Open Access Article
Open Access ArticleMismatch binding ligand upregulated back-splicing reaction producing circular RNA in a cellular model†
Lu
Ni
 ,
Takeshi
Yamada
,
Takeshi
Yamada
 ,
Asako
Murata
,
Asako
Murata
 and
Kazuhiko
Nakatani
and
Kazuhiko
Nakatani
 *
*
SANKEN (The Institute of Scientific and Industrial Research), Osaka University, 8-1 Mihogaoka, Ibaraki 567-0047, Japan. E-mail: nakatani@sanken.osaka-u.ac.jp
First published on 24th February 2022
Abstract
Circular RNA (circRNA) is a covalently closed single-stranded RNA produced from pre-mRNAs via back-splicing reaction, an alternative form of splicing. Here, we show naphthyridine carbamate dimer (NCD) upregulating the production of a circRNA from a pre-mRNA containing NCD-binding site UGGAA/UGGAA in cells, demonstrating the feasibility of small-molecule mediated circRNA production.
CircRNAs are covalently closed single-stranded RNA, without 5′ cap or 3′ polyA tail.1,2 Although circRNAs were initially considered by-products of aberrant splicing,3,4 they have recently received a surge of interest due to their ubiquitous presence in various cell types and tissues across many eukaryotic organisms.5–10 CircRNAs can act as miRNA sponges,10–12 competitive endogenous RNAs for RNA-binding proteins,13,14 competitors to canonical splicing,13 and translation templates.15 Consequently, circRNA dysregulation has become a major focus for the study of disease pathology, as they are implicated in various diseases, especially cancers11,16,17 and neurological disorders.10,18,19
CircRNAs are produced by a unique biogenesis mechanism: back-splicing. Mature mRNAs are produced from pre-mRNA by linear splicing initiated by the attack of the 2′-hydroxyl group of the branch-point adenosine (BPA) to the phosphodiester of 5′ splice site (5′ss) in the same intron (Fig. 1A, dotted line).20 The majority of circRNAs, in contrast, is produced by back-splicing, where the initial attack of BPA's 2′-OH occurs at the 5′ss of the downstream intron (Fig. 1A, solid line, step a). The produced 3′-OH group of the exon then attacks the 3′ss in the upstream intron (step b), forming a covalently closed circRNA.21 Most notably, this back-splicing process is facilitated by base pairing between reverse complementary matches (RCMs, Fig. 1A, yellow arrows) in introns flanking the circularizing exon.22,23 RCMs are highly complementary sequences acting as a cis-regulatory element, forming base pairs between themselves to bring the BPA and the 5′ss of the downstream intron close together.16,24 Bioinformatic analysis further revealed that distinct complementary repeat sequences play the role of RCMs as they are commonly found flanking circularizing exons.25–27
We have developed small molecules binding to mismatched base pairs in RNAs as mismatch binding ligands (MBLs) to regulate the functions of RNAs by facilitating the formation of ligand-stabilized structures.28,29 In this study, we hypothesized that MBLs could upregulate circRNA production by bringing the BPA's 2′-OH in close vicinity of the downstream 5′ss.
We recently reported that one of the MBLs, NCD (Fig. 1B), forms a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex with 5′-UGGAA-3′/5′-UGGAA-3′ motif (hereafter as UGGAA/UGGAA) in duplex RNAs.30 The complex structure was determined by NMR, which revealed the binding of two NCD molecules to four guanines of the internal loop present in the UGGAA/UGGAA with a concomitant flipping out of the adenine bases (Fig. 1B). Here, we report that NCD binding to the engineered RCMs involving a UGGAA/UGGAA motif increased the production of circRNA from exons flanked by those RCMs in HeLa cells. The effect of NCD is sensitively dependent on the presence of the UGGAA/UGGAA motif and NCD concentration, showing the increased formation of circRNA is due to the proximal location of BPA to the 5′ss in back-splicing. We show that circRNA could be regulated by small molecules, offering new opportunities to study their biological functions.
1 complex with 5′-UGGAA-3′/5′-UGGAA-3′ motif (hereafter as UGGAA/UGGAA) in duplex RNAs.30 The complex structure was determined by NMR, which revealed the binding of two NCD molecules to four guanines of the internal loop present in the UGGAA/UGGAA with a concomitant flipping out of the adenine bases (Fig. 1B). Here, we report that NCD binding to the engineered RCMs involving a UGGAA/UGGAA motif increased the production of circRNA from exons flanked by those RCMs in HeLa cells. The effect of NCD is sensitively dependent on the presence of the UGGAA/UGGAA motif and NCD concentration, showing the increased formation of circRNA is due to the proximal location of BPA to the 5′ss in back-splicing. We show that circRNA could be regulated by small molecules, offering new opportunities to study their biological functions.
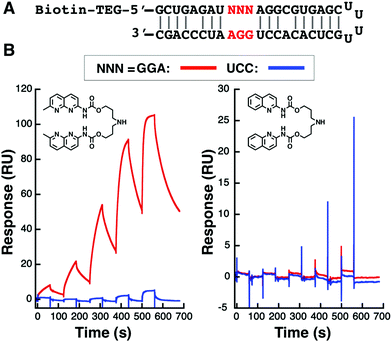
Before starting cellular model experiments, a surface plasmon resonance (SPR) assay was conducted to confirm NCD binding to GGA/GGA. 5′-Biotinylated hairpin RNA containing either a GGA/GGA mismatch or a control full match (UCC/GGA) was separately immobilized on a streptavidin-coated sensor chip. (Fig. 2A) The SPR responses were then measured at different concentrations of NCD. In addition to NCD, QCD, an NCD analog lacking binding ability towards GGA/GGA, was also studied. As shown in Fig. 2B, NCD showed a dose-dependent response to GGA/GGA-containing RNA (red line), which yielded an apparent equilibrium dissociation constant (Kdapp) of 1.29 μM. In contrast, NCD showed a weak response to UCC/GGA-containing RNA (blue line) (Fig. 2B left). On the other hand, QCD showed negligible responses to both hairpin RNA (Fig. 2B right). These results showed the selective and robust binding of NCD to RNA containing the GGA/GGA mismatch site and the lack of binding of QCD to either mismatch or fully matched sequence.
The plasmids used for cellular experiments were designed according to Scheme 1. We used a circRNA expression construct (hereon denoted as p-UAC) coding for a pre-mRNA (pre-mUAC in Fig. 1C. The whole sequence is shown in Fig. S1B, ESI†) previously reported by Wilusz et al. to produce a well-studied circRNA, circZKSCAN1 (stemming from exon 2 and 3 of host gene: ZKSCAN1).17,22Pre-mUAC contains a pair of 36-nt RCMs (shown as yellow arrows in Fig. 1A and C), which was determined to be the minimum length necessary for producing circZKSCAN1 via back-splicing reaction. We then introduced the NCD-binding site in the RCMs of pre-mUAC by changing an original UAC/GUA site in the RCM of pre-mUAC to GGA/GGA, resulting in a construct coding for NCD-responsive pre-mRNA, denoted as pre-mGGA. (Fig. 1C) In addition, pre-mUCC, where a full-match sequence UCC/GGA was introduced into the same position, was designed as a control sequence of pre-mGGA (Fig. 1C and Fig. S1A, ESI†). Hereon, plasmids coding pre-mGGA and pre-mUCC would be denoted as p-GGA and p-UCC, respectively (sequences in Fig. S1C and D and table showing the corresponding transcribed RNA is shown in Fig. S1A, ESI†). The introduced mismatch in pre-mGGA is expected to reduce the thermal stability of the base-paired complementary sequences, resulting in reduced back-splicing efficiency.22 However, binding of NCD to the mismatch site would stabilize the hairpin structure and thus can recover the efficiency loss in back-splicing and circZKSCAN1 production.
To investigate the effect of NCD on the upregulation of circZKSCAN1 production from cells expressing either pre-mGGA or pre-mUCC in the cellular environment. HeLa cells transfected with either p-GGA or p-UCC were treated in the presence of various concentrations of NCD or QCD. The expression levels of circZKSCAN1 were then determined by reverse transcription-quantitative PCR (RT-qPCR)1,31 using a “divergent” primer which amplifies across the back-splice junction (shown as a triangle in Fig. 3A) to detect the circZKSCAN1.
We first confirmed the selectivity of our primer design in detecting circZKSCAN1 stemming from either p-GGA or p-UCC in the presence of endogenous circZKSCAN1. We took advantage of a single nucleotide difference present in the ZKSCAN1 sequence of p-GGA and p-UCC (Shown in orange in Fig. 3A), where thymine in exon 3 of endogenous ZKSCAN1 gene was cytosine (The C is highlighted in red in Fig. S1, ESI†). The 3′ terminus of the forward primer (circZKSCAN1_Fw in Table S1, ESI†) was set to end at the 1-nt difference to selectively amplify circZKSCAN1 stemming from these plasmids (Fig. 3A and Fig. S1, ESI†).32 The specificity of the divergent primer towards circZKSCAN1 stemming from these plasmids was confirmed by comparing the RT-qPCR amplification curves of the non-transfected HeLa cells against HeLa cells transfected with p-GGA treated with 0 or 5 μM NCD (Fig. 3B and Table 1). While the difference in cycle threshold (Ct) values for the internal standard, beta-actin: a housekeeping gene, showed similar values for both non-transfected (black line) and p-GGA transfected cells (dotted line) (19.5, 20.0, respectively), confirming that the same amount of total RNAs were used for analysis. The non-transfected HeLa cells (green line) showed a 7.2 cycle difference in circZKSCAN1 Ct value compared to those transfected with p-GGA (red line). In summary, PCR amplification of circZKSCAN1 stemming from p-GGA is approximately 147-fold more efficient (2−7.2 = 1/147) than endogenous circZKSCAN1. Hereon all descriptions regarding circZKSCAN1 refer to circZKSCAN1 stemming from p-GGA or p-UCC.
| Samples | Average Ct | Std. Dev. |
|---|---|---|
| p-GGA transfected HeLa beta-actin | 19.5 | 0.16 |
| HeLa only beta-actin | 20.0 | 0.14 |
| p-GGA transfected HeLa circZKSCAN1 (5 μM NCD) | 24.6 | 0.38 |
| p-GGA transfected HeLa circZKSCAN1 (0 μM NCD) | 27.2 | 0.30 |
| Non-transfected HeLa circZKSCAN1 | 34.4 | 1.1 |
The specificity of the divergent primer towards circZKSCAN1 was further confirmed by polyacrylamide gel electrophoresis (PAGE) of the amplified products obtained from p-GGA transfected cells treated with 0 and 5 μM NCD, and 5 μM QCD (Fig. S2A, ESI†). Only a single DNA band of the expected size (approx. 140 bp) was detected on the gel under each condition. Moreover, the sequencing of the PCR products, obtained from the cells transfected with p-GGA or p-UCC, showed the back-splicing junction sequence (Fig. S2B, ESI†), corroborating the suitability of our divergent primer for detecting circZKSCAN1.
Additionally, to confirm the circular form of the detected circZKSCAN1, total RNA was treated by Ribonuclease R (RNase R) prior to the RT-qPCR experiment (Fig. S3, ESI†).25 The RQ of circZKSCAN1 showed a significantly higher resistance to RNase R digestion than beta-actin: a linear mRNA, further confirming the circular form of the detected circZKSCAN1.
The comparative Ct (ΔΔCt) method33 was used to determine the relative quantity (RQ) of circZKSCAN1 in the presence of different concentrations of ligands. The raw Ct of circZKSCAN1 were first normalized against mRNA of beta-actin to give a ΔCt value. The ΔCt values of treated samples were then further normalized against those in the absence of ligands (NCD = 0 μM) to provide ΔΔCt. Then the RQ value was calculated from the obtained ΔΔCt value (RQ = 2−ΔΔCt). (All Ct and ΔΔCt values used for calculations of RQ are shown in ESI†).
Cells transfected with p-GGA, expressing pre-mGGA, showed a concentration-dependent increase in the RQ of circZKSCAN1 in the presence of NCD. Where cells treated with 3 μM NCD showed an apparent 3.1-fold increase in RQ of circZKSCAN1 (p < 1 × 10−6), while at the highest concentration of 5 μM NCD, a 5.6-fold increase was observed (p < 1 × 10−6, Fig. 4A). In contrast, no statistically significant change in RQ was observed for cells treated with 5 μM QCD. On the other hand, the cells transfected with p-UCC, expressing pre-mUCC, the fully matched counterpart to pre-mGGA, were also treated similarly with the ligands (Fig. 4B). However, no statistically significant change in RQ of circZKSCAN1 was observed either upon NCD- or QCD- treatment of the cells expressing p-UCC.
Additionally, the changes in endogenous circRNAs in NCD presence were also studied. Here, the expression changes of four circRNAs (circHIKP3, circEPHB4, circFOXO3, and circPVT1) were investigated in the presence of 3 or 5 μM NCD concentration and 5 μM QCD. As shown in Fig. S4 (ESI†), we did not observe the induced circRNA formation of the above four endogenous circRNAs for both p-GGA and p-UCC transfected cells. However, we observed down-regulation of circEPHB4 under 5 μM NCD in both cases. While we searched for potential NCD binding sites within 1000 bp up/downstream intron of the circEPHB4 transcript within its host pre-mRNA, we could not identify any relevant features which may cause the fluctuations mentioned above in expression level.
Finally, to determine the extent of NCD's capacity to induce recovery in circZKSCAN1 expression for pre-mGGA, RQ of circZKCAN1 for cells transfected with p-GGA were calculated relative to circZKCAN1 amount in the cells transfected with the control p-UCC (NCD 0 μM, Fig. S5, ESI†). The results show that in the absence of NCD, cells transfected with p-GGA showed RQ of 0.074 (p < 3 × 10−6) relative to p-UCC transfected cells, indicating a significant reduction in the production of circZKSCAN1 with the introduction of GGA/GGA mismatches into the RCMs. However, in the presence of 5 μM NCD, the cells transfected with p-GGA showed an RQ of 0.41 (p < 0.005), suggesting that under the highest concentration of 5 μM, NCD was capable of inducing 34% (0.41–0.074 = 0.34) recovery in the expression of circZKSCAN1 for pre-mGGA, with regards to pre-mUCC.
In conclusion, the results of the in-cell experiments shown above are consistent with the SPR results shown in Fig. 2B. The apparent increase in circZKSCAN1 production likely follows the hypothesized mechanism, where the NCD binding to the UGGAA/UGGAA internal loop in RCMs facilitated the increase in circZKSCAN1 production by stabilizing RCMs and bringing the BPA to the proximity of 5′ss. In this paper, we have demonstrated that an RNA-binding small molecule can upregulate circZKSCAN1 production in the cellular model by using our rationally designed molecule NCD and engineered construct containing an NCD-binding motif in its introns. Moreover, RCMs and repeat sequences found in introns adjacent to exons forming circRNAs have many unpaired regions, such as stem-loops and bulge loops, when they form duplex structures. These unpaired regions in dsRNA provide potential small molecule binding sites. Thus, the demonstrated model provides a stepping stone in regulating the expression of circRNA in response to the externally added small molecules, presenting one possible strategy to regulate circRNA biogenesis. Recent progress in molecular designs that target particular RNA motifs should accelerate the discovery of RNA-binding small molecules that can regulate the biogenesis of biologically important circRNAs.
Conflicts of interest
There are no conflicts to declare.Notes and references
- W. R. Jeck and N. E. Sharpless, Nat. Biotechnol., 2014, 32, 453–461 CrossRef CAS PubMed.
- L. S. Kristensen, M. S. Andersen, L. V. W. Stagsted, K. K. Ebbesen, T. B. Hansen and J. Kjems, Nat. Rev. Genet., 2019, 20, 675–691 CrossRef CAS PubMed.
- C. Cocquerelle, B. Mascrez, D. Hétuin and B. Bailleul, FASEB J., 1993, 7, 155–160 CrossRef CAS PubMed.
- Z. Pasman, M. D. Been and M. A. Garcia-Blanco, RNA, 1996, 2, 603–610 CAS.
- S. Memczak, M. Jens, A. Elefsinioti, F. Torti, J. Krueger, A. Rybak, L. Maier, S. D. Mackowiak, L. H. Gregersen, M. Munschauer, A. Loewer, U. Ziebold, M. Landthaler, C. Kocks, F. Le Noble and N. Rajewsky, Nature, 2013, 495, 333–338 CrossRef CAS PubMed.
- A. Rybak-Wolf, C. Stottmeister, P. Glažar, M. Jens, N. Pino, S. Giusti, M. Behm, O. Bartok, R. Ashwal-Fluss, M. Herzog, L. Schreyer, P. Papavasileiou, A. Ivanov, M. Öhman, D. Refojo, S. Kadener and N. Rajewsky, Mol. Cell, 2014, 58, 870–885 CrossRef PubMed.
- E. W. Ottesen, D. Luo, J. Seo, N. N. Singh and R. N. Singh, Nucleic Acids Res., 2019, 47, 2884–2905 CrossRef CAS PubMed.
- J. Salzman, R. E. Chen, M. N. Olsen, P. L. Wang and P. O. Brown, PLoS Genet., 2013, 9, e1003777 CrossRef CAS PubMed.
- P. G. Maass, P. Glažar, S. Memczak, G. Dittmar, I. Hollfinger, L. Schreyer, A. V. Sauer, O. Toka, A. Aiuti, F. C. Luft and N. Rajewsky, J. Mol. Med., 2017, 95, 1179–1189 CrossRef CAS PubMed.
- M. Piwecka, P. Glažar, L. R. Hernandez-Miranda, S. Memczak, S. A. Wolf, A. Rybak-Wolf, A. Filipchyk, F. Klironomos, C. A. Cerda Jara, P. Fenske, T. Trimbuch, V. Zywitza, M. Plass, L. Schreyer, S. Ayoub, C. Kocks, R. Kühn, C. Rosenmund, C. Birchmeier and N. Rajewsky, Science, 2017, 357, 6357 CrossRef PubMed.
- Q. Zheng, C. Bao, W. Guo, S. Li, J. Chen, B. Chen, Y. Luo, D. Lyu, Y. Li, G. Shi, L. Liang, J. Gu, X. He and S. Huang, Nat. Commun., 2016, 7, 11215 CrossRef CAS PubMed.
- T. B. Hansen, T. I. Jensen, B. H. Clausen, J. B. Bramsen, B. Finsen, C. K. Damgaard and J. Kjems, Nature, 2013, 495, 384–388 CrossRef CAS PubMed.
- R. Ashwal-Fluss, M. Meyer, N. R. Pamudurti, A. Ivanov, O. Bartok, M. Hanan, N. Evantal, S. Memczak, N. Rajewsky and S. Kadener, Mol. Cell, 2014, 56, 55–66 CrossRef CAS PubMed.
- C. X. Liu, X. Li, F. Nan, S. Jiang, X. Gao, S. K. Guo, W. Xue, Y. Cui, K. Dong, H. Ding, B. Qu, Z. Zhou, N. Shen, L. Yang and L. L. Chen, Cell, 2019, 177, 865 CrossRef CAS PubMed.
- N. R. Pamudurti, O. Bartok, M. Jens, R. Ashwal-Fluss, C. Stottmeister, L. Ruhe, M. Hanan, E. Wyler, D. Perez-Hernandez, E. Ramberger, S. Shenzis, M. Samson, G. Dittmar, M. Landthaler, M. Chekulaeva, N. Rajewsky and S. Kadener, Mol. Cell, 2017, 66, 9–21 CrossRef CAS PubMed.
- J. N. Vo, M. Cieslik, Y. Zhang, S. Shukla, L. Xiao, Y. Zhang, Y.-M. Wu, S. M. Dhanasekaran, C. G. Engelke, X. Cao, D. R. Robinson, A. I. Nesvizhskii and A. M. Chinnaiyan, Cell, 2019, 176, 869 CrossRef CAS PubMed.
- J. Bi, H. Liu, W. Dong, W. Xie, Q. He, Z. Cai, J. Huang and T. Lin, Mol. Cancer, 2019, 18, 1–14 CrossRef CAS PubMed.
- U. Dube, J. L. Del-Aguila, Z. Li, J. P. Budde, S. Jiang, S. Hsu, L. Ibanez, M. V. Fernandez, F. Farias, J. Norton, J. Gentsch, F. Wang and S. Salloway, et al. , Nat. Neurosci., 2019, 22, 1903–1912 CrossRef CAS PubMed.
- L. Kumar, Shamsuzzama, R. Haque, T. Baghel and A. Nazir, Mol. Neurobiol., 2017, 54, 7224–7234 CrossRef CAS PubMed.
- Y. Lee and D. C. Rio, Annu. Rev. Biochem., 2015, 84, 291–323 CrossRef CAS PubMed.
- X. Li, S. Liu, L. Zhang, A. Issaian, R. C. Hill, S. Espinosa, S. Shi, Y. Cui, K. Kappel, R. Das, K. C. Hansen, Z. H. Zhou and R. Zhao, Nature, 2019, 573, 375–380 CrossRef CAS PubMed.
- D. Liang and J. E. Wilusz, Genes Dev., 2014, 28, 2233–2247 CrossRef PubMed.
- X. O. Zhang, H. Bin Wang, Y. Zhang, X. Lu, L. L. Chen and L. Yang, Cell, 2014, 159, 134–147 CrossRef CAS PubMed.
- L.-L. Chen, Nat. Rev. Mol. Cell Biol., 2016, 17, 205–211 CrossRef CAS PubMed.
- W. R. Jeck, J. A. Sorrentino, K. Wang, M. K. Slevin, C. E. Burd, J. Liu, W. F. Marzluff and N. E. Sharpless, RNA, 2013, 19, 141–157 CrossRef CAS PubMed.
- M. Wang, J. Hou, M. Müller-McNicoll, W. Chen and E. M. Schuman, iScience, 2019, 20, 237–247 CrossRef CAS PubMed.
- R. Yoshimoto, K. Rahimi, T. B. Hansen, J. Kjems and A. Mayeda, iScience, 2020, 23, 101345 CrossRef CAS PubMed.
- T. Peng and K. Nakatani, Angew. Chem., Int. Ed., 2005, 44, 7280–7283 CrossRef CAS PubMed.
- T. Peng, C. Dohno and K. Nakatani, Angew. Chem., Int. Ed., 2006, 45, 5623–5626 CrossRef CAS PubMed.
- T. Shibata, K. Nagano, M. Ueyama, K. Ninomiya, T. Hirose, Y. Nagai, K. Ishikawa, G. Kawai and K. Nakatani, Nat. Commun., 2021, 12, 236 CrossRef CAS PubMed.
- A. C. Panda, K. Abdelmohsen and M. Gorospe, Methods Mol. Biol., 2017, 1534, 79–87 CrossRef CAS PubMed.
- S. Lefever, F. Pattyn, J. Hellemans and J. Vandesompele, Clin. Chem., 2013, 59, 1470–1480 CrossRef CAS PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc06936e |
| This journal is © The Royal Society of Chemistry 2022 |