Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A biodegradable amphiphilic poly(aminoester) dendrimer for safe and effective siRNA delivery†
Chi
Ma‡
a,
Dandan
Zhu‡
 a,
Wenyi
Lin
a,
Ying
Li
a,
Yuanzheng
Huang
a,
Huiling
Zhu
a,
Mengyun
Ye
a,
Yang
Wang
bc,
Ling
Peng
a,
Wenyi
Lin
a,
Ying
Li
a,
Yuanzheng
Huang
a,
Huiling
Zhu
a,
Mengyun
Ye
a,
Yang
Wang
bc,
Ling
Peng
 *b and
Xiaoxuan
Liu
*b and
Xiaoxuan
Liu
 *a
*a
aState Key Laboratory of Natural Medicines and Jiangsu Key Laboratory of Drug Discovery for Metabolic Diseases, Center of Advanced Pharmaceuticals and Biomaterials, China Pharmaceutical University, Nanjing 210009, P. R. China. E-mail: xiaoxuanliu@cpu.edu.cn
bAix Marseille Univ, CNRS, Center Interdisciplinaire de Nanoscience de Marseille, UMR 7325, Equipe Labellisée Ligue Contre le Cancer, Marseille, France. E-mail: ling.peng@univ-amu.fr
cHubei Gedian Humanwell Pharmaceutical Co. Ltd., E-zhou, P. R. China
First published on 15th January 2022
Abstract
Small interfering RNA (siRNA)-based therapeutics represent a novel and compelling drug modality, provided that safe and competent vectors are available for their delivery. Here, we report a biodegradable amphiphilic poly(aminoester) dendrimer for effective siRNA delivery. This dendrimer is readily biodegradable upon enzyme action, and harnesses the delivery features of both lipid and polymer vectors thanks to its lipid/dendrimer hybrid structure. This study opens new perspectives for developing biodegradable and biocompatible vectors for siRNA therapeutics.
RNA interference (RNAi)-based small interfering RNA (siRNA) is emerging as a promising therapeutic modality for treating various diseases, in particular those considered as being “undruggable”.1 This is because siRNA can sequence-specifically and efficiently down-regulate any disease-associated genes, leading to potent therapeutic effects. Nevertheless, the chemical composition and anionic nature of siRNA molecules make them vulnerable to various enzymes in the body, and hinder their crossing biomembranes to enter into cells, altogether severely hampering their translation into the clinical setting.2–4 Indeed, in this context, measures to ensure safe and effective siRNA delivery are of paramount importance.
Tremendous efforts have been made in exploring various vectors for siRNA delivery and the most advanced non-viral vectors are lipids and polymers.5–9 Notably, lipid vectors mainly exploit the membrane-fusion mechanism, and polymer vectors primarily induce endocytosis-mediated delivery and endosomal release via the proton-sponge effect. Among the polymeric vectors, dendrimers, a special family of polymers, are particularly attractive for siRNA delivery by virtue of their precise chemical structures, their high degree of molecular uniformity, and their multivalent cooperativity.10,11
In order to harness the delivery advantages of both lipid and polymer vectors, while retaining the unique features of dendrimers, we have recently developed a series of small amphiphilic dendrimers for siRNA delivery.12–16 These dendrimers are lipid/dendrimer hybrids, and harbour hydrophobic lipid chains and hydrophilic poly(amidoamine) (PAMAM) dendrons. They effectively combine the self-assembling nature of lipids with the multivalency feature of dendrimers, integrating the delivery advantages of both for efficient siRNA delivery.12–16
Motivated by the excellent performance of amphiphilic dendrimer vectors for siRNA delivery, we wanted to further improve their delivery efficacy and safety profile by imparting biodegradability. We anticipated that biodegradable vectors, upon breakdown, would promote both cargo release for better delivery and rapid clearance for preventing side effects. We therefore constructed a biodegradable amphiphilic poly(aminoester) dendrimer I for siRNA delivery (Scheme 1). This dendrimer differs from the conventional PAMAM dendrimer II in that it possesses ester backbones instead of amide bonds (Scheme 1). Ester linkage is commonly used to construct biodegradable materials,17–20 as it is acid- and base- as well as enzyme-labile. Indeed, I readily disintegrated upon exposure to esterase, and benefited the delivery features of both lipid and polymer vectors. It demonstrated superior performance for siRNA delivery with much less toxicity when compared to the corresponding PAMAM dendrimer II.
 | ||
| Scheme 1 (A) Amphiphilic dendrimers I and II studied in this work. (B) Illustration of I-mediated siRNA delivery and gene silencing. | ||
We first synthesized the amphiphilic dendrimer I according to the plan illustrated in Scheme 2. The dendron building units were prepared by following the strategy we previously established for synthesizing poly(aminoester) dendrimers.21,22 Briefly, propargylamine was used as the starting material to build 1via Michael addition, followed by a three-step reaction sequence comprising deprotection of the tert-butyl ester (tBu) groups, activation of the acid terminals via cyanomethyl esters, and transesterification with excess alcohol 4 to generate 5. Conversion of 5 to 6 was achieved using the same three-step reaction sequence of deprotection, activation and trans-esterification. The obtained 6 was then coupled with the azido-bearing alkyl chain 7 to deliver the dendrimer conjugate 8via a copper-catalysed azide–alkyne cycloaddition (CuAAC) click reaction. The conjugate 8 was further treated with trifluoroacetic acid (TFA) to deprotect the Boc groups, offering the desired poly(aminoester) dendrimer I at a yield exceeding 90% (Scheme 2 and S1, ESI†). Also, the poly(aminoester) dendron 9 was prepared via deprotection of 6 using TFA. The chemical composition and structural integrity of I was characterized and confirmed using 1H NMR, 13C NMR, and high-resolution mass spectrometry (HRMS) analysis (Fig. S1, ESI†).
We next wished to examine the biodegradability of our synthesized I in the presence of an enzyme, such as pig liver esterase (PLE) (Fig. 1 and Fig. S2, ESI†). HRMS analysis showed an intense molecular peak of I at m/z 1041.7246 prior to enzymatic treatment (Fig. S1C, ESI† and inset in Fig. 1A). This peak rapidly disappeared following exposure to PLE (Fig. 1A), highlighting the efficacious enzymatic disintegration of I. The degraded products were detected as illustrated by the appearance of numerous new mass signals in HRMS (Fig. 1A and Fig. S2, ESI†), and further analysed according to accurate mass calculations. Almost all the degradation products are issued from the breakdown of the ester linkages (Fig. 1B and Table S1, ESI†). Collectively, these results demonstrate that I is readily degradable upon enzymatic hydrolysis, and thus it is endowed with biodegradable properties.
At physiological pH, I is positively charged because of the numerous amine functionalities, and hence readily interacts with the negatively charged siRNA molecules to form an electrostatic complex. As shown in Fig. 2A, I completely retarded the migration of siRNA in agarose gel at the N/P ratio > 5, indicating the formation of a stable complex between I and siRNA. In addition, I could effectively protect siRNA from enzymatic degradation (Fig. S3, ESI†). Furthermore, dynamic light scattering (DLS) analysis revealed that the siRNA/I complex was small in size, measuring around 30 nm, and had a surface potential of +31 mV (Fig. 2B). The small size and positive surface charge are advantageous features for effective cellular uptake.
We next studied the cell uptake of the siRNA/I complex with the fluorescent probe Cy5-labelled siRNA using FACS flow. We observed rapid and efficient cellular uptake of the Cy5-siRNA/I complex, and almost 100% internalization was attained within 15 min (Fig. 2C and Fig. S4, ESI†). Importantly, these complexes effectively escaped from endosomes, as detected by the dispersion and the separation of the red fluorescence signals of Cy5-siRNA from the green fluorescence signals of lysotracker labelling endosomes in the cytoplasm (Fig. 2D). Endosomal escape represents a critical issue for successful siRNA delivery.
Encouraged by the promising properties shown by the siRNA/I complex for cell uptake and endosome release, we then evaluated siRNA-mediated gene silencing using siRNA targeting protein kinase B (siAKT2). Protein kinase B (AKT2) is an oncogene that has been linked with cancer cell proliferation and cancer progression, and is acknowledged as a promising therapeutic target for cancer treatment.23 We assessed gene silencing of AKT2 in different human cancer cell lines such as pancreatic cancer PANC-1 cells, lung cancer A549 cells and ovarian cancer SKOV3 cells. As illustrated in Fig. 3A, potent and specific knockdown of AKT2 expression was observed upon treatment with the siAKT2/I complex, whereas no gene silencing was detectable with either I or siRNA alone or with the scramble siRNA/I complex (Fig. 3A and Fig. S5, ESI†). Further investigation showed that I-mediated siRNA delivery could produce a dose-dependent gene silencing effect, i.e., AKT2 knockdown started with 5 nM siRNA, and reached ca. 80% with 20 nM siRNA (Fig. 3B and Fig. S6, ESI†). Altogether, these results demonstrated that I enabled successful siRNA delivery, leading to powerful gene silencing.
It is important to mention that the amphiphilic dendrimer II bearing PAMAM dendron showed no delivery activity for siRNA (Fig. S7, ESI†), in line with our previously reported results.16 In addition, II had considerable haemolytic toxicity (Fig. S8, ESI†). This is in sharp contrast to I, which mediated functional siRNA delivery with no notable haemolytic toxicity (Fig. S7 and S8, ESI†). The only difference between I and II is the replacement of the amide backbone in II with an ester linkage in I, thus endowing I with biodegradability. Consequently, the high performance of I for siRNA delivery can at least in part be ascribed to its biodegradable properties which, by allowing the degradation of the delivery vector, facilitate the release of siRNA cargos. Indeed, siRNA dissociation from the corresponding siRNA/I complex is much easier than from the siRNA/II complex as shown by the results obtained in the heparin displacement assay (Fig. S9, ESI†). Also, the biodegradability may further contribute to the low haemolytic toxicity of Ivia its rapid disintegration in the presence of various enzymes.
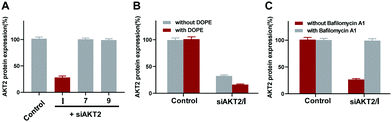
The amphiphilic dendrimer I has a lipid/dendrimer hybrid structure, and is therefore expected to exploit the delivery advantages of both lipid and dendrimer vectors. In order to confirm this, we first examined whether the dendron 9 alone or the alkyl chain 7 alone could mediate siRNA delivery. As presented in Fig. 4A, neither the alkyl chain 7 nor the dendron 9 showed any siRNA delivery activity to induce gene silencing. These results highlight that indeed it is the peculiar amphiphilic structure of I that is crucial for siRNA delivery. Importantly, I-mediated siRNA delivery and gene silencing was significantly improved in the presence of the fusogenic lipid dioleoyl-phosphatidyl ethanolamine (DOPE), which is frequently used to promote lipid-mediated delivery via a membrane fusion mechanism.24 The improved siRNA delivery and gene silencing in the presence of DOPE indicates that I effectively used the membrane fusion delivery feature of lipid vectors (Fig. 4B).
We next verified whether I exploited the delivery features of dendrimer vectors. Most effective polymer or dendrimer vectors often use the “proton-sponge” effect for endosome escape, because these vectors usually have numerous tertiary amines that allow their taking up of protons found enriched in the endosomes, leading to an ionic imbalance and hence endosome lysis and cargo release.25,26 We assessed the gene silencing efficiency in the presence of bafilomycin A1 (Fig. 4C), a proton pump inhibitor that prevents the acidification of endosomes. As shown in Fig. 4C, the presence of bafilomycin A1 significantly abolished the gene silencing effect, indicating that the I-mediated siRNA delivery was indeed dependent on the endosomal acidification, and that the proton sponge effect played a crucial role in the delivery mediated by I.
Aside delivery efficacy, the safety profile is also an essential concern for siRNA delivery. We therefore studied the biocompatibility and toxicity of I and its siRNA complex. Specifically, we evaluated the serum haemolysis, metabolic cytotoxicity and membrane disintegration of I and its siRNA complex (Fig. S10, ESI†). Neither I nor its siRNA complex exhibited any significant influence on the cell growth of various cells (PANC-1 cells, A549 cells, SKOV-3 cells, mouse fibroblast L929 cells, canine kidney MDCK cells, and human Liver L02 cells) (Fig. S10A, ESI†). In addition, neither I nor its siRNA complex caused any cell membrane damage (Fig. S10B, ESI†). Also, no haemolytic toxicity was observed with either I or its siRNA complex (Fig. S8 and S10C, ESI†). Further in vivo toxicity assessments in healthy mice also demonstrated the lack of any notable inflammatory responses or acute toxicity relating to I or its siRNA complex (Fig. S11, ESI†). Collectively, these results argue in favour of a good safety profile for I and promote its use in biomedical applications.
In conclusion, we successfully established a novel biodegradable amphiphilic poly(aminoester) dendrimer I for functional siRNA delivery. This dendrimer is a lipid/dendrimer hybrid featuring a hydrophobic alkyl chain and a small hydrophilic poly(aminoester) dendron. It shows excellent biodegradability and biocompatibility, while being devoid of notable toxicity. Most importantly, this biodegradable dendrimer has demonstrated superior siRNA delivery and gene silencing performance by harnessing not only biodegradability but also the integrated delivery advantages of both lipid and polymer vectors. It therefore holds great promise for siRNA delivery in future biomedical applications. This study also provides a new perspective in the search for biodegradable vectors for siRNA delivery, which we hope will accelerate and extend the clinical implementation of siRNA therapeutics.
X. L., L. P.: project conception and supervision; C. M., D. Z., W. L., Y. L., H. Z., M. Y., Y. H.: methodology, investigation, data analysis; C. M., D. Z., X. L., L. P. wrote the paper, all approved the manuscript.
We thank Dr Tintaru Aura (Aix-Marseille University) for her assistance with MS analysis. This study was supported by the Key Program for International S&T Cooperation Projects of China (2018YFE0117800), the National Natural Science Foundation of China (No. 51773227, and 81701815), the Natural Science Foundation of Jiangsu Province (BK20170735), the Youth Thousand-Talents Program of China, the Jiangsu Specially-Appointed Professors Program, the State Key Laboratory of Natural Medicines at China Pharmaceutical University (No. SKLNMZZ202007), Ligue Nationale Contre le Cancer (EL2021 LNCCLiP) and EU H2020 NMBP “SAFE-N-MEDTECH” (No. 814607).
Conflicts of interest
There are no conflicts to declare.Notes and references
- R. L. Setten, J. J. Rossi and S. P. Han, Nat. Rev. Drug Discovery, 2019, 18, 421 CrossRef CAS PubMed.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967 CrossRef CAS PubMed.
- S. F. Dowdy, Nat. Biotechnol., 2017, 35, 222 CrossRef CAS PubMed.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16, 630 CrossRef CAS PubMed.
- Y. Zhang, C. Sun, C. Wang, K. E. Jankovic and Y. Dong, Chem. Rev., 2021, 121, 12181 CrossRef CAS PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. V. Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527 CrossRef CAS PubMed.
- M. E. Davis, J. E. Zuckerman, C. H. Choi, D. Seligson, A. Tolcher, C. A. Alabi, Y. Yen, J. D. Heidel and A. Ribas, Nature, 2010, 464, 1067 CrossRef CAS PubMed.
- H. Ledford, Nature, 2018, 560, 291 CrossRef CAS PubMed.
- A. Mullard, Nat. Rev. Drug Discovery, 2021, 20, 728 Search PubMed.
- C. C. Lee, J. A. MacKay, J. M. Frechet and F. C. Szoka, Nat. Biotechnol., 2005, 23, 1517 CrossRef CAS PubMed.
- R. M. Kannan, E. Nance, S. Kannan and D. A. Tomalia, J. Intern. Med., 2014, 276, 579 CrossRef CAS PubMed.
- J. Chen, A. Ellert-Miklaszewska, S. Garofalo, A. K. Dey, J. Tang, Y. Jiang, F. Clement, P. N. Marche, X. Liu, B. Kaminska, A. Santoni, C. Limatola, J. J. Rossi, J. Zhou and L. Peng, Nat. Protoc., 2021, 16, 327 CrossRef CAS PubMed.
- Y. Dong, T. Yu, L. Ding, E. Laurini, Y. Huang, M. Zhang, Y. Weng, S. Lin, P. Chen, D. Marson, Y. Jiang, S. Giorgio, S. Pricl, X. Liu, P. Rocchi and L. Peng, J. Am. Chem. Soc., 2018, 140, 16264 CrossRef CAS PubMed.
- X. Liu, Y. Wang, C. Chen, A. Tintaru, Y. Cao, J. Liu, F. Ziarelli, J. J. Tang, H. B. Guo, R. Rosas, S. Giorgio, L. Charles, P. Rocchi and L. Peng, Adv. Funct. Mater., 2016, 26, 8594 CrossRef CAS.
- X. Liu, J. Zhou, T. Yu, C. Chen, Q. Cheng, K. Sengupta, Y. Huang, H. Li, C. Liu, Y. Wang, P. Posocco, M. Wang, Q. Cui, S. Giorgio, M. Fermeglia, F. Qu, S. Pricl, Y. Shi, Z. Liang, P. Rocchi, J. J. Rossi and L. Peng, Angew. Chem., Int. Ed., 2014, 53, 11822 CrossRef CAS PubMed.
- T. Yu, X. Liu, A. L. Bolcato-Bellemin, Y. Wang, C. Liu, P. Erbacher, F. Qu, P. Rocchi, J. P. Behr and L. Peng, Angew. Chem., Int. Ed., 2012, 51, 8478 CrossRef CAS PubMed.
- I. D. Kim, C. M. Lim, J. B. Kim, H. Y. Nam, K. Nam, S. W. Kim, J. S. Park and J. K. Lee, J. Controlled Release, 2010, 142, 422 CrossRef CAS PubMed.
- D. Wu, Y. Liu, X. Jiang, C. He, S. H. Goh and K. W. Leong, Biomacromolecules, 2006, 7, 1879 CrossRef CAS PubMed.
- H. J. Kim, M. S. Kwon, J. S. Choi, B. H. Kim, J. K. Yoon, K. Kim and J. S. Park, Bioorg. Med. Chem., 2007, 15, 1708 CrossRef CAS PubMed.
- A. Lancelot, R. González, R. Clavería-Gimeno, P. Romero, O. Abian, P. Martin-Duque, J. L. Serrano and T. Sierra, J. Mater. Chem. B, 2018, 6, 3956 RSC.
- C. Bouillon, G. Quelever and L. Peng, Tetrahedron Lett., 2009, 50, 4346 CrossRef CAS.
- P. Moreno, G. Quelever and L. Peng, Tetrahedron Lett., 2015, 56, 4043 CrossRef CAS.
- B. D. Manning and A. Toker, Cell, 2017, 169, 381 CrossRef CAS PubMed.
- H. Farhood, N. Serbina and L. Huang, Biochim. Biophys. Acta, 1995, 1235, 289 CrossRef.
- J. P. Behr, Chimia, 1997, 51, 34 CAS.
- N. D. Sonawane, F. C. Szoka, Jr. and A. S. Verkman, J. Biol. Chem., 2003, 278, 44826 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc06655b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |




![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: