Open Access Article
Open Access ArticleRapid and sensitive multiplex detection of COVID-19 antigens and antibody using electrochemical immunosensor-/aptasensor-enabled biochips†
Fuze
Jiang‡
ab,
Zhen
Xiao‡
cd,
Ting
Wang
e,
Jiajia
Wang
cdf,
Lihua
Bie
g,
Lanja
Saleh
h,
Kathrin
Frey
h,
Lianjun
Zhang
*cd and
Jing
Wang
 *ab
*ab
aInstitute of Environmental Engineering, ETH Zürich, Zürich, Switzerland. E-mail: jing.wang@ifu.baug.ethz.ch
bAdvanced Analytical Technologies, Empa, Dübendorf, Switzerland
cCAMS Key Laboratory of Synthetic Biology Regulatory Element, Institute of Systems Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing 100005, China. E-mail: zlj@ism.cams.cn
dSuzhou Institute of Systems Medicine, Suzhou 215123, China
eSchool of Automation, Hangzhou Dianzi University, Hangzhou 310018, China
fSchool of Engineering, China Pharmaceutical University, Nanjing, China
gCollege of Informatics, Huazhong Agricultural University, Wuhan 430070, Hubei, China
hInstitute of Clinical Chemistry, University Hospital Zurich, University of Zurich, Zurich 8091, Switzerland
First published on 26th May 2022
Abstract
We report protein- and aptamer-based electrochemical biochips for low-cost, one-step, sensitive and accurate multiplex detection of SARS-CoV-2 spike (S) and nucleocapsid (N) proteins, and IgG antibody in unprocessed clinical samples, allowing citizens to achieve rapid diagnosis at home or in community settings.
The COVID-19 pandemic has claimed millions of lives and put health systems under enormous strain. Safe reopening of the public spaces requires a series of measures to mitigate the risk of infections, including widely available vaccines and a large-scale testing capacity to rapidly confirm and isolate infected individuals.1 Commercial diagnostic approaches for COVID-19 include quantitative real-time PCR with reverse transcription (RT-qPCR) assay for nucleic acid detection and serologic-/immuno-assays for rapid antigen and antibody tests.2 RT-qPCR based nucleic acid detection can be time-consuming, and requires expensive devices and trained medical personnel to perform the analysis, thus limiting detection capacities.3 COVID-19 serologic tests can rapidly detect human SARS-CoV-2 antibodies (IgM or IgG) to determine whether a person has ever been infected and possesses a certain degree of immunity to SARS-CoV-2.4,5 Immunological tests, such as the lateral flow assay (LFA) and enzyme-linked immunosorbent assay (ELISA), utilize SARS-CoV-2 neutralizing antibodies as the probes to detect SARS-CoV-2 antigens and are less expensive than RT-qPCR tests.4 However, the serologic- or immuno-assay tests demonstrate poor sensitivity and modest limit-of-detection (LoD).6
The major SARS-CoV-2 biomarkers detected through rapid SARS-CoV-2 viral tests are the spike (S) and nucleocapsid (N) proteins.7 The S protein contains two subunits, S1 and S2. The S1 protein comprises a unique receptor-binding domain (SRBD) that can specifically interact with human angiotensin-converting enzyme 2 (ACE2) and mediate host cell recognition.8 This SRBD is an ideal target for developing sensing probes in the specific recognition of SARS-CoV-2. The N protein is the most abundant structural protein of the SARS-CoV-2 virus.9 It shows strong immunogenicity and often serves as the diagnostic biomarker for COVID-19 detection. Aptamers are short nucleic acid sequences that form distinctive three-dimensional structures that bind to target proteins (SRBD) with comparable specificity and binding affinity to antibodies.10 SARS-CoV-2 antigen-specific and high-affinity DNA aptamers have been developed from single-stranded DNA (ssDNA) libraries through systematic evolution of ligands by exponential enrichment (SELEX).11 Of note, these aptamers exhibit several advantages compared to antibodies in COVID-19 diagnostic and therapeutic applications, including better physicochemical stability, much cheaper and easier processes to produce, lower denaturation susceptibility, simpler modification steps to provide covalent bonds to other material surfaces, and lower steric hindrance due to the favourably small size (2–3 nm). Therefore, aptamers are considered as an alternative to antibodies and are capable of being utilized to establish a point of care (POC) test for rapid COVID-19 antigen detection.
We reported protein- and aptamer-initiated electrochemical biochips that allow for low-cost, one-step, sensitive, and rapid multiplex detection of SARS-CoV-2 S and N proteins, and IgG antibody. Our sensing biochips demonstrated a competitive LoD, fast turnaround time (10 min) and good selectivity, allowing for early-stage screening and tracing of suspected cases at home or in community settings.
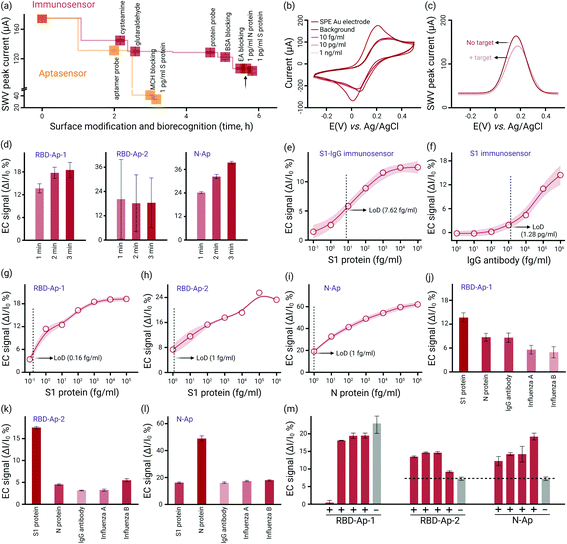
Our proposed COVID-19 test strip array consisted of five independent sensing biochips. Based on the categories of biomarkers, the test strip arrays were classified into protein-initiated immunosensors and DNA-enabled aptasensors (Fig. 1a). For immunosensors, S1-IgG antibody and S1 protein were chosen as the sensing ligands to recognize S1 protein in the swab samples and S1-IgG antibody in the blood, respectively. For the aptasensors, two S1-specific aptamers (RBD-Ap-1 and RBD-Ap-2) and one N-specific (N-Ap) aptamer, screened via the competition-based SELEX method, were anchored onto the biochips to capture S1 and N proteins (Table S1, ESI†). Nucleic Acid Package (NUPACK) predicted the minimum free energy required to form the secondary structures of the aptamers. The estimated dissociation constants (Kd) (5.8 ± 0.8 nM for RBD-Ap-1, 44.78 ± 9.97 nM for RBD-Ap-2, and 0.7 ± 0.1 nM for N-Ap) were comparable to commercial neutralizing antibodies developed to bind S1 and N proteins (Fig. 1b).10–12 According to the biorecognition tests, the highest ELISA signal response was assigned to the N-Ap aptamer, indicating the best binding affinity among the tested probes and consistent with the minimal Kd value (the lower the Kd value, the higher the binding capability) (Fig. 1b). The aptamers bound to the target proteins immediately after mixing and reached their surface saturation within 6 minutes, demonstrating a rapid SARS-CoV-2 antigen recognition. To evaluate the specificity, we investigated whether the screened aptamers could also bind non-specific proteins by incubating the biotinylated aptamers into PBS buffer (pH 7.4) (blank), bovine serum albumin (BSA), S1, S2 and N protein solutions. No obvious BSA, S2 or N binding for RBD-Ap-1 and RBD-Ap-2 aptamers was observed, even at the target concentration of 10 ng mL−1 (Fig. 1c). N-Ap aptamer did not bind to BSA, S1 or S2. Thus, the used aptamers targeted SARS-CoV-2 S1 and N with high specificity.
We prepared the COVID-19 immunosensor- and aptasensor-based biochips by immobilizing SARS-CoV-2 specific probes (proteins and aptamers) onto gold nanoparticle (∼154 nm in diameter) (Fig. S1, ESI†) modified screen-printed electrodes (AuNP-SPE) (Fig. S2 and S3, ESI†). The electrochemical property of AuNP-SPE was characterized by interrogating cyclic voltammetry (CV) scanning in 0.5 M H2SO4 and K3[Fe(CN)6]/K4[Fe(CN)6] redox indicator solution. An apparent reduction peak of gold oxide (∼0.9 V vs. Ag/AgCl pseudo-reference electrode) and a well-defined duck-shaped voltammogram (ΔEp ∼ 92.5 mV) were obtained, indicating successful electroplating of gold nanoparticles onto the SPE and the good electrochemical performance of the AuNP-SPE (Fig. S4, ESI†). The consistent CV diagrams indicated that our manufacturing process can uniformly prepare sensing electrodes. The process of immobilizing sensing ligands onto the COVID-19 biochip is critical for acquiring high-quality signals. For immunosensor-based biochips, the S1-IgG or S1 protein probes were covalently immobilized onto the AuNP-SPE through the amine coupling process. The treatment of the AuNP-SPEs with cysteamine introduced –NH2 groups onto their surface via the thiol group (S–Au bond). The S1-IgG or S1 probe was anchored on the amino-functionalized AuNP-SPEs through cysteamine-glutaraldehyde (GA) coupling (as illustrated in Fig. S5, ESI†).
The AuNP-SPE surface was activated by reacting its free –NH2 groups with –CHO of bi-functional GA at its one end. The other free –CHO group reacted with –NH2 in S1-IgG or S1 proteins, forming the Schiff base between the GA-activated AuNP-SPE and the protein probes. We used ethanolamine to deactivate remaining GA sites and introduced BSA proteins to the surface for antifouling treatment, ensuring that all non-active sites were blocked. The sensorgram produced from square wave voltammetry (SWV) measurement at various stages of surface modification (S1-IgG immunosensor) is shown in Fig. 2a. The SWV peak currents decreased with respect to the surface modification step by step, indicating that S1-IgG antibodies adsorbed onto AuNP-SPEs. A distinct N-element peak in the X-ray photoelectron spectroscopy (XPS) spectrum confirmed the successful surface modification of the protein probe (Fig. S6, ESI†). The control of 1 pg mL−1 N (non-specific protein) did not produce any measurable response, which indicated that the surface of the S1-IgG immunosensor-based biochip was well blocked, and the response to S1 was highly specific (Fig. 2a). The surface modification of the immunosensor necessitated 5.5 hours and 5 steps, yet it only required 3 hours and 2 stages to immobilize aptamer probes and blocking agents (Fig. 1a). We utilized mercaptoethanol (MCH) as the blocking agent to help the thiolated-aptamer probe to stay upright at the surface and remove non-specific bonds from the chip surface. As shown in Fig. 1a, the decrease of the SWV signal after anchoring the MCH layer indicated the successful surface antifouling blocking.
We further validated the feasibility of detecting SARS-CoV-2 antigens and antibodies using our COVID-19 test biochips. COVID-19 S1-IgG and S1 modified on the biochips allowed the target biomarkers to recognize and combine, thus introducing an extra protein layer onto the chip surface. Here, we used a surface-sensitive technique based on electrochemical cyclic voltammetry (CV) and SWV measurement to in situ probe the electrode's surface resistance changes, allowing for monitoring the biorecognition process in a highly sensitive way. Taking the RBD-AP-1 aptasensor-based biochip as an example (Fig. 2b and c), upon capturing S1 protein, bonded targets formed an additional physical barrier on the surface that hindered the redox indicator transferring to the electrode, leading to a decrease in the peak current (0.19 V) of CV and SWV scans. This peak current change extrapolated from the SWV scan was defined as the electrochemical sensing signal.
The binding as a function of time interpreted with EC signals was approximately consistent with that of the ELISA test, and thus a 10 min duration was selected for the sensor's turnaround time (Fig. 2d). We determined the biochips’ LoD by conducting S1, N and S1-IgG detection at concentrations ranging from fg mL−1 to ng mL−1. The S1-IgG and S1 modified biochips showed an LoD of 7.62 fg mL−1 and 1.28 pg mL−1 (Fig. 2d–f) for S1 protein and S1-IgG antibody detection, respectively (Fig. S7, ESI†). The N-Ap-based aptasensor demonstrated the best sensitivity in terms of the highest EC signal (ΔI/I0 = 18.7%) at the 1 fg mL−1 target concentration, significantly surpassing the RBD-Ap-1, and RBD-Ap-2 biochips (11.3% and 7.3%, respectively) (Fig. 2–i). The signal was positively correlated with the binding affinity, indicating that the N-Ap-based aptasensor possessed the highest binding capacity to capture SARS-CoV-2 viral antigens. To verify that our biochips were able to distinguish biological interferents, we proceeded with the aptasensor to detect artificially prepared samples containing non-specific viral proteins (Influenza A and Influenza B). Fig. 2–l shows that a discernible EC signal specific to the target biomarker was observed concerning the three aptasensors. However, RBD-Ap-1 demonstrated a lower specificity (minor EC signal differences) than RBD-Ap-2 and N-Ap. In the first two weeks after the onset of symptoms, the concentration of SARS-CoV-2 RNA in clinical samples ranged from 104 to 1010 copies mL−1 (corresponding 0.13 fg mL−1 to 1.30 ng mL−1 S1 proteins).13 Thus, our biochips may provide valuable early-stage detection of antigens from suspected COVID-19 patients in a clinically relevant scenario.
Finally, we explored the possibility of one-step and specific detection of SARS-CoV-2 virus from unprocessed clinical samples (Fig. S8, ESI†). Blind testing of 5 clinical swab samples showed that the analysis results of RBD-Ap-2 were in accordance with that of N-Ap, indicating the accuracy of 100% to discriminate between positive and negative samples (Fig. 2m and Table S3, ESI†). In comparison, RBD-Ap-1 could not correctly identify all the samples due to its relatively low selectivity (Fig. 2j).
In summary, our biochips exhibited a low LoD, a broad dynamic range, and excellent selectivity concerning SARS-CoV-2 S and N proteins, and IgG antibody diagnosis, which were highly competitive compared to published data (Table S4, ESI†). Our biochip array provides one-step, low-cost (2.845 $), convenient and reliable COVID-19 testing, which allows citizens, even in the poorest and most remote locations, to achieve self-testing at home or rapid diagnosis (10 min) in community settings (Table S4, ESI†).
This work was supported by the National Research Program (NRP 78 Covid-19, 198258) of the Swiss National Science Foundation (SNSF). L. Z. was in part supported by the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-061). Fuze Jiang thanks China Scholarship Council for the financial support.
Conflicts of interest
There are no conflicts to declare.References
- H. Lukas, C. Xu, Y. Yu and W. Gao, ACS Nano, 2020, 14, 16180–16193 CrossRef CAS PubMed.
- A. Tong, T. C. Sorrell, A. J. Black, C. Caillaud, W. Chrzanowski, E. Li, D. Martinez-Martin, A. McEwan, R. Wang and A. Motion, Nat. Biotechnol., 2021, 39, 144–147 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- F. Mahmoudinobar, D. Britton and J. K. Montclare, Protein Eng., Des. Sel., 2021, 34, 1–10 CrossRef PubMed.
- J. D. Whitman, J. Hiatt, C. T. Mowery, B. R. Shy, R. Yu, T. N. Yamamoto, U. Rathore, G. M. Goldgof, C. Whitty and J. M. Woo, Nat. Biotechnol., 2020, 38, 1174–1183 CrossRef CAS PubMed.
- K. Ramdas, A. Darzi and S. Jain, Nat. Med., 2020, 26, 810–811 CrossRef CAS PubMed.
- S. Campuzano, M. Pedrero, P. Yanez-Sedeno and J. M. Pingarron, Sens. Actuators, B, 2021, 345, 130349 CrossRef CAS.
- D. L. Bugembe, M. V.-T. Phan, I. Ssewanyana, P. Semanda, H. Nansumba, B. Dhaala, S. Nabadda, A. N. O’Toole, A. Rambaut, P. Kaleebu and M. Cotten, Nat. Microbiol., 2021, 6, 1094–1101 CrossRef CAS PubMed.
- A. S. Peinetti, R. J. Lake, W. Cong, L. Cooper, Y. Wu, Y. Ma, G. T. Pawel, M. E. Toimil-Molares, C. Trautmann, L. Rong, B. Marinas, O. Azzaroni and Y. Lu, Sci. Adv., 2021, 7, eabh2848 CrossRef CAS PubMed.
- L. Zhang, X. Fang, X. Liu, H. Ou, H. Zhang, J. Wang, Q. Li, H. Cheng, W. Zhang and Z. Luo, Chem. Commun., 2020, 56, 10235–10238 RSC.
- Y. Song, J. Song, X. Wei, M. Huang, M. Sun, L. Zhu, B. Lin, H. Shen, Z. Zhu and C. Yang, Anal. Chem., 2020, 92, 9895–9900 CrossRef CAS PubMed.
- M. Sun, S. Liu, X. Wei, S. Wan, M. Huang, T. Song, Y. Lu, X. Weng, Z. Lin, H. Chen, Y. Song and C. Yang, Angew. Chem., Int. Ed., 2021, 60, 10266–10272 CrossRef CAS PubMed.
- M. Norman, T. Gilboa, A. F. Ogata, A. M. Maley, L. Cohen, E. L. Busch, R. Lazarovits, C. P. Mao, Y. Cai, J. Zhang, J. E. Feldman, B. M. Hauser, T. M. Caradonna, B. Chen, A. G. Schmidt, G. Alter, R. C. Charles, E. T. Ryan and D. R. Walt, Nat. Biomed. Eng., 2020, 4, 1180–1187 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc01598f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |