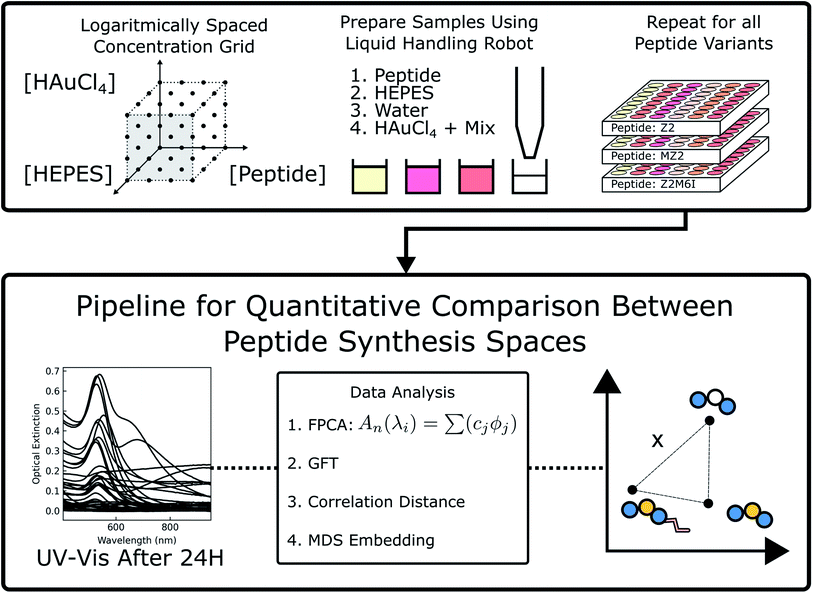
Open Access Article
Open Access ArticleMultivariate analysis of peptide-driven nucleation and growth of Au nanoparticles†
Kacper J.
Lachowski
 *ac,
Kiran
Vaddi
*ac,
Kiran
Vaddi
 a,
Nada Y.
Naser
a,
Nada Y.
Naser
 a,
François
Baneyx
a,
François
Baneyx
 a and
Lilo D.
Pozzo
a and
Lilo D.
Pozzo
 *abc
*abc
aDepartment of Chemical Engineering, University of Washington, Seattle, WA, USA. E-mail: lachok@uw.edu; dpozzo@uw.edu
bDepartment of Material Science and Engineering, University of Washington, Seattle, WA, USA
cMolecular Engineering and Sciences Institute, University of Washington, Seattle, WA, USA
First published on 1st June 2022
Abstract
In this work we synthesize gold nanostructures using a liquid-handling robot and present new data analysis methods to quantitatively compare the effect of peptides and peptide modifications on gold nanoparticle synthesis outcomes. The peptides used were gold binding peptides Z2 and AG3, as well as five Z2 variants obtained by sequence modification or conjugation of a lipid tail. Four different concentrations of peptide (0, 0.04, 0.0894, and 0.2 mM), reducing agent (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, HEPES; 0, 0.01, 0.1, and 1 mM), and precursor (HAuCl4; 0.04, 0.0684, 0.117, and 0.2 mM) were used to synthesize 64 different reagent combinations for each of the seven peptides. Each sample was characterized using UV-Vis spectroscopy, which serves as a proxy for changes in nanoparticle structure, and enabled comparisons of how peptide modifications and reagent conditions affect synthesis outcomes. We then used functional data analysis to extract a pairwise signal correlation distance between each of the peptides. The signal correlation distance quantified how different the set of 64 spectra, and therefore the synthesis outcomes, was from one peptide to the other. Spherical nanoparticles synthesized in the presence of peptides were typically 10 to 50 nm in diameter, but the size and shape of nanoparticles varied with peptide properties and reagent conditions. We show that substitution of methionine for isoleucine in Z2 has a profound impact on synthesis outcomes when compared with conjugation of a lipid tail. Electron microscopy and ultra-small angle X-ray scattering were used to corroborate our conclusions from spectroscopy experiments by directly characterizing the structure of a smaller set of samples. Scanning electron microscopy revealed interconnected plate-like structures in samples prepared with a Z2 variant with all methionines substituted by isoleucine. Z2 peptides modified with a lipid tail on the other hand, formed nanoparticles which were more colloidally stable than those prepared with non-lipidated peptides, contained no plate-like particles, and whose size depended on the ratio of peptide to precursor.
1 Introduction
The structure and composition of inorganic colloidal materials enable several unique properties and functions. Structure encompasses the morphology, characteristic dimensions, polydispersity, and crystal structure of nanoscale materials. Control of thermodynamic and kinetic processes during synthesis is required to achieve desired structural features. Particles which are composed of metals (e.g. Au, Ag, Pt, Cu) and have nanoscale structures exhibit localized surface plasmon resonance (LSPR) in response to light.1 LSPR has been used in chemical sensing, enhanced Raman spectroscopy, light harvesting, catalysis, medical imaging, and other applications.2 The particle's structure is directly related to its LSPR response. Therefore, simple optical extinction measurements can provide information about particle structure within a sample. In the simplest case of a spherical nanoparticle, the magnitude and wavelength of the LSPR band is related to the diameter of the particle.1 Different particle morphologies such as nanorods or nanotriangles allow for an additional mode of plasmonic resonance. Proximity to other particles can also modify the optical response through plasmonic coupling. As the size of features directly influences the optical properties of plasmonic particles, monodispersity is a desirable trait to obtain maximum control over the optical response. The crystal structure of metallic nanoparticles may be generally described as face-centered cubic (fcc), but the exact structural order at the surface can vary depending on the method of synthesis. This is important because the order at the nanoparticle surface is related to its catalytic properties.3Efforts at improving control over nanoparticle structure and spatial organization have often taken inspiration from biomineralization. Specifically, researchers seek to emulate the mild synthesis conditions, use of molecules with molecular recognition, and structural complexity that nature has achieved. Sequence-defined molecules such as DNA, peptides, and peptoids are attractive choices for bio-inspired synthesis approaches and have been used to control the nucleation, growth, and self-assembly of inorganic materials.4–6 Modifying the sequence of a molecule often has a significant impact on how the molecule interacts with inorganic precursors and particle surfaces.7,8 In turn, these modifications can lead to changes in nanoparticle formation mechanisms, colloidal stability, reaction kinetics, and ultimately the function of the synthesized material.3
Research into understanding molecular design rules for peptide mediated Au nanoparticle research has continued to receive significant attention due to extensive potential applications for plasmonic nanoparticles. Au nanoparticle synthesis also serves as a good model system for method development. Following several discoveries of peptides with strong binding affinities to Au surfaces using biopanning methods, Tan et al.9 performed a systematic study of each naturally occurring amino acid's contribution to Au binding and reduction. The authors synthesized peptides with each of the amino acids interdigitated with histidine. The interdigitated peptides were then used in experiments to assign scores for each amino acid's Au binding and reducing ability. However, one of the limitations of this work is related to one of the motivations for undertaking it in the first place – the order and identity of amino acids in the sequence can lead to surprising changes in peptide interaction with particle surfaces and precursors. Moreover, as was shown by Munro et al.,10 varying solution and reagent conditions will modify how a peptide performs in nanoparticle synthesis. Therefore, design rules obtained using a particular and narrow set of experimental conditions may be limited in their applicability to other experimental conditions. Munro et al. studied the effects of pH, ionic strength, and ionic composition on the formation of Au nanoparticles using the peptide AuBP1. However, unlike AuBP1, not all peptides which have a strong binding affinity towards gold surfaces also have an appreciable capability of reducing Au(III), and therefore require the use of a reducing agent. Insights into molecular design rules can be generated by studying sequence rearrangement, amino acid substitutions or additions, and by comparison of known peptides to one another. One example of the latter is the work of Li et al., who compared the use of several Au binding peptides on the formation of Au nanoparticles at fixed molar concentrations of peptide, HAuCl4, and a reducing agent.11 The researchers noted that each peptide's theoretical isoelectric point was a good predictor of zeta potential sign, but no peptide feature was identified as a good predictor of gold nanoparticle size or polydispersity. Finally, there are also several examples in which conjugation of a lipid tail to the peptide N-terminus resulted in the peptide modifying the synthesis mechanism by acting as a template for nanoparticle growth and assembly.12–17
In this paper, we seek to address the lack of systematic experimental and data analysis methods for studying connections between peptide modifications (amino acid content, sequence order, lipid tail conjugation etc.) and synthesis outcomes within a large scope of experimental conditions. There has been a surge of interest and breakthroughs in applying high-throughput experimentation, design of experiments, and new data analysis methods to accelerate colloidal synthesis research. Often, the pursued objective is an optimization of a particular nanoparticle structure or characterization of phases in the experimental design space.18,19 In contrast, our work is concerned with using high-throughput experimentation and data analysis to understand the role of peptide modifications on Au nanoparticle synthesis. We demonstrated our approach by using the Au binding peptide Z2 (RMRMKMK)20 and the mild reducing and buffering agent HEPES. Five additional Z2 variants were obtained to study the influence of different modifications to the peptide. The modifications chosen included substitution of methionine residues for isoleucine, the addition of lipid tails with two different lengths, and the addition of a lipid tail accompanied by a reversal of the sequence. A different Au binding peptide, AG3 (AYSSGAPPMPPF), was also included for comparison. Each of the peptides was used to synthesize a set of 64 samples with varying concentrations of peptide, HAuCl4, and HEPES. Each sample was then characterized using UV-Vis spectroscopy. The spectra serve as a fast proxy method for characterizing the nanoparticle structure. That is, although a given spectrum is not a unique representation of a particular nanoparticle structure, we can identify how spectra and therefore structures change as a function of reagent concentrations and peptide identity. We propose an application of functional data analysis methods to the spectral characterizations of plasmonic structures to analyze the high-throughput experimentation data obtained. Finally, we corroborate these differences in nanoparticle synthesis outcomes for each Z2 variant by using a combination of electron microscopy and small-angle scattering, and discuss the significance of changes to peptide chemistry in the context of a large experimental design space.
2 Experimental synthesis and characterization
2.1 Materials
Gold(III) chloride trihydrate (HAuCl4·3H O3H2O) (≥99.9%) was obtained from Sigma-Aldrich (St. Louis, MO, USA), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) was obtained from EMD Chemicals (Gibbstown, NJ, USA), and all peptides were acquired from Biomatik Corporation (Cambridge, ON, Canada). Peptides were purified using HPLC to >95% purity by the manufacturer and the lyophilized trifluoroacetate salt was used for experiments without further purification.2.2 Sample preparation
Stocks of HEPES, HAuCl4, and peptide were prepared by diluting the reagents with ultra pure water obtained from a Direct-Q 3 UV water purification system with a resistivity of 18.2 MΩ (Millipore Corporation, Bedford, MA, USA). The concentrations of peptide, HEPES, and HAuCl4 stock used were: 5 × 10−2 wt% (ca. 0.5 mM), 1 × 10−1 wt% or 1 × 10−2 wt% (ca. 4.18 or 0.418 mM), and 1 × 10−1 wt% (ca. 2.93 mM), respectively. Synthesis of gold nanoparticles was carried out by pipetting the stocks of peptide, HEPES, additional water, and finally chloroauric acid into 48-well Falcon microplates (Corning, Corning, NY, USA) using an Opentrons (Brooklyn, NY, USA) OT-2 liquid handling robot. Each solution was pipetted up and down, the microplates were covered, and the samples were left to react at room temperature. The Python script used to control the liquid handling robot has been shared online.222.3 UV-Vis sample characterization
Samples were measured 24 hours after synthesis using an Epoch 2 microplate spectrophotometer (BioTek, Winooski, VT, USA). Background correction was performed by subtracting the absorbance of water. Details of functional data analysis are available in the ESI.†2.4 Electron microscopy
For imaging on the scanning electron microscope (SEM), 5 μL aliquots of samples were deposited on a silicon wafer and air dried overnight. Images were acquired on a FEI (Hillsboro, OR, USA) Sirion XL30 SEM which was typically operated at 5 kV and with a spot size of 3 using through-lens detection and ultra-high resolution mode. Transmission electron microscopy (TEM) samples were prepared by diluting the sample ten-fold and then casting 7 μL on a lacey carbon coated film, 200 mesh, copper grids purchased from Electron Microscopy Sciences. Grids were dried in air and imaged on a FEI Tecnai F20 at 200 kV.2.5 USAXS measurements and data analysis
Samples were loaded in a custom-made 48-well plate assembly between two Kapton windows for a total sample thickness of 10 mm.23 USAXS data were collected on the 9-ID-C beamline at the Advanced Photon Source at Argonne National Laboratory using 21 keV X-rays.24 The USAXS data collection time was 90 seconds, beam size was 0.8 mm by 0.8 mm, and the flux density was approximately 5 × 1012 photons per second mm−2. Data were reduced using the standard data reduction protocols and software provided by the beamline (Indra).Mathematical desmearing based on the Lake method25 was performed using the provided Igor macros26 to remove instrumental slit smearing effects in preparation for data fitting. The flat background at high-q was then averaged and subtracted from the data before performing automated Guinier analysis. The latter is an estimate of the radius of gyration Rg using the Guinier approximation 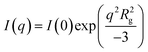 . Automated Guinier analysis was performed using AUTORG.27
. Automated Guinier analysis was performed using AUTORG.27
3 Results and discussion
3.1 Effects of peptide on Au nanoparticle synthesis
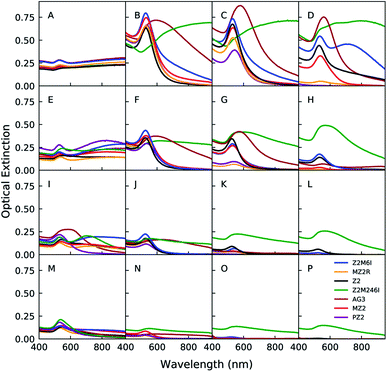
Au nanoparticles were synthesized in the presence of one of the 7 peptides listed in Table 1 at 64 different reaction conditions represented by the grid of points in Fig. 1. The concentrations of peptide, HAuCl4, and HEPES were sampled at logarithmically spaced intervals. Sample conditions with no peptide or HEPES were also included in this space. We present a portion of the UV-Vis spectra in Fig. 2 in which each row from top to bottom corresponds to a decrease in the HAuCl4 concentration level, each column from left to right represents an increasing level of peptide concentration, and the HEPES concentration is fixed at 1 mM (see (ESI) Fig. S1–S3† for other HEPES levels). These spectra can be qualitatively analyzed as a proxy for nanoparticle structure as a function of reagent conditions and peptide identity. Additionally, absorption at 400 nm can be used to determine the concentration of Au(0) in situ.28 However, as the cited work points out, the extinction coefficient of Au(0) at 400 nm depends on particle shape, size distribution, and the particle's surface chemistry (e.g. peptide or HEPES). Other contributions to optical extinction at this wavelength may include Au clusters and turbidity. Nevertheless, optical extinction at 400 nm is useful for identifying major differences in Au reduction when analyzing UV-Vis spectra. The leftmost column in Fig. 2 corresponds to the zero peptide condition and is color coded to denote the peptide that was used in the same batch of reactions. HEPES is a mild reducing and buffering agent which forms nanoplates, spherical nanoparticles, or nanostars in the presence of HAuCl4.29–31 HEPES stabilizes nanoparticles against aggregation through its adsorption to the particle surface.32,33 As the HAuCl4 concentration increases from 40 μM to 200 μM in Fig. 2(M and A), the ratio of HEPES![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HAuCl4 decreases. This results in the appearance of a second peak and broad extinction at higher wavelengths. This is consistent with formation of nanoplates and particle aggregates as seen in Fig. 7 D.
HAuCl4 decreases. This results in the appearance of a second peak and broad extinction at higher wavelengths. This is consistent with formation of nanoplates and particle aggregates as seen in Fig. 7 D.
The addition of peptide causes significant changes in the spectra for all HAuCl4 levels relative to the no peptide condition. However, the exact changes in spectra vary depending on the peptide identity and concentration. In order to discuss the potential effects of peptides on Au nanoparticle synthesis outcomes, we conceptualized the different ways that peptides can interact with Au precursors and nanoparticles as peptide presentation. Peptide presentation includes, but is not limited to, peptide sequence order, amino acid identity, lipid tail conjugation, charge of residues, solution conditions (pH, ionic strength and identity), proximity of peptide to other peptides, and peptide self-assembly. Peptide presentation in the context of peptide mediated Au nanoparticle synthesis has often been studied by modifying the peptide chemistry – i.e. sequence order, identity, chemical modification, or conjugation of lipid tails to the N-terminus.8,12,13 However, we believe that additional insights and molecular design rules for peptide mediated inorganic nanoparticle synthesis may be identified by sampling the effects of modifications to peptides in a large set of reaction conditions with the aid of high-throughput experimentation and new data analysis approaches. We identified Z2 (RMRMKMK) as a good candidate peptide for developing methods to study how peptide presentation affects nanoparticle synthesis. We selected Z2 because it has a small number of unique amino acids and lacks secondary structure. Unlike many Au binding peptides in the literature which are identified through biopanning methods, Z2 was rationally designed by combining residues with high binding affinities to Au surfaces.20 Later molecular dynamics simulations concluded that the methionine residues in Z2 act as anchor residues, which the authors define as having a strong and persistent affinity for the simulated Au(111) surface.34 Additionally, methionine is known to complex with AuCl−4 which would decrease the reduction rate of Au(III).9,35 We also chose to include AG3 (AYSSGAPPMPPF) in our study in order investigate whether our approach can yield new insights into the similarities and differences of Z2 and AG3 in the context of Au nanoparticle synthesis and stabilization beyond those that have been discussed.11
We observed that at a fixed concentration of HAuCl4 and HEPES, high concentrations of Z2 and lipidated Z2 variants led to a decrease in optical extinction at the LSPR peak and at 400 nm. Both of these observations suggest that the concentration of Au(0) decreases as the concentration of Z2 and lipidated Z2 variants is increased.28 We hypothesized that the complexation of methionine with HAuCl4 is responsible for the decrease in formation of Au nanoparticles after 24 hours. Therefore, we synthesized Z2M6I (RMRMKIK) and Z2M246I (RIRIKIK) to study the effect of partially or completely substituting methionine for isoleucine on peptide-mediated growth of Au nanoparticles. The rationale for choosing isoleucine is that it has similar physicochemical properties to methionine,36 but is not expected to bind to Au surfaces9,20 nor complex with HAuCl4. With the exception of Z2M246I, a low concentration of any Z2 variant generally led to more prominent LSPR peaks and lower optical extinction at higher wavelengths when compared to the no peptide condition at the same concentration of HEPES and HAuCl4. An increase in prominence of the LSPR peak (ca. 540 nm) is associated with increased formation of spherical nanoparticles and a decrease in extinction at higher wavelengths is consistent with a decrease in aggregation and formation of nanoplates. All samples synthesized in the presence of Z2M246I on the other hand, had significant and broad optical extinction at wavelengths higher than the typical peak position of an LSPR peak associated with spherical Au nanoparticles. Additionally, samples prepared with Z2M246I were visually distinct from samples prepared with the other Z2 variants in this study. The former sedimented and had a green or black hue, which indicated the formation of large particles or aggregates. Whereas the latter were typically pink, purple, or red and remained dispersed. We also observed that increasing the concentration of Z2M6I or Z2M246I did not lead to a significant decrease in optical extinction at 400 nm. Therefore we concluded that the structures formed in the presence of Z2M246I should be most distinct from others in the study, and that complexation of HAuCl4 with methionine was an important factor in controlling the proportion of Au(III) that was reduced to Au(0) after 24 hours.
Several previous works have investigated the use of a lipid tail conjugated to AG3 to control the in situ formation of Au nanoparticle decorated peptide nanostructures (e.g. micelles, fibrils).12–17 The addition of an alkyl tail to a water soluble peptide makes it amphiphilic, and therefore capable of self-assembling in water. Once self-assembled, peptide nanostructures may act as a template for nucleation or adsorption of Au nanoparticles. Most uses of amphiphilic peptides in the synthesis of Au nanoparticles use just one or a limited set of reaction conditions to draw conclusions about the effect of peptide modifications to Au nanoparticle synthesis outcomes. We were interested in gaining a deeper understanding of the role of peptide lipidation on Au nanoparticle synthesis by using amphiphilic peptides in a large range of reaction conditions. Increasing the length of lipidated peptide tails is one method for decreasing the critical micelle concentration. However, peptide self-assembly can also be affected by the ratio of precursor to peptide due to interactions between peptide residues and the precursor. Ionic interactions between AuCl−4 and positively charged residues and complexation may lead to changes in the critical micelle concentration and the structure of the peptide–precursor assembly. Therefore, in addition to making substitutions to the Z2 sequence, we also obtained three variants which were modified at the N-terminus with either a palmitic acid tail (PZ2), myristic acid tail (MZ2), or a myristic acid tail with a reversed sequence (MZ2R). SAXS studies of MZ2 dissolved at a concentration of 4.2 mM revealed extended structures which could be fit with a cylindrical model, and are therefore likely to be cylindrical micelles. However, this concentration is much larger than 0.2 mM, the highest concentration used in Au nanoparticle synthesis experiments. We used pyrene fluorescence to estimate the critical micelle concentration of MZ2, MZ2R, and PZ2 to be 0.47 mM, 0.33 mM, and 0.24 mM, respectively (see ESI Fig. S4†).37 Note that the conditions used in this work are all below the critical micelle concentration. Therefore, the study is limited to the interaction of dissolved peptides with particle surfaces, and does not include the growth of Au over self-assembled peptides. In the case of Z2, reversing the sequence maintains the position of the methionines but swaps the positions of lysines and arginines with one another. The physicochemical similarity of arginine and lysine may suggest that swapping their positions in a sequence should lead to negligible changes in the peptide's affinity to Au surfaces and Au nanoparticle synthesis outcomes. However, unlike lysine, arginine has been classified as an anchor residue in sequences other than Z2, and the first arginine in the Z2 sequence was close to being classified as a contact residue by Tang et al.34 Therefore, we were interested in whether changing the position of lysine and arginine relative to the lipid tail would lead to an appreciable change in Au nanoparticle synthesis outcomes.
The spectra of samples prepared in the presence of lipidated peptides were generally similar to that of the unmodified Z2. However, unlike Z2, the lipidated variants led to a more significant decrease in overall optical extinction as the peptide concentration increased. Additionally, unlike the non-lipidated peptides in this study, an increase in optical extinction at higher wavelengths for high peptide concentrations was not observed for the lipidated Z2 variants. Had the lipidated peptides led to structural organization of nanoparticles, a red-shift and peak broadening should be observed due to plasmonic coupling.1 Therefore, we interpreted the lack of optical extinction at higher wavelengths as an indication that the lipidated Z2 variants were unlikely to be organizing Au nanoparticles. However, we expected that the presence of HAuCl4 should reduce the concentration of lipidated peptide required for its self assembly. As will be discussed in a later section, peptide–precursor structures do exist in the sample, but these do not lead to organized peptide nanostructures decorated by nanoparticles. The decrease in optical extinction for lipidated peptides relative to non-lipidated peptides is likely due to a decrease in the Au reduction rate. A decrease in Au reduction rate could be caused by the peptide–precursor interactions forming structures that inhibit access of HEPES to Au species. This explanation is supported by the fact that samples prepared with PZ2 had lower optical extinction than those prepared with MZ2. The longer length of the palmitic acid tail should lead to stronger association between peptides, and therefore more strongly inhibit reactions of HEPES with Au species. The spectra of Au nanoparticles formed in the presence of MZ2R were comparable to those formed with either MZ2 or PZ2, with the distinction that MZ2R inhibited Au nanoparticle formation to a greater extent than MZ2, and less than PZ2. This observation is in line with the order of critical micelle concentrations estimated for the three peptides. Overall, changing the positions of arginine and lysine in the sequence did not lead to a significant change in the peptide's function in modulating Au nanoparticle synthesis. Therefore, the weak impact of this modification should be reflected in later analyses which use the full synthesis space to make pairwise comparisons between peptides.
The spectra from samples synthesized in the presence of AG3 show clear LSPR peaks in most conditions, but these are red-shifted and broadened relative to those synthesized in the presence of Z2. These spectral changes are consistent with formation of larger particles and aggregation. Colloidal instability of AG3 capped particles has been correlated with acidic conditions, the ratio of reducing agent to precursor, and the ratio of peptide to precursor.8 In contrast to the conditions used in this work, other publications which use HEPES, the AG3 sequence, and modified AG3 variants, utilize higher HEPES concentrations such as 0.1 M HEPES.15 The differences between synthesis conditions in which Z2 and AG3 form colloidally stable nanoparticles reveal high dissimilarity between the two sequences.
The present work is focused on measurement of spectra at just one time point (24 h) in order to connect peptide properties and reaction conditions with nanoparticle structure. Nevertheless, we also include measurements of the optical extinction at 400 nm over time in nanoparticle syntheses carried out in the presence of Z2, Z2M246I, or MZ2. These measurements demonstrate how peptide modifications affected the Au reduction rate (see ESI Fig. S5†). The slower reduction rate in the presence of MZ2 and the decrease in LSPR features in spectra prepared with high peptide concentrations after 24 hours (e.g.Fig. 2D) both demonstrate that peptide lipidation has a significant impact on nanoparticle growth dynamics. These differences in reduction rates are interesting given the similarity of spectra obtained with these reagent concentrations after 24 hours in the case of Z2 and MZ2 (Fig. 2B). The similarity in reduction rates between Z2 and Z2M246I on the other hand show that, at least in these conditions, the presence of methionine in the peptide does not appear to have an effect on the Au reduction rate. However, the connection between peptide properties, reaction conditions, and nanoparticle growth dynamics will require extensive in situ UV-Vis spectroscopy and SAXS studies in the future.
3.2 Comparison of peptide mediated nanostructure formation using the full parameter space
Each peptide's synthesis space, consisting of the 4 × 4 × 4 set of reagent concentrations and their respective spectra, is a rich dataset of nanoparticle formation outcomes. We posit that using the full dataset would strengthen our analysis of each peptide's effect on nanoparticle formation. To accomplish this, we use each peptide's synthesis space in an unsupervised learning approach to study correlations between each peptide. Comparing the synthesis space of peptides would then require comparing the set of spectra along with their sample concentrations. One crucial aspect of this comparison is the way one represents the synthesis space for data analysis purposes. To correlate different synthesis spaces of peptide sequences, we first parameterize the sampled synthesis space by a graph (i.e. a network of first-order connections) using the different component concentrations as nodes and edges obtained using a k-nearest neighbors (k-NN) approach. The above parameterization results in a natural data representation for the synthesis space via signals on the graphs where the signal corresponds to the UV-Vis characterization of the structure of Au nanoparticles. The graph signal approach also allows us to further compress the signals and delineate regions of synthesis spaces for comparison using a Fourier transform38,39 that has been well developed for graph signals with readily available open-source code.40 For example, for a synthesis space comprising of concentration of peptide, buffer, and HAuCl4, each sample (a node in the graph) in the synthesis space can be considered as a point in a three-dimensional space. Euclidean distance between the sample concentrations can then be used to obtain edges in the graph using a k-nearest neighbors construction. For each sample in the synthesis space, its characteristic UV-Vis response is collected between the wavelength region of 400–900 nm with 5 nm increments. Given that response of each sample (i.e. the signal at each node of the graph) is a function x(λ) (mapping a given wavelength to an absorption intensity) rather than an array of values typically considered in data analysis, we use a principal component analysis (PCA) adapted for functional data to obtain vector-like descriptors for each sample.41,42 This choice of representation allows us to: (a) encode the dependency between responses at different wavelengths via an inner product using integrals; (b) learn underlying basis functions to represent the functional variations such as peak position and broadening. The approach referred to as FPCA and the graph based signal processing used in this paper are described in the ESI.† In the next two subsections, we describe the application of these methods to UV-Vis spectroscopy data collected from the high-throughput sampling described earlier.3.3 Application of FPCA to UV-vis spectroscopy
In this section, we demonstrate application of functional data analysis (FDA) to spectral data collected from UV-Vis spectroscopy (see Section 1.1 in ESI† for more details). FDA and related models therein (such as FPCA) are widely used in the (applied) statistics community41,42 for analyzing time series data, shape analysis of curves and data of input/output pair type. However, to our knowledge, the techniques of FDA including the choice of function spaces as representations have not been previously applied for spectral data such as UV-Vis. Following the notation in the ESI,† the UV-Vis spectroscopy data is denoted as x(λ) where λ is a wavelength. Given a set of UV-Vis spectroscopy data, we obtain a matrix xij (where i is a sample and j is a wavelength), and compute its principal components highlighting types of variations present. For seven peptides with 64 samples in each synthesis space, we have a total of N = 448 samples. We apply FPCA to all the UV-Vis samples collected for our study and observe that ≈95% of the functional variance is captured using only 8-components. A qualitative understanding of each FPCA component can be obtained by generating spectra while varying the component using linear generative model described in Section 1.1 of ESI.† In Fig. 3, we plot spectra generated from each one of four components where we sample one standard deviation away from the mean (dotted black curve) in positive (blue curve) and negative direction (red curve). We observe that the obtained FPCA components are informative in the following sense: (i) moving one standard deviation away from the mean along PC1 direction changes the peak from being non-existent to a sharp peak thus signifying the primary signal regarding formation of gold nanoparticles. (ii) Similarly, PC2 can be associated with both a peak shift and a change in broad optical extinction at higher wavelengths, which could indicate changes in particle aggregation. (iii) PC3 corresponds to a shift along the intensity axis which we associate with changes in HAuCl4 concentration under the conditions that forms nanoparticles. (iv) We also observe that rest of components are redundant in terms of the information they encode at the spectra level. For example, PC4 can be considered as mirroring the features encoded in PC2 while PC5 is a scaled copy of PC3. Further more, PC6 can be associated with a combination of PC2 and PC3; PC7 and 8 provide minimal changes to the spectra when generated from the model. The above qualitative method is used to further decrease the components to just four given by the top four eigenvalues. For the rest of this paper, we represent each UV-Vis spectra x(λ) using its first four principal component scores fi, i = [1, 2, 3, 4] resulting in a vector representation of a signal on graph parameterization of grid-like synthesis space (see Section 1.2 in ESI† for more details on this parameterization).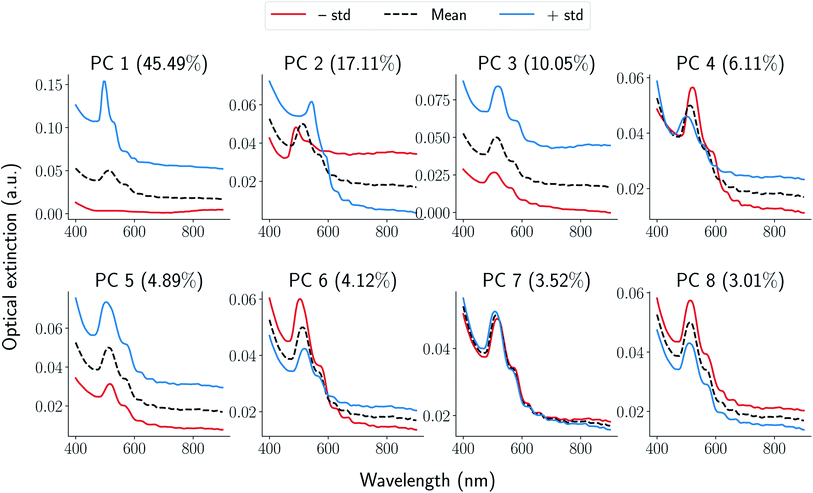 | ||
| Fig. 3 Generated samples from eqn (1) in ESI† Each panel corresponds to a principal component annotated on the top along with corresponding explained variance. For each component, the mean of the functions is denoted using a black-dotted curve while positive and negative standard deviations along the component are denoted using blue and red curves respectively. | ||
We compare synthesis space of peptides using a notion of similarity between the signals obtained over the synthesis space. For UV-Vis spectra collected over the synthesis space, the signals can now be replaced by their corresponding FPCA representation resulting in a vector descriptor for each node in the graph parameterization. Vector signals on graphs are easier to process mathematically using the linear algebra techniques extensively developed for graphs.38,39 However, it is not straightforward as to how to define a similarity function between two graph signals. Defining a graph G = (V, E) as set of vertices V, edges E associated with a signal 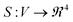 , we need a way to compare two signals Si, Sj. One approach to achieve this is to compress the graph signal into a vector by a change of basis using a graph Fourier transform (GFT) described in Section 1.2 of ESI.† GFT allows us to (a) compress the graph signal into a vector; (b) remove noise in the signal; making it a robust choice of data representation.
, we need a way to compare two signals Si, Sj. One approach to achieve this is to compress the graph signal into a vector by a change of basis using a graph Fourier transform (GFT) described in Section 1.2 of ESI.† GFT allows us to (a) compress the graph signal into a vector; (b) remove noise in the signal; making it a robust choice of data representation.
3.4 Application of GFT to high-throughput experimentation data
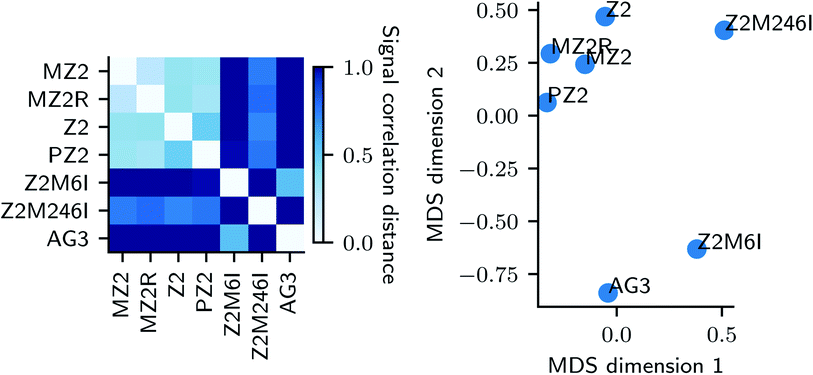
In this section, we describe an application of GFT to represent the data collected for each peptide. Suppose G = (V, E) is a graph constructed from k-NN approach described earlier. We represent each sample's UV-Vis spectra using a vector of FPCA score values and obtain a graph signal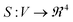 at each node. We then use GFT on S and retain the lowest 70% of frequencies resulting in Ŝ. Given a pair of peptides i, j, we compare their synthesis spaces by computing distance between Ŝi,Ŝj. A similarity measure such as the correlation distance (see eqn (3) in ESI†) is used to quantify similarities between synthesis space i.e. types of Au nanoparticles formed when we vary the concentrations in synthesis space. Fig. 4 is the result of application of GFT represented signals Ŝ for multi-variate analysis of peptide data.
at each node. We then use GFT on S and retain the lowest 70% of frequencies resulting in Ŝ. Given a pair of peptides i, j, we compare their synthesis spaces by computing distance between Ŝi,Ŝj. A similarity measure such as the correlation distance (see eqn (3) in ESI†) is used to quantify similarities between synthesis space i.e. types of Au nanoparticles formed when we vary the concentrations in synthesis space. Fig. 4 is the result of application of GFT represented signals Ŝ for multi-variate analysis of peptide data.
On the left panel of Fig. 4, we used a correlation distance (eqn (3) in ESI†) between compressed graph signal of pairs of peptides and represented the similarities using a heatmap plot. The similarity between synthesis spaces is depicted using a continuous color gradient light blue (highly similar) and dark blue (highly dissimilar). A metric embedding using multi-dimensional scaling43,44 of the correlation distance is shown on right. We observe that changes to our base peptide Z2 result in two distinct modes of synthesis space. Namely, changes to the type of alkyl chain (MZ2, PZ2) and reversing the sequencing (MZ2R) result in very similar synthesis space while changes to actual peptide sequence (AG3, Z2M6I, Z2M246I) result in a diverse synthesis space as quantified by distances in the metric embedding shown in the right panel of Fig. 4 where the two groups are clearly separated.
3.5 Structural characterization using electron microscopy and USAXS
The correlation distances describe how each pair of peptides differs in the effect it has on Au nanoparticle synthesis across the whole synthesis space. However, it is important to validate the conclusions we drew from UV-Vis spectra by more direct structural characterization methods.We repeated the synthesis of Au nanoparticles in the presence of each of the Z2 variants in a slightly modified synthesis space (6 × 4 × 2, peptide × HAuCl4 × HEPES). The bounds of this synthesis space were the same as those used for UV-Vis characterization, but the number of peptide concentrations studied was increased to more finely assess the impact of peptides on synthesis. The HEPES concentrations was 0 or 1 mM to study peptide–precursor structure on its own and because this concentration of HEPES led to more colloidally stable samples than those formed at lower concentrations, respectively. The USAXS scattering profiles presented in Fig. 5 show the results for all peptide Z2 variants. Even in the absence of HEPES, most of the scattering profiles show a Guinier region. This is consistent with the formation of hypothesized co-assembled peptide–precursor structures.
 | ||
| Fig. 5 A subset of smeared USAXS data. Remaining data is available in the ESI (Fig S10 and S11†). Concentrations of peptide, HAuCl4, and HEPES: (A) 0.04, 0.2, 0 mM, (B) 0.2, 0.2, 0 mM, (C) 0.04, 0.2, 1 mM, (D) 0.2, 0.2, 1 mM. Note that while most samples show a Guinier region in the absence of HEPES, non-lipidated peptides in the presence of HEPES and HAuCl4 do not have a Guinier region and may be colloidally unstable. | ||
The scattering profiles of all Z2 variants in the presence of HAuCl4 and no HEPES are presented in Fig. 5A and B. The scattering profiles of Z2M246I were similar to that of other Z2 variants which had methionine. Therefore, we concluded that the studied peptide variants co-assemble with the precursor due to ionic interactions between the positively charged residues (arginine and lysine) and AuCl−4, and not due to methionine-AuCl−4 complexation.
In the presence of 1 mM HEPES the scattering profiles of lipidated peptides more closely resembled one another than any of the non-lipidated variants. This can be seen in Fig. 5C and D, in which samples which were lipidated show a Guinier region and do not have signs of colloidal instability, unlike samples prepared with non-lipidated peptides. The scattering profiles of non-lipidated peptides in Fig. 5D indicate aggregation or the presence of large particles due to the upturn in scattering at low-q. The Guinier region of samples prepared with lipidated peptides in 1 mM HEPES was fit using AUTORG.27 The results are presented using a log–log plot of Rg as a function of the peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HAuCl4 molar ratio in Fig. 6. Samples which could not be fit with a Guinier region are shown as black markers with a Rg of 10 Å. All three lipidated Z2 variants show an increase in Rg as a function of peptide
HAuCl4 molar ratio in Fig. 6. Samples which could not be fit with a Guinier region are shown as black markers with a Rg of 10 Å. All three lipidated Z2 variants show an increase in Rg as a function of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HAuCl4 ratio. The majority of samples prepared with non-lipidated peptides and 1 mM HEPES did not contain a Guinier region due to aggregation or sedimentation of larger particles and could therefore not be analyzed using AUTORG. The similarity of the scattering profiles of samples prepared with lipidated peptides is in line with the analysis of the signal correlation distance of peptide synthesis spaces. Drawing conclusions from how non-lipidated peptides affected the formation of Au nanoparticles across the synthesis space is precluded by colloidal instability. Nevertheless, future high-throughput experimentation studies of nanoparticle synthesis may utilize small-angle scattering for systems which lack an easy to measure proxy of structure (i.e. LSPR). Fitting of scattering profiles provides a low-dimensional descriptor of the structure (the fit parameters) which can be used to optimize a particular structure as a function of experimental conditions.45 Alternatively, these model fitting results could be used to compare how sequence-defined molecules or other additives impact nanoparticle synthesis. However, small-angle scattering data fitting is an ill-posed problem and challenges with automated data analysis are continuing to be studied. Challenges that preclude us from applying more complex and automated data fitting to our dataset are the presence of mixed particle morphologies and sample sedimentation during longer than normal measurement times when queuing large numbers of samples.
HAuCl4 ratio. The majority of samples prepared with non-lipidated peptides and 1 mM HEPES did not contain a Guinier region due to aggregation or sedimentation of larger particles and could therefore not be analyzed using AUTORG. The similarity of the scattering profiles of samples prepared with lipidated peptides is in line with the analysis of the signal correlation distance of peptide synthesis spaces. Drawing conclusions from how non-lipidated peptides affected the formation of Au nanoparticles across the synthesis space is precluded by colloidal instability. Nevertheless, future high-throughput experimentation studies of nanoparticle synthesis may utilize small-angle scattering for systems which lack an easy to measure proxy of structure (i.e. LSPR). Fitting of scattering profiles provides a low-dimensional descriptor of the structure (the fit parameters) which can be used to optimize a particular structure as a function of experimental conditions.45 Alternatively, these model fitting results could be used to compare how sequence-defined molecules or other additives impact nanoparticle synthesis. However, small-angle scattering data fitting is an ill-posed problem and challenges with automated data analysis are continuing to be studied. Challenges that preclude us from applying more complex and automated data fitting to our dataset are the presence of mixed particle morphologies and sample sedimentation during longer than normal measurement times when queuing large numbers of samples.
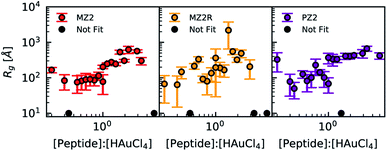 | ||
Fig. 6 Radius of gyration results obtained from AUTORG plotted as a function of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HAuCl4 molar ratio for all lipidated peptides and a HEPES concentration of 1 mM. Error bars indicate standard deviation. Profiles which did not contain a Guinier region that could be fitted were marked using black and plotted at a fixed Rg of 10 Å. Rg fit results for all peptides in the absence of HEPES are available in the ESI (Fig. S12 and S13†). Data of non-lipidated peptides in the presence of 1 mM HEPES is not presented because the majority of samples did not have a Guinier region. HAuCl4 molar ratio for all lipidated peptides and a HEPES concentration of 1 mM. Error bars indicate standard deviation. Profiles which did not contain a Guinier region that could be fitted were marked using black and plotted at a fixed Rg of 10 Å. Rg fit results for all peptides in the absence of HEPES are available in the ESI (Fig. S12 and S13†). Data of non-lipidated peptides in the presence of 1 mM HEPES is not presented because the majority of samples did not have a Guinier region. | ||
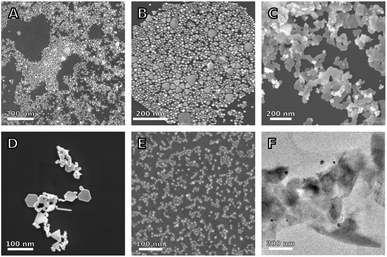
Next, we used SEM and TEM to study the structure of a small subset of samples formed in the presence of different Z2 variants. The SEM image in Fig. 7A shows that the sample synthesized in the presence of Z2 contains both spherical nanoparticles and nanoplates. However, the sample contains fewer nanoplates and is less aggregated than the sample in Fig. 7D, which corresponds to the same reagent concentration but with no peptide. Samples prepared with lipidated peptides such as MZ2R contained no plate like particles as seen in Fig. 7E and F. Additionally, the particles formed in the presence of MZ2R appear to be more monodisperse relative to the particles formed in the presence of Z2. The spectra which correspond to the reagent concentrations with and without peptides in these micrographs are shown in Fig. 2A and B, respectively. These SEM images support our previous interpretation of the spectra that the addition of Z2 leads to less aggregation and fewer nanoplates. Although the presence of nanoplates in the sample prepared with Z2 is not apparent from Fig. 2B, higher concentrations of Z2 and Z2M6I did show an increase in optical extinction at higher wavelengths and are likely to contain more plate like structures.
Samples prepared with the lipidated peptide MZ2R in Fig. 7E, on the other hand, contain no plate like particles. However, as was previously discussed, lipidation of Z2 was not sufficient to lead to formation of nanoparticle decorated peptide nanostructures. This is due to the fact the peptide concentrations in this study are below the critical micelle concentration. Additionally, Z2 has several charged amino acid groups and is therefore more likely to assemble into less extended or even spherical micelles.46 Future work into using Z2 to control the assembly of Au nanoparticles could focus on the use of higher peptide concentrations, a lipid tail that is longer than palmitic acid, or the addition of hydrophobic amino acids to the sequence.12 Using a higher peptide concentration of 0.2 mM caused sample charging in SEM. Therefore, TEM was used to characterize samples formed in the presence of MZ2R at this higher concentration. The image in Fig. 7F shows what appears to be a large amorphous aggregate with Au nanoparticles adsorbed on its surface. We interpret this as further evidence of peptide–precursor aggregation. However, the size of these amorphous aggregates is larger than what is to be expected from the fits in Fig. 6, and the structures are therefore likely to be affected by drying.
The sample prepared in the same conditions as Fig. 7A, but with the Z2M6I and Z2M246I peptides instead of Z2, are shown in Fig. 7B and C, respectively. The sample prepared with Z2M6I is similar to that prepared with Z2, with the exception that it contains a higher fraction of plate-like particles. This increase in plate-like particles is consistent with Z2M6I having higher extinction than Z2 at wavelengths above the main LSPR peak, as can be seen in Fig. 2B. The SEM image of the sample prepared with Z2M246I primarily contains flat, and interconnected structures. These interconnected structures also have smaller particles deposited on top, but the shape of these smaller particles is not clear. The large structures are similar to other reported flat and branching structures whose formation mechanism was attributed to molecular confinement.47 However, the reason for why methionine substitution for isoleucine in Z2 leads to promotion of anisotropic structures requires further study. We expected that removal of methionine would decrease the affinity of the peptide towards Au surfaces, and that this could result in structures that more closely resembled those obtained in the absence of peptide. Whereas the rigid aromatic structure of a molecule was implicated as important to achieving anisotropic particle growth,47 we do not expect the Z2M246I variant to be rigid. However, the piperazine ring in HEPES does possess this rigidity and HEPES also leads to formation of plate like structures (Fig. 7D). Therefore, we hypothesize that the observed anisotropic growth in the presence of Z2M246I is related to a modification in how HEPES interacts with Au structures, and that a different reducing agent (e.g. citrate) would not result in similar structures. The differences in particle morphology observed in samples containing Z2M6I and Z2M246I support the high signal correlation distances between these peptides and the other Z2 variants presented in Fig. 4.
We summarize our main findings in Fig. 8, where we show that each of the Z2 peptide variants formed peptide–precursor aggregates in the absence of HEPES. Given that HAuCl4 is added last to the solution, we infer that peptide–precursor aggregation is a process that happens in conjunction with Au reduction. High concentrations of peptide variants which contained methionine showed a significant decrease in the amount of Au(0) formation after 24 hours based on comparison of optical extinction at 400 nm relative to lower peptide concentrations. We note that this change cannot be explained by an increase in the amount of peptide adsorbed to the surface given that adsorption of organic molecules to citrate capped nanoparticles resulted in an increase in optical extinction.28 In the case of the lipidated peptides in this study, the difference in to what extent Au(0) formation was suppressed was inversely proportional to the values of critical micelle concentrations. Therefore, the differences in Au(0) formation in the presence of lipidated peptides are likely due to differences in modulation of HAuCl4–methionine complexation and availability of HAuCl4 to HEPES. These differences arise due to variation in the strength of lipidated peptide–precursor interactions between MZ2, MZ2R, and PZ2. Z2M6I and Z2M246I have one or all three methionine residues substituted with isoleucine, respectively. In Z2M6I, the relative proportion of plate-like particles to spherical particles appears to increase relative to Z2. Whereas for Z2M246I the structures that formed are interconnected and branching plate-like structures. Finally, the optical extinction at 400 nm for Z2M246I is least sensitive to peptide concentration out of all of the peptides studied and therefore confirms that methionine is an important modulator of HAuCl4 reduction.
4 Conclusion
In this paper, we presented a data analysis pipeline for studying large synthesis spaces using high-throughput experimentation. We characterize the formation of Au nanoparticles in the presence of peptides using our high-throughput experimentation framework to sample large synthesis spaces and collect UV-Vis spectra for each sample as a proxy signal for the nanoparticle structure. We applied our framework to study the differences in nanoparticle synthesis outcomes between six variants of the Au binding peptide Z2 and a different Au binding peptide AG3. We first focus on a portion of the synthesis space (one HEPES concentration) by analyzing the differences between spectra of peptides at specific reagent concentrations, and as these concentrations are varied. This analysis demonstrated the utility of studying modifications to sequence defined molecules while systematically varying experimental conditions. However, in order to fully take advantage of this dataset we described a data analysis framework to discern the correlations between peptide synthesis spaces. Using metrics designed to quantify higher-order correlations and variations between features, we obtain a two-dimensional embedding that provides insights into the similarity between the effect that peptides have on Au nanoparticle synthesis. The framework presented here is generic and only assumes the synthesis space being sampled is continuous. We obtained a Euclidean embedding of the correlations revealing that the synthesis spaces are more affected by the peptide sequence than conjugation of a lipid tail. The conclusions of this analysis were supported by use of USAXS and electron microscopy, which are lower-throughput but more direct methods of structural characterization. We anticipate that the proposed approach will initiate further research aiming at analyzing high-throughput experimentation data with spectral characterization to explore and understand complex synthesis spaces. The hardware and software introduced in this work can also be used to accelerate the identification of phase or state diagrams, or to perform closed-loop experimentation for material retrosynthesis.48 The latter may be used to either tune experimental conditions or molecular design to achieve desired nanomaterial structures. There are two notable limitations of the current method: (i) although the principal components capture interpretable variations in Au nanoparticle spectra, the method cannot attribute these variations to physically meaningful variables (i.e. reagent concentrations or physicochemical peptide attributes). This can, for example, be improved by using the PCA features in causal models.49,50 (ii) Lower dimensional representations obtained from FPCA via principal coefficients are not disentangled. Alternatively, data representations that characterize spectra using disentangled low dimensional representations would provide a direct way of visualizing variations in the synthesis space and their similarities across.Data availability
All of the data, notebooks for reproducing figures, and code for controlling the liquid handling robot have been shared at https://zenodo.org/record/6588769. Instructions for re-creating the ultra-small-angle X-ray scattering sample holder have been shared at https://doi.org/10.5281/zenodo.5899659. Notebook for functional data analysis can be found at https://gist.github.com/kiranvad/a46af46cbb3a114b4ab2b1970ba9cfa5.Author contributions
K. J. L., K.·V., and L. D. P. contributed equally to the conceptualization, experiment design, manuscript writing, and reviews. F. B. and N.·Y. N. contributed to drafting the manuscript. K. J. L. wrote code for high-throughput experimentation and carried out all experiments except for SEM imaging, which was performed by N.·Y. N. K. V developed the theoretical and algorithmic framework for data analysis using functional and graph representations.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is based upon work supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences (BES), as part of the Energy Frontier Research Centers program: CSSAS – The Center for the Science of Synthesis Across Scales – under Award Number DE-SC0019288. Part of this work was conducted at the Molecular Analysis Facility, a National Nanotechnology Coordinated Infrastructure (NNCI) site at the University of Washington, which is supported in part by funds from the National Science Foundation (awards NNCI-2025489, NNCI-1542101), the Molecular Engineering & Sciences Institute, and the Clean Energy Institute. K. J. L and N. Y. N. thank the Clean Energy Institute for support. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility, operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Extraordinary facility operations were supported in part by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the Coronavirus CARES Act. We would like to thank Jan Ilavsky and Ivan Kuzmenko for assistance in remote experiments at APS. Part of this work was conducted with instrumentation provided by the Joint Center for Deployment and Research in Earth Abundant Materials (JCDREAM).References
- V. Amendola, R. Pilot, M. Frasconi, O. M. Maragò and M. A. Iatì, J. Phys.: Condens. Matter, 2017, 29(20), 1–48 CrossRef CAS PubMed.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, Chem. Soc. Rev., 2012, 41, 2740–2779 RSC.
- N. M. Bedford, Z. E. Hughes, Z. Tang, Y. Li, B. D. Briggs, Y. Ren, M. T. Swihart, V. G. Petkov, R. R. Naik, M. R. Knecht and T. R. Walsh, J. Am. Chem. Soc., 2016, 138, 540–548 CrossRef CAS PubMed.
- M. R. Jones, K. D. Osberg, R. J. Macfarlane, M. R. Langille and C. A. Mirkin, Chem. Rev., 2011, 111, 3736–3827 CrossRef CAS PubMed.
- Z. Li, B. Cai, W. Yang and C. L. Chen, Chem. Rev., 2021, 121, 14031–14087 CrossRef CAS PubMed.
- M. B. Dickerson, K. H. Sandhage and R. R. Naik, Chem. Rev., 2008, 108, 4935–4978 CrossRef CAS PubMed.
- J. J. M. Lenders, L. A. Bawazer, D. C. Green, H. R. Zope, P. H. H. Bomans, G. de With, A. Kros, F. C. Meldrum and N. A. J. M. Sommerdijk, Adv. Funct. Mater., 2017, 27, 1604863 CrossRef.
- M. A. Nguyen, Z. E. Hughes, Y. Liu, Y. Li, M. T. Swihart, M. R. Knecht and T. R. Walsh, J. Phys. Chem. C, 2018, 122, 11532–11542 CrossRef CAS.
- Y. N. Tan, J. Y. Lee and D. I. Wang, J. Am. Chem. Soc., 2010, 132, 5677–5686 CrossRef CAS PubMed.
- C. J. Munro and M. R. Knecht, Langmuir, 2017, 33, 13757–13765 CrossRef CAS PubMed.
- Y. Li, Z. Tang, P. N. Prasad, M. R. Knecht and M. T. Swihart, Nanoscale, 2014, 6, 3165–3172 RSC.
- C. Zhang, C. Song, H. C. Fry and N. L. Rosi, Chem.–Eur. J., 2014, 20, 941–945 CrossRef CAS PubMed.
- S. Mokashi-Punekar, T. R. Walsh and N. L. Rosi, J. Am. Chem. Soc., 2019, 141, 15710–15716 CrossRef CAS PubMed.
- S. Mokashi-Punekar, S. C. Brooks, C. D. Hogan and N. L. Rosi, Biochemistry, 2021, 60, 1044–1049 CrossRef CAS PubMed.
- C. L. Chen, P. Zhang and N. L. Rosi, J. Am. Chem. Soc., 2008, 130, 13555–13557 CrossRef CAS PubMed.
- A. D. Merg, J. Slocik, M. G. Blaber, G. C. Schatz, R. Naik and N. L. Rosi, Langmuir, 2015, 31, 9492–9501 CrossRef CAS PubMed.
- C. Song, G. Zhao, P. Zhang and N. L. Rosi, J. Am. Chem. Soc., 2010, 132, 14033–14035 CrossRef CAS PubMed.
- H. Tao, T. Wu, M. Aldeghi, T. C. Wu, A. Aspuru-Guzik and E. Kumacheva, Nat. Rev. Mater., 2021, 6(8), 701–716 CrossRef.
- J. C. Dahl, X. Wang, X. Huang, E. M. Chan and A. P. Alivisatos, J. Am. Chem. Soc., 2020, 142, 11915–11926 CrossRef CAS PubMed.
- B. R. Peelle, E. M. Krauland, K. D. Wittrup and A. M. Belcher, Langmuir, 2005, 21, 6929–6933 CrossRef CAS PubMed.
- R. R. Naik, S. J. Stringer, G. Agarwal, S. E. Jones and M. O. Stone, Nat. Mater., 2002, 1, 169–172 CrossRef CAS PubMed.
- K. J. Lachowski, K. Vaddi, N. Y. Naser, F. Baneyx and L. D. Pozzo, Gold Nanoparticles Synthesized in the Presence of Peptides - UV-Vis Spectra, Fluorescence, USAXS, Electron Microscopy (v0.1.1) [Data set], Zenodo Electron. Microsc., 2022, https://doi.org/10.5281/zenodo.6588769 Search PubMed.
- L. Pozzo, K. Lachowski, S. Scheiwiller and E. Antonio, pozzo-research-group/Automation-Hardware: v0.1.0 – Initial release for documenting liquid sample holder on Zenodo, 2022, https://doi.org/10.5281/zenodo.5899659 Search PubMed.
- J. Ilavsky, F. Zhang, R. N. Andrews, I. Kuzmenko, P. R. Jemian, L. E. Levine and A. J. Allen, J. Appl. Crystallogr., 2018, 51, 867–882 CrossRef CAS PubMed.
- J. A. Lake, Acta Crystallogr., 1967, 23, 191–194 CrossRef CAS.
- J. Ilavsky and P. R. Jemian, J. Appl. Crystallogr., 2009, 42, 347–353 CrossRef CAS.
- M. V. Petoukhov, P. V. Konarev, A. G. Kikhney and D. I. Svergun, J. Appl. Crystallogr., 2007, 40, s223–s228 CrossRef CAS.
- T. Hendel, M. Wuithschick, F. Kettemann, A. Birnbaum, K. Rademann and J. Polte, Anal. Chem., 2014, 86, 11115–11124 CrossRef CAS PubMed.
- A. Habib, M. Tabata and Y. G. Wu, Bull. Chem. Soc. Jpn., 2005, 78, 262–269 CrossRef CAS.
- K. Liu, Z. He, J. F. Curtin, H. J. Byrne and F. Tian, Sci. Rep., 2019, 9, 1–13 Search PubMed.
- J. Xie, J. Y. Lee and D. I. Wang, Chem. Mater., 2007, 19, 2823–2830 CrossRef CAS.
- W. Xi and A. J. Haes, J. Am. Chem. Soc., 2019, 141, 4034–4042 CrossRef CAS PubMed.
- P. J. Jimmy Huang, J. Yang, K. Chong, Q. Ma, M. Li, F. Zhang, W. J. Moon, G. Zhang and J. Liu, Chem. Sci., 2020, 11, 6795–6804 RSC.
- Z. Tang, J. P. Palafox-Hernandez, W. C. Law, Z. E. Hughes, M. T. Swihart, P. N. Prasad, M. R. Knecht and T. R. Walsh, ACS Nano, 2013, 7, 9632–9646 CrossRef CAS PubMed.
- J. Cao, W. Jia, J. Zhang, X. Xu, S. Yan, Y. Wang, P. Zhang, H. Y. Chen and S. Huang, Nat. Commun., 2019, 10 Search PubMed.
- R. Grantham, Science, 1974, 185, 862–864 CrossRef CAS PubMed.
- K. Kalyanasundaram and J. K. Thomas, J. Am. Chem. Soc., 1977, 99, 2039–2044 CrossRef CAS.
- B. Ricaud, P. Borgnat, N. Tremblay, P. Gonçalves and P. Vandergheynst, C. R. Phys., 2019, 20, 474–488 CrossRef CAS.
- A. Sandryhaila and J. M. Moura, IEEE International Conference on Acoustics, Speech and Signal Processing, 2013, pp. 6167–6170 Search PubMed.
- M. Defferrard, L. Martin, R. Pena and N. Perraudin, PyGSP: Graph Signal Processing in Python (v0.5.0), Zenodo, 2017, https://doi.org/10.5281/zenodo.1003158 Search PubMed.
- J. Ramsay and B. Silvermann, Functional data analysis, springer series in statistics, 1998 Search PubMed.
- A. Srivastava and E. P. Klassen, Functional and shape data analysis, Springer, 2016, vol. 1 Search PubMed.
- J. B. Kruskal, Psychometrika, 1964, 29, 1–27 CrossRef.
- J. Friedman, T. Hastie and R. Tibshirani, The elements of statistical learning, Springer series in statistics, New York, 2001, vol. 1 Search PubMed.
- A. Y. Fong, L. Pellouchoud, M. Davidson, R. C. Walroth, C. Church, E. Tcareva, L. Wu, K. Peterson, B. Meredig and C. J. Tassone, J. Chem. Phys., 2021, 154 CrossRef CAS PubMed.
- M. H. Sangji, H. Sai, S. M. Chin, S. R. Lee, I. R Sasselli, L. C. Palmer and S. I. Stupp, Nano Lett., 2021, 21, 6146–6155 CrossRef CAS PubMed.
- S. Ye, A. P. Brown, A. C. Stammers, N. H. Thomson, J. Wen, L. Roach, R. J. Bushby, P. L. Coletta, K. Critchley, S. D. Connell, A. F. Markham, R. Brydson and S. D. Evans, Adv. Sci., 2019, 6 Search PubMed.
- K. Vaddi, H. T. Chiang and L. D. Pozzo, ChemRxiv, 2022 Search PubMed , https://doi.org/10.26434/chemrxiv-2022-dmn8w, this content is a preprint and has not been peer-reviewed.
- J. Pearl, Stat. Surv., 2009, 3, 96–146 Search PubMed.
- K. Sachs, O. Perez, D. Pe’er, D. A. Lauffenburger and G. P. Nolan, Science, 2005, 308, 523–529 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2dd00017b |
| This journal is © The Royal Society of Chemistry 2022 |