Highlights from Faraday discussion 232: Peptide–membrane interactions, 8th–10th September 2021 (online)
Sreetama
Pal†

CSIR-Centre for Cellular and Molecular Biology, Uppal Road, Hyderabad 500 007, India. E-mail: sp965@cornell.edu
First published on 1st September 2022
Abstract
The Faraday discussions meeting on peptide–membrane interactions was designed with the goal of repositioning peptides as powerful model systems that are indispensable in contemporary membrane biophysics and biology research. The meeting, originally scheduled for September 2020, was finally held in a virtual format during September 8th–10th, 2021 due to the COVID-19 pandemic. The meeting saw enthusiastic participation by ∼120 scientists from 23 countries. There were 23 talks delivered during the four sessions and 25 posters presented in two poster sessions (the published volume can be accessed online). The meeting drew attention to the multitude of open questions that persist in our understanding of the behavior of membrane peptides and proteins in spite of nearly half-a-century of intensive interdisciplinary research, and was beyond successful in throwing into sharp relief the enduring relevance of exploring membrane biophysics and biology through the looking glass of peptide–membrane interactions.
Our understanding of the foundational principles of lipid–protein interactions has grown by leaps and bounds since early reports of the preferential localization of specific lipids in the immediate annulus of membrane proteins.1 The past five decades of research on biological membranes has revolutionized the ways by which we tune in to and make sense of conversations between lipids, proteins, and other chemical entities associated with the nanoscale sheet that facilitated cellular-level evolution2 to present forms of life. The behavior of model peptide systems3 in a membrane environment has contributed immensely to the construction of a Rosetta Stone for distilling the enigma of biological membranes into measurable well-defined molecular and supramolecular parameters. However, over the years, there has been a shift in perceptions regarding the physiological relevance of peptide-based studies, due in no small part due to multiple technological breakthroughs4 that have made quantitative in vivo studies of complex membrane assemblies possible. Given this backdrop, the Faraday discussions meeting on peptide–membrane interactions was designed with the goal of repositioning peptides as powerful model systems that are indispensable in contemporary membrane biophysics and biology research.
The meeting was organized by a scientific committee chaired by Paul O’Shea (Lancaster University, UK) and consisting of co-chairs John Seddon (Imperial College London, UK), Patricia Bassereau (Institut Curie, France), Amitabha Chattopadhyay (Centre for Cellular and Molecular Biology, India), Calum Drummond (RMIT University, Melbourne), and Robert Gilbert (University of Oxford, UK). The meeting, originally scheduled for September 2020 and postponed on account of the COVID-19 pandemic, was finally held in a virtual format during September 8th–10th, 2021, due to the tireless efforts of the organizing committee and the Chemistry Biology Interface Division, Royal Society of Chemistry (RSC). The meeting was attended by ∼120 scientists from 23 countries (see Fig. 1 for a map view). There were 23 talks delivered during the four sessions and 25 posters presented in two poster sessions (the published volume can be accessed online, https://pubs.rsc.org/en/journals/journalissues/fd#!issueid=fd021232). O’Shea provided the opening remarks by briefly outlining the motivation for organizing the meeting, with an emphasis on the fundamental differences and challenges associated with defining a reaction space for peptides and that for proteins in the context of their behavior in membranes. This was followed by a short introduction to the meeting format by RSC publishing editors Alice Smallwood and Ellis Crawford.
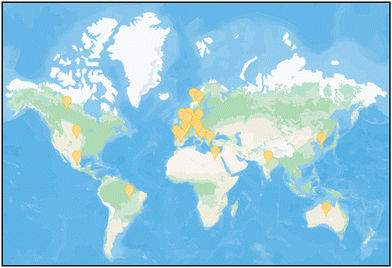 | ||
| Fig. 1 Enthusiastic participation of scientists from across the globe in the Faraday discussions meeting on peptide–membrane interactions. Image was generated by adding location symbols (yellow) to a satellite representation of a world map available https://satellites.pro/plan/world_map based on the published list of participants (DOI: https://doi.org/10.1039/D1FD90070F). | ||
Spiers memorial lecture: setting the stage
William F. DeGrado (University of California - San Francisco, USA) opened the meeting by delivering the Spiers Memorial Lecture on the design and analysis of membrane-interacting peptides (DOI: https://doi.org/10.1039/D1FD00061F). His detailed exposition provided glimpses into the evolution of our understanding of peptide–membrane interactions over the years, illustrated with representative examples from his own research. His talk spanned much of the organisational repertoire exhibited by membrane proteins, from well-structured model peptides (such as melittin, gramicidin, and the pH low insertion peptide) to intrinsically disordered ones (such as α-synuclein), all the way up to the more intricate oligomeric M2 proton channel. Further, the speaker utilized these systems as proof-of-concept models to highlight fundamental rules and requirements that govern lipid–protein interactions, and demonstrated how such knowledge could be harnessed for biotechnological and therapeutic breakthroughs. Along the way, he identified key open questions in the field and highlighted specific talks in upcoming sessions of the meeting that promised to offer novel insights into these behavioral features of membrane peptides and proteins. An important message that came out of this talk was the fact that global physicochemical properties often override local sequence information (intermolecular connectivity) in determining membrane peptide/protein function, echoing one of the fundamental tenets underlying the molecular architecture of biological membranes.5 DeGrado’s lecture was met with a host of enthusiastic questions ranging from technical clarifications to speculative and philosophical queries, which set the tone for lively and engaging discussion sessions among the attendees throughout the remainder of the meeting.Session I: theoretical and experimental comparisons of simple peptide–membrane systems; towards defining the reaction space
The six talks in Session I (on the first day of the meeting) highlighted the state-of-the-art in the choice of model systems and investigative approaches toward defining and understanding the landscape of lipid–protein interactions. Izabella Brand (University of Oldenburg, Germany) shared her team’s efforts in exploring the interaction of the antimicrobial peptide (AMP) melittin with an outer bacterial membrane mimic using in situ spectroelectrochemical characterization (DOI: https://doi.org/10.1039/D0FD00039F). Their approach yielded a detailed map of peptide–bilayer interactions in terms of global membrane potential and localized changes in lipid hydration, lipid order and peptide depth in membranes. The other two experimental talks in this session offered complementary perspectives into the membrane interactions of proteins of the Bcl-2 family (Bcl-xL, Bax, and tBid) that function as apoptosis regulators. While Francisco N. Barrera (University of Tennessee Knoxville, USA) described tunable membrane remodeling by an engineered pH-responsive fragment of the Bax protein (DOI: https://doi.org/10.1039/D0FD00070A), Ana J. Garcia-Saez (University of Cologne, Germany) presented on protein–protein interactions (between Bcl-xL and Bax or tBid) that exert a deterministic influence on the intracellular localization and apoptotic functions of these proteins (DOI: https://doi.org/10.1039/D0FD00045K). These two talks collectively highlighted the power of synthesizing insights from contrasting systems and techniques toward a comprehensive understanding of complex membrane phenomena, as reflected in the questions and comments raised during the post-talk discussions (DOI: https://doi.org/10.1039/D1FD90065J). This session further included three talks on simulation approaches from lipid and protein points-of-view, that could serve as a composite methodological blueprint for gaining dynamic insights into lipid–protein interactions. First, Gregory A. Voth (University of Chicago, USA) shared simulation studies from his group capturing lipid–protein and lipid–lipid interactions of two structural proteins of SARS-CoV-2 in a complex membrane bilayer (DOI: https://doi.org/10.1039/D1FD00031D), providing a molecular-level mechanistic understanding of viral action that could aid in developing antiviral therapeutics. D. Peter Tieleman (University of Calgary, Canada) reported on recent efforts by his laboratory toward developing a quantitative framework for the analysis of lipid dynamics in the vicinity of several representative membrane proteins (WALP23, bacterial OmpF and OmpX, aquaporin-1, and KcsA potassium channel) embedded in an asymmetric membrane bilayer (DOI: https://doi.org/10.1039/D1FD00003A). The last talk of the session was delivered by Stefano Vanni (University of Fribourg, Switzerland), who presented an important proof-of-concept study establishing the overall suitability of the MARTINI coarse-grained force field for exploring the membrane interaction of a range of peripheral membrane proteins (DOI: https://doi.org/10.1039/D0FD00058B).Session II: theoretical and experimental studies of complex peptide–membrane systems
Talks in Session II (on the second day of the meeting) focused on lipid–protein interactions in complex membranes reminiscent of the cellular environment. Ronald J. Clarke (University of Sydney, Australia) opened the session with a talk on polybasic cytoplasmic peptide domains of the integral membrane ion pump P-type ATPase (DOI: https://doi.org/10.1039/D0FD00040J). Membrane interactions of these domains are hypothesized to influence pump function by acting as “death sensors” that recognize changes in plasma membrane lipid asymmetry known to accompany apoptosis. The talk by Bart W. Hoogenboom (University College London, UK) continued on this theme of lipidic sensors and actuators by expanding on the interplay of membrane electrostatics, lipid asymmetry, and membrane domainization that underlie the highly selective action of the immune effector protein perforin on target (pathogenic) cells (DOI: https://doi.org/10.1039/D0FD00043D). Meanwhile, Paul A. Beales (University of Leeds, UK) and Durba Sengupta (National Chemical Laboratory, India) focused on the influence of membrane physical properties and lipid composition on cellular membrane remodeling events triggered by macromolecular sculptors such as the ESCRT complex and caveolin-1, respectively. Beales highlighted work from his group on the differential membrane remodeling action of the ESCRT-II/ESCRT-III complex in response to differences in membrane mechanics, curvature, and lipid localization (DOI: https://doi.org/10.1039/D0FD00042F). Sengupta presented insights on membrane remodeling and preferential lipid clustering induced by the membrane interaction of a truncated caveolin-1 construct (DOI: https://doi.org/10.1039/D0FD00062K). In an elegant counterpoint to the tour-de-force studies mentioned above, Reidar Lund (University of Oslo, Norway) demonstrated that a complex lipid and/or protein system is not an absolute requirement for accessing the elaborate reaction space underlying the various nuances in lipid–protein interactions (DOI: https://doi.org/10.1039/D0FD00046A). Indeed, even the behavior of “simple” AMPs (such as aurein 1.2, indolicidin, LL-37, and lacticin Q) in a minimalistic bilayer environment can (and often does!) evade a rigorous categorization of all possible factors at play. The post-talk discussions in this session (DOI: https://doi.org/10.1039/D1FD90066H) were particularly engaging, with several enthusiastic conversations running in parallel among the participants, much like an in-person meeting.Session III: behavior and interactions of proteins and peptides with and within membranes; from simple models to cellular membranes
The next session (Session III) on the same day was opened by John M. Sanderson (Durham University, UK), whose talk on melittin lipidation in the presence of lysophospholipids provided a fresh perspective into the membrane interaction and activity of this well-studied AMP (DOI: https://doi.org/10.1039/D1FD00030F). Boyan B. Bonev (University of Nottingham, UK) presented his team’s work on the interaction of the antibacterial peptide polymyxin-B with lipopolysaccharide-containing membranes via a pyrophosphate-mediated mechanism of molecular recognition (DOI: https://doi.org/10.1039/D1FD00036E). The last peptide-centric talk of the session was given by Sreetama Pal (Centre for Cellular and Molecular Biology, India), who presented on the sensitivity of WALP peptides toward changes in membrane interfacial electrostatics in the broader context of hydrophobic matching responses of these peptides (DOI: https://doi.org/10.1039/D0FD00065E). The remaining talks in this session represented a shift in focus toward examining complex phenomena in cellular membranes. The talk by Justin E. Molloy (The Francis Crick Institute, UK) demonstrated, by the judicious use of high-resolution single particle tracking assays, how our view of the inherent heterogeneity in plasma membrane structures could be limited by the choice of technical and analytical approaches employed on a case-by-case basis (DOI: https://doi.org/10.1039/D1FD00035G). Aurélien Roux (University of Geneva, Switzerland) made a convincing case for a unified framework for analyzing the mechanisms and consequences of surface deformation in biology, by invoking topological similarities in single lipids6 and single cells (DOI: https://doi.org/10.1039/D1FD00040C). Lastly, Shobhna Kapoor (Indian Institute of Technology Bombay, India) talked about the influence of virulent (mycobacterial) lipids on host membrane lipid abundance and localization, thereby expanding our understanding of the ways in which pathogens may stage a lipid takeover in host cells (DOI: https://doi.org/10.1039/D0FD00051E). A recurring theme in the post-talk discussions in this session (DOI: https://doi.org/10.1039/D1FD90067F) was the emphasis on advances in experimental and theoretical approaches suitable for investigations of lipid–protein interactions, which in and of itself encompass several overlapping areas of research beyond the focus of this meeting.Session IV: peptide–membrane interactions and biotechnology; enabling next-generation synthetic biology
The final session (on the third day of the meeting) consisted of talks that focused on leveraging present understanding of lipid–protein interactions toward bioengineering and biotechnological breakthroughs. The session was opened by Marie-Isabel Aguilar (Monash University, Australia), who presented on the influence of bacterial growth stages on the action of the AMP maculatin 1.1 on bacterial membranes and the implications of these observations in the context of antimicrobial drug development (DOI: https://doi.org/10.1039/D0FD00052C). Next, Burkhard Bechinger (Université de Strasbourg, France) shared work from his laboratory on the synergistic effects exerted by the lipid environment on composite AMP–membrane interactions using a mixture of two AMPs (magainin 2 and PGLa) that are secreted as part of a natural cocktail (DOI: https://doi.org/10.1039/D0FD00041H). Given that most studies on peptide–membrane interactions involve only one specific peptide at a time, such an approach could be instrumental in illustrating the extents to which insights from single peptide studies would hold true in natural systems. The talk by Georg Pabst (University of Graz, Austria) on the membrane activity of three representative AMPs (magainin 2, PGLa derivative L18W-PGLa, and lactoferricin derivative LF11-215) in asymmetric bacterial membrane mimics (DOI: https://doi.org/10.1039/D1FD00039J) shed light on yet another aspect (i.e., lipid asymmetry) of AMP–membrane interactions that remain ill-represented in the majority of studies on AMP biophysics and behavior. The last talk of this session was delivered by Franca Fraternali (King’s College London, UK) on simulation-aided design of antimicrobial nanocapsules by functionalization of a structural unit of a virus with AMP-like sequences (DOI: https://doi.org/10.1039/D1FD00041A). Interestingly, all four papers discussed in this session involved AMPs, which brings the discourse back full-circle to DeGrado’s comments on the immense application potential of AMPs during the Spiers Memorial lecture (DOI: https://doi.org/10.1039/D1FD00061F). The post-talk discussion for this session (DOI: https://doi.org/10.1039/D1FD90068D) saw a fair share of speculative and philosophical questions, reflecting the exciting vistas in the peptide–membrane interaction landscape that these talks collectively opened up. The scientific talks in this session were preceded by a talk by Laura Daly (RSC) on publishing with impact.Poster sessions
Twenty-five posters were presented on various facets of peptide–membrane interactions (major themes represented in Fig. 2) during the two poster sessions. Both sessions were well-attended, with lively discussions often extending much beyond the formal sessions, in spite of the technical challenges of a virtual presentation platform and differences in time zones for the attendees. The first poster session (on the first day of the meeting) was preceded by lightning poster presentations selected by the poster evaluation committee (headed by O’Shea and Seddon). This featured presentations by Maria Hoernke, Albert-Ludwigs-Universität Freiburg, Germany (Understanding enhanced endosomal escape: membrane permeabilization induced by pH-sensitive polycations), Elizabeth Kelley, National Institute of Standards and Technology, USA (Influence of transmembrane peptides on the mesoscale membrane dynamics), and Sujin Park, Korea Institute of Science and Technology, South Korea (Real-time visualization of phase and structural change of model lipid membranes by arginine-rich cell-penetrating peptide). Based on the recommendations of the poster evaluation committee, the Faraday Division Poster Prize was awarded to Fabian Keller, Universität Münster, Germany (Investigation of lipid bilayer asymmetry effects on transmembrane domains) and the RSC Chemical Biology Poster Prize was awarded to Olivia Hepworth, King’s College London, UK (Structural analysis of the novel fungal peptide toxin, candidalysin). The evaluation committee applauded the stellar quality and thought-provoking content of all the posters during the announcement of the poster prizes.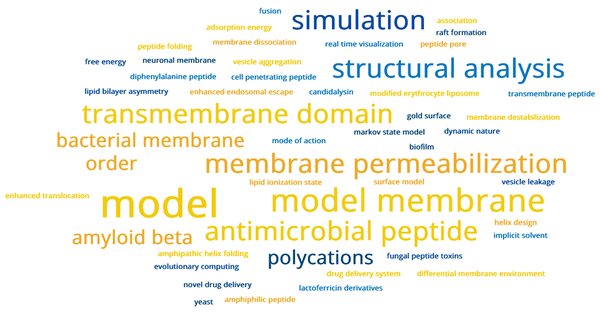 | ||
| Fig. 2 A visual representation of major research directions covered in the posters presented during the two poster sessions of the Faraday discussions meeting on peptide–membrane interactions. The word cloud representation was constructed from keywords parsed from poster titles using an https://monkeylearn.com/word-cloud, with the font size of each phrase/word scaled to the corresponding frequency of occurrence. The complete list of posters presented during the meeting is available online (DOI: https://doi.org/10.1039/D1FD90069B). | ||
Concluding remarks lecture: the road ahead
In the concluding remarks lecture (DOI: https://doi.org/10.1039/D1FD00077B), Patricia Bassereau provided a succinct summary of the very many facets of lipid–protein interactions that emerged during the talks and post-talk discussions. She started by echoing O’Shea’s introductory comments on the fundamental contrast in conceptualizing a framework for understanding the behavior of proteins vs. peptides (see above). In a stance that, for me, highlighted the near-impossibility in confining peptide–membrane interactions to narrow definitions or disciplines, the speaker moved away from a session-specific debrief to instead focus on leitmotifs recurring throughout the meeting. One such theme was the membrane interaction of AMPs or that of constituent (peptide) domains of complex multi-domain membrane proteins associated with cellular and intracellular membranes, with an emphasis on the consequences of such interactions in health and disease. Next, she commented on recent technical advancements (particularly with respect to simulation capabilities) covered during the meeting, that have been successful in providing a high-resolution window into the rich landscape of peptide–membrane interactions across different spatio-temporal scales. The talk drew attention to the multitude of open questions that persist in our understanding of the behavior of membrane peptides and proteins in spite of significant interdisciplinary research. Bassereau concluded her remarks by expressing hopes of a reunion of the participants in the next edition of a similar Faraday discussions meeting in the future.Not-the-Loving-Cup ceremony
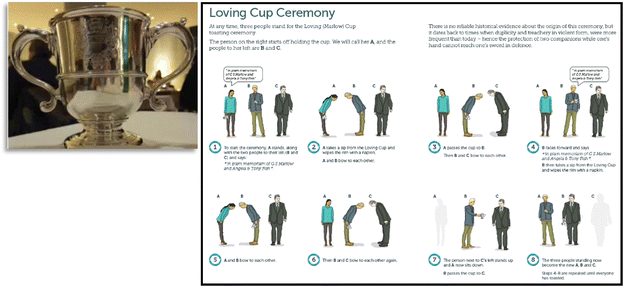
The meeting was formally concluded by Susan Perkin (University of Oxford, UK), Chair of the Faraday Standing Committee on Conferences, who introduced the participants to a brief history of the Loving Cup ceremony traditionally observed during the Conference Dinner. The Loving Cup (shown in the left panel of Fig. 3) was commissioned in the memory of Mr G. W. S. Marlow, who held the post of Secretary and Editor of the Faraday Society from 1928 to 1947. The Cup is used to toast the memory of Mr Marlow, Angela and Tony Fish, and the Faraday Division, and then passed around among the assembled (brief instructions shown in right panel of Fig. 3). Although the virtual nature of this meeting did not lend itself to a full-scale reenactment of the Loving Cup Ceremony, all the attendees enthusiastically participated in the Not-the-Loving-Cup ceremony with their choice of beverage (dictated by local time zones!).Final thoughts
Even with the flipped format of Faraday discussions meetings that emphasize discussions over monologues, this meeting was over too soon! This is a testament to the organizing committee and RSC staff who worked tirelessly to provide a welcoming virtual environment that facilitated conversations and kept participants engaged throughout the sessions. Special thanks are due to the RSC publishing editors Ellis Crawford and Alice Smallwood, who were instrumental in maintaining meticulous records of the discussion points, and to Katie Ackermann and the RSC support staff, whose superlative behind-the-scene efforts ensured an enjoyable virtual experience for everyone. The virtual platform itself deserves a special mention, since every effort was made to simulate an in-person meeting, including virtual meeting rooms mimicking the impromptu nucleation of stimulating discussions encountered in “real” meetings. The numerous debates made for an unmatched learning experience in observing seasoned scientists formulate questions, arguments, and counter-arguments in real time, and it was a privilege to participate in discussions that probed the very edge of our knowledge on peptide–membrane interactions. The Faraday discussions meeting on peptide–membrane interactions, conceptualized to reiterate the importance of peptides in understanding biological membranes (as O’Shea mentioned in his opening comments), was beyond successful in throwing into sharp relief the enduring relevance of exploring membrane biophysics and biology through the looking glass of peptide–membrane interactions.Acknowledgements
S. P. was supported as a Senior Project Associate from the SERB Distinguished Fellowship grant (Department of Science and Technology, Government of India) awarded to Amitabha Chattopadhyay (Centre for Cellular and Molecular Biology, India). S. P. thanks the organizing committee of the Faraday Discussions meeting on peptide–membrane interactions for the opportunity of putting together the conference report, and acknowledges the valuable and constructive inputs from Amitabha Chattopadhyay, Paul O’Shea (Lancaster University, UK), and John Seddon (Imperial College London, UK) during the preparation of this report.References
- P. C. Jost, O. H. Griffith, R. A. Capaldi and G. Vanderkooi, Proc. Natl. Acad. Sci. U. S. A., 1973, 70, 480–484, DOI:10.1073/pnas.70.2.480.
- I. Budin and J. W. Szostak, Annu. Rev. Biophys., 2010, 39, 245–263, DOI:10.1146/annurev.biophys.050708.133753.
- J. A. Killian and T. K. M. Nyholm, Curr. Opin. Struct. Biol., 2006, 16, 473–479, DOI:10.1016/j.sbi.2006.06.007.
- C. T. Saeui, M. P. Mathew, L. Liu, E. Urias and K. J. Yarema, J. Funct. Biomater., 2015, 6, 454–485, DOI:10.3390/jfb6020454.
- S. Pal and A. Chattopadhyay, in Membrane organization and dynamics, ed. A. Chattopadhyay, Springer, Heidelberg, 2017, pp. 1–9 DOI:10.1007/978-3-319-66601-3_1.
- J. N. Israelachvili, S. Marčelja and R. G. Horn, Q. Rev. Biophys., 1980, 13, 121–200, DOI:10.1017/s0033583500001645.
Footnote |
| † Present address: School of Chemical and Biomolecular Engineering, Cornell University, New York 14853, USA. |
| This journal is © The Royal Society of Chemistry 2022 |