Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanomaterials for virus sensing and tracking
Muqsit
Pirzada
 ab and
Zeynep
Altintas
ab and
Zeynep
Altintas
 *ab
*ab
aTechnical University of Berlin, Faculty of Natural Sciences and Maths, Straße des 17. Juni 124, Berlin 10623, Germany. E-mail: zeynep.altintas@tu-berlin.de; zeynep.altintas@tf.uni-kiel.de; Fax: +49 30 314 79552; Tel: +49 30 314 23727
bInstitute of Materials Science, Faculty of Engineering, Kiel University, Kaiserstr 2, 24143 Kiel, Germany
First published on 23rd June 2022
Abstract
The effect of the on-going COVID-19 pandemic on global healthcare systems has underlined the importance of timely and cost-effective point-of-care diagnosis of viruses. The need for ultrasensitive easy-to-use platforms has culminated in an increased interest for rapid response equipment-free alternatives to conventional diagnostic methods such as polymerase chain reaction, western-blot assay, etc. Furthermore, the poor stability and the bleaching behavior of several contemporary fluorescent reporters is a major obstacle in understanding the mechanism of viral infection thus retarding drug screening and development. Owing to their extraordinary surface-to-volume ratio as well as their quantum confinement and charge transfer properties, nanomaterials are desirable additives to sensing and imaging systems to amplify their signal response as well as temporal resolution. Their large surface area promotes biomolecular integration as well as efficacious signal transduction. Due to their hole mobility, photostability, resistance to photobleaching, and intense brightness, nanomaterials have a considerable edge over organic dyes for single virus tracking. This paper reviews the state-of-the-art of combining carbon-allotrope, inorganic and organic-based nanomaterials with virus sensing and tracking methods, starting with the impact of human pathogenic viruses on the society. We address how different nanomaterials can be used in various virus sensing platforms (e.g. lab-on-a-chip, paper, and smartphone-based point-of-care systems) as well as in virus tracking applications. We discuss the enormous potential for the use of nanomaterials as simple, versatile, and affordable tools for detecting and tracing viruses infectious to humans, animals, plants as well as bacteria. We present latest examples in this direction by emphasizing major advantages and limitations.
1. Introduction
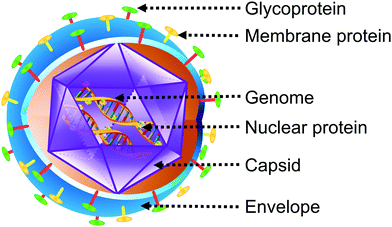
Viruses are pathogenic entities with sizes in the nanometer range.1 As the number of viruses on the planet exceeds 1031, they are considered the most cornucopian biological units on earth.2 Their ubiquity in the environment can be inferred from their concentration of 1011 virus-like particles (VLPs) per milliliter of seawater.3 Since the initial discovery of the tobacco mosaic virus in 1898 by Martinus Beijerinck,4 more than 6500 different species of viruses have been identified.5 On the basis of their genetic material, they are classified into four realms: (1) duplodnaviria which comprise of double-stranded deoxyribonucleic acid (DNA) responsible for encoding the HK97-fold, a protein essential for capsid assembly;6 (2) monodnaviria which possess single-stranded deoxyribonucleic acid (DNA) that encodes histidine-hydrophobic residue-histidine endonucleases involved in the rolling circle replication of viruses;7 (3) riboviria containing ribonucleic acid (RNA) that translates reverse transcriptase or RNA-dependent RNA polymerase which are both involved in replication of viruses such as poliovirus;8 and (4) varidnaviria that have double-stranded DNA responsible for capsid proteins involved in vertical Swiss roll fold.5,9 Viruses consist of just one type of nucleic acid encoding a corresponding protein to form a nucleic acid–protein complex. In some viruses like adenoviruses, papillomaviruses and herpesviruses, this complex is enclosed within a capsid protein. However, in the case of flavivirus, parvovirus, picornaviruses and togaviruses, a direct complex between the capsid and the nucleic acid is formed.1 In certain species such as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or human immunodeficiency virus (HIV), the capsid may be enveloped by a lipid layer (Fig. 1).Viruses can infect animals, plants, as well as microorganisms and, are considered crucial machinery for evolution since they can act as instruments of lateral gene transfer.10 In contrast to most microscopic parasites, viruses are intracellular in nature.11 Very few genes participate in the production of new viruses as reproduction is incumbent on several enzymes derived from the host cell.12 In this way, they alter cellular processes to optimize the conditions for their own reproduction. Viruses contain genes and they reproduce as well as survive following the Darwinian concept of natural selection just like living organisms.13 The defining distinction between viruses and other biological agents includes three characteristics: (1) viruses do not undergo division; (2) they are synthesized by the mere assembly of preformed building blocks; (3) the genetic information essential for the synthesis of ribosomes is absent.14
Once they infect the host, viruses may either enter a latent phase or an active replication phase. External stimuli, as well as cellular processes, can activate the repressed viral genome to encourage replication. Infected host cells may face at least one of the four possible consequences subsequently: (1) they are destroyed by various cell death mechanisms such as apoptosis (avian leukosis sarcoma virus,15 Sendai virus,16 and reovirus17), necroptosis (vaccinia18 and coxsackie19 viruses), or pyroptosis (Dengue virus20 and HIV21);22 (2) they survive with the viral genome (cytomegalovirus23) entering a phase of latency;24,25 (3) they survive with a chronic/persistent viral infection26 like in the case of varicella virus;27 (4) they become immortal and achieves the capacity to undergo uninhibited cell division28 which is often observed in the case of oncoviruses such as human T-cell lymphotropic virus (HTLV) which are responsible for causing adult T-cell lymphoma.29 Viruses are responsible for several common diseases such as influenza,30 cold,31 chickenpox,32 gastroenteritis, and pneumonia.33 Serious illnesses including smallpox,34 acquired immunodeficiency syndrome (AIDS),35 rabies,36 severe acute respiratory syndrome (SARS)37 as well as certain types of cancers are also caused by viral infections.35
Viruses have contributed to several major disease outbreaks in human history with high mortality (Fig. 2). Although it is difficult to accurately determine the cause of pre-historic epidemics, viral outbreaks had a cataclysmic impact on human society such as the end of the Bronze age due to the smallpox plague,38 the fall of Athens in the Peloponnesian war due to viral hemorrhagic fever39 as well as the rise of monotheism following the Antonine and Cyprian plagues.40 Epidemics of smallpox,41 measles,42 yellow fever,43 influenza,44 and dengue43 were rife in the middle ages due to the development of trade routes and overcrowding of cities without proportional advancement in hygiene practices. Even in modern times, in spite of the advancement in medicines, sanitation as well as an improved understanding of virology and immunology, viral diseases such as the 1918 Spanish flu,45 HIV-acquired immune deficiency syndrome (HIV)-AIDS46 pandemic as well as the ongoing coronavirus disease (COVID-19) pandemic have claimed millions of lives.47
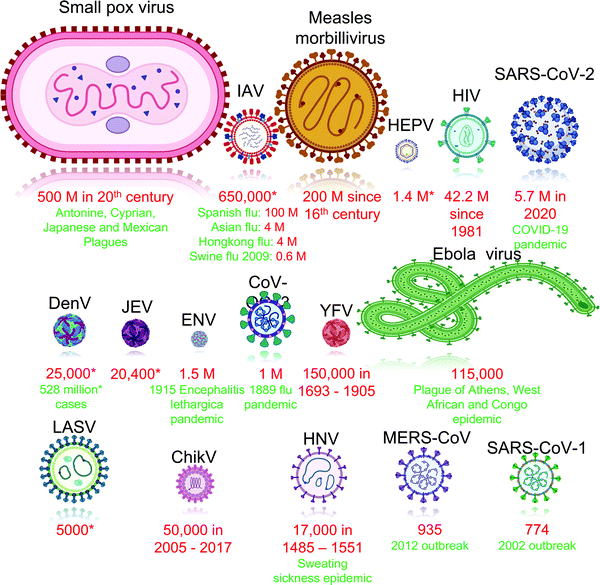 | ||
| Fig. 2 Viruses responsible for major outbreaks in human history in descending order of fatalities. Abbreviation: *: average number of cases or fatalities each year; ChikV: Chikungunya virus; CoV-OC43: human coronavirus type OC43; DenV: dengue virus; ENV: enterovirus; HEPV: hepatitis viruses; HIV: human immunodeficiency virus; HNV: hantavirus; IAV: influenza A viruses; JEV: japanese encephalitis virus; LASV: lassavirus; MERS-CoV: middle east respiratory syndrome-related coronavirus; SARS-CoV-1: severe acute respiratory syndrome coronavirus 1; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2 (Red: fatalities; green: notable outbreaks). Mortality statistics sourced from literature for: ChikV;48 CoV-OC43;49 DenV;50,51 ebola;39,52 ENV;53,54 HIV;55 HNV;56 influenza;54,57–59 LASV;60 JEV;61 measles morbillivirus;62 MERS-CoV;63 SARS-CoV-1;64 SARS-CoV-2;65 smallpox virus;66 viral hepatitis;67 yellow fever virus.68 | ||
The treatment of viral diseases and subsequent recovery hinges on the timely detection of analytes such as virions, viral proteins, nucleic acids, or antibodies synthesized as part of the body's immune response to the infection.69,70 Conventional analytical tools such as enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), serological antibody determination, and western blot demand advanced lab instruments which not only require skilled labor but also make testing expensive. Therefore, such analyses are performed in centralized testing institutes thereby prolonging the response time and delaying effective treatment.12 Such factors cripple the response to infections with high virulence such as COVID-19 in developing countries.71–73 Since biological sensors consist of receptors that exclusively bind to a chosen target analyte, they are highly sensitive and selective.74,75 As such sensors are facile to manufacture and offer rapid responses capable of real-time detection at a low cost, they are suitable for point-of-care-testing (POCT).69,76 In addition to early diagnosis, monitoring virus replication and dissemination is essential for prescribing accurate treatment.77 Tracking the mechanism of viral entry and the resultant cellular response to such cargos constitutes the foundation of antiviral drug development.78
We reviewed the integration of nanomaterials to sensing and imaging platforms in our earlier works.79–83 The typical size of a nanomaterial is lower than the wavelength of electrons which is why most of the atoms in such materials are located on the surface of the substance. These two factors in addition to their extraordinarily high surface area are responsible for the novel physicochemical properties of nanomaterials which are markedly different from the bulk form of the same material.84,85 Quantum confinement effects in these materials enhance the sensor signal and in turn increase the sensitivity.74,75 Zero-dimensional nanomaterials such as quantum dots and plasmonic nanoclusters interact with incident electromagnetic radiation resulting in their frequent use for non-invasive disease monitoring.86,87 Integration of nanomaterials to biomedical applications such as detection and imaging is therefore auspicious.88,89 Although there are some eminent reviews on nanomaterial-assisted virus recognition, they usually focus on just one type of nanomaterial without discussing viral imaging applications.84,90–92 Additionally, research works about the latest developments in nanoparticle-mediated virus detection and non-metallic nanoparticles for virus tracking from the previous half-decade are rarely discussed.93–96 Hence, we aim to present a compendious and detailed overview of the latest advancements in the field of virus recognition using various types of nanomaterials. To the best of our knowledge, this is the first review wherein nanomaterials other than quantum dots and noble metal nanoparticles are also discussed in the context of virus imaging and tracking.
2. Carbon allotrope-based nanomaterials in virus sensing and tracking
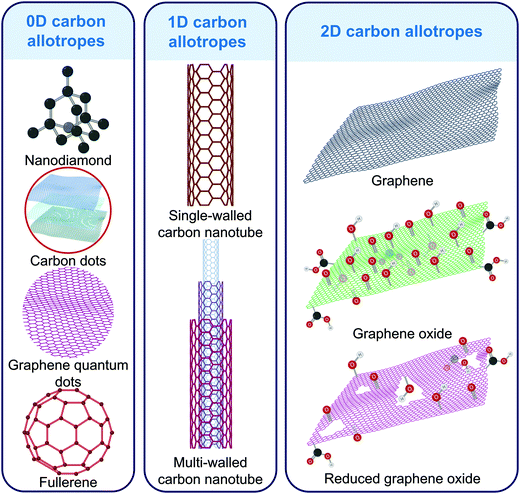
Carbon is abundantly available on earth and has therefore been frequently employed in technological and scientific areas. Elemental carbon can exist as a variety of allotropes based on its hybridization ranging between sp,97 sp2,98 and sp3.99 Furthermore, the nanoscale arrangement of these hybridized atoms allows the generation of several allotropes such as graphite, graphene, graphene quantum dots, and buckminsterfullerene all of which are sp2 hybridized (Fig. 3).84 Zero-dimensional nanomaterials such as carbon dots (CDs),100–103 graphene quantum dots (GQDs),104–106 and buckminsterfullerene (C60)107,108 are capable of undergoing dynamic as well as static quenching, electron transfer, energy transfer, and inner field effect. One-dimensional (1D) carbon nanotubes (CNTs) can be further classified into: (1) single-walled (SWCNTs) with diameters ranging from 7–14 Å; (2) multiwalled (MWCNTs) which is composed of multiple concentric graphene tubes with diameters spanning up to 1000 Å.84 Their high surface area coupled with long length facilitates the capture of various analytes in electrochemical systems.109–113 Two-dimensional (2D) carbon allotropes include graphene and its derivatives such as graphene oxide (GO) and reduced graphene oxide (rGO). Owing to the unique structure of the electronic bands of graphene, it shows extraordinary thermal, electrical, and mechanical properties.114–117 In recent years, other carbon allotropes such as nanohorns,118 nanodiamonds,119,120 and nanoparticles121,122 have also been introduced into sensing and imaging technologies due to their superior fluorescence, impedance, and energy transfer capabilities. These qualities make carbon allotrope-based nanomaterials ideal for incorporation into biosensing and bioimaging platforms.842.1 Virus sensing
| Nanomaterial | Platform | Receptor type | Target | Method | LOD | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: aCNT: aminated carbon nanotubes; AuNPs: gold nanoparticles; CNT: carbon nanotubes; DENV: dengue virus; DPV: differential pulse voltammetry; EIS: electrochemical impedance spectroscopy; FET: fluorescence energy transfer; FTO: fluorine doped tin oxide glass; GCE: glassy carbon electrode; HA: hyaluronic acid; HBV: hepatitis B virus; HCV: hepatitis C virus; HIV: human immunodeficiency virus; HPV: human papilloma virus; IDE: interdigitated electrode; IL: ionic liquid; MIP: molecularly imprinted polymer; mMWCNT: magnetic multi-walled carbon nanotube; MWCNT: multi-walled carbon nanotube; NS: non-structured protein; PFU: plaque forming units; rGO: reduced graphene oxide; SI-DPV: sequential injection differential pulse voltammetry; SPCE: screen printed carbon electrode; ssDNA: single strand deoxyribonucleic acid; SWCNT: single-walled carbon nanotube; SWV: square wave voltammetry; TCID: tissue culture infectious dose; ZNC: zeolite nanocrystal. | ||||||
| MWCNT/ZNC | FTO | ssDNA | HBV DNA | DPV, CV, EIS | 50 copies mL−1 | 130 |
| aCNT | HA/GCE | Antigen | antiHBV core protein | SWV | 34 pg mL−1 | 131 |
| MWCNT/Chitosan | GCE | MIP | HIV-p24 capsid protein | DPV | 83 fg mL−1 | 125 |
| MWCNT/Chitosan | GCE | MIP | HCV core antigen | DPV | 1.67 fg mL−1 | 124 |
| MWCNT | NH2rGO/IL | ssDNA | HPV DNA | DPV | 1.3 nM | 112 |
| Semiconducting SWCNT | Cr/Au IDE | ssDNA | H5N1 DNA | Chemiresistor | 20 pM | 132 |
| N-doped MWCNT | 2 pM | |||||
| SWCNT/1-pyrene methylamine | Au IDE | Heparin | DENV-1 | Chemiresistor | 8.4 × 102 TCID50 mL−1 | 133 |
| CNT | NiCo2O4 spinel | DNA | HIV-1 DNA | EIS | 16.7 fM | 134 |
| CNT | Cr, Pt | DNA | Influenza A DNA | FET | 1 pM | 135 |
| mMWCNT | AuNP/MNP | DNA | H1N1 DNA | LSV | 8.4 pM | 111 |
| mMWCNT | Pt/IDE | DNA | Norovirus DNA | LSV | 8.8 pM | 111 |
| MWCNT | AuNP | Antibody | H3N2 | Colorimetry | 3.4 PFU mL−1 | 136 |
| AuNP/MWCNT | Silicon chip | Antibody | DENV-2 NS1 | DPV | 300 fg mL−1 | 126 |
| AuNP/MWCNT | Ag IDE | Aptamer | H1N1 | Volymetric assay | 10 fM | 123 |
| AuNP/MWCNT | Ag IDE | Antibody | H1N1 | Volumetric assay | 1 pM | 123 |
| AuNP/CNT | AgNP/SPCE | Antibody | HBV surface antigen | SI-DPV | 860 pg mL−1 | 109 |
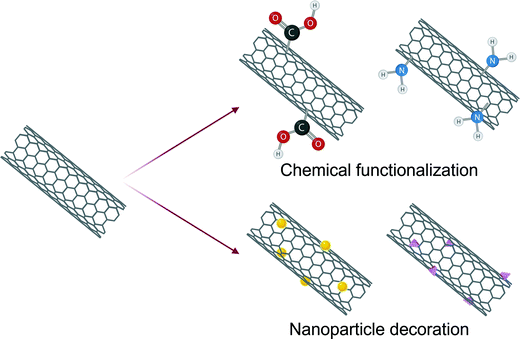
CNTs are versatile nanomaterials and chemical functionalizations can greatly alter their sensing capabilities (Fig. 4). Carboxylated CNT-based virus sensors are very specific against resemblant circulating viruses137 such as Zika virus (ZikV) which is responsible for Zika fever. Such devices allow rapid mismatch-tolerant detection for the RNA of highly variable viruses such as HIV which can progress to the life-threatening AIDS illness if left undetected and untreated.113 These CNTs exhibit excellent electrochemical stability and encourage greater electron transfer through amide bonds with -NH2 containing moieties. This concept has been harnessed for electrochemically screening ZikV and dengue virus (DenV) by evaluating the presence of nonstructural glycoprotein NS1 in serum and urine samples.138 Aminated CNTs in immunosensors amplify the charge transfer and provide a stable platform for antigen immobilization. Such platforms are essential for the detection of key serological markers for hepatitis infection such as the antibody to hepatitis B virus (HBV) core antigen.131 Chitosan functionalized MWCNTs with MIP receptors show ∼100% recovery towards viral proteins in serum samples124,125 and low relative error (−4.7 to 3.4%) against ELISA.125
Another strategy for tuning the characteristics of CNT includes decoration with metallic nanoparticles (Fig. 4). For example, gold nanoparticles (AuNPs) have been interspersed on MWCNTs to introduce peroxidase-like activity.136 These modifications have been reported to enable nearly 500 times higher sensitivity than commercial immunochromatography kits towards H3N2 detection with a 385 times lower LOD in comparison to a typical ELISA.136 AuNP-decorated CNTs exhibit exceptional specificity towards human pathogenic viruses with proteinaceous as well as nucleic acid receptors.109,111 Similarly, zinc nanoparticles intercalated on MWCNTs for the detection of Chili leaf curl betasatellite, a plant virus responsible for severe economic loss for pepper and tomato crops, promote the conjugation of single-strand DNA (ssDNA) enabling 3-fold higher specificity against non-complimentary DNA.139 A similar concept has also been developed integrating a nanocomposite of MWCNTs with zeolite nanocrystals to PCR-based amplification for genosensing HBV DNA.130 The retention of cavities in metal-organic frameworks (MOFs) embedded with CNTs encourages nucleic acid binding enabling femtomolar detection of HIV DNA.134 Incorporation of Fe3O4@SiO2 to CNT-based paper biosensor for pseudorabies virus introduced immunomagnetic properties that allows rapid detection within a few minutes without the requirement of skilled labor or expensive equipment making them suitable for POCT.127 The coexistence of nanoparticles in CNT nanocomposites offers four key benefits: (1) a stronger electrochemical response is obtained due to synergistic electronic interactions, (2) receptor conjugation via H bonding, π–π interactions and electrostatic forces is encouraged, (3) increased surface area enhances analyte adsorption, (4) the biocompatibility is improved.131,134 In addition to physical or chemical modifications, integration of computational modeling to optimize the roughness and alignment in CNT-thin films has been reported to show a higher sensitivity than the viral load observed in clinical samples by two orders of magnitude.140 Since MWCNTs are cost-effective, they find more abundant application in viral diagnostics than SWCNTs.111,112,123,128,141 However, interest in SWCNTs is increasing since they have a much larger surface area and allow ultrasensitive detection due to modulation of the Schottky barrier which is absent in MWCNTs.132,133,142,143 SWCNT-based field-effect transistors (FET) therefore demonstrate picomolar sensitivity towards influenza A virus DNA with a response time of only 1 min.142
Interest in carbon nanotube-based sensors for virus detection has risen with 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 164 research works out of 27
164 research works out of 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 916 published in the last five years alone (Web of Science). Electrochemical platforms constitute the largest class of CNT-based biosensors supporting a variety of virus detection mechanisms such as differential pulse voltammetry (DPV),137,139 electrochemical impedance spectroscopy (EIS),134 linear sweep voltammetry,111 cyclic voltammetry130 and square wave voltammetry131 (Fig. 5). CNT-based virus biosensors relying on DPV exhibit very high recovery in complex media such as serum109,125 and cells.112 CNTs have also been introduced to FET,129,135,142,144 chemiresistors,132,133 volumetric assays,123 optical sensors136 as well as conventional techniques for viral genome recognition such as PCR145,146 and loop-mediated isothermal amplification (LAMP).113 Reverse transcription LAMP-based HIV-1 DNA detection using carboxylated CNTs is even faster than fluorescent PCR and shows a 96.5% concordance rate with Roche instruments and a Nernstian response factor of 63.567 mV pH−1.113
916 published in the last five years alone (Web of Science). Electrochemical platforms constitute the largest class of CNT-based biosensors supporting a variety of virus detection mechanisms such as differential pulse voltammetry (DPV),137,139 electrochemical impedance spectroscopy (EIS),134 linear sweep voltammetry,111 cyclic voltammetry130 and square wave voltammetry131 (Fig. 5). CNT-based virus biosensors relying on DPV exhibit very high recovery in complex media such as serum109,125 and cells.112 CNTs have also been introduced to FET,129,135,142,144 chemiresistors,132,133 volumetric assays,123 optical sensors136 as well as conventional techniques for viral genome recognition such as PCR145,146 and loop-mediated isothermal amplification (LAMP).113 Reverse transcription LAMP-based HIV-1 DNA detection using carboxylated CNTs is even faster than fluorescent PCR and shows a 96.5% concordance rate with Roche instruments and a Nernstian response factor of 63.567 mV pH−1.113
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times higher sensitivity than ELISA in clinical transport medium while simultaneously being specific against the structurally similar Middle East respiratory syndrome-related coronavirus (MERS-CoV) (Fig. 6).115 Such a distinction helps in selecting a suitable therapeutic strategy since structurally similar viruses can require radically different treatment methods. Epitaxial graphene has also been employed for SARS-CoV-2 variant B.1.1.7 detection which provided a response within 0.6 s with extraordinary sensitivity (60 copies mL−1).148 Graphene possesses chiral plasmonic metasurfaces which can be harnessed for polarization-based discrimination of various types of avian influenza viruses.147 Graphene can also be composited with nanoporous silicon oxide to synthesize HBV sensitive artificial neural network.149 Such chemical modifications can significantly improve the sensitivity of graphene-based sensors and promote nucleic acid immobilization. Therefore, poly-L-lysine functionalized graphene-based FET sensors show a 113% higher sensitivity than unfunctionalized graphene platforms towards SARS-CoV-2 RNA.150 The signal-to-noise ratio of graphene FET sensors can be enhanced by ∼10% when they are nanocomposited with MXene.151 Although glutaraldehyde coupling chemistry has been reported,149 most immunosensors rely on NHS and NHS derivatives for immobilizing the antibodies on graphene.114–117
000 times higher sensitivity than ELISA in clinical transport medium while simultaneously being specific against the structurally similar Middle East respiratory syndrome-related coronavirus (MERS-CoV) (Fig. 6).115 Such a distinction helps in selecting a suitable therapeutic strategy since structurally similar viruses can require radically different treatment methods. Epitaxial graphene has also been employed for SARS-CoV-2 variant B.1.1.7 detection which provided a response within 0.6 s with extraordinary sensitivity (60 copies mL−1).148 Graphene possesses chiral plasmonic metasurfaces which can be harnessed for polarization-based discrimination of various types of avian influenza viruses.147 Graphene can also be composited with nanoporous silicon oxide to synthesize HBV sensitive artificial neural network.149 Such chemical modifications can significantly improve the sensitivity of graphene-based sensors and promote nucleic acid immobilization. Therefore, poly-L-lysine functionalized graphene-based FET sensors show a 113% higher sensitivity than unfunctionalized graphene platforms towards SARS-CoV-2 RNA.150 The signal-to-noise ratio of graphene FET sensors can be enhanced by ∼10% when they are nanocomposited with MXene.151 Although glutaraldehyde coupling chemistry has been reported,149 most immunosensors rely on NHS and NHS derivatives for immobilizing the antibodies on graphene.114–117
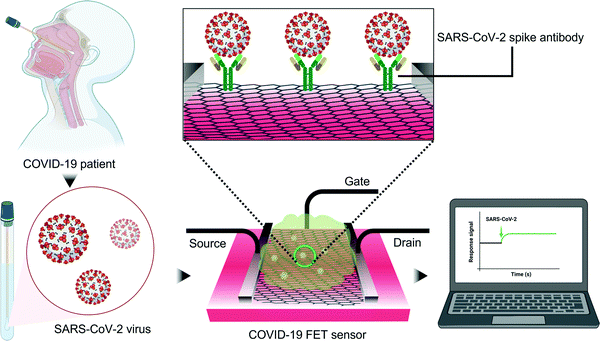 | ||
| Fig. 6 SARS-CoV-2 detection using a graphene-based FET immunosensor.115 The FET system is gated with an aqueous solution. The presence of SARS-CoV-2 alters the surface potential of the channel and results in an electrical response when the spike protein attaches to the immobilized antibody. | ||
Graphene derivatives such as GO and rGO are considerably influenced by their crystal defects as well as chemical functionalizations. GO is a product of oxidation of exfoliated graphite in which both the graphene surfaces have oxygenated functionalities. The optoelectronic and chemical properties of GO differ widely from graphene since GO has a much higher reactivity and a relatively lower electric conductivity. It also does not interact with the visible portion of the electromagnetic spectrum. Covalent, ionic, or hydrogen bonds between biomolecules and the oxygen-bearing functional groups from the edges and basal plane of GO are auspicious in the development of virus sensing platforms.84 For example, the negative charge on GO can be used for anchoring the DenV template during the molecular imprinting process.152 The quantum efficiency of GO can be quenched to avoid chemical modifications and simplify the sensing process. This fluorescence quenching property can be harvested by chemisorbing fluorescein amidate onto GO to generate a DNA-based sensor for Ebola virus (EBV) responsible for a type of viral hemorrhagic fever which is often fatal.153 Similar principles for HTLV154 and foot-and-mouth disease virus155 detection replaced fluorescein with AuNPs for quenching.
Amongst all the 2-D allotropes of carbon, rGO has been the most preferred nanomaterial in virus detection due to its crystal defects, heteroatomic impurities, and low-cost.156 It is synthesized by reducing GO via electrochemical, chemical, or thermal procedures. The reagents used for reduction determine the carbon to oxygen ratio and the crystal defects which in turn affect the electrochemical properties.84 Reduction with ascorbic acid promotes the integration of ssDNA on rGO surface by direct adsorption through π–π bonding and can thus be used for impedimetric detection of coxsackie virus which causes aseptic meningitis.157 This type of rGO can also be used as a signal enhancer for femtomolar detection of Japanese encephalitis virus (JEV) non-structural 1 protein.158 JEV is a Culex mosquito-borne virus responsible for Japanese encephalitis, a type of brain infection causing vomiting and seizures in severe cases. GO can also be reduced using porous hydrazine and subsequently conjugated with molybdenum sulfide to generate aptamer-facilitated electrochemical transducers for human papillomavirus (HPV) detection. Undetected HPV can lead to precancerous lesions and is the most common cause of cervical cancer. These hydrazine-reduced GO transducers show not only a high sensitivity (LOD: 1.75 pM) but also excellent recovery in serum (90–98%) as well as saliva (97–105%).159 Another promising approach for reduction targeted the epoxy and carboxyl groups of GO by refluxing with ethylenediamine to enable electrostatic coupling with AuNPs for the detection of HBV antibodies.160 A greater variety of biomolecular functionalizations are possible for rGO in comparison to graphene due to the presence of significantly lesser functional groups.84 These functional groups are easily hydrophilized by amination for conjugating with dendrimers to enhance the sensitivity and facilitate femtomolar detection of viral proteins.161,162 A 1.25-fold reduction in the detection limit is observed when aminated rGO is wrinkled with CdS quantum dots (QDs).163 Aminated rGO in a nanohybrid with MWCNTs and ionic liquid shows very high recovery (94–102.5%) in squamous carcinoma cells for HPV DNA.112
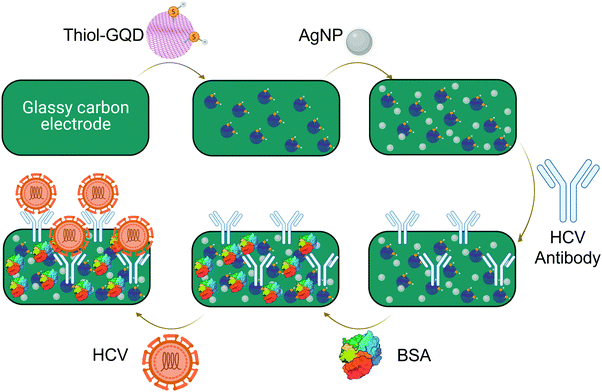 | ||
| Fig. 7 Thiolated graphene quantum dots for hepatitis virus detection.104 Abbreviations: AgNP: silver nanoparticles; BSA: bovine serum albumin; GQD: graphene quantum dots; HCV: hepatitis C virus. | ||
Crystalline allotropes of carbon are more numerous and resistant to photobleaching. High purity in addition to exquisite durability make their use in virus detection more widespread than amorphous allotropes. Nevertheless, biomolecules readily adhere to amorphous allotropes of carbon which is an integral requisite for most biosensors.122 In a recent study, nitrogen-doped porous carbon nanoparticles were synthesized from MOF to promote strong hydrogen bonds with ZikV DNA. This assembly allowed discrimination between 1–2 base mismatches as well as complimentary/non-complimentary DNA.121 As these particles show lesser residual current than glassy carbon electrodes, they act as ideal substrates for antibody physisorption.122 However, carbon nanoparticles are also suitable where chemically induced coupling is required. For example, amine-rich carbon nanoparticles derived from chitosan can be conjugated with JeV antibody through EDC-NHS chemistry.182
2.2 Virus tracking
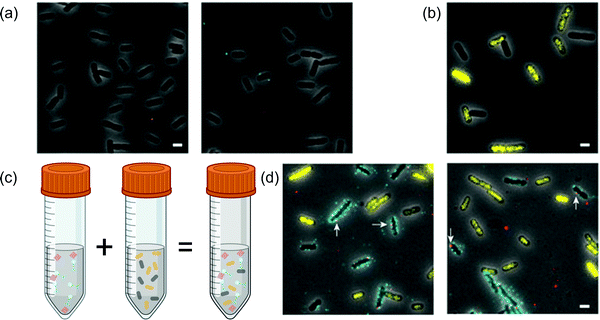 | ||
| Fig. 8 Host-specific targeting of FND-phage conjugates. (a) Cells without phage receptors do not bind to the FND conjugates; (b) a mixture of target and non-target species of bacteria; (c) scheme of FND-based imaging in the mixture of specific and non-specific bacteria; (d) epifluorescence imaging of FND-phage conjugate. Colors represent FNDs (red), phage (cyan), target bacteria (dark grey), non-target bacteria (yellow). Arrows point towards sites of FND-cell binding.184 | ||
Carbon allotrope-based virus imaging has recently been employed as a high throughput strategy for antiviral drug screening.78 In this case, an RNA-GO conjugate was developed to target RNA-dependent RNA polymerase, an essential protein present in the genome of all riboviruses such as DenV which causes hemorrhagic fever and shock syndrome. The conjugate was suitable for in vitro as well as in vivo assays. Here, the GO acted as a quencher for the fluorophore used to label RNA. The quenching-recovery mechanism followed the same approach as the previous method with one minor difference: the target nucleic acid in this case was generated in situ by the polymerase enzyme. Therefore, when a drug is successful in eliminating the virus, the fluorescence is not recovered.78 The main advantages of this strategy include convenient handling due to chemical stability of GO observed in usual room conditions. Additionally, drug screening through virus imaging exhibits high throughput and has the potential to reduce experimental burden as well as research expenditure.
3. Inorganic nanomaterials in virus sensing and tracking
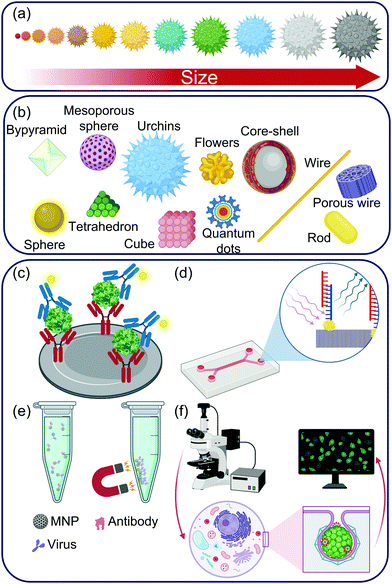
Transition and inner-transition elements have incompletely filled d and f orbitals, respectively. These orbitals contribute to the surplus of high-energy surface electrons that are responsible for the remarkable magnetic, optoelectronic and quantum properties of inorganic materials.79,187 These features can not only be easily incorporated into the sensing or imaging system as is but also be tuned to best suit the sensor requirements with the help of three strategies: (1) by manipulating the physical features of the nanomaterials such as its size or shape,188–191 (2) by introducing suitable functionalities to the nanomaterial192–196 and (3) by combining them with other nanomaterials to generate multifunctional nanocomposites (Fig. 9).177,197–199 For example, based on the distance between different nanoparticles, localized surface plasmon resonance (LSPR) can be observed. In inorganic nanomaterial-reliant biosensors, this phenomenon can be modulated by the concentration of viruses present in the ambient media.200,201 Inorganic nanomaterials include metal and metal oxide nanoparticles,92,202 quantum dots96,203 as well as silicon-based nanostructures.93 These nanomaterials come in a variety of shapes such as spheres,204 cubes,205 tetrahedrons,206 octahedrons,207 bipyramids,208 cages,207 wires,209 core–shell assemblies210 as well as rods211 and all these shapes have been harnessed for virus recognition. These nanomaterials can act as transducers,212 substrates for receptor immobilization,213 target capture probes,214 fluorophores,215 signal amplification tags,216 and as a part of labels in sandwich assays.217 Additionally, nanoparticles with magnetic characteristics can also be used to enrich viral assays and enable ultrasensitive detection.218 Some inorganic nanomaterials show exceptional fluorescent properties and can be used for tracking single virus particles.963.1 Virus sensing
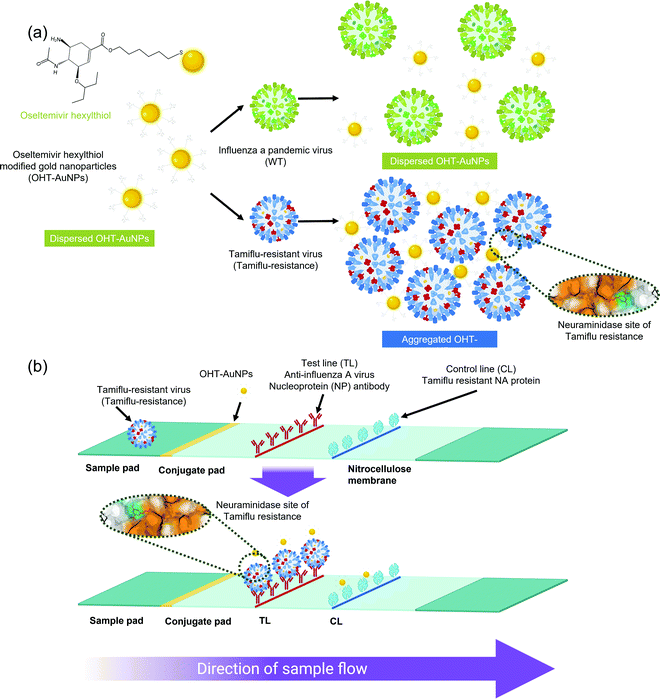 | ||
| Fig. 10 Schematic illustrations of the AuNP-assisted detection of Tamiflu-resistant influenza virus using (a) colorimetric and (b) lateral-flow assays.243https://doi.org/10.1038/s41598-018-31311-x. Used under Creative Commons CC BY 4.0 https://creativecommons.org/licenses/by/4.0/. | ||
AuNPs have also been incorporated into real-time reverse transcription PCR (nanoPCR) for the detection of foot-and-mouth disease virus which allowed the reduction of threshold cycles to 4.09 from 7.03 which was the value in the case of conventional PCR.246 Furthermore, the LOD towards the virus improved nearly 1000-fold due to the addition of GO-AuNP nanocomposites to a typical PCR system.248 NanoPCR shows 10 times higher sensitivity in half the annealing time to bovine respiratory syncytial virus in comparison to conventional PCR.249 This type of PCR can also be combined with dual-priming oligonucleotides to enable the multiplexed detection of bovine rotavirus, parvovirus and viral diarrhea virus with a 100-fold higher sensitivity than the typical PCR method.250 These 3 viruses constitute the 3 most important pathogens responsible for diarrhea in new-born calves. NanoPCR is also a promising tool for differentiation between different porcine circovirus gentoypes.251 Porcine circoviruses are responsible for the most destructive infections affecting the pig farming industry causing multisystemic wasting syndrome. Recently, the contribution of AuNP dimensions and interparticle spacing for surface plasmon resonance (SPR) sandwich immunoassay of SARS-CoV-2 was investigated.201 Gold nanorods (AuNRs) with a diameter of 10 nm and varying aspect ratios (1–4) were used as labels due to the sensitivity of their longitudinal plasmon bands to small changes in the refractive index. A 50 nm thick gold nanosheet acted as the sensing platform and the distance between AuNRs and the gold nanosheet was varied from 2–14 nm. At an aspect ratio of 4 with a distance of 14 nm, maximum plasmon enhancement was reported and incremental sensitivity of 111.11° RIU−1 was achieved. Furthermore, the optimized AuNRs enhanced the evanescent field of the basic gold nanosheet by more than 376%.201 Such virus sensors have been reported to show tremendous improvement over rapid immunoassays (up to 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold252) due to nanostructure mediated LSPR.253–256
000-fold252) due to nanostructure mediated LSPR.253–256
In addition to their size, the shape of AuNPs also determines their sensing performance.257,258 Gold nanocubes (70–140 nm) have a higher surface area than most other morphologies and therefore produce a strong electrochemical signal.240 Hexagonal AuNPs (30 nm) act as catalysts for tetramethylbenzidine oxidation by H2O2.259 Gold nanocoils (thickness: 200 nm; length: 392 nm) can be functionalized with antibody fragments and coupled with magnetic nanoparticles (MNPs) to facilitate inductive transduction.260 The superior electron transfer capabilities of nanowires (diameter: 50–70 nm) made with AuNP (6–14 nm) decorated polyaniline is responsible for the high stability of impedimetric sensors for hepatitis E virus (HEV) and white spot syndrome virus.168 Similar stability is displayed by GQD conjugated gold nanobundles (length: 700 nm; diameter 10 nm) in addition to exceptional optical confinement properties.261 Gold nanospheres (80 nm) decorated on an excessively tilted fiber grafting exhibit intrinsic LSPR.262 They magnify the effect of refractive index fluctuations on the core to co-propagating cladding and introduce the bioaffinity necessary for antibody conjugation. Gold nanobipyramids used in colorimetric sensors as substrates for silver deposition change color depending on the silver film thickness due to virus concentration-induced silver reduction.208 Chiral gold nanostars (spike length 10 nm; diameter 2 nm) allow the fabrication of circular dichroism-based sensors which show superior sensitivity in comparison to ELISA and commercial diagnostic kits.263 Similarly, gold nanofilms and nanosheets also enhance sensitivity due to the presence of multiple edges and corners that act as active sites.264,265 Nanoporous gold electrodes amplify the electrochemical response as well as encourage the covalent binding of probes to reduce baseline current signals.266,267 This concept can be integrated into PCR-based amplification to reduce the number of reaction cycles.267
The concentration of AuNPs is another critical parameter that must be optimized as a higher concentration increases not only the color intensity241 but also the possibility of nanoparticle agglomeration.268 Functionalizing the AuNPs with oligonucleotide can protect the AuNPs from aggregating with the help of the electrostatic attractions between the negatively charged nanoparticles and the positively charged nucleobases.269,270 In such genosensors, the nucleotides form an electric double layer with the negatively charged backbone phosphate groups exposed to repel other similar ssDNA–AuNP assemblies. Although not as sensitive as PCR, such assemblies facilitate naked eye detection in clinically relevant virus concentration range.269 AuNPs with peptide functionalities also tend to form large agglomerates. Such aggregates can be subjected to explosive enzymatic catalysis to observe an enhanced hypsochromic shift proportional to the concentration of the target virus (Fig. 11).271 AuNPs (15 ± 0.4 nm) can themselves be used as beacons to induce virus aggregation in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays.272 Due to their biocompatibility, they are suitable for application in enzymatic biosensors as immobilization substrates.266,273 Elemental gold can directly bond with sulfur making AuNPs ideal candidates for immobilization of thiol-activated nucleotides274–276 and cysteine-rich proteins.273,277 Therefore, AuNP-based DNA sensors are capable of femtomolar detection with simultaneous virus serotyping.172,200,278 Since they possess peroxidase-like properties, they are also used as a durable and lucrative substitute to enzymes in virus sensing assemblies that rely on signal amplification via hydrogen peroxide reduction.279–281
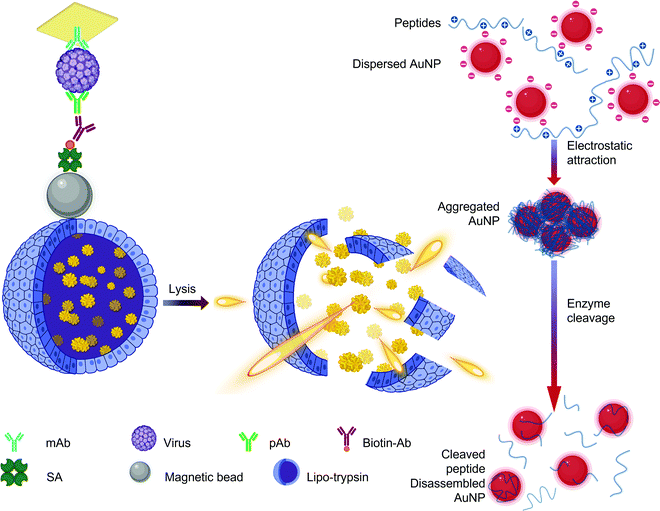 | ||
| Fig. 11 Scheme for explosive catalysis of gold nanoaggregates for virus detection.271 Abbreviations: Ab: antibody; AuNP: gold nanoparticles; mAb: monoclonal antibody; pAb: polyclonal antibody; SA: streptavidin. Reprinted with permission from (ACS Appl. Mater. Interfaces 2020, 12, 11, 12525–12532). Copyright (2020) American Chemical Society. | ||
Chemically functionalized AuNPs are emerging as a popular class of AuNPs due to their tailor-made sensing characteristics. AuNP@spherical nucleic acid core–shell assemblies enable the synthesis of reversible self-assembled structures for the picomolar quantification of HIV DNA.282 Sialic acid-functionalized AuNPs (∼20 nm) undergo target specific agglomeration due to the specific binding of the sialic acid receptor with hemagglutinin from the influenza virus.283,284 The intrinsic properties of AuNPs can be further supplemented with other nanomaterials to generate nanohybrids with extraordinary characteristics. For example, AuNP decorated graphitized carbon nitride enhances the permeability of charge transfer285 as well as photocurrent due to its crystalline structure and superior adsorption. Furthermore, AuNPs provide additional active sites thus facilitating ultrasensitive detection.286,287 Nanocomposites of AuNP and covalent organic frameworks (COFs) act as biocompatible quenchers for fluorescently labeled DNA and are suitable for in vitro and intracellular sensing.288 Different types of QDs are also common components of AuNP nanocomposites. This is because AuNPs can be employed to modulate their fluorescence.255 Fluorescent sensors using such nanocomposites have displayed 100-fold higher sensitivity than ELISA.253 Silica coated AuNR@CdSeTe QD core–shell assemblies show biocompatibility, low toxicity as well as high scattering efficiency and can be functionalized with molecular beacons for the acute recognition of as low as 1.2 copies mL−1 of norovirus (NoV) RNA.289 Norovirus infection is the most prevalent cause of pathogenic gastroenteritis involving abdominal ache, emesis, and diarrhea. The distance between QDs and AuNPs makes it possible to regulate the plasmon resonance and induce high sensitivity to fluctuations in the refractive index of the surrounding medium.200,253,255,289 It is critical for such ultrasensitive platforms to have negligible background noise to enable precise quantification. An increasingly popular technique to eliminate noise is to add MNPs or magnetic microbeads to enable the selective enrichment of the receptor-target assembly from complex media such as serum, urine, or saliva by applying a magnetic field.238,273,275,290 Due to the chemiluminescent features of such AuNP nanohybrids, attomolar sensitivity with a broad HTLV detection range of 8 orders of magnitude could be achieved in diluted serum.291 Such sensors exhibit superior tolerance than quantitative PCR in human cells and hold potential for in vivo applications.
The magnetic nature of MNPs is usually harnessed by using them to capture target molecules and then isolating such complexes under a mild magnetic field. For example, protamine-coated MNPs can be used to concentrate HAV from mussels, strawberries and green onions prior to real-time PCR.297 The particle size is a function of the method with which the MNPs were synthesized. The use of MNPs in electrochemical, optical, and conventional viral assays is therefore subject to fulfilling the following requirements: their size should never be so large that the colloidal stability is lost during sensing; they must be non-toxic and compatible with the testing matrix; the particles must retain sufficient saturation magnetization to allow their motion without the need for powerful magnets.79,83 For example, magnetic nanocubes enable the magnetic isolation and concentration of viruses in chronoamperometric immunoassays and reduce the response time to just 160 s.306 Very high serum recovery was observed when δ-FeOOH induced magnetic separation is supplemented with exonuclease III amplification.307 MNPs are compatible with a wide array of detection mechanisms as well nanomaterials and their sensing performance can be modulated to suit the required application easily. NoV immunosensors using identically sized MNPs show a higher LOD for electrical resistance measurements (1.16 pg mL−1)308 than DPV (4.1 fg mL−1) and colorimetry (340 fg mL−1)309 since each detection mechanism interacts with MNPs differently. Even when the detection mechanism, sensing medium as well as size is kept constant, MNP nanocomposites show greater sensitivity with QDs (69 NoV RNA copies mL−1)197 than AuNPs (84 NoV RNA copies mL−1)254 and liposomes (136 NoV RNA copies mL−1) (Fig. 12).187 The synergic effect of MNPs and fluorescent polymer nanoparticles in flow cytometry allows the detection of 100 fold lower concentrations of DenV than polymer nanoparticles alone.310
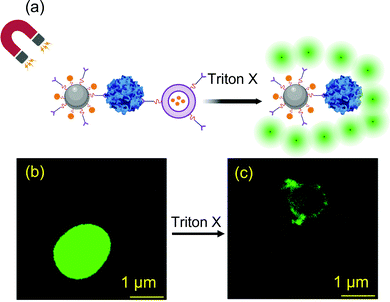 | ||
| Fig. 12 (a) Schematic illustration of norovirus detection using antibody functionalized SiO2@magnetic nanoparticles based on liposome rupture using Triton X to release calcein fluorophore; confocal microscopic images of virus bound nanoassemblies (b) before and (c) after fluorophore release.187 Reprinted with permission from (ACS Appl. Bio Mater. 2020, 3, 6, 3560–3568). Copyright (2020) American Chemical Society. | ||
Core–shell morphologies constitute the most common class of MNP nanocomposites. MNPs can act as supports for depositing different types of nanomaterials.127,192,311,312 MNP cores can be subjected to surface imprinting with whole virus templates to fabricate resonance light scattering (RLS) biosensors capable of picomolar quantification.311,313 MNP morphology plays a key role in such systems as the response time reduces by more than 7-fold when diamond-shaped particles313 are replaced with nanospheres.311 MNP supported noble metal shells are suitable for developing user-friendly lateral flow immunoassays312 and paper-based genosensors.240 MNP supports can also act as seeds for growing carbosilane dendrimers314 as well as silica nanoshells.192 The layer-by-layer deposition of γ-Fe2O3 (320–370 nm) and CdSe/ZnS QDs (7–15 nm) on poly(styrene/acrylamide) copolymer nanospheres generates fluorescent magnetic catalytic nanoparticles. These catalytic nanoparticles amplify fluorescence as well as electrochemical signals 4-fold more than commercial gold substrates315 and enable multiplex detection of various influenza viruses in chicken blood.316 Owing to their excellent catalysis, MNPs have also been employed to accelerate the recognition reactions involved in electrochemiluminescence,317 rolling circle amplification,318 RLS,311 and ELISA.314
QDs form stable links with antibodies via EDC–NHS chemistry.254 For example, CdTe QD (3.1 nm) immunoconjugates show no loss in activity even after 4 months and are capable of ZikV sensing at 100-fold lower concentrations than required for an immune response.321 They are excellent FRET acceptors329 and fluorescence enhancers.322,330 CdTe/ZnS QD nanoprobes employed in FRET-based COVID-19 detection confirm the results of PCR, thus presenting a convenient and cheaper alternative.324 CdTe QD decorations impart fluorescence to MIPs which can simultaneously detect hepatitis A virus (HAV) and HBV within 20 minutes due to virus-induced quenching.330 Zinc-doped CdTe QDs can act as supports for the immobilization of black-hole quenchers to inhibit over-valency and allow single-particle detection.331 Although CdTe QDs encourage the rapid movement of photogenerated charge carriers to improve PEC probes,286 the reproducibility improved 2-fold when cadmium-free quaternary ZnAgInS QDs (10 nm) were used instead due to higher stability of photocurrent.287 Another popular variety of cadmium-based binary QDs, CdSe (4.4 nm), has also been used in PEC-mediated genosensors with zeptomolar sensitivity towards HTLV detection.325 CdSe QDs are ultrasensitive fluorophores that show significantly higher signals than fluorescence dyes and commercial europium-chelates.192,332 CdSe QD-assisted sensors show specificity against mismatched DNA sequences332 and are capable of virus serotyping.278 They are hydrophobic and can also modulate electrochemical signals. Recently, a sensor for the chikungunya virus was developed with CdSe QD (5–7 nm) encapsulated in a liposome and MNPs (26.5 nm) as labels.193 The MNPs enabled magnetic separation following which chloroform triggered CdSe QD release facilitated dual-modal detection. Such sensors are particularly important in developing nations where chikungunya is often misdiagnosed as Zika fever or dengue due to similarity in symptoms.
The inclusion of additional elements in a QD can modify their intrinsic properties. For example, enhanced fluorescence is observed in sensors that include quaternary QDs such as CdSeTeS.197,253 In addition to alloying, other techniques such as doping,331 encapsulation,192,333 dendronization,88 intercalation,316,333etc. are also used to tune the properties of QDs. Therefore, cadmium-QDs are often coated with ZnS192,332 or ZnSeS255,278 to inhibit their cytotoxicity since the direct contact of Cd2+ ions with reactive oxygen species can cause cell death.334 ZnSeS coated QDs are also useful for tuning the LSPR performance of AuNPs by modulating the interparticle distance with peptide linkers of appropriate length (Fig. 13).255 These QDs can be supported on polystyrene nanobeads and further enveloped in SiO2 to fabricate fluorescent reporters for LFA-assisted SARS-CoV-2 detection within 18 minutes.192 CdS – dendrimer nanocomposites show higher recovery (100.11–100.33%) in serum for larger QDs (10.507 nm)88 than in the case of smaller QDs (94.2–96.45%) measuring 3–4 nm.333
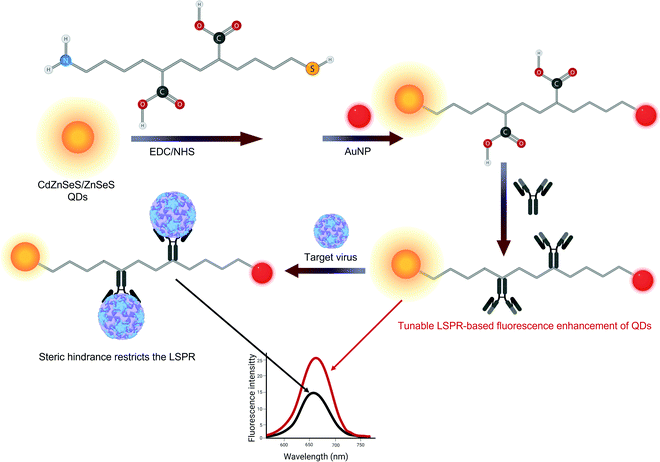 | ||
| Fig. 13 Schematic representation of the synthesis of peptide-linked quantum dot nanocomposites with gold nanoparticles and their use in norovirus detection.255 | ||
Traditionally, most QDs were cadmium-based due to their excellent photostability, bright PL, resistance to bleaching, wide excitation range in the ultraviolet spectrum, and narrow emissions.289,335 However, recent concern over the cytotoxicity of cadmium has been gradually shifting the trend towards cadmium-free QDs due to cytotoxicity issues.334 Although lead quantum dots act as ideal substituents for Cd QDs,336 additional encapsulation with biocompatible proteins is necessary to avoid toxicity issues.337 Molybdenum QDs form an important group of cadmium-free QDs. For example, MoS2 QDs can be used in dual mode sensors since they show chirality as well as PL-induced fluorescence.338 MoO3 QDs exhibit a monolayer morphology and semiconductor-based LSPR which is analogous to that of AuNPs or silver nanoparticles (AgNPs) in the NIR and visible spectrum.339 Zirconium QDs show excellent aqueous dispersibility with blue fluorescence. They readily bind with antibodies and such immunosensors can detect as low as 79.15 EID 50 μL−1 of infectious bronchitis virus in blood.198 This is the only type of coronavirus that infects chickens and the disease is extremely contagious. Kidney inflammation, respiratory distress as well as loss of internal and external egg quality include some of the most common symptoms of infection. Recently, the felicity of Si QDs for immunosensing has also been investigated for H1N1 detection.320 However, the focus of this study was only to probe the aptitude of Si QDs as substrates for immobilization and parameters like the linear response range, LOD or affinity were not reported.
AgNPs show exceptional SERS capacity with plasmon resonance near 500 nm and stable reproducible responses. SERS sensors made of AgNP-assisted LFA have enormous potential for POCT purposes since they are 2000 times more sensitive than standard colloidal gold strip and generate responses within 30 min (Fig. 14).312 With the latest advancements in nanolithography, SERS sensors can be fabricated with silver nanoislands. These nanoislands (height: 6 nm; planar size: 20 nm) readily bind with thiol functionalized aptamers to detect as few as 10 virus particles in an assay time of just 12 minutes.349 Since porous silver nanofilms synthesized using electron beam physical vapor deposition conduct SERS at their sharp edges they have been developed for size-dependent virus capture.350 A similar non-specific capturing principle has also been adopted in terahertz spectroscopy.188 Due to their extraordinary electroactivity, AgNP mediated electrochemical sensors display excellent agreement with ELISA.351 Therefore, AgNPs act as direct signaling tags in voltammetric virus detection.352,353 AgNPs are capable of improving the detection capacities of other nanomaterials. In a comparison between native AuNPs (2.7 ± 0.6 nm), Au@Ag core–shell nanoparticles (4.4 ± 0.6 nm) and alloyed Ag–AuNPs (2.9 ± 1.1 nm) for ZikV RNA detection, the respective sensitivities observed were 2.9 copies mL−1, 2.4 copies mL−1 and 1.7 copies mL−1.256 Due to their electrochemiluminescence, highly stable AgAuPt nanocubes show high sensitivity (LOD: 65 aM) and recovery (up to 98%) in human serum samples.354
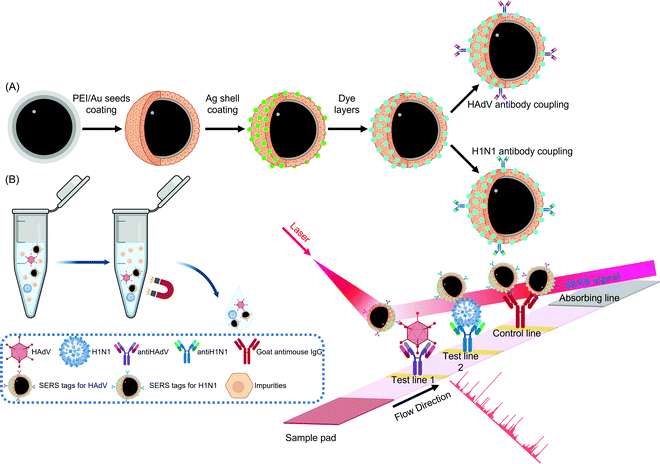 | ||
| Fig. 14 Schematic illustration of (A) the synthesis of antibody functionalized AgNP SERS tags and (B) their use in a lateral flow assay for sensing respiratory viruses312 reprinted with permission from (ACS Appl. Mater. Interfaces 2019, 11, 21, 19495–19505). Copyright 2019 American Chemical Society. Abbreviations: Ag: silver; antiHAdV: antibody for human adenovirus; antiH1N1: antibody for H1N1 virus; Au: gold; HAdV: human adenovirus; IgG: immunoglobulin G; SERS: surface-enhanced Raman spectroscopy. | ||
Spherical silica nanoparticles (SiO2 NPs) constitute the second most popular shape of silicon-based nanoparticles. Although SiO2 NPs doped with organic dyes cost much less than Au, they show satisfactory signal amplification.364 They exhibit excellent electrochemiluminescence and a 1000-fold enhancement in sensitivity is observed when SiO2 NPs are included in the sensor. SiO2 NPs are also suitable for loading a variety of substances ranging from small quantum dots to large enzymes.365 For example, pomegranate-shaped SiO2 NPs (thickness: 5 nm) show high uptake of quantum dots and they can be functionalized with antibodies for rabies lyssavirus detection.366 Timely detection of the virus is critical since rabies is almost always fatal after the onset of its symptoms such as hydrophobia, nausea, unconsciousness. Glucose oxidase-loaded SiO2 NPs (120 nm) have been used as supports for poly(acrylic) acid brushes that bind viral DNA.367 Dendritic SiO2 nanospheres loaded with red emissive CDs show up to 48.5% quantum yield in aqueous media which is auspicious for lateral flow-based ZikV detection.166 SiO2 NP supports can also be used for surface imprinting of viruses.368 The SiO2 NPs–MIP nanocomposites show more than 95% cell viability after 24 hours and are therefore ideal for in vitro virus detection.369,370 Silicon nanorods371 and fouling-based silicon nanomembranes372 have also been recently explored for virus detection although the use of these materials is not yet common due to scarce literature in the field. Silicon nanomembranes have an advantage over PCR for SARS-CoV-2 detection since they can selectively recognize intact virions thus differentiating between diseased and recently recovered individuals.372
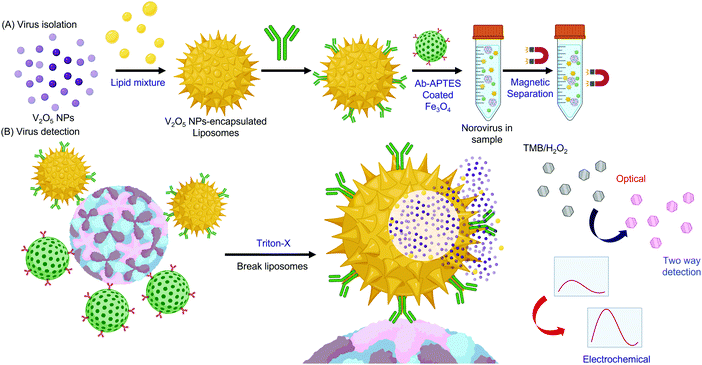 | ||
| Fig. 15 Fabrication of V2O5 liposomes and their use in virus detection.309 Abbreviation: Ab: antibody; APTES: aminopropyltriethoxy silane; TMB: tetramethylbenzidine. | ||
In tellurium doped ZnO nanowires (diameter: 50 nm), tellurium promotes the donor-acceptor emissions and simultaneously passivates the essential vacancy parameters due to isoelectronic doping.376 When such nanowires are doped with molybdenum, charge transfer is promoted and vacant sites are generated for adsorbing DNA.377 These nanowires show higher precision in comparison to PCR by demonstrating a higher sensitivity against false negatives.377 Similar nanowires of tin oxide (diameter: 25 nm; length: 3.5 μm) have been grown for EBV genosensing.378 The evolution of distance and time during SnO2 growth was described with the help of Cosmol Multiphysics and the nanowires showed high stability with 95% of initial response after 8 weeks. Similar stability is also observed when tin has been used as a dopant in heteroinjected WO3/In2O3 nanowire photoelectrodes.191,379 The tin heterogeneity intensified light absorption by the creation of exciton which allows a dynamic response. Due to this phenomenon, MoS2 nanosheets increase the electrochemiluminescence of QDs from 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 76
000 to 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (2.8 times).190 The introduction of porosity further aids in receptor immobilization due to an increase in the surface area.134,189
000 (2.8 times).190 The introduction of porosity further aids in receptor immobilization due to an increase in the surface area.134,189
Nanoflowers of metallic compounds offer a large surface area for nanoparticle immobilization thereby accelerating reaction kinetics and improving the carrier immobility as well as the efficacy of surface reaction.374,380,381 Anodic aluminum oxide nanohemispheres-based impedimetric sensors show high linear correlation (R2: 0.983) with real-time PCR detection and a low LOD of only 111 HBV DNA copieas mL−1.382 Lanthanide-based upconversion nanoparticles show comparable sensitivity to AuNPs due to the excitation of the longer wavelength of light to shorter wavelengths.383 Recently, phosphorene has been reported as a promising substitute to SARS-CoV-2 sensing due to the tunability of its band gap. Computational simulations reveal a higher Gibb's binding energy between the receptor binding domain of the virus with phosphorene (200.37 kcal mol−1) than with graphene (83.65 kcal mol−1).384 Therefore, phosphorene is expected to emerge as a promising nanomaterial for virus sensing in the future.229Table 2 includes a comprehensive list of various metallic nanoparticles that have been reported for the development of virus biosensors in the last half-decade.
| Nanomaterial | Size (nm) | Receptor | Virus | Method | LOD | Ref. |
|---|---|---|---|---|---|---|
| a PtNP dimensions. b Nanocage dimensions. c Nanocomposite film thickness. d Pore size. e Diameter of quantum dots. f Core size. g Total size including shell. h Diameter. i Length. j Petal width. k Petal length. l LOD in spiked sample. m LOD in infected oyster. n Thickness. Abbreviations: AAONH: anodic aluminum oxide nanohemisphere; Ab: antibody; Al–Mo NCM: aluminum-molybdenum nanocomposite membrane; Ap: aptamer: ArV: arbovirus; BHV: bovine herpes virus; CA: ahronoamperometry; CM: colorimetry; CNT: carbon nanotube; ConA: conconavalin A; CV: cyclic voltammetry; DenV: dengue virus; DNA: deoxyribonucleic acid; DPV: differential pulse voltammetry; DVM: differential voltage measurement; EBV: ebola virus; ECL: electrochemiluminescence; EIS: electrochemical impedance spectroscopy; FET: field-effect transistor; GO: graphene oxide; HBV: hepatitis B virus; HCV: hepatitis C virus; HEV: hepatitis E virus; HIV: human immunodeficiency virus; HNC: hollow nanocages; ITONW: indium tin oxide nanowire; LI: laser interferometry; LOD: limit of detection; NC: nanocubes; NF: nanoflowers; NNLFA: near-infrared to near-infrared lateral flow assay; NP: nanoparticles; NR: nanorod; NS: nanosheet; NSP: norovirus-specific peptide; NW: nanowire; S-BN QD: sulfur-doped boron nitrogen dots; UCNP: NaYF4:Yb,Tm@NaYF4:Ca up-conversion nanoparticles ZikV: Zika virus. | ||||||
| Pt NP | 3.9 ± 0.5 | Ab | ZikV | EIS | 10 virus particles μL−1 | 373 |
| V2O5 NP | 25 ± 1.5 | Ab | NoV | CM; DPV | 340 fg mL−1; 4.1 fg mL−1 | 309 |
| Pt-Co3O4 HNC | 4–5a; 250, 400b | Ab | HEV | CA | 61 RNA copies mL−1 | 207 |
| Pt-Pd NF; ZnO NP | 200c | DNA | DenV | CV/DPV | 43 μM | 374 |
| PtAgAu NC | 100 ± 5 | DNA | HBV | ECL | 65 aM | 354 |
| NiCo2O4 UANW | 8d | Ap | HCV | EIS | 160 ag mL−1 | 189 |
| MoS2 NS; S-BN QDs | 7e | DNA | HCV | ECL | 170 fM | 190 |
| UCNP | 30f; 45g | Ab | H5N2 | NNLFA | 102 EID50 mL−1 | 383 |
| H5N6 | 103.5 EID50 mL−1 | |||||
| ZnO NR | 300h; ∼2000i | Ab | H5N2 | CM | 8 × 103 EID50 mL−1 | 385 |
| Cu3(PO4)2-GO NF | 8000 | Ap | HBV | DPV | 1100 copies mL−1 | 380 |
| SnO2 NW | 25h; 3500i | DNA | EBV | DVM | 3.2 pM | 378 |
| Te-ZnO NW | 50h | DNA | HBV | EIS | 100 fM | 376 |
| Sn-WO3/In2O3 NW | 50h | DNA | HBV | EIS | 1 fM | 379 |
| WS2 NF | 50–180j; 500–800k; 4000–5000 | NSP | NoV | EIS | 2.37l/6.21m copies mL−1 | 381 |
| ZnO NR | 260i | Ab | HCV | CV | 0.25 mg mL−1 | 386 |
| NiCo2O4@CNT | — | DNA | HIV | EIS | 16.7 fM | 134 |
| Al–Mo NCM | 10n | Ab | BHV | LI | 90 μg mL−1 | 387 |
| AAONH | 100 | DNA | HBV | EIS | 111 copies mL−1 | 382 |
| ITONW | 50h | DNA | HBV | FET | 1 fM | 191 |
| Se NP | 42 | Ab | HIV | SERS | — | 258 |
| ZnO NP | — | ConA | ArV | EIS | 0.038–0.062 PFU mL−1 | 375 |
3.2 Virus tracking
The significance of QDs as labels for virus imaging can be attributed to the preservation of more than 90% viral infectivity that is observed in the case of single as well as dual-labeling.393 QD-based SVT has facilitated the understanding of virus infection mechanisms such as virus uncoating, replication, and transport of virus from the extracellular region to the cytoplasm. For example, the uncoating of HIV was recently investigated by labeling the RNA with a red Zn2+ doped CdTe QD and the capsid with a green organoarsenic dye as shown in Fig. 16A.331 In this work, the QDs were initially functionalized with DNA that adopted a stem-loop hairpin structure. After entering the TZM-bI cells, the DNA hybridization caused fluorescence recovery. The poor productive infection of the HIV stocks was further established when only 27 dissociations were recorded in 120 minutes from the 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 viruses that were tracked.331 The combination of QDs with varying emission spectra encourages site-specific labeling of the ribonucleoprotein complex and helps visualize the uncoating process.86 In a similar study on the internalization of Dabie bandavirus which is responsible for causing severe fever with thrombocytopenia syndrome in humans, it was discovered that the rate of membrane fusion was pH-dependent.388 In this case, multicolor imaging of streptavidin functionalized QDs attached to biotinylated viruses revealed a clathrin-mediated virus transport pathway from the cell periphery to the interior. Since the lipid-specific labeling with QDs preserves native viral infectivity, SVT demonstrates a slow peripheral movement through a dense actin network followed by rapid transport of viral load along microtubules.394 QD-based confocal imaging revealed that viruses hijack this endosomal sorting complex required for transport machinery and induce a “driver switchover” between the retrograde molecular motor proteins myosin VI and dynein.395 CdSe@ZnS QDs (8.83 ± 0.8 nm) have been used in conjunction with an inverted microscope with a confocal scanning system to interrogate respiratory syncytial virus-induced endocytosis in Hep-2 cells.396 The so called “orthopneumovirus” is responsible for bronchiolitis and pneumonia affecting all age groups.397 For such enveloped viruses, the endocytosis commenced with extensive lipid shifts and culminated with the entry into lysozymes within the perinuclear region. The dependence of such a viral entry on both actins and micropinocytosis was discovered by SVT with QD-Cas9/gRNA complex.398
000 viruses that were tracked.331 The combination of QDs with varying emission spectra encourages site-specific labeling of the ribonucleoprotein complex and helps visualize the uncoating process.86 In a similar study on the internalization of Dabie bandavirus which is responsible for causing severe fever with thrombocytopenia syndrome in humans, it was discovered that the rate of membrane fusion was pH-dependent.388 In this case, multicolor imaging of streptavidin functionalized QDs attached to biotinylated viruses revealed a clathrin-mediated virus transport pathway from the cell periphery to the interior. Since the lipid-specific labeling with QDs preserves native viral infectivity, SVT demonstrates a slow peripheral movement through a dense actin network followed by rapid transport of viral load along microtubules.394 QD-based confocal imaging revealed that viruses hijack this endosomal sorting complex required for transport machinery and induce a “driver switchover” between the retrograde molecular motor proteins myosin VI and dynein.395 CdSe@ZnS QDs (8.83 ± 0.8 nm) have been used in conjunction with an inverted microscope with a confocal scanning system to interrogate respiratory syncytial virus-induced endocytosis in Hep-2 cells.396 The so called “orthopneumovirus” is responsible for bronchiolitis and pneumonia affecting all age groups.397 For such enveloped viruses, the endocytosis commenced with extensive lipid shifts and culminated with the entry into lysozymes within the perinuclear region. The dependence of such a viral entry on both actins and micropinocytosis was discovered by SVT with QD-Cas9/gRNA complex.398
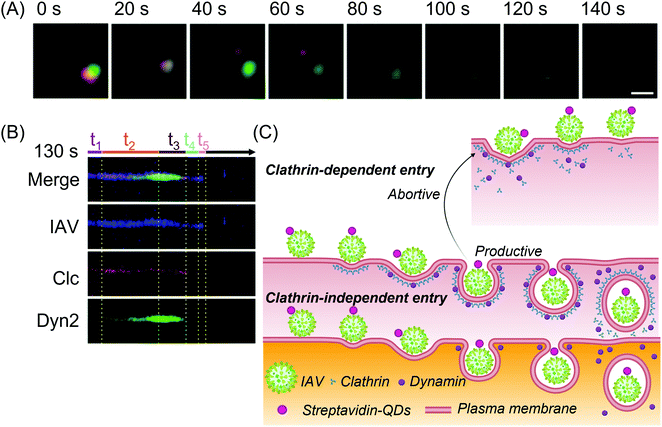 | ||
| Fig. 16 (A) Fluorescent imaging of the uncoating of the QD-labeled HIV-RNA (green) from its capsid (red)331 adapted with permission from (J. Am. Chem. Soc. 2019, 141, 34, 13454–13458). Copyright 2019 American Chemical Society. (B) Kymographs of phase-wise clathrin-dependent and independent virus entry and (C) the schematic representation of the proposed internalization mechanism399 reprinted with permission from (ACS Nano 2017, 11, 5, 4395–4406). Copyright 2017 American Chemical Society. Abbreviations: Clc: clathrin light chains; Dyn2: dynamin 2; QDs: quantum dots; t1: initiation stage; t2: dynamin assembly stage; t3: dynamin disassembly stage; t4: clathrin disassembly stage; t5: predirected stage. | ||
In addition to the common clathrin-mediated viral trafficking pathway, QDs have also been used in SVT for the real-time monitoring of virus internalization via clathrin-independent dynamin-dependent internalization (Fig. 16B and C),399 Rab5 endosomal route as well as autophagy.400 QDs have also been employed for observing other critical parameters of a viral infection such as their replication kinetics398 as well as for calculating the intracellular motility.401 Recently it was reported that certain reoviruses cross the intracellular membrane in 14.8 s and travel through the endosome-lysosome system for sorting and trafficking.402 This concept was subsequently applied to antiviral drug screening for non-enveloped viruses. Multicolor QDs at dilute concentrations do not exhibit cytotoxicity and they readily undergo budding with viral glycoproteins. Such spontaneous labeling can be used to understand the self-assembly of intraviral components.403 QD-based SVT also shows potential for differentiating between healthy and disease states in cells. Therefore, the transcription activator-like effectors of the HIV-1 provirus genomic loci can be conjugated with 2 QDs of different emission maxima to induce biorthogonal ligature reactions.404 These reactions illuminate the mechanism of individual genomic loci regulation as well as their functions within the genomic architecture thus elucidating the virus replication cycle which is a prerequisite for developing antiviral drugs.
The aggregation of AuNPs from 47.4 ± 0.8 nm particles to 300.1 ± 198.3 nm clusters enhances the fluorescence intensity enabling the detection of HBV.347 Such an enhancement is also observed when MNP@AgNP core–shell nanocomposites are bound to fluorescently labeled antibodies.312 MNPs are also employed in multiplex detection of different influenza virus species using fluorescence imaging.316 Virus stamping is one of the most interesting applications of MNPs in virus imaging.412 It is a strategy to introduce virus infection in single cells which shows high efficacy and compatibility with in vitro, ex vivo, and in vivo diagnostics using 2 photon-assisted shadow imaging. Although metallic nanoparticles show promise for virus imaging, the difficulty in sensing multiple species and the scarce availability of several labels are the two primary obstacles that should be overcome to allow widespread applications since their first recorded use for SVT in 2014.96,413
4. Organic nanomaterials in virus sensing and tracking
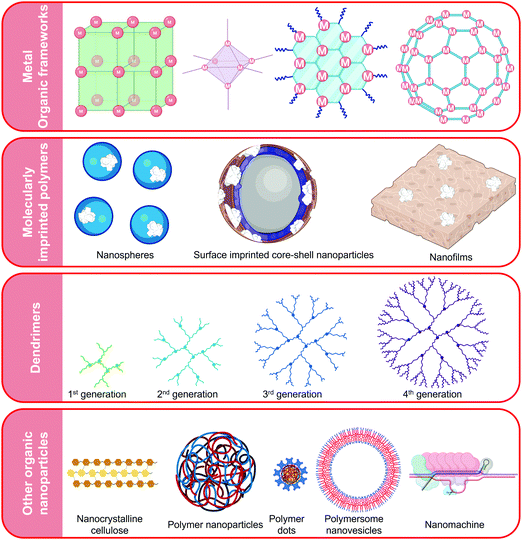
Organic nanomaterials used for virus recognition and tracking are customarily polymeric in nature (Fig. 17). Their synthesis is inexpensive and simple making these materials suitable for developing POCT devices. They are stable across wide temperature ranges and they readily co-ordinate with metals to form structurally detailed and porous frameworks with exceptional fluorescence which can act as substrates for receptor immobilization.414 Organic nanomaterials can themselves be harnessed as receptors124 or transducers415 and are the nanomaterial of choice in optical virus sensors.11,319,414,416,417 They possess intrinsic bioinertness as well as biocompatibility. Nanomaterials such as MIP nanoparticles (MIP-NPs) offer cost-effective alternatives to the delicate antibodies often used in virus detection.418 Hyper-branched polymers such as dendrimers improve the conjugation between the target and the sensing platform.88 Polymeric nanoparticles are also used in nanocomposites to tune the properties of MNPs,310 AuNPs,419 CNTs,124 and QDs.88,420 Such nanocomposites exhibit a symbiotic combination of unique morphologies with ultrasensitivity and facile fabrication processes. Such substrates effectively suppress background noise and readily bind with oligonucleotides, MIPs as well as antibodies.285,421,422 They can also act as virus mimics in the development of sensors for highly infectious viruses.423 Nanoparticles such as polymer dots have the capacity to replace cadmium-based quantum dots in virus sensing and imaging due to their exceptional fluorescent characteristics in addition to their biocompatibility.424 Apart from polymeric nanoparticles, molecular machines, so called nanomotors or nanodevices, are also emerging as promising virus receptors owing to their sub-nanomolar sensitivities for label-free recognition of viral genomic material.4254.1 Virus sensing
Biosensors use MOFs ranging from one to three dimensions or a combination of multiple varieties.428 Although 1D zinc carboxylate MOFs are water-soluble with aromatic bipyridine peripheral ligands that facilitate nanomolar sensing, they exhibit long assay times.429 In a comparative study on zwitterionic zinc carboxylate MOFs, it was observed that 2D MOF nanosheets interact with probe DNA more efficiently than 1D nanochains, 2D networks as well as 3D polymeric matrices.428 The LOD was reduced by an order of magnitude when 2D copper MOFs were used.430 3D copper MOFs (Fig. 18A) can also be harnessed as nanoreactors for copper-mediated azide-alkyne cycloaddition “click reactions” in time-resolved fluorescence immunoassays for HBV surface antigen (Fig. 18B).431
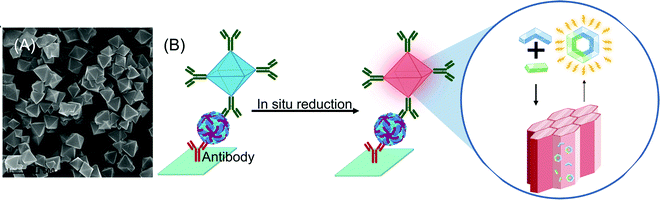 | ||
| Fig. 18 (A) SEM images of copper organic frameworks; (B) copper organic frameworks as 3D nano for sensing hepatitis B virus.431 Reprinted with permission from (Anal. Chem. 2020, 92, 4, 2972–2978), Copyright 2020 American Chemical Society. | ||
MOFs readily undergo π–π stacking interactions with viral DNA in addition to hydrogen bonds and electrostatic interactions.108,428,432–434 These interactions have also been confirmed using computational simulations and they enable the picomolar detection of Ebola RNA.434 Such simulations are also useful for understanding the binding dynamics in the case of synchronous detection of conserved viral nucleic acid sequences.435 In this study, the planar geometry of the 2D MOF exposed more functional groups on the surface thereby causing fluorescence quenching in the presence of nucleic acid and thus providing contrast for near-dark background fluorescence. These interactions are a function of the size of pores and channels in the framework which are responsible for the impressive loading efficiency of MOFs.421 For example, the low symmetry from the sp3 hybridized carbon of methylene can be compensated by metals with high coordination numbers to form MOF networks with higher dimensions and large pores.436 Chromium terephthalate MOFs are also used as support in surface imprinting for rapid (20 min) detection of viruses.414,422 A neoteric morphology of zirconium-based metal-organic gel has recently been explored in a nanocomposite with iron MOF and AuNP decorated graphitized C3N4 for electrochemiluminescence-based ZikV sensing.285 The zirconium nanogel promoted charge transfer owing to the 3D connectivity of its chromophores. It also fostered recombination in the energy rich excited state of carbon nitride.285
Dendrimer nanocomposites possess interesting features such as bright fluorescence, sensitivity in complex media, and high reproducibility. The cornucopian tertiary amine groups in PAMAM introduce water solubility and chemical stability in addition to enhancing the capacity for loading QDs.88,333,420,441 Such nanocomposites shift the resonance angle to a greater extent due to the high molecular weight of PAMAM which encourages antigen recognition.88,420 In a recent work on the detection of HBV surface antigen, it was observed that the incorporation of PAMAM multiplied the electrochemiluminescence intensity of QDs by 3.42 folds.333 AgCl nanosphere doped bis(hydroxymethyl)propionic acid dendrimers (4th generation) show low variability for enterovirus detection with intra-assay and inter-assay standard deviations of 4.15% and 6.15% respectively.439 Nanocomposites of rGO and PAMAM (4th generation) are capable of transporting bioactive agents with high efficacy. SPR sensors using the nanocomposite display high affinity (Ka: 7.6452 TM−1), sensitivity (0.2576° pM−1) along with an ultralow LOD (80 femtomolar).162
In recent years, the scope of dendrimers in virus detection has expanded beyond their role as immobilization platforms and nanomaterial carriers. For example, Au–PAMAM (5th generation) nanochains are employed for the visual paper-based detection of HIV capsid-protein p24.419 Increased levels of p24 are earlier indicators of HIV infection than the frequently targeted patient antibodies. These nanochains are 4-fold more sensitive to the target than AuNPs alone and are promising candidates in the fabrication of POCT HIV-diagnostic devices. Dendrimers derived from coumarin are excellent fluorescent materials for high output screening of viruses in human blood.442 Lately, dendrimer-based bio-bar codes for HTLV determination have been fabricated owing to the dendritic self-assembly and signal amplification features of polythymidine (Fig. 19).291 Carbosilane dendrimers (1st and 2nd generation) are compatible with ELISA as they provide the retentive potential to viral antibodies owing to their electrostatic attraction.314 Such systems present a cost-effective alternative to PCR and reduce the operational time to only 2 h in comparison to 4.5 h required for PCR.
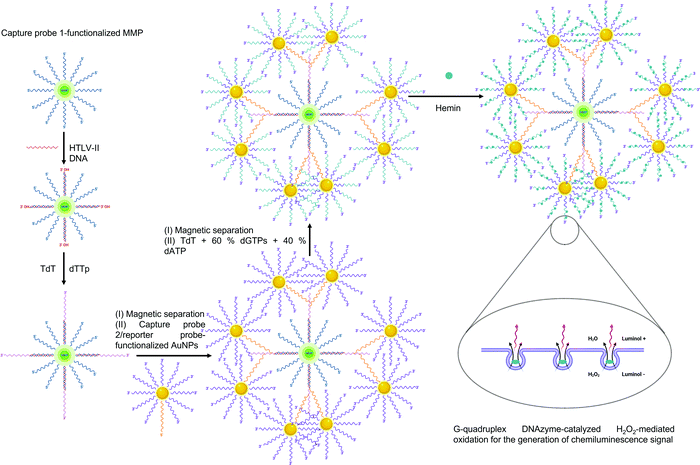 | ||
| Fig. 19 Scheme for dendrimer-mediated bio-bar codes for sensing viral DNA.291 Abbreviations: AuNP: gold nanoparticles; dATP: deoxyadenosine triphosphate; dGTP: deoxyguanosine triphosphate; DNA: deoxyribonucleic acid; dTTp: deoxythymidine triphosphate; HTLV-II: human T-lymphotropic virus II; MMP: magnetic microparticle; TdT: terminal deoxynucleotidyl transferase. Used under Creative Commons Attribution-NonCommercial 3.0 Unported Licence https://creativecommons.org/licenses/by-nc/3.0/[L. Wang, M. Ren, L. Liang and C. Zhang, Chem. Sci., 2018, 9, 4942] – Published by The Royal Society of Chemistry. | ||
The selection of suitable functional monomers is critical for developing assays with high sensitivity and rapid response (Table 3). For example, in the case of MIP-NPs for the RLS-based detection of HAV, (dimethylamino)ethyl methacrylate414 generated a pH-responsive polymer with a significantly lower LOD (100 fM) and response time (20 min) than N-isoprylacrylamide (LOD: 1.1 pM; response time: 30 min)369 or dopamine (LOD: 6.2 pM; response time: 150 min).313 With the help of computational modeling, it is possible to select the functional monomers that show the highest binding affinity with the target thus reducing experimental cost and research time significantly.444In silico designing can also be used for selecting the most stabile viral epitope as a template peptide to escape the pitfalls of template leakage during rebinding. Such MIP-NPs (65 nm) are resistant to hydrolysis enzymes and are analogous to monoclonal antibodies.416 However, unlike most anti-bodies, MIP-NPs can be easily regenerated and reused several times.33,311 The mechanism of virus detection is also a key parameter for improving sensor performance. For example, in the case of JeV imprinting with APTES, nearly 12-fold higher sensitivity was reported when RLS311 was replaced with fluorescence-based measurement.445 The detection range widened 7-fold and the imprinting factor increased 2.5 times when APTES was replaced with zinc acrylate as the functional monomer and polyethylene glycol was added as a passivating agent.422
| Functional monomer | Size (nm) | Template type | Target | Method | LOD | Ref. |
|---|---|---|---|---|---|---|
| a Size of Fe3O4@SiO2 support. b Size of diamond-shaped magnetic nanoparticle. c Size of SiO2 support. Abbreviations: AAc: acrylic acid; APTES: (3-aminopropyl)triethoxysilane; BIS: N,N′-methylenebisacrylamide; DPV: differential pulse voltammetry; EGDMA: ethylene glycol dimethacrylate; gp41: glycoprotein 41; HAV: hepatitis A virus; HIV: human immunodeficiency virus; JEV: japanese encephalitis virus; MS2: bacteriophage MS2; NIPAam: N-isopropylacrylamide; NPhAam: N-phenylacrylamide; RLS: resonance light scattering; SPR: surface plasmon resonance; TBAm: N-tert-butylacrylamide; TEOS: tetraethyl orthosilicate; TFMA: trifluoromethyl acrylic acid; TNV: tobacco necrosis virus. | ||||||
| NIPAam, BIS, TBAm, AAc | 205–238 | Virus | MS2 | SPR | 5 × 106 PFU mL−1 | 11 |
| Dimethylaminoethylmethacrylate | 13 | Virus | HAV | RLS | 100 fM | 414 |
| NIPAam, TBAm, BIS, NPhAam, TFMA | 65 | Epitope | HIV gp41 | Fluorescence | 10 nM | 416 |
| APTES, TEOS | 421 | Virus | JEV | Fluorescence | 110 fM | 445 |
| APTES, TEOS | 15 | Virus | HAV | Fluorescence | 88 pM | 76 |
| APTES, TEOS | 200a | Virus | JEV | RLS | 1.3 pM | 311 |
| Dopamine | 200b | Virus | HAV | RLS | 6.2 pM | 313 |
| Dopamine | 70c | Virus | HAV | RLS | 8.6 pM | 370 |
| NIPAam | 110c | Virus | HAV | RLS | 1.1 pM | 369 |
| Thiophene | 200 | Virus | TNV | Fluorescence | 2.2 ng L−1 | 417 |
| Zinc acrylate | 50 | Virus | JEV | Fluorescence | 13 pM | 422 |
| 2,2′:6′,6′′ terpyridine, EGDMA | <100 | Pb2+ | HIV-1 pol-gene | DPV | 300 aM | 415 |
The contribution of MIPs to virus sensors has diversified in recent years and it is not restricted to simple target-shaped cavities.444 Thermoresponsive nanostructured hydrogels synthesized from surface imprinted SiO2 nanoparticles are eligible for in vitro HAV detection.369 Such MIP-NPs (8 nm) are highly selective against viruses for diseases such as rabies, measles, and rubella370 and show high recovery in diluted serum.76 Polythiophene MIP-NPs (200 nm) emit fluorescence (λ: 410 nm) and their thickness can be optimized to obtain a response within 2 minutes.417 This year, a Pb2+ ion-imprinted terpyridine-based polymer was developed as a transducer in the genosensing of the HIV-1 pol-gene.415
 | ||
| Fig. 20 Scanning electron micrographs of the morphology of (A) polystyrene nanospheres447 and (B) polypyrrole nanowires.450 Reprinted with permission from (ACS Appl. Mater. Interfaces 2018, 10, 34, 28412–28419), Copyright 2018, American Chemical Society. | ||
In flow cytometry, fluorescence latex nanoparticles (200 nm) function as reporters and are responsible for reducing the DenV detection limit from 150 ng mL−1 to 2.5 ng mL−1.310 The intrinsically conducting polypyrrole nanowires shown in Fig. 20B (diameter: 70 nm; length: 1 μm) are conducive to aptamer loading and are capable of distinguishing attomolar concentrations of viral proteins in artificial saliva within 5 s.450 The blue-emitting polydioctylfluorene dots exhibit theranostic properties due to the synchronous detection and blocking of influenza viruses in the presence of sialyl glycans.424 Nanocrystalline cellulose (width: ∼5 nm; length: ∼200 nm) shows low toxicity and special surface chemistry that enables the antibody-mediated agglutination and detection of DenV proteins.89,451 Fluorescent probes based on carbazole form thermoreversible nanoassemblies with extraordinary solid-state fluorescence.452 These probes do not form nanoaggregates in the presence of the target virus due to electrostatic association resulting in an immediate change in color.
Molecular machines are nanoscale devices that are capable of propulsion in liquids by themselves or via an external fuel. Molecular machines used for virus detection are oligonucleotide-based. Therefore, they show ultrasensitivity to target viruses. For example, a lock-like DNA machine (7.8 nm) with a hairpin DNA and fluorescent ancient DNA opens in the presence of just 900 aM of ZikV gene and subsequently undergoes strand displacement reactions induced by a DNA-based fuel.453 Although the molecular machine enhanced the fluorescence signal by 360%, a long time (150 min) was required for DNA hybridization. DNA walkers, which combine walking and tracking particles into locked strands of DNAzyme, show good continuity and signal accumulation even though their sensitivity is lower in comparison by 5 orders of magnitude.454 3D DNA walker-based sensors show high recovery (99.3–102.0%) in human blood samples but the sensor response takes almost 2 hours.455 The cascade amplification of SPR signals by double-layer DNA tetrahedron nanodevice facilitates the label- and enzyme-free interrogation of HIV by entropy-driven strand displacement reaction.425 However, recombinant polymerase-based amplification with Cas13a nanomachine shortens the assays time (5 min–1 h) in comparison to quantitative PCR (2 h) and real-time PCR (7 days).456
4.2 Virus tracking
Organic nanoparticles have scarcely received attention for virus tracking and imaging purposes which can be attributed to the scarcity of excited electrons in these particles in comparison to quantum dots and plasmonic nanoparticles. Their large size in comparison to inorganic nanoparticles hinders their internalization into host cells as well as virus capsids. Despite these obstacles, polymer nanoparticles have recently been exploited for imaging and even single virus tracking.457 For example, latex nanobeads of 20, 100, 300, and 1000 nm elucidate the size-dependent mechanism of virus internalization.458 It has been observed that 100–300 nm beads followed a clathrin-mediated pathway whereas 1000 nm particles were internalized via a caveolae-governed route. Further investigations with stimulated emission depletion super-resolution microscopy (STED) using the 100 and 300 nm particles revealed that the clathrin coat possesses an intrinsic size limit for the synthesis of the endocytic structure and the frequency with which this machinery was recruited was a function of the virus size.458 STED is also useful for imaging the binding of adenovirus with MIP-NPs (600 nm).459 The 100 nm virus particle can be accurately resolved in the silane MIPs owing to the synthesis of a Schiff base between the lysine residues of the adenovirus and the aldehyde functionality on the MIP-NP (Fig. 21). Adenoviruses are non-enveloped viruses often responsible for mild respiratory distress in children as well as fatal organ malfunction in immunocompromised individuals. These viruses have also been imaged with polyphenylene dendrimers (1st to 5th generation) to investigate their host-guest interactions as well as manipulate their in vivo distribution.460 In this case, the theranostic features of the dendrimers can be attributed to two factors: (i) they enhanced cellular transduction capabilities of the viral gene-transfer vectors in the absence of coxsackie adenovirus receptor, and (ii) they controlled the interaction between the cargo and the Stuart-Power X enzyme which is the cornerstone of virus interactions in blood.460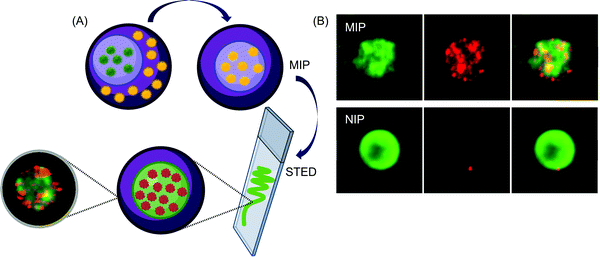 | ||
| Fig. 21 (A) Schematic illustration of molecularly imprinted polymeric nanoparticles for virus imaging; (B) dual-color imaging stimulated emission depletion (STED) microscopy of molecularly imprinted polymers (top) and non-imprinted polymers in the presence of adenovirus particles459 redrawn and reprinted with permission from (Anal. Chem. 2020, 92, 4, 3050–3057). Copyright 2020 American Chemical Society. | ||
Dye-loaded polymersome nanovesicles facilitate the study of virus replication and allow high-throughput antiviral drug screening.461 Initially, dye molecules from the polymersome are discharged to induce an endosomal escape. Subsequently, another dye of a different emission wavelength is targeted towards the Golgi complex. As a result, the budding and proliferation events in the virus replication cycle are tracked with the help of fluorescence imaging.461 Fluorescent polymer dots show excellent hole mobility, brightness as well as outstanding blue fluorescence. They are capable of suppressing the fluorescence of dye-stained cells infected with H3N2 without compromising the cell viability hinting at the scope of supramolecular nanomaterials for influenza virus treatment.424 Organic nanomaterials have also been employed as fluorescence amplifiers441 and virus substitutes to evaluate the midinfrared photothermal microscopy as a tool for virus-fingerprinting.462
5. Conclusions and future prospects
In this work, the latest developments in the role of various nanomaterials for sensing and imaging viruses were discussed. The incorporation of nanomaterials offers multifold benefits to the performance of virus sensors by improving the sensitivity, specificity, response time as well as recovery in complex media. A thorough understanding of nanomaterial interactions in these recognition systems such as nanoparticle size-dependent signal amplification, plasmonic resonance as well as charge transfer abilities could help accelerate the development of virus detection platforms with high veracity. The compatibility of nanomaterials across detection platforms including optical, electrochemical, or piezoelectric makes detection versatile and cost-effective. The spread of the global COVID-19 pandemic has incurred renewed interest in the development of alternatives to conventional virus detection systems such as PCR, ELISA, or flow cytometry. The amalgamation of nanosensors into an affordable paper strip or smartphone systems widens the scope of nanomaterials for POCT of viral pathogens in accordance with the WHO prescribed “ASSURED” (Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, Deliverable to end-users) criteria. Such POCT devices have the potential to timely assess the threat of infection at the patient level thus reducing the risk to healthcare professionals thereby helping curb the spread of epidemic viruses. Furthermore, nanomaterials are emerging to play an instrumental role in upgrading conventional techniques such as the development of nanoPCR as a highly sensitive, selective, rapid and cost-effective alternative to the traditional PCR technique which has often been dubbed as the gold standard in virus detection. Semi-synthetic and synthetic receptors such as aptamers and MIP-NPs are transforming into robust substitutes to the expensive and fragile natural receptors such as antibodies and DNA that previously dominated the field of virus detection. Another trend observed in recent years is the integration of different types of nanomaterials to generate multifunctional nanocomposites that not only positively influence the sensitivity but also improve the signal to noise ratio and make the platform suitable towards commercialization.In several nanomaterials, the particle size is smaller than the wavelength of the constituent electrons which results in quantum confinement enabling the synthesis of fluorescent nanoparticles. Since these nanoparticles are photostable and resistant to photobleaching, they have emerged as materials of interest for the non-invasive study of virus dissemination. Nanomaterial-mediated SVT helps understand virus internalization pathways and their replication mechanisms which is a prerequisite for adopting any therapeutic discourse. Fluorescent nanoparticles have emerged as promising alternatives to delicate and expensive labels such as green fluorescent protein. They also allow high throughput drug screening thus reducing development and trial costs. Nanoparticles have also been used in imaging studies to test the efficacy of emerging microscopic techniques for virus tracking. Since their size and dielectric constants are close to those of viruses, virus-mimetic nanoparticles are increasingly replacing virus-like particles in the study of intracellular host-guest interactions. Based on the recent trends in virus tracking methodologies, nanoparticles such as quantum dots, gold nanoparticles and nanodiamonds can be expected to contribute as fluorescent labels to a majority of virus imaging and internalization studies.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under the project number 428780268. Figure elements were created using BioRender.com.Notes and references
- S. Modrow, D. Falke, U. Truyen and H. Schätzl, in Molecular Virology, ed. S. Modrow, D. Falke, U. Truyen and H. Schätzl, Springer Berlin Heidelberg, Berlin, Heidelberg, 2013, pp. 17–30 Search PubMed.
- M. Breitbart and F. Rohwer, Trends Microbiol., 2005, 13, 278–284 CrossRef CAS PubMed.
- Ø. Bergh, K. Y. Børsheim, G. Bratbak and M. Heldal, Nature, 1989, 340, 467–468 CrossRef PubMed.
- M. W. Beijerinck, Verh. K. Akad. Wet. Amsterdam, Afd. Natuurkd., 1898, 6, 3–21 Search PubMed.
- International Committee on Taxonomy of Viruses, Virus Taxonomy: 2019 Release, https://talk.ictvonline.org/taxonomy/.
- R. L. Duda and C. M. Teschke, Curr. Opin. Virol., 2019, 36, 9–16 CrossRef CAS.
- K. J. Tompkins, M. Houtti, L. A. Litzau, E. J. Aird, B. A. Everett, A. T. Nelson, L. Pornschloegl, L. K. Limón-Swanson, R. L. Evans III, K. Evans, K. Shi, H. Aihara and W. R. Gordon, Nucleic Acids Res., 2021, 49, 1046–1064 CrossRef CAS PubMed.
- J. Shi, J. M. Perryman, X. Yang, X. Liu, D. M. Musser, A. K. Boehr, I. M. Moustafa, J. J. Arnold, C. E. Cameron and D. D. Boehr, Biochemistry, 2019, 58, 3735–3743 CrossRef CAS PubMed.
- C. San Martín and M. J. van Raaij, Virol. J., 2018, 15, 181 CrossRef PubMed.
- E. V. Koonin, T. G. Senkevich and V. V. Dolja, Biol. Direct, 2006, 1, 1–27 CrossRef PubMed.
- Z. Altintas, M. Gittens, A. Guerreiro, K. A. Thompson, J. Walker, S. Piletsky and I. E. Tothill, Anal. Chem., 2015, 87, 6801–6807 CrossRef CAS PubMed.
- A. Mokhtarzadeh, R. Eivazzadeh-keihan, P. Pashazadeh, M. Hejazi, N. Gharaatifar, M. Hasanzadeh, B. Baradaran and M. de la Guardia, Trends Anal. Chem., 2017, 97, 445–457 CrossRef CAS PubMed.
- F. Broecker and K. Moelling, Ann. N. Y. Acad. Sci., 2019, 1447, 53–68 CrossRef PubMed.
- A. J. Cann, Principles of molecular virology, Academic Press, Boston, 6th edn, 2016 Search PubMed.
- J. Brojatsch, J. Naughton, M. M. Rolls, K. Zingler and J. A. T. Young, Cell, 1996, 87, 845–855 CrossRef CAS PubMed.
- K. Peters, S. Chattopadhyay and G. C. Sen, J. Virol., 2008, 82, 3500 LP–3508 CrossRef PubMed.
- P. Danthi, T. Kobayashi, G. H. Holm, M. W. Hansberger, T. W. Abel and T. S. Dermody, J. Virol., 2008, 82, 161 LP–172 CrossRef PubMed.
- Y. Cho, S. Challa, D. Moquin, R. Genga, T. D. Ray, M. Guildford and F. K.-M. Chan, Cell, 2009, 137, 1112–1123 CrossRef CAS PubMed.
- K. G. Harris, S. A. Morosky, C. G. Drummond, M. Patel, C. Kim, D. B. Stolz, J. M. Bergelson, S. Cherry and C. B. Coyne, Cell Host Microbe, 2015, 18, 221–232 CrossRef CAS PubMed.
- T. Y. Tan and J. J. H. Chu, J. Gen. Virol., 2013, 94, 2215–2220 CrossRef CAS PubMed.
- K. M. Monroe, Z. Yang, J. R. Johnson, X. Geng, G. Doitsh, N. J. Krogan and W. C. Greene, Science, 2014, 343, 428 LP–432 CrossRef PubMed.
- P. Danthi, Annu. Rev. Virol., 2016, 3, 533–553 CrossRef CAS PubMed.
- D. Zhu, C. Pan, J. Sheng, H. Liang, Z. Bian, Y. Liu, P. Trang, J. Wu, F. Liu, C.-Y. Zhang and K. Zen, Nat. Microbiol., 2018, 3, 503–513 CrossRef CAS PubMed.
- J. Ribeiro, C. Oliveira, M. Malta and H. Sousa, Futur. Oncol., 2017, 13, 567–579 CrossRef CAS PubMed.
- A. M. Price, J. E. Messinger and M. A. Luftig, J. Virol., 2018, 92, e01178–17 CrossRef PubMed.
- S. Rasa, Z. Nora-Krukle, N. Henning, E. Eliassen, E. Shikova, T. Harrer, C. Scheibenbogen, M. Murovska and B. K. Prusty, J. Transl. Med., 2018, 16, 1–25 CrossRef PubMed.
- A. N. Bubak, V. Traina-Dorge, C. N. Como, B. Feia, C. M. Pearce, L. Doyle-Meyers, A. Das, J. Looper, R. Mahalingam and M. A. Nagel, J. Neurovirol., 2020, 26, 945–951 CrossRef CAS PubMed.
- J. T. Guidry, C. E. Birdwell and R. S. Scott, Oral Dis., 2018, 24, 497–508 CrossRef CAS PubMed.
- S. Ahmadi Ghezeldasht, S. A. A. Shamsian, J. Gholizadeh Navashenaq, R. Miri, F. Ashrafi, A. Mosavat and S. A. Rezaee, Rev. Med. Virol., 2021, 31, 1–15 CrossRef PubMed.
- R. E. Malosh, E. T. Martin, J. R. Ortiz and A. S. Monto, Vaccine, 2018, 36, 141–147 CrossRef PubMed.
- O. A. Khomich, S. N. Kochetkov, B. Bartosch and A. V. Ivanov, Viruses, 2018, 10, 392 CrossRef PubMed.
- A. M. Lachiewicz and M. L. Srinivas, Prev. Med. Rep., 2019, 16, 101016 CrossRef PubMed.
- Z. Altintas, J. Pocock, K. A. Thompson and I. E. Tothill, Biosens. Bioelectron., 2015, 74, 996–1004 CrossRef CAS PubMed.
- H. Meyer, R. Ehmann and L. Smith, Viruses, 2020, 12, 138 CrossRef CAS PubMed.
- Z.-Y. Jin, X. Liu, Y.-Y. Ding, Z.-F. Zhang and N. He, Sci. Rep., 2017, 7, 4890 CrossRef PubMed.
- C. R. Fisher, D. G. Streicker and M. J. Schnell, Nat. Rev. Microbiol., 2018, 16, 241–255 CrossRef CAS PubMed.
- J. A. Otter, C. Donskey, S. Yezli, S. Douthwaite, S. D. Goldenberg and D. J. Weber, J. Hosp. Infect., 2016, 92, 235–250 CrossRef CAS PubMed.
- P. Norrie, in A History of Disease in Ancient Times: More Lethal than War, ed. P. Norrie, Palgrave Macmillan, Cham, 2016, pp. 61–101 Search PubMed.
- P. E. Olson, C. S. Hames, A. S. Benenson and E. N. Genovese, Emerg. Infect. Dis., 1996, 2, 155–156 CrossRef CAS PubMed.
- S. Sabbatani and S. Fiorino, Le Infez. Med., 2009, 17, 261–275 CAS.
- A. Suzuki, Med. Hist., 2011, 55, 313–318 CrossRef PubMed.
- R. Acuna-Soto, D. W. Stahle, M. K. Cleaveland and M. D. Therrell, Emerg. Infect. Dis., 2002, 8, 360–362 CrossRef PubMed.
- D. J. Rogers, A. J. Wilson, S. I. Hay and A. J. Graham, in Advances in Parasitology, ed. S. I. Hay, A. Graham and D. J. B. T.-A. in P. Rogers, Academic Press, 2006, vol. 62, pp. 181–220 Search PubMed.
- D. M. Morens, M. North and J. K. Taubenberger, Lancet, 2010, 376, 1894–1895 CrossRef.
- T. N. Jilani, R. T. Jamil and A. H. Siddiqui, H1N1 Influenza (Swine Flu), StatPearls Publishing LLC, Treasure Island, Florida, 2019 Search PubMed.
- R. W. Eisinger and A. S. Fauci, Emerg. Infect. Dis., 2018, 24, 413–416 CrossRef PubMed.
- WHO, Weekly Epidemiological Update on COVID-19, 2021.
- A. S. Lima Neto, G. S. Sousa, O. J. Nascimento and M. C. Castro, PLoS Negl. Trop. Dis., 2019, 13, e0007575 CrossRef PubMed.
- A. King, New Sci., 2020, 246, 32–35 CrossRef CAS PubMed.
- J. E. Cogan, Dengue and severe dengue fact sheet, https://www.who.int/en/news-room/fact-sheets/detail/dengue-and-severe-dengue.
- M. Din, M. Asghar and M. Ali, J. Med. Virol., 2020, 93, 601–602 CrossRef PubMed.
- A. Aruna, P. Mbala, L. Minikulu, D. Mukadi, D. Bulemfu, F. Edidi, J. Bulabula, G. Tshapenda, J. Nsio, R. Kitenge, G. Mbuyi, C. Mwanzembe, J. Kombe, L. Lubula, J. C. Shako, M. Mossoko, F. Mulangu, A. Mutombo, E. Sana, Y. Tutu, L. Kabange, J. Makengo, F. Tshibinkufua, S. Ahuka-Mundeke, J.-J. Muyembe, E. R. Cdc and C. D. C. E. Response, MMWR Morb. Mortal. Wkly. Rep., 2019, 68, 1162–1165 CrossRef PubMed.
- R. R. Dourmashkin, G. Dunn, V. Castano and S. A. McCall, BMC Infect. Dis., 2012, 12, 136 CrossRef CAS PubMed.
- R. T. Ravenholt and W. H. Foege, Lancet, 1982, 320, 860–864 CrossRef.
- UNAIDS, Global HIV & AIDS statistics—2020 fact sheet.
- P. Heyman, L. Simons and C. Cochez, Viruses, 2014, 6, 151–171 CrossRef PubMed.
- T. Pietrasik, Up to 650 000 people die of respiratory diseases linked to seasonal flu each year, https://www.who.int/news/item/14-12-2017-up-to-650-000-people-die-of-respiratory-diseases-linked-to-seasonal-flu-each-year.
- D. M. Morens and A. S. Fauci, J. Infect. Dis., 2007, 195, 1018–1028 CrossRef PubMed.
- WHO, Report of the review committee on the functioning of the international health regulations (2005) in relation to pandemic (H1N1) 2009, 2011.
- O. Ogbu, E. Ajuluchukwu and C. J. Uneke, J. Vector Borne Dis., 2007, 44, 1–11 CAS.
- D. Kumar Pant, T. Tenzin, R. Chand, B. Kumar Sharma and P. Raj Bist, PLoS One, 2017, 12, e0180591 CrossRef PubMed.
- M. Grabowsky, JAMA Pediatr., 2014, 168, 108–109 CrossRef PubMed.
- ECDC, Geographical distribution of confirmed MERS-CoV cases by probable region of infection and exposure, from 1 January 2020 to 1 September 2020, https://www.ecdc.europa.eu/en/publications-data/geographical-distribution-confirmed-mers-cov-cases-probable-region-infection-0.
- Y. Ding, L. He, Q. Zhang, Z. Huang, X. Che, J. Hou, H. Wang, H. Shen, L. Qiu, Z. Li, J. Geng, J. Cai, H. Han, X. Li, W. Kang, D. Weng, P. Liang and S. Jiang, J. Pathol., 2004, 203, 622–630 CrossRef CAS PubMed.
- WHO, WHO coronavirus disease (COVID-19) dashboard, https://covid19.who.int/..
- G. A. Roth and E. Fee, Am. J. Public Health, 2011, 101, 1217 CrossRef PubMed.
- S. Smith, H. Harmanci, Y. Hutin, S. Hess, M. Bulterys, R. Peck, B. Rewari, A. Mozalevskis, M. Shibeshi, M. Mumba, L.-V. Le, N. Ishikawa, D. Nolna, L. Sereno, C. Gore, D. J. Goldberg and S. Hutchinson, JHEP Rep., 2019, 1, 81–89 CrossRef PubMed.
- K. D. Patterson, Soc. Sci. Med., 1992, 34, 855–865 CrossRef CAS PubMed.
- T. Ozer, B. J. Geiss and C. S. Henry, J. Electrochem. Soc., 2020, 167, 037523 CrossRef CAS PubMed.
- J. Flint, V. R. Racaniello, G. F. Rall, T. Hatziioannou and A. M. Skalka, Principles of Virology: Molecular biology, pathogenesis and control, ASM Press, Washington, DC, 5th edn, 2020 Search PubMed.
- H. H. Zalzala, New Microbes New Infect., 2020, 38, 100761 CrossRef CAS PubMed.
- M. Nagendran, J. John, K. Annamalai, M. I. Gandhi Sethuraman, N. Balamurugan, H. K. Rajendran, M. A. Deen Fakrudeen, R. Chandrasekar, S. Ranjan and V. C. Padmanaban, J. Drug Delivery Sci. Technol., 2020, 102120 Search PubMed.
- A. K. Giri and D. R. Rana, Biosaf. Heal., 2020, 2, 53–56 CrossRef.
- M. Pirzada, E. Sehit and Z. Altintas, Biosens. Bioelectron., 2020, 166, 112464 CrossRef CAS PubMed.
- E. Sehit, J. Drzazgowska, D. Buchenau, C. Yesildag, M. Lensen and Z. Altintas, Biosens. Bioelectron., 2020, 165, 112432 CrossRef CAS PubMed.
- L. Luo, W. Feng, W. Hu, C. Chen, H. Gong and C. Cai, Methods Appl. Fluoresc., 2019, 7, 015006 CrossRef CAS PubMed.
- R. Gaudin and J. G. Goetz, Trends Cell Biol., 2021, 31, 17–23 CrossRef CAS PubMed.
- S. J. Park, J. Kim, J. Kim, S. Kang, H. J. Cha, H. J. Cha, H. Shin, J. Park, Y. S. Jang, Y. S. Jang, J. S. Woo, J. S. Woo, C. Won, D. H. Min, D. H. Min and D. H. Min, Sci. Adv., 2020, 6, 1–12 Search PubMed.
- M. Pirzada and Z. Altintas, Sensors, 2019, 19, 5311 CrossRef PubMed.
- M. Kortel, B. D. Mansuriya, N. V. Santana and Z. Altintas, Micromachines, 2020, 11, 866 CrossRef PubMed.
- E. Sehit and Z. Altintas, Biosens. Bioelectron., 2020, 159, 112165 CrossRef CAS PubMed.
- B. D. Mansuriya and Z. Altintas, Sensors, 2020, 20, 1072 CrossRef CAS PubMed.
- Z. Altintas, Biosensors and Nanotechnology -Applications in Health Care Diagnostics, John Wiley & Sons Press, Hoboken, NJ, USA, 2017 Search PubMed.
- H. Ehtesabi, J. Sci. Adv. Mater. Dev., 2020, 5, 436–450 CAS.
- E. Vermisoglou, D. Panáček, K. Jayaramulu, M. Pykal, I. Frébort, M. Kolář, M. Hajdúch, R. Zbořil and M. Otyepka, Biosens. Bioelectron., 2020, 166, 112436 CrossRef CAS PubMed.
- C. Qin, W. Li, Q. Li, W. Yin, X. Zhang, Z. Zhang, X.-E. Zhang and Z. Cui, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 2577 LP–2582 CrossRef PubMed.
- Y. Cui, D. G. Grant, J. Lin, X. Yu and A. W. E. Franz, Viruses, 2019, 11 Search PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, N. A. A. Anas, N. S. M. Ramdzan and M. A. Mahdi, Results Phys., 2018, 11, 734–739 CrossRef.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, C. E. N. C. E. Chik and M. A. Mahdi, Sens. Bio-Sensing Res., 2018, 20, 16–21 CrossRef.
- Y. Wang, Y. Hu, Q. He, J. Yan, H. Xiong, N. Wen, S. Cai, D. Peng, Y. Liu and Z. Liu, Biosens. Bioelectron., 2020, 169, 112604 CrossRef CAS PubMed.
- G. Gao, Y. W. Jiang, W. Sun and F. G. Wu, Chin. Chem. Lett., 2018, 29, 1475–1485 CrossRef CAS.
- M. S. Draz and H. Shafiee, Theranostics, 2018, 8, 1985–2017 CrossRef CAS PubMed.
- J. Kang, A. Tahir, H. Wang and J. Chang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2021, 13, 1700 Search PubMed.
- B. V. Ribeiro, T. A. R. Cordeiro, G. R. Oliveira e Freitas, L. F. Ferreira and D. L. Franco, Talanta Open, 2020, 2, 100007 CrossRef PubMed.
- B. Alhalaili, I. N. Popescu, O. Kamoun, F. Alzubi, S. Alawadhia and R. Vidu, Sensors, 2020, 20 Search PubMed.
- S.-L. Liu, Z.-G. Wang, H.-Y. Xie, A.-A. Liu, D. C. Lamb and D.-W. Pang, Chem. Rev., 2020, 120, 1936–1979 CrossRef CAS PubMed.
- K. Kaiser, L. M. Scriven, F. Schulz, P. Gawel, L. Gross and H. L. Anderson, Science, 2019, 365, 1299–1301 CrossRef CAS PubMed.
- C. Chaicham, T. Tuntulani, V. Promarak and B. Tomapatanaget, Sens. Actuators, B, 2019, 282, 936–944 CrossRef CAS.
- A. Salaam, D. Dean and V. Thomas, in Drug Delivery Nanosystems for Biomedical Applications, ed. C. P. Sharma, Elsevier Inc., 2018, pp. 333–356 Search PubMed.
- A. Kurdekar, L. A. A. Chunduri, E. P. Bulagonda, M. K. Haleyurgirisetty, V. Kamisetti and I. K. Hewlett, Microfluid. Nanofluid., 2016, 20, 99 CrossRef.
- L. A. A. Chunduri, M. K. Haleyurgirisetty, S. Patnaik, P. E. Bulagonda, A. Kurdekar, J. Liu, I. K. Hewlett and V. Kamisetti, Microfluid. Nanofluid., 2016, 20, 1–10 CrossRef CAS.
- M. Zarei-Ghobadi, S.-H. Mozhgani, F. Dashtestani, A. Yadegari, F. Hakimian, M. Norouzi and H. Ghourchian, Sci. Rep., 2018, 8, 15593 CrossRef PubMed.
- L.-D. Xu, Q. Zhang, S.-N. Ding, J.-J. Xu and H.-Y. Chen, ACS Omega, 2019, 4, 21431–21438 CrossRef CAS PubMed.
- A. Valipour and M. Roushani, Biosens. Bioelectron., 2017, 89, 946–951 CrossRef CAS PubMed.
- J. Chen, M. Wang, X. Zhou, Y. Nie and X. Su, Sens. Actuators, B, 2021, 326, 128847 CrossRef CAS.
- K. Ghanbari, M. Roushani and A. Azadbakht, Anal. Biochem., 2017, 534, 64–69 CrossRef CAS PubMed.
- J. Ramos-Soriano, J. J. Reina, B. M. Illescas, N. de la Cruz, L. Rodríguez-Pérez, F. Lasala, J. Rojo, R. Delgado and N. Martín, J. Am. Chem. Soc., 2019, 141, 15403–15412 CrossRef CAS PubMed.
- C. Liu, T. Wang, J. Ji, C. Wang, H. Wang, P. Jin, W. Zhou and J. Jiang, J. Mater. Chem. C, 2019, 7, 10240–10246 RSC.
- J. Upan, P. Banet, P.-H. Aubert, K. Ounnunkad and J. Jakmunee, Electrochim. Acta, 2020, 349, 136335 CrossRef CAS.
- Z. Chaloupková, A. Balzerová, J. Bařinková, Z. Medříková, P. Šácha, P. Beneš, V. Ranc, J. Konvalinka and R. Zbořil, Anal. Chim. Acta, 2018, 997, 44–51 CrossRef PubMed.
- J. Lee, M. Morita, K. Takemura and E. Y. Park, Biosens. Bioelectron., 2018, 102, 425–431 CrossRef CAS PubMed.
- L. Farzin, S. Sadjadi, M. Shamsipur and S. Sheibani, J. Pharm. Biomed. Anal., 2020, 179, 112989 CrossRef CAS PubMed.
- H. Kong, W. Zhang, J. Yao, C. Li, R. Lu, Z. Guo, J. Li, C. Li, Y. Li, C. Zhang and L. Zhou, Sens. Actuators, B, 2020, 129118 Search PubMed.
- A. Roberts, N. Chauhan, S. Islam, S. Mahari, B. Ghawri, R. K. Gandham, S. S. Majumdar, A. Ghosh and S. Gandhi, Sci. Rep., 2020, 10, 14546 CrossRef CAS PubMed.
- G. Seo, G. Lee, M. J. Kim, S.-H. Baek, M. Choi, K. B. Ku, C.-S. Lee, S. Jun, D. Park, H. G. Kim, S.-J. Kim, J.-O. Lee, B. T. Kim, E. C. Park and S. Il Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- J. Lee, K. Takemura and E. Y. Park, Sens. Actuators, B, 2018, 276, 254–261 CrossRef CAS.
- R. M. Torrente-Rodríguez, H. Lukas, J. Tu, J. Min, Y. Yang, C. Xu, H. B. Rossiter and W. Gao, Matter, 2020, 3, 1981–1998 CrossRef PubMed.
- W. Huang, G. Xiang, D. Jiang, L. Liu, C. Liu, F. Liu and X. Pu, Electroanalysis, 2016, 28, 1126–1133 CrossRef CAS.
- K. Siuzdak, P. Niedziałkowski, M. Sobaszek, T. Łęga, M. Sawczak, E. Czaczyk, K. Dziąbowska, T. Ossowski, D. Nidzworski and R. Bogdanowicz, Sens. Actuators, B, 2019, 280, 263–271 CrossRef CAS.
- G. Z. Feuerstein, M. A. Mansfield, P. I. Lelkes, S. Alesci, C. Marcinkiewicz, N. Butlin and M. Sternberg, Int. J. Nanomedicine, 2020, 15, 7583–7599 CrossRef CAS PubMed.
- J. Li, K. Yang, Z. Wu, X. Li and Q. Duan, Talanta, 2019, 205, 120091 CrossRef CAS PubMed.
- J. Mandli, A. Attar, M. M. Ennaji and A. Amine, J. Electroanal. Chem., 2017, 799, 213–221 CrossRef CAS.
- F. Wang, S. C. B. Gopinath and T. Lakshmipriya, Int. J. Nanomedicine, 2019, 14, 8469–8481 CrossRef CAS PubMed.
- K. Ghanbari and M. Roushani, Sens. Actuators, B, 2018, 258, 1066–1071 CrossRef CAS.
- Y. Ma, X. L. Shen, Q. Zeng, H. S. Wang and L. S. Wang, Talanta, 2017, 164, 121–127 CrossRef CAS PubMed.
- Q. Palomar, X. X. Xu, C. Gondran, M. Holzinger, S. Cosnier and Z. Zhang, Microchim. Acta, 2020, 187, 1–10 CrossRef PubMed.
- X. Guo, J. Hou, Z. Yuan, H. Li and S. Sang, Nanotechnology, 2021, 32, 355102 CrossRef CAS PubMed.
- M. Sudan Guray, P. Minakshi, B. Brar, R. Rani and U. P. Lambe, J. Nanosci. Nanotechnol., 2021, 21, 3513–3523 CrossRef PubMed.
- P. G. Gopinath, V. R. Anitha and S. Aruna Mastani, Alexandria Eng. J., 2018, 57, 671–681 CrossRef.
- J. Narang, C. Singhal, N. Malhotra, S. Narang, A. K. P. N., R. Gupta, R. Kansal and C. S. Pundir, Biosens. Bioelectron., 2016, 86, 566–574 CrossRef CAS PubMed.
- D. G. A. Cabral, E. C. S. Lima, P. Moura and R. F. Dutra, Talanta, 2016, 148, 209–215 CrossRef CAS PubMed.
- Y. Fu, V. Romay, Y. Liu, B. Ibarlucea, L. Baraban, V. Khavrus, S. Oswald, A. Bachmatiuk, I. Ibrahim, M. Rümmeli, T. Gemming, V. Bezugly and G. Cuniberti, Sens. Actuators, B, 2017, 249, 691–699 CrossRef CAS.
- D. Wasik, A. Mulchandani and M. V. Yates, Biosens. Bioelectron., 2017, 91, 811–816 CrossRef CAS PubMed.
- Z. Jia, Y. Ma, L. Yang, C. Guo, N. Zhou, M. Wang, L. He and Z. Zhang, Biosens. Bioelectron., 2019, 133, 55–63 CrossRef CAS PubMed.
- T. L. Tran, T. T. Nguyen, T. T. Huyen Tran, V. T. Chu, Q. Thinh Tran and A. Tuan Mai, Phys. E, 2017, 93, 83–86 CrossRef CAS.
- S. R. Ahmed, J. Kim, T. Suzuki, J. Lee and E. Y. Park, Biosens. Bioelectron., 2016, 85, 503–508 CrossRef CAS PubMed.
- G. Cabral-Miranda, A. R. Cardoso, L. C. S. Ferreira, M. G. F. Sales and M. F. Bachmann, Biosens. Bioelectron., 2018, 113, 101–107 CrossRef CAS PubMed.
- P. D. Mendonça, L. K. B. Santos, M. V. Foguel, M. A. B. Rodrigues, M. T. Cordeiro, L. M. Gonçalves, E. T. A. Marques and R. F. Dutra, Anal. Bioanal. Chem., 2021, 413, 4873–4885 CrossRef PubMed.
- M. A. Tahir, S. Hameed, A. Munawar, I. Amin, S. Mansoor, W. S. Khan and S. Z. Bajwa, J. Virol. Methods, 2017, 249, 130–136 CrossRef CAS PubMed.
- S. Y. Kim, J.-C. Lee, G. Seo, J. H. Woo, M. Lee, J. Nam, J. Y. Sim, H.-R. Kim, E. C. Park and S. Park, Small Sci., 2021, n/a, 2100111 Search PubMed.
- Z. S. Miripour, R. Sarrami-Forooshani, H. Sanati, J. Makarem, M. S. Taheri, F. Shojaeian, A. H. Eskafi, F. Abbasvandi, N. Namdar, H. Ghafari, P. Aghaee, A. Zandi, M. Faramarzpour, M. Hoseinyazdi, M. Tayebi and M. Abdolahad, Biosens. Bioelectron., 2020, 165, 112435 CrossRef CAS PubMed.
- C. T. Xuan, N. T. Thuy, T. T. Luyen, T. T. T. Huyen and M. A. Tuan, J. Electron. Mater., 2017, 46, 3507–3511 CrossRef CAS.
- R. L. Pinals, F. Ledesma, D. Yang, N. Navarro, S. Jeong, J. E. Pak, L. Kuo, Y.-C. Chuang, Y.-W. Cheng, H.-Y. Sun and M. P. Landry, Nano Lett., 2021, 21, 2272–2280 CrossRef CAS PubMed.
- M. A. Zamzami, G. Rabbani, A. Ahmad, A. A. Basalah, W. H. Al-Sabban, S. Nate Ahn and H. Choudhry, Bioelectrochemistry, 2022, 143, 107982 CrossRef CAS PubMed.
- Y.-T. Yeh, Y. Tang, A. Sebastian, A. Dasgupta, N. Perea-Lopez, I. Albert, H. Lu, M. Terrones and S.-Y. Zheng, Sci. Adv., 2016, 2, e1601026 CrossRef PubMed.
- S. Jeong, E. González-Grandío, N. Navarro, R. L. Pinals, F. Ledesma, D. Yang and M. P. Landry, ACS Nano, 2021, 15, 10309–10317 CrossRef PubMed.
- M. Amin, O. Siddiqui, H. Abutarboush, M. Farhat and R. Ramzan, Carbon N. Y., 2021, 176, 580–591 CrossRef CAS PubMed.
- S. Kim, H. Ryu, S. Tai, M. Pedowitz, J. R. Rzasa, D. J. Pennachio, J. R. Hajzus, D. K. Milton, R. Myers-Ward and K. M. Daniels, Biosens. Bioelectron., 2022, 197, 113803 CrossRef CAS PubMed.
- J. Basu, N. Samanta, S. Jana and C. RoyChaudhuri, Microelectron. Reliab., 2018, 91, 154–159 CrossRef CAS.
- J. Gao, C. Wang, C. Wang, Y. Chu, S. Wang, M. Yuan Sun, H. Ji, Y. Gao, Y. Wang, Y. Han, F. Song, H. Liu, Y. Zhang and L. Han, Anal. Chem., 2022, 94, 1626–1636 CrossRef CAS PubMed.
- Y. Li, Z. Peng, N. J. Holl, M. R. Hassan, J. M. Pappas, C. Wei, O. H. Izadi, Y. Wang, X. Dong, C. Wang, Y.-W. Huang, D. Kim and C. Wu, ACS Omega, 2021, 6, 6643–6653 CrossRef CAS PubMed.
- K. Navakul, C. Warakulwit, P. Yenchitsomanus, A. Panya, P. A. Lieberzeit and C. Sangma, Nanomedicine, 2017, 13, 549–557 CrossRef CAS PubMed.
- J. Wen, W. Li, J. Li, B. Tao, Y. Xu, H. Li, A. Lu and S. Sun, Sens. Actuators, B, 2016, 227, 655–659 CrossRef CAS.
- M. Z. Ghobadi, S.-H. Mozhgani, F. Hakimian, M. Norouzi, S. A. Rezaee and H. Ghourchian, Heliyon, 2018, 4, e00996 CrossRef PubMed.
- J.-W. Kim, K.-W. Park, M. Kim, K. K. Lee and C.-S. Lee, Nanomaterials, 2022, 12 CAS.
- R. Tarcan, O. Todor-Boer, I. Petrovai, C. Leordean, S. Astilean and I. Botiz, J. Mater. Chem. C, 2020, 8, 1198–1224 RSC.
- B. Nagar, M. Balsells, A. de la Escosura-Muñiz, P. Gomez-Romero and A. Merkoçi, Biosens. Bioelectron., 2019, 129, 238–244 CrossRef CAS PubMed.
- A. Roberts, V. Kesarwani, R. Gupta and S. Gandhi, Biosens. Bioelectron., 2022, 198, 113837 CrossRef CAS PubMed.
- F. Chekin, K. Bagga, P. Subramanian, R. Jijie, S. K. Singh, S. Kurungot, R. Boukherroub and S. Szunerits, Sens. Actuators, B, 2018, 262, 991–1000 CrossRef CAS.
- M. F. Abd Muain, K. H. Cheo, M. N. Omar, A. S. Amir Hamzah, H. N. Lim, A. B. Salleh, W. S. Tan and A. Ahmad Tajudin, Bioelectrochemistry, 2018, 122, 199–205 CrossRef CAS PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, Y. Mustapha Kamil, W. M. E. M. M. Daniyal, A. R. Sadrolhosseini and M. A. Mahdi, Sci. Rep., 2020, 10, 2374 CrossRef CAS PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, A. R. Sadrolhosseini, Y. M. Kamil, N. I. M. Fauzi, H. S. Hashim and M. A. Mahdi, Nanomaterials, 2020, 10, 1–14 CrossRef PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, M. H. M. Zaid and M. A. Mahdi, Optik, 2018, 171, 934–940 CrossRef CAS.
- V. Georgakilas, J. A. Perman, J. Tucek and R. Zboril, Chem. Rev., 2015, 115, 4744–4822 CrossRef CAS PubMed.
- O. J. Achadu, F. Abe, F. Hossain, F. Nasrin, M. Yamazaki, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2021, 193, 113540 CrossRef CAS PubMed.
- L.-D. Xu, S.-W. Wang, J. Zhu, T. Zhou and S.-N. Ding, ChemistrySelect, 2021, 6, 9787–9793 CrossRef CAS.
- L.-D. Xu, F.-L. Du, J. Zhu and S.-N. Ding, Analyst, 2021, 146, 706–713 RSC.
- K. Takemura, J. Satoh, J. Boonyakida, S. Park, A. D. Chowdhury and E. Y. Park, J. Nanobiotechnol., 2020, 18, 152 CrossRef CAS PubMed.
- S. Savas and Z. Altintas, Materials, 2019, 12, 2189 CrossRef CAS PubMed.
- N. Li, L. Shi, X. Zou, T. Wang, D. Wang, Z. Gong and M. Fan, Microchem. J., 2022, 173, 107046 CrossRef CAS PubMed.
- Q. Xiang, J. Huang, H. Huang, W. Mao and Z. Ye, RSC Adv., 2018, 8, 1820–1825 RSC.
- A. D. Chowdhury, A. B. Ganganboina, F. Nasrin, K. Takemura, R. Doong, D. I. S. Utomo, J. Lee, I. M. Khoris and E. Y. Park, Anal. Chem., 2018, 90, 12464–12474 CrossRef PubMed.
- Y. Li, P. Ma, Q. Tao, H.-J. Krause, S. Yang, G. Ding, H. Dong and X. Xie, Sens. Actuators, B, 2021, 337, 129786 CrossRef CAS PubMed.
- J. L. Gogola, G. Martins, A. Gevaerd, L. Blanes, J. Cardoso, F. K. Marchini, C. E. Banks, M. F. Bergamini and L. H. Marcolino-Junior, Anal. Chim. Acta, 2021, 1166, 338548 CrossRef CAS PubMed.
- Y.-D. Ye, L. Xia, D.-D. Xu, X.-J. Xing, D.-W. Pang and H.-W. Tang, Biosens. Bioelectron., 2016, 85, 837–843 CrossRef CAS PubMed.
- Y. Li, J. Li, Y. Cao, P. Jiang, Y. Tang, Z. Chen and K. Han, Anal. Chim. Acta, 2022, 1203, 339693 CrossRef CAS PubMed.
- O. J. Achadu, D. X. Lioe, K. Kagawa, S. Kawahito and E. Y. Park, Microchim. Acta, 2020, 187, 466 CrossRef CAS PubMed.
- D. Nidzworski, K. Siuzdak, P. Niedziałkowski, R. Bogdanowicz, M. Sobaszek, J. Ryl, P. Weiher, M. Sawczak, E. Wnuk, W. A. Goddard, A. Jaramillo-Botero and T. Ossowski, Sci. Rep., 2017, 7, 15707 CrossRef PubMed.
- S. Ramanathan, S. C. B. Gopinath, Z. H. Ismail, M. K. Md Arshad and P. Poopalan, Biosens. Bioelectron., 2022, 197, 113735 CrossRef CAS PubMed.
- A. Valipour and M. Roushani, Microchim. Acta, 2017, 184, 2015–2022 CrossRef CAS.
- A. Pérez-García, A. Aguinaga, A. Navascués, J. Castilla and C. Ezpeleta, Int. J. Infect. Dis., 2019, 89, 131–136 CrossRef PubMed.
- H. C. Lai, S. F. Chin, S. C. Pang, M. S. Henry Sum and D. Perera, J. Nanomater., 2017, 2017, 3615707 Search PubMed.
- M. S. Purdey, J. G. Thompson, T. M. Monro, A. D. Abell and E. P. Schartner, Sensors, 2015, 15, 31904–31913 CrossRef CAS PubMed.
- J. T. Trinh, M. H. Alkahtani, I. Rampersaud, A. Rampersaud, M. Scully, R. F. Young, P. Hemmer and L. Zeng, Biotechnol. Bioeng., 2018, 115, 1427–1436 CrossRef CAS PubMed.
- Y. Wu, S. Cao, M. N. A. Alam, M. Raabe, S. Michel-Souzy, Z. Wang, M. Wagner, A. Ermakova, J. J. L. M. Cornelissen and T. Weil, J. Mater. Chem. B, 2021, 9, 5621–5627 RSC.
- R. S. Li, B. Yuan, J. H. Liu, M. L. Liu, P. F. Gao, Y. F. Li, M. Li and C. Z. Huang, J. Mater. Chem. B, 2017, 5, 8719–8724 RSC.
- A. D. Chowdhury, S. Sharmin, F. Nasrin, M. Yamazaki, F. Abe, T. Suzuki and E. Y. Park, ACS Appl. Bio Mater., 2020, 3, 3560–3568 CrossRef CAS PubMed.
- J. T. Hong, S. W. Jun, S. H. Cha, J. Y. Park, S. Lee, G. A. Shin and Y. H. Ahn, Sci. Rep., 2018, 8, 15536 CrossRef CAS PubMed.
- Z. Rahmati, M. Roushani and H. Hosseini, Surf. Interfaces, 2021, 22, 100813 CrossRef CAS.
- Y. Liu, Y. Nie, M. Wang, Q. Zhang and Q. Ma, Biosens. Bioelectron., 2020, 148, 111823 CrossRef CAS PubMed.
- M. Shariati, Biosens. Bioelectron., 2018, 105, 58–64 CrossRef CAS PubMed.
- C. Li, Z. Zou, H. Liu, Y. Jin, G. Li, C. Yuan, Z. Xiao and M. Jin, Talanta, 2021, 225, 122064 CrossRef CAS PubMed.
- F. Nasrin, A. D. Chowdhury, A. B. Ganganboina, O. J. Achadu, F. Hossain, M. Yamazaki and E. Y. Park, Microchim. Acta, 2020, 187, 674 CrossRef CAS PubMed.
- G. Eom, A. Hwang, H. Kim, S. Yang, D. K. Lee, S. Song, K. Ha, J. Jeong, J. Jung, E.-K. Lim and T. Kang, ACS Sens., 2019, 4, 2282–2287 CrossRef CAS PubMed.
- Q. Lv, Y. Wang, C. Su, T. Lakshmipriya, S. C. B. Gopinath, K. Pandian, V. Perumal and Y. Liu, Int. J. Biol. Macromol., 2019, 134, 354–360 CrossRef CAS PubMed.
- S. R. Ahmed, J. C. Corredor, É. Nagy and S. Neethirajan, Nanotheranostics, 2017, 1, 338–345 CrossRef PubMed.
- A. B. Ganganboina, A. D. Chowdhury, I. M. Khoris, R. Doong, T.-C. Li, T. Hara, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2020, 170, 112680 CrossRef CAS PubMed.
- S. R. Ahmed, S. W. Kang, S. Oh, J. Lee and S. Neethirajan, Heliyon, 2018, 4, e00766 CrossRef PubMed.
- D. H. Mohsin, M. S. Mashkour and F. Fatemi, Chem. Pap., 2021, 75, 279–295 CrossRef CAS.
- A. D. Chowdhury, K. Takemura, I. M. Khorish, F. Nasrin, M. M. Ngwe Tun, K. Morita and E. Y. Park, Nanoscale Adv., 2020, 2, 699–709 RSC.
- C. M. Das, Y. Guo, G. Yang, L. Kang, G. Xu, H.-P. Ho and K.-T. Yong, Adv. Theory Simulations, 2020, 3, 2000185 CrossRef CAS PubMed.
- J.-E. Park, K. Kim, Y. Jung, J.-H. Kim and J.-M. Nam, ChemNanoMat, 2016, 2, 927–936 CrossRef CAS.
- S. Manivannan and K. Ponnuchamy, Appl. Organomet. Chem., 2020, 34, e5887 CrossRef CAS PubMed.
- M. S. Draz, A. Vasan, A. Muthupandian, M. K. Kanakasabapathy, P. Thirumalaraju, A. Sreeram, S. Krishnakumar, V. Yogesh, W. Lin, X. G. Yu, R. T. Chung and H. Shafiee, Sci. Adv., 2020, 6, eabd5354 CrossRef CAS PubMed.
- P. Kannan, P. Subramanian, T. Maiyalagan and Z. Jiang, Anal. Chem., 2019, 91, 5824–5833 CrossRef CAS PubMed.
- X. Ma and P. Miao, J. Mater. Chem. B, 2019, 7, 2608–2612 RSC.
- A. B. Ganganboina, I. M. Khoris, A. D. Chowdhury, T.-C. Li and E. Y. Park, ACS Appl. Mater. Interfaces, 2020, 12, 50212–50221 CrossRef CAS PubMed.
- S. Xu, W. Ouyang, P. Xie, Y. Lin, B. Qiu, Z. Lin, G. Chen and L. Guo, Anal. Chem., 2017, 89, 1617–1623 CrossRef CAS PubMed.
- M. Shariati, M. Vaezjalali and M. Sadeghi, Anal. Chim. Acta, 2021, 1156, 338360 CrossRef CAS PubMed.
- Y. Zhang, G. Wu, J. Wei, Y. Ding, Y. Wei, Q. Liu and H. Chen, Microchim. Acta, 2021, 188, 3 CrossRef CAS PubMed.
- S. Yadav, S. Senapati, D. Desai, S. Gahlaut, S. Kulkarni and J. P. Singh, Colloids Surf., B, 2021, 198, 111477 CrossRef CAS PubMed.
- A. Cossettini and L. Selmi, IEEE Trans Nanobioscience, 2018, 17, 102–109 Search PubMed.
- R. Viter, M. Savchuk, N. Starodub, Z. Balevicius, S. Tumenas, A. Ramanaviciene, D. Jevdokimovs, D. Erts, I. Iatsunskyi and A. Ramanavicius, Sens. Actuators, B, 2019, 285, 601–606 CrossRef CAS.
- B. Negahdari, M. Darvishi and A. A. Saeedi, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 455–461 CrossRef CAS PubMed.
- D. Li, H. Chen, X. Gao, X. Mei and L. Yang, ACS Sens., 2021, 6, 613–627 CrossRef CAS PubMed.
- M. W. Kim, H.-J. Park, C. Y. Park, J. H. Kim, C. H. Cho, L. M. T. Phan, J. P. Park, S. K. Kailasa, C.-H. Lee and T. J. Park, RSC Adv., 2020, 10, 29759–29764 RSC.
- Z. Ghafary, R. Hallaj, A. Salimi and K. Akhtari, ACS Omega, 2019, 4, 15323–15336 CrossRef CAS PubMed.
- S.-L. Hong, N. Zhang, L. Qin, M. Tang, Z. Ai, A. Chen, S. Wang and K. Liu, Analyst, 2021, 146, 930–936 RSC.
- C. Srisomwat, A. Yakoh, N. Chuaypen, P. Tangkijvanich, T. Vilaivan and O. Chailapakul, Anal. Chem., 2021, 93, 2879–2887 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- M. Steinmetz, D. Lima, A. G. Viana, S. T. Fujiwara, C. A. Pessôa, R. M. Etto and K. Wohnrath, Biosens. Bioelectron., 2019, 141, 111351 CrossRef CAS PubMed.
- M. R. Rakhshani, IEEE Sens. J., 2022, 1 Search PubMed.
- P. Etedali, M. Behbahani, H. Mohabatkar and G. Dini, Aquaculture, 2022, 548, 737628 CrossRef CAS.
- S. Cajigas, D. Alzate and J. Orozco, Microchim. Acta, 2020, 187, 594 CrossRef CAS PubMed.
- E. Ç. Yeter, S. Şahin, M. O. Caglayan and Z. Üstündağ, Chem. Pap., 2021, 75, 77–87 CrossRef CAS PubMed.
- L. A. Layqah and S. Eissa, Microchim. Acta, 2019, 186, 224 CrossRef PubMed.
- A. S. Mohammed, R. Nagarjuna, M. N. Khaja, R. Ganesan and J. Ray Dutta, Microchim. Acta, 2019, 186, 566 CrossRef PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439 LP–444 CrossRef PubMed.
- H. Jiang, Z. Sun, C. Zhang and X. Weng, Sens. Actuators, B, 2022, 354, 131232 CrossRef CAS.
- Z. Yao, Q. Zhang, W. Zhu, M. Galluzzi, W. Zhou, J. Li, A. V. Zayats and X.-F. Yu, Nanoscale, 2021, 13, 10133–10142 RSC.
- N. Cennamo, L. Pasquardini, F. Arcadio, L. Lunelli, L. Vanzetti, V. Carafa, L. Altucci and L. Zeni, Talanta, 2021, 233, 122532 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, L. Saleh, J. Tang, T. Gui, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2021, 15, 7536–7546 CrossRef CAS PubMed.
- W. A. El-Said, A. S. Al-Bogami and W. Alshitari, Spectrochim. Acta, Part A, 2022, 264, 120237 CrossRef CAS PubMed.
- J. Lin, S. C. B. Gopinath, T. Lakshmipriya, Y. Chen, W. R. Yuan and M. Yang, Int. J. Biol. Macromol., 2019, 141, 564–569 CrossRef CAS PubMed.
- A. D. Chowdhury, K. Takemura, T.-C. Li, T. Suzuki and E. Y. Park, Nat. Commun., 2019, 10, 3737 CrossRef PubMed.
- W.-P. Lin, W.-J. Wang, C.-H. Lee, F.-J. Jan and G.-J. Wang, Sens. Actuators, B, 2022, 350, 130875 CrossRef CAS.
- F. Inci, M. G. Karaaslan, A. Mataji-Kojouri, P. A. Shah, Y. Saylan, Y. Zeng, A. Avadhani, R. Sinclair, D. T.-Y. Lau and U. Demirci, Appl. Mater. Today, 2020, 20, 100709 CrossRef.
- Y. Sun, L. Xu, F. Zhang, Z. Song, Y. Hu, Y. Ji, J. Shen, B. Li, H. Lu and H. Yang, Biosens. Bioelectron., 2017, 89, 906–912 CrossRef CAS PubMed.
- H.-W. Chu, C.-S. Lai, J.-Y. Ko, S. G. Harroun, C.-I. Chuang, R. Y. L. Wang, B. Unnikrishnan and C.-C. Huang, ACS Sens., 2019, 4, 1543–1551 CrossRef CAS PubMed.
- C. Singhal, A. Dubey, A. Mathur, C. S. Pundir and J. Narang, Process Biochem., 2018, 74, 35–42 CrossRef CAS.
- P. Brangel, A. Sobarzo, C. Parolo, B. S. Miller, P. D. Howes, S. Gelkop, J. J. Lutwama, J. M. Dye, R. A. McKendry, L. Lobel and M. M. Stevens, ACS Nano, 2018, 12, 63–73 CrossRef CAS PubMed.
- P. Teengam, W. Siangproh, S. Tontisirin, A. Jiraseree-amornkun, N. Chuaypen, P. Tangkijvanich, C. S. Henry, N. Ngamrojanavanich and O. Chailapakul, Sens. Actuators, B, 2021, 326, 128825 CrossRef CAS.
- S. G. Hwang, K. Ha, K. Guk, D. K. Lee, G. Eom, S. Song, T. Kang, H. Park, J. Jung and E.-K. Lim, Sci. Rep., 2018, 8, 12999 CrossRef PubMed.
- C. Huang, T. Wen, F.-J. Shi, X.-Y. Zeng and Y.-J. Jiao, ACS Omega, 2020, 5, 12550–12556 CrossRef CAS PubMed.
- G. Eom, A. Hwang, D. K. Lee, K. Guk, J. Moon, J. Jeong, J. Jung, B. Kim, E.-K. Lim and T. Kang, ACS Appl. Bio Mater., 2019, 2, 1233–1240 CrossRef CAS PubMed.
- M. E. Hamdy, M. Del Carlo, H. A. Hussein, T. A. Salah, A. H. El-Deeb, M. M. Emara, G. Pezzoni and D. Compagnone, J. Nanobiotechnol., 2018, 16, 48 CrossRef PubMed.
- H. H. Fakih, M. M. Itani and P. Karam, Sens. Actuators, B, 2017, 250, 446–452 CrossRef CAS.
- J.-W. Kim, M. Kim, K. K. Lee, K. H. Chung and C.-S. Lee, Nanomater., 2020, 10 Search PubMed.
- Z. Liu, J. Li, Z. Liu, J. Li, Z. Li, C. Wang, J. Wang and L. Guo, BMC Vet. Res., 2019, 15, 110 CrossRef PubMed.
- M. Wang, Y. Yan, R. Wang, L. Wang, H. Zhou, Y. Li, L. Tang, Y. Xu, Y. Jiang, W. Cui and X. Qiao, Front. Microbiol., 2019, 10, 2884 CrossRef PubMed.
- L. Zhang, Y. Luo, L. Liang, J. Li and S. Cui, Infect., Genet. Evol., 2018, 60, 1–6 CrossRef CAS PubMed.
- F. Zang, Z. Su, L. Zhou, K. Konduru, G. Kaplan and S. Y. Chou, Adv. Mater., 2019, 31, 1902331 CrossRef PubMed.
- F. Nasrin, A. D. Chowdhury, K. Takemura, J. Lee, O. Adegoke, V. K. Deo, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2018, 122, 16–24 CrossRef CAS PubMed.
- K. Takemura, J. Lee, T. Suzuki, T. Hara, F. Abe and E. Y. Park, Sens. Actuators, B, 2019, 296, 126672 CrossRef CAS.
- F. Nasrin, A. D. Chowdhury, K. Takemura, I. Kozaki, H. Honda, O. Adegoke and E. Y. Park, Anal. Chim. Acta, 2020, 1109, 148–157 CrossRef CAS PubMed.
- O. Adegoke, M. Morita, T. Kato, M. Ito, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2017, 94, 513–522 CrossRef CAS PubMed.
- M. Shariati, M. Ghorbani, P. Sasanpour and A. Karimizefreh, Anal. Chim. Acta, 2019, 1048, 31–41 CrossRef CAS PubMed.
- S. L. Manoto, A. El-Hussein, R. Malabi, L. Thobakgale, S. Ombinda-Lemboumba, Y. A. Attia, M. A. Kasem and P. Mthunzi-Kufa, Saudi J. Biol. Sci., 2021, 28, 78–89 CrossRef CAS PubMed.
- S. Oh, J. Kim, V. T. Tran, D. K. Lee, S. R. Ahmed, J. C. Hong, J. Lee, E. Y. Park and J. Lee, ACS Appl. Mater. Interfaces, 2018, 10, 12534–12543 CrossRef CAS PubMed.
- E. Alipour, S. P. Shariatpanahi, H. Ghourchian, B. Piro, M. Fathipour, S. M. Boutorabi, S. L. Znoyko and P. I. Nikitin, Microchim. Acta, 2020, 187, 463 CrossRef CAS PubMed.
- S. R. Ahmed, J. Mogus, R. Chand, E. Nagy and S. Neethirajan, Biosens. Bioelectron., 2018, 103, 45–53 CrossRef CAS PubMed.
- B. Luo, Y. Xu, S. Wu, M. Zhao, P. Jiang, S. Shi, Z. Zhang, Y. Wang, L. Wang and Y. Liu, Biosens. Bioelectron., 2018, 100, 169–175 CrossRef CAS PubMed.
- S. R. Ahmed, É. Nagy and S. Neethirajan, RSC Adv., 2017, 7, 40849–40857 RSC.
- A. Karimizefreh, F. A. Mahyari, M. VaezJalali, R. Mohammadpour and P. Sasanpour, Microchim. Acta, 2017, 184, 1729–1737 CrossRef CAS.
- S. R. Ahmed, J. Kim, V. T. Tran, T. Suzuki, S. Neethirajan, J. Lee and E. Y. Park, Sci. Rep., 2017, 7, 44495 CrossRef CAS PubMed.
- Y. Zhang, Y. Gao, X. Zhang, H. Wang, T. Xia, C. Bian, S. Liang, X. Tang and X. Wang, Sens. Actuators, B, 2019, 284, 296–304 CrossRef CAS.
- L. E. Ahangar and M. A. Mehrgardi, Bioelectrochemistry, 2017, 117, 83–88 CrossRef CAS PubMed.
- S. C. Razo, V. G. Panferov, I. V. Safenkova, Y. A. Varitsev, A. V. Zherdev, E. N. Pakina and B. B. Dzantiev, Sensors, 2018, 18 Search PubMed.
- R. Jin, L. Zhai, Q. Zhu, J. Feng and X. Pan, Aquaculture, 2020, 528, 735554 CrossRef CAS.
- A. Bosak, N. Saraf, A. Willenberg, M. W. C. Kwan, B. W. Alto, G. W. Jackson, R. H. Batchelor, T. D. Nguyen-Huu, V. Sankarapani, G. D. Parks, S. Seal and B. J. Willenberg, RSC Adv., 2019, 9, 23752–23763 RSC.
- L.-H. Xiong, S. Huang, Y. Huang, F. Yin, F. Yang, Q. Zhang, J. Cheng, R. Zhang and X. He, ACS Appl. Mater. Interfaces, 2020, 12, 12525–12532 CrossRef CAS PubMed.
- X. Mi, E. M. Lucier, D. G. Turpeinen, E. L. L. Yeo, J. C. Y. Kah and C. L. Heldt, Analyst, 2019, 144, 5486–5496 RSC.
- L. Zhan, W. B. Wu, C. M. Li and C. Z. Huang, J. Anal. Test., 2017, 1, 298–305 CrossRef.
- M. Khater, A. de la Escosura-Muñiz, D. Quesada-González and A. Merkoçi, Anal. Chim. Acta, 2019, 1046, 123–131 CrossRef CAS PubMed.
- T. Ngamdee, L. S. Yin, S. Vongpunsawad, Y. Poovorawan, W. Surareungchai and B. Lertanantawong, Anal. Chim. Acta, 2020, 1134, 10–17 CrossRef CAS PubMed.
- M. Khater, A. de la Escosura-Muñiz, L. Altet and A. Merkoçi, Anal. Chem., 2019, 91, 4790–4796 CrossRef CAS PubMed.
- J. Kim, S. Y. Oh, S. Shukla, S. B. Hong, N. S. Heo, V. K. Bajpai, H. S. Chun, C.-H. Jo, B. G. Choi, Y. S. Huh and Y.-K. Han, Biosens. Bioelectron., 2018, 107, 118–122 CrossRef CAS PubMed.
- O. Adegoke and E. Y. Park, J. Mater. Chem. B, 2017, 5, 3047–3058 RSC.
- I. M. Khoris, A. D. Chowdhury, T.-C. Li, T. Suzuki and E. Y. Park, Anal. Chim. Acta, 2020, 1110, 64–71 CrossRef CAS PubMed.
- L. Zhan, T. Yang, C. M. Li, W. B. Wu and C. Z. Huang, Sens. Actuators, B, 2018, 255, 1291–1297 CrossRef CAS.
- S. R. Ahmed, K. Takemeura, T.-C. Li, N. Kitamoto, T. Tanaka, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2017, 87, 558–565 CrossRef CAS PubMed.
- A. Karami and M. Hasani, Anal. Chim. Acta, 2020, 1102, 119–129 CrossRef CAS PubMed.
- C. Lee, P. Wang, M. A. Gaston, A. A. Weiss and P. Zhang, in Biosensors and Biodetection. Methods in Molecular Biology, ed. A. Rasooly and B. Prickril, Humana Press, New York, NY, 2017, Vol. 157, pp. 109–116 Search PubMed.
- Y. Horiguchi, T. Goda, A. Matsumoto, H. Takeuchi, S. Yamaoka and Y. Miyahara, Langmuir, 2019, 35, 1798–1806 CrossRef CAS PubMed.
- Y.-W. Zhang, W.-S. Liu, J.-S. Chen, H.-L. Niu, C.-J. Mao and B.-K. Jin, Sens. Actuators, B, 2020, 321, 128456 CrossRef CAS.
- B. Sun, J. Dong, L. Cui, T. Feng, J. Zhu, X. Liu and S. Ai, Biosens. Bioelectron., 2019, 124–125, 1–7 CrossRef CAS PubMed.
- Y. Hu, Y. Huang, Y. Wang, C. Li, W. Wong, X. Ye and D. Sun, Anal. Chim. Acta, 2018, 1035, 136–145 CrossRef CAS PubMed.
- Y. Tian, Q. Lu, X. Guo, S. Wang, Y. Gao and L. Wang, Nanoscale, 2020, 12, 7776–7781 RSC.
- Z. Han, L. Chen, Q. Weng, Y. Zhou, L. Wang, C. Li and J. Chen, Sens. Actuators, B, 2018, 258, 508–516 CrossRef CAS.
- T. A. Freitas, C. A. Proença, T. A. Baldo, E. M. Materón, A. Wong, R. F. Magnani and R. C. Faria, Talanta, 2019, 205, 120110 CrossRef CAS PubMed.
- L. Wang, M. Ren, L. Liang and C. Zhang, Chem. Sci., 2018, 9, 4942–4949 RSC.
- M. Aminul Islam and M. Ziaul Ahsan, Am. J. Nanosci., 2020, 6, 6 CrossRef.
- H. Tan, G. Tang, Z. Wang, Q. Li, J. Gao and S. Wu, Anal. Chim. Acta, 2016, 940, 136–142 CrossRef CAS PubMed.
- H. Yoo, J. Shin, J. Sim, H. Cho and S. Hong, Biosens. Bioelectron., 2020, 168, 112561 CrossRef CAS PubMed.
- S. Zhou, M. Jin, R. Tan, Z. Shen, J. Yin, Z. Qiu, Z. Chen, D. Shi, H. Li, Z. Yang, H. Wang, Z. Gao, J. Li and D. Yang, Chemosphere, 2021, 132995 Search PubMed.
- L. Ye, X. Zhang, N. Yang, S. Zhao, H. Wang and Z. Wang, J. Biomed. Nanotechnol., 2021, 17, 606–614 CrossRef CAS PubMed.
- R. Wu, B. Meng, M. Corredig and M. W. Griffiths, Food Microbiol., 2022, 102, 103921 CrossRef CAS PubMed.
- B. Yin, W. K. H. Ho, Q. Zhang, C. Li, Y. Huang, J. Yan, H. Yang, J. Hao, S. H. D. Wong and M. Yang, ACS Appl. Mater. Interfaces, 2022, 14, 4714–4724 CrossRef CAS PubMed.
- K. Wu, R. Saha, D. Su, V. D. Krishna, J. Liu, M. C.-J. Cheeran and J.-P. Wang, ACS Appl. Nano Mater., 2020, 3, 9560–9580 CrossRef CAS.
- D. Su, K. Wu, V. D. Krishna, T. Klein, J. Liu, Y. Feng, A. M. Perez, M. C.-J. Cheeran and J.-P. Wang, Front. Microbiol., 2019, 10, 1077 CrossRef PubMed.
- K. Wu, T. Klein, V. D. Krishna, D. Su, A. M. Perez and J.-P. Wang, ACS Sens., 2017, 2, 1594–1601 CrossRef CAS PubMed.
- J. Zhong, E. L. Rösch, T. Viereck, M. Schilling and F. Ludwig, ACS Sens., 2021, 6, 976–984 CrossRef CAS PubMed.
- K. Wu, J. Liu, R. Saha, D. Su, V. D. Krishna, M. C.-J. Cheeran and J.-P. Wang, ACS Appl. Mater. Interfaces, 2020, 12, 13686–13697 CrossRef CAS PubMed.
- P. P. Sharma, E. Albisetti, M. Massetti, M. Scolari, C. La Torre, M. Monticelli, M. Leone, F. Damin, G. Gervasoni, G. Ferrari, F. Salice, E. Cerquaglia, G. Falduti, M. Cretich, E. Marchisio, M. Chiari, M. Sampietro, D. Petti and R. Bertacco, Sens. Actuators, B, 2017, 242, 280–287 CrossRef CAS.
- L. Li, K.-Y. Mak and Y. Zhou, Chin. Phys. B, 2020, 29, 88701 CrossRef CAS.
- M. Sayhi, O. Ouerghi, K. Belgacem, M. Arbi, Y. Tepeli, A. Ghram, Ü. Anik, L. Österlund, D. Laouini and M. F. Diouani, Biosens. Bioelectron., 2018, 107, 170–177 CrossRef CAS PubMed.
- T. Wu, X. Li, Y. Fu, X. Ding, Z. Li, G. Zhu and J. Fan, Talanta, 2020, 209, 120550 CrossRef CAS PubMed.
- J. Lee, K. Takemura, C. N. Kato, T. Suzuki and E. Y. Park, ACS Appl. Mater. Interfaces, 2017, 9, 27298–27304 CrossRef CAS PubMed.
- A. B. Ganganboina, A. D. Chowdhury, I. M. Khoris, F. Nasrin, K. Takemura, T. Hara, F. Abe, T. Suzuki and E. Y. Park, Biosens. Bioelectron., 2020, 157, 112169 CrossRef CAS PubMed.
- K. C. Sanjaya, A. Ranzoni, J. Hung, M. A. T. Blaskovich, D. Watterson, P. R. Young and M. A. Cooper, Anal. Chim. Acta, 2020, 1107, 85–91 CrossRef CAS PubMed.
- L. Luo, J. Yang, K. Liang, C. Chen, X. Chen and C. Cai, Talanta, 2019, 202, 21–26 CrossRef CAS PubMed.
- C. Wang, C. Wang, X. Wang, K. Wang, Y. Zhu, Z. Rong, W. Wang, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 19495–19505 CrossRef CAS PubMed.
- F. Zhang, L. Luo, H. Gong, C. Chen and C. Cai, RSC Adv., 2018, 8, 32262–32268 RSC.
- A. Barrios-Gumiel, D. Sepúlveda-Crespo, J. L. Jiménez, R. Gómez, M. Á. Muñoz-Fernández and F. J. de la Mata, Colloids Surf., B, 2019, 181, 360–368 CrossRef CAS PubMed.
- X. Peng, G. Luo, Z. Wu, W. Wen, X. Zhang and S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 41148–41156 CrossRef CAS PubMed.
- Z. Wu, T. Zeng, W.-J. Guo, Y.-Y. Bai, D.-W. Pang and Z.-L. Zhang, ACS Appl. Mater. Interfaces, 2019, 11, 5762–5770 CrossRef CAS PubMed.
- F. Luo, C. Long, Z. Wu, H. Xiong, M. Chen, X. Zhang, W. Wen and S. Wang, Sens. Actuators, B, 2020, 310, 127831 CrossRef CAS.
- B. Tian, J. Fock, G. A. S. Minero and M. F. Hansen, Biosens. Bioelectron., 2020, 160, 112219 CrossRef CAS PubMed.
- M. Pirzada and Z. Altintas, Micromachines, 2020, 11, 356 CrossRef PubMed.
- H. Yanagawa, A. Inoue, H. Sugimoto, M. Shioi and M. Fujii, MRS Commun., 2019, 9, 1079–1086 CrossRef CAS.
- J. F. F. Ribeiro, M. I. A. Pereira, L. G. Assis, P. E. Cabral Filho, B. S. Santos, G. A. L. Pereira, C. R. Chaves, G. S. Campos, S. I. Sardi, G. Pereira and A. Fontes, J. Photochem. Photobiol., B, 2019, 194, 135–139 CrossRef CAS PubMed.
- A. M. J. Jimenez, A. Moulick, L. Richtera, L. Krejcova, L. Kalina, R. Datta, M. Svobodova, D. Hynek, M. Masarik, Z. Heger and V. Adam, Sens. Actuators, B, 2018, 258, 295–303 CrossRef.
- R. Q. Zhang, S. L. Hong, C. Y. Wen, D. W. Pang and Z. L. Zhang, Biosens. Bioelectron., 2018, 100, 348–354 CrossRef CAS PubMed.
- G. R. Bardajee, M. Zamani, H. Mahmoodian, H. Elmizadeh, H. Yari, L. Jouyandeh, R. Shirkavand and M. Sharifi, Spectrochim. Acta, Part A, 2022, 269, 120702 CrossRef CAS PubMed.
- L. Meng, K. Xiao, X. Zhang, C. Du and J. Chen, Sens. Actuators, B, 2020, 305, 127480 CrossRef CAS.
- H. Ashiba, Y. Sugiyama, X. Wang, H. Shirato, K. Higo-Moriguchi, K. Taniguchi, Y. Ohki and M. Fujimaki, Biosens. Bioelectron., 2017, 93, 260–266 CrossRef CAS PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, M. H. M. Zaid, W. M. E. M. M. Daniyal and M. A. Mahdi, Opt. Laser Technol., 2019, 114, 204–208 CrossRef CAS.
- Y. Zhao, J. Chen, Z. Hu, Y. Chen, Y. Tao, L. Wang, L. Li, P. Wang, H.-Y. Li, J. Zhang, J. Tang and H. Liu, Biosens. Bioelectron., 2022, 202, 113974 CrossRef CAS PubMed.
- F. Sabzehparvar, T. Rahmani Cherati, A. Mohsenifar, T. Roodbar Shojaei and M. Tabatabaei, Spectrochim. Acta, Part A, 2019, 212, 173–179 CrossRef CAS PubMed.
- L. Luo, F. Zhang, C. Chen and C. Cai, Anal. Chem., 2019, 91, 15748–15756 CrossRef CAS PubMed.
- Y. Ma, G. Mao, W. Huang, G. Wu, W. Yin, X. Ji, Z. Deng, Z. Cai, X.-E. Zhang, Z. He and Z. Cui, J. Am. Chem. Soc., 2019, 141, 13454–13458 CrossRef CAS PubMed.
- M. Bao, E. Jensen, Y. Chang, G. Korensky and K. Du, ACS Appl. Mater. Interfaces, 2020, 12, 43435–43443 CrossRef CAS PubMed.
- B. Babamiri, R. Hallaj and A. Salimi, Sens. Actuators, B, 2018, 254, 551–560 CrossRef CAS.
- Z. Ranjbar-Navazi, Y. Omidi, M. Eskandani and S. Davaran, TrAC, Trends Anal. Chem., 2019, 118, 386–400 CrossRef CAS.
- F. Zhang, D. Yi, H. Sun and H. Zhang, J. Nanosci. Nanotechnol., 2014, 14, 1409–1424 CrossRef CAS PubMed.
- D. B. Ngo, T. Chaibun, L. S. Yin, B. Lertanantawong and W. Surareungchai, Anal. Bioanal. Chem., 2021, 413, 1027–1037 CrossRef CAS PubMed.
- J. Chen, Y. Kong, W. Wang, H. Fang, Y. Wo, D. Zhou, Z. Wu, Y. Li and S. Chen, Chem. Commun., 2016, 52, 4025–4028 RSC.
- S. R. Ahmed and S. Neethirajan, Glob. Challenges, 2018, 2, 1700071 CrossRef PubMed.
- O. J. Achadu, K. Takemura, I. M. Khoris and E. Y. Park, Sens. Actuators, B, 2020, 321, 128494 CrossRef CAS PubMed.
- H. Malekzad, P. Sahandi Zangabad, H. Mirshekari, M. Karimi and M. R. Hamblin, Nanotechnol. Rev., 2017, 6, 301–329 CAS.
- Z. Bognár, M. I. De Jonge and R. E. Gyurcsányi, Nanoscale, 2022, 14, 2296–2303 RSC.
- N. Wu, H.-C. Zhang, X.-H. Sun, F.-N. Guo, L.-X. Feng, T. Yang and J.-H. Wang, Sens. Actuators, B, 2022, 352, 131041 CrossRef CAS.
- L. L. Zheng, X. J. Shuai, Y. Liu, C. M. Li, S. J. Zhen, J. J. Liu, Y. F. Li and C. Z. Huang, Chem. Eng. J., 2022, 434, 134658 CrossRef CAS.
- Y. Tao, K. Yi, H. Wang, H.-W. Kim, K. Li, X. Zhu and M. Li, J. Colloid Interface Sci., 2022, 613, 406–414 CrossRef CAS PubMed.
- Y. Tao, Y.-H. Lao, K. Yi, Y. Xu, H. Wang, D. Shao, J. Wang and M. Li, Talanta, 2021, 234, 122675 CrossRef CAS PubMed.
- N. Gao, F. Gao, S. He, Q. Zhu, J. Huang, H. Tanaka and Q. Wang, Anal. Chim. Acta, 2017, 951, 58–67 CrossRef CAS PubMed.
- F. Jin, H. Li and D. Xu, Anal. Chim. Acta, 2019, 1077, 297–304 CrossRef CAS PubMed.
- A. D. Kurdekar, L. A. A. Chunduri, S. M. Chelli, M. K. Haleyurgirisetty, E. P. Bulagonda, J. Zheng, I. K. Hewlett and V. Kamisetti, RSC Adv., 2017, 7, 19863–19877 RSC.
- V. I. Kukushkin, N. M. Ivanov, A. A. Novoseltseva, A. S. Gambaryan, I. V. Yaminsky, A. M. Kopylov and E. G. Zavyalova, PLoS One, 2019, 14, e0216247 CrossRef CAS PubMed.
- N. N. Durmanov, R. R. Guliev, A. V. Eremenko, I. A. Boginskaya, I. A. Ryzhikov, E. A. Trifonova, E. V. Putlyaev, A. N. Mukhin, S. L. Kalnov, M. V. Balandina, A. P. Tkachuk, V. A. Gushchin, A. K. Sarychev, A. N. Lagarkov, I. A. Rodionov, A. R. Gabidullin and I. N. Kurochkin, Sens. Actuators, B, 2018, 257, 37–47 CrossRef CAS.
- E. Khristunova, J. Barek, B. Kratochvil, E. Korotkova, E. Dorozhko and V. Vyskocil, Bioelectrochemistry, 2020, 135, 107576 CrossRef CAS PubMed.
- M. Awan, S. Rauf, A. Abbas, M. H. Nawaz, C. Yang, S. A. Shahid, N. Amin and A. Hayat, J. Mol. Liq., 2020, 317, 114014 CrossRef CAS.
- Y. Khristunova, E. Korotkova, B. Kratochvil, J. Barek, E. Dorozhko, V. Vyskocil, E. Plotnikov, O. Voronova and V. Sidelnikov, Sensors, 2019, 19 Search PubMed.
- L. Gou, Y. Sheng, Q. Peng, J. Ling, H. Yue, F. Chen and H. Tang, Sens. Actuators, B, 2020, 312, 127987 CrossRef CAS.
- V. M. Generalov, O. V. Naumova, B. I. Fomin, S. A. P’yankov, I. V. Khlistun, A. S. Safatov, B. N. Zaitsev, E. G. Zaitseva and A. L. Aseev, Optoelectron. Instrum. Data Process., 2019, 55, 618–622 CrossRef.
- M. Uhm, J.-M. Lee, J. Lee, J. H. Lee, S. Choi, B.-G. Park, D. M. Kim, S.-J. Choi, H.-S. Mo, Y.-J. Jeong and D. H. Kim, Sensors, 2019, 19 Search PubMed.
- S. Choi, H.-S. Mo, J. Kim, S. Kim, S. M. Lee, S.-J. Choi, D. M. Kim, D.-W. Park and D. H. Kim, Curr. Appl. Phys., 2020, 20, 828–833 CrossRef.
- K. A. Malsagova, T. O. Pleshakova, R. A. Galiullin, I. D. Shumov, A. F. Kozlov, T. S. Romanova, V. P. Popov, A. V. Glukhov, V. A. Konev, A. I. Archakov and Y. D. Ivanov, Coatings, 2020, 10 Search PubMed.
- K. A. Malsagova, T. O. Pleshakova, R. A. Galiullin, A. L. Kaysheva, I. D. Shumov, M. A. Ilnitskii, V. P. Popov, A. V. Glukhov, A. I. Archakov and Y. D. Ivanov, Anal. Methods, 2018, 10, 2740–2749 RSC.
- A. A. Leonardi, M. J. Lo Faro, S. Petralia, B. Fazio, P. Musumeci, S. Conoci, A. Irrera and F. Priolo, ACS Sens., 2018, 3, 1690–1697 CrossRef CAS PubMed.
- M. M. N. Nuzaihan, U. Hashim, M. K. Md Arshad, S. R. Kasjoo, S. F. A. Rahman, A. R. Ruslinda, M. F. M. Fathil, R. Adzhri and M. M. Shahimin, Biosens. Bioelectron., 2016, 83, 106–114 CrossRef CAS PubMed.
- K. A. Gonchar, S. N. Agafilushkina, D. V. Moiseev, I. V. Bozhev, A. A. Manykin, E. A. Kropotkina, A. S. Gambaryan and L. A. Osminkina, Mater. Res. Express, 2020, 7, 35002 CrossRef CAS.
- M. B. Gongalsky, U. A. Tsurikova, J. V. Samsonova, G. Z. Gvindzhiliiia, K. A. Gonchar, N. Y. Saushkin, A. A. Kudryavtsev, E. A. Kropotkina, A. S. Gambaryan and L. A. Osminkina, Results Mater., 2020, 6, 100084 CrossRef.
- C. Singhal, A. Ingle, D. Chakraborty, A. K. P. N., C. S. Pundir and J. Narang, Int. J. Biol. Macromol., 2017, 98, 84–93 CrossRef CAS PubMed.
- N. Wiriyachaiporn, S. Sirikaew, S. Bamrungsap, T. Limcharoen, P. Polkankosit, P. Roeksrungruang and K. Ponlamuangdee, Anal. Methods, 2021, 13, 1687–1694 RSC.
- J. Zhou, M. Ren, W. Wang, L. Huang, Z. Lu, Z. Song, M. F. Foda, L. Zhao and H. Han, Anal. Chem., 2020, 92, 8802–8809 CrossRef CAS PubMed.
- L. Ding, C. Xiang and G. Zhou, Talanta, 2018, 181, 65–72 CrossRef CAS PubMed.
- S. Chen, L. Luo, L. Wang, C. Chen, H. Gong and C. Cai, Analyst, 2021, 146, 3924–3932 RSC.
- Y. Liu, T. Shen, L. Hu, H. Gong, C. Chen, X. Chen and C. Cai, Sens. Actuators, B, 2017, 253, 1188–1193 CrossRef CAS.
- B. Yang, H. Gong, C. Chen, X. Chen and C. Cai, Biosens. Bioelectron., 2017, 87, 679–685 CrossRef CAS PubMed.
- W. S. Lee, J. Ahn, S. Jung, J. Lee, T. Kang and J. Jeong, BioChip J., 2021, 15, 260–267 CrossRef CAS PubMed.
- M. E. Klaczko, K. Lucas, A. T. Salminen, M. C. McCloskey, B. Ozgurun, B. M. Ward, J. Flax and J. L. McGrath, Analyst, 2022, 147, 213–222 RSC.
- M. S. Draz, M. Venkataramani, H. Lakshminarayanan, E. Saygili, M. Moazeni, A. Vasan, Y. Li, X. Sun, S. Hua, X. G. Yu and H. Shafiee, Nanoscale, 2018, 10, 11841–11849 RSC.
- C. Singhal, C. S. Pundir and J. Narang, Biosens. Bioelectron., 2017, 97, 75–82 CrossRef CAS PubMed.
- E. P. Simão, D. B. S. Silva, M. T. Cordeiro, L. H. V. Gil, C. A. S. Andrade and M. D. L. Oliveira, Talanta, 2020, 208, 120338 CrossRef PubMed.
- F. Khosravi-Nejad, M. Teimouri, S. Jafari Marandi and M. Shariati, Appl. Phys. A: Mater. Sci. Process., 2019, 125, 616 CrossRef.
- M. Shariati, M. Sadeghi and S. H. R. Shojaei, Anal. Chim. Acta, 2022, 1195, 339442 CrossRef CAS PubMed.
- M. H. Nguyen, H. H. Nguyen and A. T. Mai, Sens. Actuators, A, 2020, 312, 112171 CrossRef CAS.
- M. Shariati and M. Sadeghi, Anal. Bioanal. Chem., 2020, 412, 5367–5377 CrossRef CAS PubMed.
- F. Zhao, Y. Bai, L. Cao, G. Han, C. Fang, S. Wei and Z. Chen, J. Electroanal. Chem., 2020, 867, 114184 CrossRef CAS.
- S. H. Baek, C. Y. Park, T. P. Nguyen, M. W. Kim, J. P. Park, C. Choi, S. Y. Kim, S. K. Kailasa and T. J. Park, Food Control, 2020, 114, 107225 CrossRef CAS.
- C.-C. Chen, Z.-L. Lai, G.-J. Wang and C.-Y. Wu, Biosens. Bioelectron., 2016, 77, 603–608 CrossRef CAS PubMed.
- J. Kim, J. H. Kwon, J. Jang, H. Lee, S. Kim, Y. K. Hahn, S. K. Kim, K. H. Lee, S. Lee, H. Pyo, C.-S. Song and J. Lee, Biosens. Bioelectron., 2018, 112, 209–215 CrossRef CAS PubMed.
- A. Mehranfar, M. Khavani and M. Izadyar, J. Mol. Liq., 2022, 345, 117852 CrossRef CAS PubMed.
- Y. Xia, Y. Chen, Y. Tang, G. Cheng, X. Yu, H. He, G. Cao, H. Lu, Z. Liu and S.-Y. Zheng, ACS Sens., 2019, 4, 3298–3307 CrossRef CAS PubMed.
- G. Biasotto, J. P. C. Costa, P. I. Costa and M. A. Zaghete, Appl. Phys. A: Mater. Sci. Process., 2019, 125, 821 CrossRef CAS.
- R. van den Hurk, M. Baghelani, J. Chen, M. Daneshmand and S. Evoy, Sens. Actuators, A, 2019, 296, 186–191 CrossRef CAS.
- J. Liu, M. Xu, B. Tang, L. Hu, F. Deng, H. Wang, D.-W. Pang, Z. Hu, M. Wang and Y. Zhou, Small, 2019, 15, 1803788 CrossRef PubMed.
- W. Hou, Y. Li, W. Kang, X. Wang, X. Wu, S. Wang and F. Liu, Sci. Rep., 2019, 9, 1307 CrossRef PubMed.
- J. Liu, M. Su, X. Chen, Z. Li, Z. Fang and L. Yi, J. Biomater. Appl., 2022, 08853282211063298 Search PubMed.
- Q. Zhang, J. Li, Y. Li, G. Tan, M. Sun, Y. Shan, Y. Zhang, X. Wang, K. Song, R. Shi, L. Huang, F. Liu, Y. Yi and X. Wu, Biosens. Bioelectron., 2022, 113978 CrossRef CAS PubMed.
- K.-I. Joo, Y. Lei, C.-L. Lee, J. Lo, J. Xie, S. F. Hamm-Alvarez and P. Wang, ACS Nano, 2008, 2, 1553–1562 CrossRef CAS PubMed.
- Z.-Y. Hong, Z.-L. Zhang, B. Tang, J. Ao, C. Wang, C. Yu and D.-W. Pang, Anal. Chem., 2018, 90, 14020–14028 CrossRef CAS PubMed.
- L.-J. Zhang, S. Wang, L. Xia, C. Lv, H.-W. Tang, Z. Liang, G. Xiao and D.-W. Pang, mBio, 2020, 11, e00135–20 Search PubMed.
- L.-J. Zhang, L. Xia, S.-L. Liu, E.-Z. Sun, Q.-M. Wu, L. Wen, Z.-L. Zhang and D.-W. Pang, ACS Nano, 2018, 12, 474–484 CrossRef CAS PubMed.
- L. L. Zheng, C. M. Li, S. J. Zhen, Y. F. Li and C. Z. Huang, Nanoscale, 2017, 9, 7880–7887 RSC.
- J. A. Coultas, R. Smyth and P. J. Openshaw, Thorax, 2019, 74, 986–993 CrossRef PubMed.
- Y.-B. Yang, Y.-D. Tang, Y. Hu, F. Yu, J.-Y. Xiong, M.-X. Sun, C. Lyu, J.-M. Peng, Z.-J. Tian, X.-H. Cai and T.-Q. An, Nano Lett., 2020, 20, 1417–1427 CrossRef CAS PubMed.
- E.-Z. Sun, A.-A. Liu, Z.-L. Zhang, S.-L. Liu, Z.-Q. Tian and D.-W. Pang, ACS Nano, 2017, 11, 4395–4406 CrossRef CAS PubMed.
- Q.-M. Wu, S.-L. Liu, G. Chen, W. Zhang, E.-Z. Sun, G.-F. Xiao, Z.-L. Zhang and D.-W. Pang, Small, 2018, 14, 1702841 CrossRef PubMed.
- L. L. Zheng, C. M. Li, S. J. Zhen, Y. F. Li and C. Z. Huang, Nanoscale, 2016, 8, 18635–18639 RSC.
- J. Liu, C. Yu, J.-F. Gui, D.-W. Pang and Q.-Y. Zhang, Front. Microbiol., 2018, 9, 2797 CrossRef PubMed.
- L. Wen, Y. Lin, Z.-L. Zhang, W. Lu, C. Lv, Z.-L. Chen, H.-Z. Wang and D.-W. Pang, Biomaterials, 2016, 99, 24–33 CrossRef CAS PubMed.
- Y. Ma, M. Wang, W. Li, Z. Zhang, X. Zhang, T. Tan, X.-E. Zhang and Z. Cui, Nat. Commun., 2017, 8, 15318 CrossRef CAS PubMed.
- A. Pramanik, Y. Gao, S. Patibandla, D. Mitra, M. G. McCandless, L. A. Fassero, K. Gates, R. Tandon and P. C. Ray, J. Phys. Chem. Lett., 2021, 12, 2166–2171 CrossRef CAS PubMed.
- R. Cheng, F. Zhu, M. Huang, Q. Zhang, H. H. Yan, X. H. Zhao, F. K. Luo, C. M. Li, H. Liu, G. L. Liang, C. Z. Huang and J. Wang, Biosens. Bioelectron., 2022, 202, 114001 CrossRef CAS PubMed.
- H. Zhao, B. Ni, X. Jin, H. Zhang, J. J. Hou, L. Hou, J. H. Marsh, L. Dong, S. Li, X. W. Gao, D. Shi, X. Liu and J. Xiong, Appl. Opt., 2022, 61, 546–553 CrossRef CAS PubMed.
- J. C. Abrego-Martinez, M. Jafari, S. Chergui, C. Pavel, D. Che and M. Siaj, Biosens. Bioelectron., 2022, 195, 113595 CrossRef CAS PubMed.
- G. Pang, Y. Zhang, X. Wang, H. Pan, X. Zhang, Y. Li, S. Zhang, C. Yan, L. Sun, H. Wang and J. Chang, Nano Today, 2021, 40, 101264 CrossRef CAS.
- T. Wiegand, M. Fratini, F. Frey, K. Yserentant, Y. Liu, E. Weber, K. Galior, J. Ohmes, F. Braun, D.-P. Herten, S. Boulant, U. S. Schwarz, K. Salaita, E. A. Cavalcanti-Adam and J. P. Spatz, Nat. Commun., 2020, 11, 32 CrossRef CAS PubMed.
- F. Xu, A. Bandara, H. Akiyama, B. Eshaghi, D. Stelter, T. Keyes, J. E. Straub, S. Gummuluru and B. M. Reinhard, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9041 LP–E9050 Search PubMed.
- R. Schubert, S. Trenholm, K. Balint, G. Kosche, C. S. Cowan, M. A. Mohr, M. Munz, D. Martinez-Martin, G. Fläschner, R. Newton, J. Krol, B. G. Scherf, K. Yonehara, A. Wertz, A. Ponti, A. Ghanem, D. Hillier, K.-K. Conzelmann, D. J. Müller and B. Roska, Nat. Biotechnol., 2018, 36, 81–88 CrossRef CAS PubMed.
- V. Marjomäki, T. Lahtinen, M. Martikainen, J. Koivisto, S. Malola, K. Salorinne, M. Pettersson and H. Häkkinen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1277 LP–1281 CrossRef PubMed.
- L. Luo, F. Zhang, C. Chen and C. Cai, Microchim. Acta, 2020, 187, 140 CrossRef CAS PubMed.
- M. Shamsipur, L. Samandari, L. Farzin and A. Besharati-Seidani, Microchem. J., 2021, 160, 105714 CrossRef CAS.
- J. Xu, F. Merlier, B. Avalle, V. Vieillard, P. Debré, K. Haupt and B. Tse Sum Bui, ACS Appl. Mater. Interfaces, 2019, 11, 9824–9831 CrossRef CAS PubMed.
- S. Wankar, N. W. Turner and R. J. Krupadam, Biosens. Bioelectron., 2016, 82, 20–25 CrossRef CAS PubMed.
- M. Pirzada and Z. Altintas, in Molecular Imprinting for Nanosensors and Other Sensing Applications, ed. A. Denizli, Elsevier, Amsterdam, 1st edn, 2021, pp. 367–406 Search PubMed.
- A. J. Ruiz-Sanchez, C. Parolo, B. S. Miller, E. R. Gray, K. Schlegel and R. A. McKendry, J. Mater. Chem. B, 2017, 5, 7262–7266 RSC.
- N. A. S. Omar, Y. W. Fen, S. Saleviter, Y. M. Kamil, W. M. E. M. M. Daniyal, J. Abdullah and M. A. Mahdi, Sens. Actuators, A, 2020, 303, 111830 CrossRef CAS.
- C. Liu, J. Dong, S. Ning, J. Hou, G. I. N. Waterhouse, Z. Cheng and S. Ai, Microchim. Acta, 2018, 185, 423 CrossRef PubMed.
- J. Yang, W. Feng, K. Liang, C. Chen and C. Cai, Talanta, 2020, 212, 120744 CrossRef CAS PubMed.
- M. Antipchik, D. Polyakov, E. Sinitsyna, A. Dzhuzha, M. Shavlovsky, E. Korzhikova-Vlakh and T. Tennikova, Sensors, 2020, 20 Search PubMed.
- C.-Z. Wang, H.-H. Han, X.-Y. Tang, D.-M. Zhou, C. Wu, G.-R. Chen, X.-P. He and H. Tian, ACS Appl. Mater. Interfaces, 2017, 9, 25164–25170 CrossRef CAS PubMed.
- W. Diao, M. Tang, S. Ding, X. Li, W. Cheng, F. Mo, X. Yan, H. Ma and Y. Yan, Biosens. Bioelectron., 2018, 100, 228–234 CrossRef CAS PubMed.
- J. Ma, W. Wang, Y. Li, Z. Lu, X. Tan and H. Han, Anal. Chem., 2021, 93, 2090–2096 CrossRef CAS PubMed.
- Z. Su, D. Tang, X. Yang, Y. Peng, B. Wang, X. Li, J. Chen, Y. Hu and X. Qin, Microchem. J., 2022, 174, 107038 CrossRef CAS.
- H.-Q. Zhao, G.-H. Qiu, Z. Liang, M.-M. Li, B. Sun, L. Qin, S.-P. Yang, W.-H. Chen and J.-X. Chen, Anal. Chim. Acta, 2016, 922, 55–63 CrossRef CAS PubMed.
- B. Sun, H.-Q. Zhao, B.-P. Xie, L.-P. Bai, Z.-H. Jiang and J.-X. Chen, J. Inorg. Biochem., 2017, 176, 17–23 CrossRef CAS PubMed.
- H.-Q. Zhao, S.-P. Yang, N.-N. Ding, L. Qin, G.-H. Qiu, J.-X. Chen, W.-H. Zhang, W.-H. Chen and T. S. A. Hor, Dalton Trans., 2016, 45, 5092–5100 RSC.
- L. Xue, Y. Yang, S. Wu, Y. Huang, J. Li, Y. Xiang and G. Li, Anal. Chem., 2020, 92, 2972–2978 CrossRef CAS PubMed.
- B.-P. Xie, G.-H. Qiu, P.-P. Hu, Z. Liang, Y.-M. Liang, B. Sun, L.-P. Bai, Z.-H. Jiang and J.-X. Chen, Sens. Actuators, B, 2018, 254, 1133–1140 CrossRef CAS.
- Y. Pan, S. Zhan and F. Xia, Anal. Biochem., 2018, 546, 5–9 CrossRef CAS PubMed.
- G.-H. Qiu, Z.-H. Weng, P.-P. Hu, W.-J. Duan, B.-P. Xie, B. Sun, X.-Y. Tang and J.-X. Chen, Talanta, 2018, 180, 396–402 CrossRef CAS PubMed.
- B.-P. Xie, G.-H. Qiu, B. Sun, Z.-F. Yang, W.-H. Zhang, J.-X. Chen and Z.-H. Jiang, Inorg. Chem. Front., 2019, 6, 148–152 RSC.
- S.-P. Yang, W. Zhao, P.-P. Hu, K.-Y. Wu, Z.-H. Jiang, L.-P. Bai, M.-M. Li and J.-X. Chen, Inorg. Chem., 2017, 56, 14880–14887 CrossRef CAS PubMed.
- T. Matsubara, M. Ujie, T. Yamamoto, Y. Einaga, T. Daidoji, T. Nakaya and T. Sato, ACS Sens., 2020, 5, 431–439 CrossRef CAS PubMed.
- H. Que, D. Zhang, B. Guo, T. Wang, H. Wu, D. Han and Y. Yan, Biosens. Bioelectron., 2019, 143, 111610 CrossRef CAS PubMed.
- A.-L. Sun, Analyst, 2018, 143, 487–492 RSC.
- Y. Mustapha Kamil, S. H. Al-Rekabi, M. H. Yaacob, A. Syahir, H. Y. Chee, M. A. Mahdi and M. H. Abu Bakar, Sci. Rep., 2019, 9, 13483 CrossRef PubMed.
- B. Babamiri, R. Hallaj and A. Salimi, Methods Appl. Fluoresc., 2018, 6, 35013 CrossRef CAS PubMed.
- H. C. Nguyen, H. Park, H.-J. Shin, N. M. Nguyen, A. T. V. Nguyen, T.-T. T. Trinh, T. H. Y. Duong, H. T. Tuong, V. T. Hoang, G.-E. Seo, H.-J. Sohn and S.-J. Yeo, Intervirology, 2019, 62, 145–155 CrossRef CAS PubMed.
- M. Gast, S. Kühner, H. Sobek, P. Walther and B. Mizaikoff, Anal. Chem., 2018, 90, 5576–5585 CrossRef CAS PubMed.
- Z. Altintas, in Advanced Molecularly Imprinting Materials, ed. A. Tiwari and L. Uzun, 2016, pp. 389–411 Search PubMed.
- W. Feng, C. Liang, H. Gong and C. Cai, New J. Chem., 2018, 42, 3503–3508 RSC.
- M. Du, N. Li, G. Mao, Y. Liu, X. Wang, S. Tian, Q. Hu, X. Ji, Y. Liu and Z. He, Anal. Chim. Acta, 2019, 1084, 116–122 CrossRef CAS PubMed.
- Q. Yan, Y. Yang, Z. Tan, Q. Liu, H. Liu, P. Wang, L. Chen, D. Zhang, Y. Li and Y. Dong, Biosens. Bioelectron., 2018, 103, 151–157 CrossRef CAS PubMed.
- X. Zhu, X. Wang, L. Han, T. Chen, L. Wang, H. Li, S. Li, L. He, X. Fu, S. Chen, M. Xing, H. Chen and Y. Wang, Biosens. Bioelectron., 2020, 166, 112437 CrossRef CAS PubMed.
- P. Piedimonte, L. Sola, M. Cretich, A. Gori, M. Chiari, E. Marchisio, P. Borga, R. Bertacco, A. Melloni, G. Ferrari and M. Sampietro, Biosens. Bioelectron., 2022, 113996 CrossRef CAS PubMed.
- K. H. Cho, D. H. Shin, J. Oh, J. H. An, J. S. Lee and J. Jang, ACS Appl. Mater. Interfaces, 2018, 10, 28412–28419 CrossRef CAS PubMed.
- C. L. Pirich, R. A. de Freitas, R. M. Torresi, G. F. Picheth and M. R. Sierakowski, Biosens. Bioelectron., 2017, 92, 47–53 CrossRef CAS PubMed.
- N. Dey, D. Bhagat and S. Bhattacharya, ACS Sustainable Chem. Eng., 2019, 7, 7667–7675 CrossRef CAS.
- C. Song, Y. Sun, J. Zhang, T. Wang, Y. Wang, Y. Liu and L. Wang, Analyst, 2020, 145, 5475–5481 RSC.
- H. Zhang, X. Xu and W. Jiang, Chem. Sci., 2020, 11, 7415–7423 RSC.
- G.-X. Liang, S.-Y. Ye, H.-M. Yu, K.-R. Zhao, P.-F. Liu, Z.-J. Liu and L. Wang, Sens. Actuators, B, 2022, 354, 131199 CrossRef CAS.
- Y. Liu, H. Xu, C. Liu, L. Peng, H. Khan, L. Cui, R. Huang, C. Wu, S. Shen, S. Wang, W. Liang, Z. Li, B. Xu and N. He, J. Biomed. Nanotechnol., 2019, 15, 790–798 CrossRef CAS PubMed.
- T. Zhao, K. Asawa, T. Masuda, A. Honda, K. Kushiro, H. Cabral and M. Takai, J. Colloid Interface Sci., 2021, 601, 825–832 CrossRef CAS PubMed.
- M. Fratini, T. Wiegand, C. Funaya, Z. Jiang, P. N. M. Shah, J. P. Spatz, E. A. Cavalcanti-Adam and S. Boulant, ACS Infect. Dis., 2018, 4, 1585–1600 CrossRef CAS PubMed.
- M. Gast, F. Wondany, B. Raabe, J. Michaelis, H. Sobek and B. Mizaikoff, Anal. Chem., 2020, 92, 3050–3057 CrossRef CAS PubMed.
- Y. Wu, L. Li, L. Frank, J. Wagner, P. Andreozzi, B. Hammer, M. D’Alicarnasso, M. Pelliccia, W. Liu, S. Chakrabortty, S. Krol, J. Simon, K. Landfester, S. L. Kuan, F. Stellacci, K. Müllen, F. Kreppel and T. Weil, ACS Nano, 2019, 13, 8749–8759 CrossRef CAS PubMed.
- H.-O. Kim, W. Na, M. Yeom, J.-W. Lim, E.-H. Bae, G. Park, C. Park, H. Lee, H. K. Kim, D. G. Jeong, K.-S. Lyoo, V. P. Le, S. Haam and D. Song, ACS Appl. Mater. Interfaces, 2020, 12, 6876–6884 CrossRef CAS PubMed.
- Y. Zhang, C. Yurdakul, A. J. Devaux, L. Wang, X. G. Xu, J. H. Connor, M. S. Ünlü and J.-X. Cheng, Anal. Chem., 2021, 93, 4100–4107 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |