Incorporating concentration-dependent sediment microbial activity into methylmercury production kinetics modeling†
Abstract
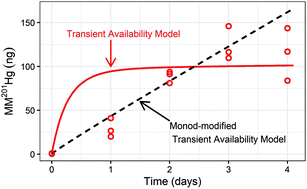
In anoxic environments, anaerobic microorganisms carrying the hgcAB gene cluster can mediate the transformation of inorganic mercury (Hg(II)) to monomethylmercury (MMHg). The kinetics of Hg(II) transformation to MMHg in periphyton from East Fork Poplar Creek (EFPC) in Oak Ridge, TN have previously been modeled using a transient availability model (TAM). The TAM for Hg(II) methylation combines methylation/demethylation kinetics with kinetic expressions for processes that decrease Hg(II) and MMHg availability for methylation and demethylation (multisite sorption of Hg(II) and MMHg, Hg(II) reduction/Hg(0) oxidation). In this study, the TAM is used for the first time to describe MMHg production in sediment. We assessed MMHg production in sediment microcosms using two different sediment types from EFPC: a relatively anoxic, carbon-rich sediment with higher microbial activity (higher CO2 production from sediment) and a relatively oxic, sandy, carbon-poor sediment with lower microbial activity (lower CO2 production from sediment). Based on 16s rRNA sequencing, the overall microbial community structure in the two sediments was retained during the incubations. However, the hgcA containing methanogenic Euryarchaeota communities differed between sediment types and their growth followed different trajectories over the course of incubations, potentially contributing to the distinct patterns of MMHg production observed. The general TAM paradigm performed well in describing MMHg production in the sediments. However, the MMHg production and ancillary data suggested the need to revise the model structure to incorporate terms for concentration-dependent microbial activity over the course of the incubations. We modified the TAM to include Monod-type kinetics for methylation and demethylation and observed an improved fit for the carbon-rich, microbially active sediment. Overall our work shows that the TAM can be applied to describe Hg(II) methylation in sediments and that including expressions accounting for concentration-dependent microbial activity can improve the accuracy of the model description of the data in some cases.
- This article is part of the themed collection: Biogeochemistry of the Trace Elements


 Please wait while we load your content...
Please wait while we load your content...