Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Native ambient mass spectrometry of intact protein assemblies directly from Escherichia coli colonies†
Yuying
Du
 ,
Robin C.
May
,
Robin C.
May
 and
Helen J.
Cooper
and
Helen J.
Cooper
 *
*
School of Biosciences, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK. E-mail: h.j.cooper@bham.ac.uk
First published on 24th May 2022
Abstract
Here, we demonstrate that by combining electroporation with native ambient mass spectrometry, it is possible to detect intact non-covalent protein complexes directly from bacterial colonies growing on agar. Homodimers HdeA and HdeB were identified, together with the 50 kDa Mn-bound superoxide dismutase homodimer, in addition to some previously undetected monomeric proteins.
Liquid extraction surface analysis (LESA)1 mass spectrometry (MS) is an ambient mass spectrometry technique which combines liquid microjunction sampling with nano-electrospray mass spectrometry, and is particularly suitable for the analysis of intact proteins. LESA MS has been demonstrated for the analysis of proteins for a range of substrates, including dried blood spots,2 thin tissue sections3 and bacterial colonies growing on agar.4–6 Compared with matrix-assisted laser desorption ionization time-of-flight (MALDI TOF) MS, which is recognized as the gold standard for routine identification of a wide range of microorganisms,7,8 LESA MS focuses on the identification of intact bacterial proteins rather than spectral matching for species identification. Other ambient MS approaches that have been applied to bacteria, albeit for the analysis of small molecules, include rapid evaporative ionization mass spectrometry (REIMS),9 laser ablation electrospray ionization (LAESI),10,11 paperspray mass spectrometry12 and nanospray desorption electrospray ionization (nano-DESI).13
In terms of protein identification, analysis can either be “top-down”, in which the intact protein ion is fragmented in the mass spectrometer, or “bottom-up”, in which proteolytic peptides are subjected to MS analysis. The advantage of top-down MS is that all information relating to primary sequence and post-translational modifications are retained, unlike the bottom-up approach where information relating to the presence of single nucleotide polymorphisms or connectivity between post-translational modifications may be lost. Our work to date on LESA MS of bacteria has made use of denaturing LESA solvents based on aqueous organic mixtures.5,14 The use of these has been necessary to breach the cell wall. A limitation of this approach is that any structural information is lost as the proteins are unfolded. As protein structure and function are intricately linked, there is a drive to develop analytical methods capable of obtaining structural information, e.g., protein assembly stoichiometry, protein-ligand binding, directly from the bacteria.
Another branch of MS, native mass spectrometry,15 allows the analysis of tertiary and quaternary protein structure. Proteins are electrosprayed from solutions designed to mimic native conditions, and intra- and intermolecular non-covalent interactions present in solution are maintained in the gas-phase. We have demonstrated that native mass spectrometry may be coupled with LESA MS and nano-DESI MS for the analysis of protein assemblies and complexes directly from tissue.16,17 Native LESA MS generally offers higher sensitivity but lower spatial resolution for mass spectrometry imaging than nano-DESI MS due to the larger sampling area. Sun and co-workers have demonstrated native mass spectrometry of E. coli lysates by integrating size exclusion chromatography and capillary zone electrophoresis.18 Huang and co-workers introduced an approach termed “in-cell” MS in which suspensions of bacteria are subjected to on-line electroporation, releasing endogenous proteins for introduction to the mass spectrometer.19 Electroporation is the process by which cell membranes are made permeable, either reversibly or irreversibly, through the application of an electric field,20–22 and has found applications in both the delivery of exogenous molecules into cells and the extraction of molecules from cells.23 For the latter, it has been shown that higher molecular weight molecules such as proteins present a greater challenge than those with lower molecular weight.24 Nevertheless, complexes of the protein calmodulin overexpressed in E. coli have been observed by the in-cell MS approach.19
We recently designed and built an electroporation device for integration with the LESA MS workflow.14 In that work, our aim was analysis of intact proteins in yeast colonies. Yeast had proved resistant to LESA MS, even with the use of harsh solvents, due to its rigid chitinous cell wall, but electroporation resulted in rapid release of intact proteins. A feature of electroporation is that it is associated with minimal heating,22 which led us to hypothesise that by combining electroporation with native-like LESA solvents, it would be possible to perform native ambient mass spectrometry, i.e., obtain structural information, directly from living bacterial colonies.
Three workflows were investigated (summarised in Fig. S1, ESI†). In the first, a small volume (2 μl) of 200 mM ammonium acetate was pipetted onto the bacterial colony, which was then subjected to electroporation followed by LESA sampling. The rationale behind addition of ammonium acetate was avoidance of air breakdown during electroporation. In the second workflow, 2 μl of 200 mM ammonium acetate was deposited onto the colony before LESA sampling, i.e., the electroporation step was omitted. In the third, the bacterial colony was subjected to LESA sampling only. The success rate, defined as the number of sampling events that led to mass spectra containing peaks corresponding to proteins in relation to the total number of sampling events, was 77% for addition of solvent and electroporation, 14% for addition of solvent with no electroporation and 31% for no solvent addition and no electroporation. See Table S1, ESI.†
Initial mass spectrometry experiments were performed in the mass range m/z 900–3000, with no source collision energy. Fig. 1 shows a representative native LESA mass spectrum obtained following electroporation of an E. coli K12 colony. Twenty-two proteins were detected, of which eleven were identified by tandem mass spectrometry (MS/MS). See Table S2, ESI† for a summary of protein assignments. Full details are provided in Protein Identification, ESI.† Eight of the proteins identified (HdeA, HdeB, CspE, Acyl carrier protein, YibT, Hpr, PsiF and YgiW) were previously undetected by LESA MS. Among those proteins, HdeA (monomer), HdeB (monomer), CspE and Hpr have previously been identified by MALDI top-down MS of cell lysates of various strains of E. coli.25 HdeA, HdeB, YibT and YgiW were identified in a bottom-up LC-MS proteomics study of outer membrane vesicles obtained from a range of E. coli strains.26 Acyl carrier protein has been identified in a bottom-up MALDI study.27
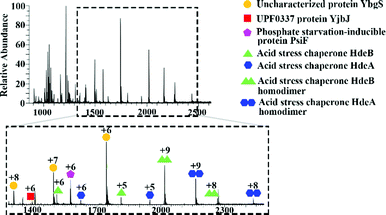 | ||
| Fig. 1 Representative mass spectrum acquired by native LESA MS following electroporation of E. coli K12 colony. | ||
Subsequent experiments were performed in a higher mass range (m/z 1500–4000) with 80% source fragmentation energy, resulting in identification of two further proteins, including the monomeric antigen 43 α chain (∼49.8 kDa, see Fig. S2 and S3, ESI†) which has not previously been detected by LESA MS, although has been identified in bottom-up studies.28–30
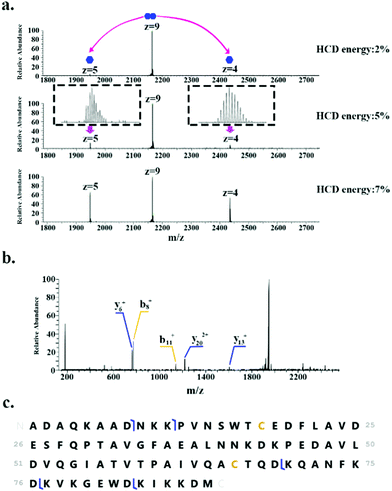
In addition to monomeric proteins, three protein assemblies were identified, corresponding to the homodimers HdeA, HdeB and Mn-bound superoxide dismutase. HdeA and HdeB are acid stress chaperones which restrict aggregation of denatured periplasmic proteins in acidic environments.31 HdeA was detected as both the monomer (∼9.7 kDa, charge states 4+ to 6+) and homodimer (∼19.5 kDa, charge states 8+ to 9+) as indicated in Fig. 1. Fig. 2 shows the identification of the intact homodimer. Tandem mass spectrometry by higher energy collision dissociation (HCD)32 of the 9+ precursor ions (m/z 2165.2) resulted in detection of monomer subunits in charge states 5+ and 4+. The abundance of the monomer subunits increases with HCD collision energy (Fig. 2a). HCD of the 5+ monomer ions (32% normalised collision energy, NCE) results in detection of sequence fragments allowing the protein identity to be confirmed (Fig. 2b and c). The homodimer of HdeA, along with the homodimer of Hpr (not observed here) have previously been identified from E. coli lysates in native proteomics experiments which coupled size exclusion chromatography with capillary zone electrophoresis MS.18
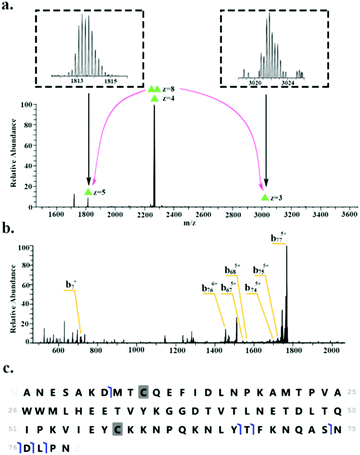
HdeB was detected as both the monomer (∼9.1 kDa, charge states 4+ to 6+) and homodimer (∼18.1 kDa, charge states 8+ to 9+). Fig. 3 shows the identification of the homodimer. HCD of the 8+ precursor ions (m/z 2266.6) results in the observation of monomer subunits in charge states 5+, 4+ and 3+ (Fig. 3a). MS/MS of the monomer was obtained for identification (Fig. 3b) and sequence fragments are shown in Fig. 3c. The fragments observed confirm the identity of the protein and suggest the presence of a disulfide bond between Cys10 and Cys58. Confirmation of the disulphide bond was obtained by electron transfer dissociation with higher-energy collision dissociation (EThcD)33,34 of the 5+ monomer ions (m/z 1813.5). See Table S3, ESI.†
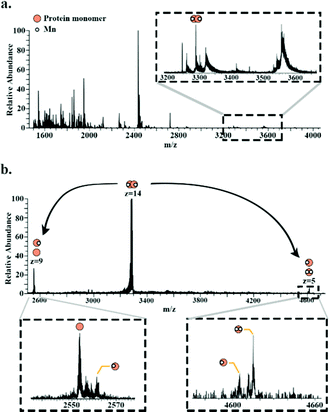
Superoxide dismutase was detected as a dimer (∼49.8 kDa) in which both subunits are bound to two Mn2+ ions. Fig. 4a shows a full scan mass spectrum with the peak corresponding to the 14+ charge state of the dimer indicated. The stoichiometry of the complex was confirmed by MS/MS (HCD at 28% NCE) which resulted in dissociation of the dimer to its monomeric subunits in charge states 5+ and 9+ (Fig. 4b). Asymmetric partitioning of the metal ions amongst the subunits was observed following HCD. Further increase in HCD energy resulted in detection of sequence fragments allowing the protein identity to be confirmed (see ESI†).
In summary, we have demonstrated that by combining electroporation with native LESA MS, it is possible to detect and identify intact protein assemblies up to 50 kDa directly from colonies of E. coli growing on a solid substrate. The stoichiometry of the protein assemblies and the identities of the proteins were confirmed by HCD MS. Although it was possible in some cases to detect proteins in the absence of electroporation, this could not be achieved reliably (success rate 14–31%).
YD is funded by a Darwin Studentship. HJC is funded by EPSRC (EP/S002979/1). The authors thank Dr Emma Sisley for helpful discussions. The Orbitrap Eclipse mass spectrometer used in this work was funded by BBSRC (BB/S019456/1). Supplementary data supporting this research is openly available from: https://doi.org/10.25500/edata.bham.00000841.
Conflicts of interest
There are no conflicts of interest to declare.Notes and references
- V. Kertesz and G. J. Van Berkel, Fully automated liquid extraction-based surface sampling and ionization using a chip-based robotic nanoelectrospray platform, J. Mass Spectrom., 2010, 45, 252–260 CrossRef CAS PubMed.
- R. L. Edwards, A. J. Creese, M. Baumert, P. Griffiths, J. Bunch and H. J. Cooper, Hemoglobin Variant Analysis via Direct Surface Sampling of Dried Blood Spots Coupled with High-Resolution Mass Spectrometry, Anal. Chem., 2011, 83, 2265–2270 CrossRef CAS PubMed.
- J. Sarsby, N. J. Martin, P. F. Lalor, J. Bunch and H. J. Cooper, Top-Down and Bottom-Up Identification of Proteins by Liquid Extraction Surface Analysis Mass Spectrometry of Healthy and Diseased Human Liver Tissue, J. Am. Soc. Mass Spectrom., 2014, 25, 1953–1961 CrossRef CAS PubMed.
- E. C. Randall, J. Bunch and H. J. Cooper, Direct Analysis of Intact Proteins from Escherichia coli Colonies by Liquid Extraction Surface Analysis Mass Spectrometry, Anal. Chem., 2014, 86, 10504–10510 CrossRef CAS PubMed.
- J. Havlikova, R. C. May, I. B. Styles and H. J. Cooper, Liquid Extraction Surface Analysis Mass Spectrometry of ESKAPE Pathogens, J. Am. Soc. Mass Spectrom., 2021, 32, 1345–1451 CrossRef CAS PubMed.
- K. I. Kocurek, L. Stones, J. Bunch, R. C. May and H. J. Cooper, Top-Down LESA Mass Spectrometry Protein Analysis of Gram-positive and Gram-negative Bacteria, J. Am. Soc. Mass Spectrom., 2017, 28, 2066–2077 CrossRef CAS PubMed.
- V. Horneffer, J. Haverkamp, H.-G. Janssen and R. Notz, MALDI-TOF-MS analysis of bacterial spores: Wet heat-treatment as a new releasing technique for biomarkers and the influence of different experimental parameters and microbiological handling, J. Am. Soc. Mass Spectrom., 2004, 15, 1444–1454 CrossRef CAS PubMed.
- L. Simon, P. J. Dufresne, S. Hafid, D. Marc-Christian, B. Sadjia, L. Brigitte, T. Cécile and C. Vishnu, A Side by Side Comparison of Bruker Biotyper and VITEK MS: Utility of MALDI-TOF MS Technology for Microorganism Identification in a Public Health Reference Laboratory, PLoS One, 2015, 10, e0144878 CrossRef PubMed.
- J. M. Kinross, L. Muirhead, J. Alexander, J. Balog, C. Guallar-Hoya, A. Speller, O. Golff, R. Goldin, A. Darzi, J. Nicholson and Z. Takats, Abstract 3977: iKnife: Rapid evaporative ionization mass spectrometry (REIMS) enables real-time chemical analysis of the mucosal lipidome for diagnostic and prognostic use in colorectal cancer, Cancer Res., 2016, 76, 3977 Search PubMed.
- G. Parsiegla, B. Shrestha, F. Carrière and A. Vertes, Direct Analysis of Phycobilisomal Antenna Proteins and Metabolites in Small Cyanobacterial Populations by Laser Ablation Electrospray Ionization Mass Spectrometry, Anal. Chem., 2012, 84, 34–38 CrossRef CAS.
- S. N. Dean, C. Walsh, H. Goodman and M. L. van Hoek, Analysis of mixed biofilm (Staphylococcus aureus and Pseudomonas aeruginosa) by laser ablation electrospray ionization mass spectrometry, Biofouling, 2015, 31, 151–161 CrossRef CAS PubMed.
- S. Chiang, W. Zhang and Z. Ouyang, Paper spray ionization mass spectrometry: recent advances and clinical applications, Expert Rev. Proteomics, 2018, 15, 781–789 CrossRef CAS PubMed.
- J. Watrous, P. Roach, B. Heath, T. Alexandrov, J. Laskin and P. C. Dorrestein, Metabolic Profiling Directly from the Petri Dish Using Nanospray Desorption Electrospray Ionization Imaging Mass Spectrometry, Anal. Chem., 2013, 85, 10385–10391 CrossRef CAS PubMed.
- K. I. Kocurek, J. Havlikova, E. Buchan, A. Tanner, R. C. May and H. J. Cooper, Electroporation and Mass Spectrometry: A New Paradigm for In Situ Analysis of Intact Proteins Direct from Living Yeast Colonies, Anal. Chem., 2020, 92, 2605–2611 CrossRef CAS PubMed.
- A. C. Leney and A. J.-R. Heck, Native Mass Spectrometry: What is in the Name?, J. Am. Soc. Mass Spectrom., 2017, 28, 5–13 CrossRef CAS PubMed.
- O. J. Hale and H. J. Cooper, Native Mass Spectrometry Imaging of Proteins and Protein Complexes by Nano-DESI, Anal. Chem., 2021, 93, 4619–4627 CrossRef CAS PubMed.
- R. L. Griffiths, E. K. Sisley, A. F. Lopez-Clavijo, A. L. Simmonds, I. B. Styles and H. J. Cooper, Native Mass Spectrometry Imaging of Intact proteins and Protein Complexes in Thin Tissue Sections, Int. J. Mass Spectrom., 2017, 437, 23–29 CrossRef.
- X. Shen, Q. Kou, R. Guo, Z. Yang, D. Chen, X. Liu, H. Hong and L. Sun, Native Proteomics in Discovery Mode Using Size-Exclusion Chromatography–Capillary Zone Electrophoresis–Tandem Mass Spectrometry, Anal. Chem., 2018, 90, 10095–10099 CrossRef CAS PubMed.
- G. Li, S. Yuan, S. Zheng, Y. Liu and G. Huang, In Situ Living Cell Protein Analysis by Single-Step Mass Spectrometry, Anal. Chem., 2018, 90, 3409–3415 CrossRef CAS PubMed.
- T. Y. J. E. Tsong, Electroporation of cell membranes, Eur. J. Cell Biol., 1989, 149–163 Search PubMed.
- R. Stampfli, Reversible electrical breakdown of the excitable membrane of a Ranvier node, An Acad. Brasil Ciens., 1958, 30, 57–61 Search PubMed.
- A. J. H. Sale and W. A. Hamilton, Effects of high electric fields on microorganisms: I. Killing of bacteria and yeasts, Biochim. Biophys. Acta, Gen. Subj., 1967, 148, 781–788 CrossRef.
- T. Geng and C. Lu, Microfluidic electroporation for cellular analysis and delivery, Lab Chip, 2013, 13, 3803–3821 RSC.
- N. Bao, J. Wang and C. Lu, Microfluidic electroporation for selective release of intracellular molecules at the single-cell level, Electrophoresis, 2008, 29, 2939–2944 CAS.
- C. K. Fagerquist and C. E. Dodd, Top-down proteomic identification of plasmid and host proteins produced by pathogenic Escherichia coli using MALDI-TOF-TOF tandem mass spectrometry, PLoS One, 2021, 16, e0260650 CrossRef CAS PubMed.
- D. J. Wurpel, D. G. Moriel, M. Totsika, D. M. Easton and M. A. Schembri, Comparative analysis of the uropathogenic Escherichia coli surface proteome by tandem mass-spectrometry of artificially induced outer membrane vesicles, J. Proteomics, 2015, 115, 93–106 CrossRef CAS PubMed.
- P. Pribil and C. Fenselau, Characterization of Enterobacteria Using MALDI-TOF Mass Spectrometry, Anal. Chem., 2005, 77, 6092–6095 CrossRef CAS PubMed.
- C. Cirulli, G. Marino and A. Amoresano, Membrane proteome in Escherichia coli probed by MS3 mass spectrometry: a preliminary report, Rapid Commun. Mass Spectrom., 2007, 21, 2389–2397 CrossRef CAS PubMed.
- M. P. Molloy, B. R. Herbert, M. B. Slade, T. Rabilloud, A. S. Nouwens, K. L. Williams and A. A. Gooley, Proteomic analysis of the Escherichia coli outer membrane, Eur. J. Biochem., 2000, 267, 2871–2881 CrossRef CAS PubMed.
- S. H. Yoon, M.-J. Han, S. Y. Lee, K. J. Jeong and J.-S. Yoo, Combined transcriptome and proteome analysis of Escherichia coli during high cell density culture, Biotech. Bioeng., 2003, 81, 753–767 CrossRef CAS PubMed.
- R. Kern, A. Malki, J. Abdallah, J. Tagourti and G. J. J. O. B. Richarme, Escherichia coli HdeB is an acid stress chaperone, J. Bacteriol., 2007, 189, 603–610 CrossRef CAS PubMed.
- J. V. Olsen, B. Macek, O. Lange, A. Makarov, S. Horning and M. Mann, Higher-energy C-trap dissociation for peptide modification analysis, Nat. Methods, 2007, 4, 709–712 CrossRef CAS PubMed.
- F. Liu, B. V. Breukelen and A. Heck, Facilitating Protein Disulfide Mapping by a Combination of Pepsin Digestion, Electron Transfer Higher Energy Dissociation (EThcD), and a Dedicated Search Algorithm SlinkS, Mol. Cell. Proteomics, 2014, 13, 2776–2786 CrossRef CAS PubMed.
- S. R. Cole, X. Ma, X. Zhang and Y. Xia, Electron Transfer Dissociation (ETD) of Peptides Containing Intrachain Disulfide Bonds, J. Am. Soc. Mass Spectrom., 2012, 23, 310–320 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, comparison of workflows, protein identification, additional MS/MS spectra. See DOI: https://doi.org/10.1039/d2cc02085h |
| This journal is © The Royal Society of Chemistry 2022 |