 Open Access Article
Open Access ArticleElucidation of the working principle of a gene-directed caged HDAC inhibitor with cell-type selectivity†
Kotoko
Sakamoto
,
Ayumi
Hirano
,
Rika
Hidaka
,
Akinobu Z.
Suzuki
,
Taro
Ueno
 and
Toshiaki
Furuta
and
Toshiaki
Furuta
 *
*
Department of Biomolecular Science, Faculty of Science, Toho University, 2-2-1 Miyama, Funabashi, 274-8510, Japan. E-mail: furuta@biomol.sci.toho-u.ac.jp
First published on 30th August 2022
Abstract
Histone deacetylases (HDACs) play crucial roles in the epigenetic regulation of gene expression. Here, we report CM-Bhc-SAHA, a novel caged HDAC inhibitor, genetically targeting cells of interest. Mammalian cells expressing porcine liver esterase led to the optochemical inhibition of endogenous HDAC activity when treated with CM-Bhc-SAHA and irradiated with 405 nm light.
Epigenetic regulation of gene expression critically regulates physiological processes such as proliferation,1 development,2 and cellular memory3 and potentially causes common diseases such as cancer.4 Histone acetylation is a primary epigenetic process that regulates transcription by loosening or tightening chromatin structure. Histone deacetylases (HDACs) are enzymes that catalyze the deacetylation of ε-acetylated lysine residues and mainly promote repression of gene expression.5,6 There are two types of HDAC families:7 Zinc-dependent Class I (HDAC 1–3, 8), IIa (HDAC 4–7, 9), IIb (HDAC 6, 10), IV (HDAC 11), and NAD-dependent Class III (SIRT1–7). Recent studies have shown that many non-histone target proteins are dynamically deacetylated by HDACs outside the nucleus, thereby demonstrating HDAC functions beyond the chromatin.8 HDAC dysregulation causes various diseases: abnormal expression of Class I HDACs in broad ranges of cancer cells,9 an association of deregulation of Class I (HDAC 1 and 2) and Class IIb (HDAC6) with cognitive impairment,10 and neurodegenerative diseases.11 Thus, HDAC inhibitors (HDACi) have emerged as chemical probes to elucidate post-transcriptional protein modification mechanisms and as drug candidates for treating diseases such as cancer12 and neurodegenerative disorders.13 The pan-selective nature of the HDACis for the HDAC family would render off-target effects upon systemic administration.
Tissue- and cell-type-selective modulation of HDAC activity is desirable for basic research and clinical applications. To achieve this goal, light-controlled modulation of HDAC activity by photoresponsive chemical probes, such as azobenzene-derived reversible HDAC inhibitors14,15 and photocaged HDACi,16–20 has been reported. Light triggering of photocaged compounds is a powerful technique that can control biological systems with high spatiotemporal resolution in experimental designs with adherent cells or tissue slices.21–23 Caged compounds are small molecular weight organic compounds that are not genetically encoded, and hence cannot target specific types of cells or tissues of interest. To overcome this problem, we recently reported the concept of gene-directed caged compounds that are photoactivated only in cells tagged with preselected genes.24 Our gene-directed caging strategy allows the use of a variety of substrate/enzyme pairs as photochemical “locks” and their “key” enzymes. In our previous paper, we used a pair of β-galactosyl moiety and E. coli β-galactosidase to prepare gene-directed caged cyclic nucleotides that function as optochemical tools with cell-type selectivity.24 This study reports a new gene-directed caged HDAC inhibitor with the 1-methyl-1-cyclopropancarboxylmethyl (CM) moiety as a chemical lock to expand the repertoire of gene-directed caged compounds and achieve cell-type selective inhibition of HDAC activity. The CM group was designed to be orthogonally hydrolyzed in mammalian cells only in the presence of externally expressed porcine liver esterase (PLE).25
For our proof-of-concept experiments, as an HDACi, we chose suberoylanilide hydroxamic acid (SAHA), also known as vorinostat, a pan-selective HDAC inhibitor effective in both Classes I and IIb HDACs.26 This hydroxamic acid coordinates to zinc ions, located inside the active site of HDAC. Therefore, we decided to mask this group with a CM-Bhc group to suppress its inhibitory activity. We hypothesized the locked caged HDAC inhibitor CM-Bhc-SAHA to be photoactivatable only in the presence of its “key” enzyme PLE (Fig. 1).
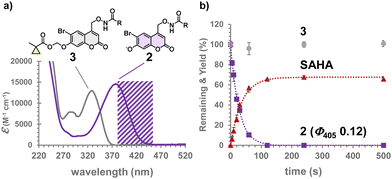
Synthesis of Bhc-caged SAHAs was performed starting from MOMBhc-CH2OH,27 as shown in Scheme 1. The 4-hydroxymethyl group was converted to alkoxyamine by Mitsunobu reaction and reduction28 to yield MOMBhc-CH2ONH2 (1). Esterification with suberanilic acid and removal of MOM protection produced Bhc-SAHA (2). Alkylation of the 7-OH group with CM moiety yielded a candidate for gene-directed caged SAHA, CM-Bhc-SAHA (3). This synthetic method can also be applied to other HDAC inhibitors with hydroxamic acids, such as Trichostatin A and Panobinostat.
The absorption maximum of 3 is at 330 nm in an aqueous solution at neutral pH, while that of 2 is at 374 nm. CM-Bhc-SAHA does not absorb light at a wavelength longer than 380 nm (purple shaded area in Fig. 2a). Therefore, we hypothesized 405 nm light irradiation to cause selective uncaging of 2 in the presence of 3. The time course of photolysis of caged SAHAs is shown in Fig. 2b where Bhc-SAHA produced SAHA (Fig. S1, ESI†) at 405 nm (3.4 mJ s−1) with a photolysis quantum yield (Φ405) of 0.12. Photolysis efficiency (εΦ) of 900 M−1 cm−1 is higher than that of previously reported diethyaminocoumarin-caged SAHA16 and lower than that of Bhc-cAMP (εΦ 2840 M−1 cm−1).24 The difference in quantum yield observed between Bhc-SAHA (0.12) and Bhc-cAMP (0.40) can be attributed to changes related to the difference in the acidity of the leaving groups. The pKa of the hydroxamic acid moiety of SAHA is 9.2, which is much more weakly acidic than that of the cyclic phosphate of cAMP (pKa 3.92). As expected from its absorption properties, no photolytic consumption of 3 was observed under the same irradiation conditions, confirming that the modification by the CM group locks the photolability of 3 (Fig. 2b).
The hydrolytic stability of caged SAHA was measured in an aqueous solution at neutral pH under dark conditions; no traceable amounts of degradation were observed after 15 days for 2 and 5 days for 3. Because SAHA is hydrolysed in the presence of plasma proteins (half-life 75–115 min),29 degradation of 3 was observed in serum containing medium at 37 °C. Since only 4% conversion from 3 to 2 was observed after 24 h, stability was not considered to be a problem in subsequent experiments.
We checked whether locked caged SAHA 3 served as a substrate of PLE and produced unlocked 2 with a practically usable reactivity. Thus, we treated 3 with PLE from the porcine liver. Analysis of the reaction mixture by HPLC confirmed the yield of 2 to be 80% (Fig. 3a and Fig. S2, ESI†). The Michaelis constant (Km) and apparent reaction rate (kcat) of 3 were 3.2 μM and 0.87 s−1, respectively. The specificity constant (kcat/Km) was determined to be 2.7 × 105 M−1 s−1, comparable to reported CM derivatives, such as fluorescein-CM2 (Km = 0.50 μM, kcat/Km = 5.1 × 104 M−1 s−1).25 The results indicated that 3 is a suitable substrate for PLE and produces 2 with a reactivity comparable to known substrates that have been successfully used in live-cell experiments.
Next, we examined the effect of mammalian internal esterases on the hydrolytic stability of the 1-methylcyclopropylcarboxy methyl group (CM). Therefore, 3 was treated with either HEK293T/PLE (PLE(+)), which constitutively expresses the PLE gene, or HEK293T (PLE(−)), which does not express PLE. The conversion from 3 to 2 was completed in 1 minute, using PLE(+) cell lysate, suggesting the practical reactivity of 3 for PLE expressed in mammalian cultured cells (Fig. 3b and Fig. S3, ESI†). Unexpectedly, the PLE(−) lysate gradually deprotected the CM group under the same reaction conditions (15% conversion in 30 min). Since 3 was hydrolytically stable at neutral pH during the experiments, the deprotection observed with PLE(−) lysates was considered a consequence of enzymatic hydrolysis catalyzed by internal esterases. To confirm that SAHA is photochemically produced from 3 in the presence of PLE(+) cells, the reaction mixtures were irradiated with 405 nm light; 3 was incubated with PLE(+) or PLE(−) cell lysates for 1 h and irradiated with 405 nm light (3.4 mJ s−1) for 120 s. HPLC analysis revealed that SAHA was produced only from PLE(+) lysate (Fig. S4, trace 2, ESI†) and not from the PLE(−) one (Fig. S4, trace 4, ESI†).
In HEK293T cells, the internal esterase-mediated enzymatic hydrolysis of the CM moiety of 3 was not entirely suppressed, although, the reactivity with PLE was much better than that with internal esterases in the test cells, suggesting 3 can be used as a gene-directed caged SAHA with PLE as a “key” enzyme.
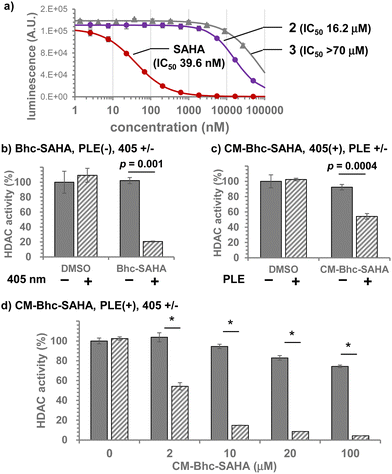
The introduction of caging groups must inhibit the HDAC inhibitory activity of caged SAHAs. Therefore, we measured the lysine deacetylation activity of HDACs from HeLa cell nuclear extracts in the presence of caged SAHAs (Fig. 4a). The half-maximal inhibitory concentrations (IC50) of caged SAHAs were 16.2 μM for 2 and over 70 μM for 3, which was 1/1000 of the inhibitory activity of unmodified SAHA (IC50 39.6 nM). Since SAHA is a nonspecific, broad-spectrum inhibitor of HDAC subtypes26 and Class I HDACs primarily involve HeLa cell nuclear extracts, the IC50 values observed with caged SAHAs must directly reflect the suppression of inhibitory activity achieved by the caging of hydroxamate moiety. The lesser residual activity of CM-Bhc, which is bulkier than Bhc, is also consistent with the caging effect. The bulkiness of the chemical lock allows caged SAHAs to suppress residual HDACi activity in non-target cells and, as a result, suppress the undesirable off-target effects.
Since the CM-Bhc caging group is converted to Bhc group when applied to PLE-expressing cells, we examined photo-induced HDAC inhibition both in a cuvette and in cells using 2. HDAC activity was quantified after incubation of HeLa cell nuclear extracts with 500 nM of 2 and irradiation with 405 nm light (3.4 mJ s−1). As shown in Fig. S5 (ESI†), photon dose-dependent inhibition of HDAC activity was observed in the presence of 2, confirming that photochemically liberated SAHA achieved HDAC inhibition.
The compounds were then tested in live cultured cells. SAHA was added to live HEK293T cells to estimate the working concentration, and the deacetylation activity of endogenous HDACs was quantified (Fig. S6, ESI†). 625 nM SAHA in culture medium achieved approximately 40% inhibition. This HDACi activity was consistent with the in-cuvette experiments shown in Fig. S5 (ESI†), in which 500 nM Bhc-SAHA caused 52% HDAC inhibition when exposed to 405 nm light (204 mJ cm−2, relative HDAC activity: DMSO 71.6%, Bhc-SAHA 37.6%). Since the expected concentration of photochemically released intracellular SAHA was likely lower than the caged SAHAs applied to the culture media, subsequent experiments were performed using caged compounds at concentrations of 2 μM or higher.
Incubation of HEK293T cells with 100 μM of 2 and irradiation with 405 nm light (204 mJ cm−2) inhibited approximately 80% of endogenous HDAC activity. Under these conditions, neither 405 nm light irradiation nor adding 2 alone affected the endogenous HDAC activity (Fig. 4b). When the effect of PLE expression was tested in HEK293T (PLE(−)) or HEK293T/PLE (PLE(+)) cells using 2 μM of 3, HDAC activity was inhibited by 40% only when PLE(+) cells were exposed to 405 nm light (Fig. 4c). Fig. 4d shows the dose dependence of HDACi activity of 3 when exposed to 405 nm light (204 mJ cm−2), confirming that the majority of endogenous HDAC activity in PLE(+) cells is almost completely suppressed photochemically by the addition of 10 μM or more of 3 to the culture media. From these results, we conclude that CM-Bhc-SAHA (3) is a membrane-permeable gene-directed caged SAHA that can selectively suppress endogenous HDAC activity in cells expressing the “key” enzyme, PLE.
In conclusion, we developed a new caged HDAC inhibitor that can genetically target cells of interest. CM-Bhc-SAHA (3) is stable in an aqueous solution at neutral pH, is inert to 405 nm irradiation, and produces Bhc-SAHA (2) with practical reactivity (kcat/Km 2.7 × 105 M−1 s−1) as a substrate for PLE. The HDAC inhibitory activities of caged SAHAs 2 and 3 were suppressed by a factor of 103 compared to SAHA alone. The target cells were tagged with an externally expressed PLE gene, and photo-induced selective inhibition of endogenous HDAC activity was achieved using a new caged SAHA 3. Thus, the CM-Bhc group is a new addition to the gene-directed photocaging groups that can be selectively uncaged by 405 nm light irradiation in the presence of its “key” enzyme. The same gene-directed caging strategy can be applied to other class-selective HDAC inhibitors having hydroxamic acid moieties.30 Gene-directed caged HDAC inhibitors would further expand the utility of methods to modulate HDACi activity in a cell- and tissue-type-selective and spatiotemporal manner. Some issues remain to be improved. Uncaging by PLE and 405 nm light should only occur in cells expressing PLE, but since the Bhc-SAHA and SAHA produced are membrane permeable, they could diffuse out of the cells and re-enter nearby cells, reducing cell type selectivity. This can be an inherent drawback of this system as long as a membrane-permeable HDACi is used.
By utilizing the concept of gene-directed caging, cell-type targetability can be incorporated into previously reported Bhc-caged compounds.31,32 Furthermore, because of their substrate specificity, it is possible to use two or more chemical lock-and-key enzyme pairs in a biologically orthogonal manner. This would allow, for example, the simultaneous targeting of two or more caged molecules and separate targeting of different types of cells or tissues.
This work was supported by JSPS KAKENHI grant number 20H02882 (TF) and a Grant-in-Aid for Scientific Research on Innovative Areas 19H05778 (TF) “MolMovies.” We thank Ms Sayaka Kado (Chiba University) for the measurement of HR-MS.
Conflicts of interest
There are no conflicts to declare.Notes and references
- N. Reichert, M.-A. Choukrallah and P. Matthias, Cell. Mol. Life Sci., 2012, 69, 2173–2187 CrossRef CAS PubMed
.
- J. C. Kiefer, Dev. Dyn., 2007, 236, 1144–1156 CrossRef CAS PubMed
.
- R. M. Barrett and M. A. Wood, Learn. Mem., 2008, 15, 460–467 CrossRef CAS PubMed
.
- M. Esteller, N. Engl. J. Med., 2008, 358, 1148–1159 CrossRef CAS PubMed
.
- A. J. M. d Ruijter, A. H. v Gennip, H. N. Caron, S. Kemp and A. B. P. v Kuilenburg, Biochem. J., 2003, 370, 737–749 CrossRef PubMed
.
- E. Seto and M. Yoshida, Cold Spring Harbor Perspect. Biol., 2014, 6, a018713–a018713 CrossRef PubMed
.
- S.-Y. Park and J.-S. Kim, Exp. Mol. Med., 2020, 52, 204–212 CrossRef CAS PubMed
.
- I. Ali, R. J. Conrad, E. Verdin and M. Ott, Chem. Rev., 2018, 118, 1216–1252 CrossRef CAS PubMed
.
- B. Barneda-Zahonero and M. Parra, Mol. Oncol., 2012, 6, 579–589 CrossRef CAS PubMed
.
- P.-C. Pao and L.-H. Tsai, ACS Chem. Neurosci., 2022, 13, 848–858 CrossRef CAS PubMed
.
- M. Lemos and N. Stefanova, Front. Synaptic Neurosci., 2020, 12, 586453 CrossRef CAS PubMed
.
- Y. Lu, Y.-T. Chan, H.-Y. Tan, S. Li, N. Wang and Y. Feng, Mol. Cancer, 2020, 19, 79 CrossRef PubMed
.
- A. M. Burns and J. Gräff, Curr. Opin. Neurobiol., 2021, 67, 75–84 CrossRef CAS PubMed
.
- S. A. Reis, B. Ghosh, J. A. Hendricks, D. M. Szantai-Kis, L. Tork, K. N. Ross, J. Lamb, W. Read-Button, B. Zheng, H. Wang, C. Salthouse, S. J. Haggarty and R. Mazitschek, Nat. Chem. Biol., 2016, 12, 317–323 CrossRef CAS PubMed
.
- W. Szymanski, M. E. Ourailidou, W. A. Velema, F. J. Dekker and B. L. Feringa, Chem. – Eur. J., 2015, 21, 16517–16524 CrossRef CAS PubMed
.
- N. Ieda, S. Yamada, M. Kawaguchi, N. Miyata and H. Nakagawa, Bioorg. Med. Chem., 2016, 24, 2789–2793 CrossRef CAS PubMed
.
- A. Leonidova, C. Mari, C. Aebersold and G. Gasser, Organometallics, 2016, 35, 851–854 CrossRef CAS
.
- B. Parasar and P. V. Chang, Chem. Sci., 2017, 8, 1450–1453 RSC
.
- L. L. Shi, L. Xu, Q. H. Guan, X. Jin, J. P. Yang and X. Y. Zhu, Bioconjugate Chem., 2018, 29, 1344–1351 CrossRef CAS PubMed
.
- G. R. Sama, H. Liu, S. Mountford, P. Thompson, A. Robinson and A. E. Dear, Bioorg. Med. Chem. Lett., 2020, 30, 127291 CrossRef CAS PubMed
.
- P. Klan, T. Solomek, C. G. Bochet, A. Blanc, R. Givens, M. Rubina, V. Popik, A. Kostikov and J. Wirz, Chem. Rev., 2013, 113, 119–191 CrossRef CAS PubMed
.
- R. Weinstain, T. Slanina, D. Kand and P. Klán, Chem. Rev., 2020, 120, 13135–13272 CrossRef CAS PubMed
.
- Y. Hou, Z. Zhou, K. Huang, H. Yang and G. Han, ChemPhotoChem, 2018, 2, 1005–1011 CrossRef CAS
.
- A. Z. Suzuki, T. Sakano, H. Sasaki, R. Watahiki, M. Sone, K. Horikawa and T. Furuta, Chem. Commun., 2021, 57, 5630–5633 RSC
.
- L. Tian, Y. L. Yang, L. M. Wysocki, A. C. Arnold, A. Hu, B. Ravichandran, S. M. Sternson, L. L. Looger and L. D. Lavis, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 4756–4761 CrossRef CAS PubMed
.
- W. K. Kelly and P. A. Marks, Nat. Clin. Pract. Oncol., 2005, 2, 150 CrossRef CAS PubMed
.
- A. Z. Suzuki, T. Watanabe, M. Kawamoto, K. Nishiyama, H. Yamashita, M. Ishii, M. Iwamura and T. Furuta, Org. Lett., 2003, 5, 4867–4870 CrossRef CAS PubMed
.
- E. Grochowski and J. Jurczak, Synth. Stuttgart, 1976, 682–684 CrossRef CAS
.
- R. Konsoula and M. Jung, Int. J. Pharm., 2008, 361, 19–25 CrossRef CAS PubMed
.
- A. V. Bieliauskas and M. K. H. Pflum, Chem. Soc. Rev., 2008, 37, 1402–1413 RSC
.
- T. Furuta, S. S. H. Wang, J. L. Dantzker, T. M. Dore, W. J. Bybee, E. M. Callaway, W. Denk and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 1193–1200 CrossRef CAS PubMed
.
- A. Z. Suzuki, R. Sekine, S. Takeda, R. Aikawa, Y. Shiraishi, T. Hamaguchi, H. Okuno, H. Tamamura and T. Furuta, Chem. Commun., 2019, 55, 451–454 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc03552a |
| This journal is © The Royal Society of Chemistry 2022 |




![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Student's
Student's