Open Access Article
Open Access ArticleMacrocyclic DNA-encoded chemical libraries: a historical perspective
Louise
Plais
 and
Jörg
Scheuermann
and
Jörg
Scheuermann
 *
*
Department of Chemistry and Applied Biosciences, ETH Zürich (Swiss Federal Institute of Technology), Vladimir-Prelog-Weg 4, CH-8093 Zürich, Switzerland. E-mail: joerg.scheuermann@pharma.ethz.ch
First published on 29th October 2021
Abstract
While macrocyclic peptides are extensively researched for therapeutically relevant protein targets, DNA-encoded chemical libraries (DELs) are developed at a quick pace to discover novel small molecule binders. The combination of both fields has been explored since 2004 and the number of macrocyclic peptide DELs is steadily increasing. Macrocycles with high affinity and potency were identified for diverse classes of proteins, revealing DEL's huge potential. By giving a historical perspective, we would like to review the methods which permitted the rise of macrocyclic peptide DELs, describe the different DELs which were created and discuss the achievements and challenges of this emerging field.
Introduction
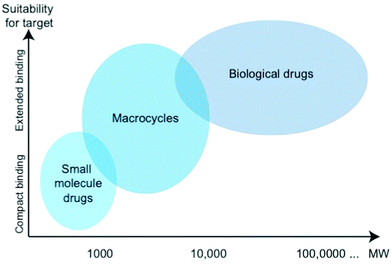
Macrocyclic peptides (MPs) essentially comprise cyclic peptidic structures with a ring size of at least twelve atoms and spanning multiple amino acid residues.1 MPs’ molecular weight may thus vary between some five hundred and several thousand Dalton, which allows them to bridge the molecular worlds of small molecules and macromolecules such as antibodies (Fig. 1).2,3 MPs therefore encompass a multitude of very diverse molecules and represent a class of promising ligands for basic and therapeutic research. While in principle small molecules may be orally available, may easily extravasate and enter cells, it is, however, very difficult to identify and develop small molecules of high affinity and specificity. On the other hand, specificity and binding affinity can be excellent for antibodies, while their pharmacokinetic properties are less favorable and oral and cellular uptake is largely hampered.4The intermediate field that is populated by MPs hence promises to amalgamate the good properties of both ends, as they possess a wide range of physical and pharmacodynamic properties with corresponding advantages and limitations.5
While usually not completely rigid, macrocycles are constrained and their residues preorganized. This eventually results in a minimized entropic loss upon binding to targets and therefore may yield exquisite binding affinities, often comparable to those of antibodies.2
As a consequence, academic and industrial research have increasingly turned towards them, in order to obtain ligands to challenging targets, and especially to tackle protein–protein interactions.6 MPs may serve as good starting points for drug discovery also because they are synthetically accessible and amenable to medicinal chemistry efforts, thus allowing to balance their pharmacokinetic and pharmacodynamic properties.7 In analogy to small molecules, MPs can potentially penetrate and diffuse into tissues, rendering them a tool of choice for targeted applications.8,9 They also tend to be less immunogenic than antibodies. The Lipinski rule of five, which describes traditional small molecules with ideal properties for drug discovery, has not been considered to apply also to MPs.10,11 Instead, adjusted rules, such as those established by Kihlberg better reflect the desired properties of MPs.12,13
The rigidity induced from cyclization provides an increased metabolic stability towards proteases. Also, the resulting physico-chemical properties may contribute to an enhanced membrane permeability, i.e. cellular uptake.14 For these reasons, chances to achieve oral bioavailability are greater for MPs compared to their linear counterparts. Finally, the peptidic backbone may confer lower toxicity in vivo, as the degradation products might easily be metabolized and excreted.15
Nature-derived macrocycles initially proved the importance of the field and provided important drugs as exemplified by the antibiotic polymyxin B, the human peptide hormone vasopressin and the immunosuppressant cyclosporine.5 Today, more than one hundred MP drugs are approved or in late-stage clinical development. On average, a new MP drug is marketed every year,1 and the pace is increasing. The remaining hurdles are set mainly by poor oral bioavailability and cell permeability, but also by metabolic stability, renal clearance and – to a lesser extent – immunogenicity.
Several methodologies are currently used to discover new macrocyclic hits. Rational design or screening of natural products are important routes yet they still require huge efforts before yielding clinical candidates.16 On the other hand, the production of peptide libraries has been rising since the discovery of phage display in 1985 by Smith and coworkers.17,18 Here, the identity of each peptide is encoded in the phage's genetic material,19,20 featuring an unambiguous link between genotype and phenotype, thus permitting the simultaneous testing of a whole library against a chosen biological target in affinity-based selections, and the subsequent decoding of the binding molecules. Also, the chemical modification of peptides allows the production of cyclic and bicyclic peptide libraries on phage, as described by Winter, Heinis and Derda.20–22 The panning of bicyclic libraries yielded drug candidates that have entered clinical trials. Another striking example in the field is the Peptidream™ MP-platform developed by Suga and coworkers. To create these libraries, the enzymatic power of specifically engineered enzymes “flexizymes” was harnessed to include also a limited number of unnatural amino acids into the peptides.23 This innovation concomitantly led to an expansion of the natural amino acid repertoire to compose the peptidic libraries and also yielded candidates which are now in clinical trials. Another biotechnological platform which allows for the synthesis of MPs relies on the SICLOPPS technology.24 The acronym stands for Split-Intein Circular Ligation of Peptides and Proteins. The library production is achieved by ribosomal protein synthesis, and as indicated by its name, is followed by an intein-like event that splices the amino acid sequence into a loop.25
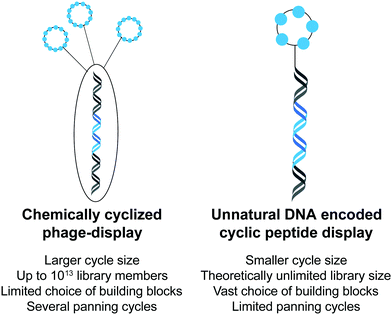
Lately, cyclic peptide display in library scale using non-natural amino acids was enabled by DNA-encoded chemical library (DEL) technology.26 DELs are collections of synthetic compounds linked to unique oligonucleotide tags.27 In analogy to phage display, phenotype and genotype are linked and allow for the production of small molecules and also MP libraries (Fig. 2). Selections of DELs for the identification of binders against a protein of interest is possible with all DEL library members present in the same experiment. Usually, the target is immobilized on a solid support and incubated with the library members.28 Non-binding library members are removed by subsequent washes while the target-bound encoded compounds are submitted to PCR amplification, to obtain enough material for high throughput DNA sequencing and hit identification.28 The libraries are typically synthesized in combinatorial fashion through alternative split-and-pool steps.29 The majority are synthesized in solution but recently, synthesis on solid support has also been explored.30,31
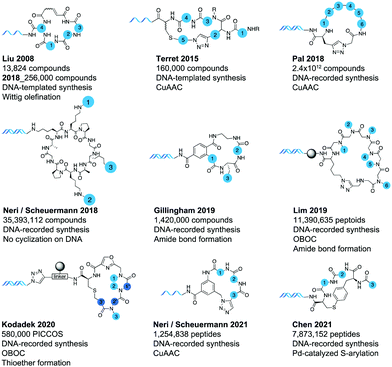
Nowadays, a large portion of DELs is dedicated to small molecules respecting Lipinski's rule of five, and such libraries delivered efficient binders to well-defined protein pockets.32 However, already in 1992, with their theoretical paper on DNA-encoded chemistry, Brenner and Lerner had envisioned peptide synthesis for DEL.33 Chemical hurdles, especially the unavailability of suitable orthogonal DNA-compatible protecting groups prevented its application for nearly a decade until, in 2004, Liu and coworkers synthetized the first macrocyclic peptide DEL (MP-DEL) based on DNA-templated synthesis (DTS).34 This pioneering work was followed-up by more DTS-derived MP-DELs.35–37 In 2018, DNA-recorded synthesis was reported for the first time for the construction of macrocyclic DELs.38,39 Since then several academic and industrial researchers published diverse DELs and methods for their synthesis.38–44 Also, DNA-encoded peptidomimetic libraries were created and successfully screened.41,45 In this review, we will describe the historical course of DNA-encoded libraries of macrocycles empowered by DNA-templated and, more recently, DNA-recorded synthesis. We will provide an overview of the published libraries, their respective setups, and the results they obtained. Finally, we will reflect on the challenges of MP-DEL technologies and comment on potential improvements.
DNA-templated synthesis of macrocycles
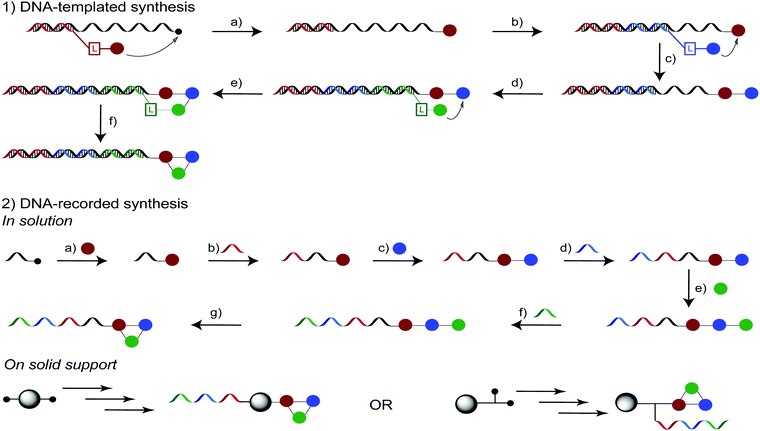
Inspired by nature, DNA-templated synthesis (DTS) was designed in 2001 by the Liu group to efficiently bring together reactants from complex mixtures through the mediation of DNA sequences.46,47DTS implementation requires a DNA template comprising the coding sequences for each final DEL library member and a chemically modifiable anchor, as well as corresponding reactants (building blocks) attached to unique oligonucleotides by cleavable linkers. According to the DNA template coding sequences, reactants and template oligonucleotides are brought together and allowed to react. The effective molarity of such reactions may be very high, thus allowing to conduct reactions which are otherwise considered difficult or impossible to implement with conventional chemistry.46
DTS has been employed to construct MP-DELs and the process to create a trimeric MP-DEL is described in Fig. 3a. Briefly, in a single vessel, the templates are annealed with a first set of code-complementary oligonucleotides bearing each a different reactant, which are then chemically attached to the template. The linker between the short oligonucleotide and the reactant is subsequently cleaved and this process can be repeated two more times leading to a trimeric linear library. Finally, the library may be cyclized and used for selection experiments against target proteins.
In 2004, nearly a decade after DEL was conceptualized, the DEL field was emerging with the concomitant synthesis of a (recorded) dual-display library in the Neri/Scheuermann lab,48 a small linear peptide DEL by the Harbury lab facilitated by “DNA routing”,49 and the first DTS-derived DEL by the Liu lab.34 The later was conceived as a pilot for future MP-DELs and it comprised 65 library members made from three sets of amino acids, eventually cyclized by Wittig olefination (see Fig. 4). Each set comprised four amino acids and cyclization was carried out with yields over 60%. It is worth noting that each step was followed by a biotin–streptavidin mediated capture step, to separate reacted from unreacted compounds. This pilot study proved the feasibility of sequence-programmed library synthesis and more generally, it displayed a successful screening of the library against carbonic anhydrase, PCR amplification and retrospective identification of the binder.34
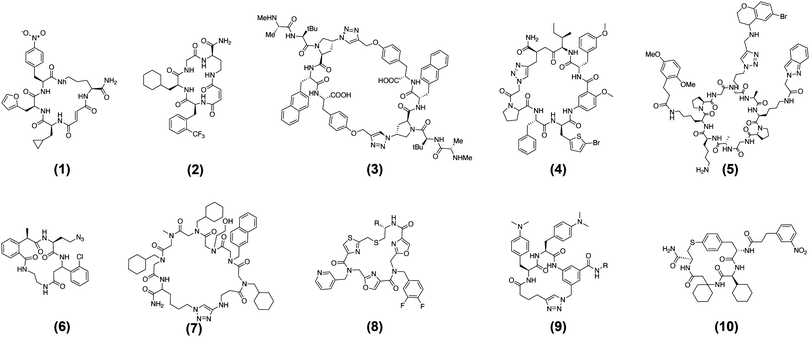
 | ||
| Fig. 4 Published MP-DELs in historical order (Last author, publication year, synthetic modalities, cyclization strategy). | ||
This first work was followed by the creation of a similar, larger library in 2008.35 Scaling-up the number of final library members demanded four key developments. First, a capping-based approach, reminiscent of solid-supported peptide synthesis, was implemented to DTS. It permitted the simplification of reagents structure and preparation, it reduced the number of required manipulations and led to increased final product yields. The number of building blocks was augmented to a total of thirty-six which were combined with eight different starting scaffolds. All chemicals were thoroughly tested for their ability to generate macrocycle products. For the DNA templates, an extended set of coding and annealing sequences was computationally designed and experimentally validated to support 1728 combinations made from three sets of building blocks. Moreover, new high-resolution LC/MS analysis methods were developed to assess the quality of larger DTS-DELs. Ultimately, these developments allowed the translation of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 824 templates into their corresponding macrocyclic structures.
824 templates into their corresponding macrocyclic structures.
The library was screened against therapeutically relevant targets in a following study.50 Hits were resynthesized without DNA tag, assayed in vitro and inhibitors with IC50 values as low as 680 nM were discovered for Src kinase (see Fig. 6, hit 1). An in-depth study of a series of enriched macrocycles showed that inhibition was depending on each building block, as well as on backbone conformation. One macrocycle proved to be activating rather than inhibiting VEGFR2, and two macrocycles were highly selective for Src compared with closely-related kinases. These two specific compounds were submitted to medicinal chemistry efforts described in a later publication.51 Macrocycles with potency reaching 4 nM were generated and characterized. Co-crystal structures revealed the molecular basis of the bi-substrate-competitive inhibition mechanism and of the selectivity for Src kinase. Modest inhibition was observed in cultured mammalian cells, which was most likely due to an insufficient membrane permeability of the inhibitor.
A similar study was published in 2014, when the library was submitted to selections against insulin degrading enzyme (IDE).52 Six macrocycles were identified, assayed and co-crystalized. Testing in mouse models showed improved glucose tolerance and slower gastric emptying. Ultimately, one compound was found to bind to a pocket away from the catalytic site of IDE, and it could be used to design an exo-site-specific screen, which revealed several inhibitors that were able to reprogram the activity of IDE.53 Among other studies, this extended investigation demonstrated the relevance of the DEL field to find modulators to enzymes of therapeutic interest and it especially established MP-DELs as useful sources of selective and potent ligands.
The Liu group further improved its DTS setup and in 2018 published a second-generation DNA-templated library of macrocycles.54 Essentially, they reviewed and improved again fundamental aspects for the design and synthesis of DNA-templated MP-DELs. Preliminary studies were carried out to computationally select the best combinations of building blocks and determine the drug-likeness of the final library members, by looking at Kihlberg rules.10,55 All DNA templates were also computationally optimized regarding the orthogonality of each annealing sequence and their assembly was improved through a polymerase-mediated strategy. In addition, methods for library isolation and purification were modified and upgraded. The integration of all these methods yielded a DNA-templated MP-DEL of 256![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 members.54
000 members.54
In vitro selections were again carried out against IDE and yielded several inhibitors. Among the hits, one macrocycle contained a surprising backbone alkene with a cis-conformation and showed high potency with an IC50 of 40 nM (see Fig. 6, hit 2).
Following on the early success of DNA-templated DELs, Ensemble Therapeutics was co-founded by Prof. Liu and continued the development of MP-DELs. In 2015, the company published a new DNA-templated MP-DEL in collaboration with Bristol Meyer Squibb.36 It contained five sets of building blocks cyclized by copper-catalyzed azide/alkyne cycloaddition (CuAAC) (see Fig. 4). In total, four libraries of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 members were generated. Each library was screened against XIAP BIR2 and BIR3 domains,56 and inhibitors with the ability to displace bound pro-apoptotic caspases were found. X-ray cocrystal structures were produced for promising compounds with XIAP BIR2 and led to structures increased in potency. Especially, it was found that dimeric macrocycles had improved affinities and inhibitory activities (see Fig. 6, hit 3). It was also shown that some dimeric macrocycles which could bind with similar affinities both XIAP BIR2 and BIR3 domains behaved as potent pro-apoptotic agents in cancer cell lines and that they could shrink tumors in a mouse xenograft model.56
000 members were generated. Each library was screened against XIAP BIR2 and BIR3 domains,56 and inhibitors with the ability to displace bound pro-apoptotic caspases were found. X-ray cocrystal structures were produced for promising compounds with XIAP BIR2 and led to structures increased in potency. Especially, it was found that dimeric macrocycles had improved affinities and inhibitory activities (see Fig. 6, hit 3). It was also shown that some dimeric macrocycles which could bind with similar affinities both XIAP BIR2 and BIR3 domains behaved as potent pro-apoptotic agents in cancer cell lines and that they could shrink tumors in a mouse xenograft model.56
The application of DTS to DEL construction was mostly supported by the efforts of the Liu group and of Ensemble Therapeutics, profiting from intrinsic advantages such as high chemical yields, access to difficult reactions, one-pot reactions with sets of building blocks and implemented purification methods.47 However, even though new strategies have been developed for the creation of “universal templates”,57 the DTS approach may be hampered by the laborious preparation of DNA templates and reagent oligonucleotides containing the chemical building blocks. Furthermore, code-specificity poses increasing difficulties with expanding library sizes, causing major limitations for the synthesis of large MP-DELs. This may explain the change of focus in the DEL field towards DNA-recorded synthesis (see below), also for the construction of MP-DELs.
DNA-recorded synthesis of macrocycles
In parallel to DTS, DNA-recorded synthesis of DELs was developed and soon became the most widely adopted method by both academia and industry to create DNA-encoded chemical libraries.27,58 This strategy makes use of split-and-mix procedures first implemented by combinatorial chemistry.59 In a first step a set of compounds is attached to the chemically modified extremity of a universal oligonucleotide. Each building block is then encoded through the ligation of a unique DNA sequence to the remote end of the universal oligonucleotide. At this point, the encoded compounds can be pooled and split again to start a new cycle of chemical modification and encoding. This process is schematically shown in Fig. 3b for a cyclized library containing three sets of building blocks, either in solution or on solid support. Nonetheless, from this general scheme many variations are possible. The resulting DEL can be either single-stranded or double-stranded.60 In the majority of DEL constructions to date double-stranded DELs are produced, e.g., the synthesis may start from a double-stranded, uniform “DNA headpiece” oligonucleotide.61,62 This oligonucleotide was designed by Praecis/GSK and exploited for DEL construction.63 The coding DNA sequences can be added to the headpiece by sticky-end ligation, allowing for a rather short final oligonucleotide. On the other hand, single-stranded libraries may feature distinct advantages, such as recently proposed selection strategies based on photocrosslinking,64,65 association with cell-penetrating peptides,66 or the affinity-maturation of ligands using a dual-display DEL setup.67 For these single-stranded libraries, the ligation process is slightly different, as it is based on adaptor-mediated ligation, whereby the used adaptors may be removed by purification or degradation.68DNA-recorded synthesis of DELs presents several advantages over DNA-templated DEL synthesis. Even though it demands more robust chemistry in order to allow for good yields, it does not require a DNA template or DNA-linked sets of building blocks.69 The choice of building blocks and the possibilities to combine them expands correspondingly, rendering this methodology more attractive for the creation of large DELs.
This is also reflected by the fact that, while the first MP-DELs were created with DTS,70 more recently the majority of MP-DELs (seven out of nine designs, described in Fig. 4) were produced by DNA-recorded synthesis. The most recent MP-DELs typically are both larger in size and in diversity of employed building blocks.
In the following, we would like to summarize the relevant MP-DELs generated with DNA-recorded synthesis and comment on their unique properties.
One of the first DNA-recorded MP-DEL was published by GSK in 2018 and nicely demonstrated the theoretical power of the split-and-pool combinatorial approach to generate very large DEL sizes. The presented MP-DEL featured 2.4 × 1012 uniquely encoded compounds.38 For the construction, six synthetic cycles where performed with amino acid building blocks and the synthesis was completed by CuAAC cyclization. In total, 276 different natural or non-natural monomeric or multimeric amino acid building blocks were used, leading to a ring size of four to twenty amino acids. Selections were carried out with the library and the respective non-cyclized control library against respiratory syncytial virus N-protein (RSV N).71 Four MP-DEL hits were resynthesized without DNA tag and their binding properties were assessed by affinity selection-mass spectrometry (AS-MS). The cyclic hit compounds were further confirmed by a time-resolved fluorescent resonance energy transfer (TR-FRET) assay disrupting the interaction between RSV N and its counterpart P-protein (see Fig. 6, hit 4).
Also in 2018, the Neri/Scheuermann lab published a completely different MP-DEL setup.39 An already cyclized decameric beta-sheet peptide mimetic was anchored to double-stranded DNA. It bore two orthogonally protected amines and an azide group, pointing towards the same face of the cycle. The functional groups were sequentially deprotected and reacted with sets of carboxylic acids and alkynes to generate a DEL of 35.4 Mio members. The YL-lib library design was inspired by the mechanism of antibody–antigen recognition. From this library specific binders with low micromolar affinity were selected against a variety of target proteins, i.a., against tumor necrosis factor (TNF),72 calmodulin (CaM) and prostate-specific antigen (PSA) (see Fig. 6, hit 5). The spatial arrangement of the library members also enabled the development of fluorescence microscopy procedures for PSA, of selective in vivo delivery of payloads to tumors for CAIX, and of the synthesis of a specific probe for CaM.
In contrast to the universal scaffold approach the Gillingham group presented a MP-DEL displaying high backbone diversity in 2019.40 Specifically inspired by polyketide and mixed peptide–polyketide structures, they aimed at producing a library rich in hydrophobic backbone elements. In total, 2142 distinct scaffolds were combined with additional side chain diversity elements to give rise to a DEL of 1.4 Mio members. The backbone diversity required important synthetic efforts to include original bifunctional building blocks. The library was tested in selections, i.a. against alpha-glycoprotein 1 (AGP),73 and provided a hit compound with a dissociation constant of 7 μM. This binder could further be improved to 4 μM by removing a side chain diversity element (see Fig. 6, hit 6).
Peptoid-like libraries are an alternative strategy to reach high backbone diversity, and such macrocyclic-peptoid DELs were conceived and synthesized. Peptoids are oligomers of N-substituted glycine and represent an interesting alternative to peptides for therapeutic approaches.74 Without peptidic amide bonds, they are less prone to metabolic degradation and can penetrate more easily into cells, while retaining the binding advantage of a cyclized structure.75 The first macrocyclic peptoid-DEL was published by the Lim group in 2019,41 followed by the Kodadek lab/Deluge Biotechnologies in 2020.45 It is worth noting that both libraries took advantage of the one-bead one-compound (OBOC) methodology for synthesis and screening.76
The respective libraries were synthesized on solid-phase using the sub-monomer approach, i.e., a succession of acylation and SN2 substitution steps. Such protocols were optimized by the groups of Paegel and Kodadek for DNA-encoded chemistry and encoding.30,77
The Lim lab produced a library counting 11.4 Mio encoded peptoids with six amide couplings and a final amide bond formation cyclization step.41 The library was tested against Skp278 in an affinity-based on-bead screening and a first set of eighty peptoids was isolated. After on-bead re-synthesis of a focused library without DNA tag, five hits were finally identified and characterized with dissociation constants between 7 and 30 μM (see Fig. 6, hit 7). Similarly, Kodadek/Deluge presented a peptoid-inspired conformationally-constrained oligomer (PICCOs) library.42 With three positions to include sets of amines and three positions to introduce backbone variations, the library comprised 580![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds. Cyclization was performed by thioether bond formation. As a proof of concept, the library was submitted to selection against streptavidin in a FACS-based screening, yielding two binders differing only by one heteroatom, with a dissociation constant in the hundred nanomolar range (see Fig. 6, hit 8).
000 compounds. Cyclization was performed by thioether bond formation. As a proof of concept, the library was submitted to selection against streptavidin in a FACS-based screening, yielding two binders differing only by one heteroatom, with a dissociation constant in the hundred nanomolar range (see Fig. 6, hit 8).
Two further MP-DELs were published in 2021. The Chen/Lu labs pursued an original way to cyclize peptides for MP-DELs.43 They investigated palladium-catalyzed intramolecular S-arylation in solution and on DNA, and constructed a corresponding four diversity elements MP-DEL comprising ca. 8 Mio. compounds. A preliminary screen against protein p30079 was performed and several compounds were identified which exhibited single digit micromolar inhibition activity (see Fig. 6, hit 10).
The Neri/Scheuermann lab constructed a MP-DEL with three variable positions using sets of natural and unnatural amino acids.44 Cyclization was achieved through CuAAC, with a collection of bifunctional carboxylic acid–alkynes. The obtained 1.3 Mio compounds were encoded by single-stranded DNA, allowing for both the screening of proteins of interest by classic affinity capture procedures, and by photo-crosslinking.65 Specific binders in the low micromolar range were enriched for several serum albumins and for NKp46,80 a marker of activated Natural Killer cells.
In spite of diverse design principles, DNA-recorded MP-DELs have demonstrated their potential for discovering ligands of interest. Different cyclization strategies, detailed below, participated in the expansion of possible scaffolds.
Cyclization strategies
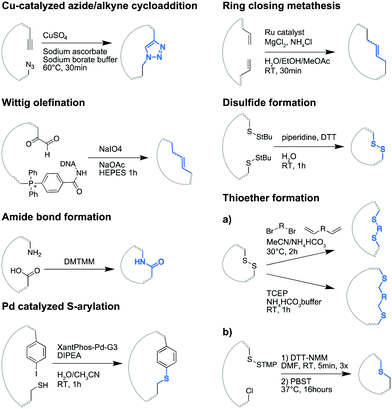
From the perspective of peptide synthesis, a crucial and challenging step in MP-DEL synthesis is macrocyclization.81 First of all, the cyclization reaction must be DNA-compatible, rendering most of the routes commonly employed in classical peptide chemistry impossible. Secondly, the reaction should be quantitative, considering that cyclization happens at the end of MP-DEL synthesis and all library compounds are pooled, meaning that they will not react uniformly and can also not be individually purified. Eventually, it would be advantageous if the progress of the cyclization reaction can be analytically monitored, e.g., by a change of molecular weight.In the macrocycle field, different cyclization options are investigated: N- to C-terminal linkages, terminal or side chain linkages, disulfide bridges and all possible combinations thereof.81 MP-DELs usually aim at rather simple structures and are thus mainly constructed using N- to C-terminal linkages. Following solid phase peptide synthesis, the traditional macrocyclization chemistry toolbox may comprise diverse sets of reactions ranging from harsh chemical setups to rather mild intein-mediated cyclization, or simply the oxidation of cysteine pairs. So far, only a small portion of these cyclization strategies has already been used for MP-DEL synthesis. In the following, we would like to shine a light on these strategies, which are also summarized in Fig. 5.
The most broadly employed strategy is cyclization by copper-catalyzed azide/alkyne cycloaddition (CuAAC). Four out of ten MP-DELs were designed using this chemistry: the DNA-templated library of Kodadek/Deluge Biotechnologies,36 the peptoid library of the Lim group,41 and the classic peptide libraries of GSK38 and of the Neri/Scheuermann group.44 CuAAC has been established as a DNA-compatible chemistry for diverse DEL setups, including non-macrocyclic DELs.39,82–84 To facilitate the access to CuAAC cyclization, Bayer recently proposed an optimized route to transform amines into azides, and surveyed the subsequent macrocyclization for a series of compounds.85 Unfortunately, an estimation of the conversion yield is analytically difficult to monitor because CuAAC does not induce a change in molecular weight. Hence, analytical understanding rather originates from investigating sets of individual representative compounds. In the case of the Neri/Scheuermann 2021 library, model compounds were synthesized and macrocyclization was detected by a shift in HPLC profiles.44 In accordance with the robustness of the well-established CuAAC reaction, the observed conversion rates typically exceeded 90%.
Further, Wittig olefination was implemented for MP-DELs by the Liu group in 2004.34,35,37 The situation here is somewhat special because the use of DNA-templated synthesis should facilitate the conversion of linear peptides into cyclic peptides.47 Nevertheless, for the final version of the library,37 each cyclizing building block was individually tested and only those resulting in at least 45% yield (typical yields were 80–90%) were selected for the library construction.
Amide bond formation is by far the most used reaction in the DEL field,82,86 to the point where it is sometimes criticized as too common to yield valuable chemical diversity. However, it had undoubtfully proven its value and robustness87,88 already before the Gilligham lab decided to use it for cyclizing a peptoid DEL.40 It is worth noting that peptoid libraries should be easier to cyclize than their peptidic equivalents, as the absence of amide bonds may allow for more rotational freedom, and preliminary studies showed that cyclization efficiency varied with the nature of the last included building block. In addition, the cyclization in pool was performed in parallel with a one-compound control, predicting the usefulness of a repetition step to achieve good overall conversion.
Recently, the Chen/Lu groups provided an interesting study on palladium-catalyzed intramolecular S-arylation as a means for cyclization.43 They first optimized the reaction in solution under mild conditions, translated it on DNA and finally constructed a MP-DEL. The reaction accepted a broad scope of reactants and exhibited greater efficiency for tetramers or larger macrocycles compared to trimers. Generally, conversion rates higher than 70% could be achieved. The formation of bicyclic structures, reminiscent of the work performed previously by the Heinis lab,21 was accomplished in solution but not yet on DNA.
The last cyclization reaction used for MP-DEL construction is thioether formation and was employed by Kodadek/Deluge Biotechnology to form the “PICCOs”-library.45 The OBOC methodology facilitated the cyclization: thanks to the solid-support, the cysteine protecting group (S-trimethoxyphenyl) could be removed in dimethylformamide, and the cyclization effectuated in aqueous solution. Moreover, cleavage of macrocycles from chosen beads allowed the analysis of individual compounds by mass spectrometry. After macrocyclization of the libraries, aliquots were stained with a thiol-reactive fluorescent dye (mBBr) to confirm that the majority of beads bore cyclized compounds.
Another study on thioether formation was published by Heinis and coworkers describing the formation of macrocycles on DNA in solution by thioether formation.89 Two tert-butylthio (S-tBu) protected cysteines were incorporated into peptides and subsequently deprotected in aqueous solution. Disulfide bond formation readily occurred, and the addition of symmetrical bis-electrophiles lead to the creation of thioether bonds via nucleophilic substitution or 1,4-addition. Conversion rates generally exceeded 70% for the described cyclization reactions, thus providing a promising basis for the creation of MP-DELs.
Ring closing metathesis was also envisioned for MP-DEL cyclization. Two preliminary studies described possible conditions for this reaction in a DNA-compatible fashion.90,91 Several ruthenium catalysts were assessed, and superiority was demonstrated for fast initiating Ru Grubbs catalysts.90 MgCl2 was employed to protect the oligonucleotides and the reactivity scope was extensively tested. A further study, published in 2019 by Simmons and coworkers, optimized the homogeneity of the reaction in a new aqueous system, implemented the use of an acidic buffer to mask problematic functional groups and developed an alternative and decomposition-resistant Ru Grubbs catalyst.91 These new conditions permitted the cyclization of an unprotected stapled peptide. However, reported conversion rates were generally around 50% indicating that further optimization might be necessary to render this challenging reaction useful for large MP-DEL synthesis.
This last example underlines the challenges of cyclization in MP-DEL construction. The value of a chosen cyclization reaction is ultimately determined by the respective screening results as DEL construction in mixtures prevents a decent quality control of the final library members. Therefore, preliminary tests on individual compounds are of great importance to assess the scope of reactivity of a given reaction, the ideal ring size and, more generally, the conversion rates that can be expected in pools.
Screening results with MP-DELs
Like in traditional DEL selections, the protein targets screened with MP-DELs originated from several pharmaceutical fields (Fig. 6) and oncology constitutes the majority of the screened proteins. Src,51 XIAP BIR2-3,56 Skp2,78 NKp4680 and p300,79 among others, are relevant tumor targets or targets of the tumor microenvironment. Similarly, inflammatory diseases were approached with selections against TNF72 and AGP.73 In addition, diabetes and viral infections were targeted through IDE52 and RSV N respectively.71 The chosen targets exemplify the capacity of MP-DELs to generate molecular binders to diverse proteins of interest.| Library | Protein | Activity/Affinity | Validation methodology | Hit | Ref. |
|---|---|---|---|---|---|
| Liu 2010 | Src | IC50 = 680 nM | Inhibition assay | (1) | 50 |
| VEGFR2 | Dose-dependent activation | Inhibition assay | 50 | ||
| Liu 2018 | IDE | IC50 = 40 nM | Inhibition assay | (2) | 37 |
| Terret 2015 | XIAP BIR2-3 | IC50 = 24 nM | Functional caspase rescue assay | (3) | 36 |
| cIAP BIR2-3 | IC50 = 3 nM | Functional caspase rescue assay | (3) | 36 | |
| Pal 2018 | RSV N | 92.3% bound | AS-MS assay | (4) | 38 |
| pIC50 = 6.98 | TR-FRET assay | ||||
| Neri 2018 | TNF | K D = 6.1 μM | Fluorescence polarization | (5) | 39 |
| CaM | K D = 0.16 μM | Fluorescence polarization | 39 | ||
| PSA | K D = 13 μM | Fluorescence polarization | 39 | ||
| Gillingham 2019 | AGP | K D = 4 μM | ITC | (6) | 40 |
| Lim 2019 | Skp2 | K D = 7.51 μM | Fluorescence anisotropy | (7) | 41 |
| Kodadek 2020 | Streptavidin | K D ∼ 30 nM | FACS-based validation | (8) | 45 |
| Neri 2021 | NKp46 | K D = 7.4 μM | ELISA | (9) | 44 |
| HSA | K D = 8.0 μM | ELISA | 44 | ||
| Chen 2021 | p300 | IC50 = 3.0 μM | Inhibition assay | (10) | 43 |
Chosen examples of discovered MP ligands are represented in Fig. 6 and picture the diversity of the MP-DELs. As originating from different laboratories, the ligands were characterized in different ways. Some were tested for affinity, reflecting directly the selection experiment, while other ligands were tested for activity. Therefore, any direct comparison between MP-DELs remains difficult. Nevertheless, all libraries reported hits exhibiting activities or affinities in the low micromolar range down to the low nanomolar range. Since peptidic structures are easily amenable to medicinal chemistry efforts, the obtained hits may constitute promising starting points to generate potent leads. Also the dimerization of a selected macrocycle permitted an immediate gain of activity.36 Alternatively, the choice of peptoidic structures allowed the Lim group to obtain hits which could penetrate HeLa cells.41
Most of the MP-DEL publications concentrate on library construction and screening, yet only a few detail examples of practical applications, such as fluorescence microscopy procedures, in vivo delivery of payloads or the development of target-specific probes.39 MP-DEL is a relatively new field in drug discovery so data can be found about affinity or activity of ligands in vitro51 and sometimes in vivo.36 While the affinity or activity of the discovered ligands usually are extensively characterized, other parameters, such as susceptibility to peptidase-mediated degradation or membrane permeability, are not generally reported.
Further progress in the field should bring a more holistic characterization of the discovered macrocycles, in order to better predict their potential for advancing to clinical stages.
Outlook
While MP-DELs share fundamental characteristics in terms of synthetic production, they embody a wide range of structurally diverse molecules. DELs based on “classical” macrocyclic peptides were described, but also beta-sheet mimicking peptides or macrocyclic peptoids. Furthermore, the macrocycle ring size is subjected to variation, depending on the number of chemical building blocks included, and on the nature of the building blocks employed: natural or unnatural amino acids, dipeptides or oligopeptides altogether creating an ultra-large repertoire.Even though the diversity of the present MP-DELs is already considerable, we expect to see it further grow as more sophisticated libraries will come up. To that aim, the combination of existing methodologies gives formidable freedom for new experimental designs. For example, one can choose between DNA-templated or DNA-recorded synthesis, the generation of single-stranded or double-stranded DELs, or a synthetic strategy in solution or on beads. Depending on the choices, one can adopt diverse screening methodologies such as library in solution,28 photo-crosslinker-65 or reversible covalent crosslinker-assisted,64 library on beads,30 micelle-based,92 on-cells,93 or even in-cells selections.66,94 Also, new combinations of natural display technologies with DEL procedures may lead to promising results.95,96
The reported sizes of MP-DELs followed the general trend in DEL, i.e., they were increasing with the DNA sequencing power.
However, theoretical library size alone is not a good indicator for library performance: DNA-templated MP-DELs counted between 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 256
000 and 256![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds and despite what is nowadays considered a “small” size, proved very successful. Later, DNA-recorded MP-DELs ranged between hundreds of thousands and tens of million compounds, and also 1012 compounds were reported.38
000 compounds and despite what is nowadays considered a “small” size, proved very successful. Later, DNA-recorded MP-DELs ranged between hundreds of thousands and tens of million compounds, and also 1012 compounds were reported.38
An important descriptor of DEL performance is the final purity of a library. Purity is affected by the total number of synthetic steps and their corresponding yields, as well as by the efficiency of encoding. Library purity depends on the method chosen for library synthesis and constitutes a central problem for DEL construction, since large compound mixtures do not allow for differential purification nor for individual quality control. The eventual quality of a MP-DEL will be determined by optimizing all chemical steps, specifically the cyclization step, and by the robustness of the encoding/decoding strategy.
MP-DELs are growing at a quick pace but they are still hampered by some limitations. It has been described that analyzing selections with very large libraries can be very challenging,97 especially with DELs of heterogeneous quality. This is in contrast with the evident wish to create larger and larger libraries from bigger and bigger collections of building blocks.
Another potential issue is the follow-up of hits generated from selection experiments. Depending on the outcome of a DEL selection, resynthesizing multiple hits off-DNA can be challenging. Following hit validation, medicinal chemistry efforts will claim resources in time, chemicals and proteins to yield leads and it would be desirable to develop tools for ranking the best hits before synthesis. For this purpose, some preliminary studies have been reported that take into account Whitty or Khilberg13,16,55 rules to maximize the probability of yielding drug-like macrocycles.
MP-DELs have proven their potential to deliver promising hits for even difficult proteins of interest, including some examples of protein–protein interactions. The field grew in popularity over the recent years, and we expect this trend to be confirmed. The increase in commercial availability of building blocks, the constant ameliorations in the DEL field, such as expansion of DNA-compatible chemistries, encoding strategies, purification techniques and deconvolution strategies together create a fertile ground for the design of ever more ambitious MP-DELs and hence the generation of beneficial compounds for basic research and medicinal applications.
Conflicts of interest
There are no conflicts to declare.References
- E. M. Driggers, S. P. Hale, J. Lee and N. K. Terrett, Nat. Rev. Drug Discovery, 2008, 7, 608–624 CrossRef CAS.
- A. A. Vinogradov, Y. Yin and H. Suga, J. Am. Chem. Soc., 2019, 141, 4167–4181 CrossRef CAS.
- N. K. Terrett, Drug Discovery Today: Technol., 2010, 7, e97–e104 CrossRef CAS.
- S. Cazzamalli, A. Dal Corso, F. Widmayer and D. Neri, J. Am. Chem. Soc., 2018, 140, 1617–1621 CrossRef CAS PubMed.
- A. Zorzi, K. Deyle and C. Heinis, Curr. Opin. Chem. Biol., 2017, 38, 24–29 CrossRef CAS PubMed.
- J. Laxio Arenas, J. Kaffy and S. Ongeri, Curr. Opin. Chem. Biol., 2019, 52, 157–167 CrossRef CAS PubMed.
- K. Fosgerau and T. Hoffmann, Drug Discovery Today, 2015, 20, 122–128 CrossRef CAS.
- V. Le Joncour and P. Laakkonen, Bioorg. Med. Chem., 2018, 26, 2797–2806 CrossRef CAS PubMed.
- P. Scodeller and E. K. Asciutto, Molecules, 2020, 25, 1–24 CrossRef.
- B. C. Doak, B. Over, F. Giordanetto and J. Kihlberg, Chem. Biol., 2014, 21, 1115–1142 CrossRef CAS PubMed.
- C. A. Lipinski, Adv. Drug Delivery Rev., 2016, 101, 34–41 CrossRef CAS.
- W. Yang, P. Gadgil, V. R. Krishnamurthy, M. Landis, P. Mallick, D. Patel, P. J. Patel, D. L. Reid and M. Sanchez-Felix, AAPS J., 2020, 22, 21 CrossRef.
- M. Tyagi, F. Begnini, V. Poongavanam, B. C. Doak and J. Kihlberg, Chem. – Eur. J., 2020, 26, 49–88 CrossRef CAS.
- P. Matsson, B. C. Doak, B. Over and J. Kihlberg, Adv. Drug Delivery Rev., 2016, 101, 42–61 CrossRef CAS.
- X. D. Kong, J. Moriya, V. Carle, F. Pojer, L. A. Abriata, K. Deyle and C. Heinis, Nat. Biomed. Eng., 2020, 4, 560–571 CrossRef CAS PubMed.
- F. Begnini, V. Poongavanam, B. Over, M. Castaldo, S. Geschwindner, P. Johansson, M. Tyagi, C. Tyrchan, L. Wissler, P. Sjö, S. Schiesser and J. Kihlberg, J. Med. Chem., 2021, 64(2), 1054–1072 CrossRef CAS.
- S. GP, Science, 1985, 228, 1315–1317 CrossRef PubMed.
- J. K. Scott and G. P. Smith, Science, 1990, 249, 386–390 CrossRef CAS.
- J. McCafferty, A. Griffiths, G. Winter and C. DJ, Nature, 1990, 348, 552 CrossRef CAS.
- C. Heinis, T. Rutherford, S. Freund and G. Winter, Nat. Chem. Biol., 2009, 5, 502–507 CrossRef CAS.
- S. S. Kale, C. Villequey, X. D. Kong, A. Zorzi, K. Deyle and C. Heinis, Nat. Chem., 2018, 10, 715–723 CrossRef CAS PubMed.
- J. Y. K. Wong, R. Mukherjee, J. Miao, O. Bilyk, V. Triana, M. Miskolzie, A. Henninot, J. J. Dwyer, S. Kharchenko, A. Iampolska, D. M. Volochnyuk, Y. S. Lin, L. M. Postovit and R. Derda, Chem. Sci., 2021, 12, 9694–9703 RSC.
- D. R. Cary, M. Ohuchi, P. C. Reid and K. Masuya, J. Synth. Org. Chem., Jpn., 2017, 75, 1171–1178 CrossRef CAS.
- C. Sohrabi, A. Foster and A. Tavassoli, Nat. Rev. Chem., 2020, 4, 90–101 CrossRef CAS.
- G. Goetz, Cyclic Peptide Design, 2019, vol. 2001 Search PubMed.
- V. Kunig, M. Potowski, A. Gohla and A. Brunschweiger, Biol. Chem., 2018, 399, 691–710 CrossRef CAS.
- D. Neri and R. A. Lerner, Annu. Rev. Biochem., 2018, 87, 479–502 CrossRef CAS PubMed.
- W. Decurtins, M. Wichert, R. M. Franzini, F. Buller, M. A. Stravs, Y. Zhang, D. Neri and J. Scheuermann, Nat. Protoc., 2016, 11, 764–780 CrossRef CAS.
- P. Dickson and T. Kodadek, Org. Biomol. Chem., 2019, 17, 4676–4688 RSC.
- A. B. MacConnell, P. J. McEnaney, V. J. Cavett and B. M. Paegel, ACS Comb. Sci., 2015, 17, 518–534 CrossRef CAS PubMed.
- N. G. Paciaroni, J. M. Ndungu and T. Kodadek, Chem. Commun., 2020, 0–3 Search PubMed.
- A. L. Satz, ACS Med. Chem. Lett., 2018, 9(5), 408–410 CrossRef CAS PubMed.
- S. Brenner and R. A. Lerner, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5381–5383 CrossRef CAS.
- Z. J. Gartner, B. N. Tse, R. Grubina, J. B. Doyon, T. M. Snyder and D. R. Liu, Science, 2004, 305, 1601–1605 CrossRef CAS.
- B. N. Tse, T. M. Snyder, Y. Shen and D. R. Liu, J. Am. Chem. Soc., 2008, 130, 15611–15626 CrossRef CAS.
- B. A. Seigal, W. H. Connors, A. Fraley, R. M. Borzilleri, P. H. Carter, S. L. Emanuel, J. Fargnoli, K. Kim, M. Lei, J. G. Naglich, M. E. Pokross, S. L. Posy, H. Shen, N. Surti, R. Talbott, Y. Zhang and N. K. Terrett, J. Med. Chem., 2015, 58, 2855–2861 CrossRef CAS.
- D. L. Usanov, A. I. Chan, J. P. Maianti and D. R. Liu, Nat. Chem., 2018, 10, 704–714 CrossRef CAS PubMed.
- Z. Zhu, A. Shaginian, L. C. Grady, T. O’Keeffe, X. E. Shi, C. P. Davie, G. L. Simpson, J. A. Messer, G. Evindar, R. N. Bream, P. P. Thansandote, N. R. Prentice, A. M. Mason and S. Pal, ACS Chem. Biol., 2018, 13, 53–59 CrossRef CAS PubMed.
- Y. Li, R. de Luca, S. Cazzamalli, F. Pretto, D. Bajic, J. Scheuermann and D. Neri, Nat. Chem., 2018, 10, 1–8 CrossRef.
- C. J. Stress, B. Sauter, L. A. Schneider, T. Sharpe and D. Gillingham, Angew. Chem., Int. Ed., 2019, 58, 9570–9574 CrossRef CAS.
- M. H. Shin, K. J. Lee and H. S. Lim, Bioconjugate Chem., 2019, 30, 2931–2938 CrossRef CAS.
- A. Roy, E. Koesema and T. Kodadek, Angew. Chem., Int. Ed., 2021, 60, 11983–11990 CrossRef CAS PubMed.
- P. Yang, X. Wang, B. Li, Y. Yang, J. Yue, Y. Suo, H. Tong, G. He, X. Lu and G. Chen, Chem. Sci., 2021, 12, 5804–5810 RSC.
- Y. Onda, G. Bassi, A. Elsayed, F. Ulrich, S. Oehler, L. Plais, J. Scheuermann and D. Neri, Chem. – Eur. J., 2021, 27, 7160–7167 CrossRef CAS.
- E. Koesema, A. Roy, N. G. Paciaroni and T. Kodadek, ChemRxiv DOI:10.26434/chemrxiv.13490151.v1.
- Z. J. Gartner and D. R. Liu, J. Am. Chem. Soc., 2001, 123, 6961–6963 CrossRef CAS.
- X. Li and D. R. Liu, Angew. Chem., Int. Ed., 2004, 43, 4848–4870 CrossRef CAS PubMed.
- S. Melkko, J. Scheuermann, C. E. Dumelin and D. Neri, Nat. Biotechnol., 2004, 22, 568–574 CrossRef CAS PubMed.
- D. R. Halpin and P. B. Harbury, PLoS Biol., 2004, 2(7), e174 CrossRef.
- R. E. Kleiner, C. E. Dumelin, G. C. Tiu, K. Sakurai and D. R. Liu, J. Am. Chem. Soc., 2010, 132, 11779–11791 CrossRef CAS.
- G. Georghiou, R. E. Kleiner, M. Pulkoski-Gross, D. R. Liu and M. A. Seeliger, Nat. Chem. Biol., 2012, 8, 366–374 CrossRef CAS PubMed.
- J. P. Maianti, A. McFedries, Z. H. Foda, R. E. Kleiner, X. Q. Du, M. A. Leissring, W. J. Tang, M. J. Charron, M. A. Seeliger, A. Saghatelian and D. R. Liu, Nature, 2014, 511, 94–98 CrossRef CAS.
- J. P. Maianti, G. A. Tan, A. Vetere, A. J. Welsh, B. K. Wagner, M. A. Seeliger and D. R. Liu, Nat. Chem. Biol., 2019, 15, 565–574 CrossRef CAS.
- D. L. Usanov, A. I. Chan, J. P. Maianti and D. R. Liu, Nat. Chem., 2018, 10, 704–714 CrossRef CAS PubMed.
- B. C. Doak, J. Zheng, D. Dobritzsch and J. Kihlberg, J. Med. Chem., 2016, 59, 2312–2327 CrossRef CAS.
- J. C. Wilkinson, E. Cepero, L. H. Boise and C. S. Duckett, Mol. Cell. Biol., 2004, 24, 7003–7014 CrossRef CAS PubMed.
- Y. Li, P. Zhao, M. Zhang, X. Zhao and X. Li, J. Am. Chem. Soc., 2013, 135, 17727–17730 CrossRef CAS.
- G. Zhao, Y. Huang, Y. Zhou, Y. Li and X. Li, Expert Opin. Drug Discovery, 2019, 14, 1–19 CrossRef PubMed.
- Á. Furka, Drug Discovery Today, 2002, 7, 1–4 CrossRef.
- G. Bassi, N. Favalli, S. Oehler, A. Martinelli, M. Catalano, J. Scheuermann and D. Neri, Biochem. Biophys. Res. Commun., 2020, 533, 223–229 CrossRef CAS.
- M. A. Clark, R. A. Acharya, C. C. Arico-Muendel, S. L. Belyanskaya, D. R. Benjamin, N. R. Carlson, P. A. Centrella, C. H. Chiu, S. P. Creaser, J. W. Cuozzo, C. P. Davie, Y. Ding, G. J. Franklin, K. D. Franzen, M. L. Gefter, S. P. Hale, N. J. V. Hansen, D. I. Israel, J. Jiang, M. J. Kavarana, M. S. Kelley, C. S. Kollmann, F. Li, K. Lind, S. Mataruse, P. F. Medeiros, J. A. Messer, P. Myers, H. O’Keefe, M. C. Oliff, C. E. Rise, A. L. Satz, S. R. Skinner, J. L. Svendsen, L. Tang, K. Van Vloten, R. W. Wagner, G. Yao, B. Zhao and B. A. Morgan, Nat. Chem. Biol., 2009, 5, 647–654 CrossRef CAS PubMed.
- P. A. Harris, B. W. King, D. Bandyopadhyay, S. B. Berger, N. Campobasso, C. A. Capriotti, J. A. Cox, L. Dare, X. Dong, J. N. Finger, L. S. C. Grady, S. J. Hoffman, J. U. Jeong, J. Kang, V. Kasparcova, A. S. Lakdawala, R. Lehr, D. E. McNulty, R. Nagilla, M. T. Ouellette, C. S. Pao, A. R. Rendina, M. C. Schaeffer, J. D. Summerfield, B. A. Swift, R. D. Totoritis, P. Ward, A. Zhang, D. Zhang, R. W. Marquis, J. Bertin and P. J. Gough, J. Med. Chem., 2016, 59, 2163–2178 CrossRef CAS PubMed.
- S. L. Belyanskaya, Y. Ding, J. F. Callahan, A. L. Lazaar and D. I. Israel, ChemBioChem, 2017, 18, 837–842 CrossRef CAS PubMed.
- A. Dal Corso, M. Catalano, A. Schmid, J. Scheuermann and D. Neri, Angew. Chem., Int. Ed., 2018, 57, 17178–17182 CrossRef CAS PubMed.
- A. Sannino, A. Gironda-martínez, E. M. D. Gorre, L. Prati, J. Scheuermann, D. Neri, E. J. Donckele and F. Samain, ACS Comb. Sci., 2020, 22(4), 204–212 CrossRef CAS PubMed.
- B. Cai, D. Kim, S. Akhand, Y. Sun, R. J. Cassell, A. Alpsoy, E. C. Dykhuizen, R. M. Van Rijn, M. K. Wendt and C. J. Krusemark, J. Am. Chem. Soc., 2019, 141, 17057–17061 CrossRef CAS.
- G. Bassi, N. Favalli, M. Vuk, M. Catalano, A. Martinelli, A. Trenner, A. Porro, S. Yang, C. L. Tham, M. Moroglu, W. W. Yue, S. J. Conway, P. K. Vogt, A. A. Sartori, J. Scheuermann and D. Neri, Adv. Sci., 2020, 7, 1–10 Search PubMed.
- M. Wichert, N. Krall, W. Decurtins, R. M. Franzini, F. Pretto, P. Schneider, D. Neri and J. Scheuermann, Nat. Chem., 2015, 7, 241–249 CrossRef CAS PubMed.
- B. Shi, Y. Zhou, Y. Huang, J. Zhang and X. Li, Bioorg. Med. Chem. Lett., 2017, 27, 361–369 CrossRef CAS PubMed.
- W. H. Connors, S. P. Hale and N. K. Terrett, Curr. Opin. Chem. Biol., 2015, 26, 42–47 CrossRef CAS.
- A. P. Oliveira, F. M. Simabuco, R. E. Tamura, M. C. Guerrero, P. G. G. Ribeiro, T. A. Libermann, L. F. Zerbini and A. M. Ventura, Virus Res., 2013, 177, 108–112 CrossRef CAS.
- F. Buller, Y. Zhang, J. Scheuermann, J. Schäfer, P. Bühlmann and D. Neri, Chem. Biol., 2009, 16, 1075–1086 CrossRef CAS PubMed.
- T. Fournier, N. Medjoubi-N and D. Porquet, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 2000, 1482, 157–171 CrossRef CAS.
- R. N. Zuckermann, Biopolymers, 2011, 96, 545–555 CrossRef CAS.
- A. M. Webster and S. L. Cobb, Chem. – Eur. J., 2018, 24, 7560–7573 CrossRef CAS.
- M. Lebl, N. F. Sepetov, S. Felder and K. S. Lam, Biopolymers, 1995, 37, 177–198 CrossRef CAS PubMed.
- K. R. Mendes, M. L. Malone, J. M. Ndungu, I. Suponitsky-Kroyter, V. J. Cavett, P. J. McEnaney, A. B. MacConnell, T. D. M. Doran, K. Ronacher, K. Stanley, O. Utset, G. Walzl, B. M. Paegel and T. Kodadek, ACS Chem. Biol., 2017, 12, 234–243 CrossRef CAS.
- J. R. Skaar, J. K. Pagan and M. Pagano, Nat. Rev. Drug Discovery, 2014, 13, 889–903 CrossRef CAS.
- N. G. Iyer, H. Özdag and C. Caldas, Oncogene, 2004, 23, 4225–4231 CrossRef CAS PubMed.
- L. Moretta and A. Moretta, Curr. Opin. Immunol., 2004, 16, 626–633 CrossRef CAS PubMed.
- R. Y. Zhang, P. Thapa, M. J. Espiritu, V. Menon and J. P. Bingham, Bioorg. Med. Chem., 2018, 26, 1135–1150 CrossRef CAS.
- A. L. Satz, J. Cai, Y. Chen, R. Goodnow, F. Gruber, A. Kowalczyk, A. Petersen, G. Naderi-Oboodi, L. Orzechowski and Q. Strebel, Bioconjugate Chem., 2015, 26(8), 1623–1632 CrossRef CAS.
- A. Litovchick, C. E. Dumelin, S. Habeshian, D. Gikunju, M. A. Guié, P. Centrella, Y. Zhang, E. A. Sigel, J. W. Cuozzo, A. D. Keefe and M. A. Clark, Sci. Rep., 2015, 5, 1–8 Search PubMed.
- N. Favalli, G. Bassi, T. Zanetti, J. Scheuermann and D. Neri, Helv. Chim. Acta, 2019, 102, e1900033 CrossRef.
- S. Ede, M. Schenk, D. Bierer, H. Weinmann and K. Graham, Molecules, 2021, 26, 1790 CrossRef CAS PubMed.
- M. L. Malone and B. M. Paegel, ACS Comb. Sci., 2016, 18, 182–187 CrossRef CAS PubMed.
- A. Tomberg and J. Boström, Drug Discovery Today, 2020, 00, 1–8 Search PubMed.
- Y. Li, E. Gabriele, F. Samain, N. Favalli, F. Sladojevich, J. Scheuermann and D. Neri, ACS Comb. Sci., 2016, 18, 438–443 CrossRef CAS PubMed.
- M. V. Pham, M. Bergeron-Brlek and C. Heinis, ChemBioChem, 2020, 21, 543–549 CrossRef CAS PubMed.
- X. Lu, L. Fan, C. B. Phelps, C. P. Davie and C. P. Donahue, Bioconjugate Chem., 2017, 28, 1625–1629 CrossRef CAS PubMed.
- O. B. C. Monty, P. Nyshadham, K. M. Bohren, M. Palaniappan, M. M. Matzuk, D. W. Young and N. Simmons, ACS Comb. Sci., 2020, 22(2), 80–88 CrossRef CAS PubMed.
- A. L. Hackler, F. G. FitzGerald, V. Q. Dang, A. L. Satz and B. M. Paegel, ACS Comb. Sci., 2020, 22, 25–34 CrossRef CAS PubMed.
- Y. Huang, L. Meng, Q. Nie, Y. Zhou, L. Chen, S. Yang, Y. M. E. Fung, X. Li, C. Huang, Y. Cao, Y. Li and X. Li, Nat. Chem., 2021, 13, 77–88 CrossRef CAS PubMed.
- L. K. Petersen, A. B. Christensen, J. Andersen, C. G. Folkesson, O. Kristensen, C. Andersen, A. Alzu, F. A. Sløk, P. Blakskjær, D. Madsen, C. Azevedo, I. Micco and N. J. V. Hansen, J. Am. Chem. Soc., 2021, 143(7), 2751–2756 CrossRef CAS.
- S. Palei, K. S. Becher, C. Nienberg, J. Jose and H. D. Mootz, ChemBioChem, 2019, 20, 72–77 CrossRef CAS PubMed.
- A. I. Ekanayake, L. Sobze, P. Kelich, J. Youk, N. J. Bennett, R. Mukherjee, A. Bhardwaj, F. Wuest, L. Vukovic and R. Derda, J. Am. Chem. Soc., 2021, 143, 5497–5507 CrossRef CAS PubMed.
- A. L. Satz, R. Hochstrasser and A. C. Petersen, ACS Comb. Sci., 2017, 19, 234–238 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |