 Open Access Article
Open Access ArticleAnalysis of the orientation of cholesterol in high-density lipoprotein nanodiscs using solid-state NMR†
Sophie
Lau
and
David A.
Middleton
 *
*
Department of Chemistry, Lancaster University, Bailrigg, Lancaster, UK. E-mail: d.middleton@lancaster.ac.uk; Tel: +44-1524-594328
First published on 7th September 2022
Abstract
Cholesterol is an essential component of eukaryotic cellular membranes that regulates the order and phase behaviour of dynamic lipid bilayers. Although cholesterol performs many vital physiological roles, hypercholesterolaemia and the accumulation of cholesterol in atherosclerotic plaques can increase the risk of coronary heart disease morbidity. The risk is mitigated by the transportation of cholesterol from peripheral tissue to the liver by high-density lipoprotein (HDL), 6–20 nm-diameter particles of lipid bilayers constrained by an annular belt of the protein apolipoprotein A-I (apoA-I). Information on the dynamics and orientation of cholesterol in HDL is pertinent to the essential role of HDL in cholesterol cycling. This work investigates whether the molecular orientation of cholesterol in HDL differs from that in the unconstrained lipid bilayers of multilamellar vesicles (MLVs). Solid-state NMR (ssNMR) measurements of dynamically-averaged 13C–13C and 13C–1H dipolar couplings were used to determine the average orientation of triple 13C-labelled cholesterol in palmitoyloleoylphosphatidylcholine (POPC) lipid bilayers in reconstituted HDL (rHDL) nanodiscs and in MLVs. Individual 13C–13C dipolar couplings were measured from [2,3,4-13C3]cholesterol in a one-dimensional NMR experiment, by using a novel application of a method to excite double quantum coherence at rotational resonance. The measured dipolar couplings were compared with average values calculated from orientational distributions of cholesterol generated using a Gaussian probability density function. The data were consistent with small differences in the average orientation of cholesterol in rHDL and MLVs, which may reflect the effects of the constrained and unconstrained lipid bilayers in the two environments. The calculated distributions of cholesterol in rHDL and MLVs that were consistent with the NMR data also agreed well with orientational distributions extracted from previous molecular dynamics simulations of HDL nanodiscs and planar POPC bilayers.
1. Introduction
Cholesterol is an important and fascinating building block of mammalian cell membranes that performs the essential roles of reinforcing membrane integrity and controlling membrane fluidity under physiological conditions and temperature.1 In plasma membranes, cholesterol represents 30–50% of total lipid content,2 but excessive accumulation in the vasculature in the form of atherosclerotic plaques can increase the risk of coronary artery disease and heart attacks. Consequently, cholesterol levels are subject to tight and complex physiological regulatory mechanisms.3 One important component of the regulatory toolkit is high-density lipoprotein (HDL), a heterogeneous ensemble of discoidal and spheroidal nanoparticles composed of bilayer phospholipids, triglycerides and cholesterol/cholesteryl esters surrounded by a belt of α-helical proteins, chiefly apolipoprotein A-I (apoA-I).Nascent, lipid-poor discoidal (pre-β) HDL particles are formed when the ATP-binding cassette transporter A1 (ABCA1) facilitates the transfer of cholesterol and lipids from peripheral tissue to circulating apoA-I. Mature spherical HDL particles bind to the hepatocyte scavenger receptor SR-B1 and transfer cholesterol and its esters to the liver for excretion.4 Whilst the behaviour of cholesterol in model phospholipid membrane systems has been studied in depth, little is known about the properties of cholesterol in HDL particles. The morphological plasticity of HDL influences their lipid-binding properties and metabolism, and the surface curvature of the lipid cargo regulates interactions with cholesterol-handling enzymes.5–8 Consequently, information on the dynamics and orientation of cholesterol in HDL is pertinent to its essential role in cholesterol cycling.4
A wealth of information on the physical properties of cholesterol and its effect on phospholipid bilayers has been gained by using solid-state NMR (ssNMR) spectroscopy with model membrane systems such as multilamellar vesicles (MLVs).9–12 Wide-line and oriented 2H NMR of deuterated cholesterol has been used extensively to study the effects of cholesterol on membrane ordering,13 membrane deformation9 and dynamics.14,15 Additionally, natural-abundance 13C–1H NMR and 31P NMR have been used to study the segmental motions of cholesterol and phospholipids16 and the effects of cholesterol on the phase behaviour of lipid bilayers.17,18 Cholesterol adopts, on average, an upright orientation approximately parallel with the lipid bilayer normal with the 3-OH group situated in the vicinity of the phospholipid glyceryl backbone and pointing toward the interface between the bilayer surface and the aqueous phase.19–21 The molecule undergoes fast axial rotation in lipid bilayers and the amplitude of motions are described by a molecular order parameter, Smol, which takes values from zero, indicating isotropic reorientation, to 1.0, indicating rotation about a single axis. NMR studies on cholesterol in model membranes typically report Smol values of between 0.80 and 0.95,11,12 dependent on temperature and physicochemical properties of the phospholipids. These values are consistent with limited random motional excursions from an average orientation.
In contrast to the detailed level of understanding of cholesterol gained from model membrane systems, there is a scarcity of information on the orientation and order of cholesterol in the lipid bilayer environment of HDL-like particles. Structural studies of discoidal, reconstituted HDL (rHDL) composed of lipids, cholesterol and apoA-I have focused largely on the topology and registration of the apoA-I double belt,22,23 but the behaviour of cholesterol in the particles has been neglected. Molecular dynamics simulations of assemblies representing nascent, discoidal HDL have been reported,24 including an atomistic simulation over a 20 μs trajectory,25 but the orientation and distribution of cholesterol in the simulations lack experimental verification. Considering the fundamental role of HDL in facilitating cholesterol uptake and recycling, it would be useful to know if the annular protein belt that surrounds the lipids in HDL, and constrains the size and shape of the particles, influences the orientational distribution of cholesterol as compared to when it diffuses freely in unconstrained lipid vesicles.
Here, we use ssNMR to examine the orientation of cholesterol in discoidal rHDL resembling nascent pre-β particles, and in MLVs. Deuterium ssNMR is a powerful tool for probing molecular orientations in lipid bilayers, and has been used study the cholesterol analogue estradiol in rHDL particles,26 but the 2H nucleus has low sensitivity and ambiguities can occur in spectral assignments of multiple-labelled molecules. We therefore use 13C cross-polarisation magic-angle spinning (CP-MAS) ssNMR methods to observe [2,3,4-13C3]cholesterol (Fig. 1a) in the rHDL and MLV preparations. Whilst the relative insensitivity of 2H NMR or 13C at natural abundance can be challenging at low sample concentrations, 13C NMR peaks for 13C-enriched cholesterol are detected rapidly and assigned unambiguously.27 Selective measurements of dynamically-averaged, residual dipolar couplings between C3 and the three bonded nuclei, C2, C4 and H3, are used to estimate the average orientation of cholesterol about the bilayer normal. The 13C–13C dipolar couplings are measured selectively in a novel application of a one-dimensional experiment that observes the mechanical excitation of zero-quantum coherence at rotational resonance.28,29
2. Results and discussion
2.1 Initial characterisation of rHDL
Nanoparticles of rHDL were prepared by detergent dialysis30 from a mixture of palmitoyloleoylphosphatidylcholine (POPC), [2,3,4-13C3]cholesterol and recombinant apoA-I in a molar ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (these proportions denote the average ratio of lipid, cholesterol and protein per particle). For NMR analysis, the particles were precipitated from aqueous solution by polyethylene glycol (PEG) as described previously.31 Visualisation by transmission electron microscopy (TEM) confirmed the expected rHDL discoidal morphology with a mean diameter of 12.6 nm (2.3 nm SD) (Fig. 1b and c). Proton-decoupled 13C CP-MAS NMR spectra of rHDL revealed peaks for the three enriched carbon sites, C2 (32.6 ppm), C3 (72.5 ppm) and C4 (43.4 ppm), distinct from the background peaks from the lipid/cholesterol 13C nuclei at natural abundance (Fig. 2a). Rapid anisotropic motions of cholesterol, combined with MAS at 9 kHz, render the peaks narrow enough to measure one-bond 13C–13C J-coupling constants of 33 Hz.
2 (these proportions denote the average ratio of lipid, cholesterol and protein per particle). For NMR analysis, the particles were precipitated from aqueous solution by polyethylene glycol (PEG) as described previously.31 Visualisation by transmission electron microscopy (TEM) confirmed the expected rHDL discoidal morphology with a mean diameter of 12.6 nm (2.3 nm SD) (Fig. 1b and c). Proton-decoupled 13C CP-MAS NMR spectra of rHDL revealed peaks for the three enriched carbon sites, C2 (32.6 ppm), C3 (72.5 ppm) and C4 (43.4 ppm), distinct from the background peaks from the lipid/cholesterol 13C nuclei at natural abundance (Fig. 2a). Rapid anisotropic motions of cholesterol, combined with MAS at 9 kHz, render the peaks narrow enough to measure one-bond 13C–13C J-coupling constants of 33 Hz.
2.2 Calculation of average dipolar couplings for cholesterol in rHDL and in MLVs
The well-known rotational resonance condition32 was exploited to observe 13C–13C dipolar couplings between C2 and C3, and C3 and C4. When the MAS frequency νR matches the difference of the C2 and C3 resonance frequencies (7010 Hz), the peaks for C2 and C3 broaden selectively; similar broadening of the peaks for C3 and C4 is observed when νR matches the frequency difference of C3 and C4 (5113 Hz) (Fig. 2b and c). This broadening occurs because the n = 1 rotational resonance conditions with respect to C2–C3 and C3–C4 are satisfied at the two MAS frequencies, whereupon the dipolar interactions between the pairs 13C nuclei are re-introduced selectively. The peak broadening is considerably less than would be expected for the rigid limit 13C–13C dipolar coupling constants of 2250 Hz calculated from the crystal structure because of scaling by the anisotropic motions of cholesterol in the bilayer. The dipolar coupling constants are therefore defined as dynamically-averaged values: b23av for C2–C3 and b34av for C3–C4. Peak fitting to the spectra at rotational resonance obtained reasonable agreement for residual 13C–13C dipolar coupling constants of |b23av| = |b34av| = 250 Hz (Fig. 2b and c). Note that absolute values are given because the sign of the averaged couplings cannot be determined experimentally.Rotational diffusion of cholesterol in lipid bilayers scales the dipolar couplings in a defined way and the observed values may be used to determine the average orientation of cholesterol in the bilayer. Although values of |b23av| and |b34av| were estimated from the NMR lineshapes at rotational resonance, measurements of weak dipolar couplings directly from NMR peaks can be prone to errors, particularly in the presence of 13C–13C J-coupling. A novel approach was therefore used for verification of the estimated values, using an experiment that mechanically (i.e., through sample rotation) excites zero quantum (ZQ) coherence between C2–C3 and C3–C4 spin pairs at n = 1 rotational resonance. The pulse sequence converts ZQ coherence to double quantum (DQ) coherence and then back to ZQ coherence, and finally observable magnetization is recovered by reversing the ZQ excitation process28,29,33 (Fig. 3a and Fig. S1a, ESI†). A series of DQ-filtered spectra was collected at different excitation intervals, τ, and the summed integrated peak intensities for the selectively recoupled spins initially increase and reach a maximum at an optimal interval given approximately by  ,28 where bijav is expressed in Hertz. The data were compared with curves simulated for different values of bijav and the zero quantum relaxation term, TZQ, to find the values in closest agreement. The simulated curves fit both sets of data closely when |b23av| = |b34av| = 230 ± 50 Hz and TZQ = 11.5 ms (Fig. 3b, left, and Table 1). These values are close to those determined by NMR line shape analysis. The build-up of DQ coherence for [2,3,4-13C3]cholesterol in POPC MLVs are consistent with |b23av| = |b34av| = 320 ± 50 Hz and TZQ = 5.5 ms (Fig. 3b, right, and Table 2). The different extent of scaling of the 13C–13C dipolar couplings in rHDL and MLVs suggest differences in the dynamic behaviour of cholesterol in the two lipid bilayer environments. This conclusion was supported by measurements of residual C3–H3 dipolar couplings using the 1H–13C DIPSHIFT NMR experiment,34 which yielded values of b33av of 3500 ± 100 Hz for cholesterol in rHDL and 4300 ± 100 Hz for cholesterol in MLVs (Fig. 3c and Tables 1, 2). In Fig. 3c it can be seen that the 13C transverse magnetization refocuses fully over one rotor period, implying that cholesterol does not undergo intermediate motions that interfere with MAS time dependence that disrupt the coherence of the time domain signal.35
,28 where bijav is expressed in Hertz. The data were compared with curves simulated for different values of bijav and the zero quantum relaxation term, TZQ, to find the values in closest agreement. The simulated curves fit both sets of data closely when |b23av| = |b34av| = 230 ± 50 Hz and TZQ = 11.5 ms (Fig. 3b, left, and Table 1). These values are close to those determined by NMR line shape analysis. The build-up of DQ coherence for [2,3,4-13C3]cholesterol in POPC MLVs are consistent with |b23av| = |b34av| = 320 ± 50 Hz and TZQ = 5.5 ms (Fig. 3b, right, and Table 2). The different extent of scaling of the 13C–13C dipolar couplings in rHDL and MLVs suggest differences in the dynamic behaviour of cholesterol in the two lipid bilayer environments. This conclusion was supported by measurements of residual C3–H3 dipolar couplings using the 1H–13C DIPSHIFT NMR experiment,34 which yielded values of b33av of 3500 ± 100 Hz for cholesterol in rHDL and 4300 ± 100 Hz for cholesterol in MLVs (Fig. 3c and Tables 1, 2). In Fig. 3c it can be seen that the 13C transverse magnetization refocuses fully over one rotor period, implying that cholesterol does not undergo intermediate motions that interfere with MAS time dependence that disrupt the coherence of the time domain signal.35
 | ||
| Fig. 3 Measurements of 13C–13C and 13C–1H dipolar couplings for cholesterol in rHDL and MLVs determined by CP-MAS ssNMR. (a) Selective excitation of C2–C3 and C3–C4 13C–13C ZQ coherences at the n = 1 rotational conditions for both spin pairs, detected by conversion of ZQ to DQ coherence and reconversion to observable magnetization. (b) Build-up and decay of C2–C3 (black circles) and C3–C4 (red circles) 13C difference intensities as a function of the ZQ excitation period τ of the NMR pulse sequence (given in the Fig. S1a, ESI†). Solid lines are the best fitting simulated ZQ build-up curves corresponding to the values of dipolar coupling constants b23av and b34av and zero-quantum relaxation time, TZQ2, given in the main text. The error bars indicate the level of the noise. (c) Evolution of the 13C transverse magnetisation of C3 over one cycle of sample rotation in the presence of C3–H3 dipolar coupling, measured using the DIPSHIFT experiment (Fig. S1b, ESI†). | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 POPC
2 POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) apoA-I nanodiscs (Fig. 6)25
apoA-I nanodiscs (Fig. 6)25
| Nuclear spin pair ij | |bijav| (Hz) | ||
|---|---|---|---|
| Experimental | Gaussian distributiona | MD simulationb | |
| a Gaussian distribution generated from αMR = 90°, αMR = 155°, αMR = 34°. b From ref. 21 and DOI: https://doi.org/10.5281/zenodo.897027. The simulations were run on the Anton 2 supercomputer, and converted to CHARMM readable files. The trajectories consisted of frames at every 4.8 ns and 10 frames were selected at random from the late stages of the 2 ms simulation. | |||
| 13C2–13C3 | 230 ± 50 | 233 | 257 |
| 13C3–13C4 | 230 ± 50 | 245 | 288 |
| 13C3–1H3 | 3500 ± 100 | 3511 | 3412 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 POPC
1 POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol MLVs (Fig. 3), calculated using the Gaussian distribution procedure (Fig. 4) and extracted from MD simulations of 200
cholesterol MLVs (Fig. 3), calculated using the Gaussian distribution procedure (Fig. 4) and extracted from MD simulations of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 POPC
22 POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol bilayers (Fig. 6)36
cholesterol bilayers (Fig. 6)36
| Nuclear spin pair ij | |bijav| (Hz) | ||
|---|---|---|---|
| Experimental | Gaussian distributiona | MD simulationb | |
| a Gaussian distribution generated from αMR = 91°, αMR = 163°, αMR = 28°. b From DOI: https://doi.org/10.5281/zenodo.3243328. Cholesterol orientations were extracted from 10 frames of a 100 ns simulation of a POPC bilayer consisting of 200 lipids (100 per leaflet) in the presence of 10% (22 molecules) cholesterol. The Slipids model (force field parameters downloaded from https://mmkluster.fos.su.se/slipids) was employed for lipids, and the tip3p model for water. | |||
| 13C2–13C3 | 320 ± 50 | 327 | 405 |
| 13C3–13C4 | 320 ± 50 | 326 | 426 |
| 13C3–1H3 | 4300 ± 100 | 4284 | 4185 |
2.3 Analysis of the average dipolar couplings to determine the orientation of cholesterol
The dynamic behaviour of cholesterol in lipid bilayers involves rapid reorientation about a principal rotational axis, R. The experimentally-determined values of b23av, b34av and b33av hold information about the average orientation of cholesterol in the rHDL and MLV lipid bilayers, relative to R and to the bilayer normal, N. The dynamically-averaged dipolar couplings, bijav, depend on the maximum, rigid-limit couplings, bijmax, according to: | (1) |
It is convenient to specify a molecular axis system within which the orientation of R can be specified by azimuthal angle αMR and polar angle βMR (Fig. 4, Steps 1 and 2). For cholesterol, we chose a molecular frame with the z-axis directed along the C3–O3 bond and y-axis in the O3–C3–H3 plane. In this molecular frame, the orientation of each internuclear vector ij, defined by angles θijDR, can be calculated for any values of αMR and βMR using equations of the form
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θijDR = xij θijDR = xij![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αMR αMR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) βMR + yij βMR + yij![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) αMR αMR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) βMR +zij βMR +zij![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) βMR βMR | (2) |
We assessed whether combinations of angles αMR and βMR could be found that are consistent with the measured values of b23av, b34av and b33av for cholesterol in rHDL and MLVs. To do so, an initial state was established in which the rotational axis R is parallel with the bilayer normal N and one particular orientation of cholesterol relative to R was specified by a pair of [αMR, βMR] angles, where each angle can take a value of 0° to 180° (Fig. 4, Step 2). A distribution function was then selected to represent the spread of orientations of R about N, defined by angles θRN. The cholesterol molecular coordinates and rotational axis R were rotated away from N by a set of n randomly-generated angles, ![[small straight theta, Greek, circumflex]](https://www.rsc.org/images/entities/i_char_e12d.gif) RN, weighted as a Gaussian distribution with
RN, weighted as a Gaussian distribution with ![[small theta, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c9.gif) RN = 0° as the fixed mean and σRN as a variable standard deviation (Fig. 4, Step 3). For each new orientation of R, a series of cholesterol rotamers was generated by revolving the molecule about R in equal increments by angle ψR, where 0° ≤ ψR < 360° (Fig. 4, Step 4). The three variables in the simulation were therefore αMR, βMR and σDR and a distribution of 5 × 105 cholesterol orientations was generated for different combinations of these variables. The values of b23av, b34av and b33av were then calculated from each distribution generated (Fig. 4, Step 5). This was done by first determining the orientations of the ij internuclear vectors with respect to the bilayer normal, given by angles θijDN, for each cholesterol orientation, where
RN = 0° as the fixed mean and σRN as a variable standard deviation (Fig. 4, Step 3). For each new orientation of R, a series of cholesterol rotamers was generated by revolving the molecule about R in equal increments by angle ψR, where 0° ≤ ψR < 360° (Fig. 4, Step 4). The three variables in the simulation were therefore αMR, βMR and σDR and a distribution of 5 × 105 cholesterol orientations was generated for different combinations of these variables. The values of b23av, b34av and b33av were then calculated from each distribution generated (Fig. 4, Step 5). This was done by first determining the orientations of the ij internuclear vectors with respect to the bilayer normal, given by angles θijDN, for each cholesterol orientation, where
 | (3) |
bijav = 〈(3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos2 cos2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θijDN(θnRN) − 1)/2〉 θijDN(θnRN) − 1)/2〉 | (4) |
An iterative process was used to calculate the values of b23av, b34av and b33av corresponding to different orientational distributions of cholesterol generated from the three variables according to Fig. 4 (see Section 5.2 for further details). Steps 2 to 6 in Fig. 4 were repeated for different combinations of αMR and βMR values within their allowed ranges (0–180°), and for σDR values of 0° to 40° in 1° increments, and values of b23av, b34av and b33av were calculated for each set of [αMR, βMR, σDR] values. Combinations of the three variables that yielded dipolar couplings that were all within the experimentally-determined ranges were captured. These experimentally-consistent values of αMR and βMR are represented by the contoured regions in Fig. 5(a and b). The [αMR, βMR] combinations are confined to only a limited range of values in both cases, being [90 ± 3°, 155 ± 3°] for cholesterol in rHDL and [91 ± 4°, 163 ± 3°] for cholesterol in MLVs. The value of ∼90° for αMR is a consequence of the near degeneracy of the dipolar couplings b23av and b34av. No other combinations of angles satisfied all three measured dipolar coupling values. For rHDL, values of αMR and βMR that were consistent with the data were only found when σRN assumed values of 34 ± 3°; for MLVs the spread of orientations was slightly lower at σRN = 28 ± 2°. Hence, the dipolar couplings measured from cholesterol in rHDL and MLVs are consistent with slightly different orientations of cholesterol relative to the principal rotational axis and suggest that the distribution of cholesterol orientations is different in the two environment.
2.4 Comparison of the experimental data with molecular dynamics simulations
To validate the interpretation of the NMR-determined dipolar coupling values, we analysed previously published atomistic molecular dynamics (MD) simulations of a 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (mol
2 (mol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol
mol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol) POPC
mol) POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol
cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) apoA-I discoidal assembly25 (analogous in composition to the rHDL formulations here) and of 200
apoA-I discoidal assembly25 (analogous in composition to the rHDL formulations here) and of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 (mol
22 (mol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol) POPC
mol) POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol in square planar bilayers of surface area ∼56 nm2 (Fig. 6a and b).36 The planar bilayers were considered to be a satisfactory approximation of MLVs, which are typically >500 nm in diameter and therefore have <5° curvature over the simulation dimensions in the outermost lamellae. We selected 10 frames at random from the later stages of the simulation trajectories (20 μs for POPC:cholesterol:apoA-I and 100 ns for POPC:cholesterol) and from these snapshots determined the distribution of cholesterol orientations relative to the bilayer normal N, defined by angles αMN and βMN (Fig. 6b and c, discrete bars). The mean αMN and βMN values and standard deviations for cholesterol in the two MD simulation environments are summarised in Table 3, and indicate that cholesterol adopts slightly different distributions of orientations in the two environments. The 13C2–13C3, 13C3–13C4 and 13C3–1H3 dipolar couplings were calculated for each cholesterol molecule in the distributions (Fig. S3, ESI†) and averaged across the ensemble of cholesterol molecules. The experimentally-determined average dipolar couplings are in excellent agreement with the values measured from cholesterol in both simulation environments (Tables 1 and 2). The different average coupling values measured from the POPC:cholesterol:apoA-I and POPC:cholesterol simulations originates from the slightly different distribution profiles of cholesterol in the two bilayer environments. Finally, we compared the cholesterol orientations in the two environments as determined from the MD simulations and as generated by the Gaussian distribution function described in Fig. 4, using the mean parameters tabulated in Fig. 5 (Fig. 6b and c, solid lines). The distribution profiles generated by the Gaussian function agree very well with the histograms calculated from the MD snapshots, with the mean values,
cholesterol in square planar bilayers of surface area ∼56 nm2 (Fig. 6a and b).36 The planar bilayers were considered to be a satisfactory approximation of MLVs, which are typically >500 nm in diameter and therefore have <5° curvature over the simulation dimensions in the outermost lamellae. We selected 10 frames at random from the later stages of the simulation trajectories (20 μs for POPC:cholesterol:apoA-I and 100 ns for POPC:cholesterol) and from these snapshots determined the distribution of cholesterol orientations relative to the bilayer normal N, defined by angles αMN and βMN (Fig. 6b and c, discrete bars). The mean αMN and βMN values and standard deviations for cholesterol in the two MD simulation environments are summarised in Table 3, and indicate that cholesterol adopts slightly different distributions of orientations in the two environments. The 13C2–13C3, 13C3–13C4 and 13C3–1H3 dipolar couplings were calculated for each cholesterol molecule in the distributions (Fig. S3, ESI†) and averaged across the ensemble of cholesterol molecules. The experimentally-determined average dipolar couplings are in excellent agreement with the values measured from cholesterol in both simulation environments (Tables 1 and 2). The different average coupling values measured from the POPC:cholesterol:apoA-I and POPC:cholesterol simulations originates from the slightly different distribution profiles of cholesterol in the two bilayer environments. Finally, we compared the cholesterol orientations in the two environments as determined from the MD simulations and as generated by the Gaussian distribution function described in Fig. 4, using the mean parameters tabulated in Fig. 5 (Fig. 6b and c, solid lines). The distribution profiles generated by the Gaussian function agree very well with the histograms calculated from the MD snapshots, with the mean values, ![[small alpha, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c2.gif) MN and
MN and ![[small beta, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c3.gif) MN and standard deviations σRN, being very similar for the two approaches (Table 3).
MN and standard deviations σRN, being very similar for the two approaches (Table 3).
 | ||
Fig. 6 Analysis of the orientational distribution of cholesterol in MD simulations of rHDL nanodiscs (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 POPC 2 POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol cholesterol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) apoA-I) and planar lipid bilayers (200 apoA-I) and planar lipid bilayers (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 POPC 22 POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cholesterol). (a and b) Single frame snapshots taken from the late stages of the simulation trajectories (2 μs for (a) and 100 ns for (b)). (c and d) Histograms of angles αMN (red) and βMN (blue) defining the distribution of orientations of the cholesterol molecular frame relative to the bilayer normal N. Solid lines represent the distribution of angles calculated using the Gaussian distribution method (step 6 in Fig. 4). The means and standard deviations of the distributions are summarised in Table 3. cholesterol). (a and b) Single frame snapshots taken from the late stages of the simulation trajectories (2 μs for (a) and 100 ns for (b)). (c and d) Histograms of angles αMN (red) and βMN (blue) defining the distribution of orientations of the cholesterol molecular frame relative to the bilayer normal N. Solid lines represent the distribution of angles calculated using the Gaussian distribution method (step 6 in Fig. 4). The means and standard deviations of the distributions are summarised in Table 3. | ||
| MD simulations | Gaussian distribution | |||
|---|---|---|---|---|
![[small alpha, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c2.gif) MN (σMN)
MN (σMN) |
![[small beta, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c3.gif) MN (σMN)
MN (σMN) |
![[small alpha, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c2.gif) MN (σMN)
MN (σMN) |
![[small beta, Greek, macron]](https://www.rsc.org/images/entities/i_char_e0c3.gif) MN (σMN)
MN (σMN) |
|
| a Gaussian distribution generated from αMR = 90°, αMR = 155°, αMR = 34°. b Gaussian distribution generated from αMR = 91°, αMR = 163°, αMR = 28°. | ||||
| rHDLa | 89° (19°) | 150° (16°) | 90° (19°) | 148° (16°) |
| MLVb | 94° (17°) | 157° (15°) | 90° (18°) | 154° (15°) |
3. Conclusion
With the aid of 13C CP-MAS ssNMR methods, new insights have been gained into the orientation of cholesterol in rHDL nanoparticles approximating the discoidal morphology of nascent pre-β HDL. By exploiting dynamically-averaged 13C–13C and 1H–13C dipolar coupling measurements, it is shown that the average orientation of cholesterol in rHDL is slightly different from that in MLV bilayers. The NMR data are also consistent with a slightly larger spread of orientations in rHDL than in MLVs. These differences in the orientational behaviour of cholesterol likely arise from the lateral and rotational diffusion of the lipid components being constrained by the diameter of the apoA-I double belt, which forces the bilayer to distort and ripple to accommodate the internal strain on the lipid components. As cholesterol is added, the overall shape and size of the cross section of the disk remains constant, but to accommodate the cholesterol the lipid bilayer core swells and this may influence the orientation of cholesterol.37 Such constraints are more relaxed in MLVs because the lipids and cholesterol molecules are free to diffuse laterally unhindered. Wide-line 2H ssNMR studies of phospholipid chain order in nanodiscs prepared with MSPΔH5, a truncated apoA-I variant, concluded that the restricted area in the nanodiscs reduce the cooperativity of lipid assembly during the gel-to-liquid crystalline phase transition and increase lipid fluidity above the transition.38 Interestingly, the same study reported an ordering effect of cholesterol, although the order/disorder of cholesterol itself was not examined. The higher orientational disorder of cholesterol presented here could contribute to the HDL particle plasticity observed in atomistic simulations.25 The particle plasticity could be required to enable the particles to interact with multiple enzymes and cell receptors in reverse cholesterol transport.4,5,39 An important outcome of this work is that the dipolar coupling measurements agree well with values calculated for cholesterol in MD simulations of HDL-like nanodiscs and in planar lipid bilayers, thereby providing hitherto unavailable experimental validation of the simulations. Unexpectedly, the cholesterol distributions generated by a Gaussian function with variables αMR, βMR and σRN, that are consistent with the NMR measurements, also agree well with the distributions extracted from the MD simulations.The methodology presented here paves the way for further investigation of cholesterol orientational order and dynamics in rHDL particles of different lipid composition and cholesterol concentration, as well as in spherical HDL in which cholesteryl esters penetrate deeper into the lipid core of the particles. In such particles, which approximate “mature” HDL, it should be possible in principle to determine the orientation of cholesterol relative to its axis of rotation.
4. Experimental procedures
4.1 Materials
[2,3,4-13C3]Cholesterol was obtained from Merck. Palmitoyloleoylphosphatidylcholine (POPC) was obtained from Avanti® Polar Lipids (Merck).4.2 Preparation of multilamellar vesicles
Aliquots of POPC (328.9 mM) and [2,3,4-13C3]cholesterol (25.7 mM) stocks in CHCl3 were combined in a round-bottom flask to a final molar ratio of POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2,3,4-13C3]cholesterol of 10
[2,3,4-13C3]cholesterol of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mol/mol). The CHCl3 was evaporated under a stream of N2, and the thin film was placed under vacuum for ≥4 hours. The film was resuspended in 0.5 mL rHDL buffer (10 mM tris base, 1 mM EDTA) pH 7.4, to form MLVs with a final POPC
1 (mol/mol). The CHCl3 was evaporated under a stream of N2, and the thin film was placed under vacuum for ≥4 hours. The film was resuspended in 0.5 mL rHDL buffer (10 mM tris base, 1 mM EDTA) pH 7.4, to form MLVs with a final POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2,3,4-13C3]cholesterol molar ratio of 26.3
[2,3,4-13C3]cholesterol molar ratio of 26.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.63 mM. The MLVs were homogenised with 10 freeze–thaw cycles, pelleted by bench-top centrifugation (10 minutes, 13.4k rpm), and transferred into a 3.2 mm diameter MAS rotor.
2.63 mM. The MLVs were homogenised with 10 freeze–thaw cycles, pelleted by bench-top centrifugation (10 minutes, 13.4k rpm), and transferred into a 3.2 mm diameter MAS rotor.
4.3 Production of rHDL particles
The rHDL particles were prepared as described previously31 using the widely established detergent mediated sodium cholate dialysis method.30 Briefly, a thin film of POPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [2,3,4-13C3]cholesterol at a 10
[2,3,4-13C3]cholesterol at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (M/M) molar ratio was prepared as above. An aliquot of sodium cholate (232.2 mM) stock in rHDL buffer, pH 7.4, was added to a final molar ratio of lipid
1 (M/M) molar ratio was prepared as above. An aliquot of sodium cholate (232.2 mM) stock in rHDL buffer, pH 7.4, was added to a final molar ratio of lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) detergent of 1
detergent of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–2 (M/M). The solution was stirred at 4 °C until clear. Recombinant WT apoA-I (2-243)40 was added to a final molar ratio of lipid
1–2 (M/M). The solution was stirred at 4 °C until clear. Recombinant WT apoA-I (2-243)40 was added to a final molar ratio of lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein of 200
protein of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (M/M) and the sample was stirred at 4 °C for ≥12 hours. The detergent was removed by dialysis against a 500-fold excess volume of rHDL buffer, pH 7.4, over 3 days at 4 °C, with at least 5 buffer changes. The dialysis tubing MWCO was 12–14 kDa (Merck).
2 (M/M) and the sample was stirred at 4 °C for ≥12 hours. The detergent was removed by dialysis against a 500-fold excess volume of rHDL buffer, pH 7.4, over 3 days at 4 °C, with at least 5 buffer changes. The dialysis tubing MWCO was 12–14 kDa (Merck).
For 13C CP-MAS ssNMR the rHDL particles were spin concentrated using an Amicon Ultra-4 device (10 kDa MWCO, Merck). The rHDL particles were precipitated in a ≥48 hour incubation at 4 °C with 24% (w/v) PEG-6000 in rHDL buffer, pH 7.4.31,41 The precipitated particles were pelleted at 4 °C by ultracentrifugation at 41k rpm for 2 hours in an SW55 Ti rotor (Beckman Coulter). The PEG-precipitated rHDL was transferred to a 3.2 mm diameter MAS rotor and stored at 4 °C. The lipid and cholesterol concentration were measured in duplicate using 96-well plate colorimetric quantification assay kits (CS0001 and MAK043, Sigma-Aldrich®). A Nanodrop™ 2000 spectrophotometer (ThermoFisher Scientific) was used to measure the protein concentration (n = 9).
4.4 Transmission electron microscopy
The rHDL particles were imaged on formvar/carbon coated 300 mesh Cu grids (agar scientific). To increase sample adhesion 10 μL of 0.1% (w/v) poly-L-lysine in dH2O was incubated on the grid for 1 minute.42 The sample (10 μL, 36 μM protein) was incubated on the grid for 2 minutes, followed by negative staining with 10 μL of 2% (w/v) PTA, pH 7.0, incubated for 1 minute. Between each step the liquid was blotted with filter paper. The reagents were kindly provided by Papatziamou. The sample grid was imaged using a JEOL JEM-1010 microscope (BLS, Lancaster University) with an 80 kV tungsten filament electron source and an AMT CCD camera at 120k× magnification. The images were processed using FIJI software.43 A total of 100 particles per window (n = 3) were measured on the TEM grid to determine the average particle diameter.4.5 NMR experiments
The 13C cross-polarization magic-angle-spinning (CP-MAS) NMR measurements were conducted on a Bruker Avance III 700 spectrometer (operating at resonance frequency 700.1 MHz for 1H and 175.9 MHz for 13C). A triple resonance (HXY) 3.2 mm MAS probe was used for all measurements, at a calibrated temperature of 298 K. In all experiments, the cross-polarization Hartmann–Hahn contact time was 2 ms at a proton field of 66 kHz, with 90° pulse lengths of 4 μs for 13C and 3 μs for 1H. To measure 1H–13C dipolar couplings, a constant-time DIPSHIFT experiment34 was used in which frequency-switched Lee Goldberg was used for homonuclear decoupling (at a field of 51 kHz) over periods τ of up to one rotor cycle (i.e., 0 ≤ τ ≤ 2π/ωr), followed by a 180° pulse and a further period of heteronuclear decoupling (tHC = 4π/ωr − tHH) to refocus the 13C chemical shifts. The spinning frequency ωr/2π was 4400 Hz. A series of free induction decays were obtained with proton decoupling for incremented periods of τ. After Fourier transformation of the FIDs, the evolution of 13C magnetisation was measured from the peak intensities for C3 at the different τ periods.For the double quantum (DQ) experiments an adaptation of the pulse sequences described by Karlsson et al. was used.28,29 This method was chosen because, unlike other one-dimensional dipolar recoupling methods, it allows for selective measurements of the C2–C3 and C3–C4 couplings at rotational resonance and so avoids ambiguities arising from multiple spin couplings involving C3. Although double quantum coherence is observed in the spectra, the rates of signal build-up (Fig. 3b) actually represent the rate of mechanical excitation of zero quantum (ZQ) coherence. The initial rate of excitation at rotational resonance is proportional to the magnitude of the dipolar coupling and hence is expected to be slower in dynamically averaged systems than in rigid solids. The experiment proceeds as follows. After cross-polarization, the transverse magnetization for the 13C–13C spin pairs of interest, S1 and S2, are converted to longitudinal difference magnetization, S1Z − S2Z. At the n = 1 rotational resonance condition the dipolar coupling between the spin pair is reintroduced and mechanically-driven excitation of ZQ coherence occurs from the initial S1Z − S2Z state over a variable interval τ. The ZQ coherence is then converted to DQ coherence and immediately back to ZQ coherence and a second τ interval regenerates the longitudinal difference magnetization. A final readout 90° pulse creates transverse magnetization with the two 13C spins having opposite phase. The receiver phase cycle selects only the magnetization that has passed through the ZQ–DQ–ZQ filter, and the Fourier transformed spectrum exhibits positive and negative peaks only from the two coupled 13C spins. The sum of the absolute values of the integrated intensities of the two peaks are plotted as a function of τ to visualise the rates of excitation and relaxation of the ZQ coherence.
5. Computational procedures
5.1 NMR simulations
Simulations of the ZQ evolution and 13C magnetization evolution under 1H dipolar coupling measured in the DIPSHIFT experiment were carried out to find the values of dynamically-averaged dipolar coupling constants providing the closest fit to the data. Simulations were carried out in the SIMPSON environment.44 Fixed parameters used in the simulations were the 13C–13C J-coupling constants (33 Hz for C2–C3 and C3–C4) and 13C chemical shift anisotropy (CSA) parameters measured from 13C NMR spectra of non-spinning samples to be approximately 1.1 kHz. The extensive scaling of the 13C CSA values meant that they had little effect on the shapes of the simulated curves. For the DIPSHIFT simulations, a series of time-domain curves representing the evolution of 13C transverse magnetization over one rotor cycle (at a MAS frequency ωr/2π = 4400 Hz) were calculated for different values of the 13C–1H dipolar coupling constant and compared with the data to find the best fit. The proton isotropic chemical shifts and chemical shift anisotropy were set to zero.For simulations of the ZQ excitation at n = 1 rotational resonance, the MAS frequency was set to the difference in the isotropic chemical shifts of the appropriate 13C spins (C2 and C3, or C3 and C4). An initial condition of 13C difference magnetization (S1z − S2z) was established and the read-out was ZQ coherence (S1+S2− + S1−S2+) as a function of excitation time. The curves generated for different 13C–13C dipolar coupling constants, bavij, were multiplied by an exponential zero-quantum relaxation term and normalised to the maximum intensity. The maximum intensity (in the absence of relaxation) occurs at close to an excitation time 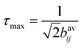 ,28 where bavij is in Hertz. This relationship was verified here from measurements on standard 13C labelled compounds for which the 13C–13C distances are known.
,28 where bavij is in Hertz. This relationship was verified here from measurements on standard 13C labelled compounds for which the 13C–13C distances are known.
5.2 Calculation of cholesterol orientations
The values of the angles αMR and βMR, defining the allowed orientations of the principal rotation axis R in the pre-defined molecular frame of cholesterol were calculated by generating a Gaussian distribution of R about the bilayer normal N, as described in the main text, and computing the average dipolar couplings bijav across the distribution. For a given value of σRN, representing the spread of orientations of R relative to N, angles αMR and βMR were each varied through 180° and for every [αMR, βMR] pair were calculated angles, θijDR, defining the orientation of each internuclear vector of interest relative to R (eqn (2)). The values of θijDR were substituted into eqn (3) and (4) to calculate θijDN and the average dipolar coupling constants bijav for the entire distribution. The allowed orientations of R were those for which αMR and βMR yielded bijav values within all three of the experimentally-measured ranges. These [αMR, βMR] pairs were saved and added to subsequent [αMR, βMR] pairs that were consistent with the measured bijav values after each incrementation of σRN. The contoured regions in the plots in Fig. 5(a and b) represent the frequency with which each captured αMR and βMR values occur across all values of σRN from 5° to 40°, with the red areas corresponding to the highest frequency. For each distribution, the individual cholesterol orientations were defined by angles αMN and βMN. By representing the bilayer normal as a unit vector![[N with combining circumflex]](https://www.rsc.org/images/entities/b_i_char_004e_0302.gif) T = (0, 0, 1) and the x- and z-axes of the cholesterol molecular frame as unit vectors
T = (0, 0, 1) and the x- and z-axes of the cholesterol molecular frame as unit vectors ![[x with combining circumflex]](https://www.rsc.org/images/entities/b_i_char_0078_0302.gif) and ẑ, the orientation of each cholesterol molecule relative to the bilayer normal may be expressed as
and ẑ, the orientation of each cholesterol molecule relative to the bilayer normal may be expressed as| αMN = cos[−x1z1z3 − x2z2z3 + x3(1 − z32)] | (5) |
βMN = a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) z3 z3 | (6) |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was funded by the British Heart Foundation Project Grant PG/21/10498. We greatly appreciate Dr Elisabeth Shaw's help in providing technical assistance with the TEM imaging and manuscript preparation. We are grateful to Dimitra Papatziamou for advice and reagents in preparing the TEM samples.References
- P. L. Yeagle, Biochim. Biophys. Acta, 1985, 822, 267–287 CrossRef CAS.
- E. Sackmann, in Structure and Dynamics of Membranes, ed. R. Lipowsky and E. Sackmann, Elsevier, Amsterdam, 1995, pp. 1–64 Search PubMed.
- A. V. Khera, M. Cuchel, M. de la Llera-Moya, A. Rodrigues, M. F. Burke, K. Jafri, B. C. French, J. A. Phillips, M. L. Mucksavage, R. L. Wilensky, E. R. Mohler, G. H. Rothblat and D. J. Rader, N. Engl. J. Med., 2011, 364, 127–135 CrossRef CAS PubMed.
- M. Ouimet, T. J. Barrett and E. A. Fisher, Circ. Res., 2019, 124, 1505–1518 CrossRef CAS PubMed.
- M. Zhang, R. Charles, H. Tong, L. Zhang, M. Patel, F. Wang, M. J. Rames, A. Ren, K.-A. Rye, X. Qiu, D. G. Johns, M. A. Charles and G. Ren, Sci. Rep., 2015, 5, 8741 CrossRef CAS PubMed.
- X. Y. Qiu, A. Mistry, M. J. Ammirati, B. A. Chrunyk, R. W. Clark, Y. Cong, J. S. Culp, D. E. Danley, T. B. Freeman, K. F. Geoghegan, M. C. Griffor, S. J. Hawrylik, C. M. Hayward, P. Hensley, L. R. Hoth, G. A. Karam, M. E. Lira, D. B. Lloyd, K. M. McGrath, K. J. Stutzman-Engwall, A. K. Subashi, T. A. Subashi, J. F. Thompson, I. K. Wang, H. L. Zhao and A. P. Seddon, Nat. Struct. Mol. Biol., 2007, 14, 106–113 CrossRef CAS PubMed.
- T. Laurenzi, C. Parravicini, L. Palazzolo, U. Guerrini, E. Gianazza, L. Calabresi and I. Eberini, J. Lipid Res., 2021, 62, 100006 CrossRef CAS.
- L. Chen, Z.-W. Zhao, P.-H. Zeng, Y.-J. Zhou and W.-J. Yin, Cell Cycle, 2022, 1–19, DOI:10.1080/15384101.2022.2042777.
- T. R. Molugu, K. J. Mallikarjunaiah, J. J. Kinnun, H. I. Petrache, M. Doktorova, G. Khelashvili, R. Ashkar and M. F. Brown, Biophys. J., 2021, 120, 41A CrossRef.
- T. R. Molugu and M. F. Brown, in Cholesterol Modulation of Protein Function: Sterol Specificity and Indirect Mechanisms, ed. A. Rosenhouse-Dantsker and A. N. Bukiya, 2019, vol. 1115, pp. 99–133 Search PubMed.
- M. P. Marsan, I. Muller, C. Ramos, F. Rodriguez, E. J. Dufourc, J. Czaplicki and A. Milon, Biophys. J., 1999, 76, 351–359 CrossRef CAS.
- E. J. Dufourc, E. J. Parish, S. Chitrakorn and I. C. P. Smith, Biochemistry, 1984, 23, 6062–6071 CrossRef CAS.
- T. Rog, M. Pasenkiewicz-Gierula, I. Vattulainen and M. Karttunen, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 97–121 CrossRef CAS PubMed.
- F. Aussenac, M. Tavares and E. J. Dufourc, Biochemistry, 2003, 42, 1383–1390 CrossRef CAS PubMed.
- E. K. Tiburu, P. C. Dave and G. A. Lorigan, Magn. Reson. Chem., 2004, 42, 132–138 CrossRef CAS.
- T. M. Ferreira, F. Coreta-Gomes, O. H. S. Ollila, M. J. Moreno, W. L. C. Vaz and D. Topgaard, Phys. Chem. Chem. Phys., 2013, 15, 1976–1989 RSC.
- J. A. Clarke, J. M. Seddon and R. V. Law, Soft Matter, 2009, 5, 369–378 RSC.
- J. A. Clarke, A. J. Heron, J. M. Seddon and R. V. Law, Biophys. J., 2006, 90, 2383–2393 CrossRef CAS PubMed.
- N. P. Franks, J. Mol. Biol., 1976, 100, 345–358 CrossRef CAS PubMed.
- D. L. Worcester and N. P. Franks, J. Mol. Biol., 1976, 100, 359–378 CrossRef CAS.
- S. Waldie, M. Moulin, L. Porcar, H. Pichler, G. A. Strohmeier, M. Skoda, V. T. Forsyth, M. Haertlein, S. Maric and M. Cardenas, Sci. Rep., 2019, 9, 5118 CrossRef PubMed.
- Z. Wu, M. A. Wagner, L. Zheng, J. S. Parks, J. M. Shy, J. D. Smith, V. Gogonea and S. L. Hazen, Nat. Struct. Mol. Biol., 2007, 14, 861–868 CrossRef CAS PubMed.
- R. Silva, G. M. Hilliard, L. Li, J. P. Segrest and W. S. Davidson, Biochemistry, 2005, 44, 8600–8607 CrossRef CAS.
- M. Pourmousa and R. W. Pastor, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 2094–2107 CrossRef CAS PubMed.
- M. Pourmousa, H. D. Song, Y. He, J. W. Heinecke, J. P. Segrest and R. W. Pastor, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5163–5168 CrossRef CAS.
- A. Vogel, H. A. Scheidt, S. E. Feller, J. Metso, R. M. Badeau, M. J. Tikkanen, K. Wahala, M. Jauhiainen and D. Huster, Biophys. J., 2014, 107, 114–125 CrossRef CAS.
- L. A. Della Ripa, Z. A. Petros, A. G. Cioffi, D. W. Piehl, J. M. Courtney, M. D. Burke and C. M. Rienstra, Methods, 2018, 138, 47–53 CrossRef PubMed.
- T. Karlsson, M. Eden, H. Luthman and M. H. Levitt, J. Magn. Reson., 2000, 145, 95–107 CrossRef CAS PubMed.
- T. Karlsson, C. E. Hughes, J. Gunne and M. H. Levitt, J. Magn. Reson., 2001, 148, 238–247 CrossRef CAS PubMed.
- A. Jonas, Methods Enzymol., 1986, 128, 553–582 CAS.
- S. Lau and D. A. Middleton, Angew. Chem., Int. Ed., 2020, 59, 18126–18130 CrossRef CAS PubMed.
- S. Dusold and A. Sebald, in Annual Reports on Nmr Spectroscopy, ed. G. A. Webb, 2000, vol. 41, pp. 185–264 Search PubMed.
- T. Karlsson, A. Brinkmann, P. J. E. Verdegem, J. Lugtenburg and M. H. Levitt, Solid State Nucl. Magn. Reson., 1999, 14, 43–58 CrossRef CAS.
- M. G. Munowitz, R. G. Griffin, G. Bodenhausen and T. H. Huang, J. Am. Chem. Soc., 1981, 103, 2529–2533 CrossRef CAS.
- E. R. Deazevedo, K. Saalwachter, O. Pascui, A. A. De Souza, T. J. Bonagamba and D. Reichert, J. Chem. Phys., 2008, 128, 104505 CrossRef PubMed.
- M. Javanainan DOI:10.5281/zenodo.640085 .
- S. R. Midtgaard, M. C. Pedersen and L. Arleth, Biophys. J., 2015, 109, 308–318 CrossRef CAS.
- D. Martinez, M. Decossas, J. Kowal, L. Frey, H. Stahlberg, E. J. Dufourc, R. Riek, B. Habenstein, S. Bibow and A. Loquet, Chem. Phys. Chem., 2017, 18, 2651–2657 CrossRef CAS PubMed.
- D. Sahoo, T. C. Trischuk, T. Chan, V. A. B. Drover, S. Ho, G. Chimini, L. B. Agellon, R. Agnihotri, G. A. Francis and R. Lehner, J. Lipid Res., 2004, 45, 1122–1131 CrossRef CAS PubMed.
- D. Townsend, E. Hughes, R. Hussain, G. Siligardi, S. Baldock, J. Madine and D. A. Middleton, Biochemistry, 2017, 56, 1632–1644 CrossRef CAS.
- K. Mors, C. Roos, F. Scholz, J. Wachtveitl, V. Dotsch, F. Bernhard and C. Glaubitz, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 1222–1229 CrossRef.
- R. S. Anjum, S. M. Bray, J. K. Blackwood, M. L. Kilkenny, M. A. Coelho, B. M. Foster, S. R. Li, J. A. Howard, L. Pellegrini, S. V. Albers, M. J. Deery and N. P. Robinson, Nat. Commun., 2015, 6, 8163 CrossRef.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- M. Bak, J. T. Rasmussen and N. C. Nielsen, J. Magn. Reson., 2000, 147, 296–330 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cp02393h |
| This journal is © the Owner Societies 2022 |




