Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A pH-responsive, endosomolytic liposome functionalized with membrane-anchoring, comb-like pseudopeptides for enhanced intracellular delivery and cancer treatment†
Siyuan
Chen
 ab,
Gabriella
Morrison
ab,
Gabriella
Morrison
 a,
Wenyuan
Liu
b,
Apanpreet
Kaur
a and
Rongjun
Chen
a,
Wenyuan
Liu
b,
Apanpreet
Kaur
a and
Rongjun
Chen
 *a
*a
aDepartment of Chemical Engineering, Imperial College London, South Kensington Campus, London, SW7 2AZ, UK. E-mail: rongjun.chen@imperial.ac.uk; Fax: +44 (0)2075945638; Tel: +44 (0)20 75942070
bResearch Institute for Biomaterials, Tech Institute for Advanced Materials, College of Materials Science and Engineering, Suqian Advanced Materials Industry Technology Innovation Center, NJTech-BARTY Joint Research Center for Innovative Medical Technology, Nanjing Tech University, Nanjing 212000, China
First published on 17th October 2022
Abstract
Low intracellular delivery efficiency and multidrug resistance are among major barriers to effective cancer therapy. Herein, we report a novel, virus-mimicking, endosomolytic liposomal drug-delivery platform to address these two key challenges. The pH-responsive, comb-like pseudopeptides were prepared by grafting relatively long alkyl side chains onto a polyamide, poly(L-lysine isophthalamide), to mimic fusogenic peptides in viral spikes. The cholesterol-containing liposome, which mimics the viral envelope, was readily coated with these pseudopeptides due to their hydrophobic side chains acting as membrane anchors. These endosomolytic pseudopeptides displayed high adsorption onto the liposomal membrane and enabled the significantly higher cellular uptake. The virus-mimicking system showed a pH-triggered content-release profile which could be manipulated by varying the structure and concentration of the adsorbed polymers. The endosomolytic ability of the multifunctional liposome and its use for efficient intracellular delivery of the widely used anticancer drug doxorubicin (DOX) were demonstrated. The virus-mimicking liposomal system with DOX encapsulation exhibited considerably higher potency against HeLa cervical cancer cells, A549 lung cancer cells, MES-SA uterus cancer cells, and MES-SA/DX5 multidrug-resistant cancer cells than DOX-loaded bare liposomes and free DOX. These results suggest its potential applications for enhanced cytoplasmic delivery and cancer treatment.
Introduction
Multidrug resistance is a major factor in the failure of many chemotherapeutic drugs in cancer treatment.1,2 A variety of multidrug resistance mechanisms have been reported, including over-expression of efflux pumps such as P-glycoprotein (also known as multidrug resistance protein 1, MDR1), breast cancer resistance protein (BCRP), and multidrug resistance associated protein 1 (MRP1).3,4 Certain cancers can develop multidrug resistance toward a broad spectrum of anticancer drugs, such as doxorubicin (DOX), vincristine, etoposide and paclitaxel.3,5 A high dose of cytotoxic drugs is usually required to overcome multidrug resistance, thus resulting in severe side effects. One attractive strategy to improve cancer treatment is utilization of intelligent nanoscale drug delivery systems which can bypass drug efflux pumps by changing the cellular entry pathway from simple diffusion to endocytosis, and can also enhance the intracellular delivery efficiency.6–9 A number of stimuli-responsive delivery systems that can achieve a sufficiently high concentration of intracellular chemotherapeutic drugs by endocytosis have demonstrated their ability to overcome multidrug resistance, such as pH/redox dual-responsive polyplexes,10 pH-sensitive micelles,11 and pH/near-infrared light dual-responsive DNA-conjugated gold nanoparticles.12–14In nature, influenza viruses have evolved as sophisticated, efficient gene delivery systems over millennia. The anionic fusogenic peptides in their protein coats can be transformed to a membrane-disruptive state upon acidification in endosomes, resulting release of the nucleic cargo from endosomes into the cytoplasm.15,16 Inspired by this, researchers have been motivated to develop pH-responsive liposomes via surface modification with fusogentic peptides.17 However, safety concerns of the viral fusogenic peptides remain a major hurdle to their clinical application. Instead, surface modification of liposomes with pH-sensitive, membrane-active, anionic synthetic polymers is regarded as a more cost-effective and safer strategy.18 Polymer chains can be anchored on the liposomal surface via electrostatic interaction or insertion of hydrophobic groups into the lipid bilayer.19 Through post-modification of already formulated liposomes, polymers could be localized mainly on the outer surface of liposomes, thus resulting in more inner core space available for loading of drugs.20
A number of membrane proteins can be immobilized on the cell membrane through the utilization of prenyl or/and alkyl chains as membrane anchors.21 Saturated alkyl chains have been reported to provide stronger binding to phospholipid membranes than unsaturated prenyl chains.22 Therefore, researchers have conjugated alkyl chains to synthetic polymers for surface modification of multifunctional liposomes.23 A variety of alkyl anchor-bearing polymers, such as poly(vinyl alcohol)-alkyl,24 poly(N-isopropylacrylamide-co-methacrylic acid-co-octadecyl acrylate)25 and alkylated derivatives of poly(α-ethylacrylic acid),26 have been demonstrated to display stronger interaction with lipid membranes compared to their unmodified counterparts. However, those synthetic vinyl polymers are not readily biodegradable, which may limit their potential clinical application.27
We have recently reported a series of pH-responsive, comb-like, anionic pseudopeptides by grafting hydrophobic decylamine (NDA) onto carboxyl groups pendant to the backbone of a biodegradable, metabolite-derived polyamide, poly(L-lysine isophthalamide) (PLP), at different degrees of substitutions.28,29 Like membrane-anchoring proteins, these comb-like pseudopeptidic polymers contain relatively long aliphatic side chains, which can act as hydrophobic anchors to enhance the polymer-membrane interaction. These pseudopeptides can display a pH-dependent coil-to-globule conformational change and consequent endosomolytic activity due to the protonation of pendant carboxyl groups upon pH reduction, which resembles pH-responsive anionic fusogenic peptides at the N-terminal region of the haemagglutinin-2 protein (HA2) subunit of an influenza virus.30 The optimal polymer containing 18 mol% NDA, PLP-NDA18, was non membrane-lytic at pH 7.4 but displayed superior membrane destabilization activity within the pH range typical of endosomes. PLP-NDA18 caused almost complete hemolysis at pH 5.5 within only 20 min, while only 0.5% hemolysis was observed for its linear counterpart PLP under the same condition. The endosomolytic pseudopeptide has been successfully applied for cytoplasmic delivery into a variety of cell types.28 All these characteristics indicate that the membrane-anchoring PLP-NDA18 is an ideal candidate for the liposomal surface modification to enhance intracellular drug delivery.
A virus-mimicking, endosomolytic liposomal system, whereby the surface of a cholesterol-containing liposome is coated with the viral-peptide-mimicking PP75 pseudopeptide bearing L-phenylalanine (Phe) pendant to the PLP backbone, has previously been developed in our group.31–33 Herein, we report a new multifunctional liposomal platform (Scheme 1) which consists of the pH-responsive, membrane-anchoring, comb-like PLP-NDA pseudopeptides mimicking the fusogenic peptides in the viral spikes, the liposomal bilayer structure mimicking the viral envelope, and the incorporated drug payload mimicking the viral genome. This new virus-mimicking liposomal system displayed more controllable and improved polymer coating and consequently enhanced intracellular delivery. Compared to the PP75-modified liposomal system, the comb-like, endosomolytic PLP-NDA pseudopeptides exhibited much higher adsorption to the liposomal surface due to the favorable membrane anchoring through their hydrophobic relatively long alkyl side chains. The pH-responsive drug release profile and liposomal size change were investigated, and the drug release mechanism was elucidated. The intracellular trafficking and endosomal escape of the multifunctional liposomes were confirmed by confocal microscopy and hemolysis assay, and their cellular uptake was quantitatively analyzed by flow cytometry. The intracellular delivery of DOX by the novel virus-mimicking liposomal system and its potency against four different cancer cell lines including a multidrug resistant cell line were assessed and compared with the DOX-loaded bare liposomes and free DOX.
 | ||
| Scheme 1 Schematic of the pH-responsive, endosomolytic liposome with the PLP-NDA pseudopeptide surface coating. | ||
Experimental
Materials
Defibrinated sheep red blood cells (RBCs) were purchased from TCS Biosciences Ltd (Buckingham, UK). Iso-phthaloyl chloride, Dulbecco's modified Eagle's medium (DMEM), cholesterol, Dulbecco's phosphate buffered saline (D-PBS), calcein, L-α-phosphatidylcholine from egg yolk (EPC), McCoy's 5A modified medium, 6-aminofluorescein, penicillin, fetal bovine serum (FBS), DOX and NDA were obtained from Sigma-Aldrich (Dorset, UK). Hoechst 33342, LysoTracker® deep red, alamar blue, N,N-dimethylformamide, 4-dimethylaminopyridine (DMAP), Texas Red® hydrazide, dimethyl sulfoxide (DMSO), Lissamine™ rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (Rh-PE), triethylamine, sodium chloride, N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (NBD-PE) were obtained from Fisher Scientific (Loughborough, UK). d6-DMSO, sodium hydroxide, anhydrous ethanol, chloroform, sodium citrate dihydrate acetone, sodium phosphate, potassium carbonate and diethyl ether were obtained from VWR (Lutterworth, UK). Triton® X-100, N,N′-dicyclohexylcarbodiimide (DCC), L-lysine methyl ester dihydrochloride and N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid) (HEPES) were obtained from Alfa Aesar (Heysham, UK). The FITC annexin V apoptosis detection kit with 7-amino-actinomycin D (7-AAD) was purchased from BioLegend (San Diego, US).Synthesis and characterization of pseudopeptidic polymers
PLP34,35 and its alkylated derivatives grafted with NDA at various degrees of substitutions, such as PLP-NDA3, PLP-NDA10, and PLP-NDA18,28 were synthesized according to the previously published methods. Briefly, was prepared through the single-phase polymerization of aqueous L-lysine methyl ester·2HCL with an equivalent amount of iso-phthaloyl chloride in acetone followed by hydrolysis in DMSO with ethanolic sodium hydroxide.34 NDA was then conjugated onto the PLP backbone at various degrees of substitution via DCC/DMAP coupling. Solid impurities were removed by vacuum filtration and the supernatant was added with 5 wt% NaOH in anhydrous ethanol. PLP-NDA polymers such as PLP-NDA3, PLP-NDA10 and PLP-NDA18 were collected after precipitation into five volumes of diethyl ether, purified and lyophilized. The numbers, 3, 10 and 18, denote to the actual degrees of grafting, i.e. molar percentages of NDA relative to the pendant carboxylic acid groups on the PLP backbone, as determined using the ratio of the integral 0.77–0.91 ppm to the integral 7.45–7.64 ppm in their 1H NMR spectra in d6-DMSO (Fig. S1†) recorded on a 400 MHz NMR spectrometer (Bruker, Germany).28 The molecular weight and polydispersity index (PDI) of PLP (Mw = 35.7 kDa, Mn = 17.9 kDa, PDI = 1.99) were determined according to our previous publication.35 The molecular weights of the comb-like pseudopeptides were calculated based on their actual degrees of grafting, e.g., PLP-NDA3 (Mw = 36.3 kDa, PDI = 1.99), PLP-NDA10 (Mw = 37.7 kDa, PDI = 1.99), and PLP-NDA18 (Mw = 39.3 kDa, PDI = 1.99). The FT-IR spectra of PLP, PLP-NDA3, PLP-NDA10 and PLP-NDA18 in acid form (Fig. S2†) were recorded on a Spectrum 100 FT-IR Spectrometer (PerkinElmer, USA), with characteristic peaks located at 3292 cm−1 (N–H str and O–H str), 1724 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O acid str), 1629 cm−1 and 1529 cm−1 (amide bands I and II).28 The fluorophore-labelled pseudopeptides were synthesized by grafting 6-aminofluorescein onto the pendant carboxylic acid groups at a grafting degree of 2 mol% via DCC/DMAP-mediated coupling chemistry.
O acid str), 1629 cm−1 and 1529 cm−1 (amide bands I and II).28 The fluorophore-labelled pseudopeptides were synthesized by grafting 6-aminofluorescein onto the pendant carboxylic acid groups at a grafting degree of 2 mol% via DCC/DMAP-mediated coupling chemistry.
Preparation of multifunctional liposomes
Cholesterol and EPC were dissolved in chloroform at a fixed ratio of 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 (mol
60 (mol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mol). The organic solvent was removed by rotary evaporation. The obtained lipid film was hydrated with HBS buffer (5 mL, 150 mM NaCl, 20 mM HEPES, pH 7.4) in the absence or presence of model drugs (e.g. 100 mM calcein or 1 mg mL−1 DOX) at 40 °C for 1 h. The size of the resulting bare liposomes was controlled to be around 100 nm by sonication (Sonicator, 120 watt, pulse mode, Fisher Scientific) for 20 min. Free calcein or DOX was removed by gel filtration chromatography using Sephadex G-50. The sample was then stored at 4 °C after filter-sterilization using a 0.22 μm filter.
mol). The organic solvent was removed by rotary evaporation. The obtained lipid film was hydrated with HBS buffer (5 mL, 150 mM NaCl, 20 mM HEPES, pH 7.4) in the absence or presence of model drugs (e.g. 100 mM calcein or 1 mg mL−1 DOX) at 40 °C for 1 h. The size of the resulting bare liposomes was controlled to be around 100 nm by sonication (Sonicator, 120 watt, pulse mode, Fisher Scientific) for 20 min. Free calcein or DOX was removed by gel filtration chromatography using Sephadex G-50. The sample was then stored at 4 °C after filter-sterilization using a 0.22 μm filter.
pH-Responsive multifunctional liposomes were prepared by incubating bare liposomes with a specific PLP-NDA polymer solution in HBS buffer (pH 7.4) at a certain concentration overnight at room temperature. Dialysis was performed using Float-A-Lyzer® (MWCO 300 kDa, Spectrumlabs, USA) against the HBS buffer at pH 7.4 for 7 h to remove the free polymer. The cholesterol proportion of 40 mol% was chosen to minimize the leakage of payloads out of the multifunctional liposomes at pH 7.4, whilst ensuring significant pH-responsive payload release behavior upon acidification of PLP-NDA polymers.33
Polymer adsorption measurement
The polymer adsorption on the liposomal surface was quantified using PLP-NDA polymers conjugated with 6-aminofluorescein at a grafting degree of 2 mol%. The fluorescence intensity of the 6-aminofluorescein labelled polymer adsorbed on the liposomal surface after purification was measured by a spectrofluorometer (GloMax®-Multi Detection System, Promega, USA) at the excitation wavelength of 490 nm and the emission wavelength of 510–570 nm. The total amount of the polymer adsorbed on the liposomal surface was calculated based on the calibration curve of the fluorescent polymer concentration versus fluorescence intensity.pH-Dependent payload release
The pH-dependent payload release behavior of the multifunctional liposomes was examined using a calcein dequenching assay.33 Calcein (100 mM, self-quenching concentration) was loaded into the liposomes. The pH of the multifunctional liposomes was adjusted to the desired value by titration with 1 wt% HCl. An aliquot was withdrawn and equilibrated at 37 °C for 20 min. To avoid the interference of pH on the fluorescence intensity, the sample pH was adjusted back to neutral and the fluorescence intensity was measured by the spectrofluorometer (GloMax®-Multi Detection System, Promega, USA) at the excitation wavelength of 490 nm and the emission wavelength of 510–570 nm. Triton® X-100 was used to totally disrupt liposomal membranes in a positive control sample. The percentage of released calcein was calculated according to the previously published method.33Particle size and morphology
Dynamic light scattering (DLS) was applied to investigate the hydrodynamic sizes of the liposomes with or without surface modification at specific pHs. The liposomal samples were diluted with specific citrate or HBS buffer to reach certain pH. The size measurement was taken after 5 min equilibration and analyzed by DLS (137°, repeated 11 times, Zetasizer Nano S, Malvern, UK) in 10 mm diameter cells. The pH-dependent reversible size change was performed by tuning the pH between 7.4 and 4.5 using 0.1 M NaOH or 1 wt% HCl for several cycles. Size measurement by DLS was performed after 5 min equilibration of each titration.Transmission electron microscopy (TEM) (JEOL JEM 2100F) was applied to observe the morphology of liposomes with or without polymer modification. The liposome samples were diluted for certain times and dropped on a carbon-coated cooper grid (Agar, 300 Mesh). 2% phosphotungstic acid was used to stain the samples for 2 min and air-dried at room temperature.
To visualize the liposomal size change, liposomes were modified with the fluorphore-labelled polymer, diluted with citrate or HBS buffer at a specific pH, and imaged by laser scanning confocal microscopy (Zeiss LSM-510 inverted laser scanning confocal microscope, Germany) at the excitation wavelength of 488 nm and the emission wavelength of 535 nm.
Lipid mixing assay
A lipid mixing assay was carried out by monitoring fluorescence resonance energy transfer (FRET)36 to investigate whether membrane fusion occurred in the pH-triggered liposomal membrane destabilization. Briefly, the liposomal membrane was incorporated with the donor lipid NBD-PE (1 mol%) and the acceptor lipid Rh-PE (1 mol%), and then coated with the PLP-NDA pseudopeptides. The resulting probe liposomes were mixed with the unlabeled liposomes at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (mol/mol) at different pHs for various time durations. The donor NBD fluorescence was detected at λex = 490 nm and λem = 510–570 nm with the spectrofluorometer. The lipid mixing percentage was calculated using the following equation.
9 (mol/mol) at different pHs for various time durations. The donor NBD fluorescence was detected at λex = 490 nm and λem = 510–570 nm with the spectrofluorometer. The lipid mixing percentage was calculated using the following equation. | (1) |
Hemolysis assay
The membrane disruptive activity of the liposomes was examined by hemolysis assay31 to investigate whether the comb-like pseudopeptides which were adsorbed on the liposomal surface retained their pH-responsive endosomolytic activity. After three times of wash with 150 mM NaCl aqueous solution, RBCs were resuspended in the liposomal solutions in citrate or phosphate buffer at specific pHs to a final concentration of 1–2 × 108 RBCs mL−1. RBCs resuspended in deionized water or buffer alone were used as positive and negative controls, respectively. The samples were incubated at 37 °C with shaking for 1 h, and then centrifuged at 4000 rpm for 4 min. The relative hemolysis was calculated based on the absorbance of the supernatant at 541 nm, measured with a UV-Vis spectrophotometer (GENESYS™ 10S UV-Vis, Thermo Scientific, USA).Cell culture
HeLa human cervical cancer cells and A549 human lung cancer cells were grown in DMEM supplemented with 100 U mL−1 penicillin and 10% (v/v) FBS unless specified otherwise. HeLa and A549 cells were trypsinized using trypsin-EDTA and maintained at 37 °C in a humidified incubator containing 5% CO2.MES-SA human uterus cancer cells and multidrug resistant cancer cells MES-SA/DX5 were cultured in McCoy's 5a medium containing 100 U mL−1 penicillin and 10% (v/v) FBS unless specified otherwise. MES-SA and MES-SA/DX5 cells were detached using EDTA solution (0.8 mM disodium EDTA, 68.5 mM NaCl, 6.7 mM sodium bicarbonate, 5.6 mM glucose and 5.4 mM KCl) and maintained at 37 °C in a humidified incubator containing 5% CO2.
Laser scanning confocal microscopy
In order to assess the effect of the liposomal coating with the PLP-NDA pseudopeptides on intracellular trafficking, fluorescent liposomes were prepared by incorporating Rh-PE in the liposomal membrane and visualized by laser scanning confocal microscopy. 2 mL of HeLa cells (1 × 105 cells per mL) were seeded into a 35 mm glass bottom petri-dish (MatTek, USA) and cultured for 24 h. The HeLa cells were treated with the serum-free DMEM containing fluorescent liposomes (1 mol% Rh-PE, 0.22 μm filter-sterilized) with or without PLP-NDA coating. After 1 h of treatment, the cells were washed with D-PBS (containing calcium and magnesium to avoid cell detachment during the washing process) for three times and further incubated with serum-free DMEM for 3 h. The cells were then stained with LysoTracker deep red (50 nM) and Hoechst 33342 (1.6 μM) and imaged using the inverted laser scanning confocal microscope (Zeiss LSM-510, Germany). Rhodamine was excited at 543 nm and the emission at 580 nm was collected. Lysotracker deep red and Hoechst were excited at 647 and 405 nm, respectively, and their respective emissions at 668 and 461 nm were collected. Colocalization of the PLP-NDA18-coated fluorescent liposomes with lysotracker was analyzed by means of the Pearson's correlation coefficient (Rr) using the “colocalization finder” plugin of the ImageJ software.37The intracellular delivery of DOX to a variety of cell lines was explored by laser scanning confocal microscopy. Cells (2 mL, 1 × 105 cells per mL) were seeded into a petri-dish (35 mm, glass bottom) and cultured for 24 h. The spent medium was replaced with 1 mL of the serum-free culture medium containing free DOX or DOX-loaded liposomal samples with or without PLP-NDA coating (0.22 μm filter-sterilized). The DOX concentration for treatment of HeLa, A549 and MES-SA cells was fixed at 2.5 μM. Due to the multidrug resistance characteristics, MES-SA/DX5 cells were treated with samples at a fixed DOX concentration of 5 μM in order to achieve a sufficient fluorescence signal. The cells were then washed with D-PBS containing calcium and magnesium after 1 h of incubation and stained with Hoechst 33342 (1.6 μM). The cells were imaged using the Zeiss LSM-510 inverted laser scanning confocal microscope (excitation wavelength at 480 nm, emission wavelength at 560 nm).
Flow cytometry
Flow cytometry was employed to quantitatively investigate the cellular uptake of the liposomes with or without PLP-NDA coating. HeLa cells were cultured in a 6-well plate (3 × 105 cells per well) for 24 h and then treated with 1 mL of the serum-free DMEM containing fluorescently labelled liposomes (1 mol% Rh-PE) with or without PLP-NDA coating (0.22 μm filter-sterilized). The cells were washed with D-PBS containing calcium and magnesium for three times after 1 h of treatment and then further incubated with serum-free DMEM medium for 3 h. The cells were trypsinized and supernatants were removed by centrifugation at 1000 rpm. The cell pellets were resuspended in serum-free DMEM and filtered through 40 μm Flowmi™ tip strainers (Bel-Art, USA) to remove aggregates. The flow cytometry measurements were carried out by a cell analyzer (LSRFortessa, BD, USA) with an excitation wavelength of 543 nm.The intracellular delivery of DOX was assessed by flow cytometry. Cells were cultured in a 6-well plate (5 × 105 cells per well) for 24 h. The spent medium was replaced with 1 mL serum-free culture medium containing free DOX or DOX-loaded liposomes with or without PLP-NDA coating (0.22 μm filter-sterilized). The DOX concentration for treatment of HeLa, A549 and MES-SA cells was fixed at 2.5 μM, whilst MES-SA/DX5 cells were incubated with samples at a fixed DOX concentration of 5 μM. The cells were washed with D-PBS containing calcium and magnesium for three times after 1 h of treatment, detached by trypsin or EDTA and the supernatants were removed by centrifugation. The cell pellets were resuspended and filtered before measurements by flow cytometry with an excitation wavelength of 488 nm.
In vitro cytotoxicity
The cytotoxicity of the multifunctional liposomes with or without DOX payload was studied by Alamar Blue assay. Cells (104 cells per well) were cultured in a 96-well plate (Corning, USA) for 24 h. The spent medium was replaced with 0.1 mL of the filter-sterilized sample solutions in serum-free culture medium. The negative control was performed by incubating the cells with 0.1 mL of serum-free culture medium and the positive control was prepared by complete cell lysis with 1% Triton® X-100. After incubation for a specific time duration, the sample solutions were completely removed, and the cells were washed three times with D-PBS containing calcium and magnesium. Only for the cells treated with DOX-containing samples, there was further incubation with the complete medium for 24 h. The plate with replenished complete medium containing alamar blue (10%, v/v) was then incubated for 4 h. The fluorescence in each well was quantified by the spectrofluorometer (excitation wavelength at 525 nm, emission wavelength at 580–640 nm). The half inhibitory concentrations (IC50) were calculated from the concentration-dependent cell viability curves.The in vitro cytotoxicity was further assessed by flow cytometry. A549 cells were cultured for 24 h, followed by 24 h of treatment with serum-free culture medium containing 2.5 μM free DOX or DOX-loaded liposomes with or without PLP-NDA coating. The cells were washed, detached and resuspended as described above and stained with Annexin V and 7-AAD for 30 min following the manufacturer's instructions. The cells were measured by flow cytometry with an excitation wavelength of 488 nm.
Statistical analysis
Results and graphical data are presented as mean ± standard deviations. Two tailed Student's t test was carried out to evaluate the statistical significance of difference. A value of P < 0.05 was considered to be statistically significant and is shown as follows: *0.01 < p ≤ 0.05; **0.001 < p ≤ 0.01; and ***p ≤ 0.001. All data points were repeated in triplicate (n = 3).Results and discussion
pH-Triggered payload release
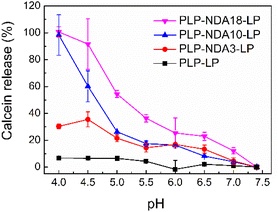
pH-Responsive liposomes can be developed by surface modification of already formulated liposomes with pH-sensitive polymers.38 Hydrophobic interaction is one major driving force controlling adsorption of polymers onto the surface of liposomes.39 Grafting relatively long hydrophobic alkyl chains, which act as membrane anchors, onto the PLP backbone can increase the hydrophobicity of the amphiphilic pseudopeptide significantly, thus leading to considerable enhancement of its hydrophobic interaction with the neutral EPC/cholesterol liposomal membrane. Cholesterol plays an important role in altering the permeability of the lipid membrane. It can tighten the alkyl chains of phospholipids in the lipid bilayer, and induce a high order degree in the membrane resulting in a pronounced thickening of the lipid bilayer, thus increase the membrane rigidity and decrease the membrane permeability.33,40 Upon pH reduction, protonation of the pendant carboxyl groups can cause the coil-to-globule conformational change of the adsorbed polymers, thus resulting in the enhanced membrane destabilization and consequent pH-dependent content release from the modified liposomes. As shown in Fig. 1, the PLP-coated liposome exhibited almost no calcein release throughout the pH range (4.0–7.4) tested. As comparison, pH-dependent release profiles were observed for all multifunctional liposomes coated with the comb-like, membrane-anchoring pseudopeptides containing different amounts of relatively long hydrophobic NDA side chains. These PLP-NDA-coated liposomes displayed essentially no calcein release at pH 7.4 whilst significantly increased payload release upon pH reduction. In addition, the pH-responsive release capacity of the multifunctional liposomes was enhanced with increasing the amount of NDA side chains pendant to the PLP backbone. The liposome coated with PLP-NDA18 displayed the superior release behavior, causing almost complete payload release at pH 4.5. This is due to the improved polymer-cell membrane interaction as a result of the enhanced hydrophobicity of the amphiphilic pseudopeptide at a higher degree of grafting.23,26 It is consistent with our previously reported finding that PLP-NDA18 exhibited the stronger cell membrane interaction and higher hemolysis at acidic pHs than PLP-NDA10, PLP-NDA3 and PLP with a lower amount or no NDA side chains as membrane anchors.28 This pH-responsive release profile is ideal for a smart drug delivery system that is stable during blood circulation but could quickly release its payload in response to the environmental pH change.41 Further increasing the degree of grafting above 18 mol% NDA could enable PLP-NDA to significantly enhance membrane activity at physiological pH and thus lead to the unwanted payload leakage at pH 7.4. Therefore, PLP-NDA18 was chosen as an optimal polymer for preparing the virus-mimicking liposomes.Compared with the amphiphilic PP polymers containing hydrophobic amino acids Phe pendant to the PLP backbone,33 a lower degree of grafting is required for the comb-like, membrane-anchoring PLP-NDA polymers with the purpose to develop the polymer-coated, pH-responsive liposomes. The virus-mimicking liposomes coated with PLP-NDA3 (molar percentage of NDA at 3% relative to the pendant carboxylic acid groups on the PLP backbone) showed 5.5 times higher calcein release than the liposomes coated with PP25 (molar percentage of Phe at 25%) at pH 4.5. With increasing the molar degree of grafting of NDA to 18%, almost all the liposomal content was released at pH 4.5, which was comparable to the pH-responsive release capacity of the virus-mimicking liposomes coated with PP75 with an actual molar degree of grafting of Phe at 63.4%. Fluorescence measurement using 6-aminofluorescein-labeled PLP-NDA18 detected 87.8 μM polymer adsorbed on the liposomal membrane, reaching an adsorption efficiency of 34.5%. This was 1.5 times more efficient than the liposomal surface coating with PP75 under the same condition. It was interesting to note that the pH-responsive release profile of the PLP-NDA18-coated liposomes remained similar with decreasing the concentration of the adsorbed PLP-NDA18 to 6.1 μM, comparable with the liposomes coated with 58.2 μM PP75.33 Thus, lower amount of PLP-NDA18 was needed for the liposomal modification in order to achieve the same level of pH-responsiveness compared to the surface coating with PP75. These results are consistent with the findings that alkyl side chains play an important role in facilitating the binding of various membrane proteins and antimicrobial peptides/polymers to lipid membranes.42 This is because that long alkyl chains could enable strong membrane anchorage through deep membrane insertion.21 The hydrophobic amino acid Phe with an aromatic group has been shown to enhance the interaction with the lipid bilayer.43,44 However, Großauer et al. have found that the hydrophobic alkyl side chains, such as octanoyl (C8) of the peptide hormone ghrelin can play a more important role than Phe in membrane binding.45
pH-Triggered size change
As shown in Fig. S3A,† the mean particle size of bare liposomes characterized by DLS was 77.1 ± 0.7 nm. After surface modification with PLP-NDA18, the DLS size increased to 89.2 ± 0.5 nm (Fig. S3B†), further confirming the successful coating of PLP-NDA18 on the outer surface of the liposomal membrane. No significant change in morphology was observed by TEM between the bare liposomes (Fig. S3A†) and PLP-NDA18 modified liposomes (Fig. S3B†). Fig. 2A shows that the particle size of the unmodified liposomes remained the same within the pH range (4.0–7.4) tested. Negligible pH-dependent size change was observed for the liposomes coated with PLP and PLP-NDA3. However, the PLP-NDA10-coated liposomes displayed a noticeable size increase at pH 4.0, whilst for the liposomes coated with PLP-NDA18, a remarkable size change was observed in the range 4.0 ≤ pH ≤ 5.5. The pH-induced size change was also confirmed by confocal microscopy. As seen in Fig. S4,† a homogeneous distribution of tiny green dots was observed at pH ranging from 6.0 to 7.4, indicative of the well-dispersed liposomes modified with FITC-labelled PLP-NDA18. Aggregates were visualized once the pH was dropped below 5.5, which was in agreement with the size change measured by DLS (Fig. 2A). We have previously reported that the increasing degree of substitution with relatively long hydrophobic NDA side chains enhanced polymer hydrophobicity,28 which has been reported to increase inter-vesicular interactions and the consequent liposomal aggregation.33,46A lipid mixing assay was then employed to investigate whether membrane fusion was involved during liposomal aggregation by measuring the FRET between the donor NBD-PE and the receptor Rh-PE upon incubation of the probe liposomes (containing 1 mol% NBD-PE and 1 mol% Rh-PE) with the unlabeled liposomes. As shown in Fig. 2B, almost no lipid mixing was detected at pH 7.4 or 4.5 for the liposomes with or without PLP-NDA18 coating, indicative of no obvious liposomal membrane fusion. This was validated by the reversible liposomal size change in response to pH. As seen in Fig. 2C, once pH was decreased from 7.4 to 4.5, a dramatic size increase was observed for the liposomes coated with PLP-NDA18. Upon titration of the pH from 4.5 back to 7.4, within only 5 min the particle decreased to its original size. This reversible size change was repeated for several titration circles, further confirming that the liposomal size increase at acidic pHs was not induced by membrane fusion.33
The polymer-bridged liposomal network model33 could be applied to interpret the pH-triggered, reversible size change of the liposomes surface-coated with the comb-like polymers. Specifically, a number of hydrophobic relatively long alkyl side chains pendant onto the backbone of PLP-NDA18 could facilitate its anchoring with more than one liposome, resulting in the formation of a polymer-bridged liposomal network. At pH 7.4 no significant liposomal size change was observed due to the expanded polymer structure as a result of electrostatic repulsion between negatively charged carboxylate groups. With decreasing pH, the comb-like polymers were neutralized and displayed a coil-to-globule conformational change.28 The formation of compact polymer structure led to aggregation of the polymer-bridged liposomal network, which resulted in a dramatic size increase.47 Once pH was changed back to 7.4, the particle size was reduced because of the expansion of polymer structure. The reversibility of the liposomal size change further confirms the successful coating of PLP-NDA18 on the liposomal surface and suggests that PLP-NDA18 facilitated efficient payload release from the multifunctional liposomes at acidic pHs (Fig. 1) without permanent damage of the lipid membrane.28 It is also worth mentioning that the significant release of liposomal content could be achieved at some pHs where no obvious size change was detected (Fig. 1 and 2A), suggesting that payload release was not necessarily dependent on the change of colloidal stability.
Cellular uptake
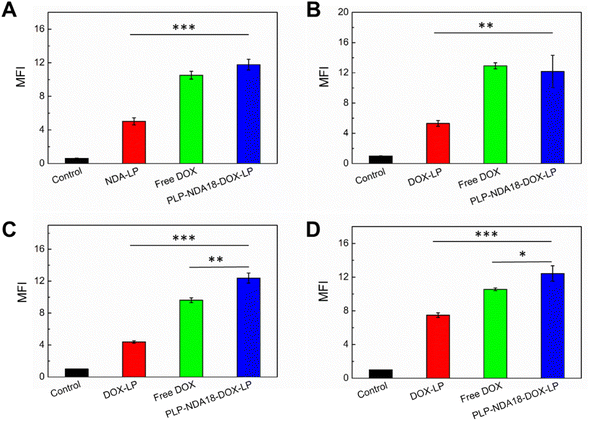
The bare liposomes and the virus-mimicking liposomes were fluorescently labelled with 1 mol% Rh-PE to investigate their cellular uptake and intracellular trafficking. As shown in Fig. 3A, both liposomes appeared as red punctate spots, with partial colocalization with the green lysotracker (leading to yellow dots), suggesting that they were internalized mainly through endocytosis. Thus, unlike many chemotherapeutic drugs which enter cells by diffusion and could be extruded by efflux pumps overexpressed on the plasma membrane,41 liposomes can shuttle loaded drugs across the cellular membrane via endocytosis and thereby avoid the drug efflux.41,48The intracellular distribution of red fluorescence, shown in Fig. 3A, suggests that the PLP-NDA18-coated liposomes displayed the significantly enhanced uptake by HeLa cells compared to the bare liposomes. The quantitative flow cytometry analysis (Fig. 3B and C) further confirmed that the mean fluorescence intensity (MFI) of the cells treated with the PLP-NDA18-coated liposomes was 4.7 ± 0.2 times higher than those treated with the bare liposomes. The improved cellular uptake of the PLP-NDA18-coated liposomes could be due to the presence of the negatively charged, comb-like pseudopeptides containing relatively long hydrophobic NDA side chains. Alkyl chains have been utilized as membrane anchors by a variety of membrane proteins to bind with the lipid membrane.49 For the liposomes coated with PLP-NDA18, accessible free NDA chains on the comb-like polymers could facilitate the hydrophobic interaction of the multifunctional liposomes with the cell membrane, thus improving cellular uptake. Furthermore, other researchers have reported that anionic polymers or delivery systems modified with anionic polymers could also display enhanced cell internalization due to their recognition by scavenger receptors that are expressed on the surface of a wide range of cell types including HeLa cells.33,46,50
Furthermore, Fig. 3D shows that some red fluorescent PLP-NDA18-coated liposomes were not colocalized with green lysotracker. The values of Pearson's correlation coefficients at zero or almost zero show no correlation between localization of the virus-mimicking liposomes and lysotracker, indicative of a successful endosomal escape.36 The endosomolytic activity of the virus-mimicking liposomes was validated by the hemolysis assay using RBCs as a model of endosomes.31 As shown in Fig. 3E, the bare liposomes and the PLP-coated liposomes displayed almost no hemolytic activity within the pH range (5.0–7.4) tested. As comparison, the PLP-NDA18-coated liposomes were almost non-hemolytic at pH 7.4, but hemolyzed 92.1 ± 1.5% RBCs at pH 6.0 characteristic of early endosomes. The pH-responsive hemolytic activity of the PLP-NDA18-coated liposomes was similar to that of PLP-NDA18,28 suggesting that the comb-like pseudopeptidic polymers coated on the liposomal surface well-retained their endosomolytic ability.
Intracellular drug delivery
The PLP-NDA18-coated, endosomolytic liposomes were then evaluated for intracellular delivery of a widely used anticancer drug DOX. Quantitative results indicate that the virus-mimicking liposomes had an encapsulation efficiency of 57.3 ± 1.1% and a loading efficiency of 14.8 ± 3.5%. Due to overexpression of drug efflux pumps on the membrane of multidrug resistant MES-SA/DX5 cancer cells,51 relatively weak red fluorescence was observed in MES-SA/DX5 cells compared to HeLa cervical cancer cells, A549 lung cancer cells and MES-SA uterus cancer cells treated with free DOX at the same concentration of 2.5 μM (Fig. 4 and S5†). Thus, higher DOX concentrations were used for treatment of MES-SA/DX5 cells in further experiments. As shown in Fig. 4, the red fluorescence intensity in HeLa cells (Fig. 4A), A549 cells (Fig. 4B), MES-SA cells (Fig. 4C), and MES-SA/DX5 multidrug resistant cancer cells (Fig. 4D) treated with the DOX-loaded bare liposomes was all significantly lower than those treated with free DOX at an equivalent drug concentration. This is because free DOX is usually internalized by cells and enters the nuclei (active site) by rapid diffusion,52 whilst the DOX-loaded liposomes are internalized mainly through endocytosis and the released DOX can enter the nuclei.53 In contrast, the DOX fluorescence intensity in all the four cell lines treated with the PLP-NDA18-coated, DOX-loaded liposomes was comparable to those treated with free DOX, while much higher compared to those treated with the DOX-loaded bare liposomes.The intracellular delivery of DOX was further quantified by flow cytometry. The DOX-loaded multifunctional liposomes enabled considerably enhanced DOX fluorescence in all the four cell lines compared to the DOX-loaded bare liposomes (Fig. S6†). As shown in Fig. 5, the MFI of HeLa, A549, MES-SA and MES-SA/DX5 cells treated with the PLP-NDA18-coated, DOX-loaded liposomes was 2.4 ± 0.3, 2.3 ± 0.5, 2.8 ± 0.1, and 1.7 ± 0.1 times, respectively, higher than those treated with the DOX-loaded bare liposomes. This could be because that the presence of the surface-coated PLP-NDA18 enhanced cellular uptake (Fig. 3A–C) and then the acidification process inside endosomes/lysosomes triggered the membrane-destabilizing activity of PLP-NDA18,28 leading to not only efficient liposomal drug release (Fig. 1) but also efficient endosomal escape (Fig. 3D). Interestingly, the PLP-NDA18-coated liposomal system was especially efficient at delivering DOX into MES-SA (Fig. 5C) and MES-SA/DX5 cancer cells (Fig. 5D), where the intracellular DOX fluorescence intensity was significantly (p < 0.05) higher than the diffusion of free DOX. These suggest that the PLP-NDA18-coated liposomes could facilitate efficient intracellular drug delivery to a variety of cell lines, including multidrug resistant cancer cells.
In vitro cytotoxicity
Alamar Blue assay was carried out to assess the effect of the pH-responsive, endosomolytic liposomes, at a fixed polymer-to-lipid ratio of 2.85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 but varying lipid concentrations up to 300 μM, on the metabolic activity of a variety of cell lines. The bare liposomes and the liposomes coated with PLP-NDA18 were well tolerated by HeLa (Fig. S7A†), A549 (Fig. S7B†) and MES-SA/DX5 (Fig. S7D†) cells after 24 h of treatment within the lipid concentration range (5–300 μM) tested. However, as shown in Fig. S7C,† MES-SA cells which were treated with the PLP-NDA18-coated liposomes at the lipid concentration ≥75 μM (i.e. the concentration of the adsorbed PLP-NDA18 ≥2.1 μM) for 24 h displayed a significantly reduced viability. As comparison, the bare liposomes were well tolerated by MES-SA cells for up to 24 h within the lipid concentration range (5–300 μM) tested. This indicates MES-SA cells which have been reported to be sensitive to various drugs, including doxorubicin, dactinomycin, mitomycin C and bleomycin,54,55 might also be sensitive to the pH-responsive, comb-like PLP-NDA pseudopeptides. Therefore, a tolerant concentration of the PLP-NDA18-coated liposomes against MES-SA cells (and kept fixed for the other cell lines tested) was chosen to investigate the potency of the DOX-loaded virus-mimicking liposomes.
100 but varying lipid concentrations up to 300 μM, on the metabolic activity of a variety of cell lines. The bare liposomes and the liposomes coated with PLP-NDA18 were well tolerated by HeLa (Fig. S7A†), A549 (Fig. S7B†) and MES-SA/DX5 (Fig. S7D†) cells after 24 h of treatment within the lipid concentration range (5–300 μM) tested. However, as shown in Fig. S7C,† MES-SA cells which were treated with the PLP-NDA18-coated liposomes at the lipid concentration ≥75 μM (i.e. the concentration of the adsorbed PLP-NDA18 ≥2.1 μM) for 24 h displayed a significantly reduced viability. As comparison, the bare liposomes were well tolerated by MES-SA cells for up to 24 h within the lipid concentration range (5–300 μM) tested. This indicates MES-SA cells which have been reported to be sensitive to various drugs, including doxorubicin, dactinomycin, mitomycin C and bleomycin,54,55 might also be sensitive to the pH-responsive, comb-like PLP-NDA pseudopeptides. Therefore, a tolerant concentration of the PLP-NDA18-coated liposomes against MES-SA cells (and kept fixed for the other cell lines tested) was chosen to investigate the potency of the DOX-loaded virus-mimicking liposomes.
The potency of the DOX-loaded liposomes against a variety of cancer cell lines was evaluated to assess the therapeutic potential of the pH-responsive, endosomolytic, PLP-NDA18-coated liposomal system. The endocytic mode of cellular uptake of DOX-loaded bare liposomes could reduce anticancer effects as result of not only the decreased internalization but also the entrapment within intracellular vesicles and possible lysosomal degradation of DOX.56 As shown in Fig. 6A, the DOX-loaded bare liposomes showed a much lower cytotoxic effect toward HeLa cells compared to free DOX at an equivalent concentration above 0.5 μM. However, the DOX-loaded, PLP-NDA18-coated liposomes displayed a higher potency against HeLa cells than the DOX-loaded bare liposomes and even free DOX at an equivalent DOX concentration ≥1 μM. The IC50 of the DOX-loaded, PLP-DNA18-coated liposomes against HeLa cells was 0.80 ± 0.01 μM, which was lower than the IC50 of the free DOX (1.25 ± 0.07 μM) and the DOX-loaded bare liposomes (5.62 ± 0.87 μM). Similarly, the DOX-loaded, PLP-NDA18-coated liposomes also displayed higher cytotoxicity toward A549 cells (Fig. 6B) than the DOX-loaded bare liposomes and even free DOX. The enhanced cytotoxicity was further confirmed by flow cytometry, as evidenced by a remarkably decreased number of A549 cells treated with the DOX-loaded, PLP-DNA18-coated liposomes (Fig. S8D†) compared with the cells treated with culture medium alone (Fig. S8A†), free DOX (Fig. S8B†) or DOX-loaded bare liposomes (Fig. S8C†), where the initial cell number in all those four groups was kept the same.
Multidrug resistance remains one of the major barriers to successful cancer therapy.3 Herein, the human uterine sarcoma MES-SA cell line and the multidrug resistant MES-SA/DX5 cell line that derived from MES-SA were employed to further evaluate the therapeutic efficacy of the PLP-NDA18-coated, DOX-loaded liposomes. It has been reported by other researchers that overexpression of P-glycoprotein on the MES-SA/DX5 plasma membrane can extrude DOX out of the cells, and thus result in multidrug resistance.55 As expected, nearly all (92.3 ± 4.2%) multidrug resistant MES-SA/DX5 cells were viable after treatment with a relatively high concentration of free DOX at 5 μM for 24 h (Fig. 6D). In contrast, 5 μM DOX killed nearly all HeLa (Fig. 6A), A549 (Fig. 6B) and MES-SA cells (Fig. 6C) after 24 h of treatment. The multidrug resistance of MES-SA/DX5 cells was also evidenced by a considerably higher IC50 of free DOX at 21.51 ± 1.02 μM (Fig. 6D), than that of free DOX at 0.78 ± 0.04 μM against non-resistant MES-SA cells (Fig. 6C). As comparison, the IC50 of the DOX-loaded, PLP-NDA18-coated liposomes against multidrug resistant MES-SA/DX5 cells was significantly reduced to 7.50 ± 0.62 μM (Fig. 6D), which was 2.9 and 6.0 times lower than that of free DOX and DOX-loaded bare liposomes, respectively. In other words, a remarkably lower dose of DOX was required to achieve the same anticancer effect once it was encapsulated inside the pH-responsive, endosomolytic liposomes coated with the comb-like polymers.
The remarkably enhanced cytotoxicity of the DOX-loaded, PLP-NDA18-coated virus-mimicking liposomes toward multidrug resistant cancer cells could be attributed to several reasons. Firstly, only approximately below 10% of drug payload was released at neutral pH (Fig. 1 and S9†). Majority of the loaded drug was stably retained in the virus-mimicking liposomes and then internalized by cells via endocytosis (Fig. 3A), thereby changing the cellular uptake mode and bypassing the drug efflux pumps.57,58 As seen in Fig. 5C and D, although the free DOX concentration for treatment of multidrug resistant cells MES-SA/DX5 was twice the concentration for MES-SA cells, no significant enhancement in the intracellular DOX MFI was observed. As compassion, when treated with DOX-LP, the MFI of MES-SA/DX5 cells was 1.7 times higher than that of MES-SA cells. This suggests that there is potential to overcome multidrug resistance by liposomal encapsulation of DOX liposomes. Secondly, lower cellular uptake of nanoscale drug delivery systems compared to free small-molecule drugs usually remains a long-standing challenge.16 As a result of surface modification with the membrane-protein-mimicking, comb-like pseudopeptides, the virus-mimicking liposomes displayed significantly improved cellular uptake (Fig. 3A–C) due to the enhanced membrane binding attributed to the “anchor-like” hydrophobic NDA side chains28,49 and the favorable recognition by scavenger receptors.33,46,50 Furthermore, the comb-like pseudopeptidic PLP-NDA18 coated on the liposomal surface well-retained its membrane activity. This enabled the virus-mimicking liposomes to have a synergy of efficient endosomal-escape ability and efficient liposomal drug release behavior in response to pH decrease (Fig. 1, 3A, D, 4 and 5), which can overcome endo-lysosomal sequestration and avoid enzymatic degradation of drugs. A higher intracellular drug delivery rate compared to the drug efflux rate has been reported by other researchers to cause the accumulation of chemotherapeutic agents in the cytoplasm and eventually kill multidrug-resistant cancer cells before the drug efflux pump can exclude all the drug.3,41 Therefore, all those taken together enabled the DOX-loaded, PLP-NDA18-coated liposomes to overcome multidrug resistance and result in high cytotoxicity. Other nanoscale drug delivery systems, such as pH-responsive endsomolytic polymeric micelles, pH-sensitive nanoporous silicon particles, and heparin/protamine/calcium carbonate hybrid nanovesicles have also been reported to be capable of combating multidrug resistance via similar mechanisms.48,51,59–61
Conclusions
A novel virus-mimicking liposomal drug delivery system was successfully developed, with the membrane-anchoring, comb-like pseudopeptides mimicking the fusogenic peptides in the viral spikes, the self-assembled liposomal bilayer structure mimicking the viral envelope, and the encapsulated drug payload mimicking the viral genome. The pH-responsive release profile of the resulting multifunctional liposomes could be manipulated by controlling the structure and concentration of the adsorbed polymers. The liposomes coated with PLP-NDA18 (degree of substitution with NDA at 18 mol%) were demonstrated to display the desired pH-responsive release profile and endosomolytic activity. The multifunctional liposomes had a reversible change of the mean hydrodynamic size in response to pH due to particle aggregation without involvement of membrane fusion and PLP-NDA18 facilitated efficient payload release at acidic pHs without permanent damage of the lipid membrane. The considerably improved intracellular delivery of DOX to HeLa cervical cancer cells, A549 lung cancer cells, MES-SA uterus cancer cells, and MES-SA/DX5 multidrug resistant cancer cells was demonstrated by confocal microscopy and flow cytometry. The potency of the PLP-NDA18-coated, DOX-loaded, virus-mimicking liposomal system against all the four cancer cell lines including the multidrug resistant cell line was shown to be remarkably higher than the DOX-loaded bare liposomes and free DOX, due to the bypass of the efflux mechanism, enhanced cellular uptake, efficient endosomal escape and efficient liposomal drug release. All these characteristics of the novel virus-mimicking liposomes are of critical importance for the applications in intracellular drug delivery and multidrug resistance treatment.Author contributions
R. C. designed and supervised the research; G. M. and A. K. performed the flow cytometry of A549 cells; W. L. carried out the TEM characterization of liposomes; S. C. completed all the other research. S. C. and R. C. analyzed data; S. C. and R. C. wrote the paper. All authors have read, commented on, and approved the final version of the paper.Data access statement
Data underlying this article can be accessed on Zenodo (https://www.zenodo.org) at 10.5281/zenodo.2653262 and is openly available under the Creative Commons CC-BY license.Conflicts of interest
R. C. and S. C. are inventors of the patent WO 2018/011580. The other authors have no conflicts to declare.Acknowledgements
The authors would like to acknowledge the Department of Chemical Engineering at Imperial College London for funding this project (to R. C.) and for providing a fully funded Ph.D. Studentship to S. C. Both G. M. and A. K. were supported by the Engineering and Physical Sciences Research Council Doctoral Training Partnership (EPSRC DTP) Studentship. The authors also acknowledge the financial support from the National Natural Science Foundation of China (No. 32101068 to S. C.) and the Start-up Funding from the College of Materials Science and Engineering, Nanjing Tech University (to S. C.). The authors thank Professor João Cabral for access to dynamic light scattering in the Department of Chemical Engineering at Imperial College London.References
- M. Ramirez, S. Rajaram, R. J. Steininger, D. Osipchuk, M. A. Roth, L. S. Morinishi, L. Evans, W. Ji, C. H. Hsu, K. Thurley, S. Wei, A. Zhou, P. R. Koduru, B. A. Posner, L. F. Wu and S. J. Altschuler, Nat. Commun., 2016, 7, 1–8 CrossRef PubMed.
- Y. G. Assaraf, A. Brozovic, A. C. Gonçalves, D. Jurkovicova, A. Linē, M. Machuqueiro, S. Saponara, A. B. Sarmento-Ribeiro, C. P. R. Xavier and M. H. Vasconcelos, Drug Resistance Updates, 2019, 46, 100645 CrossRef PubMed.
- R. W. Robey, K. M. Pluchino, M. D. Hall, A. T. Fojo, S. E. Bates and M. M. Gottesman, Nat. Rev. Cancer, 2018, 18, 452–464 CrossRef CAS PubMed.
- K. M. Hanssen, M. Haber and J. I. Fletcher, Drug Resistance Updates, 2021, 59, 100795 CrossRef CAS PubMed.
- L. Zhou, H. Wang and Y. Li, Theranostics, 2018, 8, 1059–1074 CrossRef CAS PubMed.
- G. M. Ngandeu Neubi, Y. Opoku-Damoah, X. Gu, Y. Han, J. Zhou and Y. Ding, Biomater. Sci., 2018, 6, 958–973 RSC.
- Z. Su, S. Dong, S. C. Zhao, K. Liu, Y. Tan, X. Jiang, Y. G. Assaraf, B. Qin, Z. S. Chen and C. Zou, Drug Resistance Updates, 2021, 58, 100777 CrossRef CAS PubMed.
- M. May, Nat. Med., 2022, 28, 1100–1102 CrossRef CAS PubMed.
- H. Tan, M. Zhang, Y. Wang, P. Timashev, Y. Zhang, S. Zhang, X. J. Liang and F. Li, Nanotechnology, 2021, 33, 052001 CrossRef PubMed.
- Y. Gao, L. Jia, Q. Wang, H. Hu, X. Zhao, D. Chen and M. Qiao, ACS Appl. Mater. Interfaces, 2019, 11, 16296–16310 CrossRef CAS PubMed.
- Y. Liu, C. Zhou, S. Wei, T. Yang, Y. Lan, A. Cao, J. Yang and Y. Hou, Colloids Surf., B, 2018, 170, 330–340 CrossRef CAS PubMed.
- D. Yang, T. Wang, Z. Su, L. Xue, R. Mo and C. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 22431–22441 CrossRef CAS PubMed.
- L. Song, Y. Guo, D. Roebuck, C. Chen, M. Yang, Z. Yang, S. Sreedharan, C. Glover, J. A. Thomas, D. Liu, S. Guo, R. Chen and D. Zhou, ACS Appl. Mater. Interfaces, 2015, 7, 18707–18716 CrossRef CAS PubMed.
- W. Zhang, F. Wang, Y. Wang, J. Wang, Y. Yu, S. Guo, R. Chen and D. Zhou, J. Controlled Release, 2016, 232, 9–19 CrossRef CAS PubMed.
- P. Lönn, A. D. Kacsinta, X. S. Cui, A. S. Hamil, M. Kaulich, K. Gogoi and S. F. Dowdy, Sci. Rep., 2016, 6, 1–9 CrossRef PubMed.
- M. Somiya, Q. Liu and S. Kuroda, Nanotheranostics, 2017, 1, 415–429 CrossRef PubMed.
- S. Ganta, H. Devalapally, A. Shahiwala and M. Amiji, J. Controlled Release, 2008, 126, 187–204 CrossRef CAS PubMed.
- M. Abri Aghdam, R. Bagheri, J. Mosafer, B. Baradaran, M. Hashemzaei, A. Baghbanzadeh, M. de la Guardia and A. Mokhtarzadeh, J. Controlled Release, 2019, 315, 1–22 CrossRef CAS PubMed.
- V. P. Torchilin, Nat. Rev. Drug Discovery, 2005, 4, 145–160 CrossRef CAS PubMed.
- O. K. Nag and V. Awasthi, Pharmaceutics, 2013, 5, 542–569 CrossRef CAS PubMed.
- M. D. Resh, Prog. Lipid Res., 2016, 63, 120–131 CrossRef CAS PubMed.
- H. Khandelia, J. H. Ipsen and O. G. Mouritsen, Biochim. Biophys. Acta, Biomembr., 2008, 1778, 1528–1536 CrossRef CAS PubMed.
- M. A. Yessine and J. C. Leroux, Adv. Drug Delivery Rev., 2004, 56, 999–1021 CrossRef CAS PubMed.
- Y. Teramura, Y. Kaneda, T. Totani and H. Iwata, Biomaterials, 2008, 29, 1345–1355 CrossRef CAS PubMed.
- Y. J. Hong, H. Y. Lee and J. C. Kim, Colloid Polym. Sci., 2009, 287, 1207–1214 CrossRef CAS.
- T. Chen, D. Mcintosh, Y. He, J. Kim, D. A. Tirrell, P. Scherrer, D. B. Fenske, A. P. Sandhu and P. R. Cullis, Mol. Membr. Biol., 2004, 21, 385–393 CrossRef CAS PubMed.
- R. Duncan, Nat. Rev. Cancer, 2006, 6, 688–701 CrossRef CAS PubMed.
- S. Chen, S. Wang, M. Kopytynski, M. Bachelet and R. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 8021–8029 CrossRef CAS PubMed.
- S. Chen, L. Wu, J. Ren, V. Bemmer, R. Zajicek and R. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 28941–28951 CAS.
- Y. Cheng, R. C. Yumul and S. H. Pun, Angew. Chem., 2016, 128, 12192–12196 CrossRef.
- R. Chen, S. Khormaee, M. E. Eccleston and N. K. H. Slater, Biomaterials, 2009, 30, 1954–1961 CrossRef CAS PubMed.
- S. Zhang, A. Nelson, Z. Coldrick and R. Chen, Langmuir, 2011, 27, 8530–8539 CrossRef CAS PubMed.
- S. Chen and R. Chen, ACS Appl. Mater. Interfaces, 2016, 8, 22457–22467 CrossRef CAS PubMed.
- M. E. Eccleston, S. L. Williams, Z. Yue, R. Chen, C. K. Lee, E. Anikina, C. Pawlyn, M. A. Barrand and N. K. H. Slater, Food Bioprod. Process., 2005, 83, 141–146 CrossRef CAS.
- R. Chen, S. Khormaee, M. E. Eccleston and N. K. H. Slater, Biomacromolecules, 2009, 10, 2601–2608 CrossRef CAS PubMed.
- T. F. Martens, K. Remaut, J. Demeester, S. C. De Smedt and K. Braeckmans, Nano Today, 2014, 9, 344–364 CrossRef CAS.
- F. Cardarelli, D. Pozzi, A. Bifone, C. Marchini and G. Caracciolo, Mol. Pharm., 2012, 9, 334–340 CrossRef CAS PubMed.
- A. E. Felber, M.-H. Dufresne and J.-C. Leroux, Adv. Drug Delivery Rev., 2012, 64, 979–992 CrossRef CAS PubMed.
- K. Andreev, M. W. Martynowycz, M. L. Huang, I. Kuzmenko, W. Bu, K. Kirshenbaum and D. Gidalevitz, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1414–1423 CrossRef CAS PubMed.
- F. de Meyer and B. Smit, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3654–3658 CrossRef CAS PubMed.
- M. Kanamala, W. R. Wilson, M. Yang, B. D. Palmer and Z. Wu, Biomaterials, 2016, 85, 152–167 CrossRef CAS PubMed.
- H. Sato and J. B. Feix, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 1245–1256 CrossRef CAS PubMed.
- H. Hong, S. Park, R. H. F. Jiménez, D. Rinehart and L. K. Tamm, J. Am. Chem. Soc., 2007, 129, 8320–8327 CrossRef CAS PubMed.
- J. L. MacCallum, W. F. D. Bennett and D. P. Tieleman, J. Gen. Physiol., 2007, 129, 371–377 CrossRef CAS PubMed.
- J. Großauer, S. Kosol, E. Schrank and K. Zangger, Bioorg. Med. Chem., 2010, 18, 5483–5488 CrossRef PubMed.
- E. Yuba, A. Harada, Y. Sakanishi and K. Kono, J. Controlled Release, 2011, 149, 72–80 CrossRef CAS PubMed.
- Y. Y. Chieng and S. B. Chen, J. Phys. Chem. B, 2010, 114, 4828–4835 CrossRef CAS PubMed.
- R. Li and Y. Xie, J. Controlled Release, 2017, 251, 49–67 CrossRef CAS PubMed.
- T. A. Rapoport, Nature, 2007, 450, 663–669 CrossRef CAS PubMed.
- X.-P. Yang, M. J. Amar, B. Vaisman, A. V. Bocharov, T. G. Vishnyakova, L. a. Freeman, R. J. Kurlander, A. P. Patterson, L. C. Becker and A. T. Remaley, J. Lipid Res., 2013, 54, 2450–2457 CrossRef CAS PubMed.
- H. Mostoufi, G. Yousefi, A. Tamaddon and O. Firuzi, Colloids Surf., B, 2019, 174, 17–27 CrossRef CAS PubMed.
- L. Niu, F. Zhu, B. Li, L. Zhao, H. Liang, Y. Yan and H. Tan, Mater. Chem. Front., 2018, 2, 1529–1538 RSC.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- E. Wang, M. D. Lee and K. W. Dunn, J. Cell. Physiol., 2000, 184, 263–274 CrossRef CAS PubMed.
- O. Wesolowska, M. Paprocka, J. Kozlak, N. Motohashi, D. Dus and K. Michalak, Anticancer Res., 2005, 25, 383–389 CAS.
- E. Koren, A. Apte, A. Jani and V. P. Torchilin, J. Controlled Release, 2012, 160, 264–273 CrossRef CAS PubMed.
- J. L. Markman, A. Rekechenetskiy, E. Holler and J. Y. Ljubimova, Adv. Drug Delivery Rev., 2013, 65, 1866–1879 CrossRef CAS PubMed.
- L. M. Mu, R. J. Ju, R. Liu, Y. Z. Bu, J. Y. Zhang, X. Q. Li, F. Zeng and W. L. Lu, Adv. Drug Delivery Rev., 2017, 115, 46–56 CrossRef CAS PubMed.
- A. Varela-Moreira, Y. Shi, M. H. A. M. Fens, T. Lammers, W. E. Hennink and R. M. Schiffelers, Mater. Chem. Front., 2017, 1, 1485–1501 RSC.
- R. Xu, G. Zhang, J. Mai, X. Deng, V. Segura-Ibarra, S. Wu, J. Shen, H. Liu, Z. Hu, L. Chen, Y. Huang, E. Koay, Y. Huang, J. Liu, J. E. Ensor, E. Blanco, X. Liu, M. Ferrari and H. Shen, Nat. Biotechnol., 2016, 34, 414–418 CrossRef CAS PubMed.
- M. Q. Gong, J. L. Wu, B. Chen, R. X. Zhuo and S. X. Cheng, Langmuir, 2015, 31, 5115–5122 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm01087a |
| This journal is © The Royal Society of Chemistry 2022 |