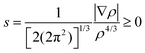Open Access Article
Open Access ArticleRight- and left-handed PPI helices in cyclic dodecapeptoids†‡
Giovanni
Pierri
 ,
Rosaria
Schettini
,
Rosaria
Schettini
 ,
Francesco F.
Summa
,
Francesco F.
Summa
 ,
Francesco
De Riccardis
,
Francesco
De Riccardis
 ,
Guglielmo
Monaco
,
Guglielmo
Monaco
 ,
Irene
Izzo
,
Irene
Izzo
 * and
Consiglia
Tedesco
* and
Consiglia
Tedesco
 *
*
Department of Chemistry and Biology “A. Zambelli”, University of Salerno, via Giovanni Paolo II, 132, I-84084 Fisciano, Italy. E-mail: iizzo@unisa.it; ctedesco@unisa.it
First published on 30th March 2022
Abstract
Enantiomorphic right- and left-handed polyproline type I helices in four cyclic dodecapeptoids with methoxyethyl and propargyl side chains are observed for the first time by single crystal X-ray diffraction. The peculiar absence of NH⋯OC hydrogen bonds in peptoids unveils the role of intramolecular backbone-to-backbone CO⋯CO interactions and CH⋯OC hydrogen bonds in the stabilization of the macrocycle conformation. Moreover, intramolecular backbone-side chain C5 CH⋯OC hydrogen bonds emerge as a stabilizing factor.
Cyclic peptoids (cyclic oligomers of N-substituted glycine residues) constitute a flexible platform to develop functional molecules with adjustable structures, easy synthesis, and excellent stability to enzymatic degradation.1 Although more than 200 members are known, the landscape of larger oligomers (with more than ten residues) is almost completely uncharted. Yet, macrocyclic dodecadepsipeptides, as valinomycin,2 are known for their ability to evoke biological activities.3 Antibiotic, antifungal, antimicrobial, antiproliferative, immunosuppressant and nematicidal activities were observed for naturally occurring dodecameric cyclic peptides.4
While larger peptides show a certain degree of conformational stability, in peptoids the isoenergetic cis/trans amide bond conformations and the absence of the Cα stereogenic centre may lead to unwanted backbone flexibility. However, cyclization and judicious incorporation of side chains represent a successful strategy to generate secondary structural motifs.5,6
In our ongoing studies devoted to the solid state assembly of cyclic peptoids,7 we designed cyclooligomers as building blocks for the accretion of solid state supramolecular architectures. Combining the appropriate side chains and ring size it was possible to induce different solid state assemblies such as columns,8 tubes,9 and layers.9b,10 Recently, we reported the synthesis of the N-propargylated cyclic dodecapeptoid cyclo-(Npa)12 (compound 1 in Fig. 1), which proved to be a useful scaffold for the preparation of the iminosugar-cyclopeptoid conjugate displaying the largest binding enhancement for α-mannosidase inhibition reported so far.11,12
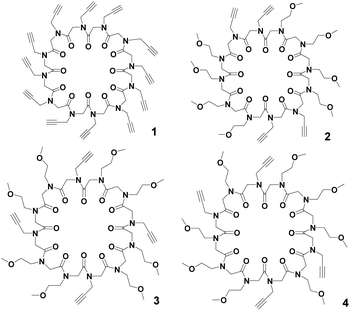 | ||
| Fig. 1 Cyclic dodecapeptoids: cyclo-(Npa)121, cyclo-[(Npa)2(Nme)4]22, cyclo-[(Npa)(Nme)2]43 and cyclo-[(Npa)(Nme)3(Npa)(Nme)]24; Npa = N-(propargyl)glycine, Nme = N-(methoxyethyl)glycine. | ||
To extend our structural analysis of cyclic peptoids to cyclododecamers, we succeeded in crystallizing cyclo-(Npa)121. We also synthesized and crystallized three further cyclic dodecamers: cyclo-[(Npa)2(Nme)4]22, cyclo-[(Npa)(Nme)2]43 and cyclo-[(Npa)(Nme)3(Npa)(Nme)]24 (Fig. 1).
The linear precursors of macrocyclic compounds were prepared on a 2-chlorotrityl resin following the sub-monomer approach.13 Repeating iteratively a two-step sequence (acylation nucleophilic substitution with propargyl or methoxyethyl amine), we obtained the linear dodecamers in quantitative yields. Cleavage from the resin and subsequent cyclization in high dilution conditions in the presence of HATU as the coupling reagent afforded the crude compounds 2–4, which were recovered after chromatography purification in 25%, 24% and 36% overall yields, respectively (see the ESI‡ for details).
For all compounds, single crystals suitable for X-ray diffraction analysis were obtained by slow evaporation, from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 methanol/toluene solution for compound 1, from a 1
1 methanol/toluene solution for compound 1, from a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ethyl acetate/toluene solution for 2, from an i-PrOH solution for 3, and from a 1
1 ethyl acetate/toluene solution for 2, from an i-PrOH solution for 3, and from a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 chloroform/diethyl ether solution for 4. Crystal data and refinement details are reported in the ESI.‡
2 chloroform/diethyl ether solution for 4. Crystal data and refinement details are reported in the ESI.‡
Interestingly, in all compounds the observed sequence of the amide group conformations is ccccctccccct, which is the largest stretch of consecutive cis peptoid junctions in cyclic peptoids, exceeding by one residue that observed in a cyclic peptoid nonamer with (S)-N-(1-phenylethyl) side chains.14
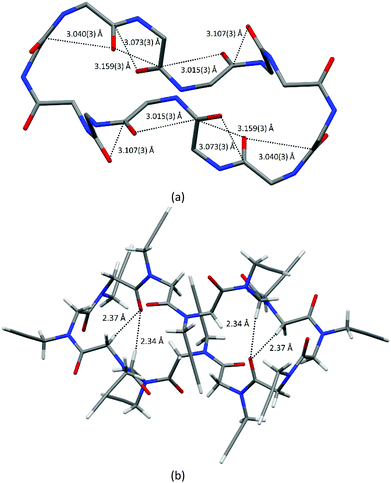
The backbone torsion angles ω, φ and ψ are reported in Table S2 (ESI‡). Since all the macrocycles possess a crystallographic inversion centre, five cis residues exhibit a typical right-handed PPI helical conformation (with φ and ψ values close to −75° and 160°, respectively), while the opposite ones exhibit a left-handed PPI helical arrangement with opposite φ and ψ values. In this way, two helices with opposite screw sense are bridged by two trans amide bonds (Fig. 2 and Fig. S2, ESI‡).
PPI helices were previously observed in linear peptoid oligomers with α-chiral side chains (including aromatic15 or alkyl16–18 groups). In our cyclododecamers both methoxyethyl and propargyl side chains can induce a cis sequence of amide bonds. On the other side, methoxyethyl side chains are associated with trans peptoid bonds in compounds 2–4.
Interestingly, the observed molecular conformation appears as an intrinsic feature of cyclic dodecapeptoids as all four crystal structures were obtained in different crystallization conditions.
Indeed, intramolecular CO⋯CO interactions and CH⋯OC hydrogen bonds, involving backbone atoms, contribute to stabilizing the observed conformation.
Both one-sided and reciprocal CO⋯CO interactions are observed in the macrocycle, as shown by short contacts below 3.22 Å between adjacent carbonyl groups (see Table S3, ESI,‡Fig. 2a and Fig. S2, ESI‡).
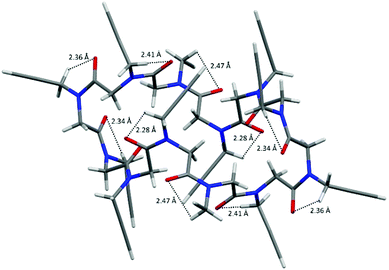
Intramolecular backbone-to-backbone CH⋯OC contacts (indicated as C > 7 CH⋯OC bonds) arise between the carbonyl groups and the opposite methylene hydrogen atoms three and two residues before (2.34 Å and 2.37 Å in 1, 2.38 Å and 2.40 Å in 2, 2.36 Å and 2.43 Å in 3, 2.40 Å and 2.46 Å in 4; see also Fig. 2b, Fig. S3 and Table S4, ESI‡). Noteworthily, analogous interactions were observed between the carbonyl groups of the ith residue and the backbone methylene hydrogen atoms up to four residues before in linear oligopeptoids.17
By further investigating the intramolecular interactions, the presence of a conspicuous number of intramolecular backbone-to-side chain C5 CH⋯OC hydrogen bonds caught our attention. C5 intraresidue NH⋯OC hydrogen bonds were proposed by Benedetti and Toniolo19 and recently discussed further by Newberry and Raines in the view of their biological importance as possible promoters of amyloid fibrils.20 They find out that C5 intraresidue NH⋯OC hydrogen bonds are energetically relevant if donor–acceptor distances are less than 2.5 Å.
As shown in Fig. 3 (see also Fig. S4 and Table S5, ESI‡), in all four molecular structures the cis carbonyl oxygen atoms are always at a distance less than 2.5 Å from one methylene hydrogen atom of the following N-linked side chains, and thus a ring of 5 atoms (namely the carbonyl O and C atoms, the N atom, the side chain methylene C and one of its H atoms) is formed and a C5 CH⋯OC hydrogen bond may be defined.
 | ||
| Fig. 3 C5 CH⋯OC hydrogen bonds in cyclo-(Npa)121: cis carbonyl oxygen atoms are always at a distance less than 2.5 Å from one methylene hydrogen atom of the following N-linked side chains. | ||
Indeed, intramolecular C5 CH⋯OC hydrogen bonds are almost ubiquitous in the X-ray molecular structures of all peptoids and could have a role in the stabilization of the cis conformation.
As for the molecular assembly in the solid state (Fig. S8, ESI‡), we observe that three out of four compounds crystallize in solvate forms, as this seems to be a peculiar feature of larger cyclic peptoids.7 For the solvate crystal structures of compounds 1, 2 and 3, we applied a solvent mask procedure for disordered guest molecules (see the ESI‡ for further details).21
In compounds 2, 3 and 4 cyclic peptoid molecules form columns by aligning on top of each other along the shortest axis. Two opposite propargyl side chains are involved in the columnar assembly forming intermolecular C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CH⋯OC hydrogen bonds (Fig. S9, ESI‡). In compound 1 methanol molecules bridge cyclic peptoid molecules along the shortest axis (a axis, Fig. S10, ESI‡). The crystal packing of compounds 1 and 2 features small cavities occupied by the guest molecules, while in 3 guest molecules occupy channel voids along the b axis (Fig. S11, ESI‡), and one water molecule bridges the cyclic peptoid molecules along the b axis.
CH⋯OC hydrogen bonds (Fig. S9, ESI‡). In compound 1 methanol molecules bridge cyclic peptoid molecules along the shortest axis (a axis, Fig. S10, ESI‡). The crystal packing of compounds 1 and 2 features small cavities occupied by the guest molecules, while in 3 guest molecules occupy channel voids along the b axis (Fig. S11, ESI‡), and one water molecule bridges the cyclic peptoid molecules along the b axis.
The nature of non-covalent (NC) intramolecular interactions in cyclo-(Npa)121 was investigated by QTAIM,22 searching for bond critical points (BCPs), using the SYSMOIC package,23 taking as inputs the wavefunction files obtained using Gaussian1624 at the B3LYP/6-311G(d,p) level, either on the experimental geometry or on a fully optimized geometry at the same level, to overcome the packing influence.
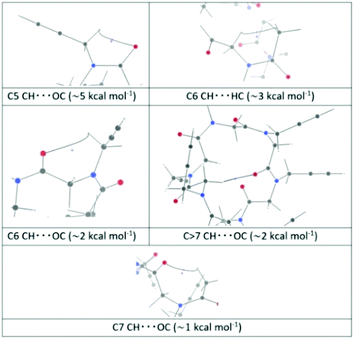
A third fully optimized B3LYP/6-311+G(d,p) computation was performed. The fully optimized B3LYP/6-311G(d,p) computation gave the richest collection of NC-BCPs (not associated to standard covalent bonds): 32 NC-BCPs as opposed to 26 for the other two computations.
A rough quantification of the corresponding interaction energies, through the Espinosa–Molins–Lecomte (EML) equation,25–27 gave 18 values exceeding 1 kcal mol−1 (see the ESI‡ for details). These interactions, listed in order of decreasing interaction energy, correspond to 4 C5 CH⋯OC bonds (∼5 kcal mol−1), 4 C6 CH⋯HC bonds (∼2.8 kcal mol−1), 4 C6 CH⋯OC bonds (∼2 kcal mol−1), 4 C > 7 CH⋯OC bonds (∼2 kcal mol−1), and 2 C7 CH⋯OC bonds (∼1.1 kcal mol−1). Representative bond paths (BPs) for these interactions are shown in Fig. 4, together with the BCP and the closest ring critical point (RCP).
The C6 CH⋯HC interactions are a sort of the much debated28–34 H–H bonding;35 in this case the bond path (BP) does not connect two hydrogen atoms, but a carbon and a hydrogen atom, as previously reported.33
In the other two BCP computations, besides other differences, the lack of the strongest C5 CH⋯OC is most striking (Fig. S12 and Table S8, ESI‡). In fact, it is known that changes in local geometry, induced by side groups (as in homologous compounds)36 or even by vibrations,37 can lead to the disappearance of BCPs. To check the genuine nature of the HB interactions revealed only by the fully optimized B3LYP/6-311G(d,p) calculation, we resorted to the NCI approach,36–38 which is rooted in the reduced density gradient
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sign(λ2) reveals a downward peak on the negative/positive semi-axis for each attractive/repulsive (BCP/RCP-like) interaction. The presence of an interaction can then be assessed even if the downward peak is not deep enough to reach the zero line for s, i.e. for a missing BCP.34,36
sign(λ2) reveals a downward peak on the negative/positive semi-axis for each attractive/repulsive (BCP/RCP-like) interaction. The presence of an interaction can then be assessed even if the downward peak is not deep enough to reach the zero line for s, i.e. for a missing BCP.34,36
Fig. S13 and S14 (ESI‡) indicate that the C5 CH⋯OC interaction is indeed associated with an attractive and a repulsive interaction (leftmost and rightmost peaks, respectively, traces of the BCP and RCP), in all three computations, although only in one of them the peak reaches the zero line for s. Thus, in this case, the lower level B3LYP/6-311G(d,p) calculation allowed visualization and characterization of the C5 CH⋯OC interaction using standard QTAIM.
In conclusion, in the present contribution we report for the first time the X-ray crystal structures of four cyclic dodecapeptoids, decorated with propargyl and methoxyethyl side chains. An unprecedented sequence of the amide group conformations ccccctccccct has been unveiled for all these compounds, regardless of crystallization conditions. Two PPI helices, right- and left-handed, are connected by trans amide bonds. Once again, our results clearly indicate that peptoids provide an extraordinary platform for evidencing the influence of weak interactions (as CO⋯CO and CH⋯OC interactions) on the stabilization of molecular conformations, otherwise hidden by the presence of stronger interactions as the NH⋯OC hydrogen bonds in peptides. In particular, intraresidue C5 CH⋯OC hydrogen bonds, similar to the intramolecular NH⋯OC observed in peptides, are a general feature of all X-ray molecular structures of cyclic peptoids and are well defined in terms of both geometry and electrons, unveiling their role in the stabilization of the macrocycle conformation.
Financial support from the University of Salerno (FARB2021) is gratefully acknowledged.
Conflicts of interest
There are no conflicts to declare.Notes and references
-
(a) F. De Riccardis, Eur. J. Org. Chem., 2020, 2981–2994 CrossRef CAS
; (b) A. M. Webster and S. L. Cobb, Chem. – Eur. J., 2018, 24, 7560–7573 CrossRef CAS PubMed
; (c) B. Yoo, S. B. Y. Shin, M. L. Huang and K. Kirshenbaum, Chem. – Eur. J., 2010, 16, 5528–5537 CrossRef CAS PubMed
.
- H. Brockmann and G. Schmidt-Kastner, Chem. Ber., 1955, 88, 57–61 CrossRef CAS
.
-
(a) P. Lauger, Science, 1972, 178, 24–30 CrossRef CAS PubMed
; (b) Z. Su, X. Ran, J. J. Leitch, A. L. Schwan, R. Faragher and J. Lipkowski, Langmuir, 2019, 35, 16935–16943 CrossRef CAS PubMed
; (c) D. Zhang, Z. Ma, H. Chen, Y. Lu and X. Chen, Biomed. J., 2020, 43, 414–423 CrossRef PubMed
.
-
(a) M. Inman, H. L. Dexter and C. J. Moody, Org. Lett., 2017, 19, 3454–3457 CrossRef CAS PubMed
; (b) S. Luo, A. Krunic, H.-S. Kang, W.-L. Chen, J. L. Woodard, J. R. Fuchs, S. M. Swanson and J. Orjala, J. Nat. Prod., 2014, 77, 1871–1880 CrossRef CAS PubMed
; (c) F. Boyaud, Z. Mahiout, C. Lenoir, S. Tang, J. Wdzieczak-Bakala, A. Witczak, I. Bonnard, B. Banaigs, T. Ye and N. Inguimbert, Org. Lett., 2013, 15, 3898–3901 CrossRef CAS PubMed
; (d) B. Thern, J. Rudolph and J. Gunther, Angew. Chem., Int. Ed., 2002, 41, 2307–2309 CrossRef CAS
; (e) J. B. MacMillan and T. F. Molinski, Org. Lett., 2002, 4, 1883–1886 CrossRef CAS PubMed
; (f) H. Morita, A. Gonda, K. Takeya, H. Itokawa, T. Hirano, K. Oka and O. Shirota, Tetrahedron, 1997, 53, 7469–7478 CrossRef CAS
.
- K. L. Bicker and S. L. Cobb, Chem. Commun., 2020, 56, 11158–11168 RSC
.
-
(a) A. D’Amato, G. Pierri, C. Tedesco, G. Della Sala, I. Izzo, C. Costabile and F. De Riccardis, J. Org. Chem., 2019, 84, 10911–10928 CrossRef PubMed
; (b) A. D’Amato, G. Pierri, C. Costabile, G. Della Sala, C. Tedesco, I. Izzo and F. De Riccardis, Org. Lett., 2018, 20, 640–643 CrossRef PubMed
; (c) S. B. Y. Shin, B. Yoo, L. J. Todaro and K. Kirshenbaum, J. Am. Chem. Soc., 2007, 129, 3218–3225 CrossRef CAS PubMed
.
- C. Tedesco, L. Erra, I. Izzo and F. De Riccardis, CrystEngComm, 2014, 16, 3667–3687 RSC
.
-
(a) E. Macedi, A. Meli, F. De Riccardis, P. Rossi, V. J. Smith, L. J. Barbour, I. Izzo and C. Tedesco, CrystEngComm, 2017, 19, 4704–4708 RSC
; (b) C. Tedesco, E. Macedi, A. Meli, G. Pierri, G. Della Sala, C. Drathen, A. N. Fitch, G. B. M. Vaughan, I. Izzo and F. De Riccardis, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2017, 73, 399–412 CrossRef CAS PubMed
; (c) A. Meli, E. Macedi, F. De Riccardis, V. J. Smith, L. J. Barbour, I. Izzo and C. Tedesco, Angew. Chem., Int. Ed., 2016, 55, 4679–4682 CrossRef CAS PubMed
; (d) I. Izzo, G. Ianniello, C. De Cola, B. Nardone, L. Erra, G. Vaughan, C. Tedesco and F. De Riccardis, Org. Lett., 2013, 15, 598–601 CrossRef CAS PubMed
; (e) G. Pierri, A. Landi, E. Macedi, I. Izzo, F. De Riccardis, R. Dinnebier and C. Tedesco, Chem. – Eur. J., 2020, 26, 14320–14323 CrossRef CAS PubMed
; (f) G. Pierri, M. Corno, E. Macedi, M. Voccia and C. Tedesco, Cryst. Growth Des., 2021, 21, 897–907 CrossRef CAS PubMed
.
-
(a) C. Tedesco, R. Schettini, V. Iuliano, G. Pierri, A. N. Fitch, F. De Riccardis and I. Izzo, Cryst. Growth Des., 2019, 19, 125–133 CrossRef CAS
; (b) C. Tedesco, A. Meli, E. Macedi, V. Iuliano, A. G. Ricciardulli, F. De Riccardis, G. B. M. Vaughan, V. J. Smith, L. J. Barbour and I. Izzo, CrystEngComm, 2016, 18, 8838–8848 RSC
; (c) S. B. L. Vollrath, C. Hu, S. Bräse and K. Kirshenbaum, Chem. Commun., 2013, 49, 2317–2319 RSC
.
- G. Pierri, R. Schettini, J. Nuss, R. E. Dinnebier, F. De Riccardis, I. Izzo and C. Tedesco, CrystEngComm, 2020, 22, 6371–6384 RSC
.
-
(a) M. L. Lepage, J. P. Schneider, A. Bodlenner, A. Meli, F. De Riccardis, M. Schmitt, C. Tarnus, N.-T. Nguyen-Huynh, Y.-N. Francois, E. Leize-Wagner, C. Birck, A. Cousido-Siah, A. Podjarny, I. Izzo and P. Compain, Chem. – Eur. J., 2016, 22, 5151–5155 CrossRef CAS PubMed
; (b) M. L. Lepage, A. Meli, A. Bodlenner, C. Tarnus, F. De Riccardis, I. Izzo and P. Compain, Beilstein J. Org. Chem., 2014, 10, 1406–1412 CrossRef PubMed
.
- E. Howard, A. Cousido-Siah, M. L. Lepage, J. P. Schneider, A. Bodlenner, A. Mitschler, A. Meli, I. Izzo, H. A. Alvarez, A. Podjarny and P. Compain, Angew. Chem., Int. Ed., 2018, 57, 8002–8006 CrossRef CAS PubMed
.
- R. N. Zuckermann, J. M. Kerr, S. B. H. Kent and W. H. Moos, J. Am. Chem. Soc., 1992, 114, 10646–10647 CrossRef CAS
.
- G. L. Butterfoss, B. Yoo, J. N. Jaworski, I. Chorny, K. A. Dill, R. N. Zuckermann, R. Bonneau, K. Kirshenbaum and V. A. Voelz, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14320–14325 CrossRef CAS PubMed
.
- J. R. Stringer, J. A. Crapster, I. A. Guzei and H. E. Blackwell, J. Am. Chem. Soc., 2011, 133, 15559–15567 CrossRef CAS PubMed
.
- C. W. Wu, K. Kirshenbaum, T. J. Sanborn, J. A. Patch, K. Huang, K. A. Dill, R. N. Zuckermann and A. E. Barron, J. Am. Chem. Soc., 2003, 125, 13525–13530 CrossRef CAS PubMed
.
- G. Angelici, N. Bhattacharjee, O. Roy, S. Faure, C. Didierjean, L. Jouffret, F. Jolibois, L. Perrin and C. Taillefumier, Chem. Commun., 2016, 52, 4573–4576 RSC
.
- O. Roy, G. Dumonteil, S. Faure, L. Jouffret, A. Kriznik and C. Taillefumier, J. Am. Chem. Soc., 2017, 139, 13533–13540 CrossRef CAS PubMed
.
-
(a) C. Toniolo and E. Benedetti, CRC Crit. Rev. Biochem., 1980, 9, 1–44 CrossRef CAS PubMed
; (b) M. Crisma, F. Formaggio, C. Alemán, J. Torras, C. Ramakrishnan, N. Kalmankar, P. Balaram and C. Toniolo, Pept. Sci., 2018, 110, e23100 CrossRef
.
- R. W. Newberry and R. T. Raines, Nat. Chem. Biol., 2016, 12, 1084–1088 CrossRef CAS PubMed
.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, J. Appl. Crystallogr., 2009, 42, 339–341 CrossRef CAS
.
-
R. F. W. Bader, Atoms in Molecules - A Quantum Theory, Oxford University Press, Oxford, 1990 Search PubMed
.
- G. Monaco, F. F. Summa and R. Zanasi, J. Chem. Inf. Model., 2021, 61, 270–283 CrossRef CAS PubMed
.
-
M. J. Frisch, et al., Gaussian16, Revision C.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed
.
- E. Espinosa, E. Molins and C. Lecomte, Chem. Phys. Lett., 1998, 285, 170–173 CrossRef CAS
.
- A. Romanova, K. Lyssenko and I. Ananyev, J. Comput. Chem., 2018, 39, 1607–1616 CrossRef CAS PubMed
.
- M. Jabłoński and G. Monaco, J. Chem. Inf. Model., 2013, 53, 1661–1675 CrossRef PubMed
.
- J. Hernández-Trujillo and C. F. Matta, Struct. Chem., 2007, 18, 849–857 CrossRef
.
- S. Grimme, C. Mück-Lichtenfeld, G. Erker, G. Kehr, H. Wang, H. Beckers and H. Willner, Angew. Chem., Int. Ed., 2009, 48, 2592–2595 CrossRef CAS PubMed
.
- A. M. Pendás and J. Hernández-Trujillo, J. Chem. Phys., 2012, 137, 134101–134109 CrossRef PubMed
.
- I. Cukrowski, J. H. de Lange, A. S. Adeyinka and P. Mangondo, Comput. Theor. Chem., 2015, 1053, 60–76 CrossRef CAS
.
- P. Della Porta, R. Zanasi and G. Monaco, J. Comput. Chem., 2015, 36, 707–716 CrossRef CAS PubMed
.
- B. Landeros-Rivera, V. Jancik, R. Moreno-Esparza, D. Martínez Otero and J. Hernández-Trujillo, Chem. – Eur. J., 2021, 27, 11912–11918 CrossRef CAS PubMed
.
- R. Laplaza, R. A. Boto, J. Contreras-García and M. M. Montero-Campillo, Phys. Chem. Chem. Phys., 2020, 22, 21251–21256 RSC
.
- C. F. Matta, J. Hernández-Trujillo, T. Ting-Hua and R. F. W. Bader, Chem. – Eur. J., 2003, 9, 1940–1951 CrossRef CAS PubMed
.
- J. R. Lane, J. Contreras-García, J.-P. Piquemal, B. J. Miller and H. G. J. Kjaergaard, J. Chem. Theory Comput., 2013, 9, 3263–3266 CrossRef CAS PubMed
.
- C. Foroutan-Nejad, S. Shahbazian and R. Marek, Chem. – Eur. J., 2014, 20, 10140–10152 CrossRef CAS PubMed
.
- E. R. Johnson, S. Keinan, P. Mori-Sánchez, J. Contreras-García, A. J. Cohen and W. Yang, J. Am. Chem. Soc., 2010, 132, 6498–6506 CrossRef CAS PubMed
.
Footnotes |
| † Dedicated to Prof. Riccardo Zanasi on occasion of his 70th birthday. |
| ‡ Electronic supplementary information (ESI) available: Synthetic procedures, characterization, crystallographic and computational details. CCDC 2117235–2117238. For ESI and crystallographic data in CIF or other electronic format see https://doi.org/10.1039/d2cc00682k |
| This journal is © The Royal Society of Chemistry 2022 |