Open Access Article
Open Access ArticleFluorescent metabolic labeling-based quick antibiotic susceptibility test for anaerobic bacteria†
Juan
Gao‡
a,
Juanxiu
Qin‡
b,
Chenling
Ding‡
c,
Yuan
Gao
c,
Junnan
Guo
d,
Min
Li
*b,
Chaoyong
Yang
 *ad and
Wei
Wang
*ad and
Wei
Wang
 *a
*a
aInstitute of Molecular Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200127, China. E-mail: cyyang@xmu.edu.cn; wwang@shsmu.edu.cn
bDepartment of Critical Care Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200127, China. E-mail: ruth_limin@126.com
cDepartment of Laboratory Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200127, China
dThe MOE Key Laboratory of Spectrochemical Analysis and Instrumentation, Key Laboratory for Chemical Biology of Fujian Province State Key Laboratory of Physical Chemistry of Solid Surfaces, Department of Chemical Biology, College of Chemistry and Chemical Engineering Xiamen University, Xiamen 361005, China
First published on 9th September 2022
Abstract
Because of the advancements in medicine and science, the numbers of patients surviving complicated diseases are continuously increasing, which in turn leads to elevated chances of anaerobic infections by endogenous bacteria. Traditional growth yield-based antibiotic susceptibility tests (ASTs) against anaerobic bacteria are very time-consuming (≥48 h) and labor intensive, which delays the timely guidance of antibiotic prescription and increases the mortality of patients. Inspired by a fluorescent D-amino acid (FDAA) labeling-based AST (FaAST) that we recently developed for quick determination of aerobic bacteria's susceptibilities, here we report an accurate and fast AST method for anaerobic pathogens. Based on flow cytometry analysis of anaerobes that have been treated with various doses of antibiotics and metabolically labeled with FDAA, the intensities of which can reflect their affected metabolic status by the drugs, the MICs of each drug can then be determined. The whole process can be completed in 5 h. After testing 40 combinations of the representative anaerobic bacteria and antibiotics, our method demonstrates a high susceptibility category accuracy of 95.0%. This FaAST-based protocol is helpful in accurately and quickly guiding antibiotic decisions when treating critical infections caused by anaerobic bacteria.
Introduction
Because of the increasing numbers of patients with complicated underlying diseases, the incidence of infections caused by endogenous anaerobic bacteria, which can be highly severe and even life-threatening, has been continuously rising.1–3 The underlying conditions associated with the development of anaerobic infections include immunosuppression conditions (organ transplantation, use of corticosteroids and cytotoxic agents, diabetes), hematological malignancies, solid tumors, gynecological or gastrointestinal surgeries, etc.3 With the rapid development of medicines and medical technologies and constant progress of the aging of the population, anaerobic infection is expected to be an important threat to human health in the near future.4Often accompanied by aerobic pathogens, anaerobic bacteria have been found in 1–20% of all bacteremia.5 These surprisingly varying numbers of incidence can be in part explained by the difficulties in diagnosing anaerobes in clinical samples (blood, cerebrospinal fluid, etc.). As the go-to method for identifying anaerobic pathogens, traditional anaerobic blood culture is still commonly performed at clinics when anaerobic infection is suspected. In clinical microbiology, bacterial diagnosis of aerobic pathogens is frequently followed by AST. However, only 21% of laboratories in the US routinely perform AST with anaerobes,1 partially because carrying out AST against anaerobes is technically challenging and very time-consuming (≥48 h for agar dilution, broth dilution and Etest methods). Empirical broad-spectrum antimicrobial therapy, therefore, is often prescribed long before the results of AST are reported and this has been proved to be associated with poor clinical responses.6 Many anaerobic bacteria possess genetically-encoded drug resistance,7,8 metronidazole-resistance in some Gram-positive rods,9 clindamycin-resistance in Fusobacterium,10 and cefoxitin-resistance in Clostridioides difficile,11 to name a few. Moreover, multidrug-resistant strains have also been increasingly prevalent in the last decade from Bacteroides/Parabacteroides, which are among the most common anaerobic pathogens found in bacteremia.12,13 This expanding drug resistance highlights the necessity of AST for anaerobes.
In the past decade, a number of new AST techniques have been developed for aerobic bacteria.14,15 These techniques can be categorized into optical/mechanical/electrical analysis-based, DNA sequencing-facilitated, and microfluidics-based ASTs, based on how they function. However, to the best of our knowledge, few of the methods have been demonstrated to be capable of analyzing anaerobes, in part because of their unsuitability under anaerobic environments. Therefore, considering the growing threat of anaerobic infections, the time-consuming process of current AST for anaerobes and the lack of new testing methods, developing a new AST strategy for anaerobic pathogens is still highly desired.
Results and discussion
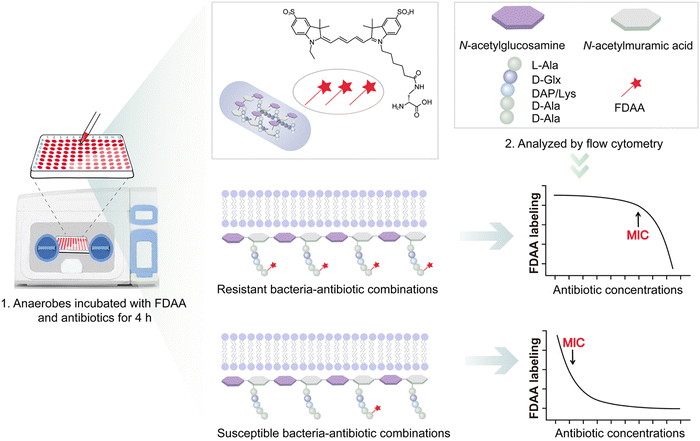
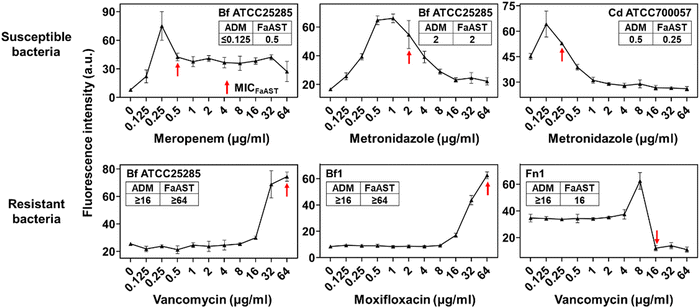
Recently, we reported a fluorescent D-amino acid (FDAA) labeling-based AST (FaAST), which could offer accurate minimum inhibitory concentrations (MICs) for aerobic bacteria in 2–3 h.16 FDAAs are metabolic labeling probes that can fluorescently tag bacteria via their peptidoglycan transpeptidases (both D, D- and L, D-),17 and we recently found that the labeling intensities of FDAA highly correlated with the bacterial metabolic status.18 Therefore, in FaAST, the resistant bacteria showed a slow decline in FDAA-labeling after antibiotic treatment and the susceptible ones demonstrated quick labeling-decline when low doses of drugs were used. MICs can then be determined according to their intensity changing trends (Fig. 1). Inspired by the successful use of FaAST against aerobes, here we tested the suitability of using FaAST in MIC analysis of anaerobic pathogens.Four important anaerobic bacterial species were first chosen to test via FaAST. Bacteroides fragilis (Bf) is currently the most prevalent anaerobic pathogen in bacteremia;12Fusobacterium nucleatum (Fn) has been intensely studied in the last decade as a pro-carcinogenic bacterium for colorectal cancer;10,19C. difficile (Cd) and Clostridium perfringens (Cp) are the most frequent bacteria causing intestinal anaerobic infections.20,21 Three susceptible combinations (Bf ATCC25285 against meropenem and metronidazole, Cd ATCC700057 against metronidazole) and three resistant combinations (Bf ATCC25285 and Fn1 against vancomycin, Bf1 against moxifloxacin) were first tested. The four antibiotics, meropenem (a penicillin-binding protein-inhibiting carbapenem drug), metronidazole (a nitroso radical-forming and DNA-disrupting nitroimidazole drug), vancomycin (functions via binding with lipid II to inhibit peptidoglycan synthesis) and moxifloxacin (a DNA gyrase-inhibiting fluoroquinolone drug), were chosen because of their clinical relevance against anaerobic infections and all four were bacteriocidal.
FaAST protocols used for aerobic bacteria were adopted here with some modifications. In brief, fresh bacterial suspensions with an optical density at 600 nm (OD600) of 0.05 were inoculated into 96-well plates loaded with different concentrations of antibiotic (0–64 μg mL−1, 2-fold serial dilution) and the FDAA probe (cyanine5-amine-D-alanine probe, Cy5ADA, 0.15 mM). After 4 h of incubation, the fluorescence intensities of the diluted anaerobic bacterial solutions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with phosphate buffered saline, PBS) were analyzed via flow cytometry. Different from the previous FaAST protocol, here, all operations were conducted in an anaerobic chamber before flow cytometry.
10 with phosphate buffered saline, PBS) were analyzed via flow cytometry. Different from the previous FaAST protocol, here, all operations were conducted in an anaerobic chamber before flow cytometry.
The results indicated that the metabolic activities of anaerobic strains in response to different doses of drugs were accurately revealed by FDAA-labeling (Fig. 2). When testing aerobic bacteria, FaAST-MIC was empirically defined as the first antibiotic concentration where a steady drop of FDAA-labeling appeared. Using this definition, the MICs of the three susceptible combinations were determined to be 0.5 μg mL−1 of meropenem, 2 μg mL−1 of metronidazole for Bf ATCC25285, and 0.25 μg mL−1 of metronidazole for Cd ATCC700057. As expected, the resistant combinations showed much higher MICs (≥64 μg mL−1 of vancomycin for Bf ATCC25285, ≥64 μg mL−1 of moxifloxacin for Bf1, and 16 μg mL−1 of vancomycin for Fn1). MICs were also determined in parallel using the standard agar dilution method (ADM, values are shown in each diagram) and the maximum concentration for all antibiotics was set to 16 μg mL−1 according to the anaerobic MIC breakpoints and clinical experience. Testing results of Bf ATCC25285 against metronidazole are shown as an example (Fig. S1, ESI†). Although the values between the two methods were not exactly identical, especially in some resistant combinations (due to the difference of the maximum testing concentrations between the two methods), their susceptibility categories were consistent in each drug-bacteria combination. Of note, the sudden fluorescence increases at lower drug concentration near their MICs (showed in both susceptible and resistant combinations), which were also observed in aerobic FaAST,16 agreed with the fact that bactericidal drugs were associated with stimulated metabolic rates at sub-MIC ranges.22
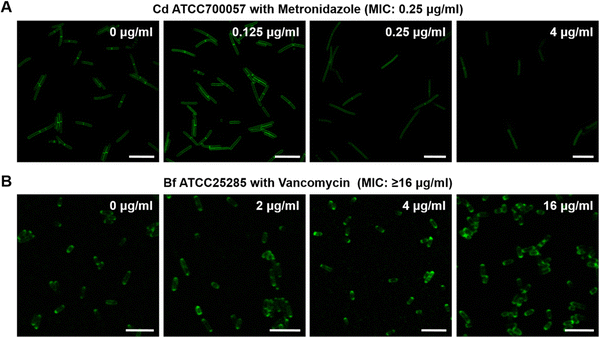
An imaging characterization of the anaerobic bacteria's responses to antibiotics was also performed. The FDAA-labeling intensity of the susceptible strain Cd ATCC700057 declined as the dose of metronidazole increased (Fig. 3A). For resistant bacteria, however, Bf ATCC25285 still showed high labeling even when treated with 16 μg mL−1 of vancomycin (Fig. 3B). These imaging results further demonstrated that the FDAA labeling could accurately reflect the anaerobic bacteria's metabolic responses to different antibiotics, which underlies a reliable AST strategy for anaerobic pathogens.
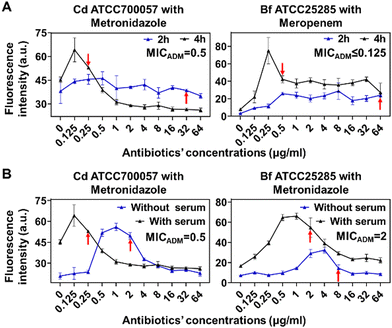
To further improve the speed and accuracy of FaAST for anaerobic bacteria, we went on to optimize the protocol. First, we tested whether we could shorten the incubation time to 2 h, which was suitable for testing aerobic bacteria, but found that a minimum of 4 h was required to accurately determine the MICs of metronidazole against Cd and meropenem against Bf (Fig. 4A). This discrepancy might be due to the slower growth rate of anaerobes in contrast to aerobic bacteria.23 In traditional AST for anaerobes, 5% of lysed horse blood is normally supplemented to the Brucella broth to stimulate the bacterial growth.24 Out of concern about the potential interference of blood cells with flow cytometry analysis, here filtered bovine serum (5%, v/v) was used instead to provide nutrients.25 As shown in Fig. 4B, compared with the tests without using serum, serum supplementation resulted in improved MIC-accuracy of Cd and Bf against metronidazole.
To test the potential of our method in testing the susceptibilities of more antibiotics, we used a model strain of E. coli ATCC25922 and tested it against a broad antibiotic panel, including two bacteriocidals (D-cycloserine, fosfomycin) and four bacteriostatics (chloramphenicol, tetracycline, linezolid, and erythromycin). These six antibiotics are also occasionally used in treating anaerobic infections. In parallel with our previous aerobic FaAST report,16 the MICs of this E. coli strain were tested under both anaerobic and aerobic conditions. Considering the high reliability of the broth microdilution method (BMD) in testing the MICs of aerobic bacteria, we used it as the standard protocol in these experiments. As shown in Table 1, FaAST showed agreeing MICs with the traditional BMD method against most antibiotics (graphical charts shown in Fig. S2, ESI†). One exception was fosfomycin, where FaAST showed quite discrepant MICs compared with the standard method. It is known that the MIC of this drug cannot be accurately determined by a broth-based but only agar-based dilution method.26 This phenomenon was also observed in our tests; compared with MIC by BMD, MIC by ADM was more consistent with the CLSI standard values (Table 1). This might explain the inaccuracy in our FaAST test (also broth-based) against this antibiotic. Taking these data together, FaAST demonstrated its capability in accurately determining the susceptibilities of a wide range of antibiotics under anaerobic conditions.
| Antibiotics/MICsa | Aerobic | Anaerobic | |||
|---|---|---|---|---|---|
| CLSI | BMD | FaAST | BMD | FaAST | |
| a MICs of antibiotics to which the bacteria are susceptible are shown in bold, resistant in italic, and those that are not categorized into susceptible or resistant by CLSI are shown in roman. Combinations mistakenly categorized by FaAST are underscored. Units, μg mL−1; NA, not available; BMD, broth microdilution method; ADM, agar dilution method. | |||||
| D-Cycloserine | NA | 32 | 64 | 64 | 64 |
| Chloroamphenicol | 2–8 | 8 | 4 | 4 | 4 |
| Tetracycline | 0.5–2 | 1 | 1 | 1 | 1 |
| Linezolid | NA | ≥64 | ≥64 | ≥64 | ≥64 |
| Erythromycin | NA | ≥64 | ≥64 | ≥64 | ≥64 |
| Fosfomycin | 0.5–2 | 16 (1 in ADM) | 64 | 16 | ≥64 |
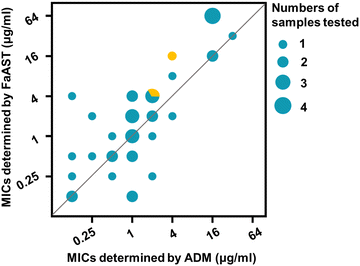
Next, to comprehensively assess the accuracy and reliability of FaAST against anaerobes, the MICs of six anaerobic isolates (2 of Bf, 2 of Cd, 1 of Fn, and 1 of Clostridium perfringens (Cf)) collected from different clinical sources at Renji Hospital (Table S1, ESI†) and a standard strain of Bf and Cd from the Clinical Laboratory Standard Institute (CLSI) were analyzed using the optimized protocol. A total of 40 antibiotic-bacterium combinations were detected in parallel by FaAST and the standard ADM method (Table 2). According to the Food and Drug Administration (FDA) guidance document,27 the ratios of minor discrepancy (min), major discrepancy (maj), very major discrepancy (vmj), and the category agreement (CA) rates were calculated. The min, maj, vmj, and CA rates of FaAST were 2.5%, 3.0%, 0, and 95.0%, respectively (a more intuitive correlation plot of the data is shown in Fig. 5). These high CA and low error rates of FaAST for anaerobic bacteria fully meet the FDA's requirements for a new AST method (min ≤ 10%, maj ≤ 3%, vmj ≤ 1.5%, and CA ≥ 90%). Taking these data together, compared with traditional AST methods, which require at least 42–48 h to report,28 FaAST-MICs can be accurately obtained in ∼5 h. This may greatly improve the efficiency of anaerobic infection treatment by saving ∼2 days in guiding antibiotic use.
| Antibiotics/a Bacteria/MICs | Gram-negative bacteria | Gram-positive bacteria | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteroides fragilis | Fusobacterium nucleatum | Clostridioides difficile | Clostridium perfringens | ||||||||||||||||
| Bf ATCC25285 | Bf1 | Bf2 | Fn1 | Cd ATCC700057 | Cd253 | Cd211 | Cp0317601 | ||||||||||||
| QC | ADM | FaAST | ADM | FaAST | ADM | FaAST | ADM | FaAST | QC | ADM | FaAST | ADM | FaAST | ADM | FaAST | ADM | FaAST | ||
| ADM | BMD | ADM | |||||||||||||||||
| a MICs of antibiotics to which the bacteria are susceptible are shown in roman; resistant in bold; intermediate in italic; the combinations mistakenly categorized by FaAST are underscored; units, μg mL−1. QC, quality control; BMD, broth microdilution method. | |||||||||||||||||||
| Vancomycin | — | — | ≥16 | ≥64 | ≥16 | 64 | ≥16 | ≥64 | ≥16 | 16 | 0.5–4 | 2 | 0.25 | 1 | 1 | 1 | 0.125 | 2 | ≥4 |
| Linezolid | 2–8 | 2–8 | 4 | 8 | 2 | 4 | 2 | 4 | 2 | 2 | 1–4 | 1 | 0.5 | 1 | 0.5 | 1 | 0.125 | 1 | 2 |
| Moxifloxacin | 0.125–0.5 | 0.125–0.5 | 0.5 | 1 | ≥16 | ≥64 | 4 | ≥16 | 0.5 | 0.5 | 1–4 | 1 | 4 | ≥32 | 32 | 16 | 16 | 0.25 | 2 |
| Meropenem | 0.03–0.25 | 0.03–0.25 | ≤0.125 | 0.5 | 1 | 2 | 0.25 | 0.5 | ≤0.125 | 4 | 0.5–4 | 1 | 2 | 4 | 2 | 0.5 | 0.5 | ≤0.125 | 0.125 |
| Metronidazole | 0.25–1 | 0.25–2 | 2 | 2 | 1 | 1 | 1 | 1 | ≤0.125 | 0.25 | 0.125–0.5 | 0.5 | 0.25 | 2 | 1 | 0.125 | 0.125 | 1 | 4 |
Conclusions
As new techniques for fast and accurate identification of bacterial taxon (the matrix-assisted laser desorption–ionization time-of-flight mass spectrometry-based protocol, for example) are quickly popularized in hospitals worldwide and the blood culture method is continuously improved, it is reasonable to expect that the numbers of anaerobic pathogen diagnoses will be continuously increasing in the future. Of note, our understanding of the physiological and pathological roles of human gut microbiota has been quickly progressing in the past two decades and the translocation of gut bacteria into circulation in clinically critical patients has also been recently proved using advanced sequencing and bioinformatical techniques.29 The recognition of gut anaerobe-induced system infections, therefore, may be paid more attention by physicians. Considering the diversity of anaerobes, more tests for different types of bacteria and antibiotics are required to further evaluate the reliability of FaAST. Due to the difficulties of handling bacteria in an anaerobic chamber, automated and integrated equipment for FaAST-based anaerobe analysis will greatly facilitate the practice of this new strategy. We expect that as our FaAST-based strategy will be continuously optimized, this new antibiotic-prescription guiding method will truly benefit patients in the foreseeable future.Ethical statement
This study was approved by the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China, protocol number KY2020-174.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We are grateful to the National Natural Science Foundation of China (Grants 22122702 and 81902118), the Science and Technology Commission Project of Shanghai Municipality (Grant 18411951100), and the Innovative Research Team of High-Level Local Universities in Shanghai for financial support (SHSMU-ZLCX20212601).References
- F. Cobo, J. Borrego, E. Gómez, I. Casanovas, E. Calatrava, C. Foronda and J. M. Navarro-MaríClinical, Antibiotics, 2020, 9, 345 CrossRef CAS PubMed.
- W. Jamal, G. Al Hashem and V. O. Rotimi, Anaerobe, 2015, 31, 25–30 CrossRef CAS PubMed.
- M. Gajdács and E. Urbán, Eur. J. Microbiol. Immunol., 2020, 10, 64–75 Search PubMed.
- M. Gajdács, G. Spengler and E. Urbán, Antibiotics, 2017, 6, 25 CrossRef PubMed.
- E. J. Goldstein, Clin. Infect. Dis., 1996, 23, S97–S101 CrossRef PubMed.
- E. J. Goldstein, J. S. Solomkin, D. M. Citron and J. D. Alder, Clin. Infect. Dis., 2011, 53, 1074–1080 CrossRef CAS PubMed.
- I. N. Rôças and J. F. Siqueira Jr., Anaerobe, 2012, 18, 576–580 CrossRef PubMed.
- I. Brook, H. M. Wexler and E. J. Goldstein, Clin. Microbiol. Rev., 2013, 26, 526–546 CrossRef CAS PubMed.
- G. A. Pankuch, M. R. Jacobs and P. C. Appelbaum, Antimicrob. Agents Chemother., 1993, 37, 1649–1654 CrossRef CAS PubMed.
- C. A. Brennan and W. S. Garrett, Nat. Rev. Microbiol., 2019, 17, 156–166 CrossRef CAS PubMed.
- Z. Peng, A. Addisu, S. Alrabaa and X. Sun, Front. Microbiol., 2017, 8, 02584 CrossRef.
- T. Y. Tan, L. S. Ng, L. L. Kwang, S. Rao and L. C. Eng, Anaerobe, 2017, 43, 69–74 CrossRef PubMed.
- J. C. Ezeji, D. K. Sarikonda, A. Hopperton, H. L. Erkkila, D. E. Cohen, S. P. Martinez, F. Cominelli, T. Kuwahara, A. E. K. Dichosa, C. E. Good, M. R. Jacobs, M. Khoretonenko, A. Veloo and A. Rodriguez-Palacios, Gut Microbes, 2021, 13, 1922241 CrossRef PubMed.
- A. van Belkum, C. D. Burnham, J. W. A. Rossen, F. Mallard, O. Rochas and W. M. Dunne, Nat. Rev. Microbiol., 2020, 18, 299–311 CrossRef CAS PubMed.
- A. van Belkum, T. T. Bachmann, G. Ludke, J. G. Lisby, G. Kahlmeter, A. Mohess, K. Becker, J. P. Hays, N. Woodford, K. Mitsakakis, J. Moran-Gilad, J. Vila, H. Peter, J. H. Rex and W. J. Dunne, Nat. Rev. Microbiol., 2019, 17, 51–62 CrossRef CAS PubMed.
- J. Gao, J. Guo, J. Chen, C. Ding, J. Wang, Q. Huang, Y. Jian, X. Zhao, M. Li, Y. Gao, C. Yang and W. Wang, Adv. Healthcare Mater., 2021, 2101736 Search PubMed.
- E. Kuru, A. Radkov, X. Meng, A. Egan, L. Alvarez, A. Dowson, G. Booher, E. Breukink, D. I. Roper, F. Cava, W. Vollmer, Y. Brun and M. S. VanNieuwenhze, ACS Chem. Biol., 2019, 14, 2745–2756 CrossRef CAS PubMed.
- L. Lin, J. Song, Y. Du, Q. Wu, J. Gao, Y. Song, C. Yang and W. Wang, Angew. Chem., Int. Ed., 2020, 59, 11923–11926 CrossRef CAS PubMed.
- T. Yu, F. Guo, Y. Yu, T. Sun, D. Ma, J. Han, Y. Qian, I. Kryczek, D. Sun, N. Nagarsheth, Y. Chen, H. Chen, J. Hong, W. Zou and J. Fang, Cell, 2017, 170, 548–563 CrossRef CAS PubMed.
- C. R. Kelly, M. Fischer, J. R. Allegretti, K. LaPlante, D. B. Stewart, B. N. Limketkai and N. H. Stollman, Am. J. Gastroenterol., 2021, 116, 1124–1147 CrossRef PubMed.
- E. C. Martens, M. Neumann and M. S. Desai, Nat. Rev. Microbiol., 2018, 16, 457–470 CrossRef CAS PubMed.
- D. N. Wilson, Nat. Rev. Microbiol., 2014, 12, 35 CrossRef CAS PubMed.
- E. Nagy, L. Boyanova and U. S. Justesen, Clin. Microbiol. Infec., 2018, 24, 1139–1148 CrossRef CAS PubMed.
- NCCLS, Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria; Approved Standard—Sixth Edition, USA, 2004.
- J. C. Lagier, S. Edouard, I. Pagnier, O. Mediannikov, M. Drancourt and D. Raoult, Clin. Microbiol. Rev., 2015, 28, 208–236 CrossRef CAS PubMed.
- M. P. Weinstein, Performance standards for antimicrobial susceptibility testing, CLSI supplement M100, Clinical and Laboratory Standards Institute, Wayne, PA, 31st edn, 2021 Search PubMed.
- H. Services, Class II Special Controls Guidance Document: Antimicrobial Susceptibility Test (AST) Systems; Guidance for Industry and FDA, USA, 2007.
- A. N. Schuetz, Clin. Infect. Dis., 2014, 59, 698–705 CrossRef PubMed.
- F. B. Tamburini, T. M. Andermann, E. Tkachenko, F. Senchyna, N. Banaei and A. S. Bhatt, Nat. Med., 2018, 24, 1809–1814 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cb00163b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |