Enhancing cellular sulfane sulfur through β-glycosidase-activated persulfide donors: mechanistic insights and oxidative stress mitigation†
Prerona
Bora
 ,
Manjima B.
Sathian
,
Manjima B.
Sathian
 and
Harinath
Chakrapani
and
Harinath
Chakrapani
 *
*
Department of Chemistry, Indian Institute of Science Education and Research Pune, Pune 411 008, Maharashtra, India. E-mail: harinath@iiserpune.ac.in
First published on 4th February 2022
Abstract
Sulfane sulfur species such as persulfides and polysulfides along with hydrogen sulfide protect cells from oxidative stress and are key members of the cellular antioxidant pool. Here, we report perthiocarbamate-based prodrugs that are cleaved by β-glycosidases to produce persulfide and relatively innocuous byproducts. The β-glucosidase-activated persulfide donor enhances cellular sulfane sulfur and protects cells against lethality induced by elevated reactive oxygen species (ROS).
Redox-active species derived from sulfur play a central role in cellular signalling and metabolism, stress response and homeostasis.1 Persulfides (RS-SH) and polysulfides (RS-(S)nH), which are members of the sulfane sulfur pool, have emerged as important mediators in stress response.2,3 Collectively, the sulfane sulfur pool together with hydrogen sulfide (H2S) is now considered a reservoir of antioxidant species that responds to oxidative stress and protects key cellular components from oxidative damage.4 Hence, in addition to thiols such as glutathione and H2S, persulfides are summoned to counter stress caused by elevated reactive oxygen species (ROS). RS-SH is more nucleophilic than RSH as evidenced by a higher HOMO (51 kJ mol−1 for CysS-SH vs. CysSH) leading to superior reactivity with electrophilic species.5 Persulfides and polysulfides are also better at sequestering reactive oxygen species (ROS) and countering oxidative stress.6 Protein persulfidation, which is an oxidative post translational modification of cysteine residues, not only contributes to redox signalling but also protects cysteines from irreversible oxidation.7 Persulfides and polysulfides are excellent persulfidating agents compared to H2S and hence new strategies to enhance cellular persulfides responsive to fluoride,8 esterase,9 pH,10 light,11 H2O212,13 and nitroreductase14 have been developed. Recently, artificial substrates for 3-mercaptopyruvate sulfurtransferase (3-MST) as enhancers of cellular persulfides was reported.15 Several of the aforementioned strategies have shown promise in mitigating oxidative stress in cellular and animal models (Fig. 1a). However, several of them produce byproducts that are electrophilic, some have poor selectivity across cell types, many have diminished aqueous solubility, and in a few cases, no therapeutic relevance was demonstrated. Here, we report the design and development of a cell permeable β-glycosidase-activated persulfide donor that protects cells from oxidative stress.
 | ||
| Fig. 1 (a) Prodrug strategies to generate sulfane sulfur in cells. (b) Present work; prodrugs of persulfides/polysulfides cleavable by β-glycosidase (1: β-glucoside, 2: β-galactoside). | ||
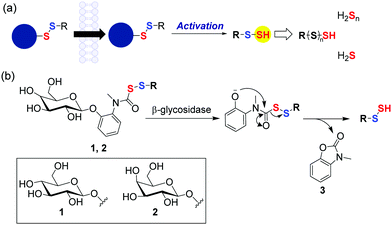
β-Glycosidases are a class of enzymes that cleave glycosidic bonds in oligo/polysaccharides. These enzymes are over-expressed in the gastrointestinal (GI) tract, especially the colon. Elevated levels of β-glycosidases are associated with certain pathophysiologies of the GI tract including inflammatory bowel disorder (IBD), Crohn's disease and ulcerative colitis and cancer.16,17 Increased reactive oxygen species (ROS) leading to collateral damage of the tissue is common in these conditions.18 Thus, enhancing antioxidants is a possible therapeutic approach for such conditions. Several colon-specific prodrug strategies that involve cleavage by β-D-glucosidase, β-D-galactosidase, β-D-xylosidase produced by the intestinal microflora are known.16,19 Hence, taking the aforementioned aspects into consideration, we designed β-glycosidase-activated persulfide prodrugs (Fig. 1b); two series of compounds were considered: one cleavable by β-glucosidase (1) and the other by β-galactosidase (2). Being sugars, these compounds are expected to have enhanced aqueous solubility and would therefore have better applicability. Lastly, unlike certain other donors, once a persulfide is generated, no electrophilic and potentially toxic byproduct is formed.20
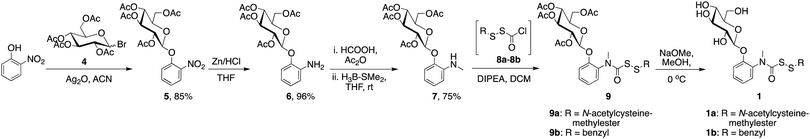
Compound 1a (R = N-acetylcysteine methylester, Scheme 1) was synthesized in 5 steps (Scheme 1). First, the reaction of 2-nitrophenol with 2,3,4,6-tetra-O-acetyl-β-D-glucopyranosyl bromide 4, using a reported protocol21 gave compound 5. This was followed by reduction of the nitro group in 5 to its corresponding aniline 6 by using zinc in HCl. Formylation of the aniline followed by its reduction using borane dimethylsulfide22 gave the N-methylaniline derivative 7. In a separate reaction, N-acetylcysteine methylester (NACMe) was treated with chlorocarbonylsulfenyl chloride to obtain the S-perthiocarbonyl chloride 8a, which was immediately reacted with compound 7 to obtain compound 9.10 Finally, de-protection of acetyl groups of 9 using sodium methoxide in methanol afforded 1a.
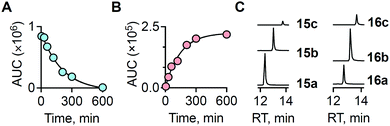
To ascertain the reactivity of the compound towards β-glucosidase, 1a was incubated with β-glucosidase in pH 7.4 phosphate buffer at 37 °C. LC/MS analysis of the reaction mixture revealed a complete decomposition of the compound within 10 h (Fig. 2a). A time course for this decomposition was obtained and curve fitting to a first order equation gave a rate constant of 5.4 × 10−3 min−1 with a half-life of 128 min (Fig. 2a and Fig. S1, ESI†).
Under these conditions, as expected, the formation of N-methyl benzoxazolone byproduct (3) was observed with m/z = 150.0551 [M + H]+ (expected m/z = 150.0555) (Fig. S2, ESI†). Curve fitting gave a rate constant of 7.1 × 10−3 min−1, which is comparable to the rate of decomposition of 1a (Fig. 2b). Together, these data suggest that the cleavage step is the rate determining and once the sugar is cleaved, the release of the persulfide is fast. The ability of 1a to generate persulfides under these conditions was next evaluated. A standard method for the characterization of persulfide species is to trap them with an electrophile to form a covalent adduct, which can be detected using HPLC or LC/MS.
N-(4-Hydroxyphenethyl)-2-iodoacetamide (HPE-IAM) has been previously reported23,24 to be a potent and efficient persulfide/polysulfide alkylating agent (Scheme 2).25 When 1a was co-incubated in the presence of β-glucosidase and HPE-IAM, the appearance of a new peak at 12.36 min with m/z = 387.1042 [M + H]+ was observed that gradually increased over time (Fig. S3, ESI†). It was attributed as the NACMe persulfide adduct, 15a (expected m/z = 387.1048), thus confirming the generation of a persulfide under these conditions (Fig. 2c). We also found evidence for the formation of polysulfide adducts, 15b and 15c under these conditions (Fig. 2c). Additionally, appreciable amounts of H2S as the bis-S-HPE-AM adduct (16a) with m/z = 389.1545 [M + H]+ (expected m/z = 389.1535) was detected along with hydrogen polysulfides (H2Sn, n = 2 and 3) (Fig. 2c). Furthermore, disulfide and trisulfide of NACMe were also detected (Fig. S4, ESI†). Together, these data demonstrate the ability of 1a to produce a gamut of reactive sulfur species, hydrogen sulfide, persulfide and polysulfide.
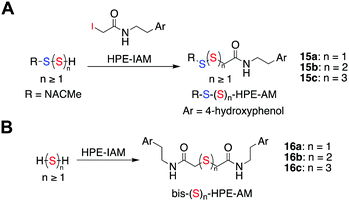 | ||
| Scheme 2 (a) Reaction scheme showing detection of persulfides/polysulfides as their HPE-AM adducts. Ar = 4-hydroxyphenol (b) hydrogen sulfide/polysulfides as their bis-S-HPE-AM adducts. | ||
Having confirmed the in vitro generation of persulfides/polysulfides from 1a, we attempted to study its cell permeability and intracellular generation of sulfane sulfur. Human cytosolic β-glucosidase is present in significant concentrations in the liver, kidney, spleen and colon.26,27 Human colon carcinoma (DLD-1) and hepatocarcinoma (HepG2) cell line were therefore used as model systems. A standard cell viability assay was conducted to assess the cytotoxicity of 1a on DLD-1 and HepG2 cells. No significant toxicity was observed up to a concentration of 100 μM (Fig. S5, ESI†). To detect intracellular sulfane sulfur, the persulfide probe SSP2 was used.28 DLD-1 cells were pre-treated with SSP2 (50 μM) in the presence of CTAB (500 μM) followed by treatment with 1a. A significant increment in the fluorescence signal corresponding to generation of sulfane sulfur was observed upon treatment with 1a (Fig. S6, ESI†). The elevated concentration needed for enhancement of cellular persulfides is likely due to the diminished stability of persulfides in the reducing environment of the cells. Nevertheless, our data suggests that 1a is cell permeable and is able to enhance the levels of intracellular sulfane sulfur pool.
Persulfides/polysulfides have previously been reported to have potent antioxidant effects and are efficient scavengers of ROS. Previously, H2S-NSAID hybrids have been reported to have potent anti-inflammatory effects on models of colitis, attributable to H2S.29 In addition, COS/H2S donors have shown to display cytoprotective effects against xenobiotic induced stress in colon cells.30 To test the protective effects of persulfides/polysulfides in the colon against oxidative stress induced lethality, MGR-1, a cell permeable ROS generator was used to induce oxidative stress (Fig. S7, ESI†).31 DLD-1 cells that were pre-treated with 1a for 12 h were next exposed to MGR-1 for 4 h, following which cell viability was determined using a standard MTT assay. The assay indicates that 1a was able to rescue cells from ROS induced lethality in a dose dependant manner. Under similar conditions, NAC failed to exhibit protective effects (Fig. S8a, ESI†). Next, to further corroborate our results, we used another cell line with an elevated expression of β-glucosidase, HepG2 cell line. When tested on HepG2 cells, similar results were obtained supporting the cytoprotective effects of the cell permeable persulfide donor 1a (Fig. S8b, ESI†).
The next series of compounds (2) were designed to be cleaved by β-galactosidase. Our attempts to synthesize the galactopyranosyl derivative with NACMe persulfide 2a (Fig. 1b; R = N-acetylcysteine methyl ester) were unsuccessful. The benzyl persulfide derivative 2b was however synthesized (Fig. 1b, R = Bn; Scheme S1, ESI†). The rate of cleavage of 2b by β-galactosidase significantly faster than that of the β-glucoside by β-glucosidase (Fig. S9a; rate constant, 6.8 × 10−2 min−1 and t1/2 = 10 min, Fig. S9b, ESI†). The observed differences in the rates of cleavage by these enzymes is similar to previous reports.32,33 The byproduct 3 was observed under these conditions and its rate of formation was calculated to be 8.6 × 10−2 min−1 (Fig. S10, ESI†). Upon co-incubation with β-galactosidase and HPE-IAM, as expected benzyl persulfide/polysulfide Bn(S)nSH (n = 1–3) along with the H2Sn (n = 1–3) adducts were detected (Fig. S11, ESI†). The compound 2b was however found to be moderately cytotoxic when compared with 1a (Fig. S12, ESI†). To verify if the benzyl persulfide contributed to cytotoxicity, the analogue 1b was next synthesized by reacting 7 with the S-perthiocarbonyl chloride generated from benzyl mercaptan (8b) (Scheme 1). The decomposition profile of 1b and persulfide/polysulfide generation profile of 1b was comparable with that of 1a (Fig. S13–S16, ESI†). The analogue 1b was also found to be moderately cytotoxic in cells suggesting the importance of the functional group that is bound to the persulfide (Fig. S17, ESI†).
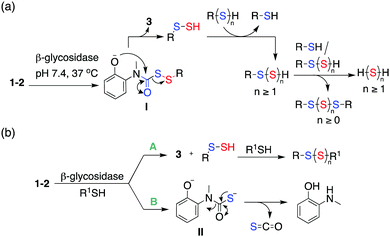
The following mechanism for the formation of persulfides/polysulfides from the glycopersulfide donors is proposed (Scheme 3a). The persulfide generators undergo decomposition in the presence of β-glycosidases to generate the intermediate phenolate I. Intramolecular cyclization of I produces a persulfide species along with the benzoxazolone byproduct 3. The persulfide species once formed can further react with itself or reduced thiols to generate hydropolysulfide (RSSnH) species. Hydropolysulfides (RSSnH) upon reduction by thiols/persulfides would result in the generation of hydrogen polysulfide (H2Sn, n ≥ 1). A similar perthiocarbamate scaffold has been reported by Khodade et al. as precursors to persulfides that reacts with thiols to generate carbonyl sulfide (COS) as well.10 COS undergoes hydrolysis to produce H2S and this reaction is accelerated by carbonic anhydrase, an enzyme that is widely prevalent in cells.34 Hence, in the presence of β-glycosidase and thiols, two possible parallel pathways A and B are possible (Scheme 3b). Pathway A indicates cleavage by β-glycosidase that forms the carbamate byproduct 3 with concomitant formation of persulfides/polysulfides. Pathway B indicates the cleavage of the disulfide bond by thiols resulting in the generation of COS, which generates H2S.34 The donor 2b was examined for its reactivity towards biologically relevant thiols like N-acetyl cysteine (NAC) and glutathione (GSH) in the absence of an electrophile. When 2b was co-incubated with β-galactosidase and NAC (5 equiv.) at 37 °C, if pathway B is significant, a reduction in the yield of 3 is expected. Indeed, when this reaction was conducted, a 25% reduction in the yield of the byproduct 3 was observed within 30 min (Fig. S18, ESI†). Mixed disulfides/polysulfides of NAC and benzyl (NAC-SS-Bn and NAC-(S)3-Bn) were also observed, which is indicative of pathway A (Scheme 3b and Fig. S19, ESI†). Whereas with GSH (10 equiv.), a 65% reduction in the formation of 3 was observed (Fig. S20, ESI†). The persulfide produced upon cleavage can also react with the prodrug via pathway B, forming COS. The derivative 1a has a slower rate of cleavage by β-glucosidase derived from almonds with a half-life of 128 mins. Given the propensity of the compounds to react with thiols, it was envisaged that an important pathway for intracellular decomposition of 1a would be pathway B. Previous reports show that human cytosolic β-glucosidases (hCBG) are more efficient at cleavage of β-glucosidic bonds compared to the enzyme derived from plants supporting the relevance of pathway A in mammalian cells.35 Hence, our overall analysis suggests that the use of prodrugs developed herein results in the enhancement of cellular sulfur species, which have a protective effect against elevated ROS.
Financial support was from the Science and Engineering Research Board (CRG/2019/002900), Department of Science and Technology (PB, DST, INSPIRE Scheme), Department of Biotechnology (HC, BH/HRD/NBM-NWB/39/2020-21) and IISER Pune. DST Fund for Improvement of S&T Infrastructure (SR/FST/LSII-043/2016) to the IISER Pune Biology Department for setting up the Biological Mass Spectrometry Facility. The manuscript was written with inputs from all authors. PB and MBS carried out all experiments.
Conflicts of interest
There are no conflicts to declare.Notes and references
- T. V. Mishanina, M. Libiad and R. Banerjee, Nat. Chem. Biol., 2015, 11, 457 CrossRef CAS PubMed.
- M. R. Filipovic, J. Zivanovic, B. Alvarez and R. Banerjee, Chem. Rev., 2018, 118, 1253–1337 CrossRef CAS PubMed.
- C. Yang, N. O. Devarie-Baez, A. Hamsath, X. Fu and M. Xian, Antioxid. Redox Signaling, 2020, 33, 1092–1114 CrossRef CAS PubMed.
- T. Zhang, H. Tsutsuki, K. Ono, T. Akaike and T. Sawa, J. Clin. Biochem. Nutr., 2021, 68, 5–8 CrossRef CAS PubMed.
- E. Cuevasanta, M. Lange, J. Bonanata, E. L. Coitiño, G. Ferrer-Sueta, M. R. Filipovic and B. Alvarez, J. Biol. Chem., 2015, 290, 26866–26880 CrossRef CAS PubMed.
- T. Ida, T. Sawa, H. Ihara, Y. Tsuchiya, Y. Watanabe, Y. Kumagai, M. Suematsu, H. Motohashi, S. Fujii, T. Matsunaga, M. Yamamoto, K. Ono, N. O. Devarie-Baez, M. Xian, J. M. Fukuto and T. Akaike, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7606–7611 CrossRef CAS PubMed.
- J. Zivanovic, E. Kouroussis, J. B. Kohl, B. Adhikari, B. Bursac, S. Schott-Roux, D. Petrovic, J. L. Miljkovic, D. Thomas-Lopez, Y. Jung, M. Miler, S. Mitchell, V. Milosevic, J. E. Gomes, M. Benhar, B. Gonzalez-Zorn, I. Ivanovic-Burmazovic, R. Torregrossa, J. R. Mitchell, M. Whiteman, G. Schwarz, S. H. Snyder, B. D. Paul, K. S. Carroll and M. R. Filipovic, Cell Metab., 2019, 30, 1152–1170 CrossRef CAS PubMed.
- J. Kang, S. Xu, M. N. Radford, W. Zhang, S. S. Kelly, J. J. Day and M. Xian, Angew. Chem., Int. Ed., 2018, 57, 5893–5897 CrossRef CAS PubMed.
- Y. Zheng, B. Yu, Z. Li, Z. Yuan, C. L. Organ, R. K. Trivedi, S. Wang, D. J. Lefer and B. Wang, Angew. Chem., Int. Ed., 2017, 56, 11749–11753 CrossRef CAS PubMed.
- V. S. Khodade, B. M. Pharoah, N. Paolocci and J. P. Toscano, J. Am. Chem. Soc., 2020, 142, 4309–4316 CrossRef CAS PubMed.
- A. Chaudhuri, Y. Venkatesh, J. Das, M. Gangopadhyay, T. K. Maiti and N. D. P. Singh, J. Org. Chem., 2019, 84, 11441–11449 CrossRef CAS PubMed.
- P. Bora, P. Chauhan, S. Manna and H. Chakrapani, Org. Lett., 2018, 20, 7916–7920 CrossRef CAS PubMed.
- C. R. Powell, K. M. Dillon, Y. Wang, R. J. Carrazzone and J. B. Matson, Angew. Chem., Int. Ed., 2018, 57, 6324 CrossRef CAS PubMed.
- Y. Wang, K. M. Dillon, Z. Li, E. W. Winckler and J. B. Matson, Angew. Chem., Int. Ed., 2020, 59, 16698–16704 CrossRef CAS PubMed.
- P. Bora, S. Manna, M. A. Nair, R. R. M. Sathe, S. Singh, V. S. Sreyas Adury, K. Gupta, A. Mukherjee, D. K. Saini, S. S. Kamat, A. B. Hazra and H. Chakrapani, Chem. Sci., 2021, 12, 12939–12949 RSC.
- D. R. Friend and G. W. Chang, J. Med. Chem., 1984, 27, 261–266 CrossRef CAS PubMed.
- H. Englyst, FEMS Microbiol. Lett., 1987, 45, 163–171 CrossRef CAS.
- H. Zhu and Y. R. Li, Exp. Biol. Med., 2012, 237, 474–480 CrossRef CAS PubMed.
- V. R. Sinha and R. Kumria, Pharm. Res., 2001, 18, 557–564 CrossRef CAS PubMed.
- The LD50 of 3 has been found as 890 mg kg−1 with no reported toxicity in mice, in Registry of Toxic Effects of Chemical Substances, ed. D. V. Sweet, US Department of Health and Human Services CDC, 1987 Search PubMed.
- X. Chen, X. Ma, Y. Zhang, G. Gao, J. Liu, X. Zhang, M. Wang and S. Hou, Anal. Chim. Acta, 2018, 1033, 193–198 CrossRef CAS PubMed.
- I. Okamoto, M. Terashima, H. Masu, M. Nabeta, K. Ono, N. Morita, K. Katagiri, I. Azumaya and O. Tamura, Tetrahedron, 2011, 67, 8536–8543 CrossRef CAS.
- H. A. Hamid, A. Tanaka, T. Ida, A. Nishimura, T. Matsunaga, S. Fujii, M. Morita, T. Sawa, J. M. Fukuto, P. Nagy, R. Tsutsumi, H. Motohashi, H. Ihara and T. Akaike, Redox Biol., 2019, 21, 101096 CrossRef PubMed.
- T. Numakura, H. Sugiura, T. Akaike, T. Ida, S. Fujii, A. Koarai, M. Yamada, K. Onodera, Y. Hashimoto, R. Tanaka, K. Sato, Y. Shishikura, T. Hirano, S. Yanagisawa, N. Fujino, T. Okazaki, T. Tamada, Y. Hoshikawa, Y. Okada and M. Ichinose, Thorax, 2017, 72, 1074–1083 CrossRef PubMed.
- T. Akaike, T. Ida, F.-Y. Wei, M. Nishida, Y. Kumagai, M. M. Alam, H. Ihara, T. Sawa, T. Matsunaga, S. Kasamatsu, A. Nishimura, M. Morita, K. Tomizawa, A. Nishimura, S. Watanabe, K. Inaba, H. Shima, N. Tanuma, M. Jung, S. Fujii, Y. Watanabe, M. Ohmuraya, P. Nagy, M. Feelisch, J. M. Fukuto and H. Motohashi, Nat. Commun., 2017, 8, 1177 CrossRef PubMed.
- B. Hultberg and P. A. Öckerman, Clin. Chim. Acta, 1970, 28, 169–174 CrossRef CAS.
- H. M. M. Arafa, Eur. J. Pharmacol., 2009, 616, 58–63 CrossRef CAS PubMed.
- W. Chen, C. Liu, B. Peng, Y. Zhao, A. Pacheco and M. Xian, Chem. Sci., 2013, 4, 2892 RSC.
- S. Fiorucci, S. Orlandi, A. Mencarelli, G. Caliendo, V. Santagada, E. Distrutti, L. Santucci, G. Cirino and J. L. Wallace, Br. J. Pharmacol., 2009, 150, 996–1002 CrossRef PubMed.
- P. Chauhan, K. Gupta, G. Ravikumar, D. K. Saini and H. Chakrapani, Chem. – Asian J., 2019, 14, 4717–4724 CrossRef CAS PubMed.
- D. S. Kelkar, G. Ravikumar, N. Mehendale, S. Singh, A. Joshi, A. K. Sharma, A. Mhetre, A. Rajendran, H. Chakrapani and S. S. Kamat, Nat. Chem. Biol., 2019, 15, 169–178 CrossRef CAS PubMed.
- T. B. Cai, D. Lu, X. Tang, Y. Zhang, M. Landerholm and P. G. Wang, J. Org. Chem., 2005, 70, 3518–3524 CrossRef CAS PubMed.
- E. Calatrava-Pérez, S. A. Bright, S. Achermann, C. Moylan, M. O. Senge, E. B. Veale, D. C. Williams, T. Gunnlaugsson and E. M. Scanlan, Chem. Commun., 2016, 52, 13086–13089 RSC.
- C. P. Chengelis and R. A. Neal, Toxicol. Appl. Pharmacol., 1980, 55, 198–202 CrossRef CAS PubMed.
- S. Tribolo, J.-G. Berrin, P. A. Kroon, M. Czjzek and N. Juge, J. Mol. Biol., 2007, 370, 964–975 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Preparative methods, assay protocols and spectral data. See DOI: 10.1039/d1cc07162a |
| This journal is © The Royal Society of Chemistry 2022 |