Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A switchable DNA origami/plasmonic hybrid device with a precisely tuneable DNA-free interparticle gap†
Michael
Erkelenz‡
a,
Richard
Kosinski‡
b,
Helene
Giesler
a,
Oliver
Sritharan
a,
Jesil
Jose
a,
Barbara
Saccà
 *b and
Sebastian
Schlücker
*b and
Sebastian
Schlücker
 *a
*a
aDepartment of Chemistry, Center for Nanointegration Duisburg-Essen (CENIDE) and Center for Medical Biotechnology (ZMB), University of Duisburg-Essen, Universitaetsstraße 5, 45141 Essen, Germany. E-mail: Sebastian.Schluecker@uni-due.de
bDepartment of Biology, Center for Nanointegration Duisburg-Essen (CENIDE) and Center for Medical Biotechnology (ZMB), University of Duisburg-Essen, Universitaetsstraße 2, 45117 Essen, Germany. E-mail: Barbara.Sacca@uni-due.de
First published on 10th November 2022
Abstract
We here show a reconfigurable DNA/plasmonic nanodevice with a precisely tunable and DNA-free interparticle gap. The nanodevice comprises two DNA boxes for the size-selective incorporation of nanoparticles in a face-to-face orientation and an underlying switchable DNA platform for the controlled and reversible adjustment of the interparticle distance.
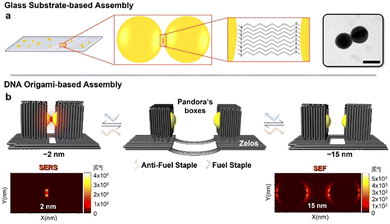
Dimers of gold nanoparticles (AuNP) are well-known for being the simplest structure exhibiting the phenomenon of plasmon coupling, which results in a very strong enhancement of the electric field in the narrow gap (hot spot) between them.1–3 The irradiation-induced field enhancement strongly depends on the gap size and enables the application of various AuNP dimers in surface-enhanced Raman scattering (SERS)4,5 and surface-enhanced fluorescence (SEF).6,7 By influencing the local enhanced electric field,8,9 the size of the gap essentially offers a means to manipulate hot spot and quenching effects. Hence, the programmable control of such a distance in a modular and reversible fashion – and especially with sub-nanometre resolution – may greatly enhance the spectrum of AuNP dimers applications in optical spectroscopy.
We have previously reported on a dimerization approach that relies on the use of alkane dithiols (Fig. 1a) as linker molecules to form ideal dimers of gold nanospheres.10 However, this strategy lacks the possibility to control gap distances larger than 3 nm and does not allow to switch from one distance to another once the dimer is assembled. Alternative designs have been also reported in the literature7,11–14 and all of them rely on the functionalization of the particle surface with DNA sequences that are complementary to the handles extruding from the DNA origami surface. All these methods share the drawback that the hot-spot is filled with linker molecules, hampering further functionalisation of the nanoparticles for single-molecule studies. A possible way to bypass this issue relies on the programmability of the DNA.15 The predictable Watson–Crick pairing of nucleobases enables nowadays to realize three-dimensional DNA objects of almost any shape and about 100 nm in size.16 Those objects, called DNA origami,11 can be further assembled into larger complexes using well-established hierarchical procedures17 and toehold-mediated single-strand displacement (TMSD) mechanisms can be finally applied to reconfigure these objects in a programmable and reversible fashion.18 Remarkably, since the structure of the double helix is known, DNA origami constructs can be addressed with nm-sized accuracy, thus enabling the placement of desired molecules in a predefined spatial arrangement. DNA origami architectures have been combined with plasmonically active nanostructures leading to hybrid nanomaterials for applications in sensing,19,20,21 spectroscopy,22–25 and biomedical detection methods,26,27 including single-molecule approaches.13,28,29
In particular, dimerisation of AuNP has been successfully achieved using origami slit4 or sheet-like designs.23 Although being suitable for spectroscopic applications, these constructs display hot spot regions that are set and filled with DNA. Conversely, in this study, we present a strategy for the formation of hot spot regions devoid of DNA or any other linker molecule.
The difference between these two approaches is schematically illustrated in Fig. 1. In a conventional dimerization approach, a glass substrate is used to bind gold nanosphere monomers electrostatically (Fig. 1a, left). Thiol-functionalized linkers are then added and incubated with additional monomeric nanospheres, leading to formation of dimers (Fig. 1a, right), the hot spot of which is filled with alkane dithiol molecules. In contrast, the DNA origami-based procedure presented here starts with the encapsulation of one gold nanosphere within the cavity of a lidless origami Pandora's Box, as previously described,30 and proceeds with the placement of two such boxes on top of a switchable platform (Zelos). The boxes are positioned at a defined intermolecular distance with the accessible (bare) regions of the nanoparticles facing each other (Fig. 1b). The platform is composed of two halves connected by a set of duplexes, the length of which can be varied in a programmable way, thereby affecting the distance between the two boxes and, in turn, the size of the interparticle gap. The main feature of our system is the absence of DNA or other linker molecules at the interface between the nanospheres, thus leading to an interparticle gap devoid of any “foreign” molecule and thus suitable for single-molecule applications. Nanometre precise distance-control of the boxes is achieved by a mechanism of single-strand displacement that switches the platform from a “flexible” state, characterized by an undefined distance between the two halves of the platform (Fig. 1b, middle panel), to one of several “fixed” states, where the distance between the two halves is instead programmable (Fig. 1b, left and right panels). Upon reaching a fixed state, the device can be switched back to its flexible form, ready for the next reconfiguration into an inter-box distance of choice. In other words, the transition from one well-defined interparticle distance to another must pass through an unstructured flexible configuration of the device.
Our design strategy is therefore modular, meaning that multiple and distinct equilibrium states can be achieved from a single structure. This feature may be advantageous to cover various types of applications that require distinct gap distances in between the nanoparticles, all achievable in a post-assembly process and from the same initial construct. We calculated the electric field enhancement originated by the plasmon coupling of gold nanoparticles placed at 2 or 15 nm distance (Fig. 1b, bottom). The calculated enhancement factors (|E4|) were determined using finite-difference time-domain (FDTD) simulations (Fig. S5, ESI†), thereby demonstrating their potential application for SERS and SEF, respectively. The dimer with a 2 nm gap size shows a drastic increase in local electric field intensity inside the gap (Fig. 1b, bottom left), enabling the use of the device for SERS spectroscopy. In contrast, the local electric field intensities for a 15 nm gap size show their maxima on the edges of the nanospheres (Fig. 1b, bottom right), with great potential for SEF applications.
Fig. 2 illustrates the strategy used to control the gap distance between the two halves of the Zelos platform. For a matter of clarity, the scheme reports the design approach used for one staple, although the same procedure is adopted identically for a total of eight staples. Essentially, the scaffold is routed back and forth along the bilayered platform leaving eight single stranded portions of the scaffold, each 63 bases-long, at the central seam (four above and four below the inner cavity, as visualised in Fig. 2a, top and Fig. S3, ESI†). The scaffold loops at the seam are those regions that will be targeted by sequential addition of fuel/antifuel strands, thus dictating the distance between the two halves of the platform. To adjust the gap distance, so-called “fuel staples” are designed to bind at both halves of the structure, thus forming eight duplexes of defined length. Each of these fuel staples can be partitioned into several domains. A first, 15 bases-long anchoring domain, hybridizes to the scaffold within the structure and tightly secures the staple at each of the two halves of Zelos (blue segments). A second domain binds to a portion of the scaffold that spans the gap region in between the two halves (red segment). Since fuel staples do not fully hybridize to the entire unpaired scaffold segment, a scaffold loop is formed. The larger the size of this loop (that is, the portion of the scaffold which is excluded from the duplex bridges), the shorter will be the gap distance between the two halves of Zelos. Finally, a third domain is represented by a toehold region at the termini of each fuel staple (green segment). This single-stranded DNA domain consists of 8 nucleobases and serves as the starting point of a toehold-mediated strand displacement reaction. Upon addition of so-called anti-fuel strands that are fully complementary to the fuel staples, these latter are displaced from the origami, thereby resulting in the two halves being connected only by eight unstructured single-stranded scaffold segments. In this flexible configuration, the distance between the two halves is undefined; nevertheless, this intermediate enables the binding of another set of fuel staples that can adjust the length of the duplexes, and thus the gap distance, to another desired value. Finally, one terminal nucleobase of the scaffold was left unbound (yellow region) in order to relieve the mechanical strain at the interface between the duplex bridges and the adjacent origami platform and favour the alignment of the Zelos’ halves on the same plane.
To test the modularity of our design, that is, the ability of the device to assume distinct and pre-defined gap distances, Zelos was assembled into five different fixed states, as shown in Fig. 2a. The five fixed states display, respectively, 0, 6, 12, 24 and 43 bp-long duplex bridges and are supposed to lead to a theoretical distance of 0, 2, 4, 8 and 15 nm between the halves of the platform (assuming 0.34 nm bp−1).31Fig. 2b shows representative TEM images of the generated structures. In the 0 bp-state, no gap was visible, and TEM analysis revealed a length of 60.7 ± 3.5 nm and a width of 45.9 ± 5.7 nm. Increased length of the fuel strands, namely to 6, 12, 24 and 43 bp, resulted in mean gap distances of, respectively, 2.8 ± 1 nm (n = 21), 4.2 ± 0.9 nm (n = 56), 8.1 ± 1.3 nm (n = 78) and 14.5 ± 1.3 nm (n = 100). In the 43 bp-state, the platform showed a mean length of 74.8 ± 3.4 nm and width of 50.4 ± 2.0 nm. Hence, in general, the platform appears well-folded in all configurational states, and the gap distances estimated by statistical analysis of the TEM images correspond exceptionally well with the expected theoretical values, indicating that a highly precise arrangement of objects (specifically, AuNPs) on top of the platform is indeed possible. Besides single-particle TEM studies, AGE and FRET studies were performed to gain deeper insights in the transition of the device between distinct states, in aqueous solution and at the ensemble level (Fig. S12, ESI†).
In the next step, two AuNPs were fixed in a face-to-face orientation on top of the Zelos platform and at well-defined positions. In this way, the precision achieved in the structural reconfiguration of the platform can be reliably translated into a predictable hot spot gap between the DNA-free nanoparticles. For this purpose, four staple strands were designed to protrude from each half of Zelos. These staples, while being anchored within the DNA platform, have a second domain (red staple sin Fig. 3a) that can incorporate into four distinct positions within the bottom face of the DNA box. Additional T3 spacers (green staples in Fig. 3a) were introduced to reduce electrostatic repulsion between the two origami structures. This ensures that the relative orientation of each DNA box on top of the Zelos platform can be fully predicted. We designed the anchoring staples such to orient two distinct DNA boxes with their lidless sides facing each other. Encapsulation of DNA boxes with single AuNPs and further positioning of these latter on top of the Zelos platform should then result in the face-to-face orientation of DNA-free metal surfaces at a predictable interparticle distance. For the nanoparticle synthesis, we used a modified seed-mediated protocol published by Park et al.32 A detailed insight into the design and formation of Pandora's Box as well as the incorporation of gold nanoparticles of several sizes and morphologies is reported by Erkelenz et al.30
The hierarchical assembly of such a large structure (about 15 MDa in MW) is clearly hampered by steric hindrance and strong electrostatic repulsions among the negatively charged DNA origami structures. For this reason, hybridization of Zelos and DNA boxes was performed over seven days at 30 °C in 1× TEMg11 buffer, using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Zelos
4 (Zelos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Box) ratio. AGE analysis of DNA–origami only structures and TEM images of products previously purified by gel-extraction indicate an almost complete binding of Zelos to at least one DNA box with about 16% of all structures showing successful attainment of the desired architecture, with two DNA boxes linked on the same side of the Zelos platform (Fig. S13, ESI†). Finally, the success of the hierarchical assembly process upon incorporation of AuNP inside Pandora's Boxes was demonstrated by TEM (n = 98) and revealed 51% of Zelos with one AuNP-containing box (Fig. 3c, bottom left) and 4% of the target superstructure consisting of a platform with two DNA boxes, each one incorporating a single gold nanosphere (Fig. 3c, bottom right).
Box) ratio. AGE analysis of DNA–origami only structures and TEM images of products previously purified by gel-extraction indicate an almost complete binding of Zelos to at least one DNA box with about 16% of all structures showing successful attainment of the desired architecture, with two DNA boxes linked on the same side of the Zelos platform (Fig. S13, ESI†). Finally, the success of the hierarchical assembly process upon incorporation of AuNP inside Pandora's Boxes was demonstrated by TEM (n = 98) and revealed 51% of Zelos with one AuNP-containing box (Fig. 3c, bottom left) and 4% of the target superstructure consisting of a platform with two DNA boxes, each one incorporating a single gold nanosphere (Fig. 3c, bottom right).
The achievement of the target device has been therefore possible, although some challenges still remain to be addressed, especially the low yield of well-formed structures and the lack of AuNP positional control within the DNA boxes. This issue indeed is fundamental to attain a predictable interparticle distance. A possible way to overcome this issue is to use larger nanoparticles than those used in this study (10 nm), to fill out the cavity and ensure interparticle distances that match the gap defined by the underlying platform. Alternatively, if nanoparticles of smaller sizes are to be used, smaller DNA cavities can be realized using a shorter scaffold. Nevertheless, the present study shows that, once experimental and design challenges are solved, the realization of a high precision hybrid device with a tuneable and DNA-free gap distance is possible.
M. E. performed assemblies of the DNA Origami structures as well as incorporation and hybridisation experiments, analysed data and prepared figures. R. K. contributed the designs of Pandora's Box and Zelos. H. G. performed assembly experiments of Pandora's Box and wrote the manuscript. O. S. carried out AuNP syntheses as well as stabilisation and ligand exchange studies. J. J. performed FDTD simulations. B. S and S. S. conceptualised the project idea, supervised the project and wrote the manuscript.
We acknowledge financial support by the German Research Foundation (DFG) within the framework of the CRC 1093 ‘‘Supramolecular Chemistry on Proteins’’, projects A06 and A09.
Conflicts of interest
There are no conflicts of interest to declare.References
- H. Wang, D. W. Brandl, P. Nordlander and N. J. Halas, Acc. Chem. Res., 2007, 40, 53–62 CrossRef CAS PubMed.
- J. H. Yoon, F. Selbach, L. Schumacher, J. Jose and S. Schlücker, ACS Photonics, 2019, 6, 642–648 CrossRef CAS.
- S. E. J. Bell, G. Charron, E. Cortes, J. Kneipp, M. L. de la Chapelle, J. Langer, M. Prochazka, V. Tran and S. Schlücker, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- V. V. Thacker, L. O. Herrmann, D. O. Sigle, T. Zhang, T. Liedl, J. J. Baumberg and U. F. Keyser, Nat. Commun., 2014, 5, 3448 CrossRef PubMed.
- E. C. Le Ru and P. G. Etchegoin, MRS Bull., 2013, 38, 631–640 CrossRef.
- A. Bek, R. Jansen, M. Ringler, S. Mayilo, T. A. Klar and J. Feldmann, Nano Lett., 2008, 8, 485–490 CrossRef CAS PubMed.
- T. Zhang, N. Gao, S. Li, M. J. Lang and Q. H. Xu, J. Phys. Chem. Lett., 2015, 6, 2043–2049 CrossRef CAS PubMed.
- P. Anger, P. Bharadwaj and L. Novotny, Phys. Rev. Lett., 2006, 96, 113002 CrossRef PubMed.
- J. F. Li, C. Y. Li and R. F. Aroca, Chem. Soc. Rev., 2017, 46, 3962–3979 RSC.
- J. H. Yoon, F. Selbach, L. Langolf and S. Schlücker, Small, 2018, 14, 1702754 CrossRef PubMed.
- G. P. Acuna, F. M. Möller, P. Holzmeister, S. Beater, B. Lalkens and P. Tinnefeld, Science, 2012, 338, 506–510 CrossRef CAS PubMed.
- V. Glembockyte, L. Grabenhorst, K. Trofymchuk and P. Tinnefeld, Acc. Chem. Res., 2021, 54, 3338–3348 CrossRef CAS PubMed.
- K. Tapio, A. Mostafa, Y. Kanehira, A. Suma, A. Dutta and I. Bald, ACS Nano, 2021, 15, 7065–7077 CrossRef CAS PubMed.
- C. Heck, K. Yuya, J. Kneipp and I. Bald, Angew. Chem., Int. Ed., 2018, 57, 7444–7447 CrossRef CAS PubMed.
- N. C. Seeman, Nature, 2003, 421, 427–431 CrossRef PubMed.
- S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. D. Nijenhuis, B. Saccà, F. C. Simmel, H. Yan and P. Zhan, Nat. Rev. Methods Primers, 2021, 1, 13 CrossRef CAS.
- W. Pfeifer and B. Saccà, ChemBioChem, 2016, 17, 1063–1080 CrossRef CAS PubMed.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- T. Funck, F. Nicoli, A. Kuzyk and T. Liedl, Angew. Chem., Int. Ed., 2018, 57, 13495–13498 CrossRef CAS PubMed.
- M. Dass, F. N. Gur, K. Kolataj, M. J. Urban and T. Liedl, J. Phys. Chem. C, 2021, 125, 5969–5981 CrossRef CAS PubMed.
- A. Kuzyk, R. Schreiber, Z. Fan, G. Pardatscher, E. M. Roller, A. Hogele, F. C. Simmel, A. O. Govorov and T. Liedl, Nature, 2012, 483, 311–314 CrossRef CAS PubMed.
- S. Kogikoski, K. Tapio, R. E. von Zander, P. Saalfrank and I. Bald, Molecules, 2021, 26, 1684 CrossRef CAS PubMed.
- P. Kuhler, E. M. Roller, R. Schreiber, T. Liedl, T. Lohmüller and J. Feldmann, Nano Lett., 2014, 14, 2914–2919 CrossRef PubMed.
- G. P. Acuna, F. M. Möller, P. Holzmeister, S. Beater, B. Lalkens and P. Tinnefeld, Science, 2012, 338, 506–510 CrossRef CAS PubMed.
- L. Xin, X. Duan and N. Liu, Nat. Commun., 2021, 12, 3207 CrossRef CAS PubMed.
- L. Dai, P. Liu, X. Hu, X. Zhao, G. Shao and Y. Tian, Analyst, 2021, 146, 1807–1819 RSC.
- Z. Ge, L. Guo, G. Wu, J. Li, Y. Sun, Y. Hou, J. Shi, S. Song, L. Wang, C. Fan, H. Lu and Q. Li, Small, 2020, 16, e1904857 CrossRef PubMed.
- S. E. Ochmann, C. Vietz, K. Trofymchuk, G. P. Acuna, B. Lalkens and P. Tinnefeld, Anal. Chem., 2017, 89, 13000–13007 CrossRef CAS PubMed.
- S. E. Ochmann, H. Joshi, E. Büber, H. G. Franquelim, P. Stegemann, B. Saccà, U. F. Keyser, A. Aksimentiev and P. Tinnefeld, Nano Lett., 2021, 21, 8634–8641 CrossRef CAS PubMed.
- M. Erkelenz, R. Kosinski, O. Sritharan, H. Giesler, B. Saccà and S. Schlücker, Chem. Commun., 2021, 57, 3151–3153 RSC.
- Y. Ke, S. M. Douglas, M. Liu, J. Sharma, A. Cheng, A. Leung, Y. Liu, W. M. Shih and H. Yan, J. Am. Chem. Soc., 2009, 131, 15903–15908 CrossRef CAS PubMed.
- J. E. Park, Y. Lee and J. M. Nam, Nano Lett., 2018, 18, 6475–6482 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc05324a |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |