Intramolecular G-quadruplex-hairpin loop structure competition of a GC-rich exon region in the TMPRSS2 gene†
Wataru
Sugimoto
 a,
Natsuki
Kinoshita
a,
Natsuki
Kinoshita
 a,
Minori
Nakata
a,
Tatsuya
Ohyama
b,
Hisae
Tateishi-Karimata
a,
Minori
Nakata
a,
Tatsuya
Ohyama
b,
Hisae
Tateishi-Karimata
 b,
Takahito
Nishikata
a,
Naoki
Sugimoto
b,
Takahito
Nishikata
a,
Naoki
Sugimoto
 ab,
Daisuke
Miyoshi
ab,
Daisuke
Miyoshi
 *a and
Keiko
Kawauchi
*a
*a and
Keiko
Kawauchi
*a
aFaculty of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-mimamimachi, Chuo-ku, Kobe 650-0047, Japan. E-mail: miyoshi@konan-u.ac.jp; kawauchi@konan-u.ac.jp
bFrontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-mimamimachi, Chuo-ku, Kobe 650-0047, Japan
First published on 16th November 2021
Abstract
We identified cytosine-rich regions adjacent to guanine-rich regions in protease genes. A typical GC-rich sequence derived from the TMPRSS2 gene showed structural competition between a G-quadruplex and a hairpin loop, and this competition significantly affected transcription efficiency. These results suggest an impact of neighboring sequences on the gene expression of guanine-rich sequences.
A guanine-rich (G-rich) sequence can form a G-quartet with Hoogsteen base pairs between four guanine bases.1–3 G4-forming G-rich sequences have been identified in the human genome at telomeric and other biologically important regions such as promoters and introns of various genes.4–10 Except for the terminus of the 3′ ends of telomeres, all G4-forming G-rich sequences in the genome have complementary sequences that form duplexes. Thus, intramolecular quadruplexes comprising a G4 of a G-rich sequence and an i-motif of a C-rich sequence compete with an intermolecular duplex, inhibiting G4 formation in the genome. This inhibition is responsible at least in part for the smaller number of genomic G4s observed in living cells compared to the number of putative G4-forming G-rich sequences identified by bioinformatic studies.11–13 It has been proposed that unwinding of the duplex during replication and transcription assists the formation of G4s, leading to a cell cycle dependency of G4 formation in living cells.14–16 Another structural competition should also be taken into consideration: that between an intramolecular duplex (hairpin loop) and intramolecular quadruplexes (G4 and i-motif). The importance of this competition is based on sequence analysis showing that regulatory regions of human protooncogenes are enriched not only in G and but also in C, generating GC-rich regions in which the C-rich regions are located either adjacent to or mixed with G-rich regions. Given this finding, it is important to further investigate DNA and RNA structures in GC-rich regions.12,13 For example, in GC-rich regions derived from some protooncogenic mRNAs, the GC-rich RNA sequences form a stable A-form duplex as a major structure essentially independent of the experimental conditions.17 In contrast, the corresponding GC-rich DNA sequences show structural polymorphisms, including duplexes, G4s, and i-motifs, depending on the environmental conditions.12 A competition between hairpin loop and G4 within an RNA strand was also reported.18 It is therefore important to consider structural competition between intramolecular hairpin loops and quadruplexes (G4 and i-motif) in GC-rich regions. Moreover, it remains unknown how such a structural switch affects the transcriptional efficiency. In this study, we analyzed sequences of a series of human protease genes and found GC-rich regions in which a C-rich region is adjacent to a G-rich region as described below, suggesting that structural competition may affect gene expression of these proteases.
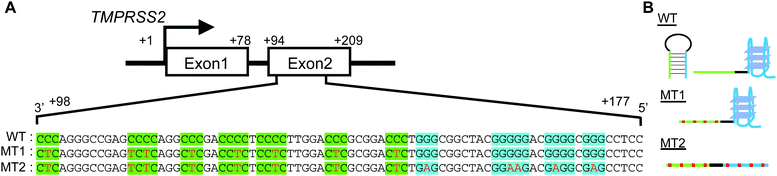
As a typical protease gene possessing GC-rich regions, we focused on the transmembrane serine protease TMPRSS2, which plays a critical role in infection by the SARS CoV-2 and influenza A viruses.19–22 A putative G4-forming sequence in the GC-rich region within exon 2 of TMPRSS2 is shown in Fig. 1A. A C-rich region at the 3′ side of the template strand comprises six cytosine stretches (highlighted in green). At the 5′ side there is a G-rich region comprising four guanine stretches (highlighted in blue) that is generally considered a putative G4-forming sequence. Each G-rich and C-rich region alone can form a G4 and an i-motif, respectively, although an i-motif cannot be formed in neutral or slightly basic solutions, as described below. Moreover, intramolecular hybridization between the G-rich and C-rich regions may result in formation of a stable hairpin loop structure (Fig. 1B). To study the structural competition and its effects on transcription, we synthesized a DNA strand corresponding to the +98 to +177 region of the TMPRSS2 gene (WT).
We further designed the two sequences MT1 and MT2 (Fig. 1A and Table S1, ESI†). MT1 was designed to reduce hairpin loop formation by introducing C-to-T mutations in the C-rich region (Fig. 1B). MT2 has G-to-A mutations in the G-rich region, in addition to the C-to-T mutations in the C-rich region. Thus, MT2 cannot fold into a G4 with these G-stretches (Fig. 1B). The values of ΔG°25 for the hairpin loop of WT, MT1 and MT2 were determined using m-fold23 to be −24.8, −12.8 and −13.1 kcal mol−1, respectively (Fig. S1, ESI†).
First, we attempted to study the structure of the DNA strands. The fluorescence intensity of thioflavin T (ThT) is enhanced upon binding to G4 in a non-sequence-specific manner.24 Fig. S2A (ESI†) shows the fluorescence spectra of 1.0 μM ThT in the presence of various concentrations of WT in 150 mM KCl, 40 mM Tris-HCl (pH 7.2) and 8 mM MgCl2 buffer. The increase in fluorescence intensity with higher concentrations of WT indicates G4 formation by WT. Fig. 2A shows the fluorescence intensity of ThT at 488 nm in the presence of various concentrations of WT, MT1 or MT2. Fluorescence spectra of ThT in the presence of MT1 and MT2 are shown in Fig. S2B and C (ESI†) respectively. The fluorescence intensity of ThT followed the order: MT1 > WT > MT2, suggesting that G4 formation by MT1 is accelerated by reducing competition with the hairpin loop. In contrast, G4 formation by MT2 is reduced by the mutations. In addition to ThT, N-methyl mesoporphyrin IX (NMM),25 which is also known as a G4 fluorescence indicator, was utilized to study G4 formation. The results were consistent with that for ThT (Fig. S2D, E and F, ESI†). Thus, the oligonucleotides designed here are suitable for studying structural competition.
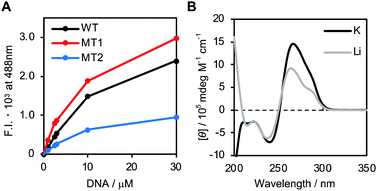 | ||
| Fig. 2 (A) Fluorescence intensity of 1 μM ThT at 488 nm in the presence of various concentrations of WT, MT1 or MT2. (B) CD spectra of WT with KCl or LiCl. | ||
The structures of the oligonucleotides were further studied using circular dichroism (CD) (Fig. 2B) in the presence of K+ or Li+. Note that at this experimental condition (pH = 7.2), the C-rich region cannot form an i-motif because this requires hemi-protonated cytosines, which have a pKa value of 6.5.26–29 The CD spectrum with K+ has positive and negative peaks around 265 nm and 240 nm, respectively, with a small shoulder around 290 nm. The positive peak and the shoulder in the presence of K+ were significantly decreased in the presence of Li+. Thermal stability of G4 in the presence of K+ is higher than that in the presence of Li+, but duplex stability is independent of cation species.30 Therefore, the differences in the CD spectra in the presence of K+ and Li+ suggest that K+ enhances the G4 component. The CD spectra of MT1 showed a similar trend (Fig. S3A, ESI†), suggesting that G4 stability is higher in the presence of K+. In contrast, the CD spectra of MT2 with K+ and Li+ were almost identical (Fig. S3B, ESI†), showing that MT2 does not form G4. These results, combined with the ThT and NMM fluorescence data, indicate that WT folds into G4 and hairpin loop structures, depending on the coexisting cation, that MT1 forms more G4 than does WT, and that MT2 does not form G4.
Furthermore, thermal denaturation curves of WT, MT1, and MT2 in the presence of K+, Li+, and K+ with PEG200 were traced by CD intensities at 265 nm and 290 nm (Fig. S4, ESI†). By comparing the thermal denaturation curves, it was further supported that WT forms G4 and hairpin loop, depending on the surrounding condition, whereas the dominant structures of MT1 and MT2 are G4 and hairpin loop, respectively, almost independent of the surrounding condition. See Fig. S4 (ESI†) for details regarding the thermal denaturation curves.
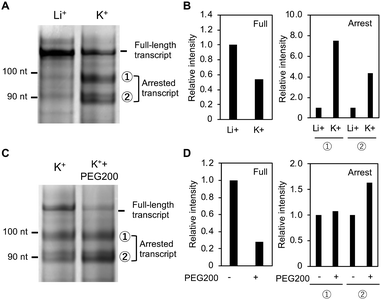
Next, to evaluate the effect of intramolecular structural competition on the transcription reaction, we designed the template DNA strands Temp-WT, Temp-MT1 and Temp-MT2, which contain WT, MT1 and MT2, respectively (Table S1 and Fig. S5, ESI†). These DNA strands have a T7 polymerase binding site which is separated from the GC-rich region by 35 nucleotides.31 Template DNAs were transcribed by T7 RNA polymerase at 37 °C in 150 mM LiCl or 150 mM KCl, 40 mM Tris-HCl (pH 7.2), 8 mM MgCl2. The transcription product was analyzed by denaturing gel electrophoresis (Fig. S6–S8, ESI†). The product band intensities increased with reaction time and nearly saturated after 120 min (Panel B in Fig. S6–S8, ESI†). Thus, the transcription efficiency was studied by analyzing the product at 120 min.
One main band and two faint bands were observed for the product from Temp-WT in the presence of Li+ (left, Fig. 3A). The top band with the slowest migration potentially corresponds to the full-length transcript, which is 115 nucleotides long. The second and the third bands from the top might be due to transcripts arrested by the G4. Indeed, the amount of these shorter transcripts increased in the presence of K+, and the amount of the longest transcript decreased (right, Fig. 3A). Fig. 3B shows the relative band intensities for the full-length transcript (left) and the arrested transcripts (right). The amount of the full-length product was halved in the presence of K+, whereas the amount of the longer and shorter arrested products increased by 7.5 and 4.4 times, respectively, indicating that Temp-WT G4 arrests transcription. It was previously reported that G4 formed in a template inhibits transcription more significantly than does a hairpin loop.32 To confirm this point, we designed additional oligonucleotides, MT3 and MT4, both of which form hairpin loop with similar with MT2 and higher thermodynamic stabilities than MT2, respectively (Fig. S9, ESI†). Structural analyses of MT3 and MT4 with the ThT assay, the NMM assay, and the CD spectroscopy were show in Fig. S10 (ESI†). The transcription reactions with temp-MT3 and temp-MT4 showed that the amounts of the full-length transcripts were similar with that of temp-MT2 (Fig. S11–S13, ESI†). These results confirm that hairpin loop has the lower inhibitory effect on the transcription than G4.
The products are approximately 85 and 95 nucleotides long and may result from arrest just before the first and the second G-tracks of G4, respectively (Fig. S5A, ESI†). The three bands were identified by comparison with transcripts from truncated Temp-WTs with lengths of 85 and 95 nucleotides (see Fig. S5B (ESI†) for details regarding identification of these bands). Since a G4 and a duplex are stabilized and destabilized, respectively, by molecular crowding with poly(ethylene glycol) with a molecular weight of 200 (PEG200),33,34 10% PEG200 was added to the reaction buffer to stabilize the G4 formed by Temp-WT. Fig. 3C shows the products at 0% and 10% PEG200 with K+. The amount of the full-length product was further decreased by PEG200, whereas the intensity of the arrested bands increased (Fig. 3D). These results demonstrate that the transcription efficiency largely depends on the thermal stability of G4 formed in the GC-rich region.
Given the critical role of G4 formation in the template strand, we investigated how intramolecular structural competition affects transcription. Fig. 4A shows the three main products from Temp-WT, Temp-MT1 and Temp-MT2 in the presence of K+. (Whole gel images are shown in Fig. S5, ESI.†) Fig. 4B shows the relative band intensities for the three products. As expected, the full-length transcript of MT1 was significantly reduced to 0.14 times that of Temp-WT, and the full-length transcript increased to 2.15 for Temp-MT2. On the other hand, the total amount of the two arrested transcripts increased to 1.79 for Temp-MT1 and decreased to 0.25 for temp-MT2. As shown above, a structural analysis showed that the degree of G4 formation followed the order MT1 > WT > MT2 (Fig. 2). Thus, these results demonstrate that G4 formed in the template strand regulates the ratio of full-length and arrested products. More importantly, the difference between Temp-WT and Temp-MT1 shows possible roles of the adjacent C-rich region. The C-rich region forms an intramolecular hairpin loop with the G-rich region, resulting in a smaller amount of G4. This smaller amount of G4 in the template strand increases and decreases the amount of arrested and full-length products, respectively. Therefore, if we focus on the G-rich region only, corresponding to MT1 in this study, the results of the structural analysis and the effects on transcription are significantly different to the results taking the C-rich region into account, here corresponding to the results obtained using WT.
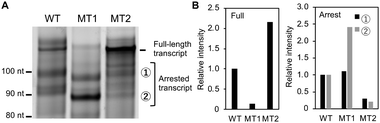 | ||
| Fig. 4 (A) Transcripts from Temp-WT, Temp-MT1 and Temp-MT2 in the presence of K+. (B) Relative amounts of full (left) and arrested (right) transcripts observed in panel (A). | ||
Structural and transcription analyses demonstrated the impact of the C-rich region adjacent to the G-rich region, and thus we further attempted to evaluate how commonly a C-rich region exists in contiguity with a G-rich region. We analysed 62 human protease genes and found 45 putative G4-forming sequences (Table S2, ESI†). We analysed the number of cytosines within 50 nucleotides upstream and downstream of these regions. A random distribution of cytosines gives an average of 12.5 cytosines (= 50/4) in each region. In contrast, we observed that the average number of cytosines within 50 nucleotides upstream and downstream was 13.8 ± 5.2 and 14.6 ± 5.1, respectively (Fig. S14, ESI†). Although the enrichment of cytosine was not significant, the number varied widely between genes. For example, a G-rich region in intron 7 of TMPRSS6 has 20 and 18 cytosines in the upstream and downstream regions, respectively, leading to a structural competition, as seen in TMPRSS2. In contrast, a G-rich region found in intron 3 of TMPRSS6 has only 5 and 9 cytosines in these regions, suggesting weak or no structural competition. Therefore, the structural competition between G4 and a hairpin loop at the intragenic region identified in this study is a fairly common phenomenon, although some G4s do not engage in competition. Thus, it is necessary to take the neighbouring nucleotide sequence into consideration when studying how an intragenic G4-forming sequence affects gene expression. Depending on the presence or absence of a C-rich region adjacent to a G-rich region, the most stable structure of the region and thus the transcription efficiency are totally different.
In conclusion, we found a C-rich region adjacent to the G-rich region in an intragenic region in a series of protease genes. Structural analysis of GC-rich regions derived from the intragenic region of the TMPRSS2 gene showed intramolecular structural competition between G4 and the hairpin loop. The T7 transcription assay demonstrated that structural competition significantly affects the transcription efficiency. These results suggest the need to take the neighbouring nucleotide sequence into account when studying how an intergenic G4-forming sequence affects gene expression. Since the hairpin loop rescues suppresses the arrest by G4, the structural competition provides a new method for controlling gene expression by regulating the stability of both hairpin loop and of G4.
K. K. and D. M. created the project. W. S., N. K. and M. N. performed the experiments. T. O. and H. T. K. contributed to sequence analysis. W. S., N. K., M. N., T. O., H. T. K., T. N., N. S., D. M. and K. K. analysed and discussed the overall data. W. S. wrote the paper.
This work was supported by JSPS KAKENHI grant numbers 21H02062, 21H05109, 20K21259, 20H02864, 18KK0164, 17H06351 (Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems”), and 19J21096, a Research Grant of the Asahi Glass Foundation, Japan, and the Hirao Taro Foundation of Konan Gakuen for Academic Research, Japan.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J. R. Williamson, Annu. Rev. Biophys. Biomol. Struct., 1994, 23, 703–730 CrossRef CAS.
- J. L. Mergny and C. Helene, Nat. Med., 1998, 4, 1366–1367 CrossRef CAS PubMed.
- S. Balasubramanian and S. Neidle, Curr. Opin. Chem. Biol., 2009, 13, 345–353 CrossRef CAS PubMed.
- J. Spiegel, S. Adhikari and S. Balasubramanian, Trends Chem., 2020, 2, 123–136 CrossRef CAS.
- D. Varshney, J. Spiegel and K. Zyner, et al. , Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- V. S. Chambers, G. Marsico and J. M. Boutell, et al. , Nat. Biotechnol., 2015, 33, 877–881 CrossRef PubMed.
- G. Marsico, V. S. Chambers and A. B. Sahakyan, et al. , Nucleic Acids Res., 2019, 47, 3862–3874 CrossRef CAS PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261–275 CrossRef CAS PubMed.
- J. Robinson, F. Raguseo and S. P. Nuccio, et al. , Nucleic Acids Res., 2021, 49, 8419–8431 CrossRef CAS.
- G. Biffi, D. Tannahill and J. McCafferty, et al. , Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- W. Li, D. Miyoshi and S. Nakano, et al. , Biochemistry, 2003, 42, 11736–11744 CrossRef CAS PubMed.
- L. Liu, L. Scott and N. Tariq, et al. , J. Phys. Chem. B, 2021, 125, 7406–7416 CrossRef CAS.
- A. L. Valton and M. N. Prioleau, Trends Genet., 2016, 32, 697–706 CrossRef.
- P. Prorok, M. Artufel and A. Aze, et al. , Nat. Commun., 2019, 10, 3274 CrossRef CAS PubMed.
- L. K. Lerner and J. E. Sale, Genes, 2019, 10, 95 CrossRef CAS.
- S. Saxena, D. Miyoshi and N. Sugimoto, Biochemistry, 2010, 49, 7190–7201 CrossRef CAS.
- A. Bugaut, P. Murat and S. Balasubramanian, J. Am. Chem. Soc., 2012, 134, 19953–19956 CrossRef CAS PubMed.
- H. Limburg, A. Harbig and D. Bestle, et al. , J. Virol., 2019, 93, e00649–19 CrossRef PubMed.
- L. W. Shen, H. J. Mao and Y. L. Wu, et al. , Biochimie, 2017, 142, 1–10 CrossRef CAS PubMed.
- M. Hoffmann, H. Kleine-Weber and S. Schroeder, et al. , Cell, 2020, 181, 271–280 CrossRef CAS.
- D. Bestle, M. R. Heindl and H. Limburg, et al. , Life Sci. Alliance, 2020, 3, e202000786 CrossRef PubMed.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS.
- V. Gabelica, R. Maeda and T. Fujimoto, et al. , Biochemistry, 2013, 52, 5620–5628 CrossRef CAS.
- A. Kreig, J. Calvert and J. Sanoica, et al. , Nucleic Acids Res., 2015, 43, 7961–7970 CrossRef CAS PubMed.
- J. L. Leroy, M. Gueron and J. L. Mergny, et al. , Nucleic Acids Res., 1994, 22, 1600–1606 CrossRef CAS.
- H. A. Day, C. Huguin and Z. A. Waller, Chem. Commun., 2013, 49, 7696–7698 RSC.
- P. Bielecka, A. Dembska and B. Juskowiak, Molecules, 2019, 24, 952 CrossRef.
- K. Gehring, J. L. Leroy and M. Gueron, Nature, 1993, 363, 561–565 CrossRef CAS.
- D. Bhattacharyya, G. Mirihana Arachchilage and S. Basu, Front. Chem., 2016, 4, 38 Search PubMed.
- H. Tateishi-Karimata, K. Kawauchi and N. Sugimoto, J. Am. Chem. Soc., 2018, 140, 642–651 CrossRef CAS.
- H. Tateishi-Karimata, N. Isono and N. Sugimoto, PLoS One, 2014, 9, e90580 CrossRef.
- S. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS.
- D. Miyoshi, H. Karimata and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 7957–7963 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc05523b |
| This journal is © The Royal Society of Chemistry 2022 |