Open Access Article
Open Access ArticleInhibition of chloroplast translation as a new target for herbicides†
Kirill V.
Sukhoverkov
 ab,
Karen J.
Breese
ab,
Karen J.
Breese
 a,
Aleksandra W.
Debowski
a,
Aleksandra W.
Debowski
 ac,
Monika W.
Murcha
ac,
Monika W.
Murcha
 ab,
Keith A.
Stubbs
ab,
Keith A.
Stubbs
 *a and
Joshua S.
Mylne
*a and
Joshua S.
Mylne
 *abd
*abd
aThe University of Western Australia, School of Molecular Sciences, 35 Stirling Highway, Crawley, Perth 6009, Australia. E-mail: keith.stubbs@uwa.edu.au
bThe ARC Centre of Excellence in Plant Energy Biology, 35 Stirling Highway, Crawley, Perth 6009, Australia
cSchool of Biomedical Sciences, 35 Stirling Highway, Crawley, Perth 6009, Australia
dCentre for Crop and Disease Management, School of Molecular and Life Sciences, Curtin University, Bentley, WA 6102, Australia. E-mail: josh.mylne@curtin.edu.au
First published on 10th November 2021
Abstract
The rise in herbicide resistance over recent decades threatens global agriculture and food security and so discovery of new modes of action is increasingly important. Here we reveal linezolid, an oxazolidinone antibiotic that inhibits microbial translation, is also herbicidal. To validate the herbicidal mode of action of linezolid we confirmed its micromolar inhibition is specific to chloroplast translation and did not affect photosynthesis directly. To assess the herbicide potential of linezolid, testing against a range of weed and crop species found it effective pre- and post-emergence. Using structure–activity analysis we identified the critical elements for herbicidal activity, but importantly also show, using antimicrobial susceptibility assays, that separation of antibacterial and herbicidal activities was possible. Overall these results validate chloroplast translation as a viable herbicidal target.
Introduction
Implementing herbicides into agricultural practice in the 1940s greatly improved crop productivity, and although this success continues today, the growth of herbicide resistance in the last few decades puts world food security under threat. The first field case of herbicide resistance, triazine-resistant Senecio vulgaris, was documented in 1968 and the number of weed species resistant to one or more herbicides has increased to 263 in 2021.1,2 Although there are strategies to manage herbicide resistance, such as herbicide rotation, the appearance of multiple-resistance weed species highlight the need for new modes of action.3,4 Despite this need, over 30 years passed with no new herbicide mode of action until the recent introduction of cyclopyrimorate that targets homogentisate solanesyltransferase5 and the upcoming release of tetflupyrolimet that targets dihydroorotate dehydrogenase.6A typical approach for discovering new herbicides is screening large and diverse compound libraries to select the most active compounds for further investigation, including a detailed analysis of its mechanism of action which might be a known or new target.3 Another approach suggested as a useful starting point for herbicidal development is to exploit the physico-chemical connections between existing pharmaceuticals and herbicides and to use the principles of drug development to develop compounds towards a biological target of interest.7 The similarities between drugs and herbicides,8,9 is highlighted by herbicides with antiprotozoal activity such as glyphosate,10 trifluralin11 and endothall,12 that are toxic to Plasmodium spp., and haloxyfop that is active against T. gondii, the causative agent of toxoplasmosis.13 Recent work has shown the converse to also be true, that many antiprotozoals, especially antimalarials, have herbicidal activity. Herbicidal antimalarial drugs include dihydrofolate reductase inhibitors,14–16 the non-mevalonate pathway inhibitor fosmidomycin17 and antimicrobial fluoroquinolones (e.g. ciprofloxacin) that act on DNA gyrase.18 Overall, these results suggest drug targets might be a source of inspiration for new herbicidal modes of action.
Unlike other eukaryotes, plants contain three genomes: nuclear, mitochondrial and plastid. The latter is present in chloroplasts, which are regarded as originating from symbiotic cyanobacteria and therefore its genome is homologous to prokaryotes.19 Although most chloroplast proteins are synthesized in the cytosol and imported by plastids, the chloroplast genome includes genes encoding essential parts of photosynthetic and translational machinery that are translated within the plastid.20 Therefore chloroplast translation is an essential process for plant viability, and its interruption interferes with embryo and seed development,21 causes chlorophyll deficiency,22 impairs cell division, root stem cell and leaf development,23,24 reduces oxygen consumption and decreases fitness,25 and eventually leads to death of a plant.21,22,26 Therefore, chloroplast translation machinery might be a viable target for herbicide development.
Despite differences in their regulatory pathways, the molecular mechanisms for chloroplast and bacterial translation are conserved27 so chloroplast translation can be inhibited by antibiotics that block microbial protein synthesis, e.g. spectinomycin and lincomycin.28 Another example is the actinonin that inhibits both bacterial and chloroplast peptide deformylase making it antibacterial and herbicidal.29 Virtually all information about chloroplast translation inhibitors such as puromycin, spectinomycin, lincomycin, chloramphenicol, streptomycin, erythromycin and anisomycin, and their effects on plants (as herbicides) is derived from studies with agar-grown plants,21,22,25,30–32 and few studies report outcomes as foliar applications or pre-emergence with soil-grown plants.33 Although chloroplast translation is an essential plant process, it is not targeted by any commercial herbicide.3 Therefore, an opportunity exists to peruse this potentially new herbicidal mode of action by starting with antimicrobials that inhibit protein synthesis. To achieve this, we used the antimicrobial linezolid 1, a known inhibitor of bacterial protein translation. Linezolid 1, is potent against Gram-positive bacteria and a member of the oxazolidinone family that share an N-aryl substituted 2-oxazolidinone structural core and contain a (S)-methylene group in the position 2 (Fig. 1). Bacterial resistance to 1 has arisen as rRNA point mutations as well as an amino acid deletion in a ribosomal protein (see references in34). Using 1 we demonstrate that its biological target possesses several advantages including the potential to control a broad spectrum of weeds.
Results and discussion
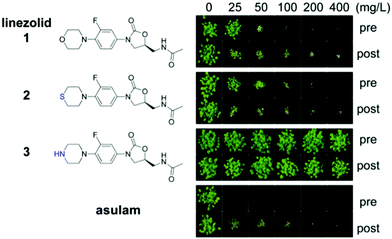
To investigate the herbicidal potential of oxazolidinones, in the first instance we used commercially available 1. This compound is highly active in vitro against S. aureus (MIC 1–8 μg mL−1) and M. tuberculosis (MIC 0.125–0.5 μg mL−1),35,36 approved for treating diseases caused by Gram-positive bacteria (e.g. E. faecium and S. aureus),37 and has potential for treating multidrug resistant tuberculosis.38 Assays against a model plant, Arabidopsis thaliana, were conducted by growing plants on soil and treating with a range of concentrations of 1 (25–400 mg L−1) either pre- or post-emergence (Fig. 1).Applying 1 as a post-emergence herbicide inhibited growth at the lowest dose 25 mg L−1 and was lethal at 100 mg L−1. Pre-emergence activity was slightly lower with growth inhibition at 50 mg L−1 and death at 100 mg L−1 (Fig. 1). The leaves of plants exposed to 1 appeared chlorotic and desiccated if the compound was applied post-emergence, whereas pre-emergence treatment resulted in plants that emerged bleached. This efficiency was less than, but comparable to the commercial herbicide asulam that was lethal at all concentrations tested pre- and post-emergence. Sutezolid 2, a thiomorpholine analogue of linezolid with similar antimicrobial effects,39 had activity similar to 1. By contrast for 1 and 2, the piperazyl analogue 3 of 1 was inactive (Fig. 1). An excellent antimicrobial, 3 displays only slightly lower activity than 140 so these results show the amino moiety of 3 plays a role either in uptake of the compound into the plant or in target binding. The known bacterial mode of action plus the observed plant chlorosis and necrosis were consistent with 1, and indeed oxazolidinones, disrupting chloroplast function.
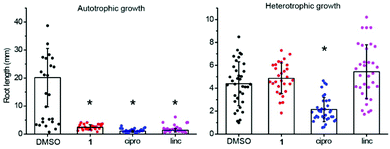
To confirm 1 interferes with chloroplast function, its effects during exclusively heterotrophic growth or exclusively autotrophic growth were compared. A. thaliana seedlings were grown on 1 (10 μM) in glucose-deficient MS-agar medium under a long day regime to stimulate photosynthesis (autotrophic growth). Exclusively heterotrophic growth was stimulated by growth on glucose-supplemented MS-agar medium in the dark. Ciprofloxacin, an inhibitor of DNA gyrase,18 was used as a control as it causes bleaching, but does not directly affect chloroplast translation. Lincomycin, an inhibitor of chloroplast translation, was also used as a control as it affects translation in chloroplasts, but not in the cytosol.32 To assess growth inhibition we measured root length. Under autotrophic conditions all compounds reduced plant growth (Fig. 2). A clear difference in phenotype was observed by the cotyledon stage for autotrophic plants with 1 causing chlorosis and a significant decrease of root length (Fig. S1, ESI†). In contrast, neither seedlings nor roots were affected significantly by 1 for plants grown in the dark on glucose (Fig. 2, Fig. S1, ESI†). The chloroplast translation inhibitor lincomycin suppressed growth in an autotrophic, but not heterotrophic growth regime. A different pattern was observed for ciprofloxacin, which inhibited growth in both regimes. Therefore, the herbicidal activity of 1 is attributable to disruption of chloroplast function, because it only affects photosynthesising plants.
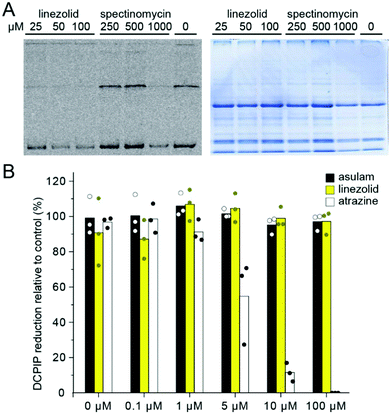
As the symptoms of injury, due to 1, (chlorosis and necrosis) can be caused by photosystem II inhibitors or inhibitors of chloroplast translation,22,41 we investigated how 1 affects electron transfer and translation using Pisum sativum (pea) chloroplasts as an in vitro model. To assess protein translation we used a [35S]-methionine in organelle protein translation assay; translation was strongly inhibited by 1 in the concentration range of 25–100 μM with an efficiency exceeding spectinomycin, a known inhibitor of chloroplast translation (Fig. 3A). To assess how linezolid affects electron transfer in thylakoid membranes we used the Hill reaction, a colorimetric assay based on reduction of dichlorophenolindophenol (DCPIP), an artificial electron acceptor used to measure electron transfer during photosynthesis.42,43 We found 1 did not affect the reduction of DCPIP (Fig. 3B), indicating 1 does not block electron transfer in the thylakoid membrane. To confirm DCPIP reduction distinguishes between inhibitors and non-inhibitors of photosynthesis we included controls atrazine (a photosystem II inhibitor, inhibits photosynthesis) and asulam (a dihydropteroate synthase inhibitor, does not inhibit photosynthesis). We found atrazine inhibited DCPIP reduction at 1 μM and completely blocked electron transfer at 100 μM, whereas asulam (like 1) did not affect the electron transfer even at the highest tested concentration, distinguishing between inhibitors of photosynthesis and those that do not affect photosynthesis. Hence, 1 inhibits protein translation in chloroplasts analogous to its mode of action in bacteria.
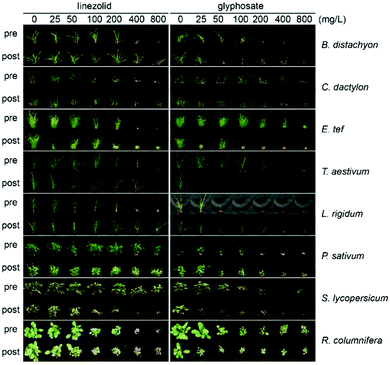
To assess the potential of 1 as a herbicidal scaffold we tested it against plant species, including monocots (Brachypodium distachyon, Cynodon dactylon, Eragrostis tef, Triticum aestivum, Lolium rigidum) and dicots (P. sativum, Solanum lycopersicum, Ratibida columnifera) at doses of 25–800 mg L−1 pre- and post-emergence (Fig. 4). We found 1 was active against all species pre- and post-emergence. Although the effective dose for 1 was ∼2–4 times greater than for A. thaliana, it had comparable activity to the glyphosate control against some species. The most susceptible species were R. columnifera and C. dactylon; these species were sensitive to 1 at 100 mg L−1 both pre- and post-emergence. The least sensitive plant was T. aestivum; visible growth inhibition was observed only at 800 mg L−1. For other species the efficient dose was 200–800 mg L−1, a use rate similar to many commercial herbicides.
With 1 having good and wide-acting activity, we explored which of its moieties were required for activity and which could be modified and retain herbicidal, but not antibacterial activity. This is crucial as herbicides should be plant-specific and 1 is a valuable antibiotic. To make sure analogues of 1 possessed physico-chemical properties suitable for herbicides, we used an interactive database of physico-chemical data for 360 commercial herbicides (used previously for herbicide discovery15,16,44,45) to choose, and then prepare analogues 4–57 (see ESI†) that varied e.g. size, shape and hydrophobicity, but overall retained favourable physico-chemical properties for herbicides (Fig. 5 and Fig. S2, ESI†).
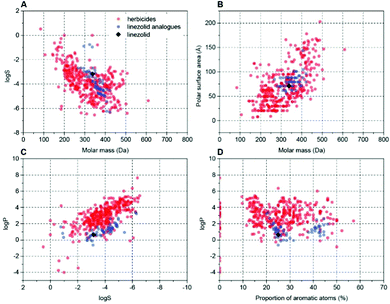 | ||
Fig. 5 Examples of cluster analysis of physicochemical properties of linezolid 1 and prepared analogues versus known herbicides. Charts were extracted from an interactive database of physicochemical properties for commercial herbicides9,45 (red dots). Data points for this work were added (blue dots, 1 shown as black rhombus) and plotted with two properties compared (A) log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S vs. molar mass; (B) polar surface area vs. molar mass; (C) log S vs. molar mass; (B) polar surface area vs. molar mass; (C) log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P vs. log P vs. log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S; (D) log S; (D) log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P vs. proportion of aromatic atoms (see also Fig. S2, ESI†). P vs. proportion of aromatic atoms (see also Fig. S2, ESI†). | ||
The overall structure of 1 contains chemically tractable motifs that were readily modified. Analogues were prepared where the core phenyloxazolidone was retained, but the piperazinyl, acetyl and fluoro groups were modified. A total of 54 analogues of 1 were synthesised (see ESI†) and then tested on soil for post-emergence activity (Fig. 6) using A. thaliana. Of the 54 analogues tested, many compounds lost herbicidal activity, but some interesting trends were observed. Extending the acetyl group of 1 with longer alkyl chains seemed to be tolerated for shorter chains (as in 5) but activity was reduced across longer chains, as in 6. Any other modification to the acetamido unit (as in 4, 7, 8 and 9) abolished activity. The morpholinyl moiety also plays an important role in activity. Substituting this unit for small groups as in 12 and 21 retained activity at 200 mg L−1, but longer and substituted acyl chains were not tolerated. Interestingly the formyl derivative 11 had no activity.
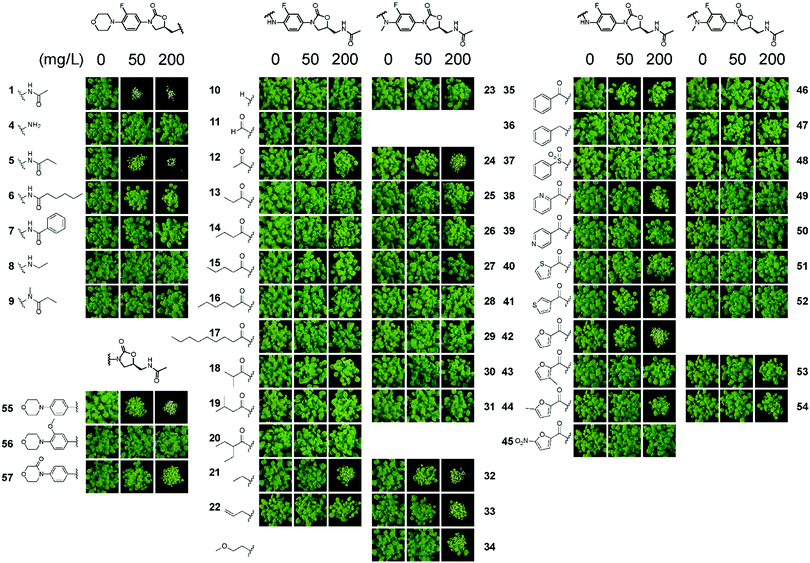 | ||
| Fig. 6 Screening of analogues 4–57 reveals insight into the post-emergent structure activity of linezolid 1. Compounds were prepared that possessed physicochemical properties as similar to commercial herbicides. Assays were performed as described in Fig. 1. | ||
In the N-methyl series 23–34, similar trends were observed with compounds 24, 32–34 all possessing good activity at 200 mg L−1. Examination of the aryl amides, which bring planarity and potential fortuitous pi-stacking interactions, showed these compounds were far less tolerated. The benzoyl 35, 2-pyridoyl 38, 2-furoyl 42 and the 5′-methylfuroyl 44 derivatives possessed some activity at 200 mg L−1 but interestingly all other molecules were inactive at the concentrations tested. Of note is that the corresponding N-methyl derivatives lost all activity. Modification of the morpholinyl ring itself as a 3-morpholinone, as in 57, also retained good activity at 200 mg L−1. A simpler structure–activity relationship was observed when modifying the fluoro group. The desfluoro analogue 55 was still quite potent at 50 mg L−1, but changing the group to a methoxy unit as in 56 abolished activity.
To assess if there was any improved selectivity gained for the prepared molecules as herbicides compared to antimicrobial activity, antibiotic susceptibility tests against Bacillus subtilis were performed for the compounds that retained herbicidal activity (Table 1). Activity against B. subtilis is relevant being that it is a common soil-borne bacterium and because 1 is more active against Gram-positive bacteria. All the synthesised analogues had zones of inhibition values less than linezolid 1 and interestingly, some compounds saw a considerable improvement in selectivity towards a herbicidal mode of action. Analogues 5 and 55, which have similar herbicidal activities compared to 1, showed a decrease in antimicrobial potency. Additionally, compounds 6, 24 and 34, despite having a slight reduction in herbicidal potency compared to 1, more importantly showed no antibacterial activity.
| Compound | Inhibition zone (mm) | Compound | Inhibition zone (mm) |
|---|---|---|---|
| Linezolid 1 | 18.6 ± 0.4 | 34 | NE |
| 5 | 15.2 ± 1.0 | 35 | NE |
| 6 | NE | 38 | 14.5 ± 1.0 |
| 12 | NE | 42 | 12.5 ± 0.5 |
| 21 | 13.7 ± 0.5 | 44 | 12.9 ± 0.4 |
| 24 | NE | 55 | 14.1 ± 0.9 |
| 32 | 15.2 ± 1.1 | 57 | NE |
| 33 | 12.7 ± 0.7 |
Conclusions
No commercial herbicide targets chloroplast protein translation. To evaluate if chloroplast translation might be a viable target we examined the oxazolidinone molecular class of microbial translation inhibitors as herbicides, using 1 as an exemplar and found this and other oxazolidinones were active against soil-grown plants (Fig. 1). Although translation in the cytosol differs from the chloroplast, some prokaryotic translation inhibitors, such as streptomycin, inhibit both cytosolic and chloroplast translation.32 Linezolid 1 and lincomycin specifically inhibit chloroplast translation as plants grow normally in a heterotrophic regime, but growth is reduced in an autotrophic regime (Fig. 2). The ability of 1 to selectively inhibit chloroplast protein translation was also confirmed using a labelled methionine incorporation assay, as 1 strongly inhibited protein translation at a concentration 1000 times lower than spectinomycin, but did not inhibit electron transfer at any tested rate of application, as confirmed by the DCPIP assay that measures electron transfer in thylakoid membranes.The utility of 1 was also demonstrated in its broad-spectrum herbicidal activity against a wide range of plant species where in all cases it was effective. Despite its promise, we cannot consider 1 itself as a potential herbicide unless its antibacterial and herbicidal activities are divorced from each other. To that end we explored which moieties of 1 are required for herbicidal activity, taking advantage of a physicochemical database9,45 to prepare analogues of 1 that retain suitable herbicidal properties. We demonstrate the critical elements that retain herbicidal activity but also show, using antimicrobial susceptibility assays, that separation of antibacterial and herbicidal activities was possible exemplified by 6, 24 and 34. Overall these results demonstrate that inhibiting chloroplast translation is a viable herbicidal mode of action and the structure of 1 could be used as a scaffold to create herbicides that target this important plant process.
Experimental procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min to pellet both intact and broken plastids. The supernatant was decanted and the plastid pellet resuspended in the residual supernatant using a soft painting brush and loaded onto 50% percoll-based density gradient prepared with grinding buffer as previously described.46 The loaded gradient was centrifuged at 12
000g for 5 min to pellet both intact and broken plastids. The supernatant was decanted and the plastid pellet resuspended in the residual supernatant using a soft painting brush and loaded onto 50% percoll-based density gradient prepared with grinding buffer as previously described.46 The loaded gradient was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100g for 10 min and the higher band of broken plastids was discarded by vacuum aspiration, whereas a lower dark-green band of intact chloroplasts was collected by a disposable Pasteur pipette, mixed with three volumes of 0.33 M sorbitol, 50 mM HEPES buffer (pH 8.0) and centrifuged at 2
100g for 10 min and the higher band of broken plastids was discarded by vacuum aspiration, whereas a lower dark-green band of intact chloroplasts was collected by a disposable Pasteur pipette, mixed with three volumes of 0.33 M sorbitol, 50 mM HEPES buffer (pH 8.0) and centrifuged at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 4 min to pellet intact plastids. The supernatant was decanted and plastids resuspended in the residual liquid. The plastid suspension was kept on ice and used immediately for in vitro in organello translation and Hill reaction assays.
000g for 4 min to pellet intact plastids. The supernatant was decanted and plastids resuspended in the residual liquid. The plastid suspension was kept on ice and used immediately for in vitro in organello translation and Hill reaction assays.
Author contributions
K. V. S., J. S. M. and K. A. S. conceived the study and analysed data. K. V. S. performed experiments validating the target of 1 and initial herbicidal analysis. K. J. B. and K. A. S. performed chemical synthesis and K. J. B. conducted herbicidal analysis of prepared compounds. K. V. S. and A. W. D. performed antimicrobial assays. M. W. M. extracted organelles and performed translation assays. K. V. S., K. A. S. and J. S. M. wrote the manuscript with help from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Philippe Hervé and Bruce Lee from Nexgen Plants for fruitful discussions. The authors are grateful to Aneta Ivanova and Mabel Gill-Hille for assistance establishing translation assays and to Maxime Corral and Joel Haywood for help establish A. thaliana herbicide assays. K. V. S. was supported by a Research Training Program Stipend and a Research Training Program Fee Offset. This work was funded in part by Nexgen Plants and in part by an Australian Research Council (ARC) Discovery Project (DP190101048) to J. S. M. and K. A. S. M. W. M. was supported by ARC Future Fellowship FT130100112.Notes and references
- G. F. Ryan, Weed Sci., 1970, 18, 614–616 CrossRef CAS.
- I. Heap, The international survey of herbicide resistant weeds, http://www.weedscience.org, (accessed 20 July 2020).
- S. O. Duke, Pest Manage. Sci., 2012, 68, 505–512 CrossRef CAS PubMed.
- P. J. Tranel, C. W. Riggins, M. S. Bell and A. G. Hager, J. Agric. Food Chem., 2011, 59, 5808–5812 CrossRef CAS PubMed.
- M. Shino, T. Hamada, Y. Shigematsu, K. Hirase and S. Banba, J. Pestic. Sci., 2018, 43, 233–239 CrossRef CAS.
- F. E. Dayan, Plants, 2019, 8, 341 CrossRef CAS.
- S. O. Duke, Weed Sci., 2010, 58, 334–339 CrossRef CAS.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Delivery Rev., 1997, 23, 3–25 CrossRef CAS.
- M. N. Gandy, M. G. Corral, J. S. Mylne and K. A. Stubbs, Org. Biomol. Chem., 2015, 13, 5586–5590 RSC.
- F. Roberts, C. W. Roberts, J. J. Johnson, D. E. Kyle, T. Krell, J. R. Coggins, G. H. Coombs, W. K. Milhous, S. Tzipori, D. J. P. Ferguson, D. Chakrabarti and R. McLeod, Nature, 1998, 393, 801 CrossRef CAS.
- T. Kaidoh, J. Nath, H. Fujioka, V. Okoye and M. Aikawa, J. Eukaryotic Microbiol., 1995, 42, 61–64 CrossRef CAS.
- J. Bajsa, K. Singh, D. Nanayakkara, S. O. Duke, A. M. Rimando, A. Evidente and B. L. Tekwani, Biol. Pharm. Bull., 2007, 30, 1740–1744 CrossRef CAS.
- E. Zuther, J. J. Johnson, R. Haselkorn, R. McLeod and P. Gornicki, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13387–13392 CrossRef CAS.
- G. Corral Maxime, J. Haywood, H. Stehl Luca, A. Stubbs Keith, W. Murcha Monika and S. Mylne Joshua, Plant J., 2018, 95, 727 CrossRef.
- M. G. Corral, J. Leroux, K. A. Stubbs and J. S. Mylne, Sci. Rep., 2017, 7, 1–9 CrossRef PubMed.
- M. G. Corral, J. Leroux, S. Tresch, T. Newton, K. A. Stubbs and J. S. Mylne, Angew. Chem., Int. Ed., 2017, 56, 9881–9885 CrossRef CAS.
- M. Witschel, F. Röhl, R. Niggeweg and T. Newton, Pest Manage. Sci., 2013, 69, 559–563 CrossRef CAS PubMed.
- M. D. Wallace, N. F. Waraich, A. W. Debowski, M. G. Corral, A. Maxwell, J. S. Mylne and K. A. Stubbs, Chem. Commun., 2018, 54, 1869–1872 RSC.
- E. H. Harris, J. E. Boynton and N. W. Gillham, Microbiol. Rev., 1994, 58, 700–754 CrossRef CAS.
- R. Zoschke and R. Bock, Plant Cell, 2018, 30, 745–770 CrossRef CAS PubMed.
- N. Parker, Y. Wang and D. Meinke, Plant Physiol., 2014, 166, 2013–2027 CrossRef.
- K. Zubko Mikhajlo and A. Day, Plant J., 1998, 15, 265–271 CrossRef.
- D. Ahlert, S. Ruf and R. Bock, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 15730–15735 CrossRef CAS.
- M. T. Nakata, M. Sato, M. Wakazaki, N. Sato, K. Kojima, A. Sekine, S. Nakamura, T. Shikanai, K. Toyooka, H. Tsukaya and G. Horiguchi, Biol. Open, 2018, 7, bio028175 CrossRef PubMed.
- P. F. Lurquin and A. Kleinhofs, Biochem. Biophys. Res. Commun., 1982, 107, 286–293 CrossRef CAS.
- B. Dudas, B. Jenes, G. B. Kiss and P. Maliga, Theor. Appl. Genet., 2012, 125, 1517–1523 CrossRef CAS.
- D. S. Stern, D. C. Higgs and J. Yang, Trends Plant Sci., 1997, 2, 308–315 CrossRef.
- R. J. Ellis, Planta, 1970, 91, 329–335 CrossRef CAS PubMed.
- C.-X. Hou and M. Williams, in Natural Products for Pest Management, ed. A. M. Rimando and S. O. Duke, American Chemical Society, Washington, DC, 2006, ch. 19, p. 319 Search PubMed.
- J. A. Sullivan and J. C. Gray, Plant Cell, 1999, 11, 901–910 CrossRef CAS.
- J. Guo, S. Wang, O. Valerius, H. Hall, Q. Zeng, J.-F. Li, D. J. Weston, B. E. Ellis and J.-G. Chen, Plant Physiol., 2011, 155, 370–383 CrossRef CAS.
- P. Mulo, S. Pursiheimo, C.-X. Hou, T. Tyystjärvi and E.-M. Aro, Funct. Plant Biol., 2003, 30, 1097–1103 CrossRef CAS PubMed.
- M. Marsoni, F. De Mattia, M. Labra, A. Bruno, M. Bracale and C. Vannini, Ecotoxicol. Environ. Saf., 2014, 108, 52–57 CrossRef CAS PubMed.
- M. J. Belousoff, Z. Eyal, M. Radjainia, T. Ahmed, R. S. Bamert, D. Matzov, A. Bashan, E. Zimmerman, S. Mishra, D. Cameron, H. Elmlund, A. Y. Peleg, S. Bhushan, T. Lithgow, A. Yonath, G. Zhang and R. A. Bonomo, mBio, 2017, 8, e00395 CrossRef CAS.
- R. E. Mendes, P. A. Hogan, J. M. Streit, R. N. Jones and R. K. Flamm, Antimicrob. Agents Chemother., 2015, 59, 2454–2457 CrossRef CAS.
- C. Yang, H. Lei, D. Wang, X. Meng, J. He, A. Tong, L. Zhu, Y. Jiang and M. Dong, Jpn. J. Infect. Dis., 2012, 65, 240–242 CrossRef CAS PubMed.
- S. M. R. Hashemian, T. Farhadi and M. Ganjparvar, Drug Des., Dev. Ther., 2018, 12, 1759–1767 CrossRef CAS.
- B. Singh, D. Cocker, H. Ryan and D. J. Sloan, Cochrane Database Syst. Rev., 2019, 3, CD012836 Search PubMed.
- S. Wen, X. Gao, W. Zhao, F. Huo, G. Jiang, L. Dong, L. Zhao, F. Wang, X. Yu and H. Huang, Int. J. Infect. Dis., 2020 DOI:10.1016/j.ijid.2021.06.055.
- W. Phetsang, M. A. T. Blaskovich, M. S. Butler, J. X. Huang, J. Zuegg, S. K. Mamidyala, S. Ramu, A. M. Kavanagh and M. A. Cooper, Bioorg. Med. Chem., 2014, 22, 4490–4498 CrossRef CAS PubMed.
- F. Dan Hess, Weed Sci., 2000, 48, 160–170 CrossRef.
- R. L. Dean and E. Miskiewicz, Biochem. Mol. Biol. Educ., 2003, 31, 410–417 CrossRef CAS.
- M. Santakumari and V. S. R. Das, Pestic. Biochem. Physiol., 1978, 9, 119–129 CrossRef CAS.
- S. Funar-Timofei, A. Borota and L. Crisan, Mol. Diversity, 2017, 21, 437–454 CrossRef CAS.
- K. V. Sukhoverkov, M. G. Corral, J. Leroux, J. Haywood, P. Johnen, T. Newton, K. A. Stubbs and J. S. Mylne, RSC Adv., 2021, 11, 8459–8467 RSC.
- K. Waegemann, J. Soll, D. W. Galbraith, D. P. Bourque and H. J. Bohnert, in Methods in Cell Biology, Academic Press, 1995, vol. 50, ch. 18, pp. 255–267 Search PubMed.
- J. E. Mullet, R. R. Klein and A. R. Grossman, Eur. J. Biochem., 1986, 155, 331–338 CrossRef CAS.
- M. F. Fisher, J. Zhang, N. L. Taylor, M. J. Howard, O. Berkowitz, A. W. Debowski, B. Behsaz, J. Whelan, P. A. Pevzner and J. S. Mylne, Plant Direct, 2018, 2, e00042 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cb00192b |
| This journal is © The Royal Society of Chemistry 2022 |