Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The C-terminus of the GKY20 antimicrobial peptide, derived from human thrombin, plays a key role in its membrane perturbation capability
Rosario
Oliva
ab,
Marco
Campanile
b,
Pompea
Del Vecchio
b,
Elio
Pizzo
c,
Andrea
Bosso
c,
Roland
Winter
 a and
Luigi
Petraccone
a and
Luigi
Petraccone
 *b
*b
aPhysical Chemistry I – Biophysical Chemistry, Department of Chemistry and Chemical Biology, TU Dortmund University, Otto-Hahn Street 4a, 44227 Dortmund, Germany
bDepartment of Chemical Sciences, University of Naples Federico II, Via Cintia 4, 80126 Naples, Italy. E-mail: luigi.petraccone@unina.it
cDepartment of Biology, University of Naples Federico II, Via Cintia 4, 80126 Naples, Italy
First published on 8th March 2022
Abstract
Previously, we characterized in detail the mechanism of action of the antimicrobial peptide GKY20, showing that it selectively perturbs the bacterial-like membrane employing peptide conformational changes, lipid segregation and domain formation as key steps in promoting membrane disruption. Here, we used a combination of biophysical techniques to similarly characterize the antimicrobial activity as well as the membrane perturbing capability of GKY10, a much shorter version of the GKY20 peptide. GKY10 is only half of the parent peptide and consists of the last 10 amino acids (starting from the C-terminus) of the full-length peptide. Despite a large difference in length, we found that GKY10, like the parent peptide, retains the ability to adopt a helical structure and to induce lipid segregation upon membrane binding. Overall, our results suggest that the amino acid sequence of GKY10 is responsible for most of the observed behaviors of GKY20. Our results shed further light on the mechanism of action of the full-length peptide and provide useful information for the design and development of new peptides that serve as antimicrobial agents.
1 Introduction
Antimicrobial peptides (AMPs) are an interesting class of bioactive peptides with a broad spectrum of activity against various microorganisms, including bacteria, fungi and even cancer cells.1–3 AMPs were first reported during the 80s of the last century,4 and, since then, an increasing number of new molecules have been identified in all life forms and even in viruses.5,6 Today, more than 3000 AMP sequences are known, which are conveniently reported in the APD3 database, which is constantly updated.7 AMPs are short peptides of variable lengths (typically from 10 up to 80 amino acid residues) and usually contain a high fraction of positively charged as well as hydrophobic residues.8,9 The exact mechanism of action of AMPs is still under debate. Most of them are membranolytic and three different putative models have been proposed to explain their action: “carpet”, “toroidal” and “barrel-stave” models.8 Despite the particular mechanism of action of a given AMP, it is believed that the primary target of AMPs is represented by the lipid bilayer of the membrane of pathogens.4,10 However, it is important to note that the mechanism depends strictly on the physicochemical properties of the peptide sequence and that a peptide does not necessarily act exclusively by only one of the mechanisms stated above. Interacting in a non-specific way (i.e., in the absence of any membrane receptor) with the lipid matrix of the membrane, AMPs can overcome the problem of resistance to antibiotics in bacteria and other microorganisms.1,11,12 Thus, understanding the molecular basis of the interaction process of AMPs with membranes is of fundamental importance for the development of new AMP-based drugs with an improved selective antimicrobial activity as well as a reduced cytotoxicity towards host cells.In a previous paper, we explored the mechanism of action of the antimicrobial peptide GKY20.13 This peptide, composed of 20 residues, is modelled on the Gly271 to Ile290 sequence of human thrombin, whose structure is shown in Fig. 1A and exhibits strong antimicrobial activity especially against Gram-negative bacteria but also a hemolytic effect on eukaryotic cells.14 Through a comprehensive biophysical characterization of its interaction with model membranes, we showed that GKY20 selectively perturbs the bacterial-like membrane via a carpet-like mechanism employing peptide conformational changes, lipid segregation and domain formation as key steps in promoting membrane disruption.13
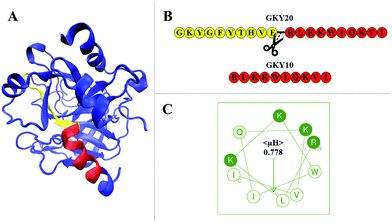 | ||
| Fig. 1 (A) Crystal structure of human thrombin (pdb: 3U69). Highlighted in red is the portion of the protein corresponding to the sequence of GKY10. The N-terminal region of the parent peptide GKY20 is highlighted in yellow. Together, the yellow and red portions constitute the full sequence of GKY20. The remaining structure of the protein is drawn in blue. The representation was prepared using VMD (visual molecular dynamics) software.18 (B) The primary sequences of the GKY20 and GKY10 peptides. The figure also depicts where GKY20 was ideally cut to obtain the sequence of the GKY10 peptide. (C) Helical wheel projection of the GKY10 peptide. The mean hydrophobic moment, 〈μH〉, is also reported. The amino acids are shown using a one letter code: hydrophobic residues in white, polar charged residues in green and polar non-charged residues in light green. The projection and the value of 〈μH〉 were obtained by means of the Heliquest software available at https://heliquest.ipmc.cnrs.fr/.19 | ||
Here, we characterized the interaction of a short version of this peptide limited to the last 10 amino acids starting from the C-terminus of GKY20 (Fig. 1B) with a bacterial-like membrane. The resulting peptide, denoted as GKY10, has a net positive charge of +4 at a physiological pH of 7.4 (one positive charge less than GKY20), a molecular weight of 1311.7 g mol−1 and no protecting groups (such as amide or acetyl) at termini. The choice of this variant of the original peptide is motivated by several considerations. First of all, a systematic study carried out on the antimicrobial potency of GKY20–analog peptides of different lengths showed that the peptide composed of the first 10 amino acids (GKYGFYTHVF sequence) shows no antimicrobial activity,14 suggesting that this portion plays a minor role in the activity of the full-length peptide. This portion showed very moderate hemolytic activity and no information about the secondary structure was available. Second, we noted that, in the full-length sequence, only the 10 amino acid residues RLKKWIQKVI on the C-terminus side of GKY20 have the possibility to form a perfect amphipathic structure as judged by the helical wheel projection (Fig. 1C) with a calculated hydrophobic moment 〈μH〉 of 0.778. Remarkably, the same region adopts an α-helical conformation in human thrombin, whereas the remaining 10 residues assume a small β-strand with turns and unstructured portions (Fig. 1A). Consistently, we experimentally found that upon binding to the membrane about half of the bound GKY20 forms an α-helical structure. Based on these observations, we speculated that the region which adopts the amphipathic α-helical structure of the peptide membrane-bound GKY20 is the C-terminus portion with the sequence RLKKWIQKVI (representing half of the peptide) and that this part is the one that plays the main role in the mechanism of action of GKY20. Indeed, it is well known that the ability to perturb the integrity of the membrane is related to the ability of the peptide to adopt amphipathic helical structures upon binding.15–17 Motivated by these considerations we characterized the interaction of GKY10 using POPC/POPG liposomes (8/2 mol/mol) as a bacterial model membrane, looking for differences compared to the full-length peptide, previously studied.13 Although we found that the antimicrobial activity of GKY10 is generally lower than that of GKY20, our results show that the truncated peptide behaves qualitatively like the parent peptide retaining the ability to adopt a helical structure and to induce lipid segregation upon surface membrane binding. Overall, our data suggest that the amino acidic sequence of GKY10 is responsible for most of the observed behavior of the full-length peptide GKY20.
2 Materials and methods
2.1 Materials
The synthetic peptide GKY10 (sequence: RLKKWIQKVI) was obtained from Primm Srl (Milano, Italy) with a purity of >95%. Lipids 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dipalmitoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (DPPG), 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPG) were purchased from Avanti Polar Lipids (Alabaster, United States). Fluorescent probes 1,6-diphenylhexatriene (DPH) and 6-dodecanoyl-2-dimethylaminonaphthalene (Laurdan), chloroform, methanol and dimethylformamide (DMF) were acquired from Merck (Darmstadt, Germany). Trifluoroethanol (TFE) was purchased from ROMIL Ltd (Cambridge, United Kingdom). Sodium phosphate buffer (10 mM pH 7.4) was prepared using deionized water.2.2 Liposome preparation
An appropriate amount of lipids was weighed in an amber vial and dissolved in a chloroform/methanol mixture (2/1 v/v). The organic solvent was removed by gentle evaporation under nitrogen flux to produce a thin film. To eliminate traces of the organic solvent, the sample was placed under vacuum for at least 4 hours. An appropriate amount of sodium phosphate buffer was then added for hydration and multilamellar vesicles (MLVs) were obtained by vigorous mixing. MLVs with embedded DPH were prepared as previously described by adding a DPH solution in chloroform to the lipid mixture at a lipid/DPH molar ratio of 150. MLVs containing Laurdan were produced likewise, adding a Laurdan solution in DMF to the mixture at a lipid/probe molar ratio of 30. Large unilamellar vesicles (LUVs) with a diameter of 200 nm were produced by extruding MLVs 21 times through a porous polycarbonate membrane using a Mini-Extruder (Avanti Polar Lipid). To check the size of the vesicles after extrusion, dynamic light scattering (DLS) measurements were performed by means of a Zetasizer Nano ZS from Malvern Instruments (Malvern, United States). The obtained hydrodynamic radius was in agreement with the size of LUVs. Liposomes with different compositions were prepared: (a) DPPC/DPPG (8/2 mol/mol), (b) DPPC/POPG (8/2 mol/mol), and (c) POPC/POPG (8/2 mol/mol). Samples in the presence of the peptide were prepared by adding an appropriate amount of peptide solution to the liposome suspension at a desired lipid-to-peptide molar ratio (L/P).2.3 Circular dichroism (CD)
Far-UV CD spectra were acquired using a Jasco J-715 spectropolarimeter from Jasco Corporation (Tokyo, Japan). All the experiments were performed in a 0.1 cm path length quartz cuvette at a scan rate of 20 nm min−1, a response time of 4 s and a bandwidth of 2 nm. Three spectra were collected for each sample to improve the signal-to-noise ratio. All the samples were prepared with a final peptide concentration of 26.6 μM. Experiments were carried out in water and solutions at TFE percentages of 20, 50 and 70. Mixtures of the peptide and POPC/POPG LUVs suspension were prepared at L/P = 10, 50, and 100 using sodium phosphate buffer (10 mM pH 7.4). For each measurement, the corresponding blank spectrum was subtracted prior to data analysis.2.4 Differential scanning calorimetry (DSC)
DSC thermograms of DPPC/DPPG and DPPC/POPG were recorded using a high sensitivity nano-DSC from TA Instruments (New Castle, USA) equipped with two twin gold capillary cells of 300 μL. MLVs were used in all the experiments in order to enhance the phase transition resolution.20 All experiments were performed in the temperature range of 25–55 °C with a scan rate of 1 °C min−1 under a pressure of 3 atm. A buffer–buffer scan was subtracted from each sample–buffer thermogram prior to baseline subtraction. Reproducibility and reversibility were verified by comparing at least four heating and four cooling scans for each sample. Lipid suspensions were diluted up to 500 μM with sodium phosphate 10 mM at pH 7.4 in the absence and presence of the peptide at L/P = 10. Data were analyzed using Nano-Analyze software provided by the manufacturer and plotted using Origin Lab software (OriginLab, Northampton, MA, USA).2.5 Steady-state fluorescence spectroscopy
Fluorescence spectroscopy experiments were performed using a Fuoromax-4 spectrofluorometer from Horiba Scientific (Edison, USA). To study the interaction between the peptide and POPC/POPG LUVs, the intrinsic fluorescence of the peptide tryptophan was monitored at an increasing concentration of lipids (up to 1 mM). All the experiments were performed at 25 °C in a 1 cm length quartz cuvette with stirring. For each sample, at least three spectra were recorded until a superimposition was obtained, indicating that an equilibrium was reached. The peptide concentration was fixed at 7 μM. The excitation wavelength was set to 280 nm and the emission was collected between 295 and 500 nm. The slits were set to 6 for the excitation monochromator and 10 for the emission monochromator. Cross-oriented polarizer configuration (Ex = 90° and Em = 0°) was adopted in order to reduce the background scattering produced by the lipid vesicles.21 The binding curve was constructed by plotting the emission intensity normalized with respect to the emission intensity of the peptide alone as a function of lipid concentration. The curve was fitted as previously described to obtain a mole-fraction partition constant Kx.13 Fluorescence anisotropy experiments were performed on DPPC/DPPG LUVs containing DPH or Laurdan at a total lipid concentration of 50 μM and at an increasing peptide concentration (up to about 50 μM). The anisotropy was recorded in a 1 cm path length quartz cuvette equipped with darkened windows to reduce the scattering interference. DPH was excited at 355 nm and its anisotropy was collected at 425 nm, while Laurdan was excited at 340 nm and its anisotropy was collected at 476 nm. Fluorescence anisotropy (r) was calculated using the equation r = (IVV − GIVH)/(IVV + 2GIVH), where IVV is the fluorescence intensity when both the excitation and emission polarizers are in a vertical orientation, IVH is the intensity when the excitation polarizer is in a vertical orientation while the emission polarizer is in a horizontal orientation, and G is a factor correcting for the detector's different responses to vertically and horizontally polarized light.132.6 Antimicrobial activity
The antimicrobial activities of GKY20 and GKY10 were evaluated by measuring their minimal inhibitory concentrations (MIC100) on the following bacterial strains: Pseudomonas aeruginosa PAO1, Salmonella enterica ATCC 14028, Klebsiella pneumonia ATCC 700603, Salmonella enteriditis 706 RIVM, Acinetobacter baumannii ATCC17878, Enterococcus faecalis ATCC 29212, methicillin resistant Staphylococcus aureus WKZ-2, Staphylococcus aureus ATCC 6538P, and Staphylococcus aureus ATCC 12600. The assay was carried out as already described.22 Briefly, bacteria were grown to the mid-logarithmic phase in Luria Bertani broth (LB) at 37 °C. Cells were then diluted to 1 × 106 CFU mL−1 in Difco 0.5× Nutrient Broth (Becton-Dickenson, Franklin Lakes, NJ) containing increasing amounts of GKY20 or GKY10 (0.75–50 μM). Following overnight incubation, MIC100 values were determined as the lowest peptide concentration responsible for no visible bacterial growth.3 Results and discussion
3.1 Interaction of GKY10 with POPC/POPG liposomes
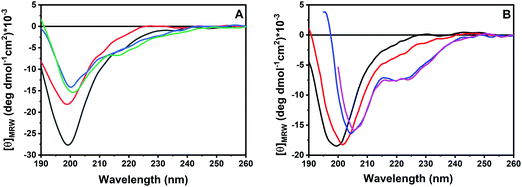
First, we evaluated the ability of GKY10 to interact with appropriate model membranes.21 Indeed, the lipid bilayer of the pathogen membrane is considered the primary target of antimicrobial peptides.8 To evaluate the interaction of the peptide with the lipid bilayer, we followed the fluorescence intensity changes of the Trp residue of GKY10 upon increasing the lipid concentration. Upon the addition of POPC/POPG liposomes to a GKY10 solution, an increase of the fluorescence emission intensity of the Trp residue and a blue shift of the maximum of emission of ∼12 nm were observed (Fig. 2), revealing that the Trp residue experiences a more hydrophobic environment.23,24 Remarkably, this result reveals that GKY10, despite its much shorter length, retains the ability to bind the lipid bilayer. We then evaluated the mole fraction partition constants (Kx) by fitting the corresponding binding isotherms, as reported in ref. 13 (see the insets in Fig. 2). The obtained value was Kx = 2.4 × 105, a value that is approximately one order of magnitude smaller than the value determined for the longer GKY20 under similar conditions.13 Although GKY10 retains the ability to interact with the lipid bilayer, this result clearly reveals that the first 10 residues of GKY20 have a non-negligible contribution in favoring the membrane–peptide interaction. This can be due to an enhanced electrostatic contribution, as the full-length peptide carries one more positive charge, and/or to additional membrane–peptide interactions established by the additional 10 N-terminus residues.In order to obtain information about the conformational behavior of the peptide in the absence (i.e., in neat buffer) and presence of the POPC/POPG model membranes, circular dichroism (CD) spectroscopy was employed. Circular dichroism is a powerful technique that allows the study of the secondary structural changes of proteins and peptides.25–28 It is known that most of the disordered, linear AMPs are able to adopt a helical conformation upon binding to membranes8,17,29 and it appears that the formation of a helical structure is related to the ability of the peptides to perturb the integrity of the membrane and exert their biological function.9,16 To this end, we first evaluated the propensity of GKY10 to adopt a helical conformation by recording its CD spectra upon increasing the concentration of the helical structure-inducer solvent 2,2,2-trifluoroethanol (TFE) in water.30–32Fig. 3, panel A, shows the CD spectra of GKY10 in the water/TFE mixture at TFE concentrations of 0, 20, 50 and 70 vol%. The peptide alone shows a negative band at about 200 nm, indicating that the peptide is mainly unstructured in solution, i.e., adopts a random coil like structure. Interestingly, on increasing the TFE concentration, only minor changes in the shape of the CD spectrum were observed, suggesting that the organic solvent is not able to induce the formation of the helical structure. This result clearly indicates that the peptide has a very low intrinsic propensity to adopt a helical conformation, as expected for such a kind of short peptide. Conversely, the addition of POPC/POPG unilamellar vesicles results in a drastic change of the CD spectra of GKY10 (Fig. 3, panel B). Particularly, at L/P = 10, a shift of the minimum from 199 nm to 202 nm was detected and a positive band starts to rise at around 190 nm. In addition, a small band at around 222 nm is also present. These observations suggest that, upon binding to lipid vesicles, the peptide changes its conformation, most likely moving towards a helical structure. According to the value of Kx reported above, under these conditions, about 50% of the total peptide should be bound to the vesicles. Thus, the spectrum at L/P = 10 can be considered a combination of the spectra of the free (random coil) and bound peptides. Conversely, in the presence of large excess of lipid vesicles (L/P = 50 and 100), the saturation has been reached and the observed spectrum corresponds to that of the bound peptide. Under these conditions, the spectrum shows two well defined minima located at about 208 nm and 220 nm and a maximum at around 195 nm, confirming that GKY10 partially adopts a helical structure upon binding. Considering the very low intrinsic helical propensity of GKY10, this result is remarkable and it strongly suggests that the α-helix is induced by the selective electrostatic and/or hydrophobic interactions with the POPC/POPG model membrane. Similar observations were reported for the full-length GKY20 peptide, which showed a pronounced α-helical conformation upon membrane binding as inferred from CD spectra. The comparison of our data with the data obtained for GKY20 supports our initial hypothesis that the C-terminus of GKY20 is strongly involved in the membrane interaction and subsequent α-helix formation.
3.2 Effect of peptide binding on the membrane structure and stability
We then explored the effect of the peptide on the stability of the lipid bilayer employing differential scanning calorimetry (DSC) measurements. Since POPC and POPG bilayers do not have a gel-to-fluid phase transition temperature suitable for DSC measurements, they were replaced by DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) and DPPG (1,2-dipalmitoyl-sn-glycero-3-phospho-1′-rac-glycerol). The DSC thermogram of pure DPPC/DPPG MLVs in the absence of peptide is characterized by two well-defined phase transitions (Fig. 4, panel A): a gel-to-gel pre-transition at 36 °C (involving the rearrangement of polar head groups) followed by the main gel-to-fluid transition centered at 42.1 °C (involving mainly the melting of the lipid chains).33,34 The addition of the peptide results in a complete disappearance of the pre-transition peak whereas the main transition peak appears to be shifted to a higher temperature. Remarkably, the enthalpy change accompanying the main transition increases slightly, indicating a more efficient lipid packing upon peptide binding (Table 1). Altogether, these observations suggest that the peptide strongly interacts with lipid polar heads at the membrane surface but does not penetrate significantly into the hydrophobic core of the membrane. Furthermore, the main peak appears to be sharper upon peptide binding, indicating a higher cooperativity of the gel-to-liquid transition. This observation suggests that the peptide reduces the lipid heterogeneity of the melting domain of lipids, maybe by means of specific lipid segregation. Unfortunately, the liposome composed of DPPC and DPPG does not allow proving unambiguously the lipid segregation phenomenon as the melting temperatures of two separate DPPG and DPPC domains are very similar and thus even complete segregation of the two lipids from their mixture should not result in a drastic change of the DSC profile. To verify the ability of GKY10 to induce the formation of lipid domains, the DSC experiment was repeated by replacing DPPG with POPG, producing multilamellar vesicles composed of DPPC/POPG (8/2 mol/mol). The substitution of DPPG with POPG has the advantage that pure POPG has a gel-to-liquid transition temperature far below (∼−5 °C) that of pure DPPG (∼41 °C); thus, its segregation in the DPPG/POPG mixture should lead to a significant modification of the overall DSC profile.13,35Fig. 4, panel B, shows the DSC thermograms of a mixture of DPPC/POPG (8/2 mol/mol) in the absence and presence of GKY10. The DSC profile of this lipid mixture in the absence of the peptide shows a single broad peak with a transition temperature for the gel-to-liquid phase transition at about 35.8 °C. After the addition of GKY10, the peak is severely affected and it is now composed of at least two transitions with a main peak centered at 38 °C and with a clear shoulder at a lower temperature. This result is consistent with a preferential interaction of the cationic peptide with negatively charged POPG lipids, leading to the formation of domains within the bilayer. Indeed, the higher temperature sharp peak should correspond to the DPPC-enriched domain. Conversely, the broad shoulder corresponds to the POPG-enriched domain that melts at a lower temperature (compared to the DPPC/DPPG 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 reference composition)36 and where the short peptide is, most likely, localized. Furthermore, as already observed for DPPC/DPPG liposomes, the total enthalpy change increases slightly, pointing towards a surface binding of the peptide without penetration into the hydrophobic core of the membrane, i.e., the lipid packing is maintained (Table 1).37 It is also important to note that the longer peptide GKY20 is able to induce the formation of lipid domains. However, comparing the data obtained for GKY10 with the DSC data previously reported for the longer GKY20,13 it seems that the ability to induce lipid segregation of GKY10 is less prominent. This could be related to the smaller net negative charge and the lower affinity of GKY10 compared to the longer peptide that can reduce the extent of the lipid segregation and the consequent formation of domains. This could have an impact on the biological activity of the peptide. Indeed, accordingly, the biological activity of GKY10 is lower compared to that of GKY20 (see Table 2).
2 reference composition)36 and where the short peptide is, most likely, localized. Furthermore, as already observed for DPPC/DPPG liposomes, the total enthalpy change increases slightly, pointing towards a surface binding of the peptide without penetration into the hydrophobic core of the membrane, i.e., the lipid packing is maintained (Table 1).37 It is also important to note that the longer peptide GKY20 is able to induce the formation of lipid domains. However, comparing the data obtained for GKY10 with the DSC data previously reported for the longer GKY20,13 it seems that the ability to induce lipid segregation of GKY10 is less prominent. This could be related to the smaller net negative charge and the lower affinity of GKY10 compared to the longer peptide that can reduce the extent of the lipid segregation and the consequent formation of domains. This could have an impact on the biological activity of the peptide. Indeed, accordingly, the biological activity of GKY10 is lower compared to that of GKY20 (see Table 2).
| System | ΔHpa/kJ mol−1 | T p/°C | ΔHma/kJ mol−1 | T m/°C |
|---|---|---|---|---|
| a ΔHp and ΔHm represent the enthalpy change, normalized by the total lipid moles, for the pre-transition and the main transition, respectively. b T p and Tm represent the temperature for the pre-transition and the main transition, respectively. | ||||
| DPPC/DPPG | 1.73 | 36.0 | 31.4 | 42.1 |
| +GKY10 | — | — | 35.6 | 43.0 |
| DPPC/POPG | — | — | 31.8 | 35.8 |
| +GKY10 | — | — | 35.2 | 38.2 |
| Bacterial strain | MIC values (μM) | ||
|---|---|---|---|
| GKY-20 | GKY-10 | ||
| Gram negative | Pseudomonas aeruginosa 01 | 3.1 | 25 |
| Salmonella enterica serovar Typhimurium ATCC 14028 | 3.1 | 25 | |
| Klebsiella pneumonia ATCC 700603 | 6.2 | 25 | |
| Salmonella enteriditis 706 RIVM | 3.1 | 25 | |
| Acinetobacter baumannii ATCC17878 | 3.1 | 6.2 | |
| Gram positive | Enterococcus faecalis ATCC 29212 | 3.1 | 25 |
| Methicillin resistant Staphylococcus aureus WKZ-2 | 3.1 | >50 | |
| Staphylococcus aureus ATCC 6538P | 1.5 | 6.2 | |
| Staphylococcus aureus ATCC 12600 | 3.1 | 12.5 | |
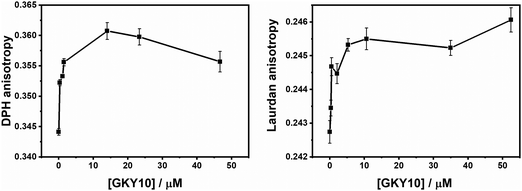
To further monitor the degree of peptide penetration into the lipid bilayer, we performed fluorescence anisotropy experiments of the probe 1,6-diphenyl-1,3,5-hexatriene (DPH), a fluorescent molecule that partitions inside the hydrophobic interior of the bilayer.38,39 The fluorescence anisotropy of the DPH embedded in the DPPC/DPPG liposome slightly increases on increasing the peptide concentration (Fig. 5, panel A), thus confirming that the peptide does not significantly perturb the lipid packing. This observation is consistent with the DSC results and reveals that the peptide does not penetrate deeply into the hydrophobic core of the membrane, but it is rather localized at the membrane surface. To directly monitor the peptide binding to the lipid head group region, we performed additional fluorescence anisotropy measurements using the Laurdan probe embedded in DPPC/DPPG vesicles (Fig. 5, panel B). The fluorescent polarity-sensitive moiety (naphthalene) of Laurdan is mainly localized close to the bilayer/water interface, thus becoming a selective probe of this bilayer region.40–42 Consistent with binding at the membrane surface, we observed an increase of the Laurdan anisotropy on increasing the peptide concentration, which indicates a reduced degree of mobility (or a higher degree of order) of the lipid head groups upon peptide binding. Interestingly, surface binding coupled with anionic lipid segregation together with a membrane rigidification was also observed for the full length GKY20 peptide.13 Taken together, these results show that the half-length GKY10 shares with the GKY20 the ability to bind at the membrane surface and to induce anionic lipid segregation which is a critical step in the GKY20 action mechanism.13
3.3 Antimicrobial activity
Finally, we compared the antibacterial activity of the full-length GKY20 and its truncated version GKY10 by evaluating their minimal inhibitory concentrations (MIC100) on different bacterial strains including both Gram-negative and Gram-positive bacteria (Table 2). An increase of the MIC100 values of the truncated version compared to the full-length of about 8 times was observed, except for Staphylococci and Acinetobacter baumannii, for which the activities of GKY20 and GKY10 are more comparable. This observed lower antimicrobial activity on the bacterial strains studied seems to be rather consistent, with few exceptions, with the observed difference in the affinity constants for the lipid bilayer (close to one order of magnitude). Taken together, our data suggest that the 10 N-terminus residues in GKY20 act to increase the peptide affinity for the membrane, whereas the 10 C-terminus residues are more involved in the membrane-perturbing capability of the peptide. Indeed, the reported data showed that the N-terminus region is more than just a flanking region and, even if it does not play a leading role, it can be involved to a lesser extent in the antimicrobial activity of the full-length peptide.It is interesting to compare the activity of GKY10 with other truncated peptides present in the literature. For example, removing 5 residues from the peptide LFampin 265–284, producing the peptide LFampin 270–284, causes a complete inactivation of the peptide.43 It is remarkable that LFampin 265–284 adopts a helical structure upon binding to model membranes. Conversely, the peptide LFampin 270–284 is not able to fold into any ordered structure upon binding to the bilayer, highlighting that conformational changes are fundamental in determining the biological activity of peptides. Instead, removing four residues, starting from the C-terminus (obtaining LFampin 265–280), causes a decrease of the antimicrobial activity against E. coli but not inactivation and the peptide retains the ability to fold into a helical structure.43 These results are consistent with those observed for GKY20 truncations: when the ability to fold into an amphipathic helical structure is preserved (as for GKY10), the antimicrobial activity is also partially preserved. However, it is important to note that LFampin 265–280 is a truncation of about 20% of the length of the parent peptide whereas GKY10 results from a 50% truncation of GKY20. In this regard, a comparable case is the one reported for the peptide dCHAT.44 The parent peptide showed a remarkable antimicrobial activity against several bacterial strains (MIC values between 2 and 8 μM). If the length of the peptide is reduced to about half of the original one (as in the case of GKY20), a complete loss of antimicrobial activity against the same bacterial strains was observed. In light of these observations, it is remarkable that GKY10, despite the strong truncation, is still active highlighting the central role played by this peptide portion in the full-length peptide. We speculate that this result can be justified by the ability of this peptide portion to participate in the amphipathic helical formation in GKY20 as well as in the full thrombin protein (see the “Introduction” section). On the other hand, when truncations represent a small percentage of the parent peptide, the effect on the antimicrobial activity can be very different. For example, in the case of peptides from marine organisms, the truncation in some cases led to peptides with an improved activity, as in the case of peptide GE (pardaxin).45 This is particularly interesting because, on removing 6 residues from the C-terminus, 4 times higher antimicrobial activity against Vibrio alginolyticus was observed. In other cases, instead, the deletion of some residues causes a reduction of activity, as observed for the peptide SALF-1.45 Removing the last 5 residues from the SALF-1 peptide, obtaining the SALF-5 peptide, causes a decrease of the antimicrobial activity by 2 times against Staphylococcus sp. and by 4 times against Vibrio harveyi.
In summary, the reported examples show that, generally, the truncation of peptides could have a strong impact on the antimicrobial activity, leading to an increase or decrease of the MIC value, depending on the physicochemical properties of the removed amino acids. Overall, this approach is very helpful in identifying the key residues responsible for the observed biological activities in order to produce shorter and more economically convenient sequences for medical applications.
4 Conclusions
In this study, we characterized the membrane interaction and the biological activity of the 10-residue peptide GKY10, which is derived from the C-terminus of the known antimicrobial peptide GKY20. Despite its much shorter length, GKY10 retains modest antimicrobial activity, albeit with higher MIC100 values in comparison with GKY20. The difference in the activity with respect to the full-length peptide appears, with some exceptions, to be consistent with the difference in the affinity constants towards the negatively charged bacterial model membrane, as measured by fluorescence titration experiments. DSC and fluorescence experiments using DPH and Laurdan as probes consistently show that the peptide binds at the membrane surface and induces anionic lipid segregation as previously observed for the GKY20 peptide. Despite the large difference in length, GKY10 retains the ability to adopt an α-helical structure upon membrane binding. Overall, our data show that GKY10 behaves qualitatively like the longer GKY20 peptide, suggesting that its sequence is the one that plays the major role in GKY20-induced membrane perturbation capability. Our results therefore shed further light on the mechanism of action of the GKY20 peptide and provide useful information for the development of new peptides that serve as antimicrobial agents.Abbreviations
| AMPs | Antimicrobial peptides |
| CD | Circular dichroism |
| DMF | Dimethylformamide |
| DPH | 1,6-Diphenylhexatriene |
| DPPC | 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine |
| DSC | Differential scanning calorimetry |
| Laurdan | 6-Dodecanoyl-2-dimethylaminonaphthalene |
| LUV | Large unilamellar vesicles |
| MLV | Multilamellar vesicles |
| POPC | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine |
| POPG | 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol |
| TFE | Trifluoroethanol. |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge generous financial support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2033 – 390677874 – RESOLV (R.W.).References
- J. Lei, L. Sun, S. Huang, C. Zhu, P. Li, J. He, V. Mackey, D. H. Coy and Q. He, The antimicrobial peptides and their potential clinical applications, Am. J. Transl. Res., 2019, 11, 3919–3931 CAS.
- M. Zasloff, Antimicrobial peptides of multicellular organisms, Nature, 2002, 415, 389–395 CrossRef CAS PubMed.
- L. Zhang and R. L. Gallo, Antimicrobial peptides, Curr. Biol., 2016, 26, R14–R19 CrossRef CAS PubMed.
- K. Matsuzaki, Antimicrobial peptides: basics for clinical application, Springer Berlin Heidelberg, New York, NY, 2019 Search PubMed.
- L. Fan, J. Sun, M. Zhou, J. Zhou, X. Lao, H. Zheng and H. Xu, DRAMP: a comprehensive data repository of antimicrobial peptides, Sci. Rep., 2016, 6, 24482 CrossRef CAS PubMed.
- X. Wang and G. Wang, Insights into Antimicrobial Peptides from Spiders and Scorpions, Protein Pept. Lett., 2016, 23, 707–721 CrossRef CAS PubMed.
- G. Wang, X. Li and Z. Wang, APD3: the antimicrobial peptide database as a tool for research and education, Nucleic Acids Res., 2016, 44, D1087–D1093 CrossRef CAS PubMed.
- V. Teixeira, M. J. Feio and M. Bastos, Role of lipids in the interaction of antimicrobial peptides with membranes, Prog. Lipid Res., 2012, 51, 149–177 CrossRef CAS PubMed.
- O. G. Travkova, H. Moehwald and G. Brezesinski, The interaction of antimicrobial peptides with membranes, Adv. Colloid Interface Sci., 2017, 247, 521–532 CrossRef CAS PubMed.
- O. Toke, Antimicrobial peptides: New candidates in the fight against bacterial infections, Biopolymers, 2005, 80, 717–735 CrossRef CAS PubMed.
- A. J. Alanis, Resistance to Antibiotics: Are We in the Post-Antibiotic Era?, Arch. Med. Res., 2005, 36, 697–705 CrossRef PubMed.
- J. Lakshmaiah Narayana and J.-Y. Chen, Antimicrobial peptides: Possible anti-infective agents, Peptides, 2015, 72, 88–94 CrossRef CAS PubMed.
- R. Oliva, P. Del Vecchio, A. Grimaldi, E. Notomista, V. Cafaro, K. Pane, V. Schuabb, R. Winter and L. Petraccone, Membrane disintegration by the antimicrobial peptide (P)GKY20: lipid segregation and domain formation, Phys. Chem. Chem. Phys., 2019, 21, 3989–3998 RSC.
- G. Kasetty, P. Papareddy, M. Kalle, V. Rydengård, M. Mörgelin, B. Albiger, M. Malmsten and A. Schmidtchen, Structure-Activity Studies and Therapeutic Potential of Host Defense Peptides of Human Thrombin, Antimicrob. Agents Chemother., 2011, 55, 2880–2890 CrossRef CAS PubMed.
- M. A. Amon, M. Ali, V. Bender, K. Hall, M.-I. Aguilar, J. Aldrich-Wright and N. Manolios, Kinetic and conformational properties of a novel T-cell antigen receptor transmembrane peptide in model membranes, J. Pept. Sci., 2008, 14, 714–724 CrossRef CAS PubMed.
- Z. Oren and Y. Shai, Selective Lysis of Bacteria but Not Mammalian Cells by Diastereomers of Melittin: Structure−Function Study†, Biochemistry, 1997, 36, 1826–1835 CrossRef CAS PubMed.
- F. Battista, R. Oliva, P. Del Vecchio, R. Winter and L. Petraccone, Insights into the Action Mechanism of the Antimicrobial Peptide Lasioglossin III, Int. J. Mol. Sci., 2021, 22, 2857 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, VMD: Visual molecular dynamics, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- R. Gautier, D. Douguet, B. Antonny and G. Drin, HELIQUEST: a web server to screen sequences with specific -helical properties, Bioinformatics, 2008, 24, 2101–2102 CrossRef CAS PubMed.
- R. L. Biltonen and D. Lichtenberg, The use of differential scanning calorimetry as a tool to characterize liposome preparations, Chem. Phys. Lipids, 1993, 64, 129–142 CrossRef CAS.
- A. S. Ladokhin, S. Jayasinghe and S. H. White, How to Measure and Analyze Tryptophan Fluorescence in Membranes Properly, and Why Bother?, Anal. Biochem., 2000, 285, 235–245 CrossRef CAS PubMed.
- A. Bosso, L. Pirone, R. Gaglione, K. Pane, A. Del Gatto, L. Zaccaro, S. Di Gaetano, D. Diana, R. Fattorusso, E. Pedone, V. Cafaro, H. P. Haagsman, A. van Dijk, M. R. Scheenstra, A. Zanfardino, O. Crescenzi, A. Arciello, M. Varcamonti, E. J. A. Veldhuizen, A. Di Donato, E. Notomista and E. Pizzo, A new cryptic host defense peptide identified in human 11-hydroxysteroid dehydrogenase-1 β-like: from in silico identification to experimental evidence, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 2342–2353 CrossRef CAS PubMed.
- Principles of Fluorescence Spectroscopy, ed. J. R. Lakowicz, Springer, US, Boston, MA, 2006 Search PubMed.
- B. Valeur and M. N. Berberan-Santos, Molecular Fluorescence: Principles and Applications, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2012 Search PubMed.
- Circular Dichroism and the Conformational Analysis of Biomolecules, ed. G. D. Fasman, Springer, US, Boston, MA, 1996 Search PubMed.
- S. M. Kelly, T. J. Jess and N. C. Price, How to study proteins by circular dichroism, Biochim. Biophys. Acta, Proteins Proteomics, 2005, 1751, 119–139 CrossRef CAS PubMed.
- R. Oliva, S. K. Mukherjee, Z. Fetahaj, S. Möbitz and R. Winter, Perturbation of liquid droplets of P-granule protein LAF-1 by the antimicrobial peptide LL-III, Chem. Commun., 2020, 56, 11577–11580 RSC.
- R. Oliva, S. K. Mukherjee, L. Ostermeier, L. A. Pazurek, S. Kriegler, V. Bader, D. Prumbaum, S. Raunser, K. F. Winklhofer, J. Tatzelt and R. H. A. Winter, Remodeling of the Fibrillation Pathway of α-Synuclein by Interaction with Antimicrobial Peptide LL-III, Chem. – Eur. J., 2021, 27, 11845–11851, DOI:10.1002/chem.202101592.
- J. Bürck, S. Roth, P. Wadhwani, S. Afonin, N. Kanithasen, E. Strandberg and A. S. Ulrich, Conformation and Membrane Orientation of Amphiphilic Helical Peptides by Oriented Circular Dichroism, Biophys. J., 2008, 95, 3872–3881 CrossRef PubMed.
- J. K. Myers, C. N. Pace and J. M. Scholtz, Trifluoroethanol effects on helix propensity and electrostatic interactions in the helical peptide from ribonuclease T1: Helix propensities in trifluoroethanol, Protein Sci., 1998, 7, 383–388 CrossRef CAS PubMed.
- L. S. Ramirez, J. Pande and A. Shekhtman, Helical Structure of Recombinant Melittin, J. Phys. Chem. B, 2019, 123, 356–368 CrossRef CAS PubMed.
- V. Srinivas, P. Santhoshkumar and K. Krishna Sharma, Effect of Trifluoroethanol on the Structural and Functional Properties of α-Crystallin, J. Protein Chem., 2002, 21, 87–95 CrossRef CAS PubMed.
- R. Oliva, M. Chino, A. Lombardi, F. Nastri, E. Notomista, L. Petraccone and P. D. Vecchio, Similarities and differences for membranotropic action of three unnatural antimicrobial peptides, J. Pept. Sci., 2020, 26, e3270, DOI:10.1002/psc.3270.
- E. Pizzo, R. Oliva, R. Morra, A. Bosso, S. Ragucci, L. Petraccone, P. Del Vecchio and A. D. Maro, Binding of a type 1 RIP and of its chimeric variant to phospholipid bilayers: evidence for a link between cytotoxicity and protein/membrane interactions, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 2106–2112 CrossRef CAS PubMed.
- T. Wiedmann, A. Salmon and V. Wong, Phase behavior of mixtures of DPPC and POPG, Biochim. Biophys. Acta, Lipids Lipid Metab., 1993, 1167, 114–120 CrossRef CAS.
- R. Oliva, M. Chino, K. Pane, V. Pistorio, A. De Santis, E. Pizzo, G. D’Errico, V. Pavone, A. Lombardi, P. Del Vecchio, E. Notomista, F. Nastri and L. Petraccone, Exploring the role of unnatural amino acids in antimicrobial peptides, Sci. Rep., 2018, 8, 8888 CrossRef PubMed.
- O. Cañadas and C. Casals, in Lipid-Protein Interactions, ed. J. H. Kleinschmidt, Humana Press, Totowa, NJ, 2013, vol. 974, pp. 55–71 Search PubMed.
- L. Loura, Interaction of peptides with binary phospholipid membranes: application of fluorescence methodologies, Chem. Phys. Lipids, 2003, 122, 77–96 CrossRef CAS PubMed.
- V. Tiriveedhi and P. Butko, A Fluorescence Spectroscopy Study on the Interactions of the TAT-PTD Peptide with Model Lipid Membranes, Biochemistry, 2007, 46, 3888–3895 CrossRef CAS PubMed.
- B. Gironi, R. Oliva, L. Petraccone, M. Paolantoni, A. Morresi, P. Del Vecchio and P. Sassi, Solvation properties of raft-like model membranes, Biochim. Biophys. Acta, Biomembr., 2019, 1861, 183052 CrossRef CAS PubMed.
- T. Parasassi, G. De Stasio, G. Ravagnan, R. M. Rusch and E. Gratton, Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence, Biophys. J., 1991, 60, 179–189 CrossRef CAS PubMed.
- A. D. Lúcio, C. C. Vequi-Suplicy, R. M. Fernandez and M. T. Lamy, Laurdan Spectrum Decomposition as a Tool for the Analysis of Surface Bilayer Structure and Polarity: a Study with DMPG, Peptides and Cholesterol, J. Fluoresc., 2010, 20, 473–482 CrossRef PubMed.
- M. Bastos, R. Adão, K. Nazmi and J. G. M. Bolscher, C- and N-truncated antimicrobial peptides from LFampin 265–284: Biophysical versus microbiology results, J. Pharm. BioAllied Sci., 2011, 3, 60 CrossRef PubMed.
- X. Feng, S. Jin, M. Wang, Q. Pang, C. Liu, R. Liu, Y. Wang, H. Yang, F. Liu and Y. Liu, The Critical Role of Tryptophan in the Antimicrobial Activity and Cell Toxicity of the Duck Antimicrobial Peptide DCATH, Front. Microbiol., 2020, 11, 1146 CrossRef PubMed.
- M.-C. Lin, C.-F. Hui, J.-Y. Chen and J.-L. Wu, Truncated antimicrobial peptides from marine organisms retain anticancer activity and antibacterial activity against multidrug-resistant Staphylococcus aureus, Peptides, 2013, 44, 139–148 CrossRef CAS PubMed.
| This journal is © the Owner Societies 2022 |