteomic approach, using beads of three different sizes (0.1, 1 and 10 μm in diameter) covering two orders of magnitude. Objects of 10 μm are in the upper range of what macrophages can internalize,38,39 and are in the microplastics range, while 1 μm objects are in the size range of bacteria and are thus well known for macrophages, and 100 nm objects are considered to be nanoplastics.40,41
Proteomic approaches have been applied with success for studying the effects of other nanomaterials on various cell types (e.g. in ref. 42–45), including macrophages (e.g. in ref. 46–48), and this approach is likely to provide interesting results when studying the effects of polystyrene on macrophages, as suggested by the results obtained on different cell types using amino-modified polystyrene.49
2. Materials and methods
2.1. Plastic particles
For the proteomic experiments and some of the validation experiments, fluorescent polystyrene particles were used. Yellow-green fluorescent particles were purchased from Polysciences (Hirschberg an der Bergstrasse, Germany) with nominal diameters of 0.1 μm (catalog number #17150), 1 μm (catalog number #17154), and 10 μm (catalog number #18140). For validation experiments with flow cytometry, such fluorescent particles cannot be used because of their excessive brightness. Thus unlabelled particles were used. They were purchased from Spherotech, (Lake Forest, IL, USA) with nominal diameters of 50–100 nm (catalog number #PP-008-10), 1–1.4 μm (catalog number #PP-10-10) and 8–12 μm (catalog number #PP-100-10). The suspensions were pasteurized overnight at 80 °C before use in cell culture. The actual size distribution of the particles was checked by optical microscopy for the 1 and 10 μm beads, and by dynamic light scattering (DLS) for the 0.1 μm beads. The zeta potential was measured on a Malvern Zetasizer instrument.2.2. Cell culture and sorting
The J774A.1 cell line (mouse macrophages) was purchased from European cell culture collection (Salisbury, UK). Cells were routinely propagated in DMEM supplemented with 10% fetal bovine serum (FBS) in non-adherent flasks (Cellstar flasks for suspension culture, Greiner Bio One, Les Ulis, France). For routine culture, the cells were seeded at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml and passaged two days later, with a cell density ranging from 800
000 cells per ml and passaged two days later, with a cell density ranging from 800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1
000 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. For exposure to plastic particles and to limit the effects of cell growth, cells were seeded at 500
000 cells per ml. For exposure to plastic particles and to limit the effects of cell growth, cells were seeded at 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml, allowed to settle and recover for 24 hours, and then exposed to the particles at 40 μg ml−1 for 24 hours before harvesting for the experiments. For cells exposed to fluorescent nanoparticles, the cells having internalized the particles were selected on the basis of fluorescence by flow cytometry using a Melody cell sorter (Becton Dickinson Biosciences, Le Pont-de-Claix, France). When nonfluorescent plastic beads were used, the cells having internalized the 10 μm particles were selected on the basis of light scattering, as this parameter was shown to be related to bead internalization using fluorescent beads. The cells were collected in 1 ml of culture medium, unless stated otherwise.
000 cells per ml, allowed to settle and recover for 24 hours, and then exposed to the particles at 40 μg ml−1 for 24 hours before harvesting for the experiments. For cells exposed to fluorescent nanoparticles, the cells having internalized the particles were selected on the basis of fluorescence by flow cytometry using a Melody cell sorter (Becton Dickinson Biosciences, Le Pont-de-Claix, France). When nonfluorescent plastic beads were used, the cells having internalized the 10 μm particles were selected on the basis of light scattering, as this parameter was shown to be related to bead internalization using fluorescent beads. The cells were collected in 1 ml of culture medium, unless stated otherwise.
2.3. Confocal microscopy
For confocal microscopy experiments, the cells were seeded on glass coverslips and allowed to adhere for 24 hours prior to the double exposure. At the end of the exposure period, the cells were washed twice for 5 min in PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. After two washes (5 min in PBS), they were permeabilized in Triton X100 (0.1% w/v) for 5 min at room temperature. After two more washes in PBS, Phalloidin-Atto 550 (Sigma) (250 nM) or Phalloidin-Atto 390 (500 nM) was added to the cells and left for 20 min at room temperature in the dark. Coverslip-attached cells were washed, and cell nuclei were colored via DAPI (1 μg ml−1, Eurobio, Les Ulis, France). The coverslips were washed twice and placed on microscope slides (Thermo Scientific, Illkirch, France) using a Vectashield mounting medium and imaged using a Zeiss LSM 800 confocal microscope. The images were processed using the ImageJ software.2.4. Neutral red assay
The 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sorted cells were recovered by centrifugation (200g, 5 minutes), resuspended in 100 μl of complete culture medium, seeded in 96 well adherent plates and allowed to settle and adhere for 24 hours. A solution containing 2 mg ml−1 neutral red in EtOH 50% was prepared. This solution was diluted 100 fold in pre-warmed complete culture medium. The medium in the 96 well plates was removed and replaced by the neutral red containing medium. After incubation at 37 °C for 2 hours, cells were washed twice with PBS and eluted with 200 μl of acid–alcohol solution (acetic acid 1% v/v, ethanol 50% v/v). The plates were incubated for 30 min at room temperature on a rotating table. The red color released was read at 540 nm.50
000 sorted cells were recovered by centrifugation (200g, 5 minutes), resuspended in 100 μl of complete culture medium, seeded in 96 well adherent plates and allowed to settle and adhere for 24 hours. A solution containing 2 mg ml−1 neutral red in EtOH 50% was prepared. This solution was diluted 100 fold in pre-warmed complete culture medium. The medium in the 96 well plates was removed and replaced by the neutral red containing medium. After incubation at 37 °C for 2 hours, cells were washed twice with PBS and eluted with 200 μl of acid–alcohol solution (acetic acid 1% v/v, ethanol 50% v/v). The plates were incubated for 30 min at room temperature on a rotating table. The red color released was read at 540 nm.50
2.5. Phagocytosis assay
For this assay, it was not necessary to perform cell sorting. The cells were first exposed to the yellow-green fluorescent particles. After 24 hours of exposure, the cells were exposed to 0.5 μm latex beads (carboxylated surface, fluorescent red labelled, catalog number #L3280 from Sigma, Saint Quentin Fallavier, France) for 2.5 hours. After this second exposure, the cells were collected, rinsed twice with PBS, and analyzed for two types of fluorescence (green and red) on a Melody flow cytometer.2.6. Cytokine release assays
The 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sorted cells were recovered by centrifugation (200g, 5 minutes), seeded in 100 μl of complete culture medium in 96 well adherent plates and allowed to settle and adhere for 24 hours. At the end of this period, the medium was collected, and tumor necrosis factor (TNFα) (catalog number 558299, BD Biosciences, Le Pont de Claix, France) and interleukin 6 (IL-6) (catalog number 558301, BD Biosciences, Le Pont de Claix) levels were measured using the Cytometric Bead Array Mouse Inflammation Kit (catalog number 558266, BD Biosciences, Le Pont de Claix), and analyzed with FCAP Array software (3.0, BD Biosciences) according to the manufacturer's instructions.
000 sorted cells were recovered by centrifugation (200g, 5 minutes), seeded in 100 μl of complete culture medium in 96 well adherent plates and allowed to settle and adhere for 24 hours. At the end of this period, the medium was collected, and tumor necrosis factor (TNFα) (catalog number 558299, BD Biosciences, Le Pont de Claix, France) and interleukin 6 (IL-6) (catalog number 558301, BD Biosciences, Le Pont de Claix) levels were measured using the Cytometric Bead Array Mouse Inflammation Kit (catalog number 558266, BD Biosciences, Le Pont de Claix), and analyzed with FCAP Array software (3.0, BD Biosciences) according to the manufacturer's instructions.
2.7. Mitochondrial transmembrane potential assay
For this assay, it was not necessary to perform cell sorting. Cells were cultivated in non-adherent 6 well plates and exposed to non-fluorescent plastic particles for 24 hours.For the mitochondrial transmembrane potential assay, Rhodamine 123 (80 nM final concentration) was added to the cultures, which were further incubated at 37 °C for 30 minutes. At the end of this period, the cells were collected, washed in cold PBS containing 0.1% glucose, resuspended in PBS glucose and analyzed for green fluorescence on a Melody flow cytometer.
2.8. Surface marker assays
After exposure to non-fluorescent polystyrene beads, the cells recovered from 6 well plates were collected, rinsed with PBS and labelled with 200 μl of 50 fold diluted FITC-labelled anti CD11b antibody (BD Pharmingen, catalog number #553310, Becton Dickinson Biosciences, Le Pont-de-Claix, France) or with 200 μl of 100 fold diluted FITC-labelled anti CD38 antibody (BD Pharmingen, catalog number #558813) in PBS for 30 minutes at room temperature in the dark. One milliliter of PBS was then added, and the cells were collected by centrifugation, resuspended in PBS and analyzed for green fluorescence on a Melody flow cytometer.2.9. Proteomics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fluorescence-positive cells in 1 ml of PBS. For the negative control, the same number of cells was collected without any selection on fluorescence. The cell suspensions were centrifuged (200g, 5 minutes) and lysed in 15 μl of extraction buffer (4 M urea, 2.5% cetyltrimethylammonium chloride, 100 mM sodium phosphate buffer pH 3, 150 μM methylene blue). The extraction was allowed to proceed at room temperature for 1 hour, after which the lysate was centrifuged (15
000 fluorescence-positive cells in 1 ml of PBS. For the negative control, the same number of cells was collected without any selection on fluorescence. The cell suspensions were centrifuged (200g, 5 minutes) and lysed in 15 μl of extraction buffer (4 M urea, 2.5% cetyltrimethylammonium chloride, 100 mM sodium phosphate buffer pH 3, 150 μM methylene blue). The extraction was allowed to proceed at room temperature for 1 hour, after which the lysate was centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 30 minutes) to pellet the nucleic acids. The supernatants were then stored at −20 °C until use.
000g, 30 minutes) to pellet the nucleic acids. The supernatants were then stored at −20 °C until use.
As mentioned above, methylene blue was included in the cell lysis buffer. The other initiator solutions consisted of a 1 M solution of sodium toluene sulfinate in water and in a saturated water solution of diphenyliodonium chloride. The ready-to-use polyacrylamide solution consisted of 1.2 ml of a commercial 40% acrylamide/bis solution (37.5/1) to which 100 μl of diphenyliodonium chloride solution, 100 μl of sodium toluene sulfinate solution and 100 μl of water were added.
To the protein samples (15 μl), 5 μl of acrylamide solution were added and mixed by pipetting in a 500 μl conical polypropylene microtube. 100 μl of water-saturated butanol were then layered on top of the samples, and polymerization was carried out under a 1500 lumen 2700 K LED lamp for 2 hours, during which the initially blue gel solution discolored. At the end of the polymerization period, butanol was removed, and the gel plugs were fixed for 2 × 1 h with 200 μl of 30% ethanol–2% phosphoric acid, followed by a 30 minute wash in 30% ethanol. The fixed gel plugs were then stored at −20 °C until use.
Gel plug processing, digestion, peptide extraction and nanoLC-MS/MS were performed as previously described,52 without the robotic protein handling system and using a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Further details are available in Methods S1.†
For protein identification, the MS/MS data were interpreted using a local Mascot server with the MASCOT 2.6.2 algorithm (Matrix Science, London, UK) against an in-house database containing all Mus musculus and Rattus norvegicus entries from UniProtKB/SwissProt (version 2019_10, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 sequences) and the corresponding 50
313 sequences) and the corresponding 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 313 reverse entries. Spectra were searched with a mass tolerance of 10 ppm for MS and 0.07 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Trypsin was specified as an enzyme. Acetylation of protein n-termini, carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Identification results were imported into Proline software version 2.1 (https://proline.profiproteomics.fr/) for validation. Peptide Spectrum Matches (PSM) with pretty rank equal to one were retained. The False Discovery Rate was then optimized to be below 1% at the PSM level using the Mascot Adjusted E-value and below 1% at the Protein Level using the Mascot Mudpit score.
313 reverse entries. Spectra were searched with a mass tolerance of 10 ppm for MS and 0.07 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Trypsin was specified as an enzyme. Acetylation of protein n-termini, carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Identification results were imported into Proline software version 2.1 (https://proline.profiproteomics.fr/) for validation. Peptide Spectrum Matches (PSM) with pretty rank equal to one were retained. The False Discovery Rate was then optimized to be below 1% at the PSM level using the Mascot Adjusted E-value and below 1% at the Protein Level using the Mascot Mudpit score.
Mass spectrometry data are available via ProteomeXchange with the identifier PXD031609.
Proteins were considered as significantly different if their p value in the Student T test was inferior to 0.05. No quantitative change threshold value was applied. However, a false discovery rate was calculated for each selected protein, using the sequential Fisher test approach.54 The selected proteins were then submitted to pathway analysis using the DAVID tool,55 with a cutoff value set at an FDR of 0.1.
3. Results
3.1. Plastic particle uptake
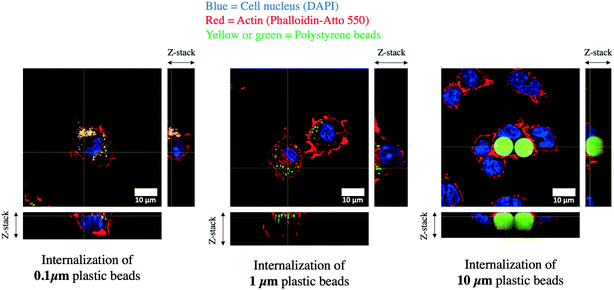
We first checked the actual size distribution of the particles. The measured diameters were 106.9 ± 3.3 nm for the 0.1 μm beads, 1.09 ± 0.16 μm for the 1 μm beads, and 9.61 ± 1.21 μm for the 10 μm beads (Fig. S1†). The 0.1 μm beads had a zeta potential of −63.30 ± 13.7 mV, the 1 μm beads had a zeta potential of −37.87 ± 74.5 mV and the 10 μm beads had a zeta potential of 68.02 ± 8.1 mV. We then checked that all the particles tested were effectively internalized by the macrophages. To this purpose, we examined cells exposed to the various plastic particles by confocal microscopy. The results, shown in Fig. 1, indicated an efficient uptake, even for 10 μm particles. These large particles induced nevertheless a deformation of the cells that was not observed for smaller particles.3.2. Analysis of the proteomic results
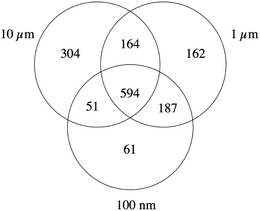
A problem that was encountered for performing proteomics is due to the fact that when the diameter of the beads increased by 10-fold, the volume and thus the mass of the beads increased by 1000-fold. This meant in turn that at equal plastic mass loaded into the cultures, the number of beads present decreased by 1000-fold. We decided to keep a constant plastic mass across the various sizes, which meant in turn that at the non-toxic dose of 40 μg ml−1, there was less than one 10 μm bead per cell present in the culture. It was then necessary for the proteomic experiments to resort to cell sorting, which allowed us to select and compare only cells that had internalized the plastic beads. Indeed, the observed percentage of fluorescence-positive cells was 98.1 ± 0.3% for the cells treated with 0.1 μm beads, 97 ± 1% for the cells treated with 1 μm beads, and 5.1 ± 0.4% for the cells treated with 10 μm beads. As the sorting process could induce cellular responses per se, which may reflect in the proteome, we submitted all cells to the sorting process prior to the proteomic analyses, even the untreated cells.Although performed on 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells, proteomics was able to detect and quantify 2855 proteins (Table S1†). Proteins modulated by the internalization of plastic particles were selected on the basis of a T test <0.05 in the comparison of plastic-treated cells compared to unexposed controls, which resulted in three separate lists, one for each plastic particle size (Tables S2–S4†). These tables indicated that the number of modulated proteins increased with the size of the plastic particles. While 893 proteins were modulated in cells treated with 0.1 μm beads, 1107 were modulated by the exposure to 1 μm beads and 1113 by the exposure to 10 μm beads. The comparison of these three lists, schematized by the Venn diagram shown in Fig. 2 and in Tables S5–S8,† showed that 594 proteins were modulated for all bead sizes. This important proportion can be viewed as the common response to polystyrene particles.
000 cells, proteomics was able to detect and quantify 2855 proteins (Table S1†). Proteins modulated by the internalization of plastic particles were selected on the basis of a T test <0.05 in the comparison of plastic-treated cells compared to unexposed controls, which resulted in three separate lists, one for each plastic particle size (Tables S2–S4†). These tables indicated that the number of modulated proteins increased with the size of the plastic particles. While 893 proteins were modulated in cells treated with 0.1 μm beads, 1107 were modulated by the exposure to 1 μm beads and 1113 by the exposure to 10 μm beads. The comparison of these three lists, schematized by the Venn diagram shown in Fig. 2 and in Tables S5–S8,† showed that 594 proteins were modulated for all bead sizes. This important proportion can be viewed as the common response to polystyrene particles.
In order to gain further insight into the significance of the observed changes, these 3 lists were used to perform pathway analyses by the DAVID software. The results of the pathway analyses are detailed in Tables S9–S11,† respectively. The main pathways highlighted by the DAVID software are summarized in Fig. 3. Some highlighted pathways indicated a global stress response (e.g. translation, proteasome, oxidoreductases, carbon metabolism) which is expected for any cellular stress, while other pathways appeared more specific of cellular internalization of particles (e.g. endocytosis, lysosomes, actin cytoskeleton, innate immunity).
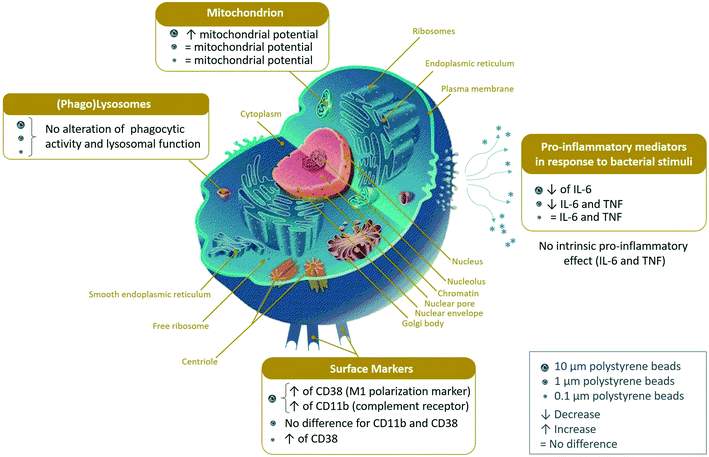 | ||
| Fig. 3 Modified pathways observed with the proteomic study (DAVID pathway analysis). Adapted from the common Wikimedia cell diagram. | ||
In addition to the results of the pathway analysis, a manual analysis of the 3 lists was also performed to retrieve isolated protein changes, which are discarded by pathway analyses but may be of interest in the frame of macrophage physiology. This analysis of the proteomic results led us to perform validation experiments on several functions.
3.3. Mitochondria
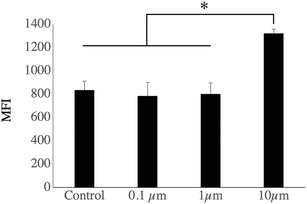
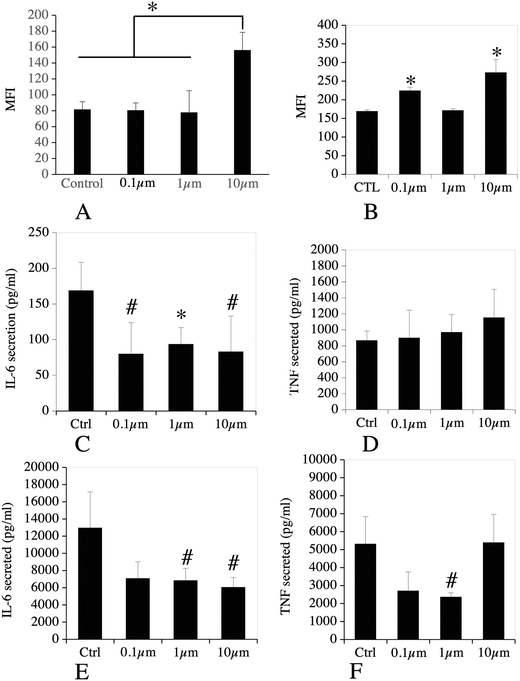
The list of the mitochondrial proteins modulated by cell treatment with one of the 3 plastic beads used is presented in Table S12,† and included 109 proteins, of which 105 showed at least one significant change. Among these, 32 were modulated by all sizes of polystyrene beads, while an additional 4 were modulated by the micrometric beads (1 and 10 μm) and an additional 8 were modulated by the small beads only (0.1 and 1 μm). Of the 52 that were modulated by only one bead, 40 were modulated by the large beads, 2 by the nanobeads only and 10 by the 1 μm beads only. The main classes of mitochondrial proteins modulated upon exposure to polystyrene were the respiratory complexes I and III (NADH–ubiquinone oxidoreductase and ubiquinol–cytochrome c oxidoreductase, respectively), ATP synthase and the MICOS complex. The MICOS complex participates in the organization of mitochondrial structure, and aberrant mitochondrial structure is found in a range of diseases.56 Although the changes observed in protein abundances were usually of low magnitude for each protein, the changes occurring on multiple subunits of the same complexes may suggest a functional alteration. To test this hypothesis, we tested the mitochondrial transmembrane potential as a proxy of mitochondrial function. The results, shown in Fig. 4, indicated that the transmembrane potential was increased after treatment with 10 μm beads, but did not differ significantly from the control cells for the whole cell population treated with 0.1 μm and 1 μm beads, as for those two bead sizes almost all the cells had internalized beads, as demonstrated with the fluorescent beads.3.4. Endosomes, lysosomes, phagocytosis
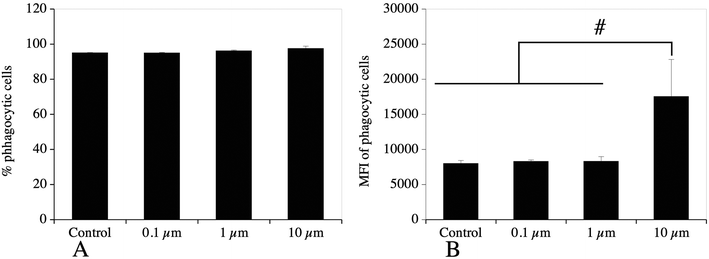
For this important macrophage function, we gathered the proteins selected under the “lysosome”, “endosome”, “autophagy” and “V-type proton ATPase”. The list of the proteins falling under these categories and whose abundance was modulated by cell treatment with one of the 3 plastic beads used is presented in Table S13,† and included 71 proteins, of which 67 showed at least one significant change. Among the 11 subunits of the V-type proton ATPase that were detected in the proteomic screen, 8 showed at least one significant change. Among the 67 modulated proteins, 64 showed significant increases after cell treatment with large beads, including the 8 modulated V-type proton ATPase subunits. The only proteins that were not modulated by the large 10 μm beads were Retreg1 (Q5FVM3), modulated by cell treatment with 1 μm beads only, and Lrp1 (Q91ZX7) and Acp2 (P20611) both modulated by cell treatment with both 0.1 μm and 1 μm beads. Of the 64 proteins modulated in response to treatment with the 10 μm beads, 28 were modulated also in response to treatment with both 0.1 μm and 1 μm beads, i.e. by all sizes tested. Another 5 proteins, i.e. Eps15 (P42567), Ankfy1 (Q810B6), Atp6v1b2 (P62814), Rufy1 (Q8BIJ7) and Ap1g1 (P22892), were modulated by cell treatment with both 10 μm and 1 μm beads. Two proteins, Lamp2 (P17047) and Slc15a3 (Q8BPX9), were modulated by cell treatment with both 10 μm and 0.1 μm beads, but not 1 μm beads.These results prompted us to carry out validation experiments. We first checked the phagocytic ability of the cells exposed to the polystyrene beads. To this purpose, we exposed the cells first to green fluorescent beads of the various sizes for 24 hours, changed the medium, and then exposed the cells to red fluorescent test beads of 0.5 μm diameter to test the phagocytic activity. These experiments were first analyzed by flow cytometry, and the results are shown in Fig. 5. A very high proportion (>90%) of the cells that have internalized green plastic beads were still able to internalize the test beads, and the results were not significantly different between the conditions tested. In order to confirm these results, the experiments were carried out again and analyzed by confocal microscopy. The results, shown in Fig. 6, showed that cells that have internalized the plastic beads are still able to internalize the test beads.
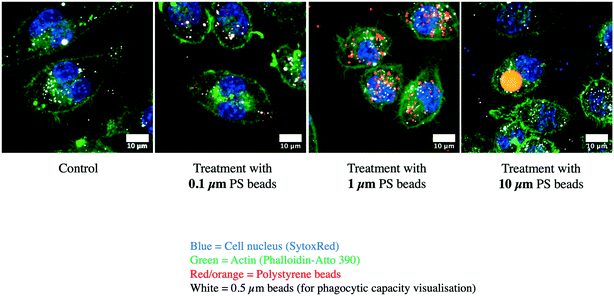 | ||
| Fig. 6 Confocal microscopy analysis of phagocytosis. Cells were treated as in Fig. 5, then fixed and analyzed by confocal microscopy. Blue: nucleus. Green: actin. Red-yellow: green-fluorescent polystyrene beads. White: test red-fluorescent carboxypolystyrene beads. | ||
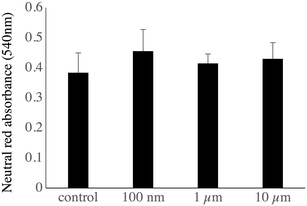
As the changes observed in the abundances of several subunits of the vacuolar proton pump (V-type proton ATPase) may indicate a change in the ability of the lysosomes to acidify their interior, we checked this parameter by two series of experiments. In the first series, the cells having internalized green fluorescent beads were sorted by flow cytometry, put again in culture for 24 hours and submitted to the neutral red assay, which requires a functional proton pump and intact lysosomes to accumulate the dye in the cells. The results, shown in Fig. 7, showed that the neutral red accumulation was not different between the control cells and those treated with any of the tested beads.
3.5. Immunity-related proteins
For defining this subset of proteins, we gathered the proteins highlighted under the “innate immunity” keyword in the pathway analysis, to which we added the Toll-Like Receptors (TLR) and CD antigens that were detected in the proteomic analysis. Furthermore, we added proteins known from their annotations to modulate the pathways of interest, e.g. proteins such as leupaxin (Q99N69) or fermitin 3 (Q8K1B8), which modulate integrin signalling57,58 or the Pycard protein (Q9EPB4, increased by 2-fold in plastic-treated cells) also known as Asc, which plays a role in the inflammasome,59 or accessory factors of the NADPH oxidase such as Ncf2 (O70145) or Ncf4 (P97369), increased in plastic-treated cells. In this particular process, the CD and TLR proteins were not selected on the basis of their changes in abundances upon cell treatment with polystyrene beads, but rather as descriptors of the immune functions of the macrophages. The list of the proteins falling under these categories is presented in Table S14.† It encompassed 47 proteins, of which 30 showed significant abundance changes upon cell treatment with at least one of the 3 plastic beads. Among these 30 proteins, only 2 were not modulated by the 10 μm beads. These were integrin alpha M-CD11b (P05555), whose abundance was changed only in response to 1 μm beads, (30% decrease) and TLR7 (P58681), whose abundance was changed only in response to 0.1 μm beads (2.5-fold increase).In order to get further insights into the modulation of the macrophages' immune functions by polystyrene beads, we tested the surface expression of CD11b, i.e. one of the subunits of Complement Receptor 3, of CD38, used as a marker of macrophage activation,60 and also the production of the proinflammatory cytokines TNF〈 and IL-6, as the proteomic data suggested a putative cell activation effect of the polystyrene beads (e.g. through the induction of Pycard, Card9, Cd2ap, Fermt3), which could however be balanced by negative regulators such as Tnfaip8l2 (ref. 61) or Nmi.62 The results, shown in Fig. 8, indicated that no change in the surface expression of CD11b was observed for the cells treated with 100 nm and 1 μm beads and that a significant increase (1.9-fold) was observed for the cells treated with 10 μm beads (Fig. 8A). A moderate but significant increase of the surface expression of CD38 was observed for the cells treated with 0.1 μm (1.3-fold) and 10 μm beads (1.6-fold), but not for cells exposed to 1 μm beads.
No significant increase of the secretion of TNF〈 and IL-6 was observed for cells treated with polystyrene beads compared to control cells (Fig. 8C & D). Oppositely, a reduction of the basal secretion of IL-6 was observed. We then investigated the secretion of TNF〈 and IL-6 in response to both beads' exposure and stimulation with LPS. The results, shown in Fig. 8E & F, showed a reduction of the secretion of TNF〈 in response to 0.1 μm and 1 μm beads, which was statistically significant only for the 1 μm beads. For IL-6, the reduction was observed in response to all beads, but was significant only in response to the 1 and 10 μm beads.
4. Discussion
Plastics are released into the environment mostly as macroplastics, which progressively degrade into microplastics (defined by a size <5 mm63) and nanoplastics (defined by a size ≤1 μm40,41). These small size particles can enter the food chain and be transferred to various species, including mammals. Besides food chain contamination, airborne contamination has also been described.16 This poses in turn the problem of their effects on cells. In this context, macrophages are a cell type of high interest, as they are the scavenger cells in charge of cleaning bodies from all particulate materials. Noteworthily, macrophages are found in all vertebrates and macrophage homologs (e.g. hemocytes) are found in invertebrates, as demonstrated in the 19th century by Metchnikoff.64,65 It is therefore not surprising that alterations in the innate immune system have been described in response to micrometric microplastics and nanoplastics in annelids,66 bivalves67 and in fish.68 This should be understood in parallel with the demonstration that plastic particles up to 10 μm can translocate from the mammalian intestine in the underlying immune tissue,69 by mechanisms that are not well understood yet.70In this general context, we aimed at bringing new information on the reactions of macrophages to internalization of plastic beads of different sizes. To this purpose, we chose a proteomic approach, which is closer to cell physiology than transcriptomics,71 although transcriptomics offers unequalled comprehensiveness and single cell capabilities.68
We also chose to work with polystyrene particles, as they are not only available in controlled dimensions but are also well represented in environmental micro and nanoplastics.3,4 We also decided to work with polystyrene particles with unmodified surfaces, as strong surface charge densities have been shown to severely modify the cellular responses.31–35 Negatively charged plastic beads are recognized by the scavenger receptors,72 as are many negatively charged polymers, and this may explain the effects induced by such strongly-charged plastics.35 Regarding the dose used for the experiments, we selected a non-toxic dose (40 μg ml−1) based on previous experiments carried out on human blood cell lines with 0.1 μm polystyrene beads22 and on the same J774 murine macrophage cell line with micrometric polystyrene beads,73 and well below the 100 μg ml−1 dose reported on human cells.33 When discussing our results, the overall striking trend is that the large beads (10 μm) have a more profound effect on macrophages than smaller beads. When taking into account the cell deformation required to accommodate such large beads, this shall not come as a surprise, and the large number of modulated proteins related to cytoskeleton argues in that direction. Furthermore, this result is in line with previously-published results, especially on invertebrates, where micrometer sized plastics induce more biological effects than smaller plastics.74
Regarding lysosomes, the neutral red assay, which relies on the active pumping of the neutral red dye in acidic, functional lysosomes,50 revealed no gross lysosomal alteration upon internalization of the plastic beads.
Regarding mitochondria, several protein modulations were observed. They encompassed mainly proteins involved in lipid metabolism (Decr1, Acad9, Acsf2, Acot2, Acot13, Ech1, Acadl, Acadm, Acsl1, Acsl4), which were all induced. This suggested an increase in lipid metabolism, probably linked to an increased energy demand for internalizing and “handling” beads intracellularly. Proteins involved in the maintenance of mitochondrial structure were also increased, such as the MICOS subunits Mic13 and Mic27,56 or the Sam50 protein.75 These changes are consistent with the perturbations likely induced by the internalization of large beads, which may require in turn adaptations to maintain mitochondrial morphology. Increases were also observed for some subunits of the respiratory complexes, such as NADH dehydrogenase complex beta 3 and beta 5, BC1 complex subunits 7 and 9, and cytochrome oxidase subunit 6C. In complement to these changes, an increase was also detected for the Lrpprc protein, which indirectly controls the synthesis of the mitochondrially-encoded cytochrome oxidase subunits.76 Mirroring these proteomic responses, we detected an increase in the mitochondrial transmembrane potential in response to exposure to 10 μm beads. While a decrease in the mitochondrial transmembrane potential is clearly a sign of mitochondrial dysfunction, an increase is not devoid of physiological consequences on cells. It has been demonstrated that a high mitochondrial transmembrane potential correlates with an increased production of reactive oxygen species.77,78 Correspondingly we observed an increase for several proteins of the mitochondrial antioxidant system (Gpx4, Prdx 3, Prdx 5, Txnrd2) in the proteomic screen, suggesting a response against an oxidative stress. Interestingly, no oxidative stress has been observed on THP1 monocytic cells23 or on Caco-2 intestinal cells79 exposed to 0.1 μm polystyrene beads, while oxidative stress has been described for RAW264.7 macrophage cells exposed to 40 nm polystyrene beads.36
We also investigated some immune functions of macrophages after exposure to the polystyrene beads. First of all, phagocytosis was unaltered after exposure to all bead sizes. As phagocytosis is quite a complex process involving energy expenditure, extensive membrane remodeling, cellular movements and so on, this indicates that the cells are quite healthy even after ingestion of the polystyrene beads.
We also evaluated in more detail some other immune functions of macrophages such as their polarization and their inflammatory reaction. We evaluated macrophage polarization through the surface expression of CD38,60 used as a marker of M1 (pro-inflammatory) activation. We observed a weak but significant activation in response to exposure to 0.1 μm and 10 μm beads, but not to 1 μm beads. This prompted us to evaluate the secretion of the two pro-inflammatory proteins TNF〈 and IL-6. We did not observe any increase in the production of pro-inflammatory cytokines in response to cell exposure to polystyrene. This was in agreement with some previously-published papers also using polystyrene nanoparticles in the 0.1 μm range,23,73 but not with other papers using smaller nanoparticles.36 The discrepant result between the activation seen through CD38 expression and the lack of induction of cytokine expression may be linked to the fact that negative modulators of the TLR signalling such as Tnfaip8l2 (ref. 61) or NMI62 are increased in response to exposure to polystyrene beads.
An increase in the levels of the Toll-like receptor TLR2 has been observed in fish intestine after exposure to polystyrene beads.68 We rather observed a small, non-significant decrease of TLR2 in our macrophage system. This suggests that the increase in the expression of TLR2 observed in total fish intestine likely arises from another cell type. Nevertheless, we observed an increase in the amounts of other Toll-like receptors, in our case TLR7 and TLR13, in response to macrophage exposure to 0.1 μm polystyrene beads, and also for TLR13 in response to cell exposure to 10 μm beads. This suggested that the responses to immune challenges may be altered in cells exposed to plastics. To check this hypothesis, we sorted plastic-exposed cells, cultured them and exposed them to an LPS challenge. A weakly significant decrease (p ≤ 0.1) was observed for the secretion of IL-6 in response to all beads. For TNF〈, a weakly significant decrease (p ≤ 0.1) was observed in response to cell exposure to 0.1 μm and 1 μm beads, but not to 10 μm beads. This suggests an altered ability of plastic-exposed macrophages to respond to a bacterial challenge.
In the general context of inflammation, the Pycard protein (also known as Asc) was found strongly increased (2-fold or more) in response to all sizes of polystyrene beads. Asc is a component of several different types of inflammasomes,80 is found in vertebrates including fish,81 and plays a role in the activation of the inflammasome. Furthermore, Asc polymers may act as a pro-inflammatory signal when released extracellularly,82 which plays a role in the observed effects of ambient particles on microglial cells,83 which are macrophages located in the brain.84 Thus, the increase in Asc expression may be a signal to further investigate for a better understanding of the possible inflammatory impacts of polystyrene particles.
In conclusion, the absence of toxicity of polystyrene beads of all sizes at the concentration used suggests that the extensive proteome changes observed are mostly adaptive changes. This hypothesis is further supported by the fact that despite the extensive proteome changes observed in response to cell exposure to 0.1 μm and 1 μm beads, no alteration of the basal cell parameters (e.g. lysosomal activity, mitochondrial transmembrane potential) was detected in the validation experiments. Nevertheless, the alterations detected in the specialized immune functions of macrophages, and especially the overexpression of the Asc protein and the decreased response to a LPS challenge suggest that polystyrene beads may affect the fine functioning of the innate immune system and have indirect adverse effects. Furthermore, these effects may be size dependent. Because macrophages are able to internalize rather large micrometric particles, they may prove more sensitive to these particles than to smaller ones, a situation that may be different for other, non-phagocytic cell types.
In addition, it should be kept in mind that these moderate effects were observed on pristine particles, and are in agreement with the literature.17,23,37 However, synergistic effects of plastic particles with adsorbed toxicants and additives have been widely described in the literature on a variety of systems,85–92 and this concept can be extended to the eco-corona for macromolecules,93 or to the still poorly-known chemicals arising from plastic degradation and especially photodegradation.94 Thus, the results described here shall not be viewed as an argument in favor of the low impact of environmental plastics.
Funding
This work used the flow cytometry facility supported by GRAL, a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003), as well as the platforms of the French Proteomic Infrastructure (ProFI) project (grant ANR-10-INBS-08-03). This work was carried out in the frame of the PlasticHeal project, which has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 965196.Author contributions
TR designed and supervised the study. VCF, HD and SC performed the proteomic experiments. BD performed the confocal microscopy experiments. BD, VCF and JD performed and interpreted the flow cytometry experiments. TR drafted the initial version of the manuscript, which was completed and amended by all co-authors, who approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors wish to thank Anaelle Torres and Marie Carrière for the measurements of the zeta potential of the particles.References
- R. Lehner, C. Weder, A. Petri-Fink and B. Rothen-Rutishauser, Emergence of Nanoplastic in the Environment and Possible Impact on Human Health, Environ. Sci. Technol., 2019, 53, 1748–1765 CrossRef CAS PubMed.
- Y. V. Fan, P. Jiang, R. R. Tan, K. B. Aviso, F. You, X. Zhao, C. T. Lee and J. J. Klemeš, Forecasting plastic waste generation and interventions for environmental hazard mitigation, J. Hazard. Mater., 2022, 424, 127330 CrossRef CAS PubMed.
- K. Enders, R. Lenz, C. A. Stedmon and T. G. Nielsen, Abundance, size and polymer composition of marine microplastics ≥ 10 μm in the Atlantic Ocean and their modelled vertical distribution, Mar. Pollut. Bull., 2015, 100, 70–81 CrossRef CAS PubMed.
- A. Ter Halle, L. Jeanneau, M. Martignac, E. Jardé, B. Pedrono, L. Brach and J. Gigault, Nanoplastic in the North Atlantic Subtropical Gyre, Environ. Sci. Technol., 2017, 51, 13689–13697 CrossRef CAS PubMed.
- J. A. Brandon, A. Freibott and L. M. Sala, Patterns of suspended and salp-ingested microplastic debris in the North Pacific investigated with epifluorescence microscopy, Limnol. Oceanogr. Lett., 2020, 5, 46–53 CrossRef.
- M. Kedzierski, M. Palazot, L. Soccalingame, M. Falcou-Préfol, G. Gorsky, F. Galgani, S. Bruzaud and M. L. Pedrotti, Chemical composition of microplastics floating on the surface of the Mediterranean Sea, Mar. Pollut. Bull., 2022, 174, 113284 CrossRef CAS PubMed.
- T. Mani, A. Hauk, U. Walter and P. Burkhardt-Holm, Microplastics profile along the Rhine River, Sci. Rep., 2015, 5, 17988 CrossRef CAS PubMed.
- C. Scherer, A. Weber, F. Stock, S. Vurusic, H. Egerci, C. Kochleus, N. Arendt, C. Foeldi, G. Dierkes, M. Wagner, N. Brennholt and G. Reifferscheid, Comparative assessment of microplastics in water and sediment of a large European river, Sci. Total Environ., 2020, 738, 139866 CrossRef CAS PubMed.
- L. Weiss, W. Ludwig, S. Heussner, M. Canals, J.-F. Ghiglione, C. Estournel, M. Constant and P. Kerhervé, The missing ocean plastic sink: Gone with the rivers, Science, 2021, 373, 107–111 CrossRef CAS PubMed.
- E. S. Jones, S. W. Ross, C. M. Robertson and C. M. Young, Distributions of microplastics and larger anthropogenic debris in Norfolk Canyon, Baltimore Canyon, and the adjacent continental slope (Western North Atlantic Margin, U.S.A.), Mar. Pollut. Bull., 2021, 174, 113047 CrossRef PubMed.
- L. Cutroneo, M. Capello, A. Domi, S. Consani, P. Lamare, P. Coyle, V. Bertin, D. Dornic, A. Reboa, I. Geneselli and M. Anghinolfi, Microplastics in the abyss: a first investigation into sediments at 2443-m depth (Toulon, France), Environ. Sci. Pollut. Res., 2022, 29, 9375–9385 CrossRef CAS PubMed.
- C. J. Weber, C. Opp, J. A. Prume, M. Koch, T. J. Andersen and P. Chifflard, Deposition and in-situ translocation of microplastics in floodplain soils, Sci. Total Environ., 2021, 152039, DOI:10.1016/j.scitotenv.2021.152039.
- H. Golwala, X. Zhang, S. M. Iskander and A. L. Smith, Solid waste: An overlooked source of microplastics to the environment, Sci. Total Environ., 2021, 769, 144581 CrossRef CAS PubMed.
- F. Collard, J. Gasperi, B. Gilbert, G. Eppe, S. Azimi, V. Rocher and B. Tassin, Anthropogenic particles in the stomach contents and liver of the freshwater fish Squalius cephalus, Sci. Total Environ., 2018, 643, 1257–1264 CrossRef CAS PubMed.
- P. Schwabl, S. Köppel, P. Königshofer, T. Bucsics, M. Trauner, T. Reiberger and B. Liebmann, Detection of Various Microplastics in Human Stool: A Prospective Case Series, Ann. Intern. Med., 2019, 171, 453 CrossRef PubMed.
- L. F. Amato-Lourenço, R. Carvalho-Oliveira, G. R. Júnior, L. Dos Santos Galvão, R. A. Ando and T. Mauad, Presence of airborne microplastics in human lung tissue, J. Hazard. Mater., 2021, 416, 126124 CrossRef PubMed.
- V. Stock, L. Böhmert, E. Lisicki, R. Block, J. Cara-Carmona, L. K. Pack, R. Selb, D. Lichtenstein, L. Voss, C. J. Henderson, E. Zabinsky, H. Sieg, A. Braeuning and A. Lampen, Uptake and effects of orally ingested polystyrene microplastic particles in vitro and in vivo, Arch. Toxicol., 2019, 93, 1817–1833 CrossRef CAS PubMed.
- J. Domenech, A. Hernández, L. Rubio, R. Marcos and C. Cortés, Interactions of polystyrene nanoplastics with in vitro models of the human intestinal barrier, Arch. Toxicol., 2020, 94, 2997–3012 CrossRef CAS PubMed.
- H. A. Leslie, M. J. M. van Velzen, S. H. Brandsma, A. D. Vethaak, J. J. Garcia-Vallejo and M. H. Lamoree, Discovery and quantification of plastic particle pollution in human blood, Environ. Int., 2022, 107199, DOI:10.1016/j.envint.2022.107199.
- A. Ragusa, A. Svelato, C. Santacroce, P. Catalano, V. Notarstefano, O. Carnevali, F. Papa, M. C. A. Rongioletti, F. Baiocco, S. Draghi, E. D'Amore, D. Rinaldo, M. Matta and E. Giorgini, Plasticenta: First evidence of microplastics in human placenta, Environ. Int., 2021, 146, 106274 CrossRef CAS PubMed.
- J. C. Prata, J. P. da Costa, I. Lopes, A. C. Duarte and T. Rocha-Santos, Environmental exposure to microplastics: An overview on possible human health effects, Sci. Total Environ., 2020, 702, 134455 CrossRef CAS PubMed.
- L. Rubio, R. Marcos and A. Hernández, Potential adverse health effects of ingested micro- and nanoplastics on humans. Lessons learned from in vivo and in vitro mammalian models, J. Toxicol. Environ. Health, Part B, 2020, 23, 51–68 CAS.
- L. Rubio, I. Barguilla, J. Domenech, R. Marcos and A. Hernández, Biological effects, including oxidative stress and genotoxic damage, of polystyrene nanoparticles in different human hematopoietic cell lines, J. Hazard. Mater., 2020, 398, 122900 CrossRef CAS PubMed.
- C. M. Rochman, C. Manzano, B. T. Hentschel, S. L. M. Simonich and E. Hoh, Polystyrene Plastic: A Source and Sink for Polycyclic Aromatic Hydrocarbons in the Marine Environment, Environ. Sci. Technol., 2013, 47, 13976–13984 CrossRef CAS PubMed.
- C. M. Rochman, C. Brookson, J. Bikker, N. Djuric, A. Earn, K. Bucci, S. Athey, A. Huntington, H. McIlwraith, K. Munno, H. De Frond, A. Kolomijeca, L. Erdle, J. Grbic, M. Bayoumi, S. B. Borrelle, T. Wu, S. Santoro, L. M. Werbowski, X. Zhu, R. K. Giles, B. M. Hamilton, C. Thaysen, A. Kaura, N. Klasios, L. Ead, J. Kim, C. Sherlock, A. Ho and C. Hung, Rethinking microplastics as a diverse contaminant suite, Environ. Toxicol. Chem., 2019, 38, 703–711 CrossRef CAS PubMed.
- H. L. De Frond, E. van Sebille, J. M. Parnis, M. L. Diamond, N. Mallos, T. Kingsbury and C. M. Rochman, Estimating the Mass of Chemicals Associated with Ocean Plastic Pollution to Inform Mitigation Efforts, Integr. Environ. Assess. Manage., 2019, 15, 596–606 CrossRef CAS PubMed.
- N. Hamid, M. Junaid and D.-S. Pei, Combined toxicity of endocrine-disrupting chemicals: A review, Ecotoxicol. Environ. Saf., 2021, 215, 112136 CrossRef CAS PubMed.
- D. M. Mitrano, A. Beltzung, S. Frehland, M. Schmiedgruber, A. Cingolani and F. Schmidt, Synthesis of metal-doped nanoplastics and their utility to investigate fate and behaviour in complex environmental systems, Nat. Nanotechnol., 2019, 14, 362–368 CrossRef CAS PubMed.
- S. Jeon, J. Clavadetscher, D.-K. Lee, S. V. Chankeshwara, M. Bradley and W.-S. Cho, Surface Charge-Dependent Cellular Uptake of Polystyrene Nanoparticles, Nanomaterials, 2018, 8, E1028 CrossRef PubMed.
- S. Lambert and M. Wagner, Characterisation of nanoplastics during the degradation of polystyrene, Chemosphere, 2016, 145, 265–268 CrossRef CAS PubMed.
- V. Paget, S. Dekali, T. Kortulewski, R. Grall, C. Gamez, K. Blazy, O. Aguerre-Chariol, S. Chevillard, A. Braun, P. Rat and G. Lacroix, Specific Uptake and Genotoxicity Induced by Polystyrene Nanobeads with Distinct Surface Chemistry on Human Lung Epithelial Cells and Macrophages, PLoS One, 2015, 10, e0123297 CrossRef PubMed.
- A.-K. Fuchs, T. Syrovets, K. A. Haas, C. Loos, A. Musyanovych, V. Mailänder, K. Landfester and T. Simmet, Carboxyl- and amino-functionalized polystyrene nanoparticles differentially affect the polarization profile of M1 and M2 macrophage subsets, Biomaterials, 2016, 85, 78–87 CrossRef CAS PubMed.
- B. Prietl, C. Meindl, E. Roblegg, T. R. Pieber, G. Lanzer and E. Fröhlich, Nano-sized and micro-sized polystyrene particles affect phagocyte function, Cell Biol. Toxicol., 2014, 30, 1–16 CrossRef CAS PubMed.
- M. Zhang, J. Li, G. Xing, R. He, W. Li, Y. Song and H. Guo, Variation in the internalization of differently sized nanoparticles induces different DNA-damaging effects on a macrophage cell line, Arch. Toxicol., 2011, 85, 1575–1588 CrossRef CAS PubMed.
- I. Florance, S. Ramasubbu, A. Mukherjee and N. Chandrasekaran, Polystyrene nanoplastics dysregulate lipid metabolism in murine macrophages in vitro, Toxicology, 2021, 458, 152850 CrossRef CAS PubMed.
- Q. Hu, H. Wang, C. He, Y. Jin and Z. Fu, Polystyrene nanoparticles trigger the activation of p38 MAPK and apoptosis via inducing oxidative stress in zebrafish and macrophage cells, Environ. Pollut., 2021, 269, 116075 CrossRef CAS PubMed.
- B. Rothen-Rutishauser, C. Mühlfeld, F. Blank, C. Musso and P. Gehr, Translocation of particles and inflammatory responses after exposure to fine particles and nanoparticles in an epithelial airway model, Part. Fibre Toxicol., 2007, 4, 9 CrossRef PubMed.
- J. A. Champion, A. Walker and S. Mitragotri, Role of Particle Size in Phagocytosis of Polymeric Microspheres, Pharm. Res., 2008, 25, 1815–1821 CrossRef CAS PubMed.
- H. Chikaura, Y. Nakashima, Y. Fujiwara, Y. Komohara, M. Takeya and Y. Nakanishi, Effect of particle size on biological response by human monocyte-derived macrophages, Biosurf. Biotribol., 2016, 2, 18–25 CrossRef.
- J. Gigault, A. T. Halle, M. Baudrimont, P.-Y. Pascal, F. Gauffre, T.-L. Phi, H. El Hadri, B. Grassl and S. Reynaud, Current opinion: What is a nanoplastic?, Environ. Pollut., 2018, 235, 1030–1034 CrossRef CAS PubMed.
- A. Ter Halle and J. F. Ghiglione, Nanoplastics: A Complex, Polluting Terra Incognita, Environ. Sci. Technol., 2021, 55, 14466–14469 CrossRef CAS PubMed.
- O. Okoturo-Evans, A. Dybowska, E. Valsami-Jones, J. Cupitt, M. Gierula, A. R. Boobis and R. J. Edwards, Elucidation of Toxicity Pathways in Lung Epithelial Cells Induced by Silicon Dioxide Nanoparticles, PLoS One, 2013, 8, e72363 CrossRef CAS PubMed.
- T. Verano-Braga, R. Miethling-Graff, K. Wojdyla, A. Rogowska-Wrzesinska, J. R. Brewer, H. Erdmann and F. Kjeldsen, Insights into the cellular response triggered by silver nanoparticles using quantitative proteomics, ACS Nano, 2014, 8, 2161–2175 CrossRef CAS PubMed.
- A. Oberemm, U. Hansen, L. Böhmert, C. Meckert, A. Braeuning, A. F. Thünemann and A. Lampen, Proteomic responses of human intestinal Caco-2 cells exposed to silver nanoparticles and ionic silver, J. Appl. Toxicol., 2016, 36, 404–413 CrossRef CAS PubMed.
- A. Braeuning, A. Oberemm, J. Görte, L. Böhmert, S. Juling and A. Lampen, Comparative proteomic analysis of silver nanoparticle effects in human liver and intestinal cells, J. Appl. Toxicol., 2018, 38, 638–648 CrossRef CAS PubMed.
- H. Karlsson, J. Lindbom, B. Ghafouri, M. Lindahl, C. Tagesson, M. Gustafsson and A. G. Ljungman, Wear Particles from Studded Tires and Granite Pavement Induce Pro-inflammatory Alterations in Human Monocyte-Derived Macrophages: A Proteomic Study, Chem. Res. Toxicol., 2011, 24, 45–53 Search PubMed.
- C. Aude-Garcia, B. Dalzon, J. L. Ravanat, V. Collin-Faure, H. Diemer, J. M. Strub, S. Cianferani, A. Van Dorsselaer, M. Carriere and T. Rabilloud, A combined proteomic and targeted analysis unravels new toxic mechanisms for zinc oxide nanoparticles in macrophages, J. Proteomics, 2016, 134, 174–185 CrossRef CAS PubMed.
- B. Dalzon, C. Aude-Garcia, H. Diemer, J. Bons, C. Marie-Desvergne, J. Pérard, M. Dubosson, V. Collin-Faure, C. Carapito, S. Cianférani, M. Carrière and T. Rabilloud, The longer the worse: a combined proteomic and targeted study of the long-term versus short-term effects of silver nanoparticles on macrophages, Environ. Sci.: Nano, 2020, 7, 2032–2046 RSC.
- L. Pietrovito, V. Cano-Cortés, T. Gamberi, F. Magherini, L. Bianchi, L. Bini, R. M. Sánchez-Martín, M. Fasano and A. Modesti, Cellular response to empty and palladium-conjugated amino-polystyrene nanospheres uptake: a proteomic study, Proteomics, 2015, 15, 34–43 CrossRef CAS PubMed.
- G. Repetto, A. del Peso and J. L. Zurita, Neutral red uptake assay for the estimation of cell viability/cytotoxicity, Nat. Protoc., 2008, 3, 1125–1131 CrossRef CAS PubMed.
- L. Muller, L. Fornecker, M. Chion, A. Van Dorsselaer, S. Cianférani, T. Rabilloud and C. Carapito, Extended investigation of tube-gel sample preparation: a versatile and simple choice for high throughput quantitative proteomics, Sci. Rep., 2018, 8, 8260 CrossRef PubMed.
- C. Cavazza, V. Collin-Faure, J. Pérard, H. Diemer, S. Cianférani, T. Rabilloud and E. Darrouzet, Proteomic analysis of Rhodospirillum rubrum after carbon monoxide exposure reveals an important effect on metallic cofactor biosynthesis, J. Proteomics, 2022, 250, 104389 CrossRef CAS PubMed.
- T. Lyubimova, S. Caglio, C. Gelfi, P. G. Righetti and T. Rabilloud, Photopolymerization of polyacrylamide gels with methylene blue, Electrophoresis, 1993, 14, 40–50 CrossRef CAS PubMed.
- A. P. Diz, A. Carvajal-Rodriguez and D. O. Skibinski, Multiple hypothesis testing in proteomics: a strategy for experimental work, Mol. Cell. Proteomics, 2011, 10, M110 004374 CrossRef PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res., 2009, 37, 1–13 CrossRef CAS PubMed.
- M. J. Eramo, V. Lisnyak, L. E. Formosa and M. T. Ryan, The ‘mitochondrial contact site and cristae organising system’ (MICOS) in health and human disease, J. Biochem., 2020, 167, 243–255 CrossRef CAS PubMed.
- T. Tanaka, K. Moriwaki, S. Murata and M. Miyasaka, LIM domain-containing adaptor, leupaxin, localizes in focal adhesion and suppresses the integrin-induced tyrosine phosphorylation of paxillin, Cancer Sci., 2010, 101, 363–368 CrossRef CAS PubMed.
- M. Moser, M. Bauer, S. Schmid, R. Ruppert, S. Schmidt, M. Sixt, H.-V. Wang, M. Sperandio and R. Fässler, Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells, Nat. Med., 2009, 15, 300–305 CrossRef CAS PubMed.
- A. Abderrazak, T. Syrovets, D. Couchie, K. El Hadri, B. Friguet, T. Simmet and M. Rouis, NLRP3 inflammasome: from a danger signal sensor to a regulatory node of oxidative stress and inflammatory diseases, Redox Biol, 2015, 4, 296–307 CrossRef CAS PubMed.
- K. A. Jablonski, S. A. Amici, L. M. Webb, J. D. D. Ruiz-Rosado, P. G. Popovich, S. Partida-Sanchez and M. Guerau-de-Arellano, Novel Markers to Delineate Murine M1 and M2 Macrophages, PLoS One, 2015, 10, e0145342 CrossRef PubMed.
- H. Sun, S. Gong, R. J. Carmody, A. Hilliard, L. Li, J. Sun, L. Kong, L. Xu, B. Hilliard, S. Hu, H. Shen, X. Yang and Y. H. Chen, TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis, Cell, 2008, 133, 415–426 CrossRef CAS PubMed.
- J. Wang, B. Yang, Y. Hu, Y. Zheng, H. Zhou, Y. Wang, Y. Ma, K. Mao, L. Yang, G. Lin, Y. Ji, X. Wu and B. Sun, Negative Regulation of Nmi on Virus-Triggered Type I IFN Production by Targeting IRF7, J. Immunol., 2013, 191, 3393–3399 CrossRef CAS PubMed.
- J. P. G. L. Frias and R. Nash, Microplastics: Finding a consensus on the definition, Mar. Pollut. Bull., 2019, 138, 145–147 CrossRef CAS PubMed.
- E. Metschnikoff, Ueber den Kampf der Zellen gegen Erysipel-kokken: Ein Beitrag zur Phagocytenlehre, Arch. Pathol. Anat. Physiol. Klin. Med., 1887, 107, 209–249 CrossRef.
- E. Metchnikoff, Lectures on the Comparative Pathology of Inflammation, translated from the French by F. A. Starling and E. H. Starling, Dover Publications, New York, 1968 Search PubMed.
- X. Jiang, Y. Chang, T. Zhang, Y. Qiao, G. Klobučar and M. Li, Toxicological effects of polystyrene microplastics on earthworm (Eisenia fetida), Environ. Pollut., 2020, 259, 113896 CrossRef CAS PubMed.
- L. Liu, H. Zheng, L. Luan, X. Luo, X. Wang, H. Lu, Y. Li, L. Wen, F. Li and J. Zhao, Functionalized polystyrene nanoplastic-induced energy homeostasis imbalance and the immunomodulation dysfunction of marine clams (Meretrix meretrix) at environmentally relevant concentrations, Environ. Sci.: Nano, 2021, 8, 2030–2048 RSC.
- W. Gu, S. Liu, L. Chen, Y. Liu, C. Gu, H.-Q. Ren and B. Wu, Single-Cell RNA Sequencing Reveals Size-Dependent Effects of Polystyrene Microplastics on Immune and Secretory Cell Populations from Zebrafish Intestines, Environ. Sci. Technol., 2020, 54, 3417–3427 CrossRef CAS PubMed.
- M. P. Desai, V. Labhasetwar, G. L. Amidon and R. J. Levy, Gastrointestinal uptake of biodegradable microparticles: effect of particle size, Pharm. Res., 1996, 13, 1838–1845 CrossRef CAS PubMed.
- M. P. Desai, V. Labhasetwar, E. Walter, R. J. Levy and G. L. Amidon, The mechanism of uptake of biodegradable microparticles in Caco-2 cells is size dependent, Pharm. Res., 1997, 14, 1568–1573 CrossRef CAS PubMed.
- K. Marcus and T. Rabilloud, How Do the Different Proteomic Strategies Cope with the Complexity of Biological Regulations in a Multi-Omic World? Critical Appraisal and Suggestions for Improvements, Proteomes, 2020, 8, 23 CrossRef CAS PubMed.
- S. Kanno, A. Furuyama and S. Hirano, A Murine Scavenger Receptor MARCO Recognizes Polystyrene Nanoparticles, Toxicol. Sci., 2007, 97, 398–406 CrossRef CAS PubMed.
- V. Olivier, J. L. Duval, M. Hindié, P. Pouletaut and M. D. Nagel, Comparative particle-induced cytotoxicity toward macrophages and fibroblasts, Cell Biol. Toxicol., 2003, 19, 145–159 CrossRef CAS PubMed.
- R. Sussarellu, M. Suquet, Y. Thomas, C. Lambert, C. Fabioux, M. E. J. Pernet, N. Le Goïc, V. Quillien, C. Mingant, Y. Epelboin, C. Corporeau, J. Guyomarch, J. Robbens, I. Paul-Pont, P. Soudant and A. Huvet, Oyster reproduction is affected by exposure to polystyrene microplastics, Proc. Natl. Acad. Sci., 2016, 113, 2430–2435 CrossRef CAS PubMed.
- C. Ott, K. Ross, S. Straub, B. Thiede, M. Gotz, C. Goosmann, M. Krischke, M. J. Mueller, G. Krohne, T. Rudel and V. Kozjak-Pavlovic, Sam50 Functions in Mitochondrial Intermembrane Space Bridging and Biogenesis of Respiratory Complexes, Mol. Cell. Biol., 2012, 32, 1173–1188 CrossRef CAS PubMed.
- F. Xu, C. Morin, G. Mitchell, C. Ackerley and B. H. Robinson, The role of the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene in cytochrome oxidase assembly: mutation causes lowered levels of COX (cytochrome c oxidase) I and COX III mRNA, Biochem. J., 2004, 382, 331–336 CrossRef CAS PubMed.
- S. S. Korshunov, V. P. Skulachev and A. A. Starkov, High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria, FEBS Lett., 1997, 416, 15–18 CrossRef CAS PubMed.
- A. A. Starkov and G. Fiskum, Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state, J. Neurochem., 2003, 86, 1101–1107 CrossRef CAS PubMed.
- C. Cortés, J. Domenech, M. Salazar, S. Pastor, R. Marcos and A. Hernández, Nanoplastics as a potential environmental health factor: effects of polystyrene nanoparticles on human intestinal epithelial Caco-2 cells, Environ. Sci.: Nano, 2020, 7, 272–285 RSC.
- M. Sharma and E. de Alba, Structure, Activation and Regulation of NLRP3 and AIM2 Inflammasomes, Int. J. Mol. Sci., 2021, 22, E872 CrossRef PubMed.
- N. Morimoto, T. Kono, M. Sakai and J.-I. Hikima, Inflammasomes in Teleosts: Structures and Mechanisms That Induce Pyroptosis during Bacterial Infection, Int. J. Mol. Sci., 2021, 22, 4389 CrossRef CAS PubMed.
- J. G. de Souza, N. Starobinas and O. C. M. Ibañez, Unknown/enigmatic functions of extracellular ASC, Immunology, 2021, 163, 377–388 CrossRef CAS PubMed.
- R. Zheng, J. Zhang, X. Han, Y. Wu, J. Yan, P. Song, Y. Wang, X. Wu and H. Zhang, Particulate matter aggravates Alzheimer's disease by activating the NLRP3 inflammasome to release ASC specks, Environ. Sci.: Nano, 2021, 8, 2177–2190 RSC.
- F. Ginhoux and M. Guilliams, Tissue-Resident Macrophage Ontogeny and Homeostasis, Immunity, 2016, 44, 439–449 CrossRef CAS PubMed.
- H. Xu, P. H. M. Hoet and B. Nemery, IN VITRO TOXICITY ASSESSMENT OF POLYVINYL CHLORIDE PARTICLES AND COMPARISON OF SIX CELLULAR SYSTEMS, J. Toxicol. Environ. Health, Part A, 2002, 65, 1141–1159 CrossRef CAS PubMed.
- W. S. Lee, H.-J. Cho, E. Kim, Y. H. Huh, H.-J. Kim, B. Kim, T. Kang, J.-S. Lee and J. Jeong, Bioaccumulation of polystyrene nanoplastics and their effect on the toxicity of Au ions in zebrafish embryos, Nanoscale, 2019, 11, 3173–3185 RSC.
- N. González-Soto, J. Hatfield, A. Katsumiti, N. Duroudier, J. M. Lacave, E. Bilbao, A. Orbea, E. Navarro and M. P. Cajaraville, Impacts of dietary exposure to different sized polystyrene microplastics alone and with sorbed benzo[a]pyrene on biomarkers and whole organism responses in mussels Mytilus galloprovincialis, Sci. Total Environ., 2019, 684, 548–566 CrossRef PubMed.
- Y. Zhang and G. G. Goss, Potentiation of polycyclic aromatic hydrocarbon uptake in zebrafish embryos by nanoplastics, Environ. Sci.: Nano, 2020, 7, 1730–1741 RSC.
- C. C. Parenti, S. Magni, A. Ghilardi, G. Caorsi, C. Della Torre, L. Del Giacco and A. Binelli, Does triclosan adsorption on polystyrene nanoplastics modify the toxicity of single contaminants?, Environ. Sci.: Nano, 2021, 8, 282–296 RSC.
- H. Abduro Ogo, N. Tang, X. Li, X. Gao and W. Xing, Combined toxicity of microplastic and lead on submerged macrophytes, Chemosphere, 2022, 295, 133956 CrossRef CAS PubMed.
- S. B. Zhou, M. J. Bailey, M. J. Dunn, V. R. Preedy and P. W. Emery, A quantitative investigation into the losses of proteins at different stages of a two-dimensional gel electrophoresis procedure, Proteomics, 2005, 5, 2739–2747 CrossRef CAS PubMed.
- R. Zhou, G. Lu, Z. Yan, R. Jiang, Y. Sun and P. Zhang, Interactive transgenerational effects of polystyrene nanoplastics and ethylhexyl salicylate on zebrafish, Environ. Sci.: Nano, 2021, 8, 146–159 RSC.
- K. E. Wheeler, A. J. Chetwynd, K. M. Fahy, B. S. Hong, J. A. Tochihuitl, L. A. Foster and I. Lynch, Environmental dimensions of the protein corona, Nat. Nanotechnol., 2021, 16, 617–629 CrossRef CAS PubMed.
- Q. Shi, J. Tang, X. Liu and R. Liu, Ultraviolet-induced photodegradation elevated the toxicity of polystyrene nanoplastics on human lung epithelial A549 cells, Environ. Sci.: Nano, 2021, 8, 2660–2675 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2en00214k |
| This journal is © The Royal Society of Chemistry 2022 |