Open Access Article
Open Access ArticlePotent and selective anticancer activity of half-sandwich ruthenium and osmium complexes with modified curcuminoid ligands†
Noemi
Pagliaricci
 a,
Riccardo
Pettinari
a,
Riccardo
Pettinari
 *a,
Fabio
Marchetti
*a,
Fabio
Marchetti
 b,
Claudio
Pettinari
b,
Claudio
Pettinari
 a,
Loredana
Cappellacci
a,
Loredana
Cappellacci
 a,
Alessia
Tombesi
a,
Alessia
Tombesi
 a,
Massimiliano
Cuccioloni
a,
Massimiliano
Cuccioloni
 c,
Mouna
Hadiji
d and
Paul J.
Dyson
c,
Mouna
Hadiji
d and
Paul J.
Dyson
 *d
*d
aSchool of Pharmacy, University of Camerino, via Madonna delle Carceri (ChIP), 62032 Camerino MC, Italy. E-mail: riccardo.pettinari@unicam.it; Tel: +39 0737402338
bSchool of Science and Technology, University of Camerino, via Madonna delle Carceri (ChIP), 62032 Camerino MC, Italy
cSchool of Biosciences and Veterinary Medicine, University of Camerino, Via Gentile III Da Varano, 62032 Camerino MC, Italy
dInstitut des Sciences et Ingénierie Chimiques, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland
First published on 15th August 2022
Abstract
We have recently reported a series of half-sandwich ruthenium(II) complexes with curcuminoid ligands showing excellent cytotoxic activities (particularly ionic derivatives containing PTA (PTA = 1,3,5-triaza-7-phosphaadamantane). In the present study, new members of this family of compounds have been prepared with the objective to investigate the effect of a long hydrophobic chain obtained by replacing the OH-groups, present in curcumin and bisdemethoxycurcumin, with the palmitic acid ester. We report the synthesis of ruthenium(II) and osmium(II) p-cymene derivatives containing palmitic acid curcumin ester ligands ((1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(2-methoxy-4,1-phenylene)dipalmitate (p-curcH) and ((1E,3Z,6E)-3-hydroxy-5-oxohepta-1,3,6-triene-1,7-diyl)bis(4,1-phenylene)dipalmitate (p-bdcurcH). Complexes [M(II)(cym)(p-curc)/(p-bdcurc)(Cl)] 1–4 (M = Ru or Os) are neutral, whereas [M(II)(cym)(p-curc)/(p-bdcurc)(PTA)][SO3CF3] 5–8 are salts obtained when the chloride ligand is replaced by the PTA ligand. Stability studies performed on 1–8 in DMSO-PBS under physiological conditions (pH = 7.4) indicate that the complexes remain intact. The complexes exhibit potent and selective cytotoxic activity against an ovarian carcinoma cell line and its cisplatin-resistant form (A2780 and A2780cis), and non-cancerous human embryonic kidney (HEK293T) cells. To define the structure–activity relationships (SAR), the compounds have been compared with other Ru(II) and Os(II) complexes with curcuminoid ligands previously reported. SAR data reveal that the bisdemethoxycurcumin complexes are generally more active and selective than analogous curcumin-containing complexes.
Introduction
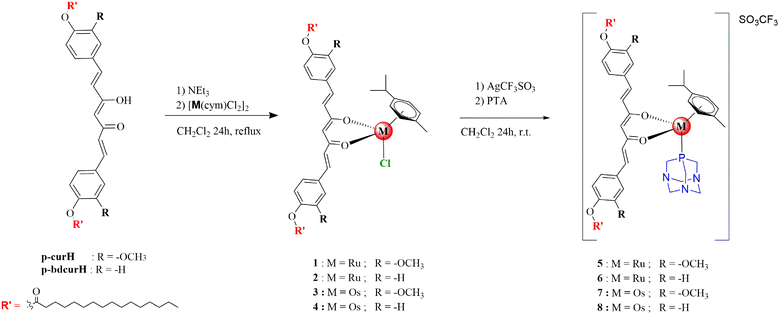
Diarylheptanoids are a relatively small class of secondary plant metabolites not directly involved in the growth and reproduction of the organism.1 However, these compounds mediate ecological interactions and produce a selective advantage for the plant by increasing its survival or fecundity. Diarylheptanoids are formed by two aromatic rings joined by a chain of seven carbon atoms, having various substituents and can be classified as cyclic or linear.2 Turmeric, the powdered rhizome of Curcuma longa, occurs in a mixture called “curcuminoids” that generally makes up approximately 1–6% of turmeric by dry weight.3 The curcuminoid extract, 1,7-bis[4-hydroxy-3-methoxyphenyl]-1,6-heptadiene-3,5-dione also called curcumin (curcH), makes up 60–70% by weight, while 1,7-bis(4-hydroxyphenyl)-1,6-heptadiene-3,5-dione, known as bisdemethoxycurcumin (bdcurcH), is the minor component (10–15%).4 Despite the interesting pharmacological properties, curcumin shows low absorption and poor bioavailability due to rapid metabolism, low water solubility and stability.5 Most recent strategies found to enhance the properties of curcumin involve modification of its structure or the application of drug delivery systems, such as nanoparticles, liposomes and micelles.6 Curcumin and bisdemethoxycurcumin have two phenolic groups which can act as potential sites for chemical variations and covalent linkage with biomolecules, such as folic acid, dipeptides, and fatty acids. Curcumin bioconjugates have been shown to enhance cellular uptake of curcumin and possess enhanced antibacterial activity against Gram-positive and Gram-negative bacteria.7,8 A curcumin bioconjugate with palmitoyl chloride (Fig. 1) was also shown to facilitate neuroprotection, preventing oligomeric Aβ40 insult8 in Alzheimer's disease. An approach to enhance bioavailability and water solubility involves the synthesis of curcumin metal complexes.9Group-8 metal complexes have been extensively explored as anticancer drugs.10–13 Research has mainly focused on iron and ruthenium complexes which are represented by several biologically active compounds evaluated in clinical trials such as NAMI-A and KP1019.14 On the other hand, studies on biologically active osmium complexes are comparatively rare,14–16 despite offering several advantageous features, such as a larger range of biologically accessible oxidation states, slower ligand exchange kinetics, a stronger π-back donation from lower oxidation states and strong spin–orbit coupling. Therefore, osmium complexes are considered interesting alternatives to ruthenium-based anticancer agents because of their stability under physiological conditions.17 Moreover, some important differences between osmium and ruthenium complexes have been observed in binding with biologically relevant targets.18
Herein, we report half-sandwich Ru(II) and Os(II) complexes with curcumin-like ligands (Scheme 1). The complexes have been fully characterized and their antiproliferative effects against several types of human cancer cell lines and non-cancerous cell lines evaluated.
Results and discussion
The bioconjugate curcumin ligands, p-curcH and p-bdcurcH, were synthesized as reported in Scheme 1, starting from the commercially available curcumin and bisdemethoxycurcumin following a modified literature procedure.7 In brief, curcumin or bisdemethoxycurcumin (1.0 mmol) were dissolved in anhydrous pyridine, DMAP was added and then palmitoyl chloride (2.2 mmol) was added to the chilled solution. The reaction mixture was stirred at room temperature overnight. Work up of the reaction mixture and purification by crystallization gave p-curcH and p-bdcurcH as yellow solids (75 and 85% yield, respectively). The p-curcH and p-bdcurcH ligands were characterized by 1H and 13C NMR spectroscopy and mass spectrometry. Complexes 1–4 were prepared from the reaction of the appropriate dimer, [Ru(cym)Cl2]2 or [Os(cym)Cl2]2, with p-curcH and p-bdcurcH and triethylamine in dichloromethane (Scheme 1). Complexes 1–4 are air-stable and soluble in acetone, acetonitrile, chlorinated solvents, DMF and DMSO. They are slightly soluble in methanol, ethanol, diethyl ether, and petroleum ether. The IR spectra of 1–4 contain the typical ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) vibrations of p-curcH and p-bdcurcH at lower wavenumbers than in the corresponding free ligands due to coordination through both the carbonyl arms to the metal. In the far-IR region, strong absorptions at 269 (for 1) and 270 cm−1 (for 2) may be assigned to ν(Ru–Cl) stretches and related strong absorptions at 273 (in 3) and 271 cm−1 (in 4), due to ν(Os–Cl) are observed. Electrospray ionization (ESI) mass spectra of 1–4 in positive ion mode, recorded in CH3CN, show the typical isotopic patterns expected and display peaks that correspond to [Ru(cym)(p-curc/p-bdcurc)]+ and [Os(cym)(p-curc/p-bdcurc)]+ respectively, arising from the dissociation of the chloride ligand. Conductivity measurements for 1–4 indicate a slight dissociation of the chloride in acetone at room temperature and 2 and 4 with the p-bdcurc ligand exhibit lower dissociation than 1 and 3 with the p-curc ligand. 1H- and 13C-NMR spectra were assigned based on the 1H–1H, and one-bond and long-range 1H–13C couplings, from {1H–1H}-COS
O) vibrations of p-curcH and p-bdcurcH at lower wavenumbers than in the corresponding free ligands due to coordination through both the carbonyl arms to the metal. In the far-IR region, strong absorptions at 269 (for 1) and 270 cm−1 (for 2) may be assigned to ν(Ru–Cl) stretches and related strong absorptions at 273 (in 3) and 271 cm−1 (in 4), due to ν(Os–Cl) are observed. Electrospray ionization (ESI) mass spectra of 1–4 in positive ion mode, recorded in CH3CN, show the typical isotopic patterns expected and display peaks that correspond to [Ru(cym)(p-curc/p-bdcurc)]+ and [Os(cym)(p-curc/p-bdcurc)]+ respectively, arising from the dissociation of the chloride ligand. Conductivity measurements for 1–4 indicate a slight dissociation of the chloride in acetone at room temperature and 2 and 4 with the p-bdcurc ligand exhibit lower dissociation than 1 and 3 with the p-curc ligand. 1H- and 13C-NMR spectra were assigned based on the 1H–1H, and one-bond and long-range 1H–13C couplings, from {1H–1H}-COS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Y, {1H–13C}-HSQC, and {1H–13C}-HMBC experiments (see ESI†). The chloride ligand in 1–4 was replaced by 1,3,5-triaza-7-phosphaadamantane (PTA), by treatment with a dichloromethane solution containing equimolar quantities of AgSO3CF3 and PTA to afford [Ru(cym)(p-curc/p-bdcurc)(PTA)][SO3CF3] (5) and (6), together with [Os(cym)(p-curc/p-bdcurc)(PTA)][SO3CF3] (7) and (8), as depicted in Scheme 1. The substitution of chloride by PTA and the formation of ionic compounds were confirmed by the absence of a ν(M–Cl) band in the IR spectra of 5–8. Moreover, a characteristic absorption pattern in the region 1000–1200 cm−1, indicative of a non-coordinated SO3CF3− anion, is observed.19
Y, {1H–13C}-HSQC, and {1H–13C}-HMBC experiments (see ESI†). The chloride ligand in 1–4 was replaced by 1,3,5-triaza-7-phosphaadamantane (PTA), by treatment with a dichloromethane solution containing equimolar quantities of AgSO3CF3 and PTA to afford [Ru(cym)(p-curc/p-bdcurc)(PTA)][SO3CF3] (5) and (6), together with [Os(cym)(p-curc/p-bdcurc)(PTA)][SO3CF3] (7) and (8), as depicted in Scheme 1. The substitution of chloride by PTA and the formation of ionic compounds were confirmed by the absence of a ν(M–Cl) band in the IR spectra of 5–8. Moreover, a characteristic absorption pattern in the region 1000–1200 cm−1, indicative of a non-coordinated SO3CF3− anion, is observed.19
The 1H-NMR spectra of 5–8 in [d6]DMSO contain only one set of resonances due to cationic [Ru(cym)(p-curc/p-bdcurc)(PTA)]+ and [Os(cym)(p-curc/p-bdcurc)(PTA)]+. The 31P-NMR resonances attributable to the PTA ligand are observed at the lower field compared to those of uncoordinated PTA, thus confirming coordination to the metal center. The conductance values in acetone confirm the existence of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 electrolyte species for 5–8 and are consistent with the conductance values of their neutral derivatives, with 6 and 8 having a lower dissociation than 5 and 7.
1 electrolyte species for 5–8 and are consistent with the conductance values of their neutral derivatives, with 6 and 8 having a lower dissociation than 5 and 7.
Stability studies
To investigate the stability of 5–8 a series of 31P-NMR spectra were recorded in DMSO-d6 solution over time. The δ values of the characteristic peaks for phosphorous in all the spectra remained unchanged over 5 days, indicating that the complexes are stable in DMSO. The 1H-NMR spectra of complexes 5–8 confirm their stability although a slight release of curcumin ligand is observed for complexes 5 and 6 after 5 days, demonstrating higher stability for the osmium complexes 7 and 8. The stability of 1 in DMSO solution was further confirmed using UV-vis spectroscopy. The spectrum remained unchanged for a period of 48 h, displaying an absorption band at about 400 nm (Fig. 2a). In order to investigate the stability profile under physiologically relevant conditions, phosphate-buffered solutions (PBS, pH = 7.4) of 1, 3 and 5 were monitored over time using UV-Visible spectroscopy. The complexes were initially dissolved in DMSO (0.7 mg L−1, 0.16 mg L−1 and 97.5 mg L−1 respectively) and then diluted to 5% DMSO with PBS. The absorbance spectra were collected after 0, 4, 18, 24, 48 and 72 h (Fig. 2b–d). | ||
| Fig. 2 UV-vis spectra of 1 in DMSO solution (a) and of 1 (b), 3 (c) and 5 (d) in 5% DMSO-PBS solution at room temperature. | ||
Over time the absorption energy of the complexes in 5% DMSO-PBS decreases, which may be attributed to precipitation, observable by clouding of the solution, probably due to the very long lipophilic chain of the palmitoyl residue. For 1 and 3 the exchange of the chloride ligand with a solvent molecule occurs immediately and the λmax remains unchanged within 72 h. In contrast, complex 5 containing PTA shows a slight decrease in the wavelength of the maximum absorption energy which might be caused by the replacement of SO3CF3− counterion with an anion present in the buffer solution. The intermolecular contacts between the anionic species with the complexes may affect the electronic structure and the structural arrangement, as previously reported by others.20,21 All the complexes showed transitions in the range 350–400 nm assignable to MLCT (metal–ligand charge transfer) from the filled 4d orbitals of Ru(II) to the empty π* ligand orbitals (4d6 Ru → π*). For complex 5 the substitution of the counterion hypothesized above is consistent with the stability studies in solution carried out using 31P NMR spectroscopy in which the phosphorus signal remains unchanged over 72 hours. The osmium complex 4 showed greater stability than the analogous ruthenium derivative 1.17
Cytotoxicity studies
The cytotoxicity of the compounds was determined on the human ovarian carcinoma cell line (A2780) and its cisplatin resistant form (A780cis) as well as non-tumorigenic human embryonic kidney (HEK293T) cells over an incubation period of 72 h using the MTT assay. The resulting IC50 values of the compounds are presented in Table 1 together with the values for cisplatin and Rapta-C used as positive and negative controls, respectively.| a Values not reproducible due to solubility issues in the biological medium. | |||
|---|---|---|---|
| Compound | A2780 | A2780cis | HEK293T |
| p-curcH | 4.2 ± 4.9 | 6.8 ± 3.0 | >50 |
| p-bdcurcH | n.aa | n.aa | n.aa |
| 1 | 43 ± 5.0 | >50 | >50 |
| 2 | 0.5 ± 0.2 | 6.3 ± 7.7 | >50 |
| 3 | 49 ± 6.0 | >50 | >50 |
| 4 | 0.4 ± 0.1 | >50 | >50 |
| 5 | 6.1 ± 1.7 | 11.2 ± 0.6 | 24 ± 80 |
| 6 | 11.8 ± 2.6 | 14.4 ± 5.7 | 21 ± 19 |
| 7 | 10.3 ± 2.9 | 14.9 ± 2.6 | 21 ± 10 |
| 8 | 3.7 ± 2.2 | 2.3 ± 0.4 | 3.7 ± 0.8 |
| Cisplatin | 1.1 ± 0.5 | 7.7 ± 0.9 | 3.4 ± 1.7 |
| Rapta-C | >100 | >100 | >100 |
Complexes 2 and 4 display the highest cytotoxicity to the A2780 cells, with IC50 values of 0.5 ± 0.2 and 0.4 ± 0.1 μM, respectively, combined with excellent selectivity profiles, with a selectivity index > 100 (IC50 > 50 μM in the HEK293T cell line). Compared to cisplatin, used as a positive control, 2 and 4 are more cytotoxic to the A2780 cells and are more selective, but do not effectively overcome acquired resistance due to cisplatin. Complexes 1 and 3 are the least cytotoxic to the A2780 cells with IC50 values of 43 ± 5 μM and 49 ± 6 μM, respectively. Previous studies on ruthenium complexes with curcumin and bisdemethoxycurcumin ligands indicated that the ionic PTA derivatives tend to be more effective.22 Here, however, the neutral Ru and Os complexes are superior to the ionic PTA derivatives. The higher cytotoxicity observed for 2 and 4 compared to previously reported compounds22 may be attributed to the presence of long aliphatic chains in the ligands that presumably favour uptake. Overall, the best results are observed for bisdemethoxycurcumin derivatives, regardless of their neutral or charged nature. SAR data revealed that bisdemethoxycurcumin complexes are generally more active and selective than the analogous curcumin-containing complexes (see Table S1†).
Binding with BSA
The interaction between serum proteins and drugs is of fundamental pharmacological importance since human serum albumin (HSA) is a major plasma protein with an established role in drug transport. The binding drug-serum albumin is known to occur mainly through the formation of non-covalent interactions at specific binding sites.23 Previous crystallographic studies reported that a wide variety of drugs and small molecule toxins targeted the deep cleft between domains I and III of HSA.24 A similar binding mode was predicted in the case of complexes 1, 2, 4 and 5. In particular, the structural analysis with Maestro R.2021-2 showed that the complexes were stabilized by hydrophobic interactions with residues Pro-113, Leu-115, Val-116, Pro-147, Tyr-148, Tyr-150, Ala-191, and by short-range polar interactions with residues Arg-114, Arg-117, Lys-190, Lys-432, Lys-436 (Fig. S57†). Given the high structural homology, bovine serum albumin (BSA) was used instead of HSA,25 in experimental binding studies of selected complexes (1, 2, 4 and 5) using a fluorometric quenching assay.26 Upon excitation of tryptophan residue at 295 nm, fluorescence emission spectra were recorded in the range 340–600 nm after addition of complexes 1, 2, 4 or 5. All compounds quenched the intrinsic fluorescence of BSA, although to different extents, based on different affinities for BSA and the distance between the fluorophore and the binding site. The biosensor analyses reported a reversible interaction, characterized by a moderate affinity in the micromolar range, with low values of both association and dissociation kinetic constants. These values indicate the slow formation of kinetically stable interactions between BSA and metal complexes 1, 2, 4 and 5, consistent with the results obtained with the docking analyses. Notably, our results showed pH-dependent affinities (Table 2), that decreases with pH. This behaviour is in line with the formation of stable protein-complexes that favour the transport in the blood (at pH = 7.3–7.5) and the promotion of the drug release at tumour sites, which are characterized by lower values of pH (6.0–7.0).27 This decrease in binding affinity at a lower pH value, is probably the cause of the ability of albumins to undergo a reversible conformational transition with changes in pH. Specifically, a significant loss of alpha helices,28 and the consequent increase in protein volume,29 causes a relaxation of the 3D BSA structure, which supports the release of the metal-complexes given a less favourable accommodation in the pocket.| Complex | pH | k ass (M−1 s−1) | k diss (s−1) | K D (mM) |
|---|---|---|---|---|
| 1 | 7.4 | 4350 ± 760 | 0.007 ± 0.002 | 1.6 ± 0.7 |
| 6.8 | 3125 ± 342 | 0.025 ± 0.009 | 7.9 ± 1.4 | |
| 2 | 7.4 | 2654 ± 784 | 0.007 ± 0.004 | 2.6 ± 0.9 |
| 6.8 | 2765 ± 433 | 0.054 ± 0.013 | 19.5 ± 4.4 | |
| 4 | 7.4 | 3534 ± 218 | 0.010 ± 0.001 | 2.8 ± 1.2 |
| 6.8 | 3876 ± 782 | 0.081 ± 0.034 | 20.9 ± 5.5 | |
| 5 | 7.4 | 2350 ± 540 | 0.008 ± 0.003 | 3.4 ± 1.3 |
| 6.8 | 3210 ± 667 | 0.036 ± 0.05 | 11.2 ± 1.6 |
Cell membrane permeability
The ability of the complexes to cross cell membrane was evaluated by monitoring the changes in cell membrane fluidity using trimethylammonium diphenylhexatriene (TMA-DPH) as a fluorescent probe. Most evidently, different behaviour was observed between the less polar (1, 2, 4) and ionic (5) complexes, which is in line with their observed cytotoxicity. Specifically, complexes 2 and 4 could easily cross cell membrane by passive transfer according to a three-stage drug internalization process (stage 1: membrane entry; stage 2: permanence in membrane; stage 3: release from membrane) in approx. 150 min. The nature of the metal centre induces significant quantitative differences only in the kinetics of membrane entry (see Fig. S53 and S54†), with the membrane entry of the Ru complex (2) being faster than the Os counterpart (4). On the other hand, complex 1 could still penetrate the cell membrane although with lower efficacy than 2 and 4, and it was retained longer in the membrane, presumably due to its higher hydrophobicity, before being released. Conversely, in a comparable time frame (180 min) no significant changes in membrane fluidity were observed upon incubation of TMA-DPH labelled cells with complex 5, the polar/ionic nature of the complex hindering its passage across cell membranes.Binding with DNA
The interactions of a representative selection of the complexes, i.e.1, 2, 4 and 5, with DNA were determined using a biosensor-based approach, with a dsDNA probe acting as the “molecular bait” for the molecules of interest. All complexes reversibly bind DNA without any apparent difference observed between the two curcuminoid ligands (p-curc or p-bdcurc). In contrast, the nature of the metal centre significantly affected the recognition event and in turn its affinity for DNA. The ruthenium derivatives (1, 2 and 5) have a higher rate of adduct formation (higher values of kass) and higher stability for the binding with DNA (lower dissociation rates, kdiss, and lower KD values) compared to the osmium derivative (4), which is consistent with the cytotoxicity studies conducted (Table 3).| Complex | k ass (M−1 s−1) | k diss (s−1) | K D (μM) |
|---|---|---|---|
| 1 | 550000 ± 37000 | 0.0076 ± 0.0045 | 0.0138 ± 0.00825 |
| 2 | 400000 ± 90000 | 0.0088 ± 0.016 | 0.022 ± 0.0091 |
| 4 | 110000 ± 20000 | 0.0103 ± 0.027 | 0.1024 ± 0.0211 |
| 5 | 416000 ± 120000 | 0.013 ± 0.009 | 0.0308 ± 0.0241 |
Although the nature of ligands does not affect the interaction with the DNA, it has shown, through competitive binding experiments, to provide evidence for a different binding mode to the biological target. These experimental results were also rationalized by molecular docking models, which showed a peculiar “hug-anchoring” mode for 1 and 5, in which the complexes wrap the ds-DNA helix and shield two contiguous major and minor grooves. The complexes containing the p-bdcurc ligands, i.e.2 and 4, showed selectivity toward the major groove of DNA (Fig. 3) as the absence of the methoxy group results in less steric hindrance.
 | ||
| Fig. 3 Comparative visualization of the best scoring complexes formed upon docking 1, 2, 4 and 5 onto dsDNA. Metal complexes and DNA are rendered as green sticks and grey solid surface, respectively. | ||
Conclusions
Here, we describe a series of novel neutral and ionic ruthenium(II) and osmium(II) arene compounds containing curcuminoid ligands obtained by replacing the OH– groups, present in curcumin and bisdemethoxycurcumin, with a long hydrophobic chain of palmitic acid ester. Two of the compounds exhibit potent antitumor activity towards the ovarian cancer cell line (A2780) and possess excellent cancer cell selectivity, i.e. they were essentially inactive against non-cancerous human embryonic kidney cells (HEK293T). The cytotoxicity values of these compounds were compared with those of previously reported Ru(II) and Os(II) complexes with curcuminoid ligands. This study highlights interesting SARs showing that bisdemethoxycurcumin complexes are generally more active and selective than complexes containing curcumin, likely due to a more efficient internalization process, rather than to the direct binding to DNA. The medicinal properties of turmeric has always been attributed to the main component, namely curcumin, but perhaps the role of bisdemethoxycurcumin, the secondary component of turmeric, has so far been underestimated and should be better investigated. We hope that our results will trigger related studies in the development of bisdemethoxycurcumin-based bioactive systems.Experimental
Materials and methods
The dimer [(cym)RuCl2]2 was purchased from Aldrich, the [(cym)OsCl2]2 was synthesized using literature methods.30 Curcumin and bisdemethoxycurcumin were purchased from TCI Europe and were used as received. All other materials were obtained from commercial sources and were used as received. IR spectra were recorded from 4000 to 600 cm−1 on a PerkinElmer Spectrum 100 FT-IR instrument. 1H, 13C-NMR, 31P-NMR, {1H–1H}-COSY NMR, {1H–13C}-HSQC and {1H–13C}-HMBC spectra were recorded on a 500 Bruker Ascend (500.1 MHz for 1H, 100 MHz for 13C and 202,4 MHz for 31P). Referencing is relative to TMS (1H) and 85% H3PO4 (31P). Positive and negative ion electrospray ionization mass spectra (ESI-MS) were obtained on a Series 1100 MSI detector HP spectrometer using methanol as the mobile phase. Solutions (3 mg mL−1) analysis were prepared using reagent-grade methanol. Masses and intensities were compared to those calculated using IsoPro Isotopic Abundance Simulator, version 2.1.28. Melting points were recorded on an STMP3 Stuart scientific instrument and a capillary apparatus. Samples for microanalysis were dried in vacuo to constant weight (20 °C, ca. 0.1 Torr) and analysed on a Fisons Instruments 1108 CHNS-O elemental analyzer. Uv-stability studies have been conducted with a Varian Caryl spectrometer. Electrical conductivity measurements (ΛM, reported as Ω−1 cm2 mol−1) of acetone solutions of the complexes were recorded using a Crison CDTM 522 conductimeter at room temperature. Binding studies were performed an IAsys + optical biosensor (Affinity Sensors – Cambridge, UK), equipped with carboxylate cuvettes (Neosensors – Crew, UK). Fluorometric assays were performed of a Shimadzu RF-5301PC fluorometer or on a SpectraMax Gemini XPS fluorescence plate reader (Molecular Device, Milan – Italy). HMGR activity assays were performed on an AKTA basic HPLC system.Cytotoxicity studies
The human ovarian carcinoma cell line and its cisplatin resistant form, A2780 and A2780cis were purchased from the European Collection of Cell Cultures (ECACC, United Kingdom). The human embryonic kidney 293T cell line (HEK293T) was kindly provided by the biological screening facility (EPFL, Switzerland). Fetal bovine serum (FBS) was obtained from (Sigma, Switzerland). RPMI 1640 GlutaMAX and DMEM GlutaMAX media were purchased from Life Technologies. The cells were cultured in RPMI 1640 GlutaMAX supplemented for the ovarian cancer cell lines A2780 and A2780cis and in DMEM GlutaMAX supplemented for HEK293T with 10% heat-inactivated FBS at 37 °C and CO2 (5%). To uphold cisplatin resistance, the A2780cis cell line was routinely treated with cisplatin at a final concentration of 2 μM in the media. MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide) assay was used to evaluate the cytotoxicity of the compounds. Stock solutions were prepared in DMSO and sequentially diluted in cell culture grade water to obtain a concentration range of 0–1 mM. 10 μL aliquots of these prepared compound solutions were added in triplicates to a 96-well plate to which 90 μL of the cell suspension (approximately 1.4 × 104 cells per well) were added (final volume 100 μL/concentrations range 0–100 μM). Cisplatin and RAPTA-C were used as positive (0–100 μM) and negative (0–100 μM) controls, respectively, and the plates were incubated for 72 h. 10 μL of an MTT solution prepared at a concentration of 5 mg mL−1 in Dulbecco's phosphate buffered saline (DPBS) was added to the cells, and the plates were incubated for additional 4 h. The culture media was carefully aspirated to preserve the purple formazan crystals that were dissolved in DMSO (100 μL per well). The absorbance of the resulting solutions, which is directly proportional to the number of surviving cells, was measured at 590 nm using SpectroMax M5e microplate reader and the data was analysed with GraphPad Prism software (version 9.3.1). The reported IC50 values are based on the means of three or two independent experiments, each comprising three tests per concentration level.BSA binding
The interaction between serum albumin and the compounds of interest was explored both fluorometrically, via quenching of BSA tryptophan fluorescence, and according to a biosensor binding assay. First, fluorescence spectra of 10 μM BSA were recorded from 300 nm to 450 nm upon excitation of tryptophan at 295 nm.26 Titrations were performed by individual additions of 5 in the range 1–10 μM at 37 °C. Next, the binding kinetics of complexes 1, 2, 4 and 5 to BSA were further evaluated on an IAsys plus biosensor. BSA sensing surface was prepared as previously reported,31 each complex being independently added at different concentrations in the range 0–2 μM and replicated at different pH values (6.8 and 7.4) at 37 °C. Raw data were globally fitted to both mono- and bi-exponential models, and the validity of each model to fit time courses was assessed by a standard F-test procedure.Fluorescence anisotropy measurements
The kinetics of transport across cell membranes were explored by monitoring the change in membrane fluidity of Caco-2 cells during the internalization phase of the complexes of interest.31 Anisotropy measurements were carried out using membrane-anchoring TMA-DPH fluorescent probe (λexc = 340 nm; λem = 460 nm) at 37 °C on a RF-5301PC Shimadzu spectrofluorometer under continuous stirring. In detail, 1.5 × 105 per mL Caco-2 cells were pre-incubated with 1 μM TMA-DPH, and individually added with 10 μM of 1–5 and kept at 37 °C. Fluorescence anisotropy (r) was calculated at 10 min intervals for 200 min using the following model:Fluorescence polarization (P) was derived using the equation:
with I| and I⊥ being the fluorescence intensities parallel (0°) and perpendicular (90°) to the excitation beam, respectively. The kinetic rate constants characterizing the main steps of the internalization event (namely, kin and kout) were derived according to a general mono-exponential model:
| rin = a(1−ekint) + c |
| rout = b(eKoutt) + d |
DNA binding
The interaction between dsDNA and the compounds of interest (namely, 1, 2, 4 and 5) was explored both according to a biosensor binding assay and spectro-fluorometrically, by exploiting the ability of the compounds of interest to compete with specific DNA binders. The dsDNA sensing surface was obtained via streptavidin cross-linking as previously described.32 Briefly, streptavidin protein anchor was covalently blocked via NHS-EDC chemistry. Next, a 5′-biotinylated dsDNA probe (sequence: 3′-CCACCCACTACCCTGGTTGGATGCTAATGT-5) was coupled to surface-blocked the streptavidin. The compounds of interest were independently added to the DNA coated surface at different concentrations, each time following binding kinetics up to equilibrium.32 Raw data were globally fitted to both mono- and bi-exponential models, and the validity of each model to fit time courses was assessed by a standard F-test procedure. Next, the binding sites on DNA for our complexes were mapped according to specific displacement assays. DNA molecules were independently labelled with DAPI (a minor groove binder) or methyl green (a major groove binder), eventually challenging individual DNA complexes with increasing concentration of the molecule of interest. Specifically, DAPI displacement was monitored from the decrease in the intensity of the emission spectra with increasing concentrations of the candidate competitors. Reaction mixtures contained different concentrations of these molecules (0–100 μM), DNA (20 μM), and DAPI (15 μM) in phosphate buffer (10 mM, pH 7.4). Likewise, methyl green displacement assay was performed by monitoring the absorbance at 630 nm.32DNA docking analysis
The predictive models of all the above-mentioned ligand-DNA complexes were computed by independently docking the crystallographic structure of the ligands onto 3′-CCACCCACTACCCTGGTTGGATGCTAATGT-5′ dsDNA oligonucleotide (target oligomer was prepared and energy minimized using Avogadro).33 Rigid geometric docking and energy refinement was performed using PatchDock34 and FireDock.35 As previously reported,311, 2, 4, or 5, and DNA being uploaded as ligand and receptor, respectively. The images of the best scoring models were rendered with PyMOL (The PyMOL Molecular Graphics System, Version 2.2.3 Schrödinger, LLC).HSA docking analysis
The molecular models of the complexes between HSA and 1, 2, 4 and 5 were obtained by flexible ligand–receptor docking using Autodock 4.2.36 Ruthenium and osmium atom parameters used manually set as “atom par Ru 2.96 0.056 12.000-0.00110 0.0 0.0 0 -1 -1 1 # Non H-bonding”, and “atom_par Os 3.12 0.120 12.000 -0.00110 0.0 0.0 0 -1 -1 1 # Non H-bonding”, respectively. The 3D structures of the metal complexes were docked onto the crystallographic structure of human serum albumin (PDB entry: 1AO637 over a grid box (90 × 90 × 50 Å) embracing the whole protein. Default settings were used throughout. The resulting best scoring models were analyzed using Maestro (Schrödinger Release 2021-2: Maestro, Schrödinger, LLC, New York, NY, USA, 2021) and PyMOL (The PyMOL Molecular Graphics System, Version 2.4 Schrödinger, LLC).General procedure for synthesis of compounds
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O); 1703 m and 1624 m ν(C
O); 1703 m and 1624 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1598 m, 1513 s, 1470 s ν(C
O), 1598 m, 1513 s, 1470 s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C).1H-NMR (DMSO-d6, 293 K): δ 0.84 (t, 6H, C(26–26′)H), 1.25 (mbr, 44H, aliphatic chain), 1.38 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.56 (m, 4H, C(12–12′)H), 3.84 (s, 6H, OCH3), 6.21 (s, 2H, C(1)H), 6.98 (d, 2H, C(3–3′)H, 3J = 16 Hz), 7.14 (d, 4H, C(9–9′)H, 3J = 8.0 Hz), 7.33 (d, 4H, C(10–10′)H, 3J = 8.0 Hz), 7.51 (s, 2H, C(6–6′)H), 7.65 (d 2H, C(4–4′)H, 3J = 16 Hz,). 1H-NMR (CDCl3, 293 K): δ 0.90 (t, 6H, C(26–26′)H), 1.28 (mbr, 44H, aliphatic chain), 1.45 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.61 (t, 4H, C(12–12′)H), 3.90 (s, 6H, OCH3), 5.88 (s, 2H, C(1)H), 6.59 (d, 2H, C(3–3′)H, 3J = 16 Hz), 7.08 (d, 4H, C(9–9′)H, 3J = 8.0 Hz), 7.14 (s, 2H, C(6–6′)H), 7.19 (d, 4H, C(10–10′)H, 3J = 8.0 Hz), 7.65 (d 2H, C(4–4′)H, 3J = 16 Hz,). 13C{1H}-NMR (CDCl3): δ 14.09 [s, C(26–26′)], 22.68 [C(13–13′)], 25.01 [C(14–14′)], 29.06, 29.27, 29.35, 29.50., 29.61, 29.65, 29.69 [from C15 to C25], 33.21 (*), 34.05 [s, C(12–12′)], 55.92 [s, OCH3], 101.70 [s, C1] 111.51 [s, C(6–6′)], 121.09 [s, C(10–10′)], 123.34 [s, (C(9–9′)], 124.22 [s, C(3–3′)], 133.85 [s, C(5–5′)], 140.02 [s, C(4–4′)], 141.54 [s, C(8–8′)], 151.51 [s, C(7–7′)], 171.63 [s, C(11–11′)], 183.12 [s, C(2–2′)
C).1H-NMR (DMSO-d6, 293 K): δ 0.84 (t, 6H, C(26–26′)H), 1.25 (mbr, 44H, aliphatic chain), 1.38 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.56 (m, 4H, C(12–12′)H), 3.84 (s, 6H, OCH3), 6.21 (s, 2H, C(1)H), 6.98 (d, 2H, C(3–3′)H, 3J = 16 Hz), 7.14 (d, 4H, C(9–9′)H, 3J = 8.0 Hz), 7.33 (d, 4H, C(10–10′)H, 3J = 8.0 Hz), 7.51 (s, 2H, C(6–6′)H), 7.65 (d 2H, C(4–4′)H, 3J = 16 Hz,). 1H-NMR (CDCl3, 293 K): δ 0.90 (t, 6H, C(26–26′)H), 1.28 (mbr, 44H, aliphatic chain), 1.45 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.61 (t, 4H, C(12–12′)H), 3.90 (s, 6H, OCH3), 5.88 (s, 2H, C(1)H), 6.59 (d, 2H, C(3–3′)H, 3J = 16 Hz), 7.08 (d, 4H, C(9–9′)H, 3J = 8.0 Hz), 7.14 (s, 2H, C(6–6′)H), 7.19 (d, 4H, C(10–10′)H, 3J = 8.0 Hz), 7.65 (d 2H, C(4–4′)H, 3J = 16 Hz,). 13C{1H}-NMR (CDCl3): δ 14.09 [s, C(26–26′)], 22.68 [C(13–13′)], 25.01 [C(14–14′)], 29.06, 29.27, 29.35, 29.50., 29.61, 29.65, 29.69 [from C15 to C25], 33.21 (*), 34.05 [s, C(12–12′)], 55.92 [s, OCH3], 101.70 [s, C1] 111.51 [s, C(6–6′)], 121.09 [s, C(10–10′)], 123.34 [s, (C(9–9′)], 124.22 [s, C(3–3′)], 133.85 [s, C(5–5′)], 140.02 [s, C(4–4′)], 141.54 [s, C(8–8′)], 151.51 [s, C(7–7′)], 171.63 [s, C(11–11′)], 183.12 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 845 [p-curcH]+.
O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 845 [p-curcH]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1701 m and 1647 m ν(C
O), 1701 m and 1647 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1598 m, 1508 m, 1463 m ν(C
O), 1598 m, 1508 m, 1463 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C).1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain), 1.44 (m, 4H, C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.59 (t, 4H, (C(12–12′)H), 5.86 (s, 2H, C(1)H), 6.61 (d, 2H, C(3–3′)H, 3J = 15.90 Hz), 7.2 (d, 4H, C(9–9′)H and C(7–7′)H, 3J = 8.5 Hz), 7.60 (d, 4H, C(10–10′)H and C(6–6′)H, 3J = 8.5 Hz), 7.67 (d, 2H, C(4–4′)H, 3J = 15.90 Hz). 13C{1H}-NMR (CDCl3): δ 14.11 [s, C(26–26′)], 22.69 [s, C(13–13′)], 24.80 [s, C(14–14′)], 29.10, 29.25, 29.36, 29.45, 29.59, 29.64, 29.66, 29.69 (from C15 to C25), 31.93 (*), 34.43 [s, C(12–12′)], 101.84 (s, C1), 122.18 [C(9–91) and C(7–7′)], 124.12 [C(3–3′)], 129.19 [C(10–10′) and C(6–6′), 132.61 [C(5–5′)], 139.61 [C(4–4′)], 152.13 [C(8–8′)], 172.02 [C(11–11′)], 183.17 [C(2–2′)
C).1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain), 1.44 (m, 4H, C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.59 (t, 4H, (C(12–12′)H), 5.86 (s, 2H, C(1)H), 6.61 (d, 2H, C(3–3′)H, 3J = 15.90 Hz), 7.2 (d, 4H, C(9–9′)H and C(7–7′)H, 3J = 8.5 Hz), 7.60 (d, 4H, C(10–10′)H and C(6–6′)H, 3J = 8.5 Hz), 7.67 (d, 2H, C(4–4′)H, 3J = 15.90 Hz). 13C{1H}-NMR (CDCl3): δ 14.11 [s, C(26–26′)], 22.69 [s, C(13–13′)], 24.80 [s, C(14–14′)], 29.10, 29.25, 29.36, 29.45, 29.59, 29.64, 29.66, 29.69 (from C15 to C25), 31.93 (*), 34.43 [s, C(12–12′)], 101.84 (s, C1), 122.18 [C(9–91) and C(7–7′)], 124.12 [C(3–3′)], 129.19 [C(10–10′) and C(6–6′), 132.61 [C(5–5′)], 139.61 [C(4–4′)], 152.13 [C(8–8′)], 172.02 [C(11–11′)], 183.17 [C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O].
O].
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-curc), 1630 m ν(C
O, of p-curc), 1630 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 w, 1525 vs., 1505 vs. ν(C
O), 1599 w, 1525 vs., 1505 vs. ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 395 m, 269 s ν(Ru–Cl). 1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-curc), 1.41 (d, 6H, –CH(CH3)2 of cym, 4J = 7 Hz), 1.45 (m, 4H, C(14–14′)H), 1.79 (m, 4H, C(13–13′)H), 2.37 (s, 3H, –CH3 of cym), 2.60 (t, 4H, C(12–12′)H), 3.00 (m, 1H, CH(CH3)2 of cym), 3.88 (s, 6H, –OCH3 of p-curc), 5.52 (s, 1H, C(1)H of p-curc), 5.33 d, 5.60 d (4H, AA′BB′ system, CH3-C6H4-CH(CH3)2 of cym, 3J = 6 Hz), 6.53 (d, 2H, C(3–3′)H of curc, 3Jtrans = 16 Hz), 7.03 (d, 2 H, C(9–9′)H of p-curc, 3Jtrans = 8 Hz), 7.11 (d, 2H, C(10–10′)H of p-curc, 3J = 8 Hz), 7.10 (sbr, 2H, C(6–6′)H of p-curc), 7.58 (d, 2H, C(4–4′)H of p-curc, 3Jtrans = 16 Hz). 13C{1H}-NMR (CDCl3, 293 K): δ 14.14 [s, C(26–26′)], 18.12 (s, –CH3 of cym), 22.44, 22.70 (s, –CH(CH3)2 of cym), 25.04 [s, C(13–13′)], 29.08 [s, C(14–14′)], 29.30, 29.37, 29.53, 29.64, 29.67, 29.71 (from C15 to C25 of p-curc), 30.88 (s, CH(CH3)2 of cym), 31.94, 34.07 [s, C(12–12′)], 55.90 (s, –OCH3 of p-curc), 79.15 [s, C(a–a′)], 83.02 [s, C(b–b′)], 97.71 (s, Ci′), 99.71 (s, Ci), 102.31 (s, C1), 111.00 [s, C(6–6′)], 120.85 [s, C(10–10′)], 123.13 [s, C(9–9′)], 127.80 [s, C(3–3′)], 134.74 [s, C(5–5′)], 138.14 [s, C(4–4′)], 140.80 [s, C(8–8′)], 151.33 [s, C(7–7′)], 171.77 [s, C(11–11′)], 178.32 [s, C(2–2′)
C); 395 m, 269 s ν(Ru–Cl). 1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-curc), 1.41 (d, 6H, –CH(CH3)2 of cym, 4J = 7 Hz), 1.45 (m, 4H, C(14–14′)H), 1.79 (m, 4H, C(13–13′)H), 2.37 (s, 3H, –CH3 of cym), 2.60 (t, 4H, C(12–12′)H), 3.00 (m, 1H, CH(CH3)2 of cym), 3.88 (s, 6H, –OCH3 of p-curc), 5.52 (s, 1H, C(1)H of p-curc), 5.33 d, 5.60 d (4H, AA′BB′ system, CH3-C6H4-CH(CH3)2 of cym, 3J = 6 Hz), 6.53 (d, 2H, C(3–3′)H of curc, 3Jtrans = 16 Hz), 7.03 (d, 2 H, C(9–9′)H of p-curc, 3Jtrans = 8 Hz), 7.11 (d, 2H, C(10–10′)H of p-curc, 3J = 8 Hz), 7.10 (sbr, 2H, C(6–6′)H of p-curc), 7.58 (d, 2H, C(4–4′)H of p-curc, 3Jtrans = 16 Hz). 13C{1H}-NMR (CDCl3, 293 K): δ 14.14 [s, C(26–26′)], 18.12 (s, –CH3 of cym), 22.44, 22.70 (s, –CH(CH3)2 of cym), 25.04 [s, C(13–13′)], 29.08 [s, C(14–14′)], 29.30, 29.37, 29.53, 29.64, 29.67, 29.71 (from C15 to C25 of p-curc), 30.88 (s, CH(CH3)2 of cym), 31.94, 34.07 [s, C(12–12′)], 55.90 (s, –OCH3 of p-curc), 79.15 [s, C(a–a′)], 83.02 [s, C(b–b′)], 97.71 (s, Ci′), 99.71 (s, Ci), 102.31 (s, C1), 111.00 [s, C(6–6′)], 120.85 [s, C(10–10′)], 123.13 [s, C(9–9′)], 127.80 [s, C(3–3′)], 134.74 [s, C(5–5′)], 138.14 [s, C(4–4′)], 140.80 [s, C(8–8′)], 151.33 [s, C(7–7′)], 171.77 [s, C(11–11′)], 178.32 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1079 [100] [Ru(cym)(p-curc)]+.
O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1079 [100] [Ru(cym)(p-curc)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-bdcurc), 1631 m ν(C
O, of p-bdcurc), 1631 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1541 s, 1520 vs, 1503 s ν(C
O), 1541 s, 1520 vs, 1503 s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 270 s ν(Ru–Cl). 1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-bdcurc), 1.42 (d, 10H, CH(CH3)2 of cym, 4J = 7 Hz, and C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.37 (s, 3H, CH3 of cym), 2.58 (t, 4H, C(12–12′)H), 3.01 (m, 1H, CH(CH3)2 of cym), 5.49 (s, 1H, C(1)H of curcumin), 5.33 d, 5.60 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.54 (d, 2H, C(3, 3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.11 (d, 4 H, C(9–9′)H and C(7, 7′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.54 (d, 4 H, C(10–10′)H and C(6–6′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.60 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz). 13C{1H}-NMR (CDCl3, 293 K): δ 14.12 [s, C(26–26′)], 18.00 (s, –CH3 of cym), 22.42, 22.69 (s, –CH(CH3)2 of cym), 24.93 [s, C(13–13′)], 29.11 [s, C(14–14′)], 29.26, 29.36, 29.46, 29.60, 29.66, 29.69 (from C15 to C25), 30.87 (s, CH(CH3)2 of cym), 31.93, 34.44 [s, C(12–12′)], 79.22 [s, C(a–a′)], 83.04 [s, C(b–b′)], 97.58 (s, Ci′), 99.68 (s, Ci), 102.48 (s, C1), 121.97 [s, C(9–9′) and C(7–7′)], 127.69 [s, C(3–3′)], 128.78 [s, C(10–10′) and C(6–6′)], 133.46 [s, C(5–5′)], 137.76 [s, C(4–4′)], 151.47 [s, C(8–8′)], 172.09 [s, C(11–11′)], 178.38 [s, C(2–2′)
C); 270 s ν(Ru–Cl). 1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-bdcurc), 1.42 (d, 10H, CH(CH3)2 of cym, 4J = 7 Hz, and C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.37 (s, 3H, CH3 of cym), 2.58 (t, 4H, C(12–12′)H), 3.01 (m, 1H, CH(CH3)2 of cym), 5.49 (s, 1H, C(1)H of curcumin), 5.33 d, 5.60 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.54 (d, 2H, C(3, 3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.11 (d, 4 H, C(9–9′)H and C(7, 7′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.54 (d, 4 H, C(10–10′)H and C(6–6′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.60 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz). 13C{1H}-NMR (CDCl3, 293 K): δ 14.12 [s, C(26–26′)], 18.00 (s, –CH3 of cym), 22.42, 22.69 (s, –CH(CH3)2 of cym), 24.93 [s, C(13–13′)], 29.11 [s, C(14–14′)], 29.26, 29.36, 29.46, 29.60, 29.66, 29.69 (from C15 to C25), 30.87 (s, CH(CH3)2 of cym), 31.93, 34.44 [s, C(12–12′)], 79.22 [s, C(a–a′)], 83.04 [s, C(b–b′)], 97.58 (s, Ci′), 99.68 (s, Ci), 102.48 (s, C1), 121.97 [s, C(9–9′) and C(7–7′)], 127.69 [s, C(3–3′)], 128.78 [s, C(10–10′) and C(6–6′)], 133.46 [s, C(5–5′)], 137.76 [s, C(4–4′)], 151.47 [s, C(8–8′)], 172.09 [s, C(11–11′)], 178.38 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1019 [5] [Ru(cym)(p-bdcurc)]+.
O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1019 [5] [Ru(cym)(p-bdcurc)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-curc), 1630 m ν(C
O, of p-curc), 1630 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 w, 1523 vs., 1505 vs. ν(C
O), 1599 w, 1523 vs., 1505 vs. ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 273 s ν(Os–Cl). 1H-NMR (CDCl3, 293 K): δ 0.90 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-curc), 1.37 (d, 6H, –CH(CH3)2 of cym, 4J = 7 Hz), 1.43 (m, 4H, C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.38 (s, 3H, –CH3 of cym), 2.60 (t, 4H, C(12–12′)H), 2.83 (m, 1H, CH(CH3)2 of cym), 3.88 (s, 6H, –OCH3 of p-curc), 5.70 (s, 1H, C(1)H of p-curc), 5.83 d, 6.06 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.51 (d, 2H, C(3–3′)H of p-curc, 3Jtrans = 16 Hz), 7.04 (d, 2 H, C(9–9′)H of p-curc, 3Jtrans = 8 Hz), 7.12 (d, 2H, C(10–10′)H, 3Jtrans = 8 Hz), 7.11 (sbr, 2H, C(6–6′)H of p-curc), 7.57 (d, 2H, C(4–4′)H of p-curc, 3Jtrans = 16 Hz). 13C{1H} NMR (CDCl3, 293 K): δ 14.11 [s, C(26–26′)], 18.29 (s, –CH3 of cym), 22.69, 22.81 (s, –CH(CH3)2 of cym), 25.03 [s, C(13–13′)], 29.07 [s, C(14–14′)], 29.28, 29.36, 29.51, 29.62, 29.66, 29.69 (from C15 to C25 of p-curc), 31.52 (s, CH(CH3)2 of cym), 31.93, 34.06 [s, C(12–12′)], 55.91 (s, -OCH3 of p-curc), 69.45 [s, C(a–a′)], 74.57 [s, C(b–b′)], 89.15 (s, Ci′), 89.80 (s, Ci), 103.71 (s, C1), 111.05 [s, C(6–6′)], 120.86 [s, C(10–10′)], 123.20 [s, C(9–9′)], 127.58 [s, C(3–3′)], 134.68 [s, C(5–5′)], 138.26 [s, C(4–4′)], 140.91 [s, C(8–8′)], 151.43 [s, C(7–7′)], 171.68 [s, C(11–11′)], 177.24 [s, C(2–2′)
C); 273 s ν(Os–Cl). 1H-NMR (CDCl3, 293 K): δ 0.90 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-curc), 1.37 (d, 6H, –CH(CH3)2 of cym, 4J = 7 Hz), 1.43 (m, 4H, C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.38 (s, 3H, –CH3 of cym), 2.60 (t, 4H, C(12–12′)H), 2.83 (m, 1H, CH(CH3)2 of cym), 3.88 (s, 6H, –OCH3 of p-curc), 5.70 (s, 1H, C(1)H of p-curc), 5.83 d, 6.06 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.51 (d, 2H, C(3–3′)H of p-curc, 3Jtrans = 16 Hz), 7.04 (d, 2 H, C(9–9′)H of p-curc, 3Jtrans = 8 Hz), 7.12 (d, 2H, C(10–10′)H, 3Jtrans = 8 Hz), 7.11 (sbr, 2H, C(6–6′)H of p-curc), 7.57 (d, 2H, C(4–4′)H of p-curc, 3Jtrans = 16 Hz). 13C{1H} NMR (CDCl3, 293 K): δ 14.11 [s, C(26–26′)], 18.29 (s, –CH3 of cym), 22.69, 22.81 (s, –CH(CH3)2 of cym), 25.03 [s, C(13–13′)], 29.07 [s, C(14–14′)], 29.28, 29.36, 29.51, 29.62, 29.66, 29.69 (from C15 to C25 of p-curc), 31.52 (s, CH(CH3)2 of cym), 31.93, 34.06 [s, C(12–12′)], 55.91 (s, -OCH3 of p-curc), 69.45 [s, C(a–a′)], 74.57 [s, C(b–b′)], 89.15 (s, Ci′), 89.80 (s, Ci), 103.71 (s, C1), 111.05 [s, C(6–6′)], 120.86 [s, C(10–10′)], 123.20 [s, C(9–9′)], 127.58 [s, C(3–3′)], 134.68 [s, C(5–5′)], 138.26 [s, C(4–4′)], 140.91 [s, C(8–8′)], 151.43 [s, C(7–7′)], 171.68 [s, C(11–11′)], 177.24 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1170 [100] [Os(cym)(p-curc)]+.
O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1170 [100] [Os(cym)(p-curc)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-curc), 1631 m ν(C
O, of p-curc), 1631 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 w, 1584 w, 1540 s, 1519 vs ν(C
O), 1599 w, 1584 w, 1540 s, 1519 vs ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 271 s ν(Os–Cl). 1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-bdcurc), 1.39 (d, 6H, CH(CH3)2 of cym, 4J = 7 Hz), 1.44 (m, 4H, C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.37 (s, 3H, CH3 of cym), 2.58 (t, 4H, C(12–12′)H), 2.84 (m, 1H, CH(CH3)2 of cym), 5.67 (s, 1H, C(1)H of curcumin), 5.82 d, 6.06 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.52 (d, 2H, C(3–3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.11 (d, 4 H, C(9–9′)H and C(7–7′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.54 (d, 4 H, C(10–10′)H and C(6–6′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.59 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz). 13C{1H}-NMR (CDCl3, 293 K): δ 14.10 [s, C(26–26′)], 18.21 (s, –CH3 of cym), 22.69, 22.81 (s, –CH(CH3)2 of cym), 24.92 [s, C(13–13′)], 29.11 [s, C(14–14′)], 29.25, 29.35, 29.46, 29.59, 29.65, 29.67, 29.69 (from C15 to C25), 31.52 (s, CH(CH3)2 of cym), 31.93, 34.44 [s, C(12–12′)], 69.53[s, C(a–a′)], 74.60 [s, C(b–b′)], 89.01 (s, Ci′), 89.73 (s, Ci), 103.89 (s, C1), 122.04 [s, C(9–9′) and C(7–7′)], 127.44 [s, C(3–3′)], 128.80 [s, C(10–10′) and C(6–6′)], 133.41 [s, C(5–5′)], 137.90 [s, C(4–4′)], 151.55 [s, C(8–8′)], 172.03 [s, C(11–11′)], 177.32 [s, C(2–2′)
C); 271 s ν(Os–Cl). 1H-NMR (CDCl3, 293 K): δ 0.91 (t, 6H, C(26–26′)H), 1.29 (mbr, 44H, aliphatic chain of p-bdcurc), 1.39 (d, 6H, CH(CH3)2 of cym, 4J = 7 Hz), 1.44 (m, 4H, C(14–14′)H), 1.78 (m, 4H, C(13–13′)H), 2.37 (s, 3H, CH3 of cym), 2.58 (t, 4H, C(12–12′)H), 2.84 (m, 1H, CH(CH3)2 of cym), 5.67 (s, 1H, C(1)H of curcumin), 5.82 d, 6.06 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.52 (d, 2H, C(3–3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.11 (d, 4 H, C(9–9′)H and C(7–7′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.54 (d, 4 H, C(10–10′)H and C(6–6′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.59 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz). 13C{1H}-NMR (CDCl3, 293 K): δ 14.10 [s, C(26–26′)], 18.21 (s, –CH3 of cym), 22.69, 22.81 (s, –CH(CH3)2 of cym), 24.92 [s, C(13–13′)], 29.11 [s, C(14–14′)], 29.25, 29.35, 29.46, 29.59, 29.65, 29.67, 29.69 (from C15 to C25), 31.52 (s, CH(CH3)2 of cym), 31.93, 34.44 [s, C(12–12′)], 69.53[s, C(a–a′)], 74.60 [s, C(b–b′)], 89.01 (s, Ci′), 89.73 (s, Ci), 103.89 (s, C1), 122.04 [s, C(9–9′) and C(7–7′)], 127.44 [s, C(3–3′)], 128.80 [s, C(10–10′) and C(6–6′)], 133.41 [s, C(5–5′)], 137.90 [s, C(4–4′)], 151.55 [s, C(8–8′)], 172.03 [s, C(11–11′)], 177.32 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1110 [100] [Os(cym)(p-bdcurc)]+.
O]. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1110 [100] [Os(cym)(p-bdcurc)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-curc), 1625 m ν(C
O, of p-curc), 1625 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 m, 1505 vs. ν(C
O), 1599 m, 1505 vs. ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 637 s ν(Ru–P). 1H-NMR (DMSO-d6, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25–1.30 (mbr, 50H, aliphatic chain of p-bdcurc and CH(CH3)2 of cym), 1.38 (m, 4H, C(14–14′)H), 1.64 (m, 4H, C(13–13′)H), 2.04 (s, 3H, CH3 of cym), 2.57 (t, 4H, C(12–12′)H), 2.67 (m, 1H, CH(CH3)2 of cym), 3.85 (s, 6H, -OCH3 of p-curc), 4.14 (s, 6H, (P–CH2–N) of PTA), 4.46 (m, 6H, (N–CH2–N) of PTA), 5.94 (s, 1H, C(1)H of curcumin), 6.10 d, 6.16 d (4H, AA′BB′ system, CH3-C6H4-CH(CH3)2 of cym, 3J = 6 Hz), 6.93 (d, 2H, C(9–9′)H of p-curc, 3J = 16 Hz), 7.15 (d, 2H, C(3, 3′)H of p-curc, 3Jtrans = 8 Hz), 7.24 (d, 2H, C(4–4′)H of p-curc, 3Jtrans = 8 Hz), 7.43 [m, 4H, C(10–10′)H and C(6–6′)H of p-curc]. 13C{1H}-NMR (DMSO-d6, 293 K): δ 14.40 [s, C(26–26′)], 16.86 (s, -CH3 of cym), 21.03, 22.23 (s, -CH(CH3)2 of cym), 24.93 [s, C(13–13′)], 28.74 [s, C(14–14′)], 29.11, 29.16, 29.35, 29.42, 29.47, 29.49 (from C15 to C25 of p-curc), 30.50 (s, CH(CH3)2 of cym), 31.75, 33.66 [s, C(12–12′)], 50.01, 51.11 [s, (P–CH2–N) of PTA, J = 13 Hz ], 56.46 (s, –OCH3 of p-curc), 72.20, 72.26 [s, (N–CH2–N) of PTA, J = 7 Hz], 88.46 [s, C(b–b′)], 90.25 [s, C(a–a′)], 96.64 (s, Ci′), 104.22 (s, Ci), 105.40 (s, C1), 111.76 [s, C(6–6′), 122.05 [s, C(4–4′)], 123.82 [s, C(3–3′)], 127.41 [s, C(9–9′)], 134.38 [s, C(5–5′)], 139.22 [s, C(10–10′)], 141.33 [s, C(8–8′)], 151.77 [s, C(7–7′)], 171.47 [s, C(11–11′)], 180.45 [s, C(2–2′)
C); 637 s ν(Ru–P). 1H-NMR (DMSO-d6, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25–1.30 (mbr, 50H, aliphatic chain of p-bdcurc and CH(CH3)2 of cym), 1.38 (m, 4H, C(14–14′)H), 1.64 (m, 4H, C(13–13′)H), 2.04 (s, 3H, CH3 of cym), 2.57 (t, 4H, C(12–12′)H), 2.67 (m, 1H, CH(CH3)2 of cym), 3.85 (s, 6H, -OCH3 of p-curc), 4.14 (s, 6H, (P–CH2–N) of PTA), 4.46 (m, 6H, (N–CH2–N) of PTA), 5.94 (s, 1H, C(1)H of curcumin), 6.10 d, 6.16 d (4H, AA′BB′ system, CH3-C6H4-CH(CH3)2 of cym, 3J = 6 Hz), 6.93 (d, 2H, C(9–9′)H of p-curc, 3J = 16 Hz), 7.15 (d, 2H, C(3, 3′)H of p-curc, 3Jtrans = 8 Hz), 7.24 (d, 2H, C(4–4′)H of p-curc, 3Jtrans = 8 Hz), 7.43 [m, 4H, C(10–10′)H and C(6–6′)H of p-curc]. 13C{1H}-NMR (DMSO-d6, 293 K): δ 14.40 [s, C(26–26′)], 16.86 (s, -CH3 of cym), 21.03, 22.23 (s, -CH(CH3)2 of cym), 24.93 [s, C(13–13′)], 28.74 [s, C(14–14′)], 29.11, 29.16, 29.35, 29.42, 29.47, 29.49 (from C15 to C25 of p-curc), 30.50 (s, CH(CH3)2 of cym), 31.75, 33.66 [s, C(12–12′)], 50.01, 51.11 [s, (P–CH2–N) of PTA, J = 13 Hz ], 56.46 (s, –OCH3 of p-curc), 72.20, 72.26 [s, (N–CH2–N) of PTA, J = 7 Hz], 88.46 [s, C(b–b′)], 90.25 [s, C(a–a′)], 96.64 (s, Ci′), 104.22 (s, Ci), 105.40 (s, C1), 111.76 [s, C(6–6′), 122.05 [s, C(4–4′)], 123.82 [s, C(3–3′)], 127.41 [s, C(9–9′)], 134.38 [s, C(5–5′)], 139.22 [s, C(10–10′)], 141.33 [s, C(8–8′)], 151.77 [s, C(7–7′)], 171.47 [s, C(11–11′)], 180.45 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O].31P-NMR (DMSO-d6, 298 K): δ −27.09. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1237 [100] [Ru(cym)(p-curc)(PTA)]+.
O].31P-NMR (DMSO-d6, 298 K): δ −27.09. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1237 [100] [Ru(cym)(p-curc)(PTA)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-bdcurc), 1623 m ν(C
O, of p-bdcurc), 1623 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1601 w, 1583 w, 1506 vs. ν(C
O), 1601 w, 1583 w, 1506 vs. ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 637 s ν(Ru-P). 1H-NMR (DMSO-d6, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25–1.30 (mbr, 50H, aliphatic chain of p-bdcurc and CH(CH3)2 of cym), 1.36 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.03 (s, 3H, CH3 of cym), 2.59 (t, 4H, C(12–12′)H), 2.66 (m, 1H, CH(CH3)2 of cym), 4.14 (s, 6H, (P–CH2–N) of PTA), 4.46 (m, 6H, (N–CH2–N) of PTA), 5.93 (s, 1H, C(1)H of p-bdcurc), 6.10 d, 6.16 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.88 (d, 2H, C(3–3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.20 (d, 4 H, C(9–9′)H and C(7–7′)H of p-bdcurc, 3Jtrans = 8 Hz), 7.45 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.74 (d, 4 H, C(10–10′)H and C(6–6′)H of p-bdcurc, 3Jtrans = 8 Hz). 13C{1H}-NMR (DMSO-d6, 293 K): δ 14.40 [s, C(26–26′)], 16.77 (s, –CH3 of cym), 22.19, 22.55, (s, –CH(CH3)2 of cym), 24.75 [s, C(13–13′)], 28.84 [s, C(14–14′)], 29.12, 29.17, 29.31, 29.42, 29.47, 29.50 (from C15 to C25 of p-bdcurc), 30.46 (s, CH(CH3)2 of cym), 31.76 (*), 33.96 [s, C(12–12′)], 50.93, 51.03 [s, (P–CH2–N) of PTA, J = 13 Hz ], 72.16, 72.22 [s, (N–CH2–N) of PTA, J = 7 Hz], 88.52 [s, C(b–b′)], 90.29 [s, C(a–a′)], 96.53 (s, Ci′), 104.09 (s, Ci), 105.30 (s, C1), 122.95 [s, C(9–9′) and C(7–7′)], 127.11 [s, C(3–3′)], 129.78 [s, C(10–10′) and C(6–6′)], 133.03 [s, C(5–5′)], 138.92 [s, C(4–4′)], 152.16 [s, C(8–8′)], 172.08 [s, C(11–11′)], 180.39 [s, C(2–2′)
C); 637 s ν(Ru-P). 1H-NMR (DMSO-d6, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25–1.30 (mbr, 50H, aliphatic chain of p-bdcurc and CH(CH3)2 of cym), 1.36 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.03 (s, 3H, CH3 of cym), 2.59 (t, 4H, C(12–12′)H), 2.66 (m, 1H, CH(CH3)2 of cym), 4.14 (s, 6H, (P–CH2–N) of PTA), 4.46 (m, 6H, (N–CH2–N) of PTA), 5.93 (s, 1H, C(1)H of p-bdcurc), 6.10 d, 6.16 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.88 (d, 2H, C(3–3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.20 (d, 4 H, C(9–9′)H and C(7–7′)H of p-bdcurc, 3Jtrans = 8 Hz), 7.45 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.74 (d, 4 H, C(10–10′)H and C(6–6′)H of p-bdcurc, 3Jtrans = 8 Hz). 13C{1H}-NMR (DMSO-d6, 293 K): δ 14.40 [s, C(26–26′)], 16.77 (s, –CH3 of cym), 22.19, 22.55, (s, –CH(CH3)2 of cym), 24.75 [s, C(13–13′)], 28.84 [s, C(14–14′)], 29.12, 29.17, 29.31, 29.42, 29.47, 29.50 (from C15 to C25 of p-bdcurc), 30.46 (s, CH(CH3)2 of cym), 31.76 (*), 33.96 [s, C(12–12′)], 50.93, 51.03 [s, (P–CH2–N) of PTA, J = 13 Hz ], 72.16, 72.22 [s, (N–CH2–N) of PTA, J = 7 Hz], 88.52 [s, C(b–b′)], 90.29 [s, C(a–a′)], 96.53 (s, Ci′), 104.09 (s, Ci), 105.30 (s, C1), 122.95 [s, C(9–9′) and C(7–7′)], 127.11 [s, C(3–3′)], 129.78 [s, C(10–10′) and C(6–6′)], 133.03 [s, C(5–5′)], 138.92 [s, C(4–4′)], 152.16 [s, C(8–8′)], 172.08 [s, C(11–11′)], 180.39 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. 31P-NMR (DMSO-d6, 298 K): δ −26.93. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1177 [100] [Ru(cym)(p-bdcurc)(PTA)]+.
O]. 31P-NMR (DMSO-d6, 298 K): δ −26.93. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1177 [100] [Ru(cym)(p-bdcurc)(PTA)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-curc), 1624 m ν(C
O, of p-curc), 1624 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1598 w, 1587 w, 1505 vs. ν(C
O), 1598 w, 1587 w, 1505 vs. ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 637 s ν(Ru-P). 1H-NMR (DMSO, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25–1.38 (mbr, 54H, aliphatic chain of p-curc, CH(CH3)2 of cym and C(14–14′)H), 1.64 (m, 4H, C(13–13′)H), 2.17 (s, 3H, CH3 of cym), 2.57 (t, 4H, C(12–12′)H), 2.67 (m, 1H, CH(CH3)2 of cym), 3.85 (s, 6H, –OCH3 of p-curc), 4.10 (s, 6H, (P–CH2–N) of PTA), 4.44 (m, 6H, (N–CH2–N) of PTA), 6.06 (s, 1H, C(1)H of p-curc), 6.18 d, 6.26 d (4H, AA′BB′ system, CH3-C6H4-CH(CH3)2 of cym, 3J = 6 Hz), 6.90 (d, 2H, C(9–9′)H of p-curc, 3J = 16 Hz), 7.15 (d, 2H, C(3–3′)H of p-curc, 3Jtrans = 8 Hz), 7.27 (d, 2H, C(4–4′)H of curc, 3Jtrans = 8 Hz), 7.46 [sbr, 2H, C(6–6′)H], 7.48 [d, 2H, C(10–10′)H of p-curc, 3J = 16 Hz]. 13C{1H} NMR (DMSO, 293 K): δ 14.40 [s, C(26–26′)], 16.89 (s, –CH3 of cym), 22.55, 22.60, (s, –CH(CH3)2 of cym), 24.93 [s, C(13–13′)], 28.74 [s, C(14–14′)], 29.11, 29.16, 29.35, 29.42, 29.47, 29.50 (from C15 to C25 of p-curc), 30.57 (s, CH(CH3)2 of cym), 31.75, 33.67 [s, C(12–12′)], 50.14, 50.29 [s, (P–CH2–N) of PTA, J = 18 Hz ], 56.48 (s, -OCH3 of p-curc), 72.18, 72.24 [s, (N–CH2–N) of PTA, J = 7 Hz], 81.44 [s, C(b–b′)], 82.56 [s, C(a–a′)], 88.20 (s, Ci’), 94.54 (s, Ci), 106.62 (s, C1), 111.74 [s, C(6–6′), 122.10 [s, C(4–4′)], 123.93 [s, C(3–3′)], 126.96 [s, C(9–9′)], 134.43 [s, C(5–5′)], 139.23 [s, C(10–10′)], 141.37 [s, C(8–8′)], 151.87 [s, C(7–7′)], 171.42 [s, C(11–11′)], 178.70 [s, C(2–2′)
C); 637 s ν(Ru-P). 1H-NMR (DMSO, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25–1.38 (mbr, 54H, aliphatic chain of p-curc, CH(CH3)2 of cym and C(14–14′)H), 1.64 (m, 4H, C(13–13′)H), 2.17 (s, 3H, CH3 of cym), 2.57 (t, 4H, C(12–12′)H), 2.67 (m, 1H, CH(CH3)2 of cym), 3.85 (s, 6H, –OCH3 of p-curc), 4.10 (s, 6H, (P–CH2–N) of PTA), 4.44 (m, 6H, (N–CH2–N) of PTA), 6.06 (s, 1H, C(1)H of p-curc), 6.18 d, 6.26 d (4H, AA′BB′ system, CH3-C6H4-CH(CH3)2 of cym, 3J = 6 Hz), 6.90 (d, 2H, C(9–9′)H of p-curc, 3J = 16 Hz), 7.15 (d, 2H, C(3–3′)H of p-curc, 3Jtrans = 8 Hz), 7.27 (d, 2H, C(4–4′)H of curc, 3Jtrans = 8 Hz), 7.46 [sbr, 2H, C(6–6′)H], 7.48 [d, 2H, C(10–10′)H of p-curc, 3J = 16 Hz]. 13C{1H} NMR (DMSO, 293 K): δ 14.40 [s, C(26–26′)], 16.89 (s, –CH3 of cym), 22.55, 22.60, (s, –CH(CH3)2 of cym), 24.93 [s, C(13–13′)], 28.74 [s, C(14–14′)], 29.11, 29.16, 29.35, 29.42, 29.47, 29.50 (from C15 to C25 of p-curc), 30.57 (s, CH(CH3)2 of cym), 31.75, 33.67 [s, C(12–12′)], 50.14, 50.29 [s, (P–CH2–N) of PTA, J = 18 Hz ], 56.48 (s, -OCH3 of p-curc), 72.18, 72.24 [s, (N–CH2–N) of PTA, J = 7 Hz], 81.44 [s, C(b–b′)], 82.56 [s, C(a–a′)], 88.20 (s, Ci’), 94.54 (s, Ci), 106.62 (s, C1), 111.74 [s, C(6–6′), 122.10 [s, C(4–4′)], 123.93 [s, C(3–3′)], 126.96 [s, C(9–9′)], 134.43 [s, C(5–5′)], 139.23 [s, C(10–10′)], 141.37 [s, C(8–8′)], 151.87 [s, C(7–7′)], 171.42 [s, C(11–11′)], 178.70 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O].31P-NMR (DMSO-d6, 298 K): δ −65.06. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1326 [100] [Os(cym)(p-curc)(PTA)]+.
O].31P-NMR (DMSO-d6, 298 K): δ −65.06. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1326 [100] [Os(cym)(p-curc)(PTA)]+.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, of p-curc), 1622 m ν(C
O, of p-curc), 1622 m ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1601 w, 1582 w,1505 vs., 1506 vs. ν(C
O), 1601 w, 1582 w,1505 vs., 1506 vs. ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C); 637 s ν(Ru-P).1H-NMR (DMSO-d6, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25 (mbr, 44 H, aliphatic chain of p-bdcurc), 1.32 (d, 6H, CH(CH3)2 of cym, 4J = 7 Hz), 1.35 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.16 (s, 3H, CH3 of cym), 2.59 (t, 4H, C(12–12′)H), 2.67 (m, 1H, CH(CH3)2 of cym), 4.09 (s, 6H, (P–CH2–N) of PTA), 4.43 (m, 6H, (N–CH2–N) of PTA), 6.05 (s, 1H, C(1)H of p-bdcurc), 6.17 d, 6.26 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.84 (d, 2H, C(3–3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.21 (d, 4 H, C(9–9′)H and C(7–7′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.50 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.76 (d, 4 H, C(10–10′)H and C(6, 6′)H of p-bdcurc, 3Jtrans = 9 Hz). 13C{1H}-NMR (DMSO-d6, 293 K): δ 14.40 [s, C(26–26′)], 16.80 (s, -CH3 of cym), 22.55, 22.57 (s, –CH(CH3)2 of cym), 24.74 [s, C(13–13′)], 28.83 [s, C(14–14′)], 29.11, 29.15, 29.30, 29.41, 29.46, 29.49 (from C15 to C25 of p-bdcurc), 30.52 (s, CH(CH3)2 of cym), 31.75, 33.96 [s, C(12–12′)], 50.11, 50.26 [s, (P–CH2–N) of PTA, J = 18 Hz], 72.17, 72.23 [s, (N–CH2–N) of PTA, J = 8 Hz], 81.57 [s, C(b–b′)], 82.67 [s, C(a–a′)], 87.97 (s, Ci′), 94.28 (s, Ci), 106.52 (s, C1), 123.06 [s, C(9–9′) and C(7–7′)], 126.69 [s, C(3–3′)], 129.81 [s, C(10–10′) and C(6–6′)], 133.10 [s, C(5–5′)], 138.92 [s, C(4–4′)], 152.20 [s, C(8–8′)], 172.04 [s, C(11–11′)], 178.62 [s, C(2–2′)
C); 637 s ν(Ru-P).1H-NMR (DMSO-d6, 293 K): δ 0.86 (t, 6H, C(26–26′)H), 1.25 (mbr, 44 H, aliphatic chain of p-bdcurc), 1.32 (d, 6H, CH(CH3)2 of cym, 4J = 7 Hz), 1.35 (m, 4H, C(14–14′)H), 1.65 (m, 4H, C(13–13′)H), 2.16 (s, 3H, CH3 of cym), 2.59 (t, 4H, C(12–12′)H), 2.67 (m, 1H, CH(CH3)2 of cym), 4.09 (s, 6H, (P–CH2–N) of PTA), 4.43 (m, 6H, (N–CH2–N) of PTA), 6.05 (s, 1H, C(1)H of p-bdcurc), 6.17 d, 6.26 d (4H, AA′BB′ system, CH3–C6H4–CH(CH3)2 of cym, 3J = 6 Hz), 6.84 (d, 2H, C(3–3′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.21 (d, 4 H, C(9–9′)H and C(7–7′)H of p-bdcurc, 3Jtrans = 9 Hz), 7.50 (d, 2H, C(4–4′)H of p-bdcurc, 3Jtrans = 16 Hz), 7.76 (d, 4 H, C(10–10′)H and C(6, 6′)H of p-bdcurc, 3Jtrans = 9 Hz). 13C{1H}-NMR (DMSO-d6, 293 K): δ 14.40 [s, C(26–26′)], 16.80 (s, -CH3 of cym), 22.55, 22.57 (s, –CH(CH3)2 of cym), 24.74 [s, C(13–13′)], 28.83 [s, C(14–14′)], 29.11, 29.15, 29.30, 29.41, 29.46, 29.49 (from C15 to C25 of p-bdcurc), 30.52 (s, CH(CH3)2 of cym), 31.75, 33.96 [s, C(12–12′)], 50.11, 50.26 [s, (P–CH2–N) of PTA, J = 18 Hz], 72.17, 72.23 [s, (N–CH2–N) of PTA, J = 8 Hz], 81.57 [s, C(b–b′)], 82.67 [s, C(a–a′)], 87.97 (s, Ci′), 94.28 (s, Ci), 106.52 (s, C1), 123.06 [s, C(9–9′) and C(7–7′)], 126.69 [s, C(3–3′)], 129.81 [s, C(10–10′) and C(6–6′)], 133.10 [s, C(5–5′)], 138.92 [s, C(4–4′)], 152.20 [s, C(8–8′)], 172.04 [s, C(11–11′)], 178.62 [s, C(2–2′)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O]. 31P-NMR (DMSO-d6, 298 K): δ −64.99. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1266.6[100] [Os(cym)(p-dbcurc)(PTA)]+.
O]. 31P-NMR (DMSO-d6, 298 K): δ −64.99. ESI-MS (+) CH3CN (m/z [relative intensity, %]): 1266.6[100] [Os(cym)(p-dbcurc)(PTA)]+.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. The authors declare no competing financial interest.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the University of Camerino (Fondo di Ateneo per la Ricerca 2018).References
- Á. Alberti, E. Riethmüller and S. Béni, J. Pharm. Biomed. Anal., 2018, 147, 13–34 CrossRef PubMed
.
- D.-J. Sun, L.-J. Zhu, Y.-Q. Zhao, Y.-Q. Zhen, L. Zhang, C.-C. Lin and L.-X. Chen, Fitoterapia, 2020, 142, 104490 CrossRef CAS PubMed
.
- A. Niranjan, S. Singh, M. Dhiman and S. K. Tewari, Anal. Lett., 2013, 46, 1069–1083 CrossRef CAS
.
- T. Esatbeyoglu, P. Huebbe, I. M. A. Ernst, D. Chin, A. E. Wagner and G. Rimbach, Angew. Chem., Int. Ed., 2012, 51, 5308–5332 CrossRef CAS PubMed
.
- K. M. Nelson, J. L. Dahlin, J. Bisson, J. Graham, G. F. Pauli and M. A. Walters, J. Med. Chem., 2017, 60, 1620–1637 CrossRef CAS PubMed
.
- A. Karthikeyan, N. Senthil and T. Min, Front. Pharmacol., 2020, 11, 487 CrossRef CAS PubMed
.
- R. K. Singh, D. Rai, D. Yadav, A. Bhargava, J. Balzarini and E. De Clercq, Eur. J. Med. Chem., 2010, 45, 1078–1086 CrossRef CAS PubMed
.
- Z. Qi, M. Wu, Y. Fu, T. Huang, T. Wang, Y. Sun, Z. Feng and C. Li, Cell. Physiol. Biochem., 2017, 44, 618–633 CrossRef PubMed
.
- S. Wanninger, V. Lorenz, A. Subhan and F. T. Edelmann, Chem. Soc. Rev., 2015, 44, 4986–5002 RSC
.
- K. M. Dale, C. I. Coleman, N. N. Henyan, J. Kluger and C. M. White, J. Am. Med. Assoc., 2006, 295, 74–80 CrossRef CAS PubMed
.
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2016, 66, 7–30 CrossRef PubMed
.
- N. J. Wheate, S. Walker, G. E. Craig and R. Oun, Dalton Trans., 2010, 39, 8113–8127 RSC
.
-
E. Alessio, Bioinorganic Medicinal Chemistry, John Wiley & Sons, Ltd, 2011 Search PubMed
.
- S. M. Meier-Menches, C. Gerner, W. Berger, C. G. Hartinger and B. K. Keppler, Chem. Soc. Rev., 2018, 47, 909–928 RSC
.
- P. Zhang and H. Huang, Dalton Trans., 2018, 47, 14841–14854 RSC
.
- R. Pettinari, F. Marchetti, C. Di Nicola and C. Pettinari, Eur. J. Inorg. Chem., 2018, 2018, 3521–3536 CrossRef CAS
.
- M. Hanif, M. V. Babak and C. G. Hartinger, Drug Discovery Today, 2014, 19, 1640–1648 CrossRef CAS PubMed
.
- A. Casini, F. Edafe, M. Erlandsson, L. Gonsalvi, A. Ciancetta, N. Re, A. Ienco, L. Messori, M. Peruzzini and P. J. Dyson, Dalton Trans., 2010, 39, 5556–5563 RSC
.
-
K. Nakamoto, 1922–2011, Infrared and Raman spectra of inorganic and coordination compounds, New York (N.Y.), Wiley, 5th edn, 1997 Search PubMed
.
- A. A. Mikhailov, V. Y. Komarov, A. S. Sukhikh, D. P. Pishchur, D. Schaniel and G. A. Kostin, New J. Chem., 2020, 44, 18014–18024 RSC
.
- V. W. W. Yam, K. H. Y. Chan, K. M. C. Wong and N. Zhu, Chem. – Eur. J., 2005, 11, 4535–4543 CrossRef PubMed
.
- R. Pettinari, F. Marchetti, F. Condello, C. Pettinari, G. Lupidi, R. Scopelliti, S. Mukhopadhyay, T. Riedel and P. J. Dyson, Organometallics, 2014, 33, 3709–3715 CrossRef CAS
.
- C. Bertucci and E. Domenici, Curr. Med. Chem., 2002, 9, 1463–1481 CrossRef CAS PubMed
.
- J. Ghuman, P. A. Zunszain, I. Petitpas, A. A. Bhattacharya, M. Otagiri and S. Curry, J. Mol. Biol., 2005, 353, 38–52 CrossRef CAS PubMed
.
- J. R. Brown, Fed. Proc., 1976, 35, 2141–2144 CAS
.
- D. E. Epps, T. J. Raub, V. Caiolfa, A. Chiari and M. Zamai, J. Pharm. Pharmacol., 1999, 51, 41–48 CrossRef CAS PubMed
.
- L. E. Gerweck, S. Vijayappa and S. Kozin, Mol. Cancer Ther., 2006, 5, 1275–1279 CrossRef CAS PubMed
.
- L. Shang, Y. Wang, J. Jiang and S. Dong, Langmuir, 2007, 23, 2714–2721 CrossRef CAS PubMed
.
- N. El Kadi, N. Taulier, J. Y. Le Huérou, M. Gindre, W. Urbach, I. Nwigwe, P. C. Kahn and M. Waks, Biophys. J., 2006, 91, 3397–3404 CrossRef CAS PubMed
.
- H. Werner and K. Zenkert, J. Organomet. Chem., 1988, 345, 151–166 CrossRef CAS
.
- M. Cuccioloni, L. Bonfili, V. Cecarini, M. Nabissi, R. Pettinari, F. Marchetti, R. Petrelli, L. Cappellacci, M. Angeletti and A. M. Eleuteri, ChemMedChem, 2020, 15, 105–113 CrossRef CAS PubMed
.
- R. Pettinari, C. Pettinari, F. Marchetti, B. W. Skelton, A. H. White, L. Bonfili, M. Cuccioloni, M. Mozzicafreddo, V. Cecarini, M. Angeletti, M. Nabissi and A. M. Eleuteri, J. Med. Chem., 2014, 57, 4532–4542 CrossRef CAS PubMed
.
- M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeerschd, E. Zurek and G. R. Hutchison, J. Cheminf., 2012, 4, 1–17 Search PubMed
.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2005, 33, W363–W367 CrossRef CAS PubMed
.
- E. Mashiach, D. Schneidman-Duhovny, N. Andrusier, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2008, 36, W229–W232 CrossRef CAS PubMed
.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed
.
- S. Sugio, A. Kashima, S. Mochizuki, M. Noda and K. Kobayashi, Protein Eng., Des. Sel., 1999, 12, 439–446 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2dt02328h |
| This journal is © The Royal Society of Chemistry 2022 |