Open Access Article
Open Access ArticleInhibition of SC4MOL and HSD17B7 shifts cellular sterol composition and promotes oligodendrocyte formation†
Matthew J
Pleshinger
a,
Ryan M.
Friedrich
 b,
Zita
Hubler
b,
Zita
Hubler
 b,
Adrianna M.
Rivera-León
b,
Adrianna M.
Rivera-León
 b,
Farrah
Gao
b,
David
Yan
b,
Joel L.
Sax
b,
Ramya
Srinivasan
b,
Ilya
Bederman
b,
H. Elizabeth
Shick
b,
Paul J.
Tesar
b and
Drew J.
Adams
b,
Farrah
Gao
b,
David
Yan
b,
Joel L.
Sax
b,
Ramya
Srinivasan
b,
Ilya
Bederman
b,
H. Elizabeth
Shick
b,
Paul J.
Tesar
b and
Drew J.
Adams
 *b
*b
aDepartment of Pharmacology, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106, USA
bDepartment of Genetics and Genome Sciences, Case Western Reserve University School of Medicine, Cleveland, Ohio 44106, USA. E-mail: drew.adams@case.edu
First published on 21st October 2021
Abstract
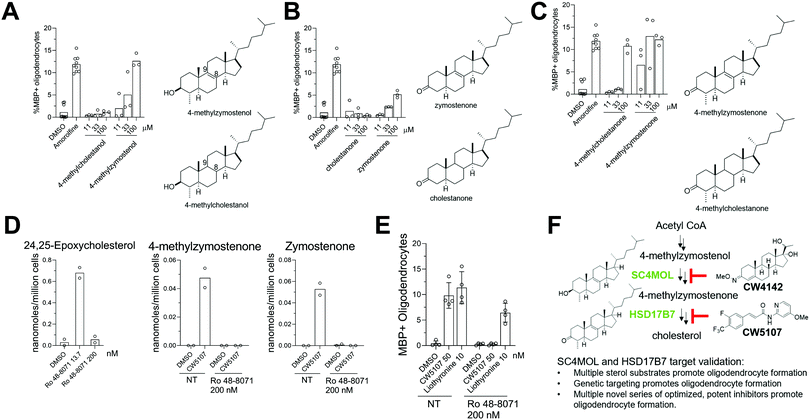
While the cholesterol biosynthesis pathway has been extensively studied, recent work has forged new links between inhibition of specific sterol pathway enzymes, accumulation of their unique sterol substrates, and biological areas as diverse as cancer, immunology, and neurodegenerative disease. We recently reported that dozens of small molecules enhance formation of oligodendrocytes, a glial cell type lost in multiple sclerosis, by inhibiting CYP51, Sterol 14-reductase, or EBP and inducing cellular accumulation of their 8,9-unsaturated sterol substrates. Several adjacent pathway enzymes also have 8,9-unsaturated sterol substrates but have not yet been evaluated as potential targets for oligodendrocyte formation or in many other biological contexts, in part due to a lack of available small-molecule probes. Here, we show that genetic suppression of SC4MOL or HSD17B7 increases the formation of oligodendrocytes. Additionally, we have identified and optimized multiple potent new series of SC4MOL and HSD17B7 inhibitors and shown that these small molecules enhance oligodendrocyte formation. SC4MOL inhibitor CW4142 induced accumulation of SC4MOL's sterol substrates in mouse brain and represents an in vivo probe of SC4MOL activity. Mechanistically, the cellular accumulation of these 8,9-unsaturated sterols represents a central driver of enhanced oligodendrocyte formation, as exogenous addition of purified SC4MOL and HSD17B7 substrates but not their 8,9-saturated analogs promotes OPC differentiation. Our work validates SC4MOL and HSD17B7 as novel targets for promoting oligodendrocyte formation, underlines a broad role for 8,9-unsaturated sterols as enhancers of oligodendrocyte formation, and establishes the first high-quality small molecules targeting SC4MOL and HSD17B7 as novel tools for probing diverse areas of biology.
Introduction
While the cholesterol biosynthesis pathway has been well characterized, recent research has indicated that modulation of pathway function can impact a wide range of disease-relevant biological processes, including remyelination. In the central nervous system, oligodendrocytes form protective, insulative myelin sheaths around axons of neurons.1,2 Multiple sclerosis (MS) and other demyelinating disorders result from loss of oligodendrocytes and their myelin sheaths, which leads to reduced action potential velocity, neuronal dysfunction, and ultimately axonal degradation.3,4 One potential therapeutic strategy for these disorders is to increase remyelination, or the formation of new oligodendrocytes and new myelin. The adult mammalian brain retains a population of oligodendrocyte precursor cells (OPCs), stem cells that can differentiate into new oligodendrocytes.5,6 While this process of adult remyelination is well-established, it is frequently insufficient to prevent disease progression.7 Future therapeutics that increase the formation of new myelinating oligodendrocytes could slow progression in MS and other demyelinating diseases.Multiple labs have reported high-throughput screens that identified novel small molecules that enhance the differentiation of cultured OPCs to oligodendrocytes.8–15 Dozens of structurally diverse hits have been validated in these screens, with many molecules shown to enhance myelination in both in vitro and in vivo models. One such drug, clemastine, significantly increased axonal conduction velocity in the optic nerve, a myelination-dependent phenotype, in MS patients.16 Although these small molecules span many diverse canonical cellular targets, we established that many enhancers of oligodendrocyte formation function in OPCs by ‘off-target’ inhibition of one of four enzymes within the cholesterol biosynthesis pathway: CYP51, Sterol 14-reductase, EBP, or LSS.13,14,17 Inhibition of these enzymes induces a sterol shift that reduces cholesterol levels and elevates atypical sterols, with LSS inducing 24,25-epoxycholesterol accumulation and the other enzymes inducing 8,9-unsaturated sterol accumulation. Addition of these purified sterols to OPCs promotes their differentiation to mature oligodendrocytes, supporting that these molecules are a central driver of enhanced oligodendrocyte formation.
Within the cholesterol biosynthesis pathway, three adjacent enzymes that have 8,9-unsaturated sterol substrates—SC4MOL, NSDHL, and HSD17B7—are underexplored and have yet to be evaluated as potential targets for enhancing oligodendrocyte formation. These enzymes comprise the C4-demethylation complex (C4DMC) and function sequentially to remove the two C4-methyl groups in T-MAS (Fig. S1, ESI†).18 To remove each methyl group, SC4MOL achieves C–H activation and further oxidations to provide a C4 carboxylic acid; NSDHL catalyzes oxidative decarboxylation to yield a C3 keto-sterol product; and HSD17B7 stereospecifically reduces the C3 ketone to provide the C4-demethylated sterol product. Two independent cycles of this sequence requiring all three enzymes are ultimately required to convert T-MAS to zymosterol. SC4MOL, NSDHL, and HSD17B7 are positioned in the cholesterol pathway between the validated targets Sterol 14-reductase and EBP, suggesting that inhibition of these three C4DMC enzymes might also influence oligodendrocyte formation (Fig. 1a). However, no small molecules identified in previous screens inhibited any of these three enzymes, and few if any high-quality small-molecule probes are available to target these enzymes.
Here, we demonstrate that exogenous addition of sterol substrates of SC4MOL and HSD17B7, but not NSDHL, increases oligodendrocyte formation, greatly expanding the structural diversity of sterols that promote this cell fate transition. Additionally, genetic targeting SC4MOL and HSD17B7 promotes oligodendrocyte formation. Further, we identified novel small molecule inhibitors of SC4MOL and HSD17B7 that also enhanced oligodendrocyte formation. Optimization of these starting points led to the first high-quality chemical probes of these enzymes, with CW4142 able to inhibit SC4MOL in the rodent brain and CW5107 inhibiting HSD17B7 with high potency (EC50 50 nM). These studies have revealed two new targets for enhancing oligodendrocyte formation, new series of optimized small-molecule inhibitors for both enzymes, and a broader mechanistic framework for how shifts in sterol accumulation in OPCs can promote remyelination.
Experimental
Small molecules and reagents
The identity and purity of key small molecules were authenticated by LC-MS. The following compounds were purchased from Cayman Chemicals: PF-1163A, APB, SB366791, Ro 48-8071. Fenpyrazamine was purchased through LGC Standards. The following sterols were purchased through Avanti Polar Lipids: T-MAS, zymosterone, MAS-414, lanosterol, and zymostenol. Cholestanone, cholestanol, 17-OHP, and (2-hydroxypropyl)-β-cyclodextrin were purchased through Sigma-Aldrich Company. 3-Hydroxy-N-(3-methyloxyphenyl)-2-naphthamide and N-(3-Methoxyphenyl) cinnamide were purchased through Synthonix US. (2E)-N,3-diphenylprop-2-enamide, (2E)-N-(3-methylphenyl)-3-phenyl-2-propenamide, N-(2,4-dimethyoxyphenyl)-3-pheynylacrylamide, N-mesityl-3-phenylacrylamide, and beta-methyl-N-phenylcinnamamide were purchased through Sigma-Aldrich Company. Z26548267, Z27782563, and Z27782075 were purchased through Enamine Store. Paraformaldehyde was purchased through Ted Pella Inc (cat 18505).Mouse OPC culture
EpiSC-derived OPCs were previously obtained using in vitro differentiation protocols and culture conditions and were a gift from Paul Tesar. Three independent derivations of OPCs generated from three separate EpiSC lines were used: OPC-5, OPC-1, and WT2.1. Data were generated using WT2.1 unless otherwise noted. OPCs were grown and expanded in flasks first coated with poly-ornithine (PO) (Sigma P3655; 100 μg ml−1) for 1 hour at 37 °C and dried then laminin-coated (Sigma L2020; 15 μg ml−1) for 1 hour at 37 °C. OPCs were grown in growth medium (DMEM/F12 (ThermoFisher, 11320-033) supplemented with N2-MAX (R&D Systems), B-27 (ThermoFisher), GlutaMax (Gibco), FGF (10 μg ml−1, R&D systems, 233-FB-025) and PDGF-AA (10 μg ml−1, R&D Systems, 233-AA-050) before harvesting and plating. Media was changed every 2 days. Cells were plated in differentiation-permissive media for experiments (DMEM/F12 supplemented with N2-MAX (R&D Systems), B-27 (ThermoFisher), GlutaMax (Gibco).OPCs in vitro phenotypic screening
OPCs were seeded in 96-well CellCarrierUltra plates (PerkinElmer) coated with poly-D-lysine (Sigma P7886; 50 μg ml−1) for 1 hour at 37 °C and washed once with 200 μl of Cell Culture grade Water then laminin for 1 hour at 37 °C (Sigma L2020; 15 μg ml−1) using multi-channel pipet. For the experiments, 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were seeded per well. Cells were incubated under standard conditions (37 °C, 5% CO2) for 3 days and fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 20 min. Fixed plates were washed with PBS (200 μl per well) twice, permeabilized with 0.1% Triton X-100 and blocked with 10% donkey serum (v/v) in PBS for 60 minutes. Then, cells were labelled with an anti-MBP antibody (Abcam, ab7349, 1
000 cells were seeded per well. Cells were incubated under standard conditions (37 °C, 5% CO2) for 3 days and fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 20 min. Fixed plates were washed with PBS (200 μl per well) twice, permeabilized with 0.1% Triton X-100 and blocked with 10% donkey serum (v/v) in PBS for 60 minutes. Then, cells were labelled with an anti-MBP antibody (Abcam, ab7349, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) overnight at 4 °C. After three PBS washes (200 μl), an Alexa Fluor-conjugated anti-rat secondary antibody (ThermoFisher, A-21208, 1
200) overnight at 4 °C. After three PBS washes (200 μl), an Alexa Fluor-conjugated anti-rat secondary antibody (ThermoFisher, A-21208, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) and DAPI (Sigma; 1 μg ml−1) were added for 45 minutes. Plates were then washed an additional four times and left in 200 μl PBS. During washing steps, PBS was added using a multi-channel pipet and aspiration was performed using Biotek EL406 washer dispenser (Biotek) equipped with a 96-well aspiration manifold.
500) and DAPI (Sigma; 1 μg ml−1) were added for 45 minutes. Plates were then washed an additional four times and left in 200 μl PBS. During washing steps, PBS was added using a multi-channel pipet and aspiration was performed using Biotek EL406 washer dispenser (Biotek) equipped with a 96-well aspiration manifold.
High-content imaging and analysis
The Operetta High Content Imaging and Analysis system (PerkinElmer) was used for plate imaging.13,14,17 Each well had confocal imaging of 6 distinct fields at 20× lens. Cells were analyzed by quantifying nuclei stained with DAPI. Live cells were analyzed by excluding nuclei whose area ranged outside of 55–250 μm2 as these are pyknotic and represent dead cells. Typically, around 1500 live cells were imaged per well. Regions traced to nuclei belonging to live cells were then expanded 3 μm beyond the live nucleus. In this perinuclear region, the intensity of the MBP stain was measured to identify mature oligodendrocytes. Positive and negative control wells on each plate were used to establish thresholds for MBP intensity that accurately identified mature oligodendrocytes.GC-MS-based sterol profiling
Cells were plated in poly-D-lysine and laminin-coated 6-well Costar plates (SigmaAldrich) at 1 million cells per ml with differentiation-permissive media. After 24 hours, media was aspirated, cells were rinsed with PBS, and stored at −80 °C. For sterol extraction and analyses, cells were incubated in hexane for 10 minutes; 100 μl of cholesterol-d7 (25,26,26,26,27,27,27-2H7-cholesterol, Cambridge Isotope Laboratories) were added as standard. Samples were then dried under nitrogen stream and derivatized with 55 μl of bis(trimethylsilyl)trifluoroacetamide/trimethylchlorosilane at 60 °C for 20 minutes to form trimethylsilyl derivatives. Following, 2 μl of each sample were analyzed by gas chromatography/mass spectrometry using an Agilent 5973 Network Mass Selective Detector equipped with a 6890-gas chromatograph system and a HP-5MS capillary column (60 m × 0.25 mm × 0.25 μm). Samples were injected in splitless mode and analyzed using electron impact ionization. Peaks of ion fragments were integrated to calculate the abundance of sterols, and quantitation was relative to the standard, cholesterol-d7. The following m/z ion fragments were used to quantitate each metabolite: cholesterol-d7 (465), cholesterol (368), zymostenol (458), zymosterol (456), desmosterol (456, 343), MAS-414 (486), T-MAS (484), 4-methylzymostenol (472), 4-methylzymosterol (470), 4-methylzymostenone (398), 4-methylzymosterone (396), zymostenone (384), zymosterone (382), 24,25-epoxycholesterol (472, 382). Calibration curves were generated by injecting varying concentrations of sterol standards and maintaining a fixed amount of cholesterol-d7.CRISPR-targeting in OPCs
Cas9 containing OPCs were generated using the lenti-CRISPRv2 backbone and virus generated from Lenti-x Hek293T cells were transfected using Lenti-x shots as per the manufacturer's protocol (Clontech cat. 631275).34 After 4 hours the media was changed to OPC media for collection of virus. 48 hours later the media was collected, supplemented with FGF, PDGF and protamine sulfate (Sigma, 8 μg ml−1), and used to transduce OPCs. 24 hours later the media was changed to non-virus containing media for 48 h. Cells underwent two 48 hours stretches of puromycin selection (ThermoFisher cat. A1113802). Cells were allowed to recover for several passages prior to being aliquoted and frozen down. Cells were thawed one passage prior to electroporation. crRNA (NTC cat. U-007501-01-05, SC4MOL cat. CM-044979-01-0002, HSD17B7 cat. CM-040937-01-0002) and tracrRNA (U-002005-05) was purchased from Dharmacon and electroporated into OPCs using the Basic Nucleofector Kit for Primary Mammalian Glial Cells and Nucleofector 2b Device by following standard manufacturer's protocol. Cells were allowed to recover for up to 8 days in growth media prior to being plated for GC-MS-based sterol profiling, differentiation experiments, and RT-qPCR.RT-qPCR analysis
OPCs where cultured in poly-D-lysine and laminin-coated 6-well NUNC plates (SigmaAldrich cat. 140685) at 1 million cells per well with differentiation-permissive media. After 48 hours, media was aspirated, cells were rinsed with PBS, and RNA was harvested by adding 1 ml of TRIzol (Zymo Research, cat. R2050-1-200). RNA isolation was then performed with the RNeasy Mini Kit (Qiagen) and cDNA synthesis was carried out using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). Exon spanning primers for GAPDH (Thermo-Fisher, Taqman, Mm99999915 _g1), SC4MOL/MSMO1 (Thermo-Fisher, Taqman, Mm00499390_m1), and HSD17B7 (Thermo-Fisher, Taqman, Mm00501703_m1) with Taqman Gene Expression Master Mix (ThermoFisher, cat. 4369016) were used using the manufacturer's protocol for detection of relative RNA levels by quantitative real time PCR (QuantStudio 7 flex system).Analysis of mouse brain sterol levels
Eight-week-old male C57BL/6 mice were intraperitoneally injected in the tail with 20 or 50 mg kg−1 CW4142 dissolved in 10% (2-hydroxypropyl)-β-cyclodextrin (SigmaAldrich cat 332607) or vehicle daily for three days. For tissue isolation, mice were anaesthetized with isoflurane and perfused with phosphate buffered saline to remove blood from the brain. Brains were collected and flash frozen using liquid nitrogen. The samples were weighed and pulverized. A modified Folch protocol was used for extraction of sterols. Briefly, samples were resuspended in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 chloroform:methanol:saline mixture and homogenized. Cell debris was removed by centrifugation at 4000g for 10 min. The chloroform-rich lower layer was extracted and cholesterol-d7 standard was added. Lipids were derivatized with 70 μl of bis(trimethylsilyl)trifluoroacetamide. 2 μl was injected and analyzed by GC-MS described above. For cholesterol measurements, the sample was diluted 1
1 chloroform:methanol:saline mixture and homogenized. Cell debris was removed by centrifugation at 4000g for 10 min. The chloroform-rich lower layer was extracted and cholesterol-d7 standard was added. Lipids were derivatized with 70 μl of bis(trimethylsilyl)trifluoroacetamide. 2 μl was injected and analyzed by GC-MS described above. For cholesterol measurements, the sample was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in bis(trimethylsilyl)trifluoroacetamide and 2 μl injected into GC-MS. All procedures were in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Case Western Reserve University Institutional Animal Care and Use Committee (IACUC).
100 in bis(trimethylsilyl)trifluoroacetamide and 2 μl injected into GC-MS. All procedures were in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Case Western Reserve University Institutional Animal Care and Use Committee (IACUC).
Statistical analysis
No statistical methods were used to predetermine sample size. Number of replicates and independent experiments performed are noted in the figure legend. P-values were calculated with tests indicated in legend. Graphpad Prism 9.0.0 (121) was used to perform all statistical analysis.Results & discussion
8,9-Unsaturated sterol substrates of SC4MOL and HSD17B7, but not NSDHL, promote oligodendrocyte formation
Previously, two SC4MOL substrate sterols (T-MAS and MAS-414) were shown to promote oligodendrocyte formation when exogenously added to OPCs, so we first evaluated whether supplying additional SC4MOL, NSDHL, or HSD17B7 substrate sterols impacted OPC differentiation (Fig. 1A and B).13 To evaluate oligodendrocyte formation, we relied on an established high-content imaging assay in which formation of mature, myelin basic protein-positive (MBP+) oligodendrocytes is quantitated three days after placing murine pluripotent stem cell-derived OPCs in differentiation-permissive media.13 We first tested the SC4MOL substrate sterol 4-methylzymostenol, which we synthesized in seven steps from 7-dehydrocholesterol (Fig. 1B). 4-Methylzymostenol was as effective as T-MAS at promoting oligodendrocyte formation, demonstrating that multiple SC4MOL substrate sterols share this ability (Fig. 1C and D). By contrast, 4-carboxyzymostenol, an NSDHL substrate sterol synthesized through a related sequence, did not enhance oligodendrocyte formation and also led to substantial reductions in cell number, suggesting that NSDHL's carboxy-sterol substrates may have cytotoxic properties (Fig. 1B–D). Further modifications to this synthesis provided the HSD17B7 keto-sterol substrates 4-methylzymostenone and zymostenone, each of which enhanced oligodendrocyte formation to a comparable extent as the SC4MOL substrate sterols (Fig. 1B–D). Together, our data indicate that supplementing OPCs with multiple SC4MOL or HSD17B7 substrate sterols, but not an NSDHL substrate, is sufficient to enhance oligodendrocyte formation. These results substantially expand the structural range of 8,9-unsaturated sterols that promote this cell fate transition and motivated us to evaluate further whether SC4MOL and HSD17B7 may be druggable targets for enhancing oligodendrocyte formation.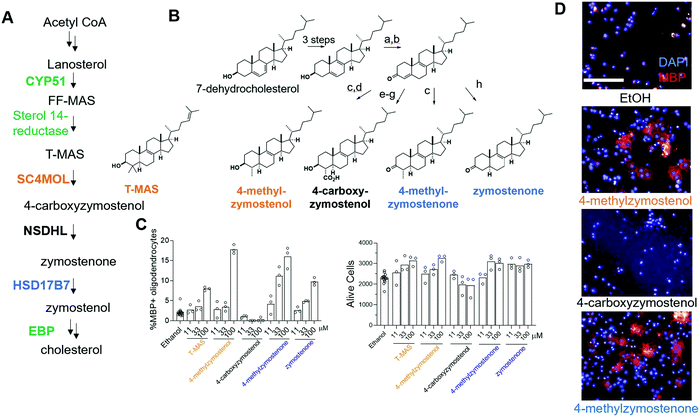 | ||
| Fig. 1 Exogenous addition of HSD17B7 and SC4MOL substrates enhance oligodendrocyte formation. (A) Simplified cholesterol biosynthesis pathway diagram. Enzymes in green have been shown to be targets for enhancing oligodendrocyte formation. A full pathway diagram showing all substrate sterols can be found in Fig. S1 (ESI†). (B) Conditions: a, oxalyl chloride, DMSO, NEt3, CH2Cl2; b, DBU, THF; c, Li0, NH3, THF; then Mel; d, LiAlH4, THF; e, Li0, NH3, THF; then methylcyanoformate; f, NaBH4, MeOH; g, KOH, MeOH, reflux; h, (BDP)CuH, PMHS, PhMe. (C) Percentage of MBP+ oligodendrocytes generated from OPCs and live cell number following treatment with the indicated 8,9-unsaturated sterol. n = 3 wells per condition. (D) Representative images of oligodendrocyte formation following sterol treatments. Blue, DAPI; Red, MBP. Scale bar, 100 μM. (C) is representative of two independent biological replicates. | ||
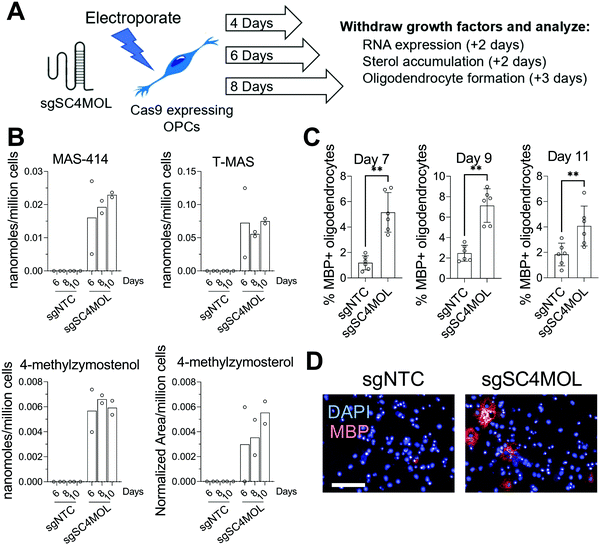
CRISPR targeting of SC4MOL and HSD17B7 promotes oligodendrocyte formation
Since supplying OPCs with SC4MOL and HSD17B7 substrates promoted OPC differentiation, we next asked if genetic suppression of these enzymes could induce endogenous accumulation of these sterols in OPCs and increase oligodendrocyte formation. To genetically target SC4MOL, we used established murine OPCs that stably express Cas9;17 electroporation then enabled delivery of synthetic CRISPR guide RNAs (gRNA) that targeted SC4MOL (Fig. 2A). We first evaluated whether targeting SC4MOL impacted cholesterol biosynthesis using GC-MS-based sterol profiling, a well-established approach that enables quantitation of cholesterol and its sterol precursors, including SC4MOL substrates.13 OPCs were transfected with guide RNA targeting SC4MOL and maintained in growth media for 4, 6, or 8 days. Cells were then re-plated for GC-MS-based sterol profiling to quantify levels of SC4MOL's substrates and additional cholesterol precursors 6, 8, and 10 days post-transfection. While SC4MOL has four substrate sterols (Fig. S1, ESI†), none of which are detected in nontreated OPCs, transfection of SC4MOL gRNA primarily induced accumulation of SC4MOL's C4-dimethyl substrates, T-MAS and MAS-414. Lower levels of 4-methylzymosterol and 4-methylzymostenol were noted as well (Fig. 2B). While elevated SC4MOL substrate sterols were detected at all times measured, we observed maximal accumulation 8 and 10 days post-transfection. No other sterol precursors to cholesterol were seen to accumulate, confirming selective inhibition of SC4MOL (Fig. S2A, ESI†). Notably, RT-qPCR analysis 6 days post-transfection revealed a 50% reduction in SC4MOL mRNA levels (Fig. S2B, ESI†).We next asked if genetic targeting of SC4MOL enhanced the formation of oligodendrocytes. In parallel with the experiments above, OPCs transfected with guide RNA targeting SC4MOL were expanded for 4, 6, or 8 days. We then re-plated these OPCs in differentiation-permissive media for an additional three days and evaluated the formation of mature, myelin basic protein-positive (MBP+) oligodendrocytes using our established high-content imaging assay 7, 9, and 11 days post-transfection. We observed a maximal increase in oligodendrocyte formation 7 and 9 days post-transfection (Fig. 2C and D), with a significant increase also seen on day 11. These data demonstrate that genetic targeting of SC4MOL in OPCs leads to accumulation of SC4MOL's substrate sterols and increases oligodendrocyte formation over multiple time points measured.
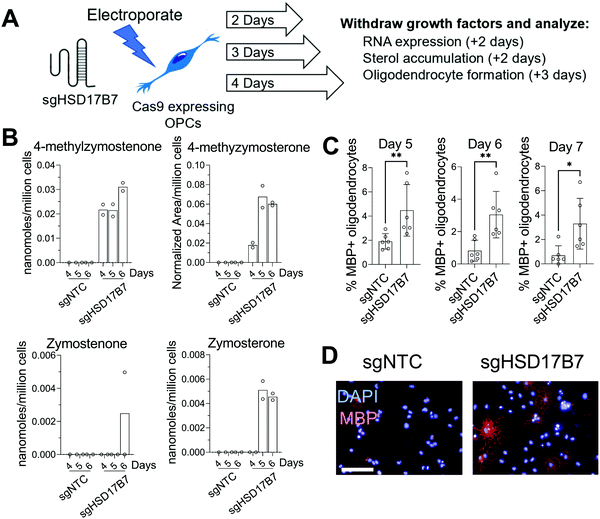
Since multiple HSD17B7 substrate sterols promote oligodendrocyte formation (Fig. 1C), we further used this CRISPR approach to target HSD17B7 in OPCs. Using OPCs that stably express Cas9, we transfected a CRISPR guide RNA targeting HSD17B7 and evaluated HSD17B7 inhibition using GC-MS-based sterol profiling. Interestingly, HSD17B7 substrate sterols, which are also undetectable in untreated OPCs, rose earlier following electroporation of HSD17B7 gRNA than observed for SC4MOL. After allowing 2, 3, or 4 days of growth post-transfection, we re-plated the OPCs for an additional 2 days and analyzed levels of HSD17B7's keto-sterol substrates and other sterols (Fig. 3A and Fig. S1, ESI†). While HSD17B7 substrates were elevated at all times measured, we observed maximal levels of HSD17B7's keto-sterol substrates 5 and 6 days after electroporation (Fig. 3B). We primarily observed accumulation of HSD17B7's C4-methyl substrates, 4-methylzymostenone and 4-methylzymosterone, with lesser levels of zymosterone also detected at later time points (Fig. S2C, ESI†). RT-qPCR analysis 4 days post-transfection showed a 25% reduction in HSD17B7 mRNA levels (Fig. S2D, ESI†).
We next asked if genetic targeting of HSD17B7 would enhance the formation of oligodendrocytes. OPCs transfected with the guide RNA targeting HSD17B7 were grown for 2, 3, and 4 days post-transfection. We then re-plated these OPCs in differentiation-permissive media for an additional three days and evaluated the formation of mature, MBP+ oligodendrocytes 5-, 6-, and 7-days post-transfection. Targeting HSD17B7 led to clear increases in oligodendrocyte formation at each time point analyzed, with a maximal response observed at 6 and 7 days after transfection (Fig. 3C and D). As observed for SC4MOL, time points with maximal sterol substrate accumulation were associated with maximal enhancement of oligodendrocyte formation. These results demonstrate that targeting HSD17B7 drives accumulation of HSD17B7's keto-sterol substrates and enhances oligodendrocyte formation from OPCs.
Small molecule inhibitors of SC4MOL promote oligodendrocyte formation
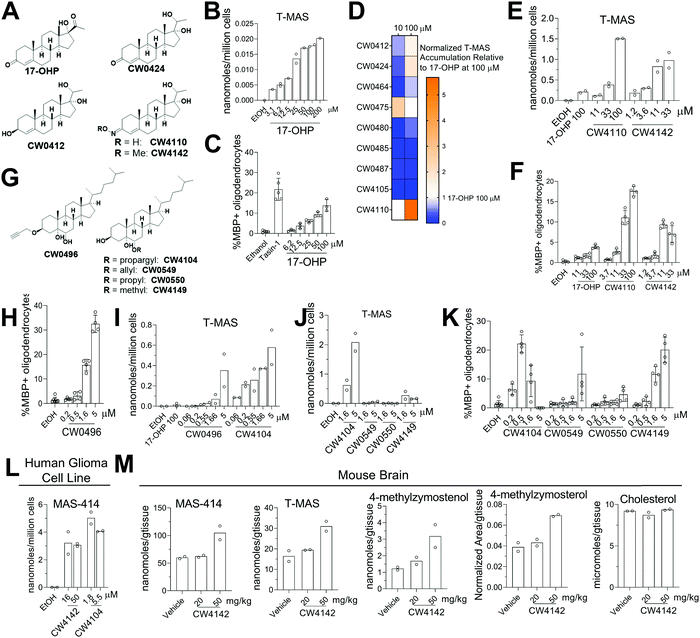
Given that genetic knockdown of SC4MOL and exogenous addition of SC4MOL substrates increase oligodendrocyte formation, we wondered whether small molecule inhibition of SC4MOL would lead to similar pro-differentiation effects on OPCs.19,20 Few inhibitors of SC4MOL are known.21–23 Among four previously reported inhibitors of yeast or mammalian SC4MOL, only treatment with 17-hydroxyprogesterone (17-OHP) or APB inhibited SC4MOL in our OPCs (Fig. S3A, ESI†). Since APB treatment led to cytotoxicity in OPCs, we focused on 17-OHP (Fig. 4A). We found that treatment of OPCs with 17-OHP in the range of 10–100 μM led to accumulation of SC4MOL's substrate sterols—primarily T-MAS, 4-methylzymosterol and 4-methylzymostenol—in a dose responsive manner (Fig. 4B and Fig. S3B, ESI†). Notably, concentrations of 17-OHP that inhibited SC4MOL in OPCs also enhanced oligodendrocyte formation, suggesting that 17-OHP enhances oligodendrocyte formation through SC4MOL inhibition (Fig. 4C). Additionally, 17-OHP increased oligodendrocyte formation to levels comparable to a previously-validated inhibitor of the pathway enzyme EBP, suggesting that inhibition of SC4MOL leads to a similar magnitude of effect as established pathway targets. To gain a preliminary understanding of the structural features responsible for 17-OHP's ability to inhibit SC4MOL, we tested three closely related steroids, androstenedione, progesterone, and testosterone, for their effects on SC4MOL. Notably, these closely related steroids did not inhibit SC4MOL in OPCs, suggesting that the 17-alpha-hydroxy functionality adds significant potency for SC4MOL (Fig. S3C and D, ESI†). Together with the CRISPR targeting of SC4MOL, these data corroborated SC4MOL as a target for enhancing oligodendrocyte formation and motivated us to identify more effective SC4MOL-inhibiting small molecules.Since 17-OHP inhibited SC4MOL and enhanced OPC differentiation to oligodendrocytes, we synthesized a set of 9 structural analogs with the goal of improving potency (for structures of all analogs synthesized, see data Table S1, ESI†). Each analog was characterized at multiple concentrations in our GC-MS-based sterol profiling assay (Fig. 4D). We identified several analogs (CW0412, CW0424, and CW4110) that improved the efficacy and potency of SC4MOL inhibition. These three molecules led to at least a 2-fold increase in accumulation of the SC4MOL substrate sterol T-MAS at 100 μM compared to 17-OHP while also increasing oligodendrocyte formation at concentrations that inhibit SC4MOL (Fig. 4D and Fig. S3E, ESI†). Further modification to CW4110 led to oxime ether CW4142, which showed additional improvement in potency for both accumulation of SC4MOL substrate sterols and enhancement of oligodendrocyte formation (Fig. 4E and F and Fig. S3F, ESI†). Relative to 17-OHP, CW4142 shows substantial improvement in potency for both inhibiting SC4MOL and enhancing oligodendrocyte formation.
While evaluating additional synthetic sterols, we identified the propargylated oxysterol CW0496 as an enhancer of oligodendrocyte formation (Fig. 4G and H). Subsequent GC-MS-based sterol profiling revealed that CW0496 surprisingly inhibited SC4MOL with improved potency and elevated SC4MOL substrate accumulation relative to 17-OHP and its derivatives (Fig. 4I and Fig. S3G, ESI†). Further, CW0496 increased oligodendrocyte formation with potency that matched its inhibition of SC4MOL. Moving the propargyl group from C3 to C6 provided CW4104, which even more potently induced accumulation of SC4MOL substrates (Fig. 4I and Fig. S3G, ESI†).
The surprising potency of these alkyne-containing oxysterols led us to synthesize a series of CW4104 analogs that contained either an allyl, propyl, or methyl ether at C6 (Fig. 4G). Among this set, CW4104 remained the most potent SC4MOL inhibitor, with allyl analog CW0550 inferior and propyl analog CW0549 inactive in our GC-MS-based sterol profiling assay (Fig. 4J and Fig. S3H, ESI†). Interestingly, methyl ether CW4149 retained moderate potency for inhibition of SC4MOL (Fig. 4J and Fig. S3H, ESI†). Evaluation of these compounds’ effects on oligodendrocyte formation from OPCs revealed a corresponding trend, with the propargyl substituent optimal, allyl inferior, propyl inactive, and methyl retaining significant pro-oligodendrocyte effects (Fig. 4K). Subsequent analogs revealed that removal of the C5 hydroxyl reduced potency for SC4MOL inhibition and oligodendrocyte formation, while removal of both the C5 and C6 hydroxyls abrogated all activity in these assays (Fig. S3J–L, ESI†). Together with our 17-OHP-derived molecules, these alkyne-containing SC4MOL inhibitors represent a suite of chemical tools that further validate SC4MOL as a target for enhancing oligodendrocyte formation and suggest structural features that lead to potent and selective cellular inhibition of SC4MOL (Fig. S3I, ESI†).
We next evaluated whether leading analogs inhibit SC4MOL in additional murine OPC derivations, human cells, and mouse brain. Using two additional derivations of OPCs obtained from two separate iPSC cell lines, we confirmed that CW4142 and CW4104 uniformly inhibited SC4MOL and promoted oligodendrocyte formation (Fig. S3M and N, ESI†). GC-MS-based sterol profiling using a human glioma cell line, GBM528, revealed that treatment with either CW4142 or CW4104 led to substantial accumulation of SC4MOL substrate sterols, led by MAS-414 (Fig. 4L). While the potency of these molecules was somewhat lower in GBM528 cells relative to murine OPCs, no other sterol intermediates accumulated upon treatment of CW4142 or CW4104, indicating a selective effect for SC4MOL (Fig. S3O, ESI†). Further, we evaluated whether CW4142 inhibits SC4MOL in mouse brain. After three daily intraperitoneal injections, mice treated with 50 mg kg−1 CW4142 showed a nearly 2-fold increase in each of four SC4MOL substrates, while cholesterol and desmosterol levels were unchanged (Fig. 4M). These results establish these SC4MOL inhibitors as leading small-molecule probes with which to explore the biological effects of SC4MOL inhibition in cultured cells (CW4142, CW4104) and in vivo (CW4142).
Novel small molecule inhibition of HSD17B7 enhances oligodendrocyte formation
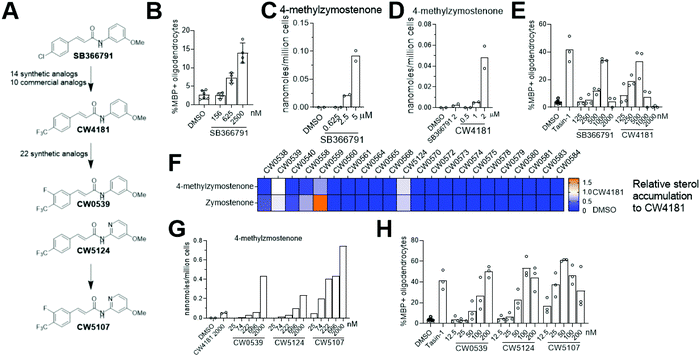
Since genetic targeting of HSD17B7 promoted oligodendrocyte formation, we also searched for inhibitors of the oxidoreductase activity of HSD17B7. While HSD17B7's role in cholesterol biosynthesis is now well-established, this enzyme was initially characterized as catalyzing the reduction of the steroid estrone at C17. Previous work identified inhibitors of HSD17B7's estrone reductase activity; however, these molecules surprisingly do not affect HSD17B7's ability to reduce keto-sterol precursors to cholesterol.24 Additionally, inhibitors of the yeast homolog of HSD17B7 are either inactive in our OPCs (fenpyrazamine, Fig. S4A and B, ESI†) or inhibit a cholesterol pathway enzyme other than HSD17B7 (fenhexmid17), leaving no known starting points for the development of inhibitors of HSD17B7's cholesterol biosynthesis activity.25–27Previously, we screened 3000 bioactive small molecules for enhancers of oligodendrocyte formation and characterized the top ten novel hits as inhibitors of CYP51, Sterol 14-reductase or EBP.13 Subsequent evaluation of molecules that fell outside the top 10 hits confirmed SB366791, a TRPV1 antagonist, as an enhancer of oligodendrocyte formation (Fig. 5A and B).28–30 Since TRPV1 and related ion channels are not expressed in OPCs,31 we evaluated whether SB366791 was functioning through an off-target mechanism. GC-MS-based sterol profiling revealed SB366791 inhibited HSD17B7 as indicated by accumulation of its keto-sterol substrates, including 4-methylzymostenone (Fig. 5C). Concentrations of SB366791 that led to the accumulation of HSD17B7 substrate sterols also increased OPC differentiation, which further suggests that SB366791 enhanced oligodendrocyte formation via ‘off-target’ inhibition of HSD17B7 (Fig. 5B and C).
We evaluated commercial and synthetic analogs of SB366791 to further correlate HSD17B7 inhibition and enhanced oligodendrocyte formation (for structures of all analogs evaluated, see Data Table S1, ESI†). We first selected 10 commercially available SB366791 analogs and evaluated each for effects on HSD17B7 substrate accumulation and oligodendrocyte formation. Surprisingly, none were able to inhibit HSD17B7 nor affect OPC differentiation at the concentrations tested (Fig. S4C and D, ESI†). We subsequently synthesized a collection of 14 analogs that made more conservative structural modifications to SB366791. Of these analogs, only CW4181, which substitutes the chloro functionality with a trifluoromethyl group, led to enhanced accumulation of HSD17B7 substrate sterols relative to SB366791 using GC-MS-based sterol profiling at 2.5 μM (Fig. 5A and Fig. S4E, ESI†). Further dose response analysis in both our GC-MS-based sterol profiling and oligodendrocyte formation assays revealed CW4181 to be 2–3-fold more potent than SB366791 in both assays (Fig. 5D and E and Fig. S4F, ESI†). Of note, maximal effects on oligodendrocyte formation are similar for SB366791, CW4181, and the EBP inhibitor Tasin-1, again suggesting that inhibition of different cholesterol pathway enzymes leads to comparable maximal increases in oligodendrocyte formation.
To identify more potent HSD17B7 inhibitors, we synthesized an additional set of 22 analogs that made modest modifications to CW4181. Remarkably, a large majority of these conservative analogs also were inactive in our cell-based sterol profiling assay at 2.5 μM (Fig. 5F). Two active analogs, CW0539 and CW5124, were selected for dose–response testing and showed enhanced potency for cellular inhibition of HSD17B7 compared to CW4181 (Fig. 5G and Fig. S4G, ESI†). These two analogs contain chemical modifications at different positions of CW4181. Combining these modifications provided compound CW5107, which as expected inhibited HSD17B7 in OPCs with enhanced potency (EC50 ca. 50 nM for 4-methylzymostenone and zymostenone accumulation, Fig. 5G and Fig. S4G, ESI†). Comparable potency trends were observed in our oligodendrocyte formation assay: CW5107 was again most potent, enhancing oligodendrocyte formation at concentrations as low as 25 nM (Fig. 5H). Similar to the parent compounds, these more potent analogs had similar maximal percentage of oligodendrocyte formation compared to EBP inhibitor. Further, CW5107 shows selectivity over other cholesterol pathway enzymes, with no other cholesterol precursors accumulating as detected by GC-MS-based sterol profiling (Fig. S4H, ESI†). CW5107 also inhibited HSD17B7 and enhanced oligodendrocyte formation in two additional derivations of OPCs (Fig. S4I and J, ESI†). Notably, CW5107 was less potent in human cells, suggesting further optimization or novel scaffolds may be needed to inhibit human HSD17B7 (Fig. S4K and L, ESI†). In total, we have identified the first inhibitors of HSD17B7's sterol synthesis activity, optimized their potency into the low nanomolar range, and demonstrated that inhibiting HSD17B7 drives oligodendrocyte formation.
Throughout these studies, we generally observed increased cell number in oligodendrocyte formation assays for both SC4MOL and HSD17B7 inhibitors that enhance oligodendrocyte formation, as seen previously for inhibitors of EBP and other pathway enzymes.13,17 However, in some oligodendrocyte formation assays, treatments for which GC/MS-based sterol profiling indicated HSD17B7 was fully inhibited led to reduced cell number at the conclusion of the 3-day assay. This phenomenon was observed for molecules with wide-ranging potency for HSD17B7 (Fig. S4M, ESI†), suggesting that full inhibition of HSD17B7 and the resulting altered sterol environment may not be tolerated by OPCs. Interestingly, we also identified one analog, CW0578, that induces submaximal levels of HSD17B7 substrate sterols over a wide concentration range. Despite its apparent partial inhibitory effect on HSD17B7, CW0578 shows strong potency, with concentrations as low as 300 nM leading to keto-sterol accumulation (Fig. S4N, ESI†). This partial inhibition of HSD17B7 strongly promotes oligodendrocyte formation without cell death across a much wider concentration range than other molecules that induce maximal accumulation of HSD17B7 sterol substrates (e.g. CW5107, Fig S4M, ESI†). While these data may further suggest that full inhibition of HSD17B7 in OPCs is not well-tolerated, supplying purified HSD17B7 substrates to OPCs was not cytotoxic (Fig. 1C). Additional chemically divergent series of HSD17B7 inhibitors may be needed to evaluate whether the observed cytotoxicity relates to HSD17B7.
Accumulation of the 8,9-unsaturated substrate sterols of SC4MOL and HSD17B7 is necessary and sufficient for enhanced oligodendrocyte formation
Previous work has provided evidence that cellular accumulation of 8,9-unsaturated sterols is the central mechanistic driver of enhanced oligodendrocyte formation following inhibition of EBP, sterol 14-reductase, and CYP51. We next evaluated whether the Δ8,9-alkene functionality itself plays a critical role for the activity of SC4MOL and HSD17B7 sterol substrates in OPCs.13 We synthesized 8,9-saturated analogs of several SC4MOL and HDS17B7 substrates and compared these head-to-head with naturally-occurring 8,9-unsaturated sterol pathway intermediates. In contrast to the SC4MOL substrate 4-methylzymostenol, its 8,9-saturated analog 4-methylcholestanol had no effect on oligodendrocyte formation in our assay (Fig. 6A). Likewise, in contrast to the HSD17B7 substrate zymostenone, the 8,9-saturated analog cholestanone was also ineffective at promoting oligodendrocyte formation (Fig. 6B). 4-Methylcholestanone was also substantially inferior in potency to its 8,9-unsaturated analog 4-methylzymostenone (Fig. 6C). These studies further highlight how inclusion of this double bond within the sterol scaffold plays a decisive role in the ability of sterols to influence the differentiation of OPCs to oligodendrocytes.To evaluate whether accumulation of 8,9-unsaturated sterols was essential for small molecule inhibitors of HSD17B7 to enhance oligodendrocyte formation, we co-treated cells with an HSD17B7 inhibitor and an LSS inhibitor, Ro 48-8071.13,17 LSS lies upstream of all 8,9-unsaturated sterol-metabolizing enzymes (Fig. S1, ESI†). As a result, full inhibition of LSS blocks flux through the cholesterol biosynthesis pathway and prevents the accumulation of downstream HSD17B7 substrates even when HSD17B7 is also inhibited. First we established, consistent with past work, that treatment of OPCs with Ro 48-8071 at 200 nM was sufficient to inhibit LSS fully. At this concentration, no 24,25-epoxycholesterol, which accumulates when LSS is inhibited partially, was detected (Fig. 6D).17 Additionally, 200 nM Ro 48-8071 was sufficient to prevent accumulation of HSD17B7 substrate sterols following co-treatment with CW5107 (Fig. 6D). Under these conditions that prevent HSD17B7 substrate sterol accumulation, CW5107 was unable to promote oligodendrocyte formation (Fig. 6E). In contrast, Liothyronine, which promotes oligodendrocyte formation via the thyroid hormone receptor and does not modulate sterol synthesis, showed only a small drop in oligodendrocyte formation when paired with Ro 48-8071 at 200 nM. These studies provide further evidence that the accumulation of HSD17B7's 8,9-unsaturated sterol substrates is necessary for CW5107 to promote oligodendrocyte formation (Fig. 6E).
Conclusions
Replenishing oligodendrocytes in the CNS may have therapeutic potential in demyelinating diseases such as MS. Previously, our work identified four enzymes within the cholesterol biosynthesis pathway whose inhibition promotes oligodendrocyte formation from OPCs.13,17 Here, we have validated two additional pathway enzymes, SC4MOL and HSD17B7. Genetic targeting, small molecule inhibition, and exogenous addition of the 8,9-unsaturated substrates of SC4MOL or HSD17B7 all enhance mouse oligodendrocyte formation, providing multiple complementary approaches validating these enzymes as targets for enhancing oligodendrocyte formation (Fig. 6F).Inhibiting any of these targets induces a sterol shift in which a portion of cholesterol pathway flux accumulates as one or more 8,9-unsaturated sterol precursors, which are undetectable in untreated OPCs but are necessary and sufficient to enhance the formation of oligodendrocytes. By synthesizing a wide range of additional complex cholesterol precursors, this work has substantially expanded the range of 8,9-unsaturated sterols that impact OPC differentiation. Notably, C3-keto-sterols are among the most potent purified sterols for enhancing oligodendrocyte formation despite lacking the C3 beta-hydroxyl functionality common to cholesterol and all its precursors. Additionally, sterols with C4 dimethyl, monomethyl, or desmethyl substitution provide comparable activity, suggesting that this position can be varied extensively. However, substitution with a C4 carboxylic acid moiety was ineffective in our OPC differentiation assay, providing initial evidence that NSDHL inhibition may not promote oligodendrocyte formation. While this result is somewhat surprising given that a wide range of other 8,9-unsaturated sterols promote oligodendrocyte formation when added exogenously, the introduction of the anionic C4 carboxylate moiety may disrupt downstream interactions needed to promote differentiation of OPCs to oligodendrocytes.
Interestingly, multiple previous screens for remyelinating small molecules did not identify any SC4MOL or HSD17B7 inhibitors among their ‘hits’; instead, inhibitors of EBP predominated. To target SC4MOL, we optimized the known low-potency inhibitor 17-OHP to generate CW4142, which showed enhanced potency in murine and human cell culture and while also inducing accumulation of SC4MOL substrates in mouse brain. Another oxime-containing sterol, the neuroprotective agent olesoxime, has been progressed clinically in ALS patients, suggesting CW4142 may be amenable for further clinical optimization.32,33 We also demonstrated that several alkyne-containing sterols, including CW4096 and CW4104, inhibited SC4MOL with enhanced potency. Intriguingly, the alkyne functionality was essential for the activity of these potent inhibitors, but the mechanistic basis for this observation is unclear.
Although TRPV1 is not expressed in OPCs in public RNA sequencing data, we identified the canonical TRPV1 antagonist SB366791 as a cellular HSD17B7 inhibitor during efforts to characterize additional molecules near the hit threshold of our previously-reported screen.13 Synthesis of more than two dozen SB366791 analogs revealed that only very conservative modifications were tolerated in our OPC assays, but our optimization efforts ultimately led to CW5107, which inhibits HSD17B7 and enhances oligodendrocyte formation in the low nanomolar range in OPCs. This series of HSD17B7 inhibitors stands in clear contrast to previously-reported inhibitors which inhibited only the distinct steroid C17 reductase activity of HSD17B7.24 As such, CW5107 and its analogs represent the first tools for probing the effects of blocking cholesterol biosynthesis at HSD17B7 in cells. We anticipate CW5107, together with our leading inhibitors of SC4MOL, will be useful small-molecule tools to elucidate additional contexts in which inhibition of these enzymes and accumulation of their 8,9-unsaturated substrates impacts cellular function.
Among the five 8,9-unsaturated sterol-metabolizing enzymes validated to date, which may be most promising for drug discovery efforts aimed at enhancing remyelination for MS patients? Notably, each of these targets appears to lead to a comparable maximal effect on oligodendrocyte formation when inhibited, consistent with a shared mechanism of inducing accumulation of 8,9-unsaturated sterols. Likewise, purified substrates of each of these five targets show similar efficacy in our OPC differentiation assay, despite significant structural variation. While inhibitors of SC4MOL and HSD17B7 appear to be rare in screening libraries relative to inhibitors of EBP and other targets, here we have demonstrated that potent inhibitors of these enzymes can be optimized for use in cells and, in some cases, in vivo. Together, these results establish that inhibition of SC4MOL and HSD17B7 shifts cholesterol biosynthesis in OPCs to favor accumulation of a diverse range of 8,9-unsaturated sterols that promote oligodendrocyte formation. Additionally, we have established high-quality chemical tools that can be used to further interrogate the roles of SC4MOL and HSD17B7 in myelin biology and other settings.
Author contributions
M. J. P., A. M. R.-L., Z. H., F. G., J. S., and D. J. A evaluated effects on oligodendrocyte formation in vitro. M. J. P., A. M. R.-L., Z. H., F. G., I. B., and D. J. A. performed and analyzed sterol profiling experiments in vitro. R. M. F., D. Y., and R. S. synthesized and characterized small molecules used in this study. M. J. P., A. M. R.-L., and D. J. A. wrote the manuscript. M. J. P., H. E. S., P. J. T., I. B., and D. J. A. performed and analyzed sterol profiling experiments in mice in vivo. All authors provided intellectual input and edited and approved the final manuscript.Conflicts of interest
The authors declare the following competing interests: D. J. A. and P. J. T. are founders, consultants, directors, and shareholders of Convelo Therapeutics, Inc., which seeks to develop remyelinating therapeutics. D. J. A., P. J. T., and Z. H. are inventors on patents and patent applications that have been licensed to Convelo.Acknowledgements
The authors thank G. Tochtrop for synthesis support, and Y. Fedorov, J. Rich, the CWRU Small-Molecule Drug Development Core, and M. Drumm for technical support. This work was supported by the following grants: NIH T32GM008803 (M. J. P.), F30MH116581 (Z. H.), NIH TL1 TR000441 (Z. H.), NIH 2R25GM075207-14 (A. M. R.-L.), the Mathers Foundation, a Mallinckrodt Foundation grant award, an NMSS Pilot grant, Thomas F. Peterson, Jr., the Mount Sinai Health Care Foundation, the CWRU School of Medicine, and the Case Comprehensive Cancer Center (P30 CA043703).Notes and references
- D. K. Hartline and D. R. Colman, Rapid Conduction and the Evolution of Giant Axons and Myelinated Fibers, Curr. Biol., 2007, 17(1), R29–R35 CrossRef CAS.
- K.-A. Nave, Myelination and the trophic support of long axons, Nat. Rev. Neurosci., 2010, 11(4), 275–283 CrossRef CAS PubMed.
- S. P. J. Fancy, M. R. Kotter, E. P. Harrington, J. K. Huang, C. Zhao and D. H. Rowitch, et al., Overcoming remyelination failure in multiple sclerosis and other myelin disorders, Exp. Neurol., 2010, 225(1), 18–23 CrossRef CAS.
- H. Lassmann, Classification of demyelinating diseases at the interface between etiology and pathogenesis, Curr. Opin. Neurol., 2001, 14(3), 253–258 CrossRef CAS PubMed.
- R. J. M. Franklin and C. Ffrench-Constant, Remyelination in the CNS: from biology to therapy, Nat. Rev. Neurosci., 2008, 9(11), 839–855 CrossRef CAS PubMed.
- H. S. Keirstead and W. F. Blakemore In: The Role of Oligodendrocytes and Oligodendrocyte Progenitors in cns Remyelination. ed. Matsas R., Tsacopoulos M., The Functional Roles of Glial Cells in Health and Disease: Dialogue between Glia and Neurons [Internet], Springer US, Boston, MA, 1999, [cited 2021 Apr 14], p. 183–197. (Advances in Experimental Medicine and Biology) DOI:10.1007/978-1-4615-4685-6_15.
- A. Chang, W. W. Tourtellotte, R. Rudick and B. D. Trapp, Premyelinating Oligodendrocytes in Chronic Lesions of Multiple Sclerosis, N. Engl. J. Med., 2002, 346(3), 165–173 CrossRef PubMed.
- J. K. Huang, A. A. Jarjour, B. Nait Oumesmar, C. Kerninon, A. Williams and W. Krezel, et al., Retinoid X receptor gamma signaling accelerates CNS remyelination, Nat. Neurosci., 2011, 14(1), 45–53 CrossRef CAS.
- V. A. Deshmukh, V. Tardif, C. A. Lyssiotis, C. C. Green, B. Kerman and H. J. Kim, et al., A regenerative approach to the treatment of multiple sclerosis, Nature, 2013, 502(7471), 327–332 CrossRef CAS PubMed.
- F. J. Najm, M. Madhavan, A. Zaremba, E. Shick, R. T. Karl and D. C. Factor, et al., Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo, Nature, 2015, 522(7555), 216–220 CrossRef CAS PubMed.
- F. Mei, S. R. Mayoral, H. Nobuta, F. Wang, C. Desponts and D. S. Lorrain, et al., Identification of the Kappa-Opioid Receptor as a Therapeutic Target for Oligodendrocyte Remyelination, J. Neurosci., 2016, 36(30), 7925–7935 CrossRef CAS PubMed.
- K. D. Lariosa-Willingham, E. S. Rosler, J. S. Tung, J. C. Dugas, T. L. Collins and D. Leonoudakis, A high throughput drug screening assay to identify compounds that promote oligodendrocyte differentiation using acutely dissociated and purified oligodendrocyte precursor cells, BMC Res. Notes, 2016, 9(1), 419 CrossRef.
- Z. Hubler, D. Allimuthu, I. Bederman, M. S. Elitt, M. Madhavan and K. C. Allan, et al., Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination, Nature, 2018, 560(7718), 372–376 CrossRef CAS PubMed.
- D. Allimuthu, Z. Hubler, F. J. Najm, H. Tang, I. Bederman and W. Seibel, et al., Diverse Chemical Scaffolds Enhance Oligodendrocyte Formation by Inhibiting CYP51, TM7SF2, or EBP, Cell Chem. Biol., 2019, 26(4), 593–599.e4 CrossRef CAS.
- G. A. Gonzalez, M. P. Hofer, Y. A. Syed, A. I. Amaral, J. Rundle and S. Rahman, et al., Tamoxifen accelerates the repair of demyelinated lesions in the central nervous system, Sci. Rep., 2016, 6, 31599 CrossRef CAS.
- A. J. Green, J. M. Gelfand, B. A. Cree, C. Bevan, W. J. Boscardin and F. Mei, et al., Clemastine fumarate as a remyelinating therapy for multiple sclerosis (ReBUILD): a randomised, controlled, double-blind, crossover trial, Lancet, 2017, 390(10111), 2481–2489 CrossRef CAS.
- Z. Hubler, R. M. Friedrich, J. L. Sax, D. Allimuthu, F. Gao and A. M. Rivera-León, et al., Modulation of lanosterol synthase drives 24,25-epoxysterol synthesis and oligodendrocyte formation, Cell Chem. Biol., 2021, 28(6), 866–875 CrossRef CAS.
- S. Darnet and H. Schaller, Metabolism and Biological Activities of 4-Methyl-Sterols, Molecules, 2019, 24(3), 451 CrossRef PubMed.
- N. M. F. S. A. Cerqueira, E. F. Oliveira, D. S. Gesto, D. Santos-Martins, C. Moreira and H. N. Moorthy, et al., Cholesterol Biosynthesis: A Mechanistic Overview, Biochemistry, 2016, 55(39), 5483–506 CrossRef CAS.
- W. D. Nes, Biosynthesis of Cholesterol and Other Sterols, Chem. Rev., 2011, 111(10), 6423–6451 CrossRef CAS.
- B. Lindenthal, A. L. Holleran, T. A. Aldaghlas, B. Ruan, G. J. Schroepfer and W. K. Wilson, et al., Progestins block cholesterol synthesis to produce meiosis-activating sterols, FASEB J., 2001, 15(3), 775–784 CrossRef CAS.
- H. Nose, H. Fushimi, A. Seki, T. Sasaki, H. Watabe and S. Hoshiko, PF1163A, a Novel Antifungal Agent, Inhibit Ergosterol Biosynthesis at C-4 Sterol Methyl Oxidase, J. Antibiot., 2002, 55(11), 969–974 CrossRef CAS.
- M. Sharma, Z. Sasvari and P. D. Nagy, Inhibition of Sterol Biosynthesis Reduces Tombusvirus Replication in Yeast and Plants, J. Virol., 2010, 84(5), 2270–2281 CrossRef CAS.
- T. Ferrante, S. Adinolfi, G. D’Arrigo, D. Poirier, M. Daga and M. L. Lolli, et al., Multiple catalytic activities of human 17β-hydroxysteroid dehydrogenase type 7 respond differently to inhibitors, Biochimie, 2020, 170, 106–117 CrossRef CAS.
- D. Debieu, J. Bach, M. Hugon, C. Malosse and P. Leroux, The hydroxyanilide fenhexamid, a new sterol biosynthesis inhibitor fungicide efficient against the plant pathogenic fungus Botryotinia fuckeliana (Botrytis cinerea), Pest Manage. Sci., 2001, 57(11), 1060–1067 CrossRef CAS PubMed.
- S. Fillinger, P. Leroux, C. Auclair, C. Barreau, C. Al Hajj and D. Debieu, Genetic analysis of fenhexamid-resistant field isolates of the phytopathogenic fungus Botrytis cinerea, Antimicrob. Agents Chemother., 2008, 52(11), 3933–3940 CrossRef CAS.
- N. Kimura, M. Hashizume, T. Kusaba and S. Tanaka, Development of the novel fungicide fenpyrazamine, J. Pestic. Sci., 2017, 42(3), 137–143 CrossRef CAS.
- M. J. Gunthorpe, H. K. Rami, J. C. Jerman, D. Smart, C. H. Gill and E. M. Soffin, et al., Identification and characterisation of SB-366791, a potent and selective vanilloid receptor (VR1/TRPV1) antagonist, Neuropharmacology, 2004, 46(1), 133–149 CrossRef CAS.
- S. C. Lappin, A. D. Randall, M. J. Gunthorpe and V. Morisset, TRPV1 antagonist, SB-366791, inhibits glutamatergic synaptic transmission in rat spinal dorsal horn following peripheral inflammation, Eur. J. Pharmacol., 2006, 540(1–3), 73–81 CrossRef CAS PubMed.
- A. Varga, J. Németh, A. Szabó, J. J. McDougall, C. Zhang and K. Elekes, et al., Effects of the novel TRPV1 receptor antagonist SB366791 in vitro and in vivo in the rat, Neurosci. Lett., 2005, 385(2), 137–142 CrossRef CAS.
- A. M. Lager, O. G. Corradin, J. M. Cregg, M. S. Elitt, H. E. Shick and B. L. L. Clayton, et al., Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells, Nat. Commun., 2018, 9(1), 3708 CrossRef PubMed.
- L. J. Martin, Olesoxime, a cholesterol-like neuroprotectant for the potential treatment of amyotrophic lateral sclerosis, IDrugs, 2010, 13(8), 568–580 CAS.
- C. Sunyach, M. Michaud, T. Arnoux, N. Bernard-Marissal, J. Aebischer and V. Latyszenok, et al., Olesoxime delays muscle denervation, astrogliosis, microglial activation and motoneuron death in an ALS mouse model, Neuropharmacology, 2012, 62(7), 2346–2352 CrossRef CAS PubMed.
- N. E. Sanjana, O. Shalem and F. Zhang, Improved vectors and genome-wide libraries for CRISPR screening, Nat. Methods, 2014, 11(8), 783–784 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cb00145k |
| This journal is © The Royal Society of Chemistry 2022 |