Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemoproteomic mapping of human milk oligosaccharide (HMO) interactions in cells†
Abdullah A.
Hassan‡
 a,
Jacob M.
Wozniak‡
b,
Zak
Vilen
ac,
Weichao
Li
a,
Jacob M.
Wozniak‡
b,
Zak
Vilen
ac,
Weichao
Li
 ab,
Appaso
Jadhav
ab,
Appaso
Jadhav
 b,
Christopher G.
Parker
b,
Christopher G.
Parker
 *bc and
Mia L.
Huang
*bc and
Mia L.
Huang
 *abc
*abc
aDepartment of Molecular Medicine, Scripps Research, 10550 N Torrey Pines Rd., La Jolla, CA 92037, USA. E-mail: miahuang@scripps.edu
bDepartment of Chemistry, Scripps Research, 10550 N Torrey Pines Rd., La Jolla, CA 92037, USA. E-mail: cparker@scripps.edu
cSkaggs Graduate School of Chemical and Biological Sciences, Scripps Research, 10550 N Torrey Pines Rd., La Jolla, CA 92037, USA
First published on 18th October 2022
Abstract
Human milk oligosaccharides (HMOs) are a family of unconjugated soluble glycans found in human breast milk that exhibit a myriad of biological activity. While recent studies have uncovered numerous biological functions for HMOs (antimicrobial, anti-inflammatory & probiotic properties), the receptors and protein binding partners involved in these processes are not well characterized. This can be attributed largely in part to the low affinity and transient nature of soluble glycan–protein interactions, precluding the use of traditional characterization techniques to survey binding partners in live cells. Here, we present the use of synthetic photoactivatable HMO probes to capture, enrich and identify HMO protein targets in live cells using mass spectrometry-based chemoproteomics. Following initial validation studies using purified lectins, we profiled the targets of HMO probes in live mouse macrophages. Using this strategy, we mapped hundreds of HMO binding partners across multiple cellular compartments, including many known glycan-binding proteins as well as numerous proteins previously not known to bind glycans. We expect our findings to inform future investigations of the diverse roles of how HMOs may regulate protein function.
Introduction
The interactions between glycans and proteins mediate a diverse range of biological processes,1 and aberrant glycosylation can result in a myriad of pathologies.2–4 Whereas the vast majority of the annotated glycome exists as either lipid- or protein-modified glycoconjugates, “free” (soluble or unconjugated) glycans represent an under-studied fraction of the glycome. From their initial discovery as plant defense signaling molecules5 and modulators of host–pathogen interactions,6 to the recent discovery of a mammalian disaccharide that induces autoimmunity,7 it is becoming increasingly conspicuous that free glycans are important biological regulators.Human milk oligosaccharides (HMOs) are a family of free glycans that are abundantly found in breast milk.8,9 While initially believed to be mere nutritional agents, increasing evidence has highlighted HMOs as important signaling molecules involved in the activation of protein receptors10 and the resolution of inflammation.9,11–13 Some HMOs also display antimicrobial activity by acting as soluble decoys for pathogenic microbes or through inhibiting biofilm formation.14,15 HMOs share a common lactose (Galβ(1-4)Glc) core structure, which is often decorated with additional modifications, such as fucose or sialic acid residues (Fig. 1A). Importantly, such variants can impart vastly different biological activities.16 Despite growing evidence demonstrating that HMOs impact diverse biology important to human health, a molecular understanding of the mechanisms and protein partners through which HMOs act remains limited.12
Similar to glycoconjugates, the interactions of free glycans with glycan-binding proteins are relatively weak (KD ∼ 10−4–10−7 M), dynamic, and prefer the physiological presentation of proteins in live cells.17 These inherent characteristics preclude the use of conventional affinity-based techniques, which require harsh sample preparation procedures. Although several methods have been developed to survey the protein-binding partners of glycoconjugates in live cells, these techniques have not been applied directly to the study of free glycans.18
Thus, we hypothesized that photoaffinity labeling could be applied towards the study of HMO interactions in live cells.19,20 Photoaffinity labeling is able to capture transient and low affinity interactions, and has been previously combined with metabolic oligosaccharide engineering to profile interactions with glycoconjugates.21–25 To enable identification of potential HMO-binding proteins, we chose to utilize a ‘fully-functionalized’ enrichment tag composed of a photoactivatable diazirine group for UV light-induced capture of HMO bound protein targets and an alkyne handle for conjugation to azide-bearing reporter tags via copper-mediated azide-alkyne (CuAAC) “click” chemistry.26,27 We synthesized four probes composed of the most abundant HMOs in human breast milk (lactose or Lac, 2’fucosyl lactose or 2FL, 3'sialyl lactose or 3SL, and 6'sialyl lactose or 6SL), and first cross-evaluated their engagement with purified glycan-binding lectins in vitro, followed by profiling their interactions with live murine macrophages. Using this strategy, we observed that the HMO probes interacted with hundreds of proteins encompassing several intracellular compartments, some of which interacted in a glycan-mediated manner. Among these interactors were galectin-1 and galectin-3, which were validated as 3SL targets in live cells.
Results and discussion
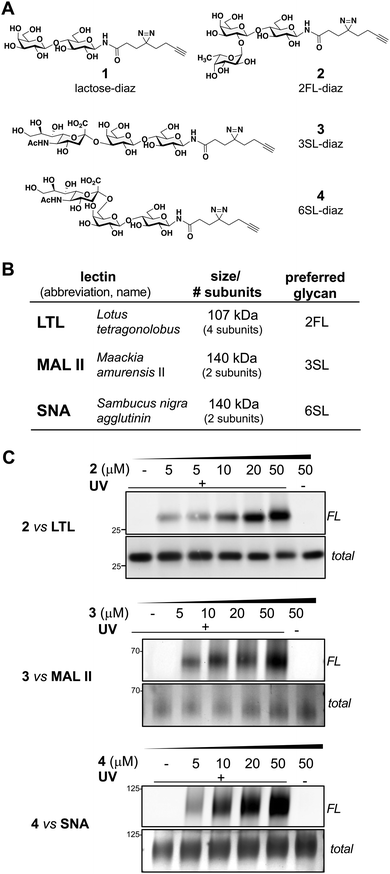
Using a similar experimental protocol previously reported by the Townsend group to prepare the 2FL probe (2),28 the chemical synthesis of our photoactivatable HMO probes commenced with the Kochetkov microwave-assisted conversion of commercially available HMO hemi-acetals into their corresponding amine derivatives,29,30 followed by amide bond coupling with a ‘fully-functionalized’ bifunctional diazirine-alkyne acid linker (see ESI†). The probes 1–4 were synthesized in 44% to 53% overall yields (Fig. 1A). The anomeric positions were chosen based on lectin crystallography studies and biological data suggesting that this site would be the most inert position for derivatization.31–33With probes in hand, we evaluated their engagement with plant lectins in vitro. Plant lectins, such as LTL, MAL II, and SNA, are commonly used reagents to describe glycan patterns in cells. Plant lectins are known to display a degree of selectivity among various glycans, but they also tolerate a wide array of substitutions (Fig. 1B).34–36 Upon incubation of the probes with the plant lectins and UV-light irradiation at 365 nm, the mixtures were reacted with tetramethylrhodamine (TAMRA) azide via CuAAC, separated using denaturing SDS-PAGE, and visualized by in-gel fluorescence scanning and silver staining (Fig. 1C).19 We observed dose-dependent (0–50 μM) photo cross-linking of probes 2–4 with the lectins (Fig. 1C and Fig. S1, ESI†), and cross-linking was dependent on UV irradiation. Consistent with the multi-subunit nature of plant lectins, we observed several bands in SDS-PAGE corresponding to lectin monomers, dimers, or oligomers (Fig. S1, ESI†).
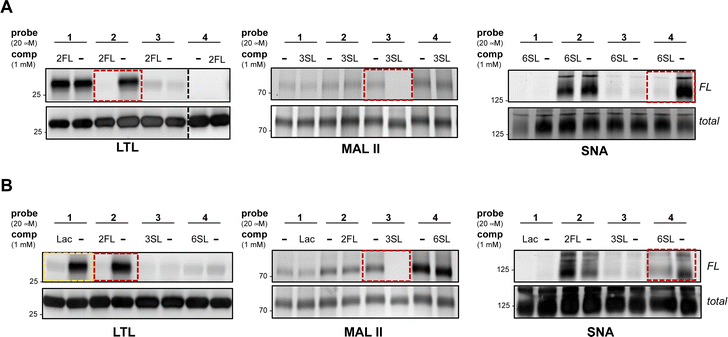
To further evaluate the selectivity of the probes and determine whether the glycan recognition domains of the lectins were engaged, we performed a cross comparison analysis of different probes against individual lectins and used an array of excess unmodified HMOs as competitors. We observed that LTL was equally captured by probes 1 and 2, and excess 2FL abrogated the interaction with 2 but not with probe 1 (Fig. 2A and Fig. S2A, ESI†). MAL II was engaged by all probes, with probe 4 showing maximum cross-linking. However, only the interaction with probe 3 was competed by excess 3SL. Although cross-linking with SNA was observed with probe 2 and probe 4, only the interaction with probe 4 was reduced by 6SL. We also evaluated the ability of unmodified soluble HMOs (Lac, 2FL, 3SL, 6SL) to reduce photo-crosslinking of the probes (Fig. 2B and Fig. S2B, ESI†), and observed that the preferred glycan of each lectin is superior as a competitor molecule compared to other HMOs. We also note, however, that the interaction of probe 1 in the presence of excess Lac reduced photo-crosslinking with LTL. Overall, these experiments demonstrate that the known preferred glycan ligands are required for successful competition, and that, while probes can cross-react with non-binding lectins, incubation with an excess of the corresponding preferred glycan competes for specific interactions.
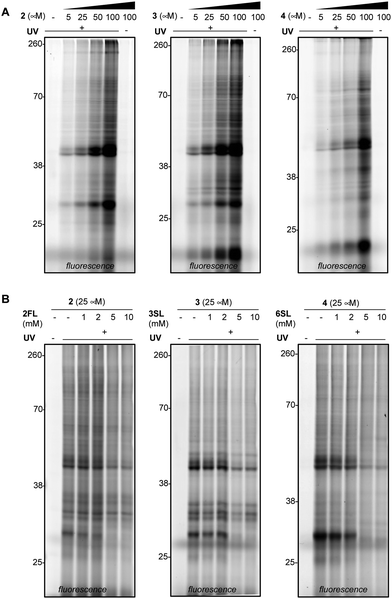
We next evaluated the ability of our probes to engage proteins in live RAW264.7 murine macrophages (Fig. 3A and Fig. S3A, ESI†), as the immunomodulatory activities of HMOs have previously been described in these cells.11,37 In addition to the previously reported immunomodulatory roles, RAW264.7 cells are widely used in immunology due to their phenotypic and functional stability.38 In brief, cells were treated with each probe, followed by exposure to UV light, harvesting, lysis, coupling of probe-modified proteins to TAMRA-azide and fluorescence visualization. We observed dose-dependent photo-crosslinking with probes 2–4 in cells, with protein targets engaged across a range of molecular weights. Photo-crosslinking was observed to be dependent on the presence of the probe and UV, and probe labeling was significantly blocked by excess soluble HMO (Fig. 3B and Fig. S3B, ESI†), suggesting the selective capture of proteins that recognize the cognate free glycan. We also observed that proteins across multiple cellular compartments, including those that are located within cells were engaged by the probes (Fig. S4A–C, ESI†).
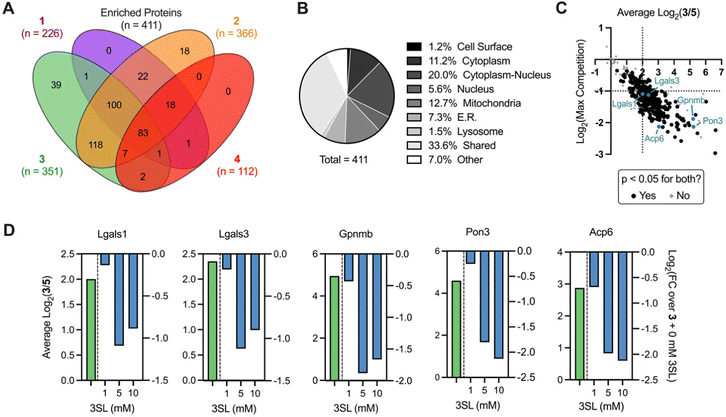
We next sought to identify the protein targets of photoactivatable HMOs using quantitative mass spectrometry-based proteomics. Briefly, following probe treatment and photo-crosslinking, we used CuAAC to append a biotin-azide enrichment handle to crosslinked proteins, which were then enriched with streptavidin beads and digested with trypsin. The resulting peptides were subsequently labeled with isobaric tandem mass tags (TMT)39,40 to facilitate multiplexed, quantitative comparisons. We also included a control probe (5; see ESI†) to account for possible background interactions dependent on the enrichment tag alone.19 Overall, we identified 512 proteins in this experiment (Appendix Table S1, ESI†), observed excellent within replicate correlation and replicates clustered together via unbiased hierarchical clustering based on Pearson correlation (Fig. S5A, ESI†). As expected, the median protein abundance (Fig. S5B, ESI†) and number of enriched protein targets (defined as >4-fold, p < 0.05 over control probe 5 across two biological replicates; Fig. S4C, ESI†) mirrored the general gel fluorescence intensity for each probe. We identified between 100–350 protein targets for each probe for a total of 411 unique targets across all probes (Fig. 4A). Targets spanned diverse cellular compartments, including the cell surface and intracellular organelles (mitochondria, ER, lysosome, Fig. 4B), consistent with our fractionation and microscopy experiments (Fig. S4A–C, ESI†). While only a fraction (3.4%) of these enriched targets are known to bind carbohydrates, >50% of them are annotated as binding to carbohydrate derivatives or other organic cyclic compounds (Fig. S5D, ESI†). With the exception of probe 4, we identified multiple proteins with robust probe-preferred interactions (Fig. S5E–G, ESI†for probes 1–3, respectively). While probe 4 did not exhibit highly preferred interactions among other probes, it did enrich proteins over one or more of the other probes (Fig. S5H, ESI†), demonstrating selectivity imparted by the different molecular recognition of each glycan moiety.
As probe 3 engaged most of the proteins detected and its cognate free glycan (3SL) displays important immunomodulatory properties,11,41,42 we pursued competition experiments to provide additional evidence that the identified targets bind 3SL. In these experiments, cells were pre-treated with excess 3SL prior to incubation with probe 3 plus 3SL and processed for proteomic analysis as described above. We observed excellent correlation between replicates (Fig. S6A, ESI†), and as expected from gel labeling experiments, we observed a strong, dose-dependent reduction of global protein abundances with co-incubation of the 3SL competitor (Fig. S6B, ESI†) and diverse competition profiles (Fig. S6C and D, ESI†).
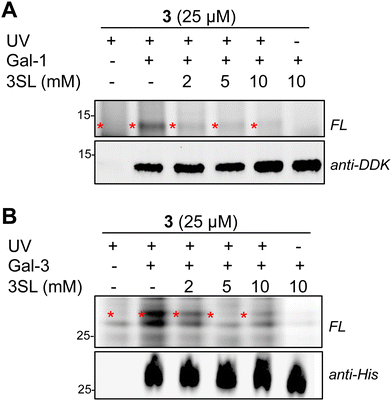
Overall, we identified 193 unique targets that were both enriched by 3 and competed with coincubation of excess free 3SL (>2-fold, p < 0.05 at maximum competition; Appendix Table S2, ESI†). Notably, this list encompasses known glycan-binding proteins including Lgals1 (galectin-1), Lgals3 (galectin-3), and Gpnmb. We also observed proteins that have not been reported to exhibit glycan-binding activities, such as Pon3 (paraoxanase 3) and Acp6 (lysophosphatidic acid phosphatase type 6). (Fig. 4C and D) Intriguingly, both proteins are thought to use small polar molecules (organophosphates, aryl esters and lactones for Pon3) or negatively charged molecules (lysophosphatidic acid for Acp6) as substrates.43–45 We selected galectin-1 and galectin-3 as targets for validation studies, which entailed over-expressing FLAG(DDK)-tagged galectin-1 or His-tagged galectin-3 in HEK293T cells (Fig. S7A–C, ESI†). We observed robust photo-crosslinking of galectin-1 (Fig. 5A) or galectin-3 (Fig. 5B) by probe 3 in live cells, which was abrogated by excess 3SL. While galectins (expressed both intra- and extracellularly)46–48 are commonly known to bind galactose-terminated glycans, they are also known to tolerate α(2,3)-sialic acid substitutions.49 Previous shotgun glycan array studies highlighted that galectins can also bind HMOs and may in some instances be receptors of HMOs.50 Together, these data pinpoint high confidence interactors of probe 3 and demonstrate the capture of known glycan binders as well as unexpected proteins with affinity for HMO probes.
Conclusions
We have demonstrated that fully-functionalized HMO probes can be used to capture and identify potential HMO protein targets in live cells. Our finding that 3SL exhibits maximal interactions with proteins found in murine macrophages is intriguing, given recent findings that it induces transcriptional activation to resolve inflammatory responses, however further investigations are needed to elucidate the underlying molecular mechanisms.11,41,42 Overall, these probes augment the existing toolkit to study of glycan–protein interactions in solution and in equilibrium, and we expect the datasets herein to facilitate our understanding HMO biological activity as well as other untapped free glycans.Methods
Detailed information regarding the availability and sources of all reagents used as well as experimental procedures used are listed in the ESI.† All raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD035729.Conflicts of interest
There are no conflicts to declareAcknowledgements
This work was supported by the NIGMS award to MLH (R35GM142462). JW is supported by NIAID T32 AI007244. We are grateful to Lars Bode and Philip Gordts for helpful discussions.References
- A. Varki, Essentials of glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 3rd edn, 2017 Search PubMed.
- A. Varki, Glycobiology, 2017, 27, 3–49 CrossRef CAS.
- B. A. H. Smith and C. R. Bertozzi, Nat. Rev. Drug Discovery, 2021, 20, 217–243 CrossRef CAS.
- C. Reily, T. J. Stewart, M. B. Renfrow and J. Novak, Nat. Rev. Nephrol., 2019, 15, 346–366 CrossRef.
- A. Darvill, C. Augur, C. Bergmann, R. W. Carlson, J. J. Cheong, S. Eberhard, M. G. Hahn, V. M. Lo, V. Marfa and B. Meyer, et al. , Glycobiology, 1992, 2, 181–198 CrossRef CAS PubMed.
- E. C. Carroll, L. Jin, A. Mori, N. Muñoz-Wolf, E. Oleszycka, H. B. T. Moran, S. Mansouri, C. P. McEntee, E. Lambe, E. M. Agger, P. Andersen, C. Cunningham, P. Hertzog, K. A. Fitzgerald, A. G. Bowie and E. C. Lavelle, Immunity, 2016, 44, 597–608 CrossRef CAS.
- C. S. Fermaintt, K. Sano, Z. Liu, N. Ishii, J. Seino, N. Dobbs, T. Suzuki, Y.-X. Fu, M. A. Lehrman, I. Matsuo and N. Yan, Nat. Commun., 2019, 10, 2377 CrossRef.
- L. Bode, Glycobiology, 2012, 22, 1147–1162 CrossRef CAS.
- L. Bode, Early Hum. Dev., 2015, 91, 619–622 CrossRef CAS.
- F. Foata, N. Sprenger, F. Rochat and S. Damak, Sci. Rep., 2020, 10, 16117 CrossRef CAS.
- A. Pessentheiner, N. Spann, C. Collins, B. Ramms, A. Chiang, Y. Wang, A. Quach, L. Booshehri, A. Hammond, C. Tognaccini, J. Latasiewicz, J. L. Witztum, H. Hoffman, N. Lewis, C. Glass, L. Bode and P. Gordts, Atherosclerosis, 2021, 331, e31 CrossRef.
- C. P. Sodhi, P. Wipf, Y. Yamaguchi, W. B. Fulton, M. Kovler, D. F. Niño, Q. Zhou, E. Banfield, A. D. Werts, M. R. Ladd, R. H. Buck, K. C. Goehring, T. Prindle, S. Wang, H. Jia, P. Lu and D. J. Hackam, Pediatr. Res., 2021, 89, 91–101 CrossRef CAS.
- W. Zhang, J. He-Yang, W. Tu and X. Zhou, Nutr. Metab., 2021, 18, 5 CrossRef CAS PubMed.
- D. L. Ackerman, R. S. Doster, J. H. Weitkamp, D. M. Aronoff, J. A. Gaddy and S. D. Townsend, ACS Infect. Dis., 2017, 3, 595–605 CrossRef CAS.
- S. K. Spicer, R. E. Moore, J. Lu, M. A. Guevara, D. R. Marshall, S. D. Manning, S. M. Damo, S. D. Townsend and J. A. Gaddy, ACS Infect. Dis., 2021, 7, 3254–3263 CrossRef CAS.
- L. Cheng, C. Kong, M. T. C. Walvoort, M. M. Faas and P. Vos, Mol. Nutr. Food Res., 2020, 64, 1900976 CrossRef CAS.
- M. Cohen, Biomolecules, 2015, 5, 2056–2072 CrossRef CAS PubMed.
- M. Critcher, A. A. Hassan and M. L. Huang, Trends Biochem. Sci., 2022, 47, 492–505 CrossRef CAS.
- C. G. Parker, A. Galmozzi, Y. Wang, B. E. Correia, K. Sasaki, C. M. Joslyn, A. S. Kim, C. L. Cavallaro, R. M. Lawrence, S. R. Johnson, I. Narvaiza, E. Saez and B. F. Cravatt, Cell, 2017, 168, 527–541.e529 CrossRef CAS.
- H. Wu and J. Kohler, Curr. Opin. Chem. Biol., 2019, 53, 173–182 CrossRef CAS PubMed.
- S. Han, B. E. Collins, P. Bengtson and J. C. Paulson, Nat. Chem. Biol., 2005, 1, 93–97 CrossRef CAS PubMed.
- Y. Tanaka and J. J. Kohler, J. Am. Chem. Soc., 2008, 130, 3278–3279 CrossRef CAS.
- H. Wu, A. Shajahan, J. Y. Yang, E. Capota, A. M. Wands, C. M. Arthur, S. R. Stowell, K. W. Moremen, P. Azadi and J. J. Kohler, Cell Chem. Biol., 2022, 29, 84 CrossRef CAS.
- S.-S. Ge, B. Chen, Y.-Y. Wu, Q.-S. Long, Y.-L. Zhao, P.-Y. Wang and S. Yang, RSC Adv., 2018, 8, 29428–29454 RSC.
- K. Sakurai, S. Ozawa and T. Yamaguchi, Bioorg. Med. Chem., 2015, 23, 5319–5325 CrossRef CAS.
- Z. Li, P. Hao, L. Li, C. Y. J. Tan, X. Cheng, G. Y. J. Chen, S. K. Sze, H.-M. Shen and S. Q. Yao, Angew. Chem., Int. Ed., 2013, 52, 8551–8556 CrossRef CAS.
- L. P. Conway, A. M. Jadhav, R. A. Homan, W. Li, J. S. Rubiano, R. Hawkins, R. M. Lawrence and C. G. Parker, Chem. Sci., 2021, 12, 7839–7847 RSC.
- S. A. Chambers and S. D. Townsend, Carbohydr. Res., 2020, 488, 107895 CrossRef CAS PubMed.
- L. M. Likhosherstov, O. S. Novikova, V. A. Derevitskaja and N. K. Kochetkov, Carbohydr. Res., 1986, 146, C1–C5 CrossRef CAS.
- M. Bejugam and S. L. Flitsch, Org. Lett., 2004, 6, 4001–4004 CrossRef CAS.
- L. Maveyraud, H. Niwa, V. Guillet, D. I. Svergun, P. V. Konarev, R. A. Palmer, W. J. Peumans, P. Rougé, E. J. M. Van Damme, C. D. Reynolds and L. Mourey, Proteins: Struct., Funct., Bioinf., 2009, 75, 89–103 CrossRef CAS.
- A. Imberty, C. Gautier, J. Lescar, S. Pérez, L. Wyns and R. Loris, J. Biol. Chem., 2000, 275, 17541–17548 CrossRef CAS.
- C. Di Carluccio, R. E. Forgione, M. Montefiori, M. Civera, S. Sattin, G. Smaldone, K. Fukase, Y. Manabe, P. R. Crocker, A. Molinaro, R. Marchetti and A. Silipo, iScience, 2021, 24, 101998 CrossRef CAS.
- A. M. Wu, E. Lisowska, M. Duk and Z. Yang, Glycoconjugate J., 2009, 26, 899–913 CrossRef CAS PubMed.
- D. Bojar, L. Meche, G. Meng, W. Eng, D. F. Smith, R. D. Cummings and L. K. Mahal, ACS Chem. Biol., 2022 DOI:10.1021/acschembio.1c00689.
- S. R. Haseley, P. Talaga, J. P. Kamerling and J. F. G. Vliegenthart, Anal. Biochem., 1999, 274, 203–210 CrossRef CAS PubMed.
- W. Y. Zhang, J. Y. Yan, L. H. Wu, Y. Yu, R. D. Ye, Y. Zhang and X. M. Liang, Carbohydr. Polym., 2019, 207, 230–238 CrossRef CAS PubMed.
- B. Taciak, M. Białasek, A. Braniewska, Z. Sas, P. Sawicka, Ł. Kiraga, T. Rygiel and M. Król, PLoS One, 2018, 13, e0198943 CrossRef PubMed.
- A. Thompson, J. Schafer, K. Kuhn, S. Kienle, J. Schwarz, G. Schmidt, T. Neumann, R. Johnstone, A. K. Mohammed and C. Hamon, Anal. Chem., 2003, 75, 1895–1904 CrossRef CAS.
- G. C. McAlister, E. L. Huttlin, W. Haas, L. Ting, M. P. Jedrychowski, J. C. Rogers, K. Kuhn, I. Pike, R. A. Grothe, J. D. Blethrow and S. P. Gygi, Anal. Chem., 2012, 84, 7469–7478 CrossRef CAS PubMed.
- L.-J. Kang, E.-S. Kwon, K. M. Lee, C. Cho, J.-I. Lee, Y. B. Ryu, T. H. Youm, J. Jeon, M. R. Cho, S.-Y. Jeong, S.-R. Lee, W. Kim and S. Yang, Br. J. Pharmacol., 2018, 175, 4295–4309 CrossRef CAS.
- L.-J. Kang, E. Oh, C. Cho, H. Kwon, C.-G. Lee, J. Jeon, H. Lee, S. Choi, S. J. Han, J. Nam, C.-U. Song, H. Jung, H. Y. Kim, E.-J. Park, E.-J. Choi, J. Kim, S.-I. Eyun and S. Yang, Sci. Rep., 2020, 10, 5603 CrossRef CAS.
- G. Manco, E. Porzio and T. M. Carusone, Antioxidants, 2021, 10, 256 CrossRef CAS.
- C. J. Ng, N. Bourquard, S. Y. Hama, D. Shih, V. R. Grijalva, M. Navab, A. M. Fogelman and S. T. Reddy, Arterioscler., Thromb., Vasc. Biol., 2007, 27, 1368–1374 CrossRef CAS.
- C. J. Ng, S. Y. Hama, N. Bourquard, M. Navab and S. T. Reddy, Mol. Genet. Metab., 2006, 89, 368–373 CrossRef CAS.
- F. T. Liu, R. J. Patterson and J. L. Wang, Biochim Biophys Acta, 2002, 1572, 263–273 CrossRef CAS.
- K. C. Haudek, K. J. Spronk, P. G. Voss, R. J. Patterson, J. L. Wang and E. J. Arnoys, Biochim. Biophys. Acta, Gen. Subj., 2010, 1800, 181–189 CrossRef CAS.
- M. I. Garin, C. C. Chu, D. Golshayan, E. Cernuda-Morollon, R. Wait and R. I. Lechler, Blood, 2007, 109, 2058–2065 CrossRef CAS.
- S. R. Stowell, C. M. Arthur, P. Mehta, K. A. Slanina, O. Blixt, H. Leffler, D. F. Smith and R. D. Cummings, J. Biol. Chem., 2008, 283, 10109–10123 CrossRef CAS PubMed.
- A. J. Noll, J.-P. Gourdine, Y. Yu, Y. Lasanajak, D. F. Smith and R. D. Cummings, Glycobiology, 2016, 26, 655–669 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cb00176d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |