 Open Access Article
Open Access ArticleCritical review of antibiotic resistance genes in the atmosphere†
David
Kormos
,
Kaisen
Lin
,
Amy
Pruden
 and
Linsey C.
Marr
and
Linsey C.
Marr
 *
*
Civil and Environmental Engineering, Virginia Tech, Blacksburg, VA, USA. E-mail: lmarr@vt.edu
First published on 25th May 2022
Abstract
We conducted a critical review to establish what is known about the sources, characteristics, and dissemination of ARGs in the atmosphere. We identified 52 papers that reported direct measurements of bacterial ARGs in air samples and met other inclusion criteria. The settings of the studies fell into the following categories: urban, rural, hospital, industrial, wastewater treatment plants (WWTPs), composting and landfill sites, and indoor environments. Certain genes were commonly studied and generally abundant: sul1, intI1, β-lactam ARGs, and tetracycline ARGs. Abundances of total ARGs varied by season and setting, with air in urban areas having higher ARG abundance than rural areas during the summer and vice versa during the winter. There was greater consistency in the types and abundances of ARGs throughout the seasons in urban areas. Human activity within indoor environments was also linked to increased ARG content (abundance, diversity, and concentration) in the air. Several studies found that human exposure to ARGs through inhalation was comparable to exposure through drinking water or ingesting soil. Detection of ARGs in air is a developing field, and differences in sampling and analysis methods reflect the many possible approaches to studying ARGs in air and make direct comparisons between studies difficult. Methodologies need to be standardized to facilitate identification of the dominant ARGs in the air, determine their major sources, and quantify the role of atmospheric transport in dissemination of ARGs in the environment. With such knowledge we can develop better policies and guidelines to limit the spread of antimicrobial resistance.
Environmental significanceThe spread of antimicrobial resistance is considered one of the greatest threats to global health. While the presence and transport of antibiotic resistance genes (ARGs) in water and soil are well documented, less is known about them in the air. Our critical review synthesizes the current state of knowledge about ARGs in the air, including their sources, temporal and spatial patterns, and potential for human exposure. We identify knowledge gaps and present a list of questions for future research. This work contributes to a more complete picture of ARGs in the total environment and will inform development of policies and guidelines to more effectively limit the spread of antimicrobial resistance. |
1. Introduction
Antimicrobial resistance (AMR) looms as a major threat to public health and is estimated to be responsible for $20 billion in direct healthcare costs and $35 billion in lost productivity per year in the US.1,2 Overuse and abuse of antibiotics in human medicine and agriculture are key drivers of increased rates of AMR among infections afflicting humans.3 Anthropogenic inputs of antimicrobials (e.g., antibiotics, antifungals, antiparasitics), antibiotic resistant bacteria, and antibiotic resistance genes (ARGs) to the environment4 have markedly augmented their occurrence in soil, sediment, and permafrost relative to background levels.5 Given that ARGs driving clinical resistance have in many cases been traced back to environmental sources,6 a US National Academies workshop identified better understanding of ARGs in the environment as a high-priority knowledge gap.7 While the behavior and fate of ARGs in soil and water have been studied extensively, along with mitigation strategies,4,8,9 far fewer studies have considered ARGs in the atmosphere. Among these, the focus is usually on short-range transport. Very few studies have addressed the potential for long-range transport of ARGs across continents.Suspension of ARGs in the air is of concern because this can potentially enhance their long-distance transport, increase transmission across environmental and ecosystem boundaries, and increase human and animal exposure via inhalation. Atmospheric transport can lead to greater dissemination of AMR in the environment beyond that facilitated by international movement of humans, animals, and goods.10 Taking advantage of a recent proliferation of studies of ARGs in the atmosphere, here we critically review the current state of knowledge regarding ARGs in the air and identify critical research needs that must be addressed in order to more holistically understand and mitigate their contribution to the spread of AMR.
2. Search strategy and data extraction
Fig. S1† summarizes the approach and criteria applied to selecting articles for inclusion in this critical review, and Table S1† summarizes the exact searches used for each of the databases. Three databases (Web of Science, PubMed, and SCOPUS) were searched on 20 January 2021 to identify publications describing ARGs in indoor and/or outdoor air. The following keywords and terms were used for the search: “air”, “airborne”, “air-borne”, “bioaerosol”, “atmosphere”, “antimicrobial”, “antibiotic”, “antibacterial”, and “resist”. No date limit was applied to the searches. A total of 2488 articles were captured through these searches (886 from WOS, 972 from PubMed, and 630 from SCOPUS). Duplicates were removed based on title by using the Excel Fuzzy Lookup add-in with a similarity of greater than 0.85, then removed by exact matching of DOI. Each set of duplicates was verified manually. There were 1529 distinct papers remaining (873 removed according to title, 86 removed according to DOI). Titles and abstracts were scrutinized to remove those that did not report direct measurement of ARGs in air samples, as described in the ESI,† reducing the total number of papers to 52 (Fig. S1†). Data were then categorized by setting (urban/suburban, rural/agricultural, industrial, hospital, WWTPs, landfills, indoor environments, or a combination of two or more) and analysis method (polymerase chain reaction (PCR), quantitative PCR (qPCR), high throughput qPCR (HT-qPCR), droplet digital PCR (ddPCR), metagenomic analysis, or a combination of methods).The settings defined for the purpose of this review are not always mutually exclusive. Urban/suburban air refers to outdoor air sampled at urban and suburban areas, away from known point sources of ARGs. For all of the studies included in the urban/suburban section, it is noted whether the sampling location was urban or suburban (generally based on population density), and our classification was based on the original studies. For the purposes of this review, rural/agricultural settings were limited to crop or livestock farms. Typically, there are several buildings on the grounds of a livestock farm, so discussion of rural air will include both indoor and outdoor settings, resulting in some overlap between indoor air and rural settings. We expect farm buildings to differ in many ways from those designed for human occupancy. Hospitals, WWTPs, and landfills are discussed within their own sections because they are known sources of ARGs to the atmosphere. These locations may be also categorized under urban or rural settings, and where applicable those broader classifications are mentioned. Sampling in residences, offices, or labs is discussed in the indoor air section, and their broader classification as either rural or urban is also addressed within the section.
3. Sample collection approaches for airborne ARG analysis
Air sampling for the purpose of microbial analysis presents many challenges.11–14 For ARGs in particular, the ideal sampling method would capture both intracellular and extracellular nucleic acids while maintaining their integrity during collection to support downstream analysis. The sampling devices utilized in the studies reviewed here collected airborne particles onto different media, including filters (n = 51) and agarose (n = 4). In some studies, total suspended particles were collected, while in others, a size-selective inlet was used to exclude larger particles and collect only coarse particulate matter 10 μm and smaller (PM10), fine particulate matter 2.5 μm and smaller (PM2.5), or other size fractions. Such size fractionation is valuable for studying transport, as size dictates how long particles can remain suspended in the atmosphere. PM2.5 can travel hundreds to thousands of kilometers, while larger particles cannot travel as far because they will be more rapidly removed from the atmosphere by gravitational settling.15 The most notable similarity among the studies was the use of 1.5 m as the sampling height, to simulate human height (n = 42).4. ARG analysis of air samples
Analytical methods for the ARGs themselves fell into four distinct categories: PCR (n = 6), qPCR (n = 34), HT-qPCR (n = 5), ddPCR (n = 2), metagenomics (n = 10), or a combination of two or more of these methods (n = 4). The PCR-based methods target pre-determined subsets of ARGs, up to 285 for HT-qPCR, 34 for qPCR, 23 for ddPCR, and 15 for PCR. Metagenomic analysis, on the other hand, affords the advantage of capturing the full profile of ARGs across the microbial assemblage (i.e., the “resistome”), without a priori selection of targets. However, metagenomics yields relative abundance quantification, has a higher detection limit and higher cost, and requires substantial bioinformatic expertise for analysis.16–19 It is also challenging to collect sufficient mass of DNA in air samples to support metagenomic analysis. Thus, it is not surprising that few air studies were found that employed metagenomics for ARG detection (n = 8). The use of metagenomics in ARG analysis is an ever-evolving process, and other reviews have covered the advances extensively.16–19The most commonly employed method for ARG analysis among the studies was qPCR, which has the advantages of high sensitivity (often as low as 1–10 gene copies per reaction can be detected), specificity, and broad quantitative range (typically six logs). The newer approach of ddPCR is thought to be more sensitive, precise, and reproducible, while also not requiring a standard curve.20 qPCR still provides a greater range of quantification than does ddPCR,20 but ddPCR will likely eclipse qPCR in application in future studies due to the other advantages.
5. Comparing airborne ARG profiles reported across studies
Existing studies of ARGs in air typically focus on either urban, rural, or indoor air, or outdoor air near specific sources, such as hospitals, WWTPs, waste sites, or industrial sites. Relevant studies are summarized in Tables S2–S9,† with their setting, sample type, list of all ARGs and other genes screened, and the dominant ARGs found. Dominance is defined here as reported by the studies reviewed, or otherwise if the gene of interest was orders of magnitude higher in concentration or abundance in relation to others. If not explicitly stated, if a gene was detected in more than 50% of samples, occurred at a relative abundance higher than 10−3 copies per 16S rRNA gene, or was at a concentration higher than 102 copies per m3, it was usually considered dominant. Genes targeted included ARGs encoding resistance to various types of antibiotics (e.g., sulfonamides, tetracyclines, β-lactams, glycopeptides, macrolides, quinolones) as well as mobile genetic elements (MGEs) involved in horizontal gene transfer (e.g., intI1, tnpA). The studies employed various sample collection methods that amassed either total suspended particles from the air or only those smaller than a given cut-off, usually PM10 or PM2.5. As an alternative to collecting samples using traditional particle sampling equipment, some researchers collected material from commercial filters used in automobiles and air conditioning units.We considered various aspects to support comparisons across studies, while noting that such comparisons were challenged by different sampling and analysis methods and ambiguous terminology. Here, we use the following common terminology: (1) relative abundance in air refers to gene copies per 16S rRNA gene (gc/16S) (2) airborne concentration refers to gene copies per cubic meter of air (gc m−3) (3) loading in airborne particles (total suspended particles, PM10, or PM2.5) refers to gene copies per mass of particulate matter (gc μg−3) and (4) diversity of samples refers to the number of unique ARGs per sample (richness). The conclusions of some of these studies are based solely on relative abundances, which provide an incomplete picture of trends in airborne ARGs. Total concentrations of bacteria, which appear in the denominator of the relative abundance calculation, may fluctuate widely during a study. Such shifts can influence the observed dynamics. For example, a large shift in relative abundance may not be as significant as it appears if a dust storm rich in bacteria but not ARGs passes through an area with a high ARG concentration.
Metagenomics inherently produces relative abundance data, typically reported as a function of total ARGs detected in the sample, normalized to total reads or another denominator. Because conventional PCR does not provide quantitation, frequency of detection is reported. In terms of ARG diversity, richness can be particularly informative when seeking to identify sources of ARGs in a given sample. An additional challenge is that studies often target different types of ARGs (e.g., tetracycline resistance versus sulfonamide resistance types and corresponding sub-types), making it impossible to compare results directly. Type of ARG refers to the class of antibiotic, such as sulfonamide, and sub-type refers to the specific gene within the type, such as sul1. A few studies compared the same ARGs in different settings,21–23 but whether differences are due to geographic location (e.g., China vs. US) or the settings of air samples collected (e.g., urban, rural, industrial) is not readily apparent. It would also be useful if studies applied the same analytical methods and targeted the same ARGs in different media (e.g., air, water, and soil) in order to make relative comparisons and to begin populating mass transfer models.
a. ARGs in urban and suburban air
Twenty-one studies were identified that sampled urban or suburban air, representing four different continents, with more than half including sampling locations in China (n = 13). In this section, we focus on samples collected away from known point sources of ARGs. The ARGs targeted and those reported to be most abundant were largely inconsistent across the studies; however, some trends were clear. A few genes were consistently dominant, including sul1, intI1, certain β-lactam ARGs, and certain tetracycline ARGs, which is not surprising because sul1, intI1, and tetW are well established as indicators of anthropogenic sources of antibiotic resistance.24 A large study of 864 publicly-available metagenomes from humans, animals, sediment, soil, water, mines, wastewater/sludge, and pharmaceutical pollution revealed that air samples from Beijing exhibited the highest diversity of ARGs compared to other samples.25 A global study of urban air found that among 19 sites surveyed, the highest total relative abundance (total gc of ARGs/16S rRNA gene) of 30 ARG sub-types detected by qPCR was in the US (San Francisco), at a level more than twice as high as in other countries.26In a city in China with longitudinal data, the relative abundance of certain ARGs grew by 30–180% over a period of 10 years, showing the increasing dominance of specific ARGs (e.g., qepA and blaTEM) in the environment.26 This finding agrees with a metagenomics study of Beijing smog, which indicated blaTEM to be one of the most abundant ARG sub-types.25 These results are somewhat inconsistent, however, with another large-scale metagenomics study in China showing that chloramphenicol exporter gene and sul1 were the most common ARG sub-types in both urban and suburban communities, followed by bacA in the urban community and aadE in the suburban community. Genes conferring resistance to tetracycline (tet) were also especially abundant in both urban and suburban communities.27 These inconsistencies could be explained by differences in analytical methods, as mentioned previously, or differences in the background environment and antibiotic usage patterns. A general challenge with such field studies is that it is difficult to isolate the factors contributing to the observed patterns, especially across multiple locations.
A study in California contrasted ARG profiles in 26 parks in four major cities (Bakersfield, San Diego, Fresno, and Los Angeles) and three different media (soil, drinking water, and air) during a single season. The samples were examined for sul1 and blaSHV, while soil samples were further examined for ermF and ermB. The largest and most significant differences in ARG abundances between the cities were observed in soil. ARG abundances in air were highly variable, even between parks within the same city.28 These results show how ARG content can vary substantially within the same region and media.
One key study found that air pollution in an urban environment correlated strongly with relative abundance of total ARGs annotated in metagenomic data (i.e., number of the hits of all ARGs types detected per one million sequencing reads, as “ppm”). Levels were higher during a severe smog event than on clean days in Beijing. Thus, emissions of anthropogenic ARGs may be correlated with emissions of traditional air pollutants. Interestingly, tetW was found to be equally abundant on both clean and polluted days, consistent with reports of this ARG being widespread in the environment,29,30 potentially related to the fact that tetracycline is a very common type of antibiotic used for treating humans and livestock. Dominant levels of tetW in urban air were confirmed in a metagenomic study of PM2.5 at Beijing University,30 but one study of PM2.5 and PM10 fractions in Harbin, China reported that concentrations of sul1, intI1, and qnrS were orders of magnitude higher than those of tetW.31 A study in Bolu, Turkey indicated similarly high abundances of sul1, intI1, and qnrS in PM10 fractions.32 Additionally, it has been shown that contaminated water sources from urban environments in Bolivia emitted bioaerosols enriched with ARGs, particularly blaTEM and intI1.33 The contrast between these findings illustrates that one study alone cannot fully characterize AMR due to the limited number of genes screened.
The total “ppm” of ARGs in the PM2.5 fraction in Beijing was as much as 7 times higher during the smog event than on clean days, and as much as 5 times higher in the PM10 fraction. The concentrations of PM2.5 and PM10 were not reported, making it difficult to define the relationship between PM and ARG abundance. The higher relative abundance of ARGs on polluted days suggests that at least some of the same sources contributing to elevated PM may also be rich sources of ARGs; tet, blaTEM, and aminoglycoside ARGs were most abundant on polluted days.29blaTEM and other β-lactamase ARGs (ampC-02, blaIMP-01, blaSHV-01, and blaCTX-M) were also dominant in a metagenomic study of PM2.5 and PM10 fractions in Beijing34 and in Tianjin, China.35 Several other studies also confirmed elevated ARGs in hazy and polluted air.31,34–38 Conversely, intI1, qnrS, and sul1 abundance measured by qPCR decreased during dust storms in urban areas.39,40
In contrast, another study in Beijing found that total relative abundances of ARGs decreased during a period of high pollution, with some specific (and more clinically-relevant) genes increasing in relative abundance. Additionally, ARG relative abundances differed greatly (up to 3-fold) between day and night. MGE genes tnpA and intI1 occurred at higher relative abundances during high PM2.5 episodes, while blaTEM and sul1 were high in relative abundances in all cases. The carbapenemase-producing blaNDM-1, which is a highly clinically-relevant ARG, was high in abundance in the early morning of polluted days. In this study, PM2.5 concentrations on the cleaner days ranged from 14 to 93 μg m−3, while on the polluted days concentrations ranged from 36 to 251 μg m−3.41 For reference, the World Health Organization's guideline value for PM2.5 is 15 μg m−3 over a 24 hours average. The changes in the distribution and makeup of ARG sub-types in the PM2.5 on polluted vs. clean days could be due to higher emissions from certain sources, varying resuspension patterns at different times of the day, or differences in atmospheric stability.
These studies illustrate that the relationship between air pollution and ARGs in the atmosphere is not straightforward. On one hand, emissions of ARG may be associated with human activities that also release traditional air pollutants, but the observation of a decrease in total ARG relative abundance under severely polluted conditions may indicate that deteriorating air quality hinders ARG dissemination in some cases. Severely polluted air could contain toxins and harmful substances that inhibit bacterial growth, driving changes in the relative abundance of ARG sub-types. This is confirmed by a study published after our literature search in Xinxiang, China, which showed that under moderate pollution levels, concentrations of sul1, sul2, intI1, ermC, and blaTEM were higher than when air quality was excellent, but at higher levels of pollution these concentrations drastically decreased. tetW concentrations started to decrease at low/moderate levels of pollution, showing how different ARGs may respond differently to varying pollution levels.42 Atmospheric transport from different regions could also play a role, as it has been shown to contribute 15–55% of PM2.5 levels during different haze events in Beijing.43 Shifts in relative abundance profiles of individual ARGs while in the air could also lead to variability throughout the day; however, these mechanisms are still not understood.44
There was also some indication that specific ARGs were associated with different regions, like blaTEM in urban areas and sul1 near the Pearl River Delta, while the ARG tetW was more widespread in both urban and rural regions in China.45tet ARGs (tetA and tetB) and ARGs conferring resistance to aminoglycoside (sat) and β-lactam were also found to be abundant in an estuary area in one study.46 This study found a relationship between the microbial community taxonomic composition and the ARG profiles utilizing a Spearman correlation heatmap, finding positive correlations between specific genes and genera, with some bacteria having intrinsic resistance, and some bacteria able to acquire resistance from MGEs. The 21 most abundant ARGs detected by qPCR and 20 of the top 30 genera detected through sequencing were included in the correlation analysis.
b. Rural and agricultural air
In rural settings, the dissemination of ARGs in the environment is heavily influenced by agricultural activity. One study found that ARG composition was similar between animal feces and airborne particles at farms, with feces having a higher relative abundance of total ARGs.47 Thus, animal feces could be one of the major sources of airborne ARGs at and downwind from animal farms. Interestingly, this study also found that some genes (especially the multidrug mexF), was higher in relative abundance in aerosols than in feces, suggesting that ARGs from other contaminants and sources were also present. Antimicrobial usage in livestock over the years may have enriched the resistomes of feces, which subsequently influence the dust and air resistomes of farms, affecting the spread and dissemination of ARGs in these environments.48,49Some of the earliest studies demonstrating the occurrence and transport of ARGs in air were conducted at rural agricultural sites.23,50,51 One rural study documented that tetracycline resistance genes (tetB, tetL, tetM, tetO, tetQ, and tetW) in air were 100–1000 times higher downwind of 10 beef cattle yards compared to upwind.23 The relative abundance of ARGs in total suspended particles was also comparable to that in manure and soils, suggesting that the latter are sources of ARGs in airborne PM.23tet ARGs, especially tetQ, tetW, and tetM, have commonly been found at high frequencies of detection and high concentrations/abundances across many different agricultural sites, including swine, turkey, poultry, and cattle farms.23,46,52–62
Different kinds of farms, livestock, and methods used for the production phase are likely to affect the ARG profile and microbial community composition of the local atmosphere. A metagenomic survey found high diversity across the ARG profiles of different swine confinement buildings on a farm in Guizhou, China, with approximately 300 sub-types detected and ARG concentrations ranging from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–70
000–70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 gene copies per type of housing.63 Type of housing refers to the different production phases of the swine, as each is housed in a separate building, such as weaning piglets and breeding boars, and occupancy ranged from 24 to 480 animals, which could affect normalization of results. In the aerosol samples, the top ten most abundant sub-types were those conferring resistance to aminoglycosides, aminocoumarin, mupirocin, elfamycin, fluoroquinolone, pleuromutilin, rifampin, and lincosamide. Finishing pigs carried the highest occurrence and diversity of ARGs, likely due to longer exposure to antibiotics. Weaning piglets was high in abundance of aminoglycoside ARGs, likely due to the use of aminoglycoside antibiotics. A smaller study of specific ARGs by use of qPCR observed lower concentrations of sul1, blaSHV, ermB, and blaTEM in air at organic beef cattle farms compared to conventional beef cattle farms, confirming how different farm practices could affect the resistome of the air.64 Another study published after our systematic literature search analyzed air samples from 333 poultry and pig farms in nine different countries for tetW, ermB, sul1, and aph(3′)-III. ARG concentrations and relative abundances were correlated with different conditions within the farm including biosecurity measures at the facility (scored by an online aggregate system). Higher biosecurity measures were associated with lower ARG relative abundance at poultry farms but higher ARG relative abundance at pig farms, showing that biosecurity measures for pathogens may not be effective for ARG control.65
000 gene copies per type of housing.63 Type of housing refers to the different production phases of the swine, as each is housed in a separate building, such as weaning piglets and breeding boars, and occupancy ranged from 24 to 480 animals, which could affect normalization of results. In the aerosol samples, the top ten most abundant sub-types were those conferring resistance to aminoglycosides, aminocoumarin, mupirocin, elfamycin, fluoroquinolone, pleuromutilin, rifampin, and lincosamide. Finishing pigs carried the highest occurrence and diversity of ARGs, likely due to longer exposure to antibiotics. Weaning piglets was high in abundance of aminoglycoside ARGs, likely due to the use of aminoglycoside antibiotics. A smaller study of specific ARGs by use of qPCR observed lower concentrations of sul1, blaSHV, ermB, and blaTEM in air at organic beef cattle farms compared to conventional beef cattle farms, confirming how different farm practices could affect the resistome of the air.64 Another study published after our systematic literature search analyzed air samples from 333 poultry and pig farms in nine different countries for tetW, ermB, sul1, and aph(3′)-III. ARG concentrations and relative abundances were correlated with different conditions within the farm including biosecurity measures at the facility (scored by an online aggregate system). Higher biosecurity measures were associated with lower ARG relative abundance at poultry farms but higher ARG relative abundance at pig farms, showing that biosecurity measures for pathogens may not be effective for ARG control.65
The finding of ARGs in rural environments, where the corresponding antibiotics have not been used, suggests that atmospheric transport may play a role in disseminating ARGs. The aminocoumarin ARG in the previously discussed metagenomics study of a large-scale swine farm in China63 was notably abundant in certain swine confinement buildings, despite the fact that novobiocin, an aminocoumarin, was not used at this farm.63 Novobiocin is commonly used for boars elsewhere in China. Another study similarly found a high frequency of the colistin resistance gene (mcr-1) in Eastern Canada, despite the restriction of colistin in Eastern Canada, indicating possible long-distance transport of this ARG.66 A study in China was conducted at a swine farm that utilized biogas digestate storage using HT-qPCR to analyze 45 ARGs.57 The most abundant ARGs detected in the airborne particulate matter at this facility belonged to the β-lactam type. MGEs (tnpA and intI1) were detected and seemed to be correlated with different ARG sub-types (tet and sul), indicating possible horizontal gene transfer.57 In horizontal gene transfer, ARGs are carried by MGEs, and since ARGs had a more significant correlation with MGEs than with bacterial communities in this study, and there were higher levels of intI1 in the winter, it is likely the main pathway from ARGs in the winter.54
Composting may also be a source of ARG emissions to the atmosphere. A study at four compositing plants found higher concentrations of IntI1 in air samples collected downwind compared to those collected upwind.67
c. Hospitals
We identified 11 studies of air in 18 hospitals, more than half of these in China (n = 10). Within urban and suburban areas, the richness and relative abundance of ARGs was higher in hospital air at the entrance of the hospital compared to that of outdoor air of surrounding communities.27 Air near the entrance to a hospital in the center of a city in China was higher in relative abundances of ARGs conferring multidrug and quinolone resistance than did urban and suburban environments 0.5 km and 10 km away, respectively.27 ARGs conferring multi-drug resistance and resistance to aminoglycoside, tetracycline, chloramphenicol and the macrolide–lincosamide–streptogramin (MLS) types of antibiotics were most abundant. These ARGs are commonly found in medicine (antibiotic use in humans) and the animal breeding industry (antibiotic use by veterinarians).27 In a hospital study in Beijing, aminoglycoside, bacitracin, β-lactam, chloramphenicol, MLS, multidrug, tetracycline, bleomycin, and sulfonamide ARGs were the most frequently detected types by metagenomics, with intI1 and tetW being the two most abundant sub-types according to qPCR, while aadD, CE, tetK, tetA, tetZ, and norA were widespread in most of the departments of the hospital.68 Slight variations in ARG profiles between the two studies may be a function of actual differences between the hospitals, such as the local pool of ARGs carried by patients, differences in antibiotic use, and differences in human activity. However, differences in sampling and detection methods may also influence the dominant ARGs identified. Other hospital studies that used qPCR69,70 or PCR71,72 for targeted ARGs also indicated a high frequency of ermA and ermB,70–72 tetracycline70,71 and methicillin ARGs,69,72 with low frequency of β-lactam69,70 and vancomycin ARGs.71,72 A metagenomics study found aminoglycoside, multidrug, and fluoroquinolone, quinolone, florfenicol, chloramphenicol, amphenicol (FCA) were the most abundant ARGs in air and confirmed that these ARGs were higher in indoor hospital samples than in dust samples and outdoor air samples.73Two studies compared the ARG abundances and concentrations in multiple environments, including hospitals.22,74 One of these studies employed qPCR on air samples collected from a WWTP (fine screens and sludge thickener), dormitory bathroom, hospital infusion room, and outdoors at Tianjin University during a haze episode.22 The highest concentrations of blaOXA-1, blaTEM, blaAmpc, ermC, tetC, and tetM were found in the hospital. It had the highest diversity and second highest concentration (behind hazy outdoors) among the environments tested, likely due to the high use of antibiotics in hospitals. The second study collected air conditioning filter samples from hospitals, farms, and residences in Ningbo City, China and detected ARGs with HT-qPCR.74 Similar to the previous study, β-lactams had one of the highest detection rates in hospitals after multidrug, followed by aminoglycoside, MLSB, and tetracycline. bl3_cpha gene was the most abundant sub-type. Total ARG relative abundance was highest in hospitals at 8.76 × 10−2 copies per 16S, followed by the farm at 3.29 × 10−2 copies per 16S, village residences at 3.29 × 10−3 copies per 16S, and city residences at 3.64 × 10−6 copies per 16S.
One metagenomic study showed how ARGs moved throughout the hospital air and were able to accumulate in air conditioning units.68 Some of the ARG-carrying bacteria within these units were found to be associated with humans through interpretation of metagenome assemblies, including bacteria known to grow in oral and nasal passages that could enter the environment via respiratory aerosols.68 Such movement patterns of ARGs in hospitals are consistent with observations of general microbial movement in hospital microbiome studies.75 Additionally a metagenomic study published after the literature search found that human sources accounted for 30% of the PM2.5 microbiome in a hospital, and that these bacteria were strongly correlated with the resistome. This study also confirmed higher total relative abundances of ARGs in hospitals versus the surrounding urban community, with an increase of 0.2 to 0.5 magnitudes for each ARG type. Some clinically relevant ARG sub-types were significantly higher in hospital vs. urban samples. These included rpoB2 (tuberculosis drugs), vanR, ugd, blaOXA, class-a, msrA, tetL, and bacA.76 These findings suggest that human activity inside of a hospital, and any indoor environment, could lead to an increase of ARGs in the air, and therefore increase the likelihood of exposure to ARGs.
d. Wastewater treatment plants (WWTPs)
Five studies were found that investigated the air around WWTPs, spanning urban and suburban areas and three different countries (Korea, China, and the US). Certain units in WWTPs are more likely to generate bioaerosols,77 especially aeration basins and sedimentation tanks, so it is not surprising that WWTPs are a source of ARG emissions to the atmosphere. A study employing PCR in Beijing illustrated that the frequency of detection of ARGs were higher downwind of WWTPs than upwind and found high frequency of detection of sul2 and intI1,78 The emission of aerosols containing ARGs from aeration of a wastewater and sludge mixture in the laboratory was confirmed by metagenomics.79 Of the 55 ARG sub-types belonging to 32 ARG types detected in the aerosols, BacA, which confers resistance to bacitracin, was the most abundant ARG detected, followed by tet (tetM, tetO, tetP, tetQ, tetS, tetT, tetW) and unspecified sulfonamide resistance genes.79 Most of the ARGs found in the aerosols in this study were confirmed to be from the wastewater and sludge mixture, based on comparison of the metagenomic analyses of the aerosols and mixture.79 Another study utilizing metagenomic analysis in the field in South Korea similarly found high abundances of sul (sul1, sul2) and tet (tetA, tetG, tetQ, tetW) in the air, along with MLS (ermB, ermG, ermF).80 The total ARG relative abundances in gene copies per 16S rRNA gene in air found in this study were comparable to levels found in activated sludge and dewatered sludge, while the diversity of ARGs in the aerosol samples was lower. This study also only detected 19 ARG types, 13 fewer than the previous study employing metagenomics.79 Again, it is difficult to discern if this is due to true differences between the settings (a WWTP located in South Korea versus a laboratory model of aerated wastewater from a WWTP located in Beijing), or if they are due to differences in metagenomic sequencing or analysis.80One study examining 84 ARG sub-types by qPCR found high similarity of ARGs (68% similar) between downwind aerosol samples and sludge samples at a WWTP in South Carolina, further confirming the aerosolization of ARGs from sludge.81 Also, the types and relative abundance of ARGs were higher in downwind samples than upwind ones, illustrating a clear effect of WWTPs. A further conclusion from these observations was that ARGs can potentially travel many miles away from the WWTP, depending on wind speed, and can also deposit onto surrounding land and water.81 A wide range of ARGs (17 out of 84 ARGs tested by qPCR) was found in the downwind samples, with blaTEM being the most abundant. Other abundant ARGs included ermB, ermC, tetA, tetB, aadA1, qnrB5, mefA, and msrA. A more recent study utilizing metagenomics compared PM2.5 samples from coastal, urban, and WWTP environments and determined that the relative abundance of ARGs were elevated at WWTPs, and comparable to the sewage and water samples from the plant. The results provided further evidence for the effect of WWTPs and human activity on the airborne resistome. The four most abundant types of ARGs in this study at Stonecutters Island Sewage Treatment Works in Hong Kong were multidrug, peptide, MLS, and aminoglycoside, followed by tetracycline, lactam, rifamycin, and glycopeptide. Some ARG sub-types, including tetA, mtrA, mexF, cpxR, bacA, and rosB, were found in much higher abundance in air samples than in the water and sewage samples, indicating that sources other than the WWTP affected the resistome.82
e. Landfills
Only one study that involved ARG detection at a landfill site was identified. Antibiotics and other selective pressures imposed by the extreme conditions (e.g., low pH, heavy metals) in landfills, could potentially contribute to enriched abundance of ARGs in the surrounding environment, including the atmosphere. High levels of ARGs, many associated with MGEs, have been found in landfills in cities.26,45 Air was sampled upwind, downwind, and directly above a landfill site in Changzhou, China. IntI1 was prevalent in all media, including air, leachate, and solid waste, along with blaTEM, followed by sul1, tetQ, and ermB.83 The estimated daily intakes of ermB, sul1, and tetA through inhalation of PM were comparable to those through drinking water, and the intake by inhalation was higher for blaTEM. These levels were lower compared to those via ingestion of vegetables (up to 5 orders of magnitude for sul1).f. Indoor environments
Five studies have sampled indoor air for ARGs in three different countries (Australia, China, US). Two studies at universities in Tianjin, China indicated that indoor aerosols was higher in relative abundances compared to outdoor samples of common ARG types and sub-types (β-lactams, multidrug, and tet) and MGEs, emphasizing the potential for higher inhalation exposure to these ARGs in such settings.35,84 In Australia, PCR conducted on dust from vacuum cleaner bags and airborne particles collected during vacuuming revealed high frequencies of tetA/C and ermB, low frequencies of vanD and tetG, and no detection of ribosomal protection protein (tetM, tetO, tetP, tetQ, tetS, tetT, tetW), erythromycin (ermA, ermB, ermF), or vatD ARGs.85 In contrast, tetW and tetX were detected by qPCR in indoor environments in Colorado in another study,86 while tetM was detected in high abundance, and tetC in low abundance, in the air in two laboratories.87 Relative abundances of the genes detected indoors in the Colorado study were much lower than those found in nearby rural environments.In a study at a wet poultry market in China (live poultry, pork, live fish, and vegetables), the highest concentrations of ARGs (tetG, tetC, sul2, and ermC) were observed in the live poultry area (indoor) and second highest in the indoor market.88 Concentrations in outdoor air up to 400 m away were measurable but much lower. It is not known whether the ARGs observed in outdoor air originated from the market or another source. There is, however, the possibility that ARG levels this far away could be influenced by sources other than the market, presenting a need for consideration of wind direction when interpreting these results.
A study of the resistomes in kindergartens in Hong Kong using qPCR documented various ARGs in indoor air, the surrounding soil, and nearby farm soil samples.89Sul1 was abundant in dust samples, which represents material that settled from the air, and urine samples. Within the kindergarten environment, intI1 co-occurred with multiple ARGs89 while plasmids co-occurred with multi-drug ARGs. Notable abundances of blaTEM and ermF might be related to high rates of use of β-lactam and macrolide antibiotics in the area, according to the authors. Possible sources of ARGs found in the kindergarten environment include nearby vegetation and soil, pollution from the surrounding community, as well as exhaled aerosols containing ARGs.70,88
In a study comparing ARGs observed in air conditioning systems, hospitals and farms were associated with the highest diversity of ARGs, followed by residences in cities and villages, respectively. The most abundant ARG types were multidrug, blaTEM, aminoglycoside, tetracycline, and MLSB across all four environments. City samples was the lowest in ARG abundance by a significant margin. Although the abundances of ARGs in air conditioning units were up to five orders of magnitude lower than those in soil, wastewater, and sludge, the units could still be a source of inhalation exposure to humans.74
6. Season and weather
Seasonal differences in atmospheric ARGs appear to be minor in urban areas and more pronounced in rural areas. One study comparing urban, rural, and industrial sites found that relative abundance of a few ARGs was higher in hot urban areas (Beijing summer) than in suburban and rural areas, while rural areas had higher abundances of ARGs during the winter. ARG abundances were comparable during the spring and fall.45 Suburban areas consistently had lower ARG levels than both urban and rural areas for the entire year. A recent study in a smaller urban area (Handan, China) reported conflicting trends, with ARG concentrations increasing in the winter as compared to summer, perhaps due to the winter haze events in the area. They also found that samples collected in winter were heavily influenced by local and inland sources while summer samples were influenced by long-range and remote sources, including the marine environment. Thus, along with urbanization, seasonal ARG trends may also be influenced by wind patterns.38Urban sites in these studies also demonstrated greater consistency in the types and abundance of ARGs throughout the seasons, demonstrating that with greater urbanization, relationships between temperature and climate and MGE and ARG abundance diminished.45 This pattern may be due to seasonally independent sources of airborne microbes in urban and industrial areas, like human activity, and lesser influence of seasonal sources such as vegetation. A metagenomics study of hospitals and urban and suburban environments in China determined that ARG abundance in the urban environment and hospital was highest in the summer, and suburban ARG abundance was highest in the winter. The hospital had the highest total abundance of ARGs in general, especially during the summer and autumn.27 Confirming this finding, a recent study showed that 11 different types of ARGs (multidrug, MLS, aminoglycoside, tetracycline, rifamycin, β-lactam, bacitracin, sulfonamide, glycopeptide, peptide, and fluoroquinolone) all increased in relative abundance from winter to summer. Interestingly, they also found that the difference between the relative abundance of ARGs in the urban and hospital settings was much more pronounced in the summertime, especially for MLS and β-lactam.76 Another hospital study in China showed that the diversity of ARGs was higher during the winter, but the abundances were comparable. In this study, samples were collected from the air filtration system in the hospital and could have been more affected by seasonal changes in operation rather than by the weather itself.68 Most other hospital studies utilized high volume samplers placed in different rooms in the hospitals, often for a period of 24 hours.22,27,69,71 The windows were open throughout the winter, and sampling periods lasted a full month. This could indicate that more ARGs came in from the surrounding urban environment during the winter, affecting the diversity more than in other studies in which sampling periods were shorter.
Rural sites showed a reduction in total bacteria but enrichment of ARGs in winter.21,65 Given that the amount and kind of vegetation changes substantially by season, this result is consistent with the role of vegetation as a main source of bacteria in the air. Further seasonal disparities were attributed to different sources, such as those of natural (soil and plants) and anthropogenic origin (livestock, composting, and WWTPs).
Humidity and precipitation may also affect the abundance of ARGs in urban areas, as evidenced by one study in Beijing where ARG concentration and loading in PM2.5 was highest the day before a heavy rain event, compared to the day after the rain event, and lowest during the rain event.30 The observed pattern could have been due to removal of PM2.5 by wet deposition (i.e., scavenging of particles from the air by rain droplets). A study conducted in Handan, China and published after our literature search was conducted, showed that rainfall can carry and transfer ARGs to soil. ARG concentrations in PM2.5 increased during rainfall and decreased afterward. ARG concentrations were higher in soil treated with rainwater than in soil treated with sterile water. intI1, cIntI1, tnpA-04, tetA, and sul2. ermB and OXA-10 were only present in air samples during rainfall, indicating the rainwater was the main mode of transport for these ARGs.90
7. Transport of ARGs in the air
While most of the studies reviewed above demonstrated short-range transport of ARGs (i.e., several meters to hundreds of meters), the aerosols sampled, and therefore the ARGs within them, may also be subject to long-range transport. We posit that there is greater potential for ARGs to spread long distances in the environment through the atmosphere compared to the hydrosphere, lithosphere, and biosphere. Long-distance atmospheric transport is responsible for distributing chemical pollutants, mineral dust, and microbes around the globe, including to remote regions.91–95 Similarly, ARGs may be carried long distances through the atmosphere and then deposit on land or water. This process introduces novel ARGs to new sites and shapes the existing resistome there.It is challenging to provide direct evidence to confirm the atmospheric transport of ARGs due to the difficulties in collecting sufficient samples and establishing effective approaches for source tracking. One study found that ARGs of apparent anthropogenic origin—clinical (aac(3) and blaIMP) and agricultural (strA and tetW)—were present in remote glaciers, possibly due to airborne bacteria and migrating birds.96 Another study found evidence that snowfall could contribute to the spread of ARGs from different point sources, especially when air pollution levels were high.37 A study published after our search of the literature found sul1, tetO, and intI1 in up to 60% of precipitation samples at a mountain site in Spain.97 The location of the sampling site above the atmospheric boundary layer means that these ARGs were present in the free troposphere and thus subject to long-range transport by global circulation patterns. Two studies in Israel found that dust storms originating in distant countries had distinct bacterial communities that could harbor pathogens. These dust storms, however, were associated with lower relative abundance of ARGs. This lower abundance could be related to the source of the dust storms, since antibiotic use in deserts would be rare in comparison to cities, or the possible unreliability of relative abundance as a measurement for events like dust storms, with large changes in bacterial load. Additionally, these studies considered only a few ARGs (intI1, sul1, and qnrS), so further analysis utilizing more primers or metagenomics is needed to better understand these trends.39,40
Knowledge about the atmospheric transport of bacteria98 could yield insight into patterns of atmospheric ARG transport, as it is likely that the bacteria are accompanied by ARGs. The correlation between ARGs in air and bacterial community composition has been investigated near typical bioaerosol emission sources.47 Results suggested that certain bacterial strains were strongly correlated with several ARGs in the air samples. In other studies, researchers compared the amount of intracellular and extracellular ARGs in water and sludge samples and found that the majority of ARGs existed in intracellular DNA.99–101 Even if the opposite were true for ARGs in PM2.5,102 there is likely to be a relationship between presence of ARGs and presence of bacteria. Direct evidence from future studies is needed to determine the magnitude of atmospheric transport of ARGs compared to other routes of dissemination in the environment.
There are several factors that can impact the extent of ARG transport in the air. First, the size of the associated particle determines its settling velocity and lifetime in the atmosphere. Smaller particles settle slowly and thus are more likely to travel farther in the atmosphere than larger particles, which settle more rapidly. Secondly, meteorological parameters, including wind speed, wind direction, and the height of the mixing layer, can affect the range of transport. Additionally, the altitude reached by these particles is a determining factor in long-range transport. Those that traverse the boundary layer and reach the free atmosphere can be readily transported continental-scale distances. Precipitation enhances removal from the atmosphere and deposition on the Earth's surface. Lastly, environmental factors that accelerate decay of bacteria and their genomic material may affect the spread of ARGs. The persistence of airborne bacteria is a function of temperature, humidity, and radiation.103–105 The inactivation of airborne bacteria may lead to damage to bacterial structures and induce the release of bacteria DNA, which will be more susceptible to degradation and deactivation via UV light exposure and other mechanisms when in extracellular form. The difference between the susceptibility of intracellular and extracellular ARGs has been previously reported in water bodies.106 Similar studies are needed for ARGs in air to advance understanding of their persistence in the atmosphere.
8. Closing thoughts and path forward
It is apparent that the atmosphere is an important and understudied reservoir of ARGs. There is a need for research aimed at building a quantitative, mechanistic understanding of ARG transport in the atmosphere. Additional studies are needed to delineate the impacts of inhaled ARGs on human health and how they might interact with the human microbiome.107 The urgency of the AMR problem and paucity of knowledge about ARGs in the atmosphere raise numerous questions, summarized in Fig. 1. Among them: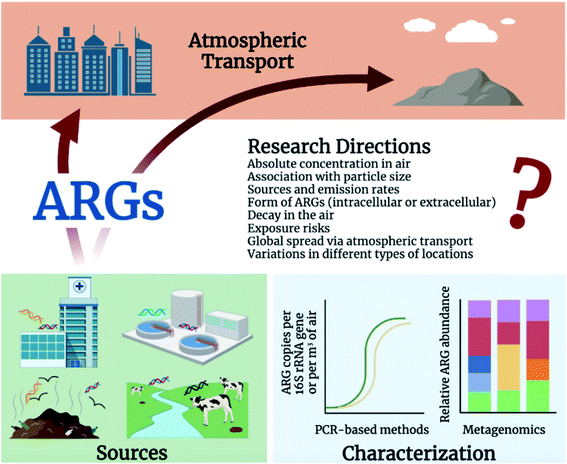 | ||
| Fig. 1 Overview of the sources, characterization methods, atmospheric transport, and future research questions of airborne ARGs. | ||
- What are the actual concentrations of ARGs in air, in terms of number per cubic meter of air, rather than just relative abundances?
- Is the abundance of ARGs correlated with the total number of bacteria in air?
- Are atmospheric ARGs primarily intracellular and confined to a few major taxa?
- How do ARG types and abundances vary among different locations, and how do they compare to profiles in potential sources, such as soil and wastewater?
- What are the major sources of ARGs to the atmosphere and at what rate do these sources emit ARGs into the atmosphere?
- Are ARGs in the air mobile and what conditions generally enhance their mobility?
- To what extent do airborne sources contribute to the global spread of antimicrobial resistance, e.g., new forms of resistance appearing in clinics in different parts of the world?
- What is the size range of particles with which airborne ARGs tend to be associated, and how far can they be transported in the atmosphere?
- Are ARGs in the atmosphere subject to decay, and if so, by what mechanisms?
- What are the exposure risks related to airborne ARGs and could epidemiological studies provide insight into this question?
To date, most studies have reported ARGs in terms of relative abundance, which is helpful for identifying “hot spots” where there is an abundant source and/or selective pressure enriching ARGs. However, concentration data (ARGs per volume of air) are needed to inform emission and exposure rates; only 19 of the studies reviewed here reported data in concentration-based units. It would be most useful to report an emission rate in gene copies per unit time, along with concentrations of ARGs per volume of air, ideally resolved by particle size. This information would enable others to predict downwind concentrations, deposition fluxes to soil and water, and exposure. Concentrations can be calculated from both qPCR and metagenomics, and both techniques have value, depending on the question being answered.
There is also a need to standardize analytical approaches and reporting of data to support reproducibility and comparability across studies. Sampling methods should be informed by prior reviews of different sampler types and filters.108–110 Collection and recovery efficiencies can vary widely by filter type. For example, one study reported that different filter materials had recovery efficiencies for the 16S rRNA gene ranging from 1% for polyvinylidene fluoride (PVDF) to 113% for polytetrafluoroethylene (PTFE), and we expect these values to apply to ARGs also.109 Recently, standardized methods for qPCR of ARGs has been proposed111 and similar efforts are needed for metagenomics.
Through interdisciplinary approaches that leverage state-of-the-science methods in air quality engineering, microbiology, and aerosol science, we can achieve a fundamental understanding about the sources, transport, and fate of ARGs in the atmosphere. Advancing knowledge about ARGs in air will provide a more complete picture of ARGs in the total environment, encompassing the hydrosphere, biosphere, lithosphere, and atmosphere, and will inform development of policies and guidelines to more effectively limit the spread of antimicrobial resistance.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Science Foundation (CBET-1936319, OISE-1545756, NRT-2125798, NNCI-2025151, and OAC-2004751).References
- CDC, Antibiotic resistance threats in the United States, US Department of Health and Human Services, 2019, pp. 1–139, DOI:10.15620/cdc:82532, https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf.
- P. Dadgostar, Antimicrobial resistance: implications and costs, Infect. Drug Resist., 2019, 12, 3903–3910 CrossRef CAS PubMed.
- Q. Chang, W. Wang, G. Regev-Yochay, M. Lipsitch and W. P. Hanage, Antibiotics in agriculture and the risk to human health: how worried should we be?, Evol. Appl., 2015, 8, 240–247 CrossRef PubMed.
- A. C. Singer, H. Shaw, V. Rhodes and A. Hart, Review of antimicrobial resistance in the environment and its relevance to environmental regulators, Front. Microbiol., 2016, 7, 01728 CrossRef PubMed.
- S. Zhang, G. Yang, S. Hou, T. Zhang, Z. Li and F. Liang, Distribution of ARGs and MGEs among glacial soil, permafrost, and sediment using metagenomic analysis, Environ. Pollut., 2018, 234, 339–346 CrossRef CAS PubMed.
- P. M. C. Huijbers, H. Blaak, M. C. M. De Jong, E. A. M. Graat, C. M. J. E. Vandenbroucke-Grauls and A. M. De Roda Husman, Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review, Environ. Sci. Technol., 2015, 49, 11993–12004 CrossRef CAS PubMed.
- V. A. Ogawa, C. M. Shah, J. M. Hughes and L. J. King, Prioritizing a One Health Approach in the Immediate Fight against Antimicrobial Resistance, Ecohealth, 2019, 16, 410–413 CrossRef PubMed.
- Y. G. Zhu, T. A. Johnson, J. Q. Su, M. Qiao, G. X. Guo, R. D. Stedtfeld, S. A. Hashsham and J. M. Tiedje, Diverse and abundant antibiotic resistance genes in Chinese swine farms, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 3435–3440 CrossRef CAS PubMed.
- P. J. Vikesland, A. Pruden, P. J. J. Alvarez, D. Aga, H. Bürgmann, X. D. Li, C. M. Manaia, I. Nambi, K. Wigginton, T. Zhang and Y. G. Zhu, Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance, Environ. Sci. Technol., 2017, 51, 13061–13069 CrossRef CAS PubMed.
- E. Castro-Sánchez, L. S. P. Moore, F. Husson and A. H. Holmes, What are the factors driving antimicrobial resistance? Perspectives from a public event in London, England, BMC Infect. Dis., 2016, 16, 465 CrossRef PubMed.
- G. Mainelis, Bioaerosol sampling: classical approaches, advances, and perspectives, Aerosol Sci. Technol., 2020, 54, 496–519 CrossRef CAS.
- C. H. Wang, B. T. Chen, B. C. Han, A. C. Y. Liu, P. C. Hung, C. Y. Chen and H. J. Chao, Field evaluation of personal sampling methods for multiple bioaerosols, PLoS One, 2015, 10, e0120308 CrossRef PubMed.
- E. Kabir, A. Azzouz, N. Raza, S. K. Bhardwaj, K. H. Kim, M. Tabatabaei and D. Kukkar, Recent Advances in Monitoring, Sampling, and Sensing Techniques for Bioaerosols in the Atmosphere, ACS Sens., 2020, 5, 1254–1267 CrossRef CAS PubMed.
- E. Franchitti, E. Pascale, E. Fea, E. Anedda and D. Traversi, Methods for bioaerosol characterization: limits and perspectives for human health risk assessment in organic waste treatment, Atmosphere, 2020, 11, 452 CrossRef CAS.
- S. Fuzzi, U. Baltensperger, K. Carslaw, S. Decesari, H. Denier Van Der Gon, M. C. Facchini, D. Fowler, I. Koren, B. Langford, U. Lohmann, E. Nemitz, S. Pandis, I. Riipinen, Y. Rudich, M. Schaap, J. G. Slowik, D. V. Spracklen, E. Vignati, M. Wild, M. Williams and S. Gilardoni, Particulate matter, air quality and climate: lessons learned and future needs, Atmos. Chem. Phys., 2015, 15, 8217–8299 CrossRef CAS.
- R. S. Hendriksen, P. Munk, P. Njage, B. van Bunnik, L. McNally, O. Lukjancenko, T. Röder, D. Nieuwenhuijse, S. K. Pedersen, J. Kjeldgaard, R. S. Kaas, P. T. L. C. Clausen, J. K. Vogt, P. Leekitcharoenphon, M. G. M. van de Schans, T. Zuidema, A. M. de Roda Husman, S. Rasmussen, B. Petersen, A. Bego, C. Rees, S. Cassar, K. Coventry, P. Collignon, F. Allerberger, T. O. Rahube, G. Oliveira, I. Ivanov, Y. Vuthy, T. Sopheak, C. K. Yost, C. Ke, H. Zheng, L. Baisheng, X. Jiao, P. Donado-Godoy, K. J. Coulibaly, M. Jergović, J. Hrenovic, R. Karpíšková, J. E. Villacis, M. Legesse, T. Eguale, A. Heikinheimo, L. Malania, A. Nitsche, A. Brinkmann, C. K. S. Saba, B. Kocsis, N. Solymosi, T. R. Thorsteinsdottir, A. M. Hatha, M. Alebouyeh, D. Morris, M. Cormican, L. O'Connor, J. Moran-Gilad, P. Alba, A. Battisti, Z. Shakenova, C. Kiiyukia, E. Ng’eno, L. Raka, J. Avsejenko, A. Bērziņš, V. Bartkevics, C. Penny, H. Rajandas, S. Parimannan, M. V. Haber, P. Pal, G. J. Jeunen, N. Gemmell, K. Fashae, R. Holmstad, R. Hasan, S. Shakoor, M. L. Z. Rojas, D. Wasyl, G. Bosevska, M. Kochubovski, C. Radu, A. Gassama, V. Radosavljevic, S. Wuertz, R. Zuniga-Montanez, M. Y. F. Tay, D. Gavačová, K. Pastuchova, P. Truska, M. Trkov, K. Esterhuyse, K. Keddy, M. Cerdà-Cuéllar, S. Pathirage, L. Norrgren, S. Örn, D. G. J. Larsson, T. Van der Heijden, H. H. Kumburu, B. Sanneh, P. Bidjada, B. M. Njanpop-Lafourcade, S. C. Nikiema-Pessinaba, B. Levent, J. S. Meschke, N. K. Beck, C. D. Van, N. Do Phuc, D. M. N. Tran, G. Kwenda, D. adjim Tabo, A. L. Wester, S. Cuadros-Orellana, C. Amid, G. Cochrane, T. Sicheritz-Ponten, H. Schmitt, J. R. M. Alvarez, A. Aidara-Kane, S. J. Pamp, O. Lund, T. Hald, M. Woolhouse, M. P. Koopmans, H. Vigre, T. N. Petersen and F. M. Aarestrup, Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage, Nat. Commun., 2019, 10, 1124 CrossRef PubMed.
- K. Pärnänen, A. Karkman, M. Tamminen, C. Lyra, J. Hultman, L. Paulin and M. Virta, Evaluating the mobility potential of antibiotic resistance genes in environmental resistomes without metagenomics, Sci. Rep., 2016, 6, 35790 CrossRef PubMed.
- C. Lal Gupta, R. Kumar Tiwari and E. Cytryn, Platforms for elucidating antibiotic resistance in single genomes and complex metagenomes, Environ. Int., 2020, 138, 105667 CrossRef PubMed.
- N. R. Noyes, M. E. Weinroth, J. K. Parker, C. J. Dean, S. M. Lakin, R. A. Raymond, P. Rovira, E. Doster, Z. Abdo, J. N. Martin, K. L. Jones, J. Ruiz, C. A. Boucher, K. E. Belk and P. S. Morley, Enrichment allows identification of diverse, rare elements in metagenomic resistome-virulome sequencing, Microbiome, 2017, 5, 142 CrossRef PubMed.
- S. C. Taylor, G. Laperriere and H. Germain, Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data, Sci. Rep., 2017, 7, 2409 CrossRef PubMed.
- J. Xie, L. Jin, X. Luo, Z. Zhao and X. Li, Seasonal Disparities in Airborne Bacteria and Associated Antibiotic Resistance Genes in PM2.5 between Urban and Rural Sites, Environ. Sci. Technol. Lett., 2018, 5, 74–79 CrossRef CAS.
- Y. Wang, C. Wang and L. Song, Distribution of antibiotic resistance genes and bacteria from six atmospheric environments: exposure risk to human, Sci. Total Environ., 2019, 694, 133750 CrossRef CAS PubMed.
- A. D. McEachran, B. R. Blackwell, J. D. Hanson, K. J. Wooten, G. D. Mayer, S. B. Cox and P. N. Smith, Antibiotics, bacteria, and antibiotic resistance genes: aerial transport from cattle feed yards via particulate matter, Environ. Health Perspect., 2015, 123, 337–343 CrossRef PubMed.
- K. Fogler, G. K. P. Guron, L. L. Wind, I. M. Keenum, W. C. Hession, L. A. Krometis, L. K. Strawn, A. Pruden and M. A. Ponder, Microbiota and Antibiotic Resistome of Lettuce Leaves and Radishes Grown in Soils Receiving Manure-Based Amendments Derived from Antibiotic-Treated Cows, Frontiers in Sustainable Food Systems, 2019, 3, e00022 CrossRef.
- C. Pal, J. Bengtsson-Palme, E. Kristiansson and D. G. J. Larsson, The structure and diversity of human, animal and environmental resistomes, Microbiome, 2016, 4, 54 CrossRef PubMed.
- J. Li, J. Cao, Y. G. Zhu, Q. L. Chen, F. Shen, Y. Wu, S. Xu, H. Fan, G. Da, R. J. Huang, J. Wang, A. L. De Jesus, L. Morawska, C. K. Chan, J. Peccia and M. Yao, Global Survey of Antibiotic Resistance Genes in Air, Environ. Sci. Technol., 2018, 52, 10975–10984 CrossRef CAS PubMed.
- P. He, Y. Wu, W. Huang, X. Wu, J. Lv, P. Liu, L. Bu, Z. Bai, S. Chen, W. Feng and Z. Yang, Characteristics of and variation in airborne ARGs among urban hospitals and adjacent urban and suburban communities: a metagenomic approach, Environ. Int., 2020, 139, 105625 CrossRef CAS PubMed.
- C. M. Echeverria-Palencia, V. Thulsiraj, N. Tran, C. A. Ericksen, I. Melendez, M. G. Sanchez, D. Walpert, T. Yuan, E. Ficara, N. Senthilkumar, F. Sun, R. Li, M. Hernandez-Cira, D. Gamboa, H. Haro, S. E. Paulson, Y. Zhu and J. A. Jay, Disparate Antibiotic Resistance Gene Quantities Revealed across 4 Major Cities in California: A Survey in Drinking Water, Air, and Soil at 24 Public Parks, ACS Omega, 2017, 2, 2255–2263 CrossRef CAS PubMed.
- J. Hu, F. Zhao, X. X. Zhang, K. Li, C. Li, L. Ye and M. Li, Metagenomic profiling of ARGs in airborne particulate matters during a severe smog event, Sci. Total Environ., 2018, 615, 1332–1340 CrossRef CAS PubMed.
- W. Ouyang, B. Gao, H. Cheng, L. Zhang, Y. Wang, C. Lin and J. Chen, Airborne bacterial communities and antibiotic resistance gene dynamics in PM2.5 during rainfall, Environ. Int., 2020, 134, 105318 CrossRef CAS PubMed.
- X. Sun, D. Li, B. Li, S. Sun, S. D. Yabo, J. Geng, L. Ma and H. Qi, Exploring the disparity of inhalable bacterial communities and antibiotic resistance genes between hazy days and non-hazy days in a cold megacity in northeast China, J. Hazard. Mater., 2020, 398, 122984 CrossRef CAS PubMed.
- N. Lang-Yona, F. Öztürk, D. Gat, M. Aktürk, E. Dikmen, P. Zarmpas, M. Tsagkaraki, N. Mihalopoulos, A. Birgül, P. B. Kurt-Karakuş and Y. Rudich, Links between airborne microbiome, meteorology, and chemical composition in northwestern Turkey, Sci. Total Environ., 2020, 725, 138227 CrossRef CAS PubMed.
- O. Ginn, D. Nichols, L. Rocha-Melogno, A. Bivins, D. Berendes, F. Soria, M. Andrade, M. A. Deshusses, M. Bergin and J. Brown, Antimicrobial resistance genes are enriched in aerosols near impacted urban surface waters in La Paz, Bolivia, Environ. Res., 2021, 194, 110730 CrossRef CAS PubMed.
- N. Qin, P. Liang, C. Wu, G. Wang, Q. Xu, X. Xiong, T. Wang, M. Zolfo, N. Segata, H. Qin, R. Knight, J. A. Gilbert and T. F. Zhu, Longitudinal survey of microbiome associated with particulate matter in a megacity, Genome Biol., 2020, 21, 55 CrossRef CAS PubMed.
- Y. Zhao, Z. Chen, J. Hou, D. Mao, H. Lin, Y. Xue and Y. Luo, Monitoring antibiotic resistomes and bacterial microbiomes in the aerosols from fine, hazy, and dusty weather in Tianjin, China using a developed high-volume tandem liquid impinging sampler, Sci. Total Environ., 2020, 731, 139242 CrossRef CAS PubMed.
- I. Gandolfi, A. Franzetti, V. Bertolini, E. Gaspari and G. Bestetti, Antibiotic resistance in bacteria associated with coarse atmospheric particulate matter in an urban area, J. Appl. Microbiol., 2011, 110, 1612–1620 CrossRef CAS PubMed.
- G. Zhu, X. Wang, T. Yang, J. Su, Y. Qin, S. Wang, M. Gillings, C. Wang, F. Ju, B. Lan, C. Liu, H. Li, X. E. Long, X. Wang, M. S. M. Jetten, Z. Wang and Y. G. Zhu, Air pollution could drive global dissemination of antibiotic resistance genes, ISME J., 2021, 15, 270–281 CrossRef CAS PubMed.
- Q. Wang, Z. Hou, L. Li, S. Guo, H. Liang, M. Li, H. Luo, L. Wang, Y. Luo and H. Ren, Seasonal disparities and source tracking of airborne antibiotic resistance genes in Handan, China, J. Hazard. Mater., 2021, 422, 126844 CrossRef PubMed.
- D. Gat, Y. Mazar, E. Cytryn and Y. Rudich, Origin-Dependent Variations in the Atmospheric Microbiome Community in Eastern Mediterranean Dust Storms, Environ. Sci. Technol., 2017, 51, 6709–6718 CrossRef CAS PubMed.
- Y. Mazar, E. Cytryn, Y. Erel and Y. Rudich, Effect of Dust Storms on the Atmospheric Microbiome in the Eastern Mediterranean, Environ. Sci. Technol., 2016, 50, 4194–4202 CrossRef CAS PubMed.
- T. Zhang, X. Li, M. Wang, H. Chen, Y. Yang, Q. lin Chen and M. Yao, Time-resolved spread of antibiotic resistance genes in highly polluted air, Environ. Int., 2019, 127, 333–339 CrossRef CAS PubMed.
- X. Yan, J. Ma, X. Chen, M. Lei, T. Li and Y. Han, Characteristics of airborne bacterial communities and antibiotic resistance genes under different air quality levels, Environ. Int., 2022, 161, 107127 CrossRef CAS PubMed.
- L. Wang, Z. Liu, Y. Sun, D. Ji and Y. Wang, Long-range transport and regional sources of PM2.5 in Beijing based on long-term observations from 2005 to 2010, Atmos. Res., 2015, 157, 37–48 CrossRef CAS.
- T. Zhang, X. Li, M. Wang, H. Chen and M. Yao, Time- and size-resolved bacterial aerosol dynamics in highly polluted air: new clues for haze formation mechanism: supplemental file, bioRxiv, 2019, 513093 Search PubMed.
- J. Xie, L. Jin, T. He, B. Chen, X. Luo, B. Feng, W. Huang, J. Li, P. Fu and X. Li, Bacteria and Antibiotic Resistance Genes (ARGs) in PM2.5 from China: Implications for Human Exposure, Environ. Sci. Technol., 2019, 53, 963–972 CrossRef CAS PubMed.
- Z. Liang, Y. Yu, Z. Ye, G. Li, W. Wang and T. An, Pollution profiles of antibiotic resistance genes associated with airborne opportunistic pathogens from typical area, Pearl River Estuary and their exposure risk to human, Environ. Int., 2020, 143, 105934 CrossRef CAS PubMed.
- Y. Yang, R. Zhou, B. Chen, T. Zhang, L. Hu and S. Zou, Characterization of airborne antibiotic resistance genes from typical bioaerosol emission sources in the urban environment using metagenomic approach, Chemosphere, 2018, 213, 463–471 CrossRef CAS PubMed.
- R. E. C. Luiken, L. Van Gompel, A. Bossers, P. Munk, P. Joosten, R. B. Hansen, B. E. Knudsen, S. García-Cobos, J. Dewulf, F. M. Aarestrup, J. A. Wagenaar, L. A. M. Smit, D. J. Mevius, D. J. J. Heederik and H. Schmitt, Farm dust resistomes and bacterial microbiomes in European poultry and pig farms, Environ. Int., 2020, 143, 105971 CrossRef PubMed.
- R. E. C. Luiken, L. Van Gompel, P. Munk, S. Sarrazin, P. Joosten, A. Dorado-García, R. Borup Hansen, B. E. Knudsen, A. Bossers, J. A. Wagenaar, F. M. Aarestrup, J. Dewulf, D. J. Mevius, D. J. J. Heederik, L. A. M. Smit, H. Schmitt, H. Graveland, A. Vanessen, B. Gonzalez-Zorn, G. Moyano, P. Sanders, C. Chauvin, J. David, A. Battisti, A. Caprioli, T. Blaha, K. Wadepohl, M. Brandt, T. Hald, A. S. Ribeiro Duarte, D. Wasyl, M. Skarzyńska, M. Zajac, H. Daskalov, H. W. Saatkamp and K. D. C. Stärk, Associations between antimicrobial use and the faecal resistome on broiler farms from nine European countries, J. Antimicrob. Chemother., 2019, 74, 2596–2604 CrossRef CAS PubMed.
- C. H. Thames, A. Pruden, R. E. James, P. P. Ray and K. F. Knowlton, Excretion of antibiotic resistance genes by dairy calves fed milk replacers with varying doses of antibiotics, Front. Microbiol., 2012, 3, 139 Search PubMed.
- L. Chambers, Y. Yang, H. Littier, P. Ray, T. Zhang, A. Pruden, M. Strickland and K. Knowlton, Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic administration of third generation cephalosporin, PLoS One, 2015, 10, e0133764 CrossRef PubMed.
- M. Gao, R. Jia, T. Qiu, M. Han and X. Wang, Size-related bacterial diversity and tetracycline resistance gene abundance in the air of concentrated poultry feeding operations, Environ. Pollut., 2017, 220, 1342–1348 CrossRef CAS PubMed.
- K. J. Wooten, G. D. Mayer and P. N. Smith, Persistence of elevated concentrations of PM, affiliated pharmaceuticals, and tetracycline resistance genes downwind of feedyards, Environ. Pollut., 2019, 247, 467–473 CrossRef CAS PubMed.
- L. Song, C. Wang, G. Jiang, J. Ma, Y. Li, H. Chen and J. Guo, Bioaerosol is an important transmission route of antibiotic resistance genes in pig farms, Environ. Int., 2021, 154, 106559 CrossRef CAS PubMed.
- M. M. T. De Rooij, G. Hoek, H. Schmitt, I. Janse, A. Swart, C. B. M. Maassen, M. Schalk, D. J. J. Heederik and I. M. Wouters, Insights into Livestock-Related Microbial Concentrations in Air at Residential Level in a Livestock Dense Area, Environ. Sci. Technol., 2019, 53, 7746–7758 CrossRef CAS PubMed.
- L. Van Gompel, W. Dohmen, R. E. C. Luiken, M. Bouwknegt, L. Heres, E. Van Heijnsbergen, B. G. M. Jongerius-Gortemaker, P. Scherpenisse, G. D. Greve, M. H. G. Tersteeg-Zijderveld, K. Wadepohl, A. S. Ribeiro Duarte, V. Muñoz-Gómez, J. Fischer, M. Skarżyńska, D. Wasyl, J. A. Wagenaar, B. A. P. Urlings, A. Dorado-García, I. M. Wouters, D. J. J. Heederik, H. Schmitt and L. A. M. Smit, Occupational exposure and carriage of antimicrobial resistance genes (tetW, ermB) in pig slaughterhouse workers, Ann. Work Exposures Health, 2020, 64, 125–137 CrossRef CAS PubMed.
- Y. Zhang, Y. Zheng, Z. Zhu, Y. Chen and H. Dong, Dispersion of antibiotic resistance genes (ARGs) from stored swine manure biogas digestate to the atmosphere, Sci. Total Environ., 2021, 761, 144108 CrossRef CAS PubMed.
- P. Y. Hong, X. Li, X. Yang, T. Shinkai, Y. Zhang, X. Wang and R. I. Mackie, Monitoring airborne biotic contaminants in the indoor environment of pig and poultry confinement buildings, Environ. Microbiol., 2012, 14, 1420–1431 CrossRef CAS PubMed.
- V. Létourneau, B. Nehmé, A. Mériaux, D. Massé, Y. Cormier and C. Duchaine, Human pathogens and tetracycline-resistant bacteria in bioaerosols of swine confinement buildings and in nasal flora of hog producers, Int. J. Hyg. Environ. Health, 2010, 213, 444–449 CrossRef PubMed.
- P. Kumari and H. L. Choi, Manure removal system influences the abundance and composition of airborne biotic contaminants in swine confinement buildings, Environ. Monit. Assess., 2015, 187, 537 CrossRef PubMed.
- P. Kumari and H. L. Choi, Seasonal variability in airborne biotic contaminants in swine confinement buildings, PLoS One, 2014, 9, e112897 CrossRef PubMed.
- N. A. Just, V. Létourneau, S. P. Kirychuk, B. Singh and C. Duchaine, Potentially pathogenic bacteria and antimicrobial resistance in bioaerosols from cage-housed and floor-housed poultry operations, Ann. Occup. Hyg., 2012, 56, 440–449 CAS.
- H. Yan, L. Zhang, Z. Guo, H. Zhang and J. Liu, Production phase affects the bioaerosol microbial composition and functional potential in swine confinement buildings, Animals, 2019, 9, 90 CrossRef PubMed.
- H. M. Sancheza, C. Echeverria, V. Thulsiraj, A. Zimmer-Faust, A. Flores, M. Laitz, G. Healy, S. Mahendra, S. E. Paulson, Y. Zhu and J. A. Jay, Antibiotic Resistance in Airborne Bacteria Near Conventional and Organic Beef Cattle Farms in California, USA, Water, Air, Soil Pollut., 2016, 227, 280 CrossRef.
- R. E. Luiken, D. J. Heederik, P. Scherpenisse, L. Van Gompel, E. van Heijnsbergen, G. D. Greve, B. G. Jongerius-Gortemaker, M. H. Tersteeg-Zijderveld, J. Fischer, K. Juraschek, M. Skarżyńska, M. Zając, D. Wasyl, J. A. Wagenaar, L. A. Smit, I. M. Wouters, D. J. Mevius and H. Schmitt, Determinants for antimicrobial resistance genes in farm dust on 333 poultry and pig farms in nine European countries, Environ. Res., 2022, 208, 112715 CrossRef CAS PubMed.
- J. Pilote, V. Létourneau, M. Girard and C. Duchaine, Quantification of airborne dust, endotoxins, human pathogens and antibiotic and metal resistance genes in Eastern Canadian swine confinement buildings, Aerobiologia, 2019, 35, 283–296 CrossRef.
- M. Gao, T. Qiu, Y. Sun and X. Wang, The abundance and diversity of antibiotic resistance genes in the atmospheric environment of composting plants, Environ. Int., 2018, 116, 229–238 CrossRef CAS PubMed.
- X. Li, Z. Wu, C. Dang, M. Zhang, B. Zhao, Z. Cheng, L. Chen, Z. Zhong, Y. Ye and Y. Xia, A metagenomic-based method to study hospital air dust resistome, Chem. Eng. J., 2021, 406, 126854 CrossRef CAS PubMed.
- X. L. Gao, M. F. Shao, Q. Wang, L. T. Wang, W. Y. Fang, F. Ouyang and J. Li, Airborne microbial communities in the atmospheric environment of urban hospitals in China, J. Hazard. Mater., 2018, 349, 10–17 CrossRef CAS PubMed.
- M. Kennedy, M. Y. Ramsheh, C. M. L. Williams, J. Auty, K. Haldar, M. Abdulwhhab, C. E. Brightling and M. R. Barer, Face mask sampling reveals antimicrobial resistance genes in exhaled aerosols from patients with chronic obstructive pulmonary disease and healthy volunteers, BMJ Open Respiratory Research, 2018, 5, e000321 CrossRef PubMed.
- Y. Gilbert, M. Veillette and C. Duchaine, Airborne bacteria and antibiotic resistance genes in hospital rooms, Aerobiologia, 2010, 26, 185–194 CrossRef.
- C. N. Drudge, S. Krajden, R. C. Summerbell and J. A. Scott, Detection of antibiotic resistance genes associated with methicillin-resistant Staphylococcus aureus (MRSA) and coagulase-negative staphylococci in hospital air filter dust by PCR, Aerobiologia, 2012, 28, 285–289 CrossRef.
- Z. C. Zhou, Y. Liu, Z. J. Lin, X. Y. Shuai, L. Zhu, L. Xu, L. X. Meng, Y. J. Sun and H. Chen, Spread of antibiotic resistance genes and microbiota in airborne particulate matter, dust, and human airways in the urban hospital, Environ. Int., 2021, 153, 106501 CrossRef CAS PubMed.
- Y. Li, H. Liao and H. Yao, Prevalence of antibiotic resistance genes in air-conditioning systems in hospitals, farms, and residences, Int. J. Environ. Res. Public Health, 2019, 16, 683 CrossRef CAS PubMed.
- R. Dhand and J. Li, Coughs and Sneezes: Their Role in Transmission of Respiratory Viral Infections, including SARS-CoV-2, Am. J. Respir. Crit. Care Med., 2020, 202, 651–659 CrossRef CAS PubMed.
- D. Wu, L. Jin, J. Xie, H. Liu, J. Zhao, D. Ye and X. dong Li, Inhalable antibiotic resistomes emitted from hospitals: metagenomic insights into bacterial hosts, clinical relevance, and environmental risks, Microbiome, 2022, 10, 19 CrossRef CAS PubMed.
- E. Mirskaya and I. E. Agranovski, Sources and mechanisms of bioaerosol generation in occupational environments, Crit. Rev. Microbiol., 2018, 44, 739–758 CrossRef CAS PubMed.
- J. Li, L. Zhou, X. Zhang, C. Xu, L. Dong and M. Yao, Bioaerosol emissions and detection of airborne antibiotic resistance genes from a wastewater treatment plant, Atmos. Environ., 2016, 124, 404–412 CrossRef CAS.
- Y. Han, T. Yang, T. Chen, L. Li and J. Liu, Characteristics of submicron aerosols produced during aeration in wastewater treatment, Sci. Total Environ., 2019, 696, 134019 CrossRef CAS PubMed.
- I. Han and K. Yoo, Metagenomic profiles of antibiotic resistance genes in activated sludge, dewatered sludge and bioaerosols, Water, 2020, 12, 1516 CrossRef CAS.
- A. Gaviria-Figueroa, E. C. Preisner, S. Hoque, C. E. Feigley and R. S. Norman, Emission and dispersal of antibiotic resistance genes through bioaerosols generated during the treatment of municipal sewage, Sci. Total Environ., 2019, 686, 402–412 CrossRef CAS PubMed.
- J. Xie, L. Jin, D. Wu, A. Pruden and X. Li, Inhalable Antibiotic Resistome from Wastewater Treatment Plants to Urban Areas: Bacterial Hosts, Dissemination Risks, and Source Contributions, Environ. Sci. Technol., 2022, 1c07023 CrossRef PubMed.
- L. Li, Q. Wang, W. Bi, J. Hou, Y. Xue, D. Mao, R. Das, Y. Luo and X. Li, Municipal Solid Waste Treatment System Increases Ambient Airborne Bacteria and Antibiotic Resistance Genes, Environ. Sci. Technol., 2020, 54, 3900–3908 CrossRef CAS PubMed.
- Y. Zhao, Q. Wang, Z. Chen, D. Mao and Y. Luo, Significant higher airborne antibiotic resistance genes and the associated inhalation risk in the indoor than the outdoor, Environ. Pollut., 2021, 268, 115620 CrossRef CAS PubMed.
- M. Veillette, L. D. Knibbs, A. Pelletier, R. Charlebois, P. B. Lecours, C. He, L. Morawska and C. Duchaine, Microbial contents of vacuum cleaner bag dust and emitted bioaerosols and their implications for human exposure indoors, Appl. Environ. Microbiol., 2013, 79, 6331–6336 CrossRef CAS PubMed.
- A. L. Ling, N. R. Pace, M. T. Hernandez and T. M. Lapara, Tetracycline resistance and class 1 integron genes associated with indoor and outdoor aerosols, Environ. Sci. Technol., 2013, 47, 4046–4052 CrossRef CAS PubMed.
- L. Song, C. Wang and Y. Wang, Optimized determination of airborne tetracycline resistance genes in laboratory atmosphere, Front. Environ. Sci. Eng., 2020, 14, 95 CrossRef CAS.
- X. L. Gao, M. F. Shao, Y. Luo, Y. F. Dong, F. Ouyang, W. Y. Dong and J. Li, Airborne bacterial contaminations in typical Chinese wet market with live poultry trade, Sci. Total Environ., 2016, 572, 681–687 CrossRef CAS PubMed.
- N. Li, Y. Chai, G. G. Ying, K. C. Jones and W. J. Deng, Airborne antibiotic resistance genes in Hong Kong kindergartens, Environ. Pollut., 2020, 260, 114009 CrossRef CAS PubMed.
- Q. Wang, S. Guo, Z. Hou, H. Lin, H. Liang, L. Wang, Y. Luo and H. Ren, Rainfall facilitates the transmission and proliferation of antibiotic resistance genes from ambient air to soil, Sci. Total Environ., 2021, 799, 149260 CrossRef CAS PubMed.
- S. Hatakeyama, A. Takami, F. Sakamaki, H. Mukai, N. Sugimoto, A. Shimizu and H. Bandow, Aerial measurement of air pollutants and aerosols during 20-22 March 2001 over the East China Sea, J. Geophys. Res.: Atmos., 2004, 109, 304 CrossRef.
- C. A. Kellogg and D. W. Griffin, Aerobiology and the global transport of desert dust, Trends Ecol. Evol., 2006, 21, 638–644 CrossRef PubMed.
- H. Maring, D. L. Savoie, M. A. Izaguirre, L. Custals and J. S. Reid, Mineral dust aerosol size distribution change during atmospheric transport, J. Geophys. Res.: Atmos., 2003, 108, 8592 CrossRef.
- D. W. Griffin, Atmospheric movement of microorganisms in clouds of desert dust and implications for human health, Clin. Microbiol. Rev., 2007, 20, 459–477 CrossRef PubMed.
- M. Scheringer, Long-range transport of organic chemicals in the environment, Environ. Toxicol. Chem., 2009, 28, 677–690 CrossRef CAS PubMed.
- T. Segawa, N. Takeuchi, A. Rivera, A. Yamada, Y. Yoshimura, G. Barcaza, K. Shinbori, H. Motoyama, S. Kohshima and K. Ushida, Distribution of antibiotic resistance genes in glacier environments, Environ. Microbiol. Rep., 2013, 5, 127–134 CrossRef CAS PubMed.
- J. Cáliz, J. Subirats, X. Triadó-Margarit, C. M. Borrego and E. O. Casamayor, Global dispersal and potential sources of antibiotic resistance genes in atmospheric remote depositions, Environ. Int., 2022, 160, 107077 CrossRef PubMed.
- C. E. Morris, C. Leyronas and P. C. Nicot, in Aerosol Science: Technology and Applications, 2014, vol. 9781119977, pp. 393–415 Search PubMed.
- P. Dong, H. Wang, T. Fang, Y. Wang and Q. Ye, Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG, Environ. Int., 2019, 125, 90–96 CrossRef CAS PubMed.
- Q. Sui, Y. Chen, D. Yu, T. Wang, Y. Hai, J. Zhang, M. Chen and Y. Wei, Fates of intracellular and extracellular antibiotic resistance genes and microbial community structures in typical swine wastewater treatment processes, Environ. Int., 2019, 133, 105183 CrossRef CAS PubMed.
- Y. Zhang, D. D. Snow, D. Parker, Z. Zhou and X. Li, Intracellular and extracellular antimicrobial resistance genes in the sludge of livestock waste management structures, Environ. Sci. Technol., 2013, 47, 10206–10213 CrossRef CAS PubMed.
- T. He, L. Jin, J. Xie, S. Yue, P. Fu and X. Li, Intracellular and Extracellular Antibiotic Resistance Genes in Airborne PM2.5 for Respiratory Exposure in Urban Areas, Environ. Sci. Technol. Lett., 2021, 8, 128–134 CrossRef CAS.
- K. Lin and L. C. Marr, Humidity-Dependent Decay of Viruses, but Not Bacteria, in Aerosols and Droplets Follows Disinfection Kinetics, Environ. Sci. Technol., 2020, 54, 1024–1032 CrossRef CAS PubMed.
- R. Ehrlich, S. Miller and R. L. Walker, Relationship between atmospheric temperature and survival of airborne bacteria, Appl. Microbiol., 1970, 19, 245–249 CrossRef CAS PubMed.
- C. Wang, S. Lu and Z. Zhang, Inactivation of airborne bacteria using different UV sources: performance modeling, energy utilization, and endotoxin degradation, Sci. Total Environ., 2019, 655, 787–795 CrossRef CAS PubMed.
- C. W. McKinney and A. Pruden, Ultraviolet disinfection of antibiotic resistant bacteria and their antibiotic resistance genes in water and wastewater, Environ. Sci. Technol., 2012, 46, 13393–13400 CrossRef CAS PubMed.
- L. Jin, J. Xie, T. He, D. Wu and X. Li, Airborne transmission as an integral environmental dimension of antimicrobial resistance through the “One Health” lens, Crit. Rev. Environ. Sci. Technol., 2021, 2006537 Search PubMed.
- J. Li, A. Leavey, Y. Wang, C. O'Neil, M. A. Wallace, C. A. D. Burnham, A. C. Boon, H. Babcock and P. Biswas, Comparing the performance of 3 bioaerosol samplers for influenza virus, J. Aerosol Sci., 2018, 115, 133–145 CrossRef CAS PubMed.
- S. Y. Jeong and T. G. Kim, Comparison of five membrane filters to collect bioaerosols for airborne microbiome analysis, J. Appl. Microbiol., 2021, 131, 780–790 CrossRef CAS PubMed.
- N. C. Burton, S. A. Grinshpun and T. Reponen, Physical collection efficiency of filter materials for bacteria and viruses, Ann. Occup. Hyg., 2007, 51, 143–151 CAS.
- I. Keenum, K. Liguori, J. Calarco, B. C. Davis, E. Milligan, V. J. Harwood and A. Pruden, A framework for standardized qPCR-targets and protocols for quantifying antibiotic resistance in surface water, recycled water and wastewater, Crit. Rev. Environ. Sci. Technol., 2021, 2024739 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2em00091a |
| This journal is © The Royal Society of Chemistry 2022 |
