Open Access Article
Open Access ArticleLabeling and sequencing nucleic acid modifications using bio-orthogonal tools
Hui
Liu
 *a,
Yafen
Wang
b and
Xiang
Zhou
*a,
Yafen
Wang
b and
Xiang
Zhou
 a
a
aCollege of Chemistry and Molecular Sciences, Wuhan University, Wuhan 430072, China. E-mail: HuiLiuwhu@whu.edu.cn
bSchool of Public Health, Wuhan University, Wuhan 430071, China
First published on 22nd June 2022
Abstract
The bio-orthogonal reaction is a type of reaction that can occur within a cell without interfering with the active components of the cell. Bio-orthogonal reaction techniques have been used to label and track the synthesis, metabolism, and interactions of distinct biomacromolecules in cells. Thus, it is a handy tool for analyzing biological macromolecules within cells. Nucleic acid modifications are widely distributed in DNA and RNA in cells and play a critical role in regulating physiological and pathological cellular activities. Utilizing bio-orthogonal tools to study modified bases is a critical and worthwhile research direction. The development of bio-orthogonal reactions focusing on nucleic acid modifications has enabled the mapping of nucleic acid modifications in DNA and RNA. This review discusses the recent advances in bio-orthogonal labeling and sequencing nucleic acid modifications in DNA and RNA.
1 Introduction
As the primary genetic material in cells, nucleic acids regulate various complex biological functions.1 Following extensive detection and analysis of the nucleic acid composition, it was discovered that there are numerous chemically modified bases in addition to the primary constituent units (ATCG in DNA, AUCG in RNA).2 The discovery of nucleic acid modifications in DNA or RNA opened up new avenues for studying the relationship between genetic information and cellular biological processes.2–4 These so-called epigenetic modifications are associated with normal cellular functions and the onset and progression of various diseases involving gene expression regulation via interactions with related proteins.5 DNA cytosine methylation is a well-known example of DNA modified bases, and it plays a complex role in regulating a variety of physiological processes within the cell.6 RNA contains significantly more modified bases than DNA,4 and its most abundant modified base, m6A, has been implicated in intracellular processes such as RNA transcription and metabolism, as well as immune regulation.7,8 Thus, labeling and analyzing modified nucleic acids can be a handy research tool for gaining a better understanding of how modified nucleic acids function within the cell.9,10 Numerous methods have been developed for detecting and mapping nucleic acid modifications, including liquid chromatography, immunostaining-based detection, and chemically labeled sequencing technologies.11,12 Due to the selectivity, biocompatibility, and biological inertness of the bio-orthogonal reaction, it has been used to label distinct biomacromolecules within cells and then track their synthesis, metabolism, and interactions with other biomacromolecules within cells.13–15 Click chemistry plays a critical role in bio-orthogonal reactions such as Diels–Alder reactions, copper(I)-catalyzed azide–alkyne cycloadditions, and strain-promoted azide–alkyne cycloadditions.16–18 By labeling the modified nucleic acids with bio-orthogonal tools, subsequent analysis such as sequencing and imaging can be performed easily (Fig. 1).19–21 This review provides a systematic overview of recent research efforts aimed at improving the intuitive understanding of the application of bio-orthogonal techniques to the labeling and sequencing of nucleic acid modifications.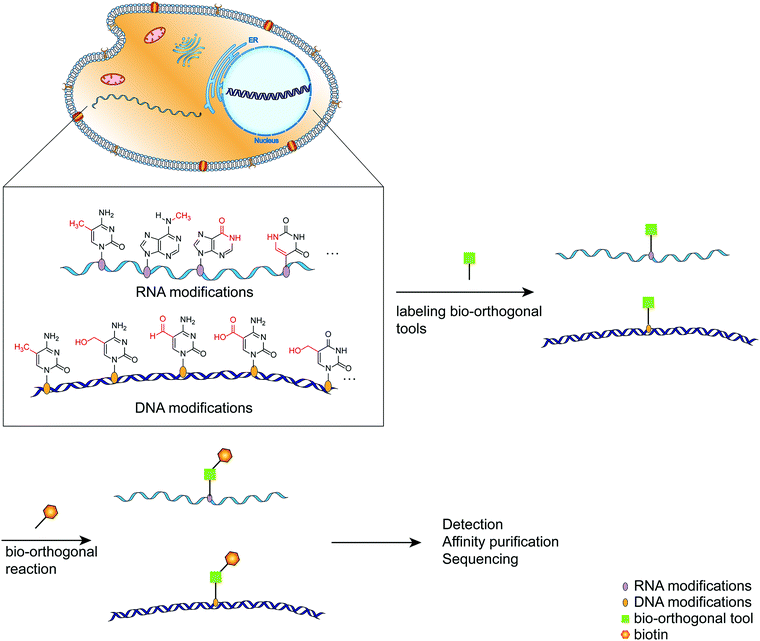 | ||
| Fig. 1 A schematic diagram of the simplified process for labeling modified bases using bio-orthogonal tools. | ||
2 Labeling nucleic acid modifications with the functional group
Due to the critical nature of nucleic acid modifications, research on them is advancing rapidly, and preliminary findings indicate over 170 distinct types of nucleic acid modifications (including DNA and RNA).10 Since the reactive activity of nucleic acid modifications varies considerably, they are frequently labeled using various methods. The methods for labeling bio-orthogonal reactive groups on modified bases fall into two broad categories: chemical labeling and enzyme-assisted labeling.2.1 Chemical labeling methods
Using selected chemical compounds, the chemical labeling approach inserts a functional group with bio-orthogonal activity into the target nucleic acid modifications. The following sections discuss labeling DNA and RNA modifications using various chemical modification methods.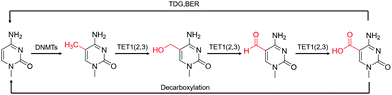 | ||
| Fig. 2 The transition of various cytosine modifications.31 | ||
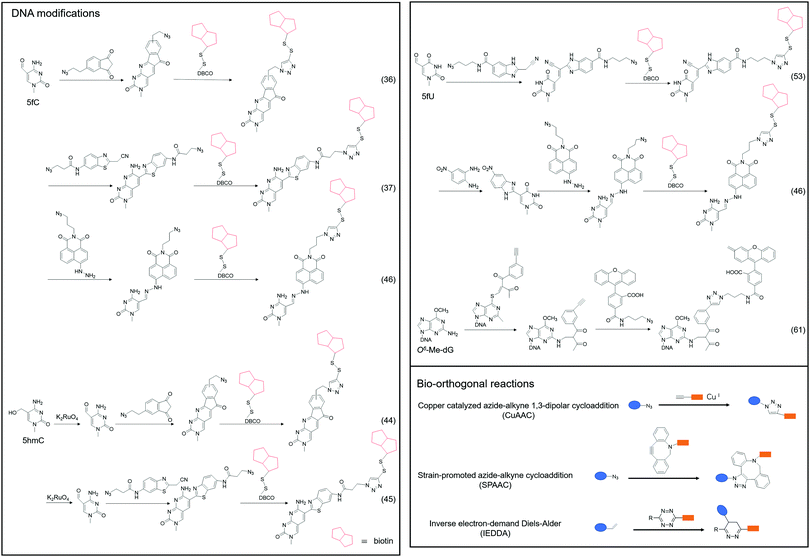
Although 5fC has been widely discovered as the oxidized derivative of 5mC, its content is extremely low, ranging between 2/10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 2/100
000 and 2/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 of cytosine.35 Chengqi Yi et al. cyclized 5fC and azido-modified 1,3-indandione (AI) intermolecularly.36 With the assistance of AI, the azide was attached to 5fC, and then a bio-orthogonal reaction with DBCO-SS-PEG3-biotin was catalyzed by strain-promoted azide–alkyne cycloaddition (SPAAC) to modify functional molecules such as biotin on 5fC for subsequent separation and purification. Our group developed a novel chemical labeling method for connecting azide molecules to 5fC37via the intermolecular cycloaddition reaction. This procedure utilizes direct condensation of aminobenzaldehydes and cyano reagents to attach 3-azido-N-(2-(cyanomethyl)benzo[d]thiazol-6-yl) propanamide (azi-BP) to 5fC, thereby introducing the azide group onto 5fC. SPAAC was then used to enrich the target sequence with DBCO-S-S-PEG3-biotin. 5hmC is a more stable form of DNA modification that occurs in greater abundance and dispersion in cells.38 It is a biomarker for various tumor types.39,40 Alternative detection methods for 5hmC have included antibody enrichment.12,41–43 Direct chemical labeling of bio-orthogonal small molecules on 5hmC is more complicated than labeling 5fC. However, by selective oxidation of 5hmC, the 5fC modification approach can be used to complete the selective labeling of 5hmC.44,45 Chengqi Yi et al. used potassium ruthenate (K2RuO4) to complete the specific oxidation of 5hmC's hydroxymethyl, oxidized 5hmC to 5fC and then used 5fC to react with AI, thereby achieving the 5hmC selection marker. Additionally, our group used K2RuO4 and intermolecular cycloaddition with azi-BP to achieve specific oxidation and labeling to modify 5hmC. Apart from intermolecular cycloaddition, our group labeled 5fC using the reaction between hydrazine and the 5-formyl group of 5fC. 5fC enables covalent attachment to the azide group via novel azide and hydrazine-tethered naphthylimide analog labeling.46 Simultaneously, this reaction can be used to modify the 5-formyl group of 5fU. 5fC and 5fU were labeled with azide under various conditions, followed by a bio-orthogonal reaction, and functional molecules such as biotin were connected. 5fU is a thymine oxidative modification product abundant in mammalian cells, bacteria, and other organisms.47 It is the byproduct of cells that have been damaged by external agents such as ultraviolet light, particle radiation, or free radicals produced by chemical substances.48–51 Since the previous method can label both 5fC and 5fU, our group developed a method for labeling 5fU specifically.52,53 The experimental results demonstrated that by using the specific reaction of (2-benzimidazolyl) acetonitrile (azi-BIAN) with 5fU, interference from 5fC can be avoided, and 5fU can be explicitly labeled.
000 of cytosine.35 Chengqi Yi et al. cyclized 5fC and azido-modified 1,3-indandione (AI) intermolecularly.36 With the assistance of AI, the azide was attached to 5fC, and then a bio-orthogonal reaction with DBCO-SS-PEG3-biotin was catalyzed by strain-promoted azide–alkyne cycloaddition (SPAAC) to modify functional molecules such as biotin on 5fC for subsequent separation and purification. Our group developed a novel chemical labeling method for connecting azide molecules to 5fC37via the intermolecular cycloaddition reaction. This procedure utilizes direct condensation of aminobenzaldehydes and cyano reagents to attach 3-azido-N-(2-(cyanomethyl)benzo[d]thiazol-6-yl) propanamide (azi-BP) to 5fC, thereby introducing the azide group onto 5fC. SPAAC was then used to enrich the target sequence with DBCO-S-S-PEG3-biotin. 5hmC is a more stable form of DNA modification that occurs in greater abundance and dispersion in cells.38 It is a biomarker for various tumor types.39,40 Alternative detection methods for 5hmC have included antibody enrichment.12,41–43 Direct chemical labeling of bio-orthogonal small molecules on 5hmC is more complicated than labeling 5fC. However, by selective oxidation of 5hmC, the 5fC modification approach can be used to complete the selective labeling of 5hmC.44,45 Chengqi Yi et al. used potassium ruthenate (K2RuO4) to complete the specific oxidation of 5hmC's hydroxymethyl, oxidized 5hmC to 5fC and then used 5fC to react with AI, thereby achieving the 5hmC selection marker. Additionally, our group used K2RuO4 and intermolecular cycloaddition with azi-BP to achieve specific oxidation and labeling to modify 5hmC. Apart from intermolecular cycloaddition, our group labeled 5fC using the reaction between hydrazine and the 5-formyl group of 5fC. 5fC enables covalent attachment to the azide group via novel azide and hydrazine-tethered naphthylimide analog labeling.46 Simultaneously, this reaction can be used to modify the 5-formyl group of 5fU. 5fC and 5fU were labeled with azide under various conditions, followed by a bio-orthogonal reaction, and functional molecules such as biotin were connected. 5fU is a thymine oxidative modification product abundant in mammalian cells, bacteria, and other organisms.47 It is the byproduct of cells that have been damaged by external agents such as ultraviolet light, particle radiation, or free radicals produced by chemical substances.48–51 Since the previous method can label both 5fC and 5fU, our group developed a method for labeling 5fU specifically.52,53 The experimental results demonstrated that by using the specific reaction of (2-benzimidazolyl) acetonitrile (azi-BIAN) with 5fU, interference from 5fC can be avoided, and 5fU can be explicitly labeled.
A portion of nucleic acid modifications occurs in both DNA and RNA, including O6-methylation of guanosine.54,55 This DNA modification, O6-Me-dG, is primarily caused by alkylating agents.56,57 It has been shown to result in GC to AT mutations during DNA replication, causing genotoxicity.58,59 Shigeki Sasaki et al. labeled O6-Me-dG in DNA60 differently from previous small molecule reaction methods.61 They modified O6-Me-dG via a functional group transfer reaction experiment. They previously discovered that by inserting 2′-deoxy-6-thioguanosine modified with a 2-methyliden-1,3-diketone group into the complementary RNA sequence, they could achieve the transfer reaction of the target base in the target RNA using the modified nucleobase and the target base's amino group. They then designed a sequence containing an alkynyl transfer group and used the functional group transfer reaction to label the 4-amino group of O6-Me-dG selectively. The alkyne-containing transfer group reacts with azide via a copper-catalyzed azide–alkyne cycloaddition reaction (click reaction), allowing for additional modification.
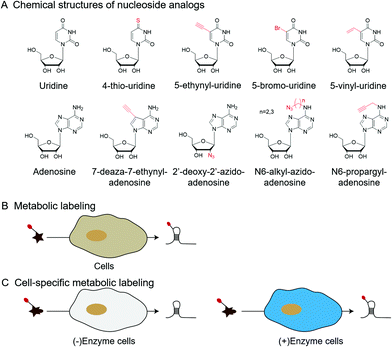 | ||
| Fig. 6 Metabolic labeling by nucleoside analogs. (A) The chemical structures of nucleoside analogs. (B) Metabolic labeling of RNA. (C) Cell-specific metabolic labeling. | ||
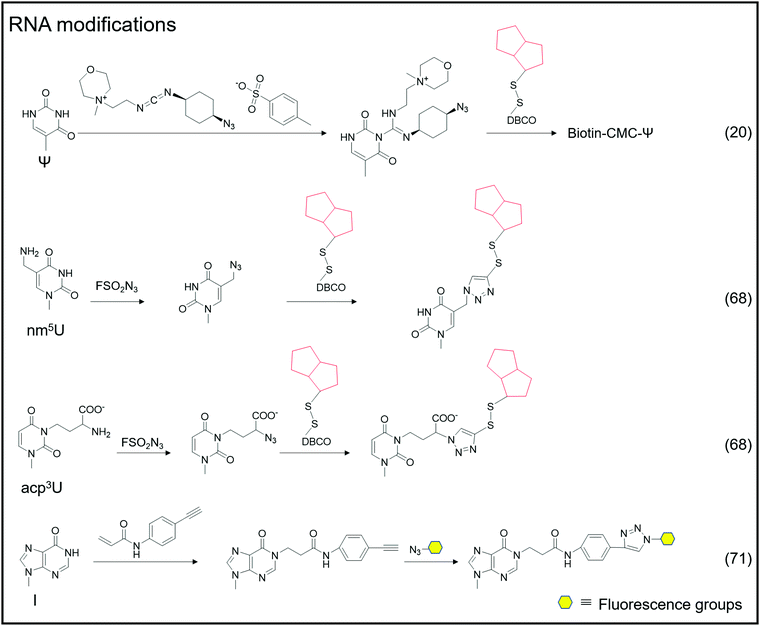
Adenosine-to-inosine (A-to-I) RNA editing frequently occurs as a post-transcriptional modification in worms to humans.69 The conversion of adenine to inosine is completed by adenosine deaminases acting on RNA (ADARs). Tsutomu Suzuki et al. developed the inosine chemical erasing (ICE) method to cyanoethylate inosine via Michael addition with acrylonitrile to understand the adenosine-to-inosine editing processes better.70 Jennifer M. Heemstra et al. used bio-orthogonal chemistry to synthesize N-(4-ethynylphenyl) acrylamide to label inosine with an alkyne group, and then inosine was subjected to secondary functionalization with azide-containing functional probes.71
2.2 Enzyme-assisted labeling methods
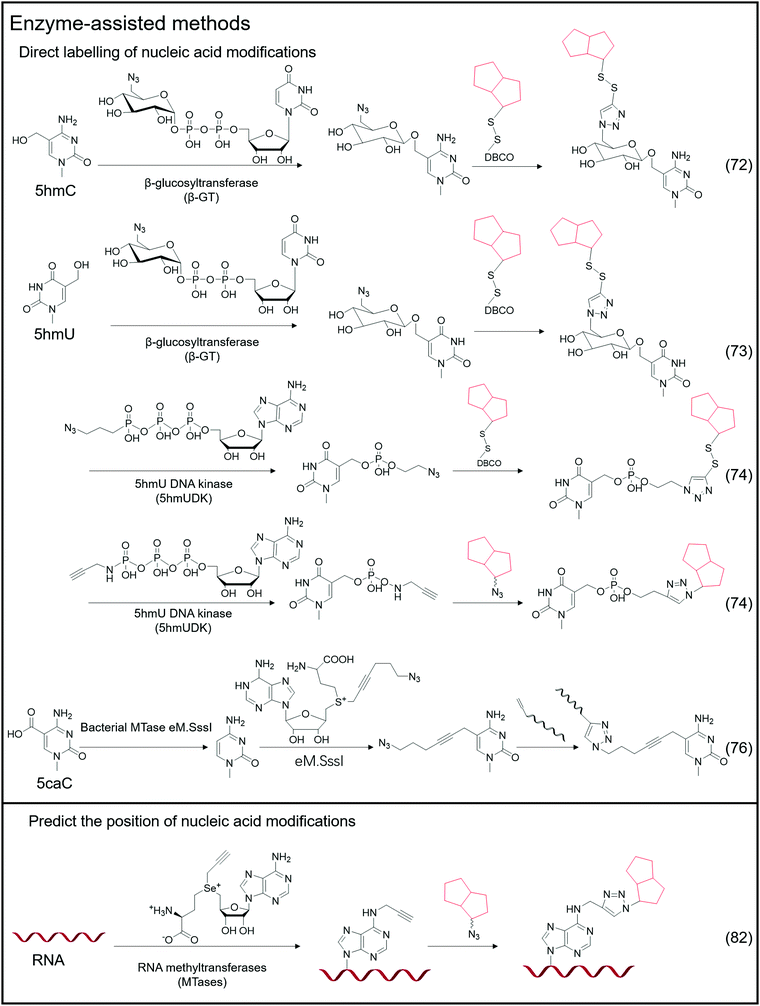
Certain enzymes are capable of specifically recognizing and labeling specific bases, and a typical example is the labeling of 5hmC-labeled bio-orthogonal small molecules via enzyme-assisted modification (Fig. 5).72 Instead of oxidizing 5hmC to 5fC and then labeling, Chuan He et al. used β-glucosyltransferase (β-GT) to enable selective labeling of 5hmC in the double-strand. This labeling method considers that β-GT can transfer the glucose moiety from uridine diphosphoglucose (UDP-Glu) to the hydroxyl group of 5hmC; hence, they further modified the glucose moiety to attach an azide group, 6-N3-glucose, which can undergo a bio-orthogonal reaction, thereby realizing the transfer of azide-modified glucose to 5hmC under β-GT catalysis. Further modification of functional small molecules such as biotin can be accomplished via the bio-orthogonal reaction with the azide group, followed by enrichment and detection. Simultaneously, Chuan He's group used this method to detect 5hmU in DNA.73 They began by oxidizing 5hmC, altering the reaction's specificity by converting 5hmC to 5caC while leaving the hydroxyl group in 5hmU unoxidized. Then, by adding β-GT and UDP-6-N3-Glc, 5hmU can be glycosylated to obtain N3-5gmU. Subsequently, biotin can be enriched through bio-orthogonal reactions. However, β-GT recognizes and modifies the paired position of 5hmU and G in the double-strand, which still contains 5hmU; β-GT is not sensitive to the mismatching of 5hmU and A. Another type of enzyme, 5hmU DNA kinase (5-hmUDK), transfers the phosphate group to 5hmU using adenosine-5-O-thiotriphosphate as a substrate, resulting in phosphorylated 5hmU, 5-thiophosphomethyluracil (5psmU). In this case, Bi-Feng Yuan et al. attached the azide or alkynyl groups to ATP-γ-S using this enzyme-assisted modification procedure, thereby connecting the azide or alkynyl group to 5hmU.74 With the phosphate group transferred 5hmU, the bio-orthogonal reaction between azide and DBCO or alkyl with azide can occur.Nucleic acid modifications and unmodified specific bases can be explicitly labeled using the enzyme-assisted labeling method. Saulius Klimasauskas et al. developed a method for selectively labeling unmodified CG dinucleotides.75 They identified CG dinucleotide sites using synthetic SAM cofactor analogs as substrates for SssI methyltransferase (eM.Sssl). The enzyme transfers the azide group on the AdoMet cofactor to the cytosine of the unmodified CG site, and the alkynyl-modified oligonucleotide sequence connects to the azide-modified cytosine via a bio-orthogonal process, allowing for the completion of further amplification operations. Based on this method, Edita Kriukienė et al. then developed an identification approach for the specific detection of 5caC in DNA.76 They employed T4 phase β-GT to glycosylate the 5hmC hydroxyl site, preventing 5hmC from interfering with DNA. To protect the CG site from further reactions, it was also methylated with MTase SssI and AdoMet. Next is decarboxylating the 5caC site in the sequence using the eM.SssI mutant enzyme, converting it to a new unmodified CG site. Then, the unmodified CG site can be tagged with the azide group, and the following amplification reaction can be carried out using the previously disclosed approach for bio-orthogonal reaction. NAD-capped RNAs were collected from total RNA using an enzyme-assisted method and their significance in gene regulation was identified.77–81 Andres Jäschke et al. catalyzed the transglycosylation of NAD with pentynol, 4-pentyn-1-ol, using adenosine diphosphate-ribosylcyclase (ADPRC).77 Following adaptor ligation, the sequence information of NAD-RNA is revealed using PCR and sequencing.
In addition to direct labeling of nucleic acid modifications, enzymes can be employed to label non-modified sites and to predict the location of nucleic acid modification sites in combination with sequencing and other approaches. Andrea Rentmeister et al. developed a method for assigning the methylation sites in RNA using the METTL3-METTL14 assist method.82 They considered that METTL3-METTL14 uses S-adenosyl-l-methionine as a substrate to catalyze the N6-propargylation of adenine sites in target RNAs. They used alkynyl-modified AdoMet analogs as substrates, determined that the METTL3-METTL14 interaction sites were alkynyl-modified, and then defined these reaction sites using bio-orthogonal chemistry. According to their findings, the modified sites obtained through metabolic labeling enabled the direct and precise assignment of m6A sites. AdoMet analogs transferred functional groups (methyl groups or groups including vinyl, alkynyl, or azide groups) into target macromolecular substrates catalysed by methyltransferases (MTs).83–87 The transferred functional groups on DNA or RNA serve as a substrate for subsequent bio-orthogonal procedures that modify fluorescent groups or other functional groups. This issue will not be discussed in depth because there is already a review on MT-mediated bio-orthogonal reactions.83
2.3 Metabolic labeling methods
In addition to labeling nucleic acid modifications, bio-orthogonal reactions can also be used to label and analyse intracellular RNA production and metabolism in combination with nucleoside analogs. Detection of RNA in cells is not only a fundamental study, but is also relevant to disease research.88 Nucleoside analogs are widely used for detecting nascent RNA and RNA-binding proteins, and bio-orthogonal methods combined with relative nucleoside analogs enrich target RNA from total RNA (Fig. 6).89–93 4-Thio-uridine (4SU) has been used as a nucleoside analog since 1978,94 and Mark Helm et al. attached an alkyne group to target RNA, based on the selective reaction between 4SU and 4-bromomethyl-7-propargyloxycoumarin (PBC).89 Based on the reaction of 4SU and PBC, the alkyne group was modified to 4SU, which is convenient for following conjugation. The nucleoside analogues containing alkynyl, alkenyl or vinyl groups allow the labeled RNA to undergo bio-orthogonal reactions upon insertion into the target RNA. These types of nucleoside analogs, such as 5-ethynyluridine (EU), are widely used in the detection of RNA synthesis.90,91 Adrian Salic et al. incorporated 5-ethynyluridine (EU) into newly transcribed RNA.90 The CuAAC reaction between the alkyne group in EU and the azide modified fluorophore was applied to assay the transcription rates of different tissues and different cell types. Xichen Bao et al. developed a method to capture the newly transcribed RNA interactome using click chemistry (RICK) by EU treatment.91 Based on RICK, proteins which bind nascent RNAs and nonpolyadenylated RNAs were captured and analysed. To overcome the cytotoxic response caused by copper ions, bio-orthogonal reactions without toxicity were used to label RNAs by nucleoside analogs, including SPAAC and inverse electron-demand Diels–Alder (IEDDA) reaction.95,96 In combination with cell-specific expression of the enzyme and the corresponding protected substrate molecule, cell-specific RNA metabolic labeling methods can be achieved in the target cell.92,93 Robert C. Spitale et al. realized cell-specific bio-orthogonal metabolic labeling.92 The 5EU was incorporated into nascent RNA by uracil phosphoribosyltransferase (UPRT) in the presence of phosphoribosyl pyrophosphate (PRPP). They further developed another small molecule–enzyme pair, penicillin G-amidase (PGA) and N6-phenylacetyl-2′-azidoadenosine (N-PAC-2′AzA). To reduce the off-target effects, Robert C. Spitale et al. developed the pair of uridine/cytidine kinase 2 and 2′-azidouridine.93 Uridine/cytidine kinase 2 (UCK2) is a rarely expressed isoform of ubiquitously expressed uridine/cytidine kinase-1 (UCK1). The cell-specific labeling realized by the cell overexpressed UCK2 selectively incorporated 2′-azidouridine into nascent RNAs. As stated above, the bio-orthogonal reactions combined with the corresponding nucleoside analogs are convenient for the detection of nascent RNA and RNA-binding proteins and the labeling of cell-specific RNA.3 Enhancing and detecting bio-orthogonal labeled nucleic acid modifications
After labeling nucleic acid modifications with various methods, subsequent detection and sequencing provide information on the amount, location, etc. of the labeled nucleic acid modifications in DNA or RNA. The sequences containing modified bases are enriched using the specific labeling method, and the enriched sequence information is compared to the unenriched sequence information.3.1 Sequencing method
To detect DNA modifications in the genome, many different methods developed realized quantification and mapping of DNA modifications. Those methods can be classified into overall detection and mapping analysis. The overall detection methods analyse DNA modifications in digested DNA by techniques such as liquid chromatography, mass spectrometry, gas chromatography and so on. The LOD value of those detection results can reach fmol levels.97,98 Combined with specific chemical labeling, some low-level DNA modifications also can be detected by LC/MS analysis.99 The overall detection methods provide a relatively simple and widely applied way to detect DNA modifications in the genome. However, those methods can’t provide the position of DNA modifications in the genome. Sequencing analysis is now a widely used technique for determining nucleic acid modifications in sequences. Chemical techniques, antibodies, or other methods recognize the nucleic acid modifications in the sequence, attach the recognition unit, and enrich the recognition unit's sequence using solid phase separation. Both enriched and unenriched sequences are sequenced and the sequencing results are compared to determine the position of the modified base target in the sequence (Fig. 7A). With the help of sequencing technology, a variety of methods can be applied to map genome-wide DNA modifications. The third-generation sequencing technology is a promising and straight way for DNA sequencing. The nanopore sequencing and single-molecule real-time sequencing have been applied for mapping DNA modifications in the genome, and enable the simultaneous detection of multiple DNA modifications.100,101 However, the high cost of the third-generation sequencing limits its application. The next-generation DNA sequencing combined with antibodies, specific affinity binding proteins or chemical modifications is used to distinguish DNA modifications from normal bases. The bisulfite sequencing (BS-seq) provides a single-base resolution way for genome-wide mapping of 5mC without enrichment,42 and the bisulfite sequencing combined with other chemical labeling methods can be expanded to the detection of 5hmC, 5fC, and 5caC.102–106 Although BS-seq is a gold standard method for 5mC mapping, the treatment condition is too harsh to degrade part of input DNA. The immunoprecipitation enrichment strategies are very useful ways for mapping DNA modifications. Based on antibodies or specific affinity binding proteins, DNA fragments which contain 5mC, 5hmC, 5fC or 6mA are isolated and then analysed by sequencing,107–110 providing straightforward mapping information of target DNA modifications in the genome. However, immunoprecipitation-based sequencing can’t offer base-resolution information. Chemical labeling-based methods offer available options for DNA modification enrichment and sequencing. Chemical labeling methods are specific for target DNA modifications, and the sequencing results are used to analyse the labeled DNA modification. As the chemical labeling approach induces base conversion of the DNA modifications, base resolution sequencing information can be obtained by sequencing,12,111 based on bio-orthogonal chemistry based labeling methods for DNA modification mapping, such as fC-CET, hmC-CATCH, Click-code-seq, caCLEAR and CeU-Seq.20,36,44,76,112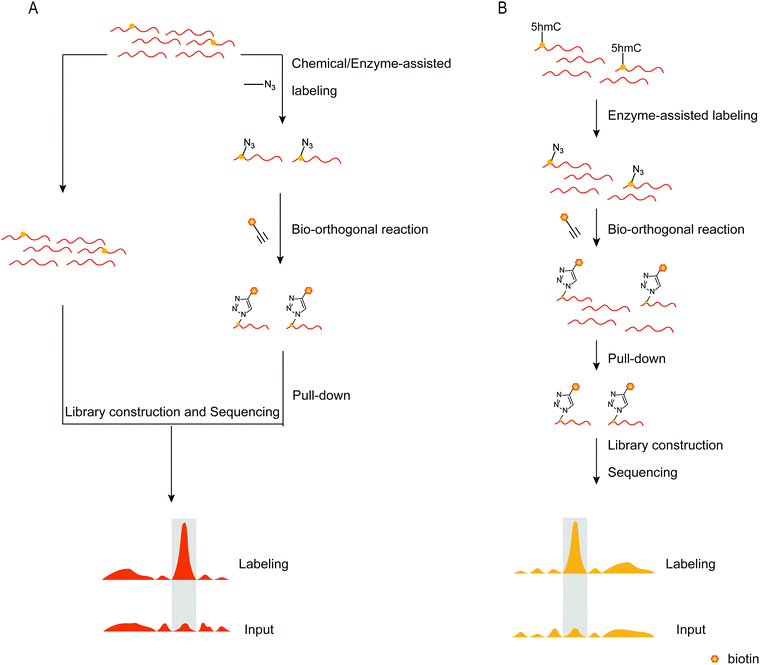 | ||
| Fig. 7 Mapping nucleic acid modifications based on bio-orthogonal tools. (A) The workflow of mapping. (B) Mapping 5hmC's position in the genome.72 | ||
The combination of bio-orthogonal tools and sequence technology enables the detection of nucleic acid modifications to be effective. Chuan He et al. labeled 5hmC using an enzyme-assisted method, in which β-GT catalyzed the transfer of the glucose moiety to 5hmC, converting it into β-6-azide-glucosyl-5-hydroxymethyl-cytosine.72 Biotin was attached to 5hmC via the bio-orthogonal reaction of azide and DBCO. The sequence is easily enhanced with 5hmC using biotin, and subsequent sequencing determines the location of 5hmC in genomic DNA (Fig. 7B). Additionally, they established a link between the presence of 5hmC in specific gene bodies and neurodegenerative disorders. They then improved the nano-hmC-Seal method, which was like the previous method in principle but detected 5hmC using nanograms of genetic DNA.113 The nano-hmC-Seal method was used to determine the differences between Tet2-mutant acute myeloid leukemia and normal stem cells. The sequencing results established that 5hmC plays a critical role in acute myeloid leukemia and regulates transcription. They extended this method to detect 5fC, dubbed the fC-Seal approach.105 5fC could not be directly recognized and labeled by β-GT, so they converted the 5fC locus to a 5hmC locus via sodium borohydride reduction and then detected 5fC using the previously used method to label 5hmC. To avoid interference from the original 5hmC in the sequence, the unmodified glycosyl group must be transferred into the hydroxyl site of 5hmC via β-GT catalysis, rendering 5hmC inactive in the subsequent bio-orthogonal reaction. The bio-orthogonal reaction between azide and alkyne provides an easy route to enrich the sequence containing 5fC. Additionally, it provides a convenient method for examining the role of 5fC.
Sequencing analysis can reveal the distribution of nucleic acid modifications in the genome or RNA and provide the position information of nucleic acid modifications at the base-resolution level, which can then be combined with subsequent PCR amplification and sequencing. The concept of mismatches following chemical modification, such as changing 5fC from C to T following modification, is a fairly standard approach for detecting base modifications in nucleic acids.36,37 Due to the exocyclic amino group of 5fC being involved in the cyclization, the reacted 5fC no longer forms a complementary base pairing with dG. Instead, the AI or azi-BP group is transferred to 5fC, and the bio-orthogonal reaction between azide and DBCO-biotin is used to enhance sequences containing 5fC (Fig. 8A). While unmodified 5fC in the input will be read as C during sequencing, modified 5fC in enhanced sequences will be read as T, allowing for single-base identification of 5fC in the genome. The detection method for 5fC is also applicable to 5hmC.44,45 By first protecting the preexisting 5fC in the sequence, oxidizing 5hmC into newly generated 5fC, and then labeling and sequencing, the single base level of 5hmC was detected and analyzed (Fig. 8B).
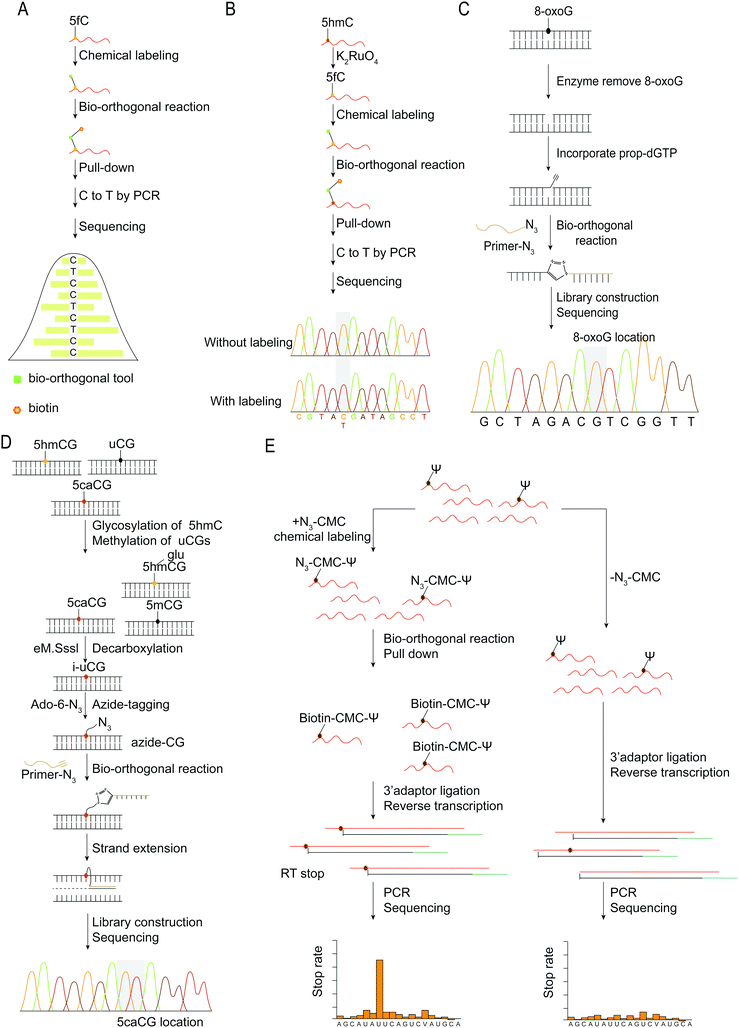 | ||
| Fig. 8 Base resolution sequencing methods for nucleic acid modifications. (A) fC-CET.36 (B) hmC-CATCH.44 (C) Click-code-seq for labeling 8-oxoG.112 (D) caCLEAR.76 (E) CeU-Seq.20 | ||
An oligonucleotide code was used to detect the 8-oxoG location in genomes using click-code-seq.112 The base excision repair (BER) proteins, formamidopyrimidine DNA glycosylase (Fpg), and human apurinic/apyrimidinic endonuclease (APE1) removed 8-oxoG, resulting in a 1 nt gap in the DNA. Furthermore, the O-3′-propargyl modified dGTP filled the gap and modified an alkynyl group in the DNA (Fig. 8C). Following ligation of the azide-modified primer sequence using a bio-orthogonal process, the sequence was amplified using PCR and evaluated via subsequent sequencing. The 8-oxoG site is the G base following the target primer sequence (5′-TCGGAA-3′). The adaptor ligation method for determining the location of nucleic acid modifications in DNA is also applicable to profiling genomic 5caCGs (Fig. 8D), referred to as caCLEAR.76 Before the 5caCG site in the genome can be labeled, the sequence's 5hmC must be glycosylated, and the sequence's unmodified CG site must also be methylated. The sequence's 5caCG site was then decarboxylated with eM.SssI, yielding a new unaltered CG site. While the eM.SssI mutant enzyme and Ado-6-N3 coexisted, the azide labeling of the new unmodified CG site was continued. After attaching the adaptor at the azide site, the sequence is dA-tailed at the 3′-end. PCR amplifies the linked sequence, which can then be sequenced. Following that, the associated adapter sequence is used to determine the sequence's location of the 5caCG site. Chengqi Yi et al. used ‘CeU-Seq’ to detect Ψ in RNA.20 This method is based on ‘stop rate’ and ‘CMC sensitivity’ to determine the site of Ψ in RNA (Fig. 8E). First, after RNA was fragmented, N3-CMC was used to label Ψ chemically and form an N3-CMC-Ψ site. Then, azide and DBCO-biotin were used for bio-orthogonal reaction, biotin modification was performed at the Ψ site, and biotin was used to enrich the sequence containing Ψ. The 3′ ends of the enriched sequences were connected with adapters and then reverse transcribed to obtain cDNA, followed by circularization, linearization, and PCR amplification. Finally, they were sequenced and compared with the control group without chemical modification. The ‘stop rate’ is gained by the equation Nistop/(Nistop + Nireadthrough) that provides the position of Ψ.20
3.3 Cellular visualization
While cell imaging does not provide precise location information for nucleic acid modifications in the target sequence, it mitigates the impact of certain experimental operations by not requiring DNA/RNA extraction from cells for detection. Location information for nucleic acid modifications can be provided at the cellular and subcellular organelle levels. However, because cells contain many reactive biomolecules, cell imaging of nucleic acid modifications faces numerous difficulties, limiting the number of nucleic acid modifications detected in cells. Yongxi Zhao et al. utilized bio-orthogonal reactions to image nucleic acid modifications in fixed cells.114,115 One example is the simultaneous imaging of 5fC and 5hmC.115 After labeling 5fC with AI, ligation with primer sequences linked to azide and DBCO was performed. Intracellular 5fC imaging was then accomplished using the ligated sequence to hybridize with the circularized DNA barcode and the barcode worked as the template for RCA. The RCA product contains repetitive sequences that capture the fluorescent probe for signal amplification. For 5hmC imaging, T4-βGT was used to catalyze UDP-N-Glu glycosylation, followed by bio-orthogonal reaction with DBCO, hybridization with another barcode, and RCA amplification to achieve intracellular simultaneous imaging analysis for 5fC and 5hmC. The imaging of 5hmU and 5fU in cells was carried out comparably.114 The 5hmU site was modified with specific primers by modifying it with an alkynyl group using 5-HMUDK and ATP-γ-alkyne as described previously, and then by linking it to an azide-modified sequence via a bio-orthogonal reaction. After reducing the 5fU site with NaBH4 to create a new 5hmU site, the modification and ligation steps were repeated, and a different primer sequence was connected to the 5fU site. RCA amplification and imaging sequence labeling are used to achieve simultaneous imaging of 5hmU and 5fU. The bio-orthogonal reactions are convenient for imaging metabolically labeled RNA. Adrian Salic et al. modified RNA by EU, and then imaged RNA synthesis, transport, and localization by using CuAAC with fluorescent azides.90 RNA polymerases I, II, and III incorporated EU into the RNA transcript. The fluorescent azides react with EU-labeled RNA fast and specifically. Their results revealed that by incubating EU with cells and tissues, they were able to achieve precise marker imaging of RNA rather than DNA. The Robert C. Spitale group produced three small molecule-enzyme pairings (EU-UPRT, N-PAC-2′AzA-PGA, and 2′AzUd-UCK2) to achieve cell-selective metabolic labeling.92,93 After reacting with enzymes, those small molecules integrated into RNA after being incubated with cells. Cell-specific incorporation of small molecule was imaged with fluorescent molecules containing azide or alkyne groups using bio-orthogonal reactions. Based on the small molecule-enzyme pairings, cell-selective imaging is achieved.4 Conclusions and outlook
This review summarizes the current state-of-the-art in labeling and detecting nucleic acid modification via bio-orthogonal reactions. Bio-orthogonal reactions are widely used in the study of intracellular macromolecules due to their low toxicity, high specificity, and rapidity. In the study of intracellular nucleic acid modifications, bio-orthogonal reactions provide an extremely convenient method for labeling and detecting nucleic acid modifications. Along with direct labeling with small molecules, these methods can be aided by enzymes with specific recognition and catalytic activities, such as labeling 5fC via intermolecular cyclization or transferring an azide-containing glucose moiety to 5hmC via β-GT. In addition, the labeling of nucleic acid modifications with bio-orthogonal reactive small molecules provides tools for subsequent sequencing, imaging, and other applications. For example, after being modified by β-GT, 5hmC allows for subsequent molecular sequencing as well as intracellular imaging via bio-orthogonal reactions;44,115 metabolic labeling approaches combined with bio-orthogonal reactions allow for accurate RNA detection and imaging analysis.89–93 Even though these bio-orthogonal reactions labeled a wide variety of nucleic acid modifications, some limitations remain. To begin, compared to the types of modified nucleic acids discovered thus far, there are still far too few modified nucleic acids that can be labeled using bio-orthogonal tools. Therefore, additional alternative methods for labeling nucleic acid modifications should be investigated. Second, while most currently available labeling methods apply to extracellular labeling, only a few apply to intracellular labeling. The fact that groups with bio-orthogonal reactivity do not react with active molecules in vivo is one of their significant advantages. Thus, the advantages of bio-orthogonal reactions can be maximized by employing them for in situ labeling of modified nucleic acids in cells. However, most labeling methods require many samples, and single-cell analysis is currently difficult. As a result, more research methods capable of carrying out bio-orthogonal reactions should be developed in the future, which can be used to specifically identify more and diverse types of nucleic acid modifications, and increase reaction efficiency and sensitivity, and we also anticipate the development of additional bio-orthogonal reactions for in situ labeling.Bio-orthogonal modifications of nucleic acids are a promising avenue and a helpful research tool for in situ labeling. Simultaneously, numerous methods for investigating nucleic acid modifications exist, including mass spectrometry analysis and antibody-based specific recognition. Each analytic approach has unique advantages and disadvantages, allowing researchers to choose the most appropriate tool. We believe that by developing additional methods in combination with bio-orthogonal reactions applicable to nucleic acid modifications, additional methods for labeling various nucleic acid modifications will become available and eventually be developed for intracellular and even in vivo applications. In addition, the bio-orthogonal reaction will be a valuable tool for examining the relationship between nucleic acid modification and various biological processes and diseases and may be applied to disease diagnosis and new drug development.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We are grateful to the National Natural Science Foundation of China (91753201, 21721005, and 22037004 to X. Z.; 210900178 to H. L.) and China Postdoctoral Science Foundation (211000157 to H. L.).References
- R. Dahm, Hum. Genet., 2007, 122, 565–581 CrossRef PubMed.
- R. Jaenisch and A. Bird, Nat. Genet., 2003, 33, 245–254 CrossRef CAS PubMed.
- I. A. Roundtree, M. E. Evans, T. Pan and C. He, Cell, 2017, 169, 1187–1200 CrossRef CAS PubMed.
- S. Kumar and T. Mohapatra, Front. Cell Dev. Biol., 2021, 9, 628415 CrossRef PubMed.
- I. V. Bure and M. V. Nemtsova, Int. J. Mol. Sci., 2021, 22, 5683 CrossRef CAS PubMed.
- A. Martisova, J. Holcakova, N. Izadi, R. Sebuyoya, R. Hrstka and M. Bartosik, Int. J. Mol. Sci., 2021, 22, 4247 CrossRef CAS PubMed.
- X. Fang, M. Li, T. Yu, G. Liu and J. Wang, Genes Dis., 2020, 7, 585–597 CrossRef CAS PubMed.
- Z. Shulman and N. Stern-Ginossar, Nat. Immunol., 2020, 21, 501–512 CrossRef CAS PubMed.
- Y. Chen, T. Hong, S. Wang, J. Mo, T. Tian and X. Zhou, Chem. Soc. Rev., 2017, 46, 2844–2872 RSC.
- Y. Wang, X. Zhang, H. Liu and X. Zhou, Chem. Soc. Rev., 2021, 50, 13481–13497 RSC.
- T. Wang, C. E. Loo and R. M. Kohli, Mol. Metab., 2022, 57, 101314 CrossRef CAS PubMed.
- Y. Dai, B. F. Yuan and Y. Q. Feng, RSC Chem. Biol., 2021, 2, 1096–1114 RSC.
- S. Y. Jang, D. P. Murale, A. D. Kim and J. S. Lee, ChemBioChem, 2019, 20, 1498–1507 CrossRef CAS PubMed.
- K. J. Bruemmer, S. W. M. Crossley and C. J. Chang, Angew. Chem., Int. Ed., 2020, 59, 13734–13762 CrossRef CAS PubMed.
- N. Z. Fantoni, A. H. El-Sagheer and T. Brown, Chem. Rev., 2021, 121, 7122–7154 CrossRef CAS PubMed.
- J. Kaur, M. Saxena and N. Rishi, Bioconjugate Chem., 2021, 32, 1455–1471 CrossRef CAS PubMed.
- J. Wang, Y. Liu, Y. Liu, S. Zheng, X. Wang, J. Zhao, F. Yang, G. Zhang, C. Wang and P. R. Chen, Nature, 2019, 569, 509–513 CrossRef CAS PubMed.
- Y. Chen, F. Wu, Z. Chen, Z. He, Q. Wei, W. Zeng, K. Chen, F. Xiao, Y. Yuan, X. Weng, Y. Zhou and X. Zhou, Adv. Sci., 2020, 7, 1900997 CrossRef CAS PubMed.
- A. Ovcharenko, F. P. Weissenboeck and A. Rentmeister, Angew. Chem., Int. Ed., 2021, 60, 4098–4103 CrossRef CAS PubMed.
- X. Li, P. Zhu, S. Ma, J. Song, J. Bai, F. Sun and C. Yi, Nat. Chem. Biol., 2015, 11, 592–597 CrossRef CAS PubMed.
- J. Xue, F. Chen, L. Su, X. Cao, M. Bai, Y. Zhao, C. Fan and Y. Zhao, Angew. Chem., Int. Ed., 2021, 60, 3428–3432 CrossRef CAS PubMed.
- O. Taryma-Lesniak, K. E. Sokolowska and T. K. Wojdacz, J. Clin. Pathol., 2021, 74, 692–696 CrossRef CAS PubMed.
- H. Wheeler, J. Am. Chem. Soc., 1904, 31, 591–606 CAS.
- T. B. Johnson and R. D. Coghill, J. Am. Chem. Soc., 1925, 47, 2838–2844 CrossRef CAS.
- R. D. Hotchkiss, J. Biol. Chem., 1948, 175, 315–332 CrossRef CAS PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu and L. Aravind, Science, 2009, 324, 930–935 CrossRef CAS PubMed.
- Y.-F. He, B.-Z. Li, Z. Li, P. Liu, Y. Wang, Q. Tang, J. Ding, Y. Jia, Z. Chen and L. Li, Science, 2011, 333, 1303–1307 CrossRef CAS PubMed.
- S. Ito, L. Shen, Q. Dai, S. C. Wu, L. B. Collins, J. A. Swenberg, C. He and Y. Zhang, Science, 2011, 333, 1300–1303 CrossRef CAS PubMed.
- S. Cortellino, J. Xu, M. Sannai, R. Moore, E. Caretti, A. Cigliano, M. Le Coz, K. Devarajan, A. Wessels and D. Soprano, Cell, 2011, 146, 67–79 CrossRef CAS PubMed.
- J. U. Guo, Y. Su, C. Zhong, G.-L. Ming and H. Song, Cell, 2011, 145, 423–434 CrossRef CAS PubMed.
- C. S. Nabel, H. Jia, Y. Ye, L. Shen, H. L. Goldschmidt, J. T. Stivers, Y. Zhang and R. M. Kohli, Nat. Chem. Biol., 2012, 8, 751–758 CrossRef CAS PubMed.
- P. A. Jones, Nat. Rev. Genet., 2012, 13, 484–492 CrossRef CAS PubMed.
- A. Parry, S. Rulands and W. Reik, Nat. Rev. Genet., 2020, 22, 59–66 CrossRef PubMed.
- M. V. C. Greenberg and D. Bourc’his, Nat. Rev. Mol. Cell Biol., 2019, 20, 590–607 CrossRef CAS PubMed.
- W. Huang, M.-D. Lan, C.-B. Qi, S.-J. Zheng, S.-Z. Wei, B.-F. Yuan and Y.-Q. Feng, Chem. Sci., 2016, 7, 5495–5502 RSC.
- B. Xia, D. Han, X. Lu, Z. Sun, A. Zhou, Q. Yin, H. Zeng, M. Liu, X. Jiang, W. Xie, C. He and C. Yi, Nat. Methods, 2015, 12, 1047–1050 CrossRef CAS PubMed.
- Y. Wang, C. Liu, X. Zhang, W. Yang, F. Wu, G. Zou, X. Weng and X. Zhou, Chem. Sci., 2018, 9, 3723–3728 RSC.
- J. Madzo, H. Liu, A. Rodriguez, A. Vasanthakumar, S. Sundaravel, D. B. D. Caces, T. J. Looney, L. Zhang, J. B. Lepore, T. Macrae, R. Duszynski, A. H. Shih, C.-X. Song, M. Yu, Y. Yu, R. Grossman, B. Raumann, A. Verma, C. He, R. L. Levine, D. Lavelle, B. T. Lahn, A. Wickrema and L. A. Godley, Cell Rep., 2014, 6, 231–244 CrossRef CAS PubMed.
- A. Vasanthakumar and L. A. Godley, Cancer Genet., 2015, 208, 167–177 CrossRef CAS PubMed.
- X. Lu, B. S. Zhao and C. He, Chem. Rev., 2015, 115, 2225–2239 CrossRef CAS PubMed.
- A. Inoue and Y. Zhang, Science, 2011, 334, 194 CrossRef CAS PubMed.
- M. J. Booth, M. R. Branco, G. Ficz, D. Oxley, F. Krueger, W. Reik and S. Balasubramanian, Science, 2012, 336, 934–937 CrossRef CAS PubMed.
- G. Hayashi, K. Koyama, H. Shiota, A. Kamio, T. Umeda, G. Nagae, H. Aburatani and A. Okamoto, J. Am. Chem. Soc., 2016, 138, 14178–14181 CrossRef CAS PubMed.
- H. Zeng, B. He, B. Xia, D. Bai, X. Lu, J. Cai, L. Chen, A. Zhou, C. Zhu, H. Meng, Y. Gao, H. Guo, C. He, Q. Dai and C. Yi, J. Am. Chem. Soc., 2018, 140, 13190–13194 CrossRef CAS PubMed.
- Y. Wang, X. Zhang, F. Wu, Z. Chen and X. Zhou, Chem. Sci., 2019, 10, 447–452 RSC.
- Y. Wang, C. Liu, W. Yang, G. Zou, X. Zhang, F. Wu, S. Yu, X. Luo and X. Zhou, Chem. Commun., 2018, 54, 1497–1500 RSC.
- L. D. Thornburg, M. T. Lai, J. S. Wishnok and J. Stubbe, Biochemistry, 1993, 32, 14023–14033 CrossRef CAS PubMed.
- C. Decarroz, J. Wagner, J. Van Lier, C. M. Krishna, P. Riesz and J. Cadet, Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med., 1986, 50, 491–505 CrossRef CAS PubMed.
- H. Hong, H. Cao, Y. Wang and Y. Wang, Chem. Res. Toxicol., 2006, 19, 614–621 Search PubMed.
- H. Kasai, A. Iida, Z. Yamaizumi, S. Nishimura and H. Tanooka, Mutat. Res., 1990, 243, 249–253 CAS.
- E. J. Privat and L. C. Sowers, Mutat. Res., 1996, 354, 151–156 CrossRef PubMed.
- W. Yang, S. Han, X. Zhang, Y. Wang, G. Zou, C. Liu, M. Xu and X. Zhou, Anal. Chem., 2021, 93, 15445–15451 CrossRef CAS PubMed.
- Y. Wang, C. Liu, F. Wu, X. Zhang, S. Liu, Z. Chen, W. Zeng, W. Yang, X. Zhang, Y. Zhou, X. Weng, Z. Wu and X. Zhou, iScience, 2018, 9, 423–432 CrossRef CAS PubMed.
- J. A. Burns, K. Dreij, L. Cartularo and D. A. Scicchitano, Nucleic Acids Res., 2010, 38, 8178–8187 CrossRef CAS PubMed.
- K. B. Altshuler, C. S. Hodes and J. M. Essigmann, Chem. Res. Toxicol., 1996, 9, 980–987 Search PubMed.
- D. T. Beranek, Mutat. Res., 1990, 231, 11–30 CrossRef CAS PubMed.
- N. Shrivastav, D. Li and J. M. Essigmann, Carcinogenesis, 2010, 31, 59–70 CrossRef CAS PubMed.
- G. P. Margison, M. F. Santibáñez Koref and A. C. Povey, Mutagenesis, 2002, 17, 483–487 CrossRef CAS PubMed.
- M. Ezerskyte, J. A. Paredes, S. Malvezzi, J. A. Burns, G. P. Margison, M. Olsson, D. A. Scicchitano and K. Dreij, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4731–4736 CrossRef CAS PubMed.
- K. Onizuka, T. Nishioka, Z. Li, D. Jitsuzaki, Y. Taniguchi and S. Sasaki, Chem. Commun., 2012, 48, 3969–3971 RSC.
- K. Onizuka, A. Shibata, Y. Taniguchi and S. Sasaki, Chem. Commun., 2011, 47, 5004–5006 RSC.
- M. Charette and M. W. Gray, IUBMB Life, 2000, 49, 341–352 CrossRef CAS PubMed.
- P. Morais, H. Adachi, J. L. Chen and Y.-T. Yu, in Epitranscriptomics RNA Technologies, ed. S. Jurga and J. Barciszewski, Springer, Cham, 2021, pp. 505–526 DOI:10.1007/978-3-030-71612-7_19.
- A. T. Yu, J. Ge and Y.-T. Yu, Protein Cell, 2011, 2, 712–725 CrossRef CAS PubMed.
- R. Sousa-Luís and M. Carmo-Fonseca, Mol. Cell, 2022, 82, 495–496 CrossRef PubMed.
- N. W. Ho and P. Gilham, Biochemistry, 1967, 6, 3632–3639 CrossRef CAS PubMed.
- A. Bakin and J. Ofengand, Biochemistry, 1993, 32, 9754–9762 CrossRef CAS PubMed.
- O. A. Krasheninina, J. Thaler, M. D. Erlacher and R. Micura, Angew. Chem., Int. Ed., 2021, 60, 6970–6974 CrossRef CAS PubMed.
- B. L. Bass, Annu. Rev. Biochem., 2002, 71, 817–846 CrossRef CAS PubMed.
- M. Sakurai, T. Yano, H. Kawabata, H. Ueda and T. Suzuki, Nat. Chem. Biol., 2010, 6, 733–740 CrossRef CAS PubMed.
- S. D. Knutson, M. M. Korn, R. P. Johnson, L. R. Monteleone, D. M. Dailey, C. S. Swenson, P. A. Beal and J. M. Heemstra, Chemistry, 2020, 26, 9874–9878 CrossRef CAS PubMed.
- C.-X. Song, K. E. Szulwach, Y. Fu, Q. Dai, C. Yi, X. Li, Y. Li, C.-H. Chen, W. Zhang, X. Jian, J. Wang, L. Zhang, T. J. Looney, B. Zhang, L. A. Godley, L. M. Hicks, B. T. Lahn, P. Jin and C. He, Nat. Biotechnol., 2011, 29, 68–72 CrossRef CAS PubMed.
- M. Yu, C.-X. Song and C. He, Methods, 2015, 72, 16–20 CrossRef CAS PubMed.
- C.-J. Ma, L. Li, W.-X. Shao, J.-H. Ding, X.-L. Cai, Z.-R. Lun, B.-F. Yuan and Y.-Q. Feng, Chem. Sci., 2021, 12, 14126–14132 RSC.
- Z. Staševskij, P. Gibas, J. Gordevičius, E. Kriukienė and S. Klimašauskas, Mol. Cell, 2017, 65, 554–564 CrossRef PubMed .e6.
- J. Ličytė, P. Gibas, K. Skardžiūtė, V. Stankevičius, A. Rukšėnaitė and E. Kriukienė, Cell Rep., 2020, 32, 108155 CrossRef PubMed.
- H. Cahova, M.-L. Winz, K. Hoefer, G. Nuebel and A. Jaeschke, Nature, 2015, 519, 374–377 CrossRef CAS PubMed.
- M.-L. Winz, H. Cahova, G. Nuebel, J. Frindert, K. Hoefer and A. Jaeschke, Nat. Protoc., 2017, 12, 122–149 CrossRef CAS PubMed.
- H. Zhang, H. Zhong, S. Zhang, X. Shao, M. Ni, Z. Cai, X. Chen and Y. Xia, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 12072–12077 CrossRef CAS PubMed.
- H. Hu, N. Flynn, H. Zhang, C. You, R. Hang, X. Wang, H. Zhong, Z. Chan, Y. Xia and X. Chen, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2025595118 CrossRef CAS PubMed.
- H. Zhang, H. Zhong, X. Wang, S. Zhang, X. Shao, H. Hu, Z. Yu, Z. Cai, X. Chen and Y. Xia, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2026183118 CrossRef CAS PubMed.
- K. Hartstock, B. S. Nilges, A. Ovcharenko, N. V. Cornelissen, N. Püllen, A. M. Lawrence-Dörner, S. A. Leidel and A. Rentmeister, Angew. Chem., Int. Ed., 2018, 57, 6342–6346 CrossRef CAS PubMed.
- E. Jalali and J. S. Thorson, Curr. Opin. Biotechnol, 2021, 69, 290–298 CrossRef CAS PubMed.
- A. A. Wilkinson, E. Jagu, K. Ubych, S. Coulthard, A. E. Rushton, J. Kennefick, Q. Su, R. K. Neely and P. Fernandez-Trillo, ACS Cent. Sci., 2020, 6, 525–534 CrossRef CAS PubMed.
- F. Muttach and A. Rentmeister, Angew. Chem., Int. Ed., 2016, 55, 1917–1920 CrossRef CAS PubMed.
- F. Muttach, N. Muthmann, D. Reichert, L. Anhaeuser and A. Rentmeister, Chem. Sci., 2017, 8, 7947–7953 RSC.
- J. M. Holstein, L. Anhaeuser and A. Rentmeister, Angew. Chem., Int. Ed., 2016, 55, 10899–10903 CrossRef CAS PubMed.
- M. Singha, L. Spitalny, K. Nguyen, A. Vandewalle and R. C. Spitale, Wiley Interdiscip. Rev.: RNA, 2021, 12, e1650 CAS.
- K. Schmid, M. Adobes-Vidal and M. Helm, Bioconjugate Chem., 2017, 28, 1123–1134 CrossRef CAS PubMed.
- C. Y. Jao and A. Salic, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15779–15784 CrossRef CAS.
- X. Bao, X. Guo, M. Yin, M. Tariq, Y. Lai, S. Kanwal, J. Zhou, N. Li, Y. Lv, C. Pulido-Quetglas, X. Wang, L. Ji, M. J. Khan, X. Zhu, Z. Luo, C. Shao, D.-H. Lim, X. Liu, N. Li, W. Wang, M. He, Y.-L. Liu, C. Ward, T. Wang, G. Zhang, D. Wang, J. Yang, Y. Chen, C. Zhang, R. Jauch, Y.-G. Yang, Y. Wang, B. Qin, M.-L. Anko, A. P. Hutchins, H. Sun, H. Wang, X.-D. Fu, B. Zhang and M. A. Esteban, Nat. Methods, 2018, 15, 213–220 CrossRef CAS PubMed.
- K. Nguyen, M. Fazio, M. Kubota, S. Nainar, C. Feng, X. Li, S. X. Atwood, T. W. Bredy and R. C. Spitale, J. Am. Chem. Soc., 2017, 139, 2148–2151 CrossRef CAS PubMed.
- S. Nainar, B. J. Cuthbert, N. M. Lim, W. E. England, K. Ke, K. Sophal, R. Quechol, D. L. Mobley, C. W. Goulding and R. C. Spitale, Nat. Methods, 2020, 17, 311–318 CrossRef CAS PubMed.
- W. T. Melvin, H. B. Milne, A. A. Slater, H. J. Allen and H. M. Keir, Eur. J. Biochem., 1978, 92, 373–379 CrossRef CAS PubMed.
- C. Feng, Y. Li and R. C. Spitale, Org. Biomol. Chem., 2017, 15, 5117–5120 RSC.
- F. Eggert and S. Kath-Schorr, Chem. Commun., 2016, 52, 7284–7287 RSC.
- R. Yin, J. Mo, M. Lu and H. Wang, Anal. Chem., 2015, 87, 1846–1852 CrossRef CAS PubMed.
- Y. Tang, X.-D. Gao, Y. Wang, B.-F. Yuan and Y.-Q. Feng, Anal. Chem., 2012, 84, 7249–7255 CrossRef CAS PubMed.
- Y. Tang, S.-J. Zheng, C.-B. Qi, Y.-Q. Feng and B.-F. Yuan, Anal. Chem., 2015, 87, 3445–3452 CrossRef CAS PubMed.
- B. A. Flusberg, D. R. Webster, J. H. Lee, K. J. Travers, E. C. Olivares, T. A. Clark, J. Korlach and S. W. Turner, Nat. Methods, 2010, 7, 461–465 CrossRef CAS PubMed.
- Z. L. Wescoe, J. Schreiber and M. Akeson, J. Am. Chem. Soc., 2014, 136, 16582–16587 CrossRef CAS PubMed.
- M. Yu, G. C. Hon, K. E. Szulwach, C.-X. Song, L. Zhang, A. Kim, X. Li, Q. Dai, Y. Shen, B. Park, J.-H. Min, P. Jin, B. Ren and C. He, Cell, 2012, 149, 1368–1380 CrossRef CAS PubMed.
- D. Tan, T. H. Chung, X. Sun and X.-Y. Jia, Anal. Chem., 2018, 90, 13200–13206 CrossRef CAS PubMed.
- M. J. Booth, G. Marsico, M. Bachman, D. Beraldi and S. Balasubramanian, Nat. Chem., 2014, 6, 435–440 CrossRef CAS PubMed.
- C.-X. Song, K. E. Szulwach, Q. Dai, Y. Fu, S.-Q. Mao, L. Lin, C. Street, Y. Li, M. Poidevin, H. Wu, J. Gao, P. Liu, L. Li, G.-L. Xu, P. Jin and C. He, Cell, 2013, 153, 678–691 CrossRef CAS PubMed.
- X. Lu, C.-X. Song, K. Szulwach, Z. Wang, P. Weidenbacher, P. Jin and C. He, J. Am. Chem. Soc., 2013, 135, 9315–9317 CrossRef CAS PubMed.
- M. Weber, J. J. Davies, D. Wittig, E. J. Oakeley, M. Haase, W. L. Lam and D. Schubeler, Nat. Genet., 2005, 37, 853–862 CrossRef CAS PubMed.
- J. P. Thomson and R. R. Meehan, Methods Mol. Biol., 2018, 1708, 679–696 CrossRef CAS PubMed.
- E.-A. Raiber, D. Beraldi, G. Ficz, H. E. Burgess, M. R. Branco, P. Murat, D. Oxley, M. J. Booth, W. Reik and S. Balasubramanian, Genome Biol., 2012, 13, R69 CrossRef PubMed.
- B. Yao, Y. Li, Z. Wang, L. Chen, M. Poidevin, C. Zhang, L. Lin, F. Wang, H. Bao, B. Jiao, J. Lim, Y. Cheng, L. Huang, B. L. Phillips, T. Xu, R. Duan, K. H. Moberg, H. Wu and P. Jin, Mol. Cell, 2018, 71, 848–857 CrossRef CAS PubMed.
- Y. Mahdavi-Amiri, K. C. K. Chung and R. Hili, Chem. Sci., 2021, 12, 606–612 RSC.
- J. Wu, M. McKeague and S. J. Sturla, J. Am. Chem. Soc., 2018, 140, 9783–9787 CrossRef CAS PubMed.
- D. Han, X. Lu, A. H. Shih, J. Nie, Q. You, M. M. Xu, A. M. Melnick, R. L. Levine and C. He, Mol. Cell, 2016, 63, 711–719 CrossRef CAS PubMed.
- M. Bai, X. Cao, F. Chen, J. Xue, Y. Zhao and Y. Zhao, Anal. Chem., 2021, 93, 10495–10501 CrossRef CAS PubMed.
- J. Xue, F. Chen, L. Su, X. Cao, M. Bai, Y. Zhao, C. Fan and Y. Zhao, Angew. Chem., Int. Ed., 2020, 60, 3428–3432 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |