Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein binding on acutely toxic and non-toxic polystyrene nanoparticles during filtration by Daphnia magna†
Egle
Kelpsiene
 ab,
Irene
Brandts
ab,
Irene
Brandts
 cd,
Katja
Bernfur
a,
Mikael T.
Ekvall
be,
Martin
Lundqvist
cd,
Katja
Bernfur
a,
Mikael T.
Ekvall
be,
Martin
Lundqvist
 ab,
Mariana
Teles
cd and
Tommy
Cedervall
ab,
Mariana
Teles
cd and
Tommy
Cedervall
 *ab
*ab
aDepartment of Biochemistry and Structural Biology, Lund University, P.O. Box 124, SE-221 00 Lund, Sweden. E-mail: tommy.cedervall@biochemistry.lu.se
bNanoLund, Lund University, Box 118, SE-221 00 Lund, Sweden
cDepartment of Cell Biology, Physiology and Immunology, Universitat Autònoma de Barcelona, 08193 Barcelona, Spain
dInstitute of Biotechnology and Biomedicine, Universitat Autònoma de Barcelona, 08193 Barcelona, Spain
eDepartment of Biology, Aquatic Ecology Unit, Lund University, Lund, Sweden
First published on 10th June 2022
Abstract
Nanomaterials can adsorb biomolecules to their surface and form a protein corona. Here we investigated the protein profile bound to different sizes of aminated and carboxylated polystyrene (PS) nanoparticles after passing through the digestive tract of the freshwater zooplankter Daphnia magna. We found that acutely toxic aminated 53 nm PS nanoparticles bind a different set of proteins compared to other non-toxic PS nanoparticles. The aminated PS nanoparticles bind a higher number of proteins, which are smaller and more acidic, compared to the proteins which bind to the PS nanoparticles that are non-toxic in acute toxicity tests. The proteins bound to toxic nanoparticles can be divided into two groups. One group of proteins which function is related to the digestive system, whereas the other group of proteins can be related to the epithelium, intracellular structures and processes. Finally, we observed that not only proteins bind to the surfaces of the nanoparticles. Triglycerides effectively bind to 200 nm carboxylated PS nanoparticles but not to the other tested nanoparticles. These results provide information about the composition of the corona formed on surfaces of nanoparticles after a short-term incubation with D. magna and give insights to what underlies the acute toxicity caused by nanoparticles.
Environmental significanceThe increasing use of nanomaterial may lead to increasing exposure in the environment. Another source of nano-sized particles are breakdown particles from plastics and rubbers. Altogether nano-sized particles are a growing environmental concern. Although the toxicity of particles is known in many cases, surprisingly little is known about the mechanisms behind the toxicity. Detailed knowledge on how toxic and non-toxic nano-sized particles interact with filtrating organisms can not only explain the mechanisms behind the toxicity but also be used as an early guide to evaluate the expected toxicity of new material and products under development. This will facilitate the evaluation of the environmental impact of already present materials as well as future materials. |
Introduction
Nanomaterials bring many revolutionary advantages to our modern way of living. Due to their small size (1–100 nm), shape and surface,1 nanomaterials are widely used in biomedical applications, personal care items and engineering technologies.2,3 However, as the use of nanomaterials increases, so does the need for careful investigations of the potential toxicity and interaction with biological matter,4 especially when nanomaterials enter the natural environment.5Once nanomaterials enter the environment or biological fluids, nanoparticles, including polystyrene (PS) nanoparticles, can adsorb biomolecules to their surfaces,6,7 and form a layer, a so-called protein corona.8–10 Protein corona formation and composition depends on several parameters, such as, nanoparticle size, surface characteristics,11,12 medium conditions,13 and time.14,15 The characteristics of these protein corona are particularly relevant, as both the interaction with cell membranes and the cellular uptake are influenced by the adsorbed proteins.16 Therefore, the corona becomes highly important when the cytotoxicity17,18 or body distribution19–21 of nanoparticles is evaluated. Even though corona mainly contain proteins, the presence of other biomolecules such as sugars, nucleic acids and lipids is also expected,20,22,23 and should be further investigated.
Daphnia magna, used in the present study, is a well-studied freshwater filter feeder. The genome of D. magna is fully sequenced, making it a suitable model organism to monitor biological responses to changes in the environment.24 Moreover, D. magna is specifically appropriate to study nanoparticles, as they typically feed on particles ranging between 240 to 640 nm in size,25 although they can also ingest particles up to 1000 nm.26 During the last years, the toxicity of PS nanoparticles has been widely studied. In general, aminated PS (PS-NH2) are more toxic than carboxylated PS (PS-COOH) due to the positive surface charge.27 It has previously been shown that 50 nm PS-NH2 are toxic to D. magna after 24 h exposure, whereas 200 nm PS-NH2, and 60 nm and 200 nm PS-COOH are not acutely toxic.28 More recently the importance of the surface charge on PS nanoparticles29 and the size of PS-NH2 (ref. 30) has been studied in detail. The weathering of PS nanoparticles and the eco-corona formation have been seen to influence nanoparticle, suggesting that biomolecules on the surface are important for the toxicity.31,32 The importance of considering the eco-corona, including the protein corona, in testing nanomaterial has recently been reviewed,7 but the current knowledge on the molecular events that initiate the toxic response is still limited.
A few previous studies have identified proteins bound to nanoparticles after being incubated with D. magna. For example, 25 nm gold nanoparticles filtrated by D. magna bound different proteins, compared to nanoparticles incubated in conditioned water, i.e. water that has been filtrated by D. magna without nanoparticles.33 Interestingly, the protein corona formed on aminated PS particles in conditioned water was shown to depended on the presence of other organic molecules.34 Another study showed that the protein corona formed after filtration by D. magna was different on freshly dispersed or medium-aged silver (Ag) and titanium dioxide (TiO2) nanoparticles.35 In the present study we show that toxic and non-toxic (as seen in acute toxicity tests) PS-NH2 and PS-COOH nanoparticles binds different sets of proteins after passing through the digestive system of D. magna. This suggest that the most abundant proteins on the particles can be linked to their toxicity and encourage future studies to understand the links between particle toxicity and protein binding.
Material and methods
Nanoparticle preparation and characterization
Aminated PS nanoparticles, PS-NH2 (53 and 200 nm), and carboxylated PS nanoparticles, PS-COOH (62 and 200 nm), were purchased from Bangs Laboratories Inc. (https://www.bangslabs.com). Before the experiments, nanoparticles were diluted to 10 mg mL−1 and dialyzed in a Standard RC Tubing, dialysis membrane (MWCO: 3.5 kD) for 72 h at 4 °C in 10 L of MilliQ water. The water was changed after 4 h the first day and once a day on the following days. To confirm the size of nanoparticles after dialysis, and ensure that there was no particle aggregation, nanoparticles were measured using both dynamic light scattering (DLS, DynaPro Plate Reader II, Wyatt instruments, USA) and differential centrifugal sedimentation (DCS, DC24000 UHR Disc Centrifuge, CPS Instruments Europe, Oosterhout, Netherlands).The z-potential, performed at 25 °C using a Zetasizer Nano ZS instrument (Malvern Instruments, Worcestershire, UK), for the particles were determined both for a particle in MilliQ H2O and four different buffers; 0.1 M glycine pH 2.2, 3.6, 8.6, and 10.6 (adjusted to the right pH with either HCL or NaOH). 2 μL of the 10% (w/v) stock solution was mixed with 98 μL MilliQ H2O whereafter 1000 μL of the buffer was added and the sample was mixed. The final concentrations in the sample were ∼0.9 M glycine and ∼0.02% (w/v) particles.
The FTIR spectra were recorded using a Perkin Elmer Spectrum Two equipped with a UATR HR unit. Each sample was recorded with 64 scans between 450–4000 cm−1 and with 4 cm−1 nominal resolution. 2 μL of the stock solution (10% w/v) was added to the diamond surface and was left drying for 30 min before the spectrum was recorded.
Incubation with D. magna
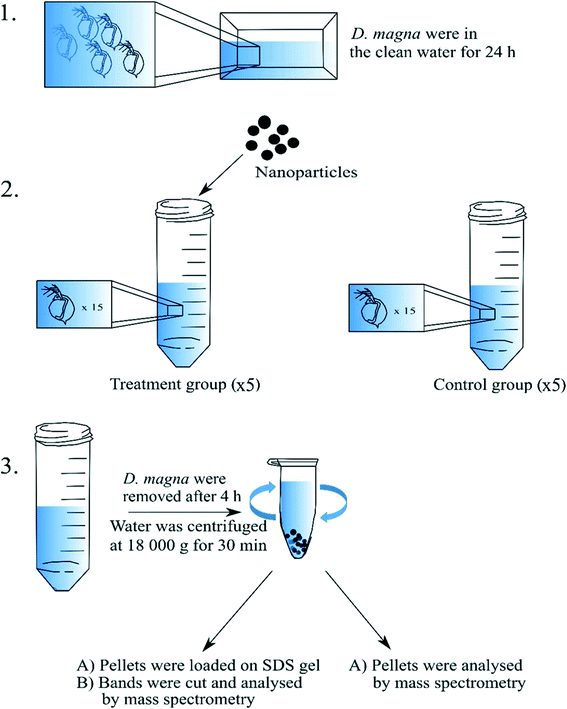
Adult, at least one week old and that had not yet reproduced, D. magna individuals that came from the same population were used in the present study. The culture originates from Lake Bysjön, southern Sweden (55°40′31.3′′N 13°32′41.9′′E) and has been kept in a laboratory environment for several hundred generations. All cultures and experiments were maintained at 18 °C under an 8:16 h light/dark photoperiod. Before the incubation with nanoparticles, D. magna adults were kept in clean tap water for 24 h to allow evacuation of remaining algal cells from the gut (Fig. 1). During the exposure, D. magna (n = 15 individuals per tube) were placed into 15 mL tubes (in total there were four replicates for each group) containing a total volume of 5 mL tap water and the following exposure concentrations: 0 mg L−1 (control group), 16 mg L−1 (for 53 and 62 nm sized nanoparticles), and 224 mg L−1 (for 200 nm sized nanoparticles) of PS nanoparticles. Different concentrations were chosen due to the equivalent surface area for the different sizes of PS nanoparticles. Individuals were allowed to filter the water containing nanoparticles or water alone for 4 h. During the incubation, immobilized D. magna were removed. All D. magna individuals were removed from the tubes after the incubation period. The nanoparticle–protein complexes were recovered by centrifugation in Eppendorf tubes at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g and 4 °C for 30 min with 1 mL total volume. After each centrifugation, 900 μL of supernatant was removed gently in order not to disturb the pellet. More exposure media, 900 μL was added to the same tube and centrifuged again. The procedure was repeated until all the 5 mL of the sample had been centrifuged in the same tube.
000g and 4 °C for 30 min with 1 mL total volume. After each centrifugation, 900 μL of supernatant was removed gently in order not to disturb the pellet. More exposure media, 900 μL was added to the same tube and centrifuged again. The procedure was repeated until all the 5 mL of the sample had been centrifuged in the same tube.
Digestion of protein samples
Proteins were digested in solution as follow: the pH of the samples was adjusted to 7.8 by adding ammonium bicarbonate (ABC) to a final concentration of 50 mM. The proteins were reduced by the addition of DL-dithiothreitol (DTT, Sigma-Aldrich) in 50 mM ABC to a final concentration of 5 mM and incubated at 37 °C for 30 min, followed by alkylation using iodoacetamide (IAA, Sigma-Aldrich) with a final concentration of 12 mM and incubation in the dark for 20 min. Finally sequencing-grade modified trypsin (Promega, Madison, WI, USA) was added to a final concentration of 2 ng μL−1 and the samples were digested overnight at 37 °C. The next day formic acid (FA) was added to a final concentration of 0.5%. The samples were centrifuged at 15 kRCF for 10 min before the peptide solutions were extracted and transferred into new tubes. The peptides were cleaned up by C18 reversed-phase micro columns using a 2% ACN, 0.1% FA equilibration buffer and an 80% ACN, 0.1% FA elution buffer. The collected samples were dried in a fume hood and resuspended in 15 μL 2% ACN, 0.1% FA.Proteins were also in gel digested as follow: from each experimental group (treatments and control), samples were prepared by adding 20 μl of SDS-PAGE loading buffer to the particle pellets or control tubes and 10 μL was loaded on a 4 to 12% premade SDS-PAGE (Bio-Rad). SDS has been shown to effectively desorb proteins from nanoparticle surfaces.36 Protein bands were visualised, using Pierce™ Silver Stain Kit (Thermo Scientific) according to the manufacturer's protocol, then immediately cut into 1 × 1 mm gel pieces and destained. The gel pieces were washed twice with 50% acetonitrile (ACN, Sigma-Aldrich)/50 mM ABC and incubated for 30 min each time. After washing, the gel pieces were dehydrated using 100% ACN before the proteins were reduced with 25 μL 10 mM DTT in 50 mM ABC for 30 min at 37 °C. The DTT was removed, and the gel pieces were dehydrated using 100% ACN before the proteins were alkylated with 25 μL 55 mM iodacetamide in 50 mM ABC for 30 min in the dark at room temperature. The gel pieces were dehydrated one last time with 100% ACN before the proteins were digested by adding 25 μL 12 ng μL−1 trypsin (sequence grade modified trypsin porcine, Promega, Fitchburg, WI, USA) in 50 mM ABC and incubated on ice for 4 hours before 20 μL 50 mM ABC were added, and the protein was incubated overnight at 37 °C. The following day 10% (FA) was added to a final concentration of 0.5%, to get a pH of 2–3 to stop the digestion, before the peptide solutions were extracted and transferred into new tubes.
Peptide separation and mass spectrometry
Peptides were subjected to a reversed phase nano-LC source (Proxeon Biosystems, Odense, Denmark) coupled to an LTQ-Orbitrap Velos Pro mass spectrometer equipped with a nano Easy spray ion source (Thermo Fisher Scientific, Stockholm, Sweden). The chromatographic separation was performed on a 2 cm C18 Acclaim PepMap precolumn (75 mm i.d.) and a 15 cm C18 EASY-Spray LC capillary separation column (75 mm i.d., packed with 3 μm resin, column temperature 45 °C) from Thermo Fisher. The gradient was created by solvent A (1% ACN, 0.1% FA in water) and solvent B (100% ACN, 0.1% FA). A flow rate of 300 nL min−1 was used throughout the whole gradient, (0–30% B for 40 minutes, 30–50% 20 minutes, 50–95% for 10 minutes and 95% for 10 minutes). One full MS scan (resolution 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at 400 m/z; mass range 400–1400 m/z) was followed by MS/MS scans of the 4 most abundant ion signals. Charge state screening was enabled where singly charged and unassigned ions were rejected. The precursor ions were isolated with 3 m/z isolation width and fragmented using collision induced dissociation (CID) at a normalized collision energy of 35. The dynamic exclusion window was limited to 500 and set to 30 s. The intensity threshold for precursor ions was set to 2500. The automatic gain control was set to 1 × 106 for both MS and MS/MS with ion accumulation times 100 ms.
000 at 400 m/z; mass range 400–1400 m/z) was followed by MS/MS scans of the 4 most abundant ion signals. Charge state screening was enabled where singly charged and unassigned ions were rejected. The precursor ions were isolated with 3 m/z isolation width and fragmented using collision induced dissociation (CID) at a normalized collision energy of 35. The dynamic exclusion window was limited to 500 and set to 30 s. The intensity threshold for precursor ions was set to 2500. The automatic gain control was set to 1 × 106 for both MS and MS/MS with ion accumulation times 100 ms.
Data analysis and protein identification
Raw files were converted to mgf-format by Mascot Distiller (version 2.6, Matrix Science) and identification of proteins were carried out with the Mascot Daemon software (version 2.4, Matrix Science). The following search settings were used: trypsin as protease, one allowed missed cleavage site, 5 ppm MS accuracy for peptides and 0.015 Da MS/MS accuracy, variable modifications: oxidation (M) and carbamidomethylation (C). The files were searched against an in-house created database containing all Daphnia protein sequences. To be considered a true protein identification all individual ion scores must have a higher score than the score given when using a significant threshold of p < 0.005. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE1 partner repository with the dataset identifier PXD033695. The isoelectric point was calculated using the site https://web.expasy.org/compute_pi.Determination of total triglyceride concentration
In order to obtain a detectible level of total triglycerides, 3 replicates representing each treatment were pooled together and freeze dried. Analysis of total triglyceride concentration of each fraction was performed according to the manufacturer's instructions (Sigma-Aldrich, USA). Absorbance intensity was recorded at 540 nm using a ProbeDrum spectrophotometer (Probation Labs, Sweden).Statistical analysis
Data were analysed using Student's t-test. All analysis was performed using the statistical computing software GraphPad Prism version 8.0.0 (224) for Windows (GraphPad Software, Inc., https://www.graphpad.com).Results and discussion
The size of PS nanoparticles was determined by DLS before and after incubation with D. magna, Table S1.† There was a large increase of the size for the smaller particles after filtration by D. magna indicating that the particles aggregated.In the present study, we incubated D. magna individuals with model PS nanoparticles for 4 h to identify which proteins were bound to the nanoparticles during filtration. The experimental outline is shown in Fig. 1. The nanoparticle concentrations were optimised to allow for proteins on the nanoparticles to be identified, maintaining similar surface areas among the different nanoparticle sizes. At these concentrations the 53 nm PS-NH2 are acutely toxic to D. magna. Therefore, immobilized D. magna individuals were regularly observed and immediately removed. No death was, as expected, observed in the other nanoparticle treatments during the incubation time. After completed incubation, all zooplankters were removed in all experimental groups. DLS measurements showed that after 4 h of incubation with D. magna, the size of 53 nm PS-NH2 nanoparticles increased 6.2 times, whereas 62 nm PS-COOH nanoparticles increased 1.5 times.
The first set of experiments compared which proteins bound to the acutely toxic 53 nm PS-NH2 and the non-toxic 200 nm PS-NH2. After the incubation with D. magna the protein–nanoparticles complexes were collected by centrifugation. The proteins were desorbed from the particles with SDS and separated using gel electrophoresis (Fig. 2). There is a considerable variation in the amount of proteins, but the pattern of proteins is the same. It appeared that a larger number of proteins were bound to 53 nm PS-NH2 (Fig. 2A) compared to 200 nm PS-NH2 (Fig. 2B). Interestingly, the molecular sizes of the proteins appeared to be smaller for 53 nm PS-NH2 compared to 200 nm PS-NH2. A density scan of the gels further visualized these size differences, see Fig. 2C.
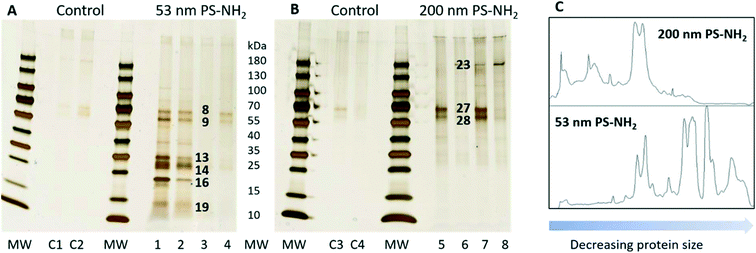 | ||
| Fig. 2 Silver-stained SDS gels after 53 nm (A) and 200 nm (B) PS-NH2 incubation with D. magna in comparison with a control group. Lane C1–C4, 1 to 4, and 5 to 8, represent four replicates for control, 53 nm PS-NH2, and 200 nm PS-NH2. The identity of proteins in bands marked with black numbers are listed in Table 1. The visualization of the size differences in proteins found in both gels (C). | ||
The visible protein bands were labelled with numbers (Fig. S1†), cut out from the gels and the proteins were identified using mass spectrometry. Only proteins that were identified based on at least two peptides with a significance threshold p < 0.0005 for the individual ion score were reported as true protein identifications. Under these conditions a total of 41 proteins were identified from 24 bands (Table S2†). The proteins were unequally distributed on the particles as 53 nm PS-NH2 nanoparticles bound more proteins compared with 200 nm PS-NH2 (27 vs. 6 proteins, respectively), confirming the conclusion from the visual observation. Only four proteins were bound to both types of nanoparticles (Fig. 3).
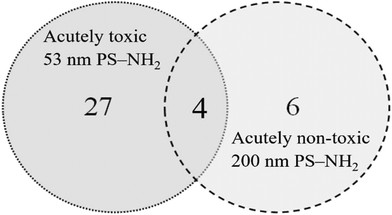 | ||
| Fig. 3 Venn diagram compares the number proteins identified on 53 and 200 nm PS-NH2 after identification using mass spectrometry after passing through the digestive system of D. magna. | ||
The silver staining of SDS-PAGE is a semi-quantitative method. Therefore, it is reasonable to assume that the most abundant proteins on the particles are represented in the most intensely stained protein bands in Fig. 2A and B. The identity of the proteins is presented in Table 1. The analysis of the most abundant proteins shows that the 53 nm and 200 nm PS-NH2 bind distinctly different proteins. The 53 nm PS-NH2 appears to bind two groups of proteins. One that can be related to the digestive system37 as carboxypeptidase B, serine protease and chymotrypsin elastase family member 2A are among the most abundant proteins. The second group of proteins can be related to the epithelium, and intracellular structures and processes, for example, beta-klotho, actin, tubulin, elongation factors and histones. Klotho protein, which is a membrane-bound protein, plays an important role for a proper function of many organs.38–40 It has previously been shown that disturbances of the klotho gene expression in mice might interfere with the lifespan or fertility.38
| Treatment | Band | MW (kDa) | Protein name | Accession number | MW (kDa) | pI |
|---|---|---|---|---|---|---|
| 53 nm PS-NH2 | 8 | 70 | Actin, alpha skeletal muscle (fragment) | A0A0N7ZDU7 | 35 | 5.4 |
| Tubulin beta chain | A0A0N7ZH16 | 52 | 4.75 | |||
| Tubulin alpha chain | A0A0P5ZYN7 | 50 | 4.94 | |||
| Fructose-bisphosphate aldolase | A0A0N8DTE5 | 39 | 8.30 | |||
| Heat shock 70 kDa protein cognate | A0A0P6DCT5 | 70 | 5.37 | |||
| ATP synthase subunit beta | A0A0P6CBB6 | 56 | 5.19 | |||
| Eukaryotic initiation factor 4A-II | A0A0P6CFN1 | 46 | 5.32 | |||
| 9 | 60 | Beta-klotho | A0A0N7ZV82 | 48 | NA | |
| 13 | 35 | Carboxypeptidase B | A0A0N8AEM3 | 5.1 | ||
| Putative carboxypeptidase | A0A0N7ZFK6 | 31 | 6.2 | |||
| A0A0N7ZNY1 | 28 | 4.7 | ||||
| A0A0N8A474 | 34 | 4.5 | ||||
| Zinc carboxypeptidase | A0A0N8B2M1 | 45 | 6.5 | |||
| Poly(U)-specific endoribonuclease | A0A0N8AU82 | 41 | 4.5 | |||
| Serine protease | A0A0N7ZYN7 | 32 | 4.9 | |||
| Serine protease (fragment) | A0A0N7ZFA7 | 27 | 4.3 | |||
| Putative serine protease (fragment) | A0A0N8C2S1 | 17 | 4.5 | |||
| Trypsin serine protease | A0A0N8AZL9 | 32 | 4.8 | |||
| 14 | 30 | Serine protease | A0A0N7ZYN7 | 32 | 4.9 | |
| Serine protease (fragment) | A0A0N7ZFA7 | 27 | 4.3 | |||
| 16 | 22 | Chymotrypsin elastase family member 2A | A0A0N7ZRR8 | 20 | 4.6 | |
| 19 | 15 | Uncharcterized protein | A0A0N8AVI2 | 40 | 5.3 | |
| C-type lectin domain family 6 member A | A0A0N7ZMI0 | 17 | 4.9 | |||
| Heat shock 70 kDa protein | A0A0N8AGX4 | 67 | 5.3 | |||
| A0A0P6A337 | 72 | 5.1 | ||||
| Chymotrypsin elastase family member 2A | A0A0N7ZRR8 | 20 | 4.6 | |||
| Tubulin beta-4B chain | A0A0N8ERR7 | 34 | 4.8 | |||
| Tubulin beta chain | A0A0P6A486_9CRUS | 57 | NA | |||
| Elongation factor 1-alpha | A0A0N7ZYV1 | 50 | 9.0 | |||
| ADP-ribosylation factor (fragment) | A0A0N8BN21 | 19 | 6.4 | |||
| Niemann-pick type C-2d | A0A0N7ZXW1 | 18 | 4.4 | |||
| Actin, alpha skeletal muscle (fragment) | A0A0N7ZDU7 | 35 | 5.7 | |||
| ATP synthase subunit beta | A0A0N7ZMA0 | 57 | 5.1 | |||
| Histone H4 | A0A0N8AML8 | 11 | 11.3 | |||
| 200 nm PS-NH2 | 23 | 190 | Vitellogenin-1 | A0A0N8ERH4 | 171 | 9.0 |
| 27 | 70 | Putative hemocyte protein-glutamine gamma-glutamyltransferase | A0A0N8AYD5 | 68 | 6.6 | |
| A0A0P5BFI8 | 58 | 5.9 | ||||
| 28 | 60 | Hemocyte protein-glutamine gamma-glutamyltransferase | A0A0N7ZEH2 | 105 | 7.9 | |
| A0A0P4ZFG8 | 89 | NA | ||||
| Lamin-A | A0A0P6IZ96 | 53 | NA |
Likewise, some proteins were uniquely bound to 200 nm PS-NH2 nanoparticles. The most prominent are the following ones, vitellogenin-1, hemocyte protein-glutamine gamma-glutamyltransferase, and putative hemocyte protein-glutamine gamma-glutamyltransferase. Vitellogenin-1, is a precursor to egg yolk, plays a critical role in oogenesis,41 and is especially highly expressed in female fish.42 Interestingly, only 4 proteins, actin, chymotrypsin elastase family member 2A, elongation factor 1-alpha and histone H4 bound to both 53 nm and 200 nm PS-NH2.
The protein corona has previously been determined on aminated PS particles in condition water, i.e., water that has been filtered by D. magna excreting proteins and other biomolecules before the D. magna are removed and the particles added.34 Some proteins found after filtration are also found on particles in conditioned water which is not surprising as the filtrated particles are exposed to both the environment in the intestine and the surrounding water.
To explain the striking difference in the number and identity of proteins bound to 53 nm and 200 nm PS-NH2 we calculated the isoelectric point (pI) for each of the proteins (Tables 1 and S2†). The pI of proteins bound to 53 nm PS-NH2 was, with a few exceptions, below 6, whereas, surprisingly, the pI of proteins bound to 200 nm PS-NH2 was in general above 6 (Fig. S2†). This means that, in the media, the proteins bound to 53 nm PS-NH2 generally had a negative net charge, whereas the proteins bound to 200 nm PS-NH2 had a positive net charge. Although charge attractions can explain why proteins with a net negative charge bind to 53 nm PS-NH2 it cannot explain why positively charged proteins, for example vitellogenin-1, with a pI of 9 and, therefore, a positive net charge in the media, bind to the 200 nm PS-NH2. However, measuring the Z-potential of the two particles in tap water revealed that 53 nm PS-NH2 had, as expected, a net positive charge (+30 mV), but the z-potential for 200 nm PS-NH2, unexpectedly, was found to be negative (−28 mV). To further characterize the surface charge, the particle size and z-potential of four PS-NH2 particles, including the 200 nm PS-NH2, were measured at different pH 10.6, 8.7, 3.5, and 2.0 (Table 2). The rationale behind the choice of pH is that the PS-NH2 amine groups, added as the functionalization and sulphone groups left from the original particle can be titrated and thereby change the stability and surface charge of the particles. As seen in Table 2 the z-potential decrease at high pH for the two small PS-NH2 and the 180 nm PS-NH2, whereas for the 200 nm PS-NH2 the z-potential is close to neutral at low pH probably as a result of deprotonation and protonation, respectively. The changes in z-potential were reflected in the stability of the dispersion especially for the 200 nm PS-NH2 that aggregated at low pH. A possible explanation of the different behaviour of the 200 nm PS-NH2 is that there is a lower number of amine groups on the surface of the 200 nm PS-NH2. Therefore, the surface chemistry was further characterized by FTIR spectrometry (Fig. S3–S5†). The areas with wavenumber ∼1570 cm−1, which have been assigned to the stretching vibrations of –NH2,43 and wavenumber ∼1005 cm−1 which have been assigned to stretching vibrations of C–N bonds,44 are present in all spectra for all PS-NH2 particles indicating that there are amine groups present although in different amounts. Furthermore, in the spectra for 180 nm PS-NH2 there is a broad signal between 1000 and 1300 cm−1, indicating sulphone groups. These signals are less prominent in the other particles. Overall, the characterization shows that there are amine groups on all particles but the number and the ratio between amine and sulfone groups vary among the different particles.
| Nominal sizes (nm) | Diameter (nm)/polydispersity (%) | z-Potential (mV) with standard deviation | ||||||
|---|---|---|---|---|---|---|---|---|
| pH 10.6 | pH 8.7 | pH 3.5 | pH 2.0 | pH 10.6 | pH 8.7 | pH 3.5 | pH 2.0 | |
| a In this condition the particle dispersion has two peaks. | ||||||||
| 50 | 60/23 | 51/23 | 48/20 | 50/21 | 26 ± 1 | 28 ± 0.7 | 30 ± 0.8 | 32 ± 2 |
| 304/19a | ||||||||
| 53 | 67/24 | 58/25 | 56/12 | 58/31 | 21 ± 0.4 | 27 ± 1 | 35 ± 0.9 | 34 ± 2 |
| 180 | 165/13 | 167/17 | 168/13 | 176/19 | −37 ± 0.8 | −31 ± 0.6 | 33 ± 0.7 | 25 ± 4 |
| 200 | 232/20 | 222/17 | 444/24 | 413/32 | −23 ± 0.7 | −29 ± 0.6 | −10 ± 0.3 | −2 ± 0.5 |
Next, we wanted to expand the number of particles in the analyses with the non-toxic 62 nm and 200 nm PS-COOH, as they have a different surface chemistry from aminated PS nanoparticles that could influence the protein binding. Despite several attempts we were not able to visualize any proteins using the above approach. This could be due to lower protein binding to the negatively charged particles or that less proteins could be desorbed from the particles. Therefore, we tried another approach in which the digestion of the proteins was made on the particles before identification with mass spectrometry. The identified proteins, using the same criteria for selection as above are reported in Table S3.† There were fewer proteins identified when the proteins were digested on the particles and only a few overlapping proteins between the different preparations of PS-NH2 were observed (Tables S2 and S3†). This may be explained by that PS-NH2 are able to strongly affect the structure of bound proteins45 and thereby what parts of the proteins are accessible for the enzyme and what peptides will be released.
The main proteins found to be bound on the 200 nm PS-NH2 are the same, i.e., vitellogenin-1 and hemocyte protein-glutamine gamma-glutamyltransferase, regardless of the preparation method. Interestingly, these proteins are also among the main proteins identified on the 62 and 200 nm PS-COOH. Furthermore, vitellogenin and hemocyte protein-glutamine gamma-glutamyltransferase are also found on the pellets of 53 nm PS-NH2. Vittelogenin-1 is one of the main proteins identified to bind to 25 nm gold particles filtered by D. magna,33 indicating that this protein may in general bind to nanoparticles. Serine protease and actin were commonly bound to all nanoparticles when the proteins were digested in the pellet. Actin plays a major role in the structure and motility of cells in both muscle and non-muscle cells,46 and changes in its expression can lead to toxicity.47
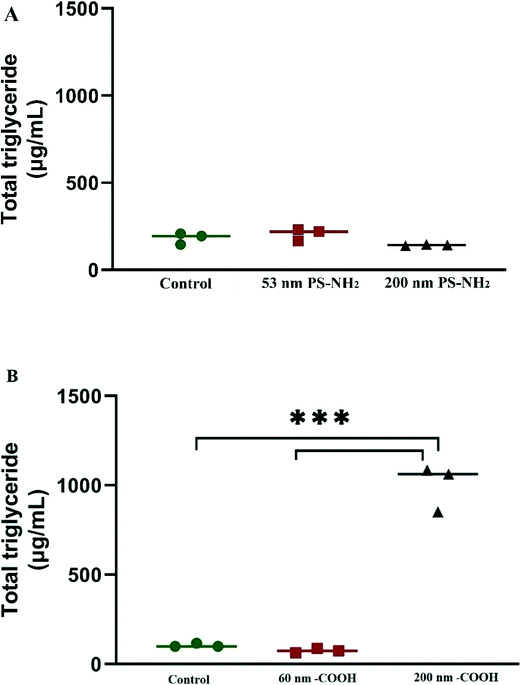
The bio-corona formed around PS nanoparticles can be composed of both proteins and lipids.23 Therefore, we measured the total triglyceride concentration of each experimental fraction. Triglyceride concentration between 53 and 200 nm PS-NH2 after incubation with D. magna was similar to the control group (Fig. 4A). However, triglyceride concentration in 200 nm PS-COOH nanoparticles after incubation with D. magna showed higher concentration compared to both control and 62 nm PS-COOH (Fig. 4B). Similarly, Lima et al.23 showed that higher levels of total triglycerides were observed in lipid corona of 200 nm PS-COOH compared to smaller size particles (80 nm PS-COOH) after 1 h incubation in mouse serum.
Conclusions
There is a remarkable difference in proteins bound to PS nanoparticles that have shown to be toxic (53 nm PS-NH2) and non-toxic (62 nm PS-COOH, 200 nm PS-COOH and 200 nm PS-NH2) in acute toxicity tests. There is a size and charge difference as smaller acetic proteins bind to the toxic 53 nm PS-NH2. The proteins which bind to acutely toxic 53 nm PS-NH2 can be divided into two groups, the ones that are functionally related to the digestive system and the ones involved in various other functions. Furthermore, 53 nm PS-NH2 bound the highest number of total and unique proteins compared to the other tested PS nanoparticles. There were several proteins, for example actin, carboxypeptidase B, chymotrypsin elastase family member 2A or beta-klotho, which were absent from the control group. It can be speculated that 53 nm PS-NH2 toxicity could be due to the binding and depleting proteins that are important for the longevity of D. magna and/or damage the integrity of cells or tissues in the digestive system. On the other hand, non-toxic nanoparticles (200 nm PS-NH2, 62 nm and 200 nm PS-COOH) appeared to bind to similar proteins. These proteins are mainly related to the epithelium and intracellular structures and processes. Even though the latter nanoparticles are shown to be non-toxic after acute exposure,28 there were a couple of proteins (serine protease and vitellogenin-1), that bound to all nanoparticles, which partly could explain the toxicity of 62 nm PS-COOH after a long-term exposure.47We found that neither 53 nm nor 200 nm PS-NH2 bound to lipids, this could be due to the electrostatic repulsion between the positively charged parts of the lipid head and the positive group on the particle surface.48 However, negatively charged 200 nm PS-COOH appeared to exhibit greater binding affinity compared to 62 nm PS-COOH. This suggests that the amount of lipids that binds to nanoparticles depends on the hydrophobicity of the nanoparticle surface, as well as nanoparticle size and/or curvature.20
The results presented in this study provide knowledge regarding what is happening after a short (4 h) incubation of PS nanoparticles together with D. magna. However, despite not being acutely toxic, 62 nm PS-COOH is shown to be toxic after long-term exposure.47 We therefore conclude that including protein corona characterization at later time points in further studies can provide a better understanding regarding the toxicity of these, as well as other, nanoparticles.
In this study we have targeted the most abundant proteins on the PS particles after filtration as we believe these are the most relevant to explain the acute toxicity. Possible explanations for the toxicity are protein depletion and tissue rupture. Future experiments using the most abundant proteins, after cloning and expression, to cover the particle surface with a single protein could elucidate if the absorption of specific proteins affect the particle toxicity.
Author contributions
Egle Kelpsiene: investigation, methodology, writing – review & editing. Irene Brandts: investigation, methodology, writing – review & editing. Katja Bernfur: formal analysis, investigation, writing – review & editing. Mikael T. Ekvall: conceptualization, formal analysis, writing – review & editing. Martin Lundqvist: investigation, writing – review & editing. Mariana Teles: conceptualization, investigation, methodology, writing – review & editing. Tommy Cedervall: conceptualization, funding acquisition, methodology, supervision, writing – original draft, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding for the present study was provided by the Swedish Environmental Protection Agency and the MISTRA Environmental NanoSafety program.References
- P.-C. Lin, S. Lin, P. C. Wang and R. Sridhar, Techniques for physicochemical characterization of nanomaterials, Biotechnol. Adv., 2014, 32(4), 711–726 CrossRef PubMed.
- K. McNamara and S. A. Tofail, Nanoparticles in biomedical applications, Adv. Phys.: X, 2017, 2(1), 54–88 CAS.
- R. Gupta and H. Xie, Nanoparticles in Daily Life: Applications, Toxicity and Regulations, J. Environ. Pathol., Toxicol. Oncol., 2018, 37(3), 209–230 CrossRef PubMed.
- K. L. Aillon, Y. Xie, N. El-Gendy, C. J. Berkland and M. L. Forrest, Effects of nanomaterial physicochemical properties on in vivo toxicity, Adv. Drug Delivery Rev., 2009, 61(6), 457–466 CrossRef CAS PubMed.
- A. R. Petosa, D. P. Jaisi, I. R. Quevedo, M. Elimelech and N. Tufenkji, Aggregation and deposition of engineered nanomaterials in aquatic environments: role of physicochemical interactions, Environ. Sci. Technol., 2010, 44(17), 6532–6549 CrossRef CAS PubMed.
- M. Markiewicz, J. Kumirska, I. Lynch, M. Matzke, J. Koser and S. Bemowsky, et al., Changing environments and biomolecule coronas: consequeences and challenges for the design of environmentally acceptable engineered nanoparticles, Green Chem., 2018, 20(18), 4133–4168 RSC.
- F. Nasser, J. Constantinou and I. Lynch, Nanomaterials in the Environment Acquire an “Eco-Corona” Impacting their Toxicity to Daphnia magna - a Call for Updating Toxicity Testing Policies, Proteomics, 2020, 20(9), e1800412 CrossRef PubMed.
- M. P. Monopoli, C. Aberg, A. Salvati and K. A. Dawson, Biomolecular coronas provide the biological identity of nanosized materials, Nat. Nanotechnol., 2012, 7(12), 779–786 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. Hoek and P. Somasundaran, et al., Understanding biophysicochemical interactions at the nano-bio interface, Nat. Mater., 2009, 8(7), 543–557 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin and H. Nilsson, et al., Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2050–2055 CrossRef CAS PubMed.
- I. Lynch and K. A. Dawson, Protein-nanoparticle interactions, Nano Today, 2008, 3(1), 40–47 CrossRef CAS.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(38), 14265–14270 CrossRef CAS PubMed.
- I. Lynch, K. A. Dawson, J. R. Lead and E. Valsami-Jones, Macromolecular coronas and their importance in nanotoxicology and nanoecotoxicology, Frontiers of nanoscience, Elsevier, 2014, vol. 7, pp. 127–156 Search PubMed.
- D. Dell'Orco, M. Lundqvist, C. Oslakovic, T. Cedervall and S. Linse, Modeling the time evolution of the nanoparticle-protein corona in a body fluid, PLoS One, 2010, 5(6), e10949 CrossRef PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz and R. Hecht, et al., Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology, Nat. Nanotechnol., 2013, 8(10), 772–781 CrossRef CAS PubMed.
- S. Ritz, S. Schöttler, N. Kotman, G. Baier, A. Musyanovych, J. Kuharev, K. Landfester, H. Schild, O. Jahn, S. Tenzer and V. Mailänder, et al., Protein corona of nanoparticles: distinct proteins regulate the cellular uptake, Biomacromolecules, 2015, 16(4), 1311–1321 CrossRef CAS PubMed.
- J. A. Kim, A. Salvati, C. Åberg and K. A. Dawson, Suppression of nanoparticle cytotoxicity approaching in vivo serum concentrations: limitations of in vitro testing for nanosafety, Nanoscale, 2014, 6(23), 14180–14184 RSC.
- K. Saha, D. F. Moyano and V. M. Rotello, Protein coronas suppress the hemolytic activity of hydrophilic and hydrophobic nanoparticles, Mater. Horiz., 2014, 1(1), 102–105 RSC.
- C. D. Walkey and W. C. Chan, Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment, Chem. Soc. Rev., 2012, 41(7), 2780–2799 RSC.
- E. Hellstrand, I. Lynch, A. Andersson, T. Drakenberg, B. Dahlbäck and K. A. Dawson, et al., Complete high-density lipoproteins in nanoparticle corona, FEBS J., 2009, 276(12), 3372–3381 CrossRef CAS PubMed.
- C. C. Fleischer and C. K. Payne, Nanoparticle–cell interactions: molecular structure of the protein corona and cellular outcomes, Acc. Chem. Res., 2014, 47(8), 2651–2659 CrossRef CAS PubMed.
- S. Wan, P. M. Kelly, E. Mahon, H. Stockmann, P. M. Rudd and F. Caruso, et al., The “sweet” side of the protein corona: effects of glycosylation on nanoparticle–cell interactions, ACS Nano, 2015, 9(2), 2157–2166 CrossRef CAS PubMed.
- T. Lima, K. Bernfur, M. Vilanova and T. Cedervall, Understanding the lipid and protein corona formation on different sized polymeric nanoparticles, Sci. Rep., 2020, 10(1), 1–9 CrossRef PubMed.
- T. Vandenbrouck, O. A. H. Jones, N. Dom, J. L. Griffin and W. De Coen, Mixtures of similarly acting compounds in Daphnia magna: From gene to metabolite and beyond, Environ. Int., 2010, 36(3), 254–268 CrossRef CAS PubMed.
- W. Geller and H. Müller, The filtration apparatus of Cladocera: filter mesh-sizes and their implications on food selectivity, Oecologia, 1981, 49(3), 316–321 CrossRef PubMed.
- P. Rosenkranz, Q. Chaudhry, V. Stone and T. F. Fernandes, A comparison of nanoparticle and fine particle uptake by Daphnia magna, Environ. Toxicol. Chem., 2009, 28(10), 2142–2149 CrossRef CAS PubMed.
- A. Sukhanova, S. Bozrova, P. Sokolov, M. Berestovoy, A. Karaulov and I. Nabiev, Dependence of Nanoparticle Toxicity on Their Physical and Chemical Properties, Nanoscale Res. Lett., 2018, 13(1), 44 CrossRef PubMed.
- K. Mattsson, E. V. Johnson, A. Malmendal, S. Linse, L.-A. Hansson and T. Cedervall, Brain damage and behavioural disorders in fish induced by plastic nanoparticles delivered through the food chain, Sci. Rep., 2017, 7(1), 11452 CrossRef PubMed.
- J. Saavedra, S. Stoll and V. I. Slaveykova, Influence of nanoplastic surface charge on eco-corona formation, aggregation and toxicity to freshwater zooplankton, Environ. Pollut., 2019, 252, 715–722 CrossRef CAS PubMed.
- A. Pochelon, S. Stoll and V. I. Slaveykova, Polystyrene Nanoplastic Behavior and Toxicity on Crustacean Daphnia magna: Media Composition, Size, and Surface Charge Effects, Environments, 2021, 8(10), 101 CrossRef.
- E. Besseling, B. Wang, M. Lurling and A. A. Koelmans, Nanoplastic Affects Growth of S. obliquus and Reproduction of D. magna, Environ. Sci. Technol., 2014, 48(20), 12336–12343 CrossRef CAS PubMed.
- F. Nasser and I. Lynch, Secreted protein eco-corona mediates uptake and impacts of polystyrene nanoparticles on Daphnia magna, J. Proteomics, 2016, 137, 45–51 CrossRef CAS PubMed.
- K. Mattsson, R. Aguilar, O. Torstensson, D. Perry, K. Bernfur and S. Linse, et al., Disaggregation of gold nanoparticles by Daphnia magna, Nanotoxicology, 2018, 12(8), 885–900 CrossRef CAS PubMed.
- O. O. Fadare, B. Wan, K. Y. Liu, Y. Yang, L. X. Zhao and L. H. Guo, Eco-Corona vs Protein Corona: Effects of Humic Substances on Corona Formation and Nanoplastic Particle Toxicity in Daphnia magna, Environ. Sci. Technol., 2020, 54(13), 8001–8009 CrossRef CAS PubMed.
- L.-J. A. Ellis and I. Lynch, Mechanistic insights into toxicity pathways induced by nanomaterials in Daphnia magna from analysis of the composition of the acquired protein corona, Environ. Sci.: Nano, 2020, 7(11), 3343–3359 RSC.
- M. Lundqvist, T. Berggard, E. Hellstrand, I. Lynch, K. A. Dawson, S. Linse and T. Cedervall, Rapid and facile purification of apolipoprotein AI from human plasma using thermoresponsive nanoparticles, J. Biomater. Nanobiotechnol., 2011, 2(03), 258–266 CrossRef CAS.
- F. L. Carboxypeptidases, in xPharm: The Comprehensive Pharmacology Reference, ed. S. J. Enna and D. B. Bylund, Elsevier, New York, 2007, pp. 1–4 Search PubMed.
- M. Kuro-o, Y. Matsumura, H. Aizawa, H. Kawaguchi, T. Suga and T. Utsugi, et al., Mutation of the mouse klotho gene leads to a syndrome resembling ageing, Nature, 1997, 390(6655), 45–51 CrossRef CAS PubMed.
- S. A. Li, M. Watanabe, H. Yamada, A. Nagai, M. Kinuta and K. Takei, Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice, Cell Struct. Funct., 2004, 29(4), 91–99 CrossRef CAS PubMed.
- T. Shiraki-Iida, H. Aizawa, Y. Matsumura, S. Sekine, A. Iida and H. Anazawa, et al., Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein, FEBS Lett., 1998, 424(1–2), 6–10 CrossRef CAS PubMed.
- A. Hara, N. Hiramatsu and T. Fujita, Vitellogenesis and choriogenesis in fishes, Fish. Sci., 2016, 82(2), 187–202 CrossRef CAS.
- J. Gao, L. Lin, A. Wei and M. S. Sepúlveda, Protein corona analysis of silver nanoparticles exposed to fish plasma, Environ. Sci. Technol. Lett., 2017, 4(5), 174–179 CrossRef CAS PubMed.
- C. Ling, X. Y. Li, Z. Y. Zhang, F. Q. Liu, Y. Q. Deng and X. P. Zhang, et al., High Adsorption of Sulfamethoxazole by an Amine-Modified Polystyrene-Divinylbenzene Resin and Its Mechanistic Insight, Environ. Sci. Technol., 2016, 50(18), 10015–10023 CrossRef CAS PubMed.
- A. Hernandez-Gordillo, S. Oros-Ruiz and R. Gomez, Preparation of efficient cadmium sulfide nanofibers for hydrogen production using ethylenediamine (NH2CH2CH2NH2) as template, J. Colloid Interface Sci., 2015, 451, 40–45 CrossRef CAS PubMed.
- R. Cukalevski, S. A. Ferreira, C. J. Dunning, T. Berggård and T. Cedervall, IgG and fibrinogen driven nanoparticle aggregation, Nano Res., 2015, 8(8), 2733–2743 CrossRef CAS.
- P. W. Gunning, U. Ghoshdastider, S. Whitaker, D. Popp and R. C. Robinson, The evolution of compositionally and functionally distinct actin filaments, J. Cell Sci., 2015, 128(11), 2009–2019 CrossRef CAS PubMed.
- E. Kelpsiene, O. Torstensson, M. T. Ekvall, L.-A. Hansson and T. Cedervall, Long-term exposure to nanoplastics reduces life-time in Daphnia magna, Sci. Rep., 2020, 10(1), 5979 CrossRef CAS PubMed.
- B. Munteanu, F. Harb, J. P. Rieu, Y. Berthier, B. Tinland and A. M. Trunfio-Sfarghiu, Charged particles interacting with a mixed supported lipid bilayer as a biomimetic pulmonary surfactant, Eur. Phys. J. E: Soft Matter Biol. Phys., 2014, 37(8), 72 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2en00125j |
| This journal is © The Royal Society of Chemistry 2022 |