Open Access Article
Open Access ArticleInterconversion between [2Fe–2S] and [4Fe–4S] cluster glutathione complexes†
Michele
Invernici
 ab,
Giulia
Selvolini
ab,
Giulia
Selvolini
 c,
José Malanho
Silva
c,
José Malanho
Silva
 ac,
Giovanna
Marrazza
ac,
Giovanna
Marrazza
 c,
Simone
Ciofi-Baffoni
c,
Simone
Ciofi-Baffoni
 ac and
Mario
Piccioli
ac and
Mario
Piccioli
 *abc
*abc
aMagnetic Resonance Center (CERM), University of Florence, Via L. Sacconi 6, Sesto Fiorentino 50019, Italy. E-mail: piccioli@cerm.unifi.it
bConsorzio Interuniversitario Risonanze Magnetiche di Metalloproteine (CIRMMP), Via L. Sacconi 6, Sesto Fiorentino 50019, Italy
cDepartment of Chemistry “Ugo Schiff”, University of Florence, Via della Lastruccia 3, Sesto Fiorentino 50019, Italy
First published on 15th February 2022
Abstract
We present here how different iron–sulfide–glutathione ratios, applied in in vitro conditions comparable to those present in the mitochondrial matrix, affect the speciation of iron–sulfur cluster glutathione complexes. An excess of sulfide with respect to iron ions promotes the formation of a tetranuclear [FeII2FeIII2S4(GS)4]2− complex, while an excess of iron ions favors the formation of a dinuclear [FeIIFeIIIS2(GS)4]3− complex. These two complexes establish an interconversion equilibrium. The latter might play a role in the composition of the mitochondrial labile iron pool potentially contributing to the regulation of cellular iron homeostasis.
The mitochondrial matrix is a major site for iron–sulfur ([Fe–S]) cluster biosynthesis in eukaryotes.1,2 Moreover, the [2Fe–2S] clusters synthesized in the mitochondrial matrix were proposed to be exported to the cytoplasm3–5 where they can play a crucial role in the maturation of cytosolic and nuclear [Fe–S] proteins. In particular, among the latter there are crucial proteins involved in the regulation of cellular iron homeostasis.6 Because of this basic cellular demand, eukaryotic cells have a pool of accessible iron in the mitochondrial matrix, named the labile iron pool (LIP),7–9 which is exploited to assemble [Fe–S] clusters for all cellular proteins that need these cofactors for their function. The redox state of the iron ions in the mitochondrial LIP comprises both Fe2+ and Fe3+ species, with Fe2+ being the largely predominant redox state.8–10 In the mitochondrial LIP, iron exists as non-proteinacious low-molecular-mass (LMM) Fe2+ complexes rather than as inorganic free Fe2+ ions.11 Among the possible Fe2+ mitochondrial chelators, the most quoted, because of its very high concentration in the mitochondrial matrix, given in the range of 10–14
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM, is glutathione (GSH).12 It was proposed that a Fe2+-glutathione adduct ([FeII(H2O)5GS]+) is the major non-proteinacious LMM Fe complex in the mitochondrial matrix and that a glutathione-coordinated [Fe2S2(GS)4]2− complex might be also part of the mitochondrial LIP10,11a to be used as reservoir for the mitochondrial [Fe–S] cluster assembly machinery13 as well as to be transferred to the cytosol.5 However, the exact composition of the non-proteinacious LMM complexes detected in mitochondria still needs to be addressed. This has prompted us to investigate in vitro how different iron–sulfide–glutathione molar ratios bring to different glutathione-coordinated [Fe–S] cluster complexes and to explore their possible interconversion phenomena.
mM, is glutathione (GSH).12 It was proposed that a Fe2+-glutathione adduct ([FeII(H2O)5GS]+) is the major non-proteinacious LMM Fe complex in the mitochondrial matrix and that a glutathione-coordinated [Fe2S2(GS)4]2− complex might be also part of the mitochondrial LIP10,11a to be used as reservoir for the mitochondrial [Fe–S] cluster assembly machinery13 as well as to be transferred to the cytosol.5 However, the exact composition of the non-proteinacious LMM complexes detected in mitochondria still needs to be addressed. This has prompted us to investigate in vitro how different iron–sulfide–glutathione molar ratios bring to different glutathione-coordinated [Fe–S] cluster complexes and to explore their possible interconversion phenomena.
The synthesis of glutathione-coordinated [Fe–S] clusters in aqueous solution has been accomplished by mixing, under anaerobic conditions, FeCl3 and Na2S at different molar ratios in the presence of a large excess of GSH. The pH was adjusted to the pKa of GSH (pKa = 8.6), exploiting the thiol as buffering agent. A high concentration of GSH (240 mM) was used in order to mimick the GSH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe ratio present in LIP of the mitochondrial matrix (Fe and GSH concentrations in the mitochondrial matrix are 10–150 μM10,14 and 10–14
Fe ratio present in LIP of the mitochondrial matrix (Fe and GSH concentrations in the mitochondrial matrix are 10–150 μM10,14 and 10–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mM,12 respectively).
mM,12 respectively).
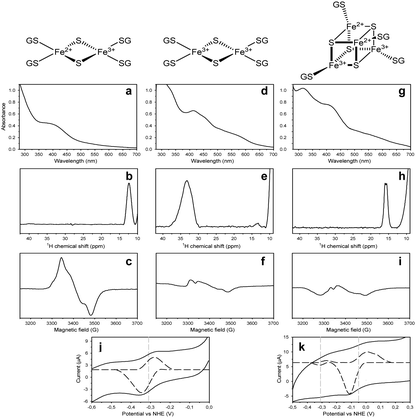
A solution containing 240 mM GSH, 2 mM Na2S and 10 mM FeCl3, i.e. with a large excess of iron ions (solution 1, hereafter), gives rise to UV-vis, paramagnetic 1H NMR and EPR spectra (Fig. 1a–c) similar to the spectra of ferredoxin-like proteins/peptides containing a reduced, [2Fe–2S]+, cluster.15–17 This result supports the formation of a dinuclear [FeIIFeIIIS2(GS)4]3− complex. The UV-vis spectrum of solution 1 reproduces, indeed, that previously obtained when an oxidized [FeIII2(GS)4]2− complex was reduced by dithionite.17 The EPR spectrum of solution 1 recorded at 45 K showed principal g values of 2.00, 1.96, and 1.92. The average g value (gav = 1.96) as well as its anisotropy are characteristic of four-cysteine-ligated reduced [2Fe–2S]+ clusters.15 The paramagnetic 1H NMR spectrum of solution 1 at 298 K shows only one hyperfine signal at 12.5 ppm, with longitudinal relaxation rate (R1) of 3500 s−1 and a Curie temperature dependence. The signal at 12.5 ppm, having a small paramagnetic shift but a large paramagnetic relaxation, can be attributed to αCH of the cysteine of GS− bound to the iron in a reduced [2Fe–2S]+ cluster with a S = 1/2 ground state.18 By exposing solution 1 containing the dinuclear [FeIIFeIIIS2(GS)4]3− complex to air for 10 min, the cluster of the complex was oxidized to obtain a [2Fe–2S]2+ cluster (Fig. 1d–f). Indeed, the UV-vis spectrum shows a near ultraviolet peak at 330 nm and visible absorption peaks at 414 and 455 nm with a shoulder at 580 nm, all typical of ferredoxin-like proteins containing [2Fe–2S]2+ oxidized clusters bound to cysteine ligands (Fig. 1d).19 The paramagnetic 1H NMR spectrum shows a broad signal, at 33 ppm, having an anti-Curie temperature dependence, typical of βCH2 of cysteines bound to an oxidized [2Fe–2S]2+ cluster, with two antiferromagnetically coupled FeIII at a S = 0 ground state (Fig. 1e).15,20 The EPR signal almost disappears, in agreement with the formation of a EPR-silent [2Fe–2S]2+ species (Fig. 1f).
A different situation was observed when the concentration of Na2S and FeCl3 are swapped, i.e. when there is a large excess of sulfide ions. The solution containing 240 mM GSH, 20 mM Na2S and 2 mM FeCl3 (solution 2, hereafter), gives rise to UV-vis, paramagnetic 1H NMR and EPR spectra (Fig. 1g–i) similar to those of systems having a [4Fe–4S]2+ bound cluster.16,21 These results support the presence of a tetranuclear [FeII2FeIII2S4(GS)4]2− complex. The UV-vis spectrum of solution 2 shows a broad absorption centered at 410 nm with a A(410 nm)/A(280 nm) absorbance ratio higher than 0.7, all typical features observed in [4Fe–4S]2+ proteins/peptides (Fig. 1g).21 The paramagnetic 1H NMR spectrum of solution 2 at 298 K shows two barely resolved signals centered at ∼16 ppm, with R1 of 730 s−1 and an anti-Curie temperature dependence, typical of Cys βCH2 bound to a [4Fe–4S]2+ cluster.15 The frozen solution at 10 K does not show any strong EPR signal, in agreement with the presence, as the major component of the mixture, of a EPR silent [4Fe–4S]2+ cluster. By exposing the solution to air for 60 min, the UV-vis, EPR and paramagnetic 1H NMR spectra do not change, showing that tetranuclear [FeII2FeIII2S4(GS)4]2− cluster behaves differently than the dinuclear [FeIIFeIIIS2(GS)4]3− complex.
Upon decreasing the pH, both reduced and oxidized forms of the dinuclear [FeIIFeIIIS2(GS)4] complex as well as the tetranuclear [FeII2FeIII2S4(GS)4]2− complex remain stable up to pH values of 7.8 and 7.3, respectively (Fig. S1, ESI†), in support of their possible existence in the mitochondrial matrix, reported to have a pH in the range 7.8–8.0.22,23
Cyclic voltammograms (CVs), recorded, respectively, on solutions 1 and 2 (Fig. 1j and k), allowed us to measure the redox potentials of the [2Fe–2S] and [4Fe–4S] clusters bound to the two complexes (Fig. S2, ESI† shows the full recorded CVs). Fig. 1j shows a redox event in solution 1 with a formal potential of E1/2 = −310 mV related to the reduction/oxidation of the [Fe2S2(GS)4]3−/2− couple, in agreement with literature values of [2Fe–2S] ferredoxins, which are in the range −240/−350 mV.24Fig. 1k from solution 2 shows a main cathodic and anodic current with E1/2 = −50 mV for the reduction peak. The low intensity current at −310 mV shows that a small amount of the dinuclear [Fe2S2(GS)4]3−/2− complex is still present in solution.
Literature data show redox potentials ranging between +30 and +480 mV and between −300 and −700 mV, respectively for [4Fe–4S] HiPIP and [4Fe–4S] ferredoxin proteins.16,25 The redox potential at −50 mV suggests that the [4Fe–4S] cluster is in a HiPIP-like situation and therefore can be attributed to the reduction/oxidation of a [Fe4S4(GS)4]2−/− couple. This high redox potential is also consistent with the relative high stability of the [Fe4S4(GS)4]2− complex observed upon air exposure, which is typical of [4Fe–4S] HiPIP proteins.25 To exclude the presence of a GSH-bound mononuclear iron complex, the cyclic voltammogram obtained by mixing 240 mM GSH and 2 mM FeCl3 at pH = 8.6 was recorded and it gives rise to a cathodic and anodic current with E1/2 = +240 mV (Fig. S3, ESI†).
The conversion from the dinuclear [FeIIFeIIIS2(GS)4]3− complex to the tetranuclear [FeII2FeIII2S4(GS)4]2− complex was observed by paramagnetic 1H NMR, by titrating a solution of 240 mM GSH and 6.0 mM FeCl3 with increasing amounts of Na2S, as shown in Fig. 2a. In order to mimick the pH of mitochondrial matrix that has been reported to be in the range 7.8–8.0,22,23 we performed this titration at pH 7.9. When FeCl3 is in excess with respect to Na2S, the dinuclear complex predominates, but, upon increasing the amount of Na2S, the broad signal belonging to the dinuclear [FeIIFeIIIS2(GS)4]3− complex decreases in intensity whereas the two signals belonging to the tetranuclear [FeII2FeIII2S4(GS)4]2− complex increase. Fig. 2b, which shows the absolute peak areas of the signals at 16–15.5 ppm (black circles) and 11.6 ppm (blue circles) vs. iron/sulfide ratio, clearly identifies a conversion from the dinuclear complex to the tetranuclear complex. Thus, we can conclude that an excess of sulfide in solution with respect to Fe3+ ions promotes the formation of the tetranuclear [FeII2FeIII2S4(GS)4]2− complex at the expenses of dinuclear [FeIIFeIIIS2(GS)4]3−complex, while the excess of Fe3+ ions favors the formation of the dinuclear [FeIIFeIIIS2(GS)4]3− complex. Upon a complete conversion, the signals of the two complexes are expected to have the same area because the concentration of the dinuclear complex is twice that of the tetranuclear complex and thus the Cys αCH signal in the dinuclear complex will have the same area of those of the Cys βCH2 in the tetranucelar complex. The behavior reported in Fig. 2b is fully consistent with what expected, thus showing a full conversion from the dinuclear to the tetranuclear complex. At an iron/sulfide ratio around 0.70, the ratio between the two complexes is around 1 and signals are broadened due to chemical exchange. To characterize the reverse conversion from tetranuclear to dinuclear complex, a solution containing 6 mM of Na2S was titrated, at pH 7.9, with increasing amounts of FeCl3. As shown in Fig. 2c, complete conversion from [FeII2FeIII2S4(GS)4]2− to [FeIIFeIIIS2(GS)4]3− was observed. When the titration is performed by the addition of increasing amounts of Fe3+ and constant sulfide content, the intensities of all signals (of both dinuclear and tetranuclear complexes) depend on the amount of iron, at variance of what it occurs in the titration monitoring the dinuclear to tetranuclear conversion, where the maximum amount of [Fe–S] cluster is already formed since the first point of the titration. Moreover, during the dinuclear to tetranuclear conversion (Fig. 2c), the excess of Fe3+ present in solution, from the first point of the titration, is reduced by GSH to mononuclear, Fe2+–thiolate complexes.21 According to Bonfio, reduced Fe2+ complexes fail to give further [Fe2S2]2+ cluster upon the following additions of Na2S, because of the reduction of ferric ions prior to the availability of sulfide ions needed for cluster synthesis.17 This is not the case for the reverse situation, where the sulfide ions, mixed with GSH prior to the additions of Fe3+ ions, give the formation of larger amount of [Fe–S] clusters and more intense NMR signal intensity of the dinuclear complex (Fig. 2c), because the rate of cluster synthesis was faster than the reduction of Fe3+ ions.17 When signal areas are normalized with respect to Fe3+ concentration (Fig. 2d), a full conversion from tetranuclear to dinuclear complex is observed, with the same Fe/S dependency observed in the conversion from dinuclear to tetranuclear complex. The two titrations have been repeated also at pH 8.6–8.9 (Fig. S4, ESI†), providing the same results. This is in agreement with the fact that a longer exposure to UV light of a solution of Fe3+–glutathione leads to increased sulfide concentrations determining a conversion from [2Fe–2S] to [4Fe–4S] cluster.17,21
 | ||
| Fig. 2 Interconversion between dinuclear and tetranuclear complexes. (a) Paramagnetic 1H NMR spectra of a solution containing: 240 mM GSH and 6.0 mM FeCl3, with increasing amounts of Na2S, at pH 7.9. Initial and final spectra of the titration are represented, respectively, as solid blue and solid black lines. (b) Peak area for the signals at 16 ppm (black circles) and 11.6 ppm (blue circles) observed in titration (a). (c) Paramagnetic 1H NMR spectra of a solution containing 240 mM GSH and 6.0 mM Na2S, with increasing amounts of FeCl3, at pH 7.9. The initial spectrum is in solid black line and the final spectrum is in solid blue line. (d) Peak area for the signals at 16 ppm (black circles) and 11.6 ppm (blue circles) observed in titration (c). In panel (d), all areas are normalized according to the amount of FeCl3 added, i.e. (Area/[Fe]added) × (normalization factor), while in panel (b) the original signal intensity is shown. Concentrations are reported in Tables S1 and S2 (ESI†). | ||
The formation of the two complexes can be interpreted through the following equilibria:
 | (1) |
 | (2) |
The glutathione-coordinated complex bearing a reduced [2Fe–2S]+ cluster is formed viareaction (1), where the two electrons required to reduce two Fe3+ to Fe2+ ions are provided by free GSH. Then, a coupling reaction (2) between two dinuclear [FeIIFeIIIS2(GS)4]3− complexes occurs to form the tetranuclear [FeII2FeIII2S4(GS)4]2− complex, establishing a dynamic interconversion equilibrium. No electrons are required in this step, since the clusters are stabilized in the appropriate redox state by the reducing environment. The formation of the tetranuclear cluster via the coupling of two dinuclear centers is well documented in literature.26 This suggests that, at low Fe/S ratios, the dinuclear complex acts as transient intermediate for the predominant formation of the tetranuclear complex, while higher Fe/S ratios stabilize the dinuclear cluster and therefore the tetranuclear is not observed.
Recently, Lindahl et al. showed that yeast mitochondria contain non-proteinacious LMM complexes with estimated masses of 580 Da (Fe580) and 1100 Da (Fe1100), which can interconvert.11b The masses of Fe580 and Fe1100 complexes were speculated to correspond to [Fe(GS)(OH2)5] and [Fe2S2(GS)4] complexes, respectively. In mammalian mitochondria, a further LMM complex was detected with a mass of 1500 Da (Fe1500), whose function and chemical composition remains unknown.11a Our in vitro study that mimicks conditions comparable to those present in the mitochondrial matrix22,23 suggests that the dinuclear [Fe2S2(GS)4]3− complex having a reduced [2Fe–2S] cluster might be present in the mitochondrial LIP and that this dinuclear complex might interconvert in the mitochondrial LIP to the tetranuclear [FeII2FeIII2S4(GS)4]2− complex with no electrons required in this process. Thus, we speculate that the Fe1100 complex reported by Lindahl et al.11 might be the dinuclear reduced [FeIIFeIIIS2(GS)4]3− complex, while the Fe1500 complex, observed only in mammalian mitochondria, might be the tetranuclear [FeII2FeIII2S4(GS)4]2− complex. The possible existence of a [Fe–S] cluster pool in the mitochondrial LIP is also supported by a recent study, which showed that yeast mitochondria generate, in the initial step of mitochondrial [Fe–S] cluster assembly machinery, a [Fe–S] cluster intermediate that is absolutely required for the cytoplasmic [Fe–S] cluster assembly machinery (CIA machinery).3 This [Fe–S] cluster intermediate is indeed exported in the cytosol via a mitochondrial inner membrane ATP-binding cassette (ABC) transporter, where it results essential for the maturation of cytosolic and nuclear [Fe–S] proteins.3 It was also proposed5,27,28 that the [Fe–S] cluster intermediate, substrate of the ABC transporter, is the dinuclear [Fe2S2(GS)4]3− complex. In this view, the exported [Fe2S2(GS)4]2− complex plays a role in the regulation of the cellular iron homeostasis, being indeed used by the CIA machinery to mature IRP1 to aconitase. The dynamic interconversion here observed between dinuclear and tetranuclear complexes might thus tune the amount of the dinuclear complex exported as the substrate of the ABC transporter in mammals. Hence, the equilibrium (2) in the mitochondrial LIP might have an effect on the action of the CIA machinery, so possibly contributing to the regulation of the iron homeostasis in mammals.
This research has been supported by Timb3, grant no. 810856, funded by the Horizon 2020 research and innovation program of the European Commission. S. C. B. thanks the Fondazione Cassa di Risparmio di Firenze for partial funding (grant no. CRF2018.0920).
Conflicts of interest
There are no conflicts to declare.Notes and references
- R. Lill and S. A. Freibert, Annu. Rev. Biochem., 2020, 89, 471–499 CrossRef CAS PubMed
.
- S. Ciofi-Baffoni, V. Nasta and L. Banci, Metallomics, 2018, 10, 49–72 CrossRef CAS PubMed
.
- A. K. Pandey, J. Pain, A. Dancis and D. Pain, J. Biol. Chem., 2019, 294, 9489–9502 CrossRef CAS PubMed
.
- S. A. Pearson and J. A. Cowan, Metallomics, 2021, 13 Search PubMed
.
- J. Li and J. A. Cowan, Chem. Commun., 2015, 51, 2253–2255 RSC
.
-
(a) U. Mühlenhoff, B. Hoffmann, N. Richter, N. Rietzschel, F. Spantgar, O. Stehling, M. A. Uzarska and R. Lill, Eur. J. Cell Biol., 2015, 94, 292–308 CrossRef PubMed
; (b) T. A. Rouault, Biometals, 2019, 32, 343–353 CrossRef CAS PubMed
.
- U. Rauen, A. Springer, D. Weisheit, F. Petrat, H. G. Korth, H. de Groot and R. Sustmann, ChemBioChem, 2007, 8, 341–352 CrossRef CAS PubMed
.
- J. Garber Morales, G. P. Holmes-Hampton, R. Miao, Y. Guo, E. Münck and P. A. Lindahl, Biochemistry, 2010, 49, 5436–5444 CrossRef CAS PubMed
.
- G. P. Holmes-Hampton, R. Miao, J. Garber Morales, Y. Guo, E. Münck and P. A. Lindahl, Biochemistry, 2010, 49, 4227–4234 CrossRef CAS PubMed
.
- R. C. Hider and X. Kong, Dalton Trans., 2013, 42, 3220–3229 RSC
.
-
(a) P. A. Lindahl and M. J. Moore, Biochemistry, 2016, 55, 4140–4153 CrossRef CAS PubMed
; (b) S. P. McCormick, M. J. Moore and P. A. Lindahl, Biochemistry, 2015, 54, 3442–3453 CrossRef CAS PubMed
.
- M. Marí, A. Morales, A. Colell, C. García-Ruiz and J. C. Fernández-Checa, Antioxid. Redox Signaling, 2009, 11, 2685–2700 CrossRef PubMed
.
-
(a) S. Sen, B. Rao, C. Wachnowsky and J. A. Cowan, Metallomics, 2018, 10, 1282–1290 CrossRef CAS PubMed
; (b) S. Sen, A. L. Hendricks and J. A. Cowan, FEBS J., 2021, 288, 920–929 CrossRef CAS PubMed
.
- F. Petrat, D. Weisheit, M. Lensen, H. de Groot, R. Sustmann and U. Rauen, Biochem. J., 2002, 362, 137–147 CrossRef CAS
.
- L. Banci, F. Camponeschi, S. Ciofi-Baffoni and M. Piccioli, J. Biol. Inorg. Chem., 2018, 23, 687 CrossRef CAS PubMed
.
- J. Liu, S. Chakraborty, P. Hosseinzadeh, Y. Yu, S. L. Tian, I. Petrik, A. Bhagi and Y. Lu, Chem. Rev., 2014, 114, 4366–4469 CrossRef CAS PubMed
.
-
C. Bonfio, PhD, University of Trento, 2017
.
- L. Banci, S. Ciofi-Baffoni, M. Mikolajczyk, J. Winkelmann, E. Bill and M. E. Pandelia, J. Biol. Inorg. Chem., 2013, 18, 883–893 CrossRef CAS PubMed
.
- H. A. Dailey, M. G. Finnegan and M. K. Johnson, Biochemistry, 1994, 33, 403–407 CrossRef CAS PubMed
.
- M. Piccioli, Magnetochemistry, 2020, 6(4), 46 CrossRef CAS
.
- C. Bonfio, L. Valer, S. Scintilla, S. Shah, D. J. Evans, L. Jin, J. W. Szostak, D. D. Sasselov, J. D. Sutherland and S. S. Mansy, Nat. Chem., 2017, 9, 1229–1234 CrossRef CAS PubMed
.
- A. M. Porcelli, A. Ghelli, C. Zanna, P. Pinton, R. Rizzuto and M. Rugolo, Biochem. Biophys. Res. Commun., 2005, 326, 799–804 CrossRef CAS PubMed
.
- J. Llopis, J. M. McCaffery, A. Miyawaki, M. G. Farquhar and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 6803–6808 CrossRef CAS PubMed
.
- P. Zanello, J. Struct. Biol., 2019, 205, 103–120 CrossRef CAS PubMed
.
- D. W. Bak and S. J. Elliott, Curr. Opin. Chem. Biol., 2014, 19, 50–58 CrossRef CAS PubMed
.
-
(a) K. S. Hagen, J. G. Reynolds and R. H. Holm, J. Am. Chem. Soc., 1981, 103, 4054–4063 CrossRef CAS
; (b) R. H. Holm and W. Lo, Chem. Rev., 2016, 116, 13685–13713 CrossRef CAS PubMed
.
- S. A. Pearson, C. Wachnowsky and J. A. Cowan, Metallomics, 2020, 12, 902–915 CrossRef CAS PubMed
.
- S. A. Pearson and J. A. Cowan, Arch. Biochem. Biophys., 2021, 697, 108661 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed experimental procedures, NMR spectra and cyclic voltammograms. See DOI: 10.1039/d1cc03566e |
| This journal is © The Royal Society of Chemistry 2022 |