Open Access Article
Open Access ArticleStatic and photoresponsive dynamic materials to dissect physical regulation of cellular functions
Jun
Nakanishi
 *abc and
Shota
Yamamoto
*abc and
Shota
Yamamoto
 ad
ad
aResearch Center for Functional Materials, National Institute for Materials Science, Japan. E-mail: NAKANISHI.Jun@nims.go.jp
bGraduate School of Advanced Science and Engineering, Waseda University, Japan
cGraduate School of Advanced Engineering, Tokyo University of Science, Japan
dGraduate School of Arts and Sciences, The University of Tokyo, Japan
First published on 12th September 2022
Abstract
Recent progress in mechanobiology has highlighted the importance of physical cues, such as mechanics, geometry (size), topography, and porosity, in the determination of cellular activities and fates, in addition to biochemical factors derived from their surroundings. In this review, we will first provide an overview of how such fundamental insights are identified by synchronizing the hierarchical nature of biological systems and static materials with tunable physical cues. Thereafter, we will explain the photoresponsive dynamic biomaterials to dissect the spatiotemporal aspects of the dependence of biological functions on physical cues.
1. Introduction
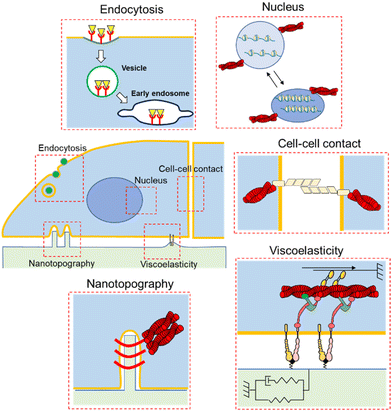
Biological systems regularly encounter artificial materials, such as body implants, blood bags, cell culture dishes, and drug delivery systems (DDSs).1,2 Each scenario evokes a wide range of reactions at the biomaterial interface. At the molecular level, protein adsorption on the material surface is the primary event that determines subsequent refusal responses, such as immune responses and blood coagulation.3 In addition, proteins chemically or physically adsorbed to the material surface can have salutary effects by interacting with cell-surface receptors, evoking intracellular signaling and gene expression regulation.4 Therefore, scientists have sought a wide variety of biomaterials not only to prevent protein adsorption to eliminate undesired rejection reactions (bioinert materials) but also to introduce biological functionalities to accelerate regeneration and healing (bioactive materials).5 In addition to these biochemical or molecular biological aspects, the physical properties of materials, such as mechanics, geometry (size), topography, and porosity, are key features for biomaterial design.6,7 Porous structures of materials are critical in tissue engineering scaffolds for efficient intercommunication of cells and nutrient/waste transport between implants and hosts; this determines the efficiency of functional tissue formation and vascularization. In contrast, depending on the size of DDS nanoparticles, they can travel throughout the body via blood circulation and can enter the cells’ interior via endocytosis and follow intrinsic intracellular trafficking routes.8 Moreover, recent progress in mechanobiology has shed light on the significance of tissue/material mechanics not only for structural robustness and flexibility but also for the direct regulation of cellular signaling and fates depending on their viscoelasticity.9 Therefore, it is crucial to focus on the physical characteristics of materials and hierarchical nature of biological systems to optimize their therapeutic and diagnostic capabilities. Furthermore, biological reactions are dynamic and mediated by the everlasting assembly and disassembly of various molecular players in broad time windows. This ranges from the time scale of signal transduction to much longer ones, like disease progression and aging. In this regard, in addition to precise spatial control of physical cues, stimulus-responsive dynamic materials are useful to replicate time-dependent changes in crosslinking and mechanics of matrices. Synchronization of the material-derived physical cues with such intrinsic spatiotemporal dynamics and time-evolving nature of biological systems is another important criterion for effective biomaterial design.10 In this regard, photoresponsive dynamic materials are the most promising tools for controlling the physical properties of materials at high spatiotemporal resolutions due to the precise and remote-controllable nature of “light”. Especially, the time-resolving nature of the photoresponsive materials can be used to mimic dynamic changes of physical cues occurring in physiological and pathological processes. Whereas the spatially resolving nature of them are useful to replicate heterogenous nature of biological systems, such as the asymmetry, gradients, and hierarchical features. Such engineered materials, either statically or dynamically, are useful not only for biomedical applications but also to further deepening of our understanding of the nanoarchitectonics and dynamics of biological systems.Most earlier reviews mainly focused on how such biomaterials can be used for biomedical applications.11–13 Whereas biomaterials for the fundamental understanding of biological systems are rather limited.14,15 Therefore, in this review, we will focus on recent trends in using materials to gain fundamental biological insights. More specifically, we introduce static/dynamic materials to replicate the spatiotemporally sophisticated architecture of biological systems by confining or manipulating physical cues exposed to the cells, such as cell geometry, extracellular matrix (ECM) topography and viscoelasticity (Fig. 1). These cues can not only manipulate cellular shape and contacts with surrounding cells, but also induce endocytosis processes or nuclear shape, either directly or indirectly, eventually altering cellular responses and phenotypes. Static biomaterials were chronologically developed first, therefore, we begin by overviewing them, due to the conceptual significance before discussing photoresponsive dynamic materials which were developed later. As photoresponsive dynamic materials, we introduce not only synthetic interfacial and hydrogel materials bearing photoresponsive functionalities, but also those based on state-of-the-art and highly versatile optogenetic techniques, although their examples are still limited. From the methodological perspective, we believe such comprehensive discussion of static and dynamic materials, together with the cutting-edge biotechnologies, will be useful for future biomaterials research and applications.
2. Static materials
2.1. Controlling ECM mechanics
It has been perceived that the stiffness of the substrates and ECM is related to cell fate and activities.16 For example, normal cells exhibit an anchorage-dependent proliferative nature, however, this is not the case for cancer cells.17,18 Polyacrylamide (PAAm) and poly(ethylene glycol) (PEG) hydrogels with various crosslinked levels are the most commonly used platforms to investigate the impact of substrate stiffness. Compared to naturally derived ECMs, such as collagen and Matrigel, these synthetic scaffolds are useful for decoupling the biochemical and mechanical regulation of cellular phenotypes. The first direct experimental evidence was obtained from Pelham and Wang, who demonstrated the impact of substrate elasticity on spreading and migration behaviours.19 Later, Discher et al. demonstrated the effect of substrate elasticity on the differentiation of mesenchymal stem cells (MSCs). The lineages changed from neuronal to muscle and osteoblast by manipulating the elastic modulus of the substrate from soft (0.1–1 kPa) to semi-stiff (8–17 kPa) or stiff (25–40 kPa) ones. Furthermore, the trend corresponded to the native stiffness of each tissue, indicating the possible role of tissue stiffness in stem cell differentiation in vivo (Fig. 2A).20 These findings have revitalized research in the field of mechanobiology or mechanotransduction, and researchers have elucidated the impact of stiffness on various biological functions, such as migration21 and epithelial–mesenchymal transition (EMT).22,23 These activities are involved in the progression of diseases, such as cancer, fibrosis, and atherosclerosis, which are associated with tissue stiffening.24 Therefore, it is physiologically relevant to investigate how ECM mechanics directly regulate those cellular activities. In fact, Chen and co-workers demonstrated changes in transforming growth factor (TGF)-β activity from apoptosis- to EMT-inducing by increasing the stiffness of the substrate (Fig. 2B).25 The role of TGF-β has been controversial; it serves as a tumor suppressor at the early stage of cancer, while it promotes tumor progression and metastasis at a later stage.26 This study demonstrated that the paradox may be caused by mechanobiological change in cellular functional responses against TGF-β, depending on substrate elasticity.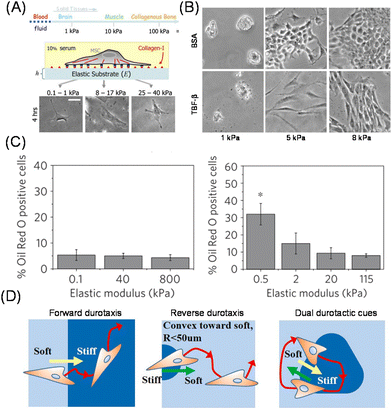 | ||
| Fig. 2 Impact of matrix elasticity on cellular functions. (A) Determination of differentiation lineages of MSCs in response to substrate elasticity. Morphologies of MSCs on the stiffness-tuned gel are shown. (B) Elasticity-dependent change in the activity of TGF beta between EMT- and apoptosis-inducing activity. Morphologies of BSA- (control) or TGF-treated MDCK cells are shown. (C) Different cellular mechanosensing behaviors on PDMS (left) and PAAm hydrogel (right) substrates. (D) Cellular durotaxis in response to photopatterned substrate elasticity. (A and D) Reproduced from ref. 20 and 35 with permission from Elsevier, Copyright 2006 and 2021. (B) Adapted from ref. 25, https://doi.org/10.1091/mbc.e11-06-0537, under the terms of the CC BY-NC-SA 3.0 license. (C) Reproduced from ref. 28 with permission from Springer Nature, Copyright 2012. | ||
In addition to hydrogel platforms, polydimethylsiloxane (PDMS) is another candidate commonly used to manipulate substrate elasticity even though its non-permeable nature against biomolecules is different from and sometimes disadvantageous compared to hydrogel matrices. In addition, another important difference between silicon elastomers and hydrogel-based platforms is their non-swellable nature. This makes the bulk mechanics stable without variation through time-dependent swelling/de-swelling. In addition, the surface is more hydrophobic, which affects the assembled structures27 and reactivity28 of ECM proteins, eventually altering cellular mechanosensing behaviors. For example, Trappmann et al. demonstrated the loss of sensitivity to substrate elasticity in keratinocytes and MSCs in terms of their spreading and differentiation behaviors, respectively (Fig. 2C).28 The authors attributed this loss to the significant alteration of porosity by changing the stiffness in PAAm but not in PDMS, thereby resulting in altered tethering of collagen to the substrate. The effect of ligand mobility is important in viscoelastic mechanobiology, as discussed below.
High-throughput platforms have been developed for the comprehensive analysis of the impact of ECM mechanics on cellular phenotypes or drug responses.29,30 Using automated robotics, combinatorial hydrogel arrays with serially controlled ECM stiffness and ligand densities can be prepared in a multi-well format.31 This allows for the study of synergistic regulation of cellular phenotypes via biochemical and biomechanical cues using morphological and/or gene expression analysis.
In addition to the impact of homogenously elastic surfaces on cellular behavior, cells can feel matrix stiffness heterogeneity. Lo et al. first demonstrated that cells can migrate along the gradient of matrix stiffness at the boundary of soft (14 kPa) and stiff (30 kPa) regions; this capability of cells to sense and migrate across the elasticity difference is later called durotaxis.32 In a similar fashion to cell migration along the gradient of chemoattractant molecules in solution or immobilized on the matrices, how cells feel and respond to the gradient of substrate mechanics is one of the most important topics in mechanobiology. This is because the cells need to respond differently in the subcellular resolution, polarize them, and direct themselves to move toward the gradient of matrix mechanics. Kidoaki et al. used photocurable styrenated gelatin to systematically investigate the cellular migration behaviors on the microelastic gradients.33 Due to its photo-crosslinkable feature, precise stiffness patterns were possible, and various unique behaviors were identified, such as the dependence of curvature of stiffness boundaries and reverse durotaxis (Fig. 2D).34,35 Additionally, aged vessels exhibit mechanical heterogeneity at subcellular levels, resulting in cell proliferation and local traction stresses.36,37 To replicate this issue, Reinhart-King et al. developed photopatterned hydrogels based on methacylated hyaluronic acid in the range of 2.7–10.3 kPa and directly demonstrated the disruption of cell–cell junctions by increasing matrix heterogeneity.38
However, biological tissues also exhibit “visco”-elastic responses, rather than simple elastic responses, against load or deformation; therefore, researchers have begun to develop viscoelastic artificial ECMs to study cellular mechanobiological responses in a more physiologically relevant manner. Cameron et al. prepared PAAm-based hydrogel substrates with varied loss modulus (G′′), maintaining a constant storage modulus (G′), and investigated the effect of loss modulus (viscous component) on the adhesion, proliferation, and differentiation potentials of MSCs (Fig. 3A).39 Their study identified increased cell spreading area and smaller focal adhesions on the substrate with a larger loss modulus (higher creep), resulting in smooth muscular differentiation via the activation of small GTPase Rac.40 Matsudaira manipulated viscosity using PDMS and demonstrated enhanced collective migration41 and coalescence42 on viscoelastic substrates. To systematically manipulate the viscous term, Mooney et al. used ionically crosslinked alginate hydrogels and investigated the effect of stress relaxation, while maintaining constant elasticity, on adhesion behaviors of epithelial U2OS cells and 3T3 fibroblasts (Fig. 3B).43 In contrast with the elastic counterpart, the stress-relaxing hydrogels allowed for larger cell spreading with enhanced stress fiber formation, together with higher YAP nuclear expression. The authors further demonstrated enhanced osteoblast differentiation of MSCs on stress-relaxing hydrogels.44 These results indicate that stress-relaxing properties are different from a mere decrease in matrix elasticity. Later, Shenoy et al. explained these phenomena using the modified molecular clutch model and theoretically demonstrated that matching cellular and material timescales are critical to maximizing cellular spreading behaviors.45 Chaudhuri et al. further identified emergent filopodia-mediated migration phenotypes on the stress relaxing hydrogels, which are mechanistically different from those observed in 3D matrices.46
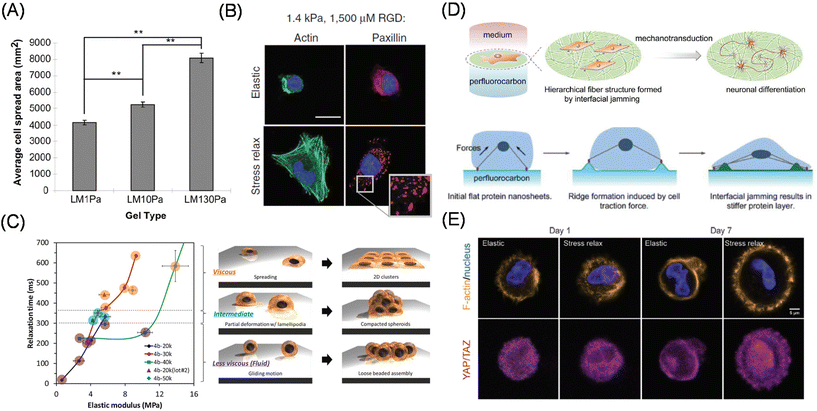 | ||
| Fig. 3 Viscoelastic mechanobiology. (A, B) Effect of viscous term on cell adhesion and differentiation behaviors using (A) PAAm-based hydrogels with varied G′′ (at constant G′) and (B) alginate hydrogel with controllable stress relaxation (at constant elasticity). Green, actin; blue, nucleus; red, paxillin. (C) Interfacial relaxation dominant epithelial cell adhesion and assembly behaviors identified by precise viscoelasticity manipulation and characterization. (D) Liquid–liquid interface induces neuronal differentiation without differentiation inducing factor via cell traction force-mediated interfacial jamming of protein layers. (E) Use of 3D hydrogel based on dynamic covalent bonding for the organoid culture. Immunofluorescence images show time-dependent changes in YAP/TAZ localization in elastic and stress relaxing 3D gels. Yellow, F-actin; blue, nucleus; pink, YAP/TAZ. (A and C) Reproduced from ref. 39 and 51 with permission from Elsevier, Copyright 2011 and 2021. (B) Reproduced from ref. 43 with permission from Springer Nature, Copyright 2015. (D) Reproduced from ref. 60 with permission from John Wiley & Sons, Copyright 2020. (E) Reproduced from ref. 65, https://doi.org/10.1002/advs.201800638, under the terms of the CC BY license. | ||
In addition to the bulk viscosity of hydrogel matrices, increasing the mobility of the material interface can also alter cellular responses. Yui et al. have used supramolecular polyrotaxane, a topological complex with cyclodextrin (CD) and PEG.47 By changing the number of CD molecules complexing with PEG, the authors succeeded in controlling the mobility of the molecules adsorbed at the interface, resulting in changes in the cell morphologies48 and effective cardiomyocyte differentiation of pluripotent stem cells.49 Alternatively, Wei et al. manipulated the diffusion of cell adhesive ligands on the surface by controlling their hydrophobic interactions with the substrates.50 The peptide ligand has a hydrophobic domain at the other end, and its adsorption strength changes depending on the chain length of alkylsiloxane layers introduced at the substrate surface. The highly diffusible ligand surface allowed the selective accumulation of specific integrin subtypes (α5β1 over αvβ3) and activated Rac and RhoA signaling, resulting in a lower degree of osteogenic differentiation of MSCs.
In contrast, Nakanishi et al. succeeded in decoupling the interfacial viscosity and elasticity of polymeric scaffolds and investigated the effect on adhesion, migration, and assembly behaviors of epithelial MDCK (Madin-Darby Canine Kidney) cells (Fig. 3C)51 by appropriately choosing the molecular weights of copolymers of ε-caplolactone and D,L-lactide and photo-crosslinking time. The authors reported that cellular mechanosensing was mainly determined by interfacial stress relaxation rather than interfacial elasticity and identified enhanced cellular aggregate formation at the intermediate interfacial relaxation. Later, this group applied the substrates to demonstrate different controls of EMT-related genes (E- and N-cadherins) by interfacial viscoelasticity.52 Notably, although an extremely fluid air–water interface has been used for the formation of spheroids, this study indicated the possible existence of more suitable interfacial mechanics (intermediate level relaxation) for the efficient production of 3D cellular structures.
Another approach is to make intrinsically highly fluid and adaptive nature of (hydrophobic) molecular liquids by hardening the liquid–liquid interfaces capable to sustain the cellular traction force. In the mid-20th century, Rosenberg has witnessed, for the first time, cellular adhesion and expansion at the interface between water and perfluorocarbons or silicon oils.53 Twenty years later, Keese and Giaever reviewed the phenomena54 and demonstrated the protein layer formation at the interface, which was further strengthened by trace amounts of surface-active compounds.55 Due to recent trends in mechanobiology and mechanotransduction, more findings have come to light.56,57 Using interfacial rheology, Gautrot et al. precisely characterized the mechanics of the protein layers and demonstrate their capability to expand stem cells in the suspension culture of oil-in-water emulsions.58,59 In contrast, Jia, Nakanishi, Ariga and coworkers focused on different mechanical evolutions and cellular mechanical sensing of protein layers depending on the types of perfluorocarbons.60 By using perfluorooctane as the underlayer, the authors found spontaneous neuronal differentiation of MSCs without the addition of differentiation-inducing factors (Fig. 3D). The authors attributed the adaptive nature of the interface to the fibrillar formation of fibronectin molecules through cell traction-induced interfacial protein jamming, which is critical for the neuronal differentiation.
Considering native tissues, elasticity manipulation of 3D culture scaffolds is more physiologically important.61 However, sometimes, the same chemically crosslinked hydrogels cannot be applied to 3D scaffolds since the frozen chemical linkages block cellular movement, thereby inducing apoptosis.62 Therefore, naturally derived collagen, gelatin, or Matrigel at various concentrations have been conventionally used to study cellular mechanosensing in 3D.63 Another commonly used approach is to crosslink gelatin PEG diacrylate; with the commercially available compound, we can take advantage of the physically crosslinkable nature of the native ECMs. The effect of ECM stiffness on the reprogramming of normal cells into tumor precursor cells has also been reported using these 3D scaffolds.64 Alternatively, artificial hydrogels bearing enzymatically cleavable and physical crosslinkers have been designed to maintain cellular viability in 3D matrices via proteolytic degradation and cell traction-driven reconfiguration of the crosslinking points. Recently, hydrogels based on dynamic covalent bonds have allowed for the manipulation of viscoelasticity in 3D.65 These include Diels–Alder reactions,66 reversible hydrazone bonds,67 and phenylboronic acids.68,69 For example, by utilizing the adaptive hydrogels, Anseth et al. demonstrated YAP/TAZ subcellular localization is directly correlated with cellular capability to remodel surrounding environments (Fig. 3E).69
2.2. Controlling the shape and geometry of cells
It is well known that there are strong correlations between shape and function in biological systems; the change in shape of cells and cell clusters is essential in developmental processes, pattern formation, and epithelial tube formation.70 On the other hand, matrix mechanics not only changes the functions as discussed in the previous section but also the shape of cells. Therefore, it is reasonable curiosity to investigate what happens if cellular shape as well as geometry was decoupled from matrix mechanics. It was only in the late 20th century that direct experimental evidence to support this was obtained at the single-cell level by taking advantage of microcontact printing (Fig. 4A).71,72 In this study, Whitesides, Ingber, and co-workers prepared micrometer-sized fibronectin-adsorbed cell adhesion regions surrounded by non-adhesive PEG-conjugated regions. On this micropatterned substrate, they demonstrated a strong correlation between cellular shape and cell proliferation; cell growth was enhanced by increasing the cell spreading area (Fig. 4A). The important point of this study is that they controlled cellular geometry not by changing the substrate mechanics or ECM ligands (fibronectin here) because they kept these parameters unchanged. Therefore, the different cellular growth rate is solely attributed to the cell geometry. Later, their effects on cell polarity and anisotropic patterns were also studied, in terms of lamellipodia extension73 and the orientation of cell division axis.74,75 It was also elucidated that the cell spreading area altered the differentiation of human MSCs into adipocyte and osteoblast lineages on small and large adhesion areas, respectively, through RhoA activity and cytoskeletal tension (Fig. 4B).76 By further changing the aspect ratio and subcellular curvatures of the cellular outlines, Mrksich et al. demonstrated the significance of geometrical cues in directing MSCs into appropriate fates by regulating the activities of kinases and wingless type (Wnt) signaling.77 Later, Piccolo et al. identified the essential role of the nuclear localization of Hippo pathway transcription cofactor YAP/TAZ in the cellular geometry sensing processes controlled by shuttling the proteins between the cytosol and nucleus, depending on the spreading degree (Fig. 4C).78 More recently, the impact of single-cell geometry on plasma membrane tension and order was observed, showing transient Ca2+ influx through mechanosensitive PIEZO-1 channels79 and stem cell differentiation via the Akt pathway (Fig. 4D),80 respectively. These studies have a strong correlation with the cellular mechanosensing of ECM mechanics discussed in the previous section.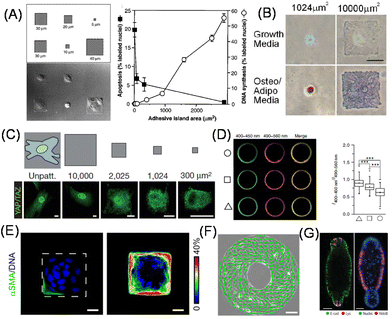 | ||
| Fig. 4 Two-dimensional cell shape control for fundamental biology. (A–D) Cell spreading area determines (A) cell proliferative activities, (B) MSC differentiation lineages (red: lipids stain; blue: alkaline phosphatase stain), (C) YAP nuclear shuttling and (D) membrane order. Left: Fluorescence images of C-laudan-stained plasma membrane vesicles isolated from different geometry cells. Right: Ratio of fluorescence intensity. (E) Geometric control of cell clusters affects EMT progression therein. Left, Immunofluorescence images of αSMA (green) and nucleus (blue); right, frequency map of αSMA (pseudocolor). (F) Emergent left-right asymmetry in collectively migrating cells confined in a specific geometry. Green lines show the direction of cellular alignment. (G) Immunofluorescence staining of reproducibly generated organoids by shape control. Left, E-cadherin (green) and lysozyme (red); right, nuclei (blue) and aldolase B (red). MSC, mesenchymal stem cell; EMT, epithelial–mesenchymal transition, αSMA, α-smooth muscle actin. (A and G) Reproduced from ref. 71 and 88 with permission from AAAS, Copyright 1997 and 2022. (B) Reproduced from ref. 76 with permission from Elsevier, Copyright 2004. (C and D) Reproduced from ref. 78 and 80 with permission from Springer Nature, Copyright 2011 and 2018. (E) Reproduced from ref. 82 with permission from John Wiley & Sons, Copyright 2010. (F) Reproduced from ref. 83 with permission from National Academy of Science. | ||
In addition to single-cell manipulation, the same concept can be applied to study the impact of the geometry of multiple cell clusters on cellular phenotypic symmetry breaks within them. For example, Nelson, Chen and coworkers demonstrated emergent patterns of cell growth within cell clusters formed on micropatterned geometries.81 The authors correlated highly proliferative foci with high traction stress regions within the clusters, which was confirmed theoretically via finite element analysis of myosin ATPase-driven cellular intrinsic contractility and experimentally using PDMS micropillars. The authors further demonstrated that epithelial cells located in the outer regions of the cluster exhibited high susceptibility to undergo EMT against TGF-β stimulation (Fig. 4E).82 Furthermore, by patterning cell clusters in a donut shape, an intrinsic chirality of the direction of collective cell migration was identified (Fig. 4F).83 Such biased motion in the direction of collective cell migration on 2D substrates is considered the origin of the left-right asymmetry generation in embryonic development.84
By extending this concept to a three-dimensional (3D) space, the impact of geometrical cues on cellular functions and fate becomes much closer to that of in vivo environments. Nelson and Bissel demonstrated the impact of tissue geometry on in vivo mammary branching morphogenesis by geometrically mimicking tubular structures in organoid cultures via a micropatterning approach.85 From a reductionist perspective, the impact of 3D geometry can also be studied to decouple from the ECM mechanical cue.86 Thanks to the advancements in additive manufacturing techniques, such as 3D printing, a more detailed analysis of the impact of geometry in more physiologically or pathologically relevant scaffold-free (or less) environments has become available. For example, Kilian et al. demonstrated the impact of geometrical cues on histone state, thereby inducing reprogramming in melanoma.87 Lutolf et al. reported a methodology for reproducible organoid formation by combining the template fabrication technique, which elucidated shape-oriented deterministic regulation of YAP and Notch signaling (Fig. 4G).88 This research provides clear evidence as to the origin of the form of our living bodies. It is ripe for more sophisticated analysis by combining cutting-edge biological techniques, such as stem cell biology and genome-editing technologies, and material science.
2.3. Manipulating subcellular structures
In the last two sections, we discussed static biomaterials to elucidate how the entire form and surrounding mechanics regulate cellular and tissue activities and fate. However, cells can sense external cues smaller than their size. Therefore, from now on, we will introduce static materials to manipulate subcellular structures. Actually, Harrison reported that even in the early 20th century, cells were elongated along the fibrous matrices.89 Later, the concept of topographical (contact) guidance in outgrowing nerve fibers was developed by Weiss,90 following which the role of surface topographies in cellular function regulation and the mechanisms underlying contact guidance have since gained attention.91 In particular, the native ECM is composed of fibrous collagen, laminin, fibronectin, and proteoglycans, and hence it is reasonable to investigate the mechanisms by which cells can sense such subcellular physical cues and feed them back to their own phenotypes. To replicate such cellular fibrous structures, electrospinning and rotary jet spinning92 are powerful techniques for biomedical applications.93 On the other hand, photolithography and multiphoton fabrication techniques,94 together with soft lithography, are more common in fundamental studies, because these allow precise manipulation of the matrix topography and studies on the effects on cellular functions.95 Dalby et al. demonstrated that MSCs cultured on surfaces bearing submicrometer-sized pits with a certain level of disorder exhibited enhanced osteoblast differentiation comparable with dexamethasone-induced stimulus.96 Subsequent studies by this group have identified reduced adhesion formation on the nontopographic surfaces, thereby inducing multipotency in MSCs cultured on the ordered arrangement of nanoscale pits (Fig. 5A)97 which has been elucidated via metabolomic analysis.98 Recently, Jia and Ariga developed large-area aligned nanopatterned surfaces by self-assembly of fullerene nanowhiskers to maintain multipotency of MSCs, which is suitable for tissue engineering and biochemical analysis.99 The impact of surface micro/nanotopography on cardiac and skeletal muscles has been intensively studied, since these cells are known to align in a specific orientation and exhibit improved performance to couple electrical stimulus into mechanical output.100–102 Therefore, it is important, even from tissue engineering perspectives, to understand the underlying mechanisms.103 Moreover, the mechanosensitive responses of fibroblasts against the gradient of nanotopography have been elegantly elucidated by Levchenko et al. using ultraviolet (UV)-assisted capillary force lithography.104 Apart from studies focusing on the impact of nanotopography on cellular fate or phenotypes, recent studies have discussed the mechanisms underlying dynamic sensing of nanotopography in contact guidance (Fig. 5B).105,106 For example, Kurniawan and Bouten showed the emergence of cellular contact guidance from energetically favorable cellular elongation and gap avoidance,107 while Provenzano et al. demonstrated the significant role of local anisotropic forces in constrained focal adhesion for contact guidance.108 A review by Barakat et al. has further elaborated on this topic.109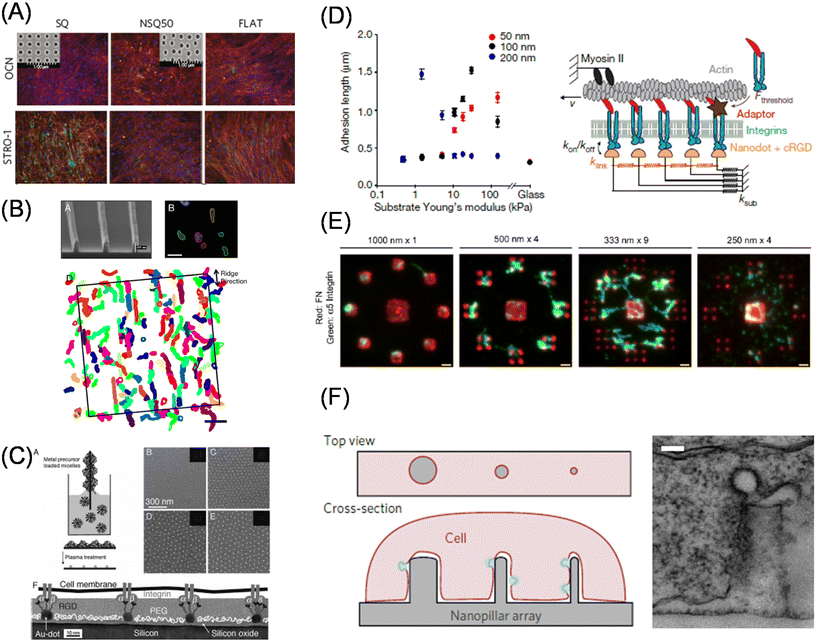 | ||
| Fig. 5 Effect of subcellular topography and adhesion geometry on cellular functions. (A) Enhanced multipotency in MSCs cultured on the substrate bearing nanoscale pits confirmed by immunofluorescence staining of osteoblast (OCN, osteocalcin) and stemness (STRO-1) markers. Inset shows ordered and semi-disordered nano-pits. (B) FA-based model of contact guidance. Top left, a Scanning electron microscopic image of nanoridges; top right, amoebae cells migration; bottom, overlapped extracted cell shapes for 22 min. (C) Block copolymer nanolithography for the control of nanoscopic separations of gold nanoparticle arrays. (D) Left, synergistic regulation of ligand spacing (red, 50 nm; black, 100 nm; blue, 200 nm) and substrate elasticity for FA maturation. Right, molecular clutch model. (E) The minimum ECM area required for focal adhesion assembly and force transmission is determined by subtractive contact printing. Immunofluorescence images of printed fibronectin (red) and bound integrin (green) on different geometrical configurations. (F) Vertical nanostructure-induced endocytosis. Left, schematic illustration of cell adhesion on the nanostructures; right, a transmission electron microscopic image of a single nanopillar wrapped with plasma membranes. (A, D and F) Reproduced from ref. 97, 113 and 122 with permission from Springer Nature, Copyright 2011, 2017, and 2017. (B) Reproduced from ref. 105, https://doi.org/10.1021/nn406637c, under the terms of the CC-BY-NC-ND license. (C) Reproduced from ref. 111 with permission from Elsevier, Copyright 2007. (E) Reproduced from ref. 114, https://doi.org/10.1242/jcs.108035, with permission from Journal of Cell Science. | ||
Focal adhesion is a micrometer-size protein assembled structure that mediates biochemical and mechanical cell–matrix interactions. Nanopatterned surfaces are useful for understanding focal adhesion maturation at a molecular level. In particular, block copolymer nanolithography developed by Spatz et al. insights for understanding integrin clustering in focal adhesion maturation.110 Gold nanoparticle arrays in a hexagonal arrangement prepared using this method enable fine tuning of interparticle distance on the order of tens of nanometers (Fig. 5C).111 Moreover, the size of the gold nanoparticles was comparable to that of integrin heterodimers. Using these features, the authors identified a universal threshold of 50–70 nm for focal adhesion maturation; above this threshold, the distance between neighboring integrins is too sparse to accumulate adaptor molecules needed for downstream signaling. The nanopatterning technique can be adapted to hydrogel surfaces.112 By exploiting this feature, Roca-Cusachs et al. explained counter-intuitive observations of enhanced cell spreading on the surfaces with sparser ligand density and softer bulk mechanics under the framework of a molecular clutch model (Fig. 5D).113 Garcia et al. focused on a larger area and identified another threshold for ECM area on a larger scale, with a submicron area (0.11 μm2) for focal adhesion assembly and force transmission using subtractive contact printing (Fig. 5E).114 Meanwhile, Sheetz et al. examined cell adhesion and focal adhesion maturation behaviors on nanolines at 10 nm width with a given separation and found that the capability of integrin clusters across the parallel nanolines (within 110 nm) was similar to that observed with homogenous surfaces.115
Subcellular patterns can be used to understand the formation of cellular polarity and in-cell structural organization. Thery et al. have used PDMS soft lithography to determine how subcellular adhesive spots affect cell division axis orientation74,75 and cell orientation polarity.116 Further detailed analyses of actin network architecture demonstrated a significant role for α-actinin in integrating the network and adapting to subcellular geometrical cues.117 Intracellular signaling induced by vertical nanostructures, such as silicone nanowires, nanopillars, nanoneedles, and nanocones, have recently become trending research topics.118–120 These nanotopographic surfaces not only induce focal adhesion and cytoskeletal rearrangements121 but also endocytosis by accumulating membrane curvature sensors122 and endocytosis-related proteins, such as caveolae and clathrin (Fig. 5F).123 Moreover, lipid-anchored K-Ras proteins change their activity depending on membrane curvature.124 In this way, subcellular-size manipulation technologies contribute to increasing the fundamental understanding of structure–function relationships in cellular functions.
3. Dynamic photoresponsive materials
3.1. Photoresponsive tools
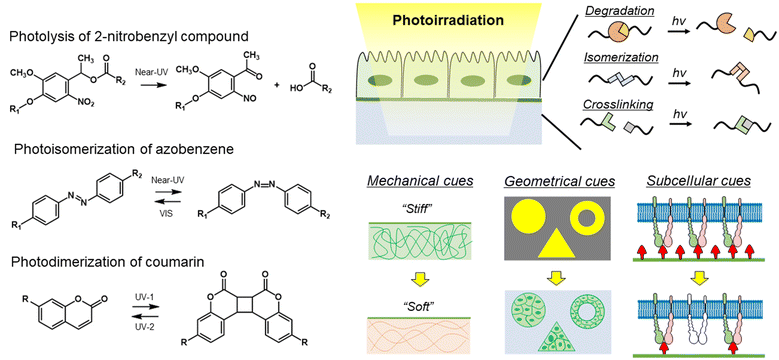
In the wake of the above-mentioned successful contribution of static biomaterials in dissecting the hierarchical nature of biological systems, dynamic materials have been developed to address the dynamic aspects of biological processes.125,126 By introducing photoresponsive functional groups to materials, photoresponsive materials allow researchers to manipulate the physical properties of materials, such as mechanical, geometrical, and subcellular cues, at high spatiotemporal resolutions (Fig. 6).127 As photoresponsive functional groups, 2-nitorobenzyl compounds, azobenzenes, spiropyrans, and coumarins are frequently used to develop photoresponsive dynamic materials. 2-Nitrobenzyl ester and its derivatives undergo photolysis under near-UV light.128,129 Azobenzene undergoes cis isomerization upon near-UV irradiation and turns into a trans-isomer when irradiated with visible light.130 The photochromic spiropyran derivatives undergo reversible chemical structural as well as electrostatic changes during the isomerization reaction.131 Coumarin and anthracene derivatives can photodimerize under near-UV irradiation, while the dimer can be divided into two coumarins under deeper UV irradiation.132,133 The coumarin group can also undergo photocleavage reaction via near UV irradiation, in a similar fashion to the 2-nitrobenzyl groups, with higher cross-sections for two-photon excitation with near IR light.134 In addition, cytocompatible photo-induced radical polymerization reactions are an alternative tool for in situ manipulations of the crosslinking density of matrices.135 Conversely, the allyl sulfide moiety cleaves via a radical addition–fragmentation chain transfer (AFCT) process.136 Combining light-harvesting nanomaterials to induce phase-transition of polymers is another approach for photocontrolling polymeric properties.137 Today, photo-switchable proteins are popular tools in molecular biology studies, such as gene editing,138 nuclear-cytosolic translocation139,140 and liquid–liquid protein phase separation.141,142 Due to the wide wavelength range, high responsivity, and reversibility,143,144 biomaterials based on photo-switchable proteins are a new trend for getting insights in fundamental biology.145Since each photoresponsive functionality has a different photoreaction mechanism, such as degradation, cis–trans isomerization, dimerization, polymerization/depolymerization, and conformation changes, the selection of these molecules is important for controlling the desired material properties. Moreover, there is accumulating evidence of substituent effects for these photoresponsive functionalities,146–148 which allows for the introduction of more than two switches in one system.149,150 From now on, we will discuss their utilization to dynamically control physical cues exposed to cells and recent trends in fundamental biological studies using photoresponsive biomaterials.
3.2. Photoresponsive matrices with switchable mechanics
As discussed in the sections of static biomaterials, cells utilize myosin-derived intrinsic contractility to sense the mechanical properties of their surroundings. Dynamic materials enable us to determine how changes in the mechanical properties can affect cellular functions, thereby these materials are considered power tools for mechanobiology studies. Dynamic biomaterials based on photoresponsive molecules such as 2-nitrobenzyl groups, azobenzene, and coumarin can change the stiffness and viscoelasticity, upon photoirradiation. To elucidate the spatial and temporal mechanisms of mechanosensing, PAAm or PEG gels bearing the 2-nitrobenzyl group as a crosslinker have been developed to modulate mechanical properties (Fig. 7A).151 The stiffness of PAAm gels can be altered from 7.2 to 5.5 kPa using photoirradiation, while soft hydrogel inhibited the spreading of NIH3T3 fibroblasts. Furthermore, changes in stiffness due to localized irradiation revealed that the cell had stiffness sensors located at the front. In addition, the concept of mechanical memory was proposed from research using photoresponsive hydrogels.152 MSCs can memorize and recall their past experiences of their mechanical environments as plastic changes in signaling and protein expression. MSCs were initially cultured in a growth medium at the stiff state on the photoactivatable hydrogels, whose stiffness was reduced by photoirradiation after a different delay time (Fig. 7B). Depending on the past period of culturing on the stiff substrate, the efficiency of MSCs differentiation changed despite the medium not being changed to differentiation one after the substrates have been softened. A recent study applied the same concept to investigate the impact of matrix softening on the phenotype of myofibroblasts, changing from transient to persistent owing to condensation of chromatin structures.153 Interestingly, transient myofibroblasts increased their condensation significantly after dynamically changing the scaffold from stiff to soft, whereas persistent myofibroblasts remained constant. In addition, myofibroblasts cultured for 7 d on stiff hydrogels during treatment with actin inhibitors showed the same level of smooth muscle α-actin (αSMA) expression as non-treated cells but chromatin condensation was inhibited in persistent myofibroblasts. Furthermore, by softening hydrogels, the αSMA signal disappeared in the inhibitor-treated myofibroblasts. These results indicate that actin stabilization is not required for initial myofibroblast activation but may be involved in the mechanical memory for myofibroblast persistence (Fig. 7C).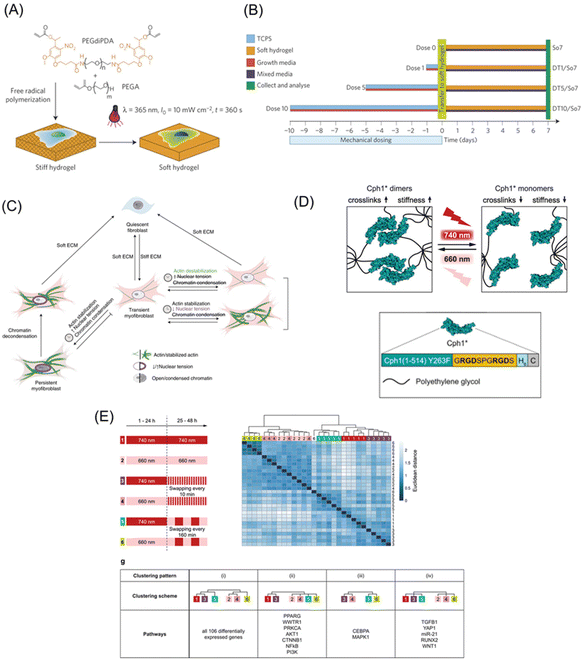 | ||
| Fig. 7 Photoresponsive matrices with switchable mechanics. (A) The PEG hydrogels with photodegradable crosslinker were prepared by free-radical polymerization. The photoresponsive hydrogels gel changes from stiff to soft in response to UV light. (B) Experimental methods for understanding mechanical memory. (C) The mechanism for stiffness-induced myofibroblast persistence using photo-softening hydrogels. (D) Design of phytochrome-based hydrogels based on the Cph1 variant. (E) Mechanical memory behavior of the 106 differentially expressed genes as a reference of Ingenuity Pathway analysis or literature in MSCs. A and B: Reproduced from ref. 152 with permission from Springer Nature, Copyright 2014. C: Reproduced from ref. 153 with permission from Springer Nature, Copyright 2021. D and E: Reproduced from ref. 164 with permission from John Wiley & Sons, Copyright 2019. | ||
Compared to photo-induced degradation of matrices, photo-induced stiffening is more physiologically relevant since the increase in elasticity is often observed not only in the development process but also in disease progressions such as fibrosis and cancer progression.154 For this purpose, the selection of photoinitiators is critical for radical polymerization cytocompatibility.135 Burdick et al. first succeeded in dynamic stiffening of methacrylated-hyaluronic acid-based hydrogels using Irgacure 2949 as a photoinitiator.155 The materials can be used to detect short- and long-term cellular responses in terms of changes in traction force and differentiation lineages in MSCs, respectively. Moreover, the authors have identified that already differentiating MSCs are less sensitive to the change in the mechanical properties. Engler et al. also utilized a similar system to demonstrate the cooperative effect of paracrine and mechanosensing pathways in stiffness-induced EMT in mammary epithelial cells.156 By combining the photopolymerization reaction and the photocleavable 2-nitrobenzyl crosslinker, Rosales, Burdick, and Anseth have succeeded in a reversible mechanics change.157
Conversely, the azobenzene group has a reversible switchable nature by itself; therefore, azobenzene-containing hydrogels have the potential to address reversible changes in mechanical properties upon light irradiation. An azobenzene-containing acrylamide gel was softened with UV irradiation and hardened using blue light irradiation.158 The substrates were shown to be noncytotoxic, while cell morphology showed characteristic cell spreading and increased aspect ratios in response to greater substrate stiffness. A combination of azobenzene and β-cyclodextrin has also been developed to control the elastic modulus of hyaluronan-based scaffolds.159 A recent study reported the development of azobenzene-based hydrogel with switchable mechanics based on photo-induced phase-transition of the polymer chains within the hydrogel.160 Utilizing this platform, the authors identified cellular adaptive nature, in terms of an immediate increase in E-cadherin mRNA expression in breast cancer MCF-7 cells in response to dynamic stiffening of the hydrogel, followed by gradual restoration to the original level.
The light-induced dimerization characteristics of coumarin can be used for stiffness and viscoelasticity control of 3D matrices. The hydrogels based on coumarin ABA-type triblock copolymers can convert from physically crosslinked viscoelastic hydrogel to chemically crosslinked elastic hydrogel.161 Using this switchable hydrogel, Ueki, Yoshida and coworkers have successfully demonstrated the impact of cell confinement on proliferation activity. Anseth et al. developed anthracene-containing hydrogels, whose stiffness can be increased by photoinduced dimerization of anthracene molecules.162 The two platforms have reversibly switchable potential through photo-induced cleavage of the dimer into monomer, but it is not suited for physiological conditions as it requires deep UV irradiation and/or heat.
In this regard, photoswitchable proteins allow for more physiologically compatible reversible switches of hydrogel mechanics in fast speed and high spatial resolution with long-wavelength light. For example, DeForest et al. developed a stiffness-controllable hydrogel-based on a light-oxygen-voltage-sensing domain 2 (LOV2).163 A blue light irradiation induced a conformation change in LOV2, thereby softening the gel, whereas the dark conditions return it to the original stiffness to return the gel stiffer. Dynamic control of hydrogel mechanical properties increased αSMA and periostin (Postn) gene expression in fibroblast. In order to make the photo-induced reaction milder, the cyanobacterial photoreceptor Cph1 was utilized for the design of PEG hydrogels with switchable mechanical properties.164 This system allows for high spatiotemporal control of material stiffness using cell-compatible tissue-penetrating red/far-red irradiation due to the chromophore of Cph1 (Fig. 7D). The crosslinking density of the hydrogel increased via the dimerization of Cph1 via 660 nm irradiation to increase its stiffness. Whereas the irradiation of 740 nm converted the Cph1 dimer into a monomer to soften the matrix. When light responsive hydrogels are cyclically alternated between these wavelengths, different signaling molecules are expressed depending on the interval. For example, downstream genes such as YAP and RUNX2 were highly dependent on the initial stiffness. Meanwhile, the upstream signaling molecules, including AKT1 and PI3K, are less sensitive to short (10 min) interval stiffness changes, although their expression levels changed significantly in response to longer interval (160 min) stiffness switching (Fig. 7E).
Some studies have introduced both mechanical and chemical cues using different wavelength photoresponsive functionalities to assess at the two effects on the single platform. For instance, del Campo et al. developed UV and Vis light-responsive hydrogel, which can control the biochemical and mechanical properties.165 Biochemical property is induced with the caged-cRGD (arginine–glycine–aspartate) upon UV irradiation, while the mechanical property is regulated by the photopolymerization of methacrylate groups in the presence of the photoinitiator eosinY upon Vis irradiation. In this hydrogel, directed and hindered migration of L929 fibroblast spheroids can be induced by UV light and Vis light, respectively. In photoresponsive matrices with switchable mechanics, the response and fate of cells can be controlled by mechanical stimuli through photoirradiation, hence these biomaterials are expected to contribute not only to fundamental biology but also medical fields such as regenerative medicine and disease prediction.
3.3. Photoresponsive materials for controlling cell shape and geometry
There are three conceptually different approaches to dynamically control the shape and geometry of cells and cell collectives: one to apply external force for deforming cells via elastic substrates,166 seconds to use additive manufacturing approach, like 3D bioprinting,167 and the third to manipulate the cell–ECM interactions such as integrin and RGD peptide by using functionalized (photoresponsive) materials.125,126,168–170 Due to the scope of this review, we herein only focus on the last approach. Pioneering photoresponsive materials are, reported by Nakanishi et al., based on dynamic substrates functionalized with photocleavable 2-nitrobenzyl groups physically or chemically adsorbed with cell-repellent protein or polymers.170–172 This initial surface passivation step is critical and indispensable for the following photocontrolling procedure, otherwise the surface permits protein adsorption and hence cell adhesion becomes uncontrollable. The illumination of near-UV light to the surface induces the photocleavage reaction of the 2-nitronbenzyl ester and release of the cell-repellent molecules, changing the surface cell-adhesive region-selectively. Moreover, by controlling the timing of irradiation, we are able to control onset of cell migration.172,173 Photoirradiation through photomasks can easily create geometrically controlled cell patterning, such as circles, triangles, and stripes on the surface. By manipulating the migration path widths via controlled photoirradiation, MDCK cells displayed unique migration responses to blebbistatin treatment depending on the migration regions, which highlights the involvement of dynamic geometrical changes in cellular drug responses (Fig. 8A and B).174 Additionally, by applying this concept to substrates with controlled chemical cues175 or stiffness-controlled polyacrylamide gel,176 or both,177 the interplay of chemical, mechanical, and geometrical regulation of collective migration in epithelial cells have been comprehensively studied.168 For example, biphasic responses of leader cell appearance against reducing interfacial cyclic RGD peptide density have been reported.178 Considering that the RGD peptide is an essential motif existing in ECM proteins that mediate the interaction with cell-surface integrin molecules, this result indicates positive regulation of mechanical and biochemical coupling in general epithelial collective behaviors.179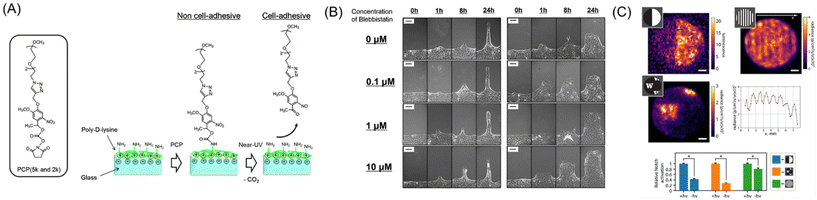 | ||
| Fig. 8 Dynamic control of cell–ECM interactions on/in photoresponsive materials. (A) Schematic illustration of a photoresponsive biomaterial based on a photocleavable PEG that changes from non-cell-adhesive to cell-adhesive upon photoirradiation. (B) Cell migration behavior of different widths after adding blebbistatin (0–10 mM). (C) Enhancement of Notch signal within the patterned delta-1 regions in collagen gels. A and B: Reproduced from ref. 174 with permission from the PCCP Owner Societies. C: Reproduced from ref. 184 with permission from National Academy of Science. | ||
In addition to the application of 2-nitrobenzyl ester as a photocleavable linker, this functionality has been used to produce so-called caged compounds to switch ON the biological activities of amino acids and proteins by utilizing the 2-nitrobenzyl ester as a photocleavable protecting group. One of the biggest successes in terms of manipulating cell–ECM interaction is the development of photoresponsive molecules based on caged RGD peptides. The activity of the peptide can be blocked (caged) by chemical modification of the aspartate (D) residue with a 2-nitrobenzyl group but recovered by photocleaving the protecting group. By introducing the caged RGD or higher affinity cyclic RGD peptide to materials, cell adhesion and migration can be controlled via photoirradiation.180,181 For example, cell migration from the endothelial monolayers using scanning lasers was observed on fibrillar adhesion tracks (3–15 μm in width).182 The frequency of escape increased monotonically with the width of the fibril and the density of the photoactivated adhesion ligand.
By intruding the concept of caged compounds into hydrogel matrices, the 3D control of cell adhesion becomes possible. Especially, the two-photon excitation is critical for precise three-dimensional drawing of cell adhesive proteins and peptides. In this regard, the coumarin molecule is a suitable caging group due to its high cross-section against two-photon excitation. Considering the chemical properties, a method to spatially control the immobilization of different growth factors in distinct volumes in 3D hydrogels was developed to specifically guide the differentiation signaling of stem cells.183 An agarose hydrogel modified with coumarin-caged thiols was uncaged to yield reactive thiol groups using two-photon excitation. Using this strategy, the differentiation factor sonic hedgehog (SHH), and ciliary neurotrophic factor (CNTF) with cell-adhesion peptide GRGDS were immobilized via the uncaged thiol group in the hydrogels. Retinal precursor cells (RPCs) were successfully attached and activated in the irradiated regions. Using photochemically patterning technologies, 3D cell migration of RPCs has been reported along an SHH gradient in agarose gels. Moreover, chemoattractant molecules and an adhesive peptide have been co-immobilized to facilitate the 3D penetration of RPCs in the hydrogel. Recent studies have extended this approach to further define the orientation of immobilized proteins. DeForest et al. developed collagen gels with an oxylamine protected by 2-nitrobenzyl molecules, which liberated the bioorthogonally reactive functional group following photolysis. This functional group formed covalent bonding with an aldehyde-modified delta-1 protein, enabling its photopatterning in the micrometer order.184 Using this method, the Notch signal was enhanced region-selectively (Fig. 8C). In this study, the two-photon excitation of the 2-nitrobenzyl group has been also demonstrated; this fact indicates that two-photon excitation stays in a practical level despite the efficiency being less efficient than coumarin. One of the most important applications of the photoresponsive matrices is 4D cell patterning in artificial biomaterials, which allows rational creation of tissue mimics in vitro. These are useful not only for fundamental understanding the development and disease progressing processes but also for physiologically relevant drug screening platform.
3.4. Subcellular-to-molecular control by photoresponsive biomaterials
Cellular activities are highly dependent on molecular-to-subcellular structures of ECM due to their cytoskeletal structures and the presence of intracellular membranous and non-membrane organelles.185,186 To this end, photoresponsive dynamic materials are useful to resolve spatiotemporal aspects of the cellular hierarchical characteristics. However, direct control of ECM cues on a molecular resolution by photoirradiation is extremely challenging due to the diffraction limit. Instead, the impact of dynamic aspects of subcellular-to-molecular cues can be illustrated by functionalizing nanostructured materials with photoresponsive molecules. For example, gold nanoparticle arrays prepared by block copolymer nanolithography become dynamic by functionalizing gold nanoparticles with photocleavable PEG and cRGD ligand (Fig. 9A).187 On this surface, the impact of nanoscopic as well as cRGD spacing on cell adhesion and migration can be discussed in the nanometer order. HeLa cells on nanoarrays showed unique phenotypes, such as detachment of cell–cell cohesion and loss of collective features, due to hampering FAK phosphorylation signaling between Y397 and Y861 by reduced clustering of integrin molecules (Fig. 9B).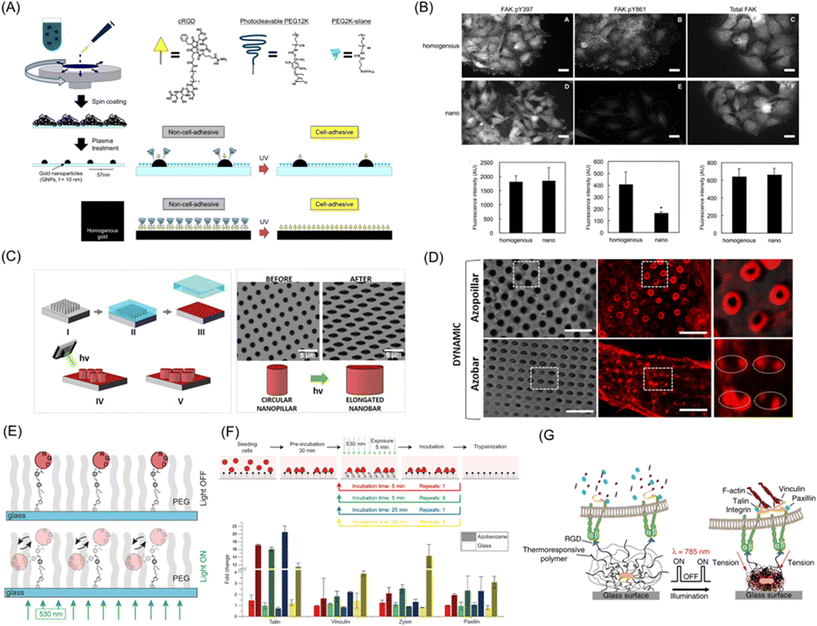 | ||
| Fig. 9 Subcellular-to-molecular control by photoresponsive biomaterials. (A) Preparation of photoactivatable nanopatterned substrates on gold nanoparticle array based on block copolymer nanolithography. (B) Immunofluorescence images (up) and average fluorescence intensities (down) of pY397, pY861, and total FAK in cells migrating on the photoresponsive homogenous and nanopatterned surfaces. (C) Schematic illustration of a nanostructure with poly(dispersed red 1 methacrylate) (pDR1m) azopolymer that changed from a vertical pillar to an elongated vertical bar using azobenzene isomerization upon photoirradiation. (D) Brightfield and fluorescence images of Lifeact-transfected U2OS cells on photoresponsive nanostructure before and after photoirradiation. (E) Schematic illustration of RGD-coupled azobenzene oscillation behavior toward biomaterials via light irradiation. (F) Change of gene expression levels to RGD-functionalized azobenzene oscillation. (G) The strategy of cRGD-functionalized optomechanical actuator (OMA) nanoparticles based on pNIPAAm embedded with Au-nanorods (AuNRs) to mechanically stimulate cell surface receptor forces. A and B: Reproduced from ref. 187, Copyright 2014. C and D: Reproduced from ref. 189 with permission from the American Chemical Society, Copyright 2020. E and F: Reproduced from ref. 191 with permission from John Wiley & Sons, Copyright 2017. G: Reproduced from ref. 192 with permission from Springer Nature, Copyright 2015. | ||
Meanwhile, azobenzene-containing biomaterials can control the shape of the material at the subcellular level by utilizing structural changes of the molecules through photo-induced surface relief (PSR) mechanisms.188 Santoro, Cui and coworkers developed nanostructures with poly(dispersed red 1 methacrylate) (pDR1m) azopolymer that changed from a vertical pillar to an elongated vertical bar shape upon photoirradiation (Fig. 9C).189 Static nanostructure substrates show that vertical structures can induce well-defined curvatures on the cell membrane and cytoskeleton factors. By dynamically changing this nanostructure using azobenzene isomerization, U2OS cells responded rapidly to changes in the nanostructure via the actin fiber and actin nucleation factor Arp2/3 complex (Fig. 9D). Netti et al. developed azopolymeric photoactive interfaces to control cellular decisions and fate. The surface topography changes from ordered parallel patterning to flat or grid by a laser beam, inducing cyclic cellular and nuclear stretches and determining mesenchymal stem cell fate.190
Photoresponsive materials, especially reversible ones, can also be used to investigate cellular responses to oscillatory or dynamic mechanical forces applied to the cell–ECM interactions. Selhuber-Unkel et al. reported on cRGD-modified azobenzene interfaces (Fig. 9E).191 Due to the extremely rapid thermally relaxing nature (reverse-isomerization) of this specific push–pull type azobenzene molecule, continuous irradiation of this molecule results in oscillatory motion and forces exertion to the cells. A high-frequency molecular oscillation upregulated the expression levels of adhesion-associated genes, such as paxillin, talin, vinculin, and zyxin (Fig. 9F). These results suggest that cells can sense small oscillatory forces at the photoisomerization rate and molecular-level strain of azobenzene. Salaita et al. developed cRGD-functionalized optomechanical actuator (OMA) nanoparticles based on pNIPAAm embedded with Au-nanorods (AuNRs) to mechanically stimulate cell surface receptor forces.192 The photothermal transductor of AuNRs converts near-infrared (NIR) illumination to localized heat, causing phase transition of the pNIPPAm and gel shrinking, delivering piconewton forces to cells through immobilized cRGD peptides. This method enables the control of T-cell adhesion, migration, and activation of T-cell receptors (Fig. 9G). Additionally, C2C12 myoblasts cultured on OMA nanoparticles enhanced nuclear YAP1 accumulation and ERK phosphorylation.193 Recently, del Campo et al. developed light-driven rotary motors based on over-crowded alkene molecules bearing terminal cell adhesive ligands, such as RGD and anti-CD3.194 The uni-directional motion of the molecular rotor increased the entanglements of the PEG chain connecting the motor and the substrate surface, therefore a tensional force is applied to membrane receptors and induce focal adhesion maturation and T cell activation.
Meanwhile, Haag et al. took advantage of spontaneous thermal reverse isomerization property, rather than photo-induced isomerization reaction itself, of a spiropyran compound to manipulate the dynamics of tethered RGD ligand.195 The concept is similar to the above mentioned oscillating azobenzene molecule-tethered surface,191 where the photochemical reaction is used to produce metastable state of the photoresponsive moiety and their time-dependent relaxing processes were used to study dynamic nature of cellular responses. In the UV-irradiated hydrophilic charged merocyanine (MC) state, the ligand diffusion increases while cellular traction force is dissipated. The spontaneous and gradual thermal reverse isomerization into a hydrophobic spiropyran (SP) state makes the adhesive force of the ligand to the substrate with the self-strengthening feature. Using this surface, the authors have identified selective activation of α5β1 integrin in the earlier stage of adhesion and conversion into αvβ3-mediated adhesion.
The use of photoresponsive proteins is mostly limited to proof-of-concept experiments; however, some studies have utilized the photoresponsive proteins for a fundamental understanding of cell–ECM interactions. Wegner et al. developed a reversibly controllable cell adhesion scaffold based on the blue light switchable protein LOV2.196 The RGD sequence inserted within the LOV2 is initially hidden in the dimerized state under dark conditions but becomes exposed upon blue light irradiation. This group also innovated another approach to control cell adhesion using green light-responsive CarH protein.197 Wang, Bian, and Cao have utilized a light-responsive protein, pdDronpa, as a linkage between a cell-adhesive ligand to the substrate for manipulating the ligand tethering.198 Owing to the photo-reversibly dimerizing property of pdDronpa, the author have successfully controlled YAP nuclear translocation in response to blue/green irradiation, resulting in changes in adipogenic/osteogenic differentiation in MSCs.198
In this way, the development of new tools as well as seeking adequate biological targets is crucial for the contribution of dynamic photoresponsive materials to fundamental biology.
4. Conclusions and future scope
Before closing this review, we discuss the future prospects of biomaterials for fundamental biological studies with reference to current limitations and bottlenecks.Compared to static materials, limited number of studies elegantly used dynamic biomaterials for obtaining biological insights. One reason is that these materials are not readily available to biological scientist. Therefore, materials scientists need to propose ideally one-pot synthesis methods or simple protocols for preparing dynamic materials. The integration of materials science and biology is expected to advance the understanding of cell behavior and responsiveness occurring within living organisms. From a perspective of time, dynamic materials, in principle, provide a useful platform for mimicking time-evolving biological events over various time ranges. To address rapid biological processes, the speed of response of materials to light (or other external stimuli) is crucial. However, most matrix changes in development and disease occur on a day-to-year scale. Therefore, the primary goal is to reduce these long processes to an experimentally accessible time range in laboratory. However, it remains unclear whether such time shortening could bias important biological processes, thereby resulting in incorrect outcomes. Furthermore, we must consider phototoxicity to discuss longer time-scale events. Therefore, the concept of bioadaptive materials, materials that sense biological signals and responses autonomously, would be useful. Also, it is difficult to manipulate targets located at distal sites, such as nuclear shape and chromatin structures in the cell interior, cell–cell junction biology, and cells located in living bodies. The use of nanoparticles, not discussed here, is one solution, although their precise manipulation is not easy at high spatiotemporal resolution. In this sense, the combination of advanced devices and optical technologies, or radical changes in material design strategy, such as in situ synthesis of materials via self-assembly, are required. Finally, the utilization of cutting-edge biological and engineering technologies is highly recommended, not only for material designs but also for their applications. For example, biomaterial design can be accelerated by adopting artificial intelligence and material informatics. In addition, patient-derived iPS cells and disease-mimetic cells/tissues reconstructed using genome editing have become easily available. Use of these new technologies will further accelerate material design and ensure that studies are more physiologically and pathologically significant.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by the Japan Society for Promotion of Science, Kakenhi (20K20645, 22H00596, and 22K14705).References
- H. K. Raut, R. Das, Z. Liu, X. Liu and S. Ramakrishna, Biotechnol. J., 2020, 15, 2000160 CrossRef CAS PubMed.
- O. S. Fenton, K. N. Olafson, P. S. Pillai, M. J. Mitchell and R. Langer, Adv. Mater., 2018, 30, 1705328 CrossRef PubMed.
- M. Weber, H. Steinle, S. Golombek, L. Hann, C. Schlensak, H. P. Wendel and M. Avci-Adali, Front. Bioeng. Biotechnol., 2018, 6, 99 CrossRef PubMed.
- K. S. Masters, Macromol. Biosci., 2011, 11, 1149–1163 CrossRef CAS PubMed.
- D. F. Williams, Bioact. Mater., 2022, 10, 306–322 CrossRef CAS PubMed.
- S. Di Cio and J. E. Gautrot, Acta Biomater., 2016, 30, 26–48 CrossRef CAS PubMed.
- B. Cheng, M. Lin, G. Huang, Y. Li, B. Ji, G. M. Genin, V. S. Deshpande, T. J. Lu and F. Xu, Phys. Life Rev., 2017, 22–23, 88–119 CrossRef PubMed.
- N. D. Donahue, H. Acar and S. Wilhelm, Adv. Drug Delivery Rev., 2019, 143, 68–96 CrossRef CAS PubMed.
- O. Chaudhuri, J. Cooper-White, P. A. Janmey, D. J. Mooney and V. B. Shenoy, Nature, 2020, 584, 535–546 CrossRef CAS.
- T. L. Rapp and C. A. DeForest, Adv. Healthcare Mater., 2020, 9, 1901553 CrossRef CAS.
- T. E. Brown and K. S. Anseth, Chem. Soc. Rev., 2017, 46, 6532–6552 RSC.
- B. R. Seo and D. J. Mooney, ACS Biomater. Sci. Eng., 2022 DOI:10.1021/acsbiomaterials.1c01477.
- P. Cai, W. R. Leow, X. Wang, Y.-L. Wu and X. Chen, Adv. Mater., 2017, 29, 1605529 CrossRef PubMed.
- B. N. Narasimhan, M. S. Horrocks and J. Malmström, Adv. NanoBiomed. Res., 2021, 1, 2100059 CrossRef.
- J. Nakanishi and K. Uto, Material-based Mechanobiology, Royal Society of Chemistry, Cambridge, UK, 2022 Search PubMed.
- D. E. Discher, P. Janmey and Y.-L. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS.
- K. K. Sanford, G. D. Likely and W. R. Earle, J. Natl. Cancer Inst., 1954, 15, 215–237 CAS.
- H. M. Temin and H. Rubin, Virology, 1958, 6, 669–688 CrossRef CAS PubMed.
- R. J. Pelham and Y.-L. Wang, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 13661–13665 CrossRef CAS PubMed.
- A. J. Engler, S. Sen, H. L. Sweeney and D. E. Discher, Cell, 2006, 126, 677–689 CrossRef CAS PubMed.
- M. R. Ng, A. Besser, G. Danuser and J. S. Brugge, J. Cell Biol., 2012, 199, 545–563 CrossRef CAS PubMed.
- S. C. Wei, L. Fattet, J. H. Tsai, Y. R. Guo, V. H. Pai, H. E. Majeski, A. C. Chen, R. L. Sah, S. S. Taylor, A. J. Engler and J. Yang, Nat. Cell Biol., 2015, 17, 678–688 CrossRef CAS PubMed.
- B. F. Matte, A. Kumar, J. K. Placone, V. G. Zanella, M. D. Martins, A. J. Engler and M. L. Lamers, J. Cell Sci., 2019, 132, jcs224360 Search PubMed.
- F. Kai, H. Laklai and V. M. Weaver, Trends Cell Biol., 2016, 26, 486–497 CrossRef CAS.
- J. L. Leight, M. A. Wozniak, S. Chen, M. L. Lynch and C. S. Chen, Mol. Biol. Cell, 2012, 23, 781–791 CrossRef CAS PubMed.
- B. Hinz, Matrix Biol., 2015, 47, 54–65 CrossRef CAS PubMed.
- T. Razafiarison, C. N. Holenstein, T. Stauber, M. Jovic, E. Vertudes, M. Loparic, M. Kawecki, L. Bernard, U. Silvan and J. G. Snedeker, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4631–4636 CrossRef CAS.
- B. Trappmann, J. E. Gautrot, J. T. Connelly, D. G. T. Strange, Y. Li, M. L. Oyen, M. A. Cohen Stuart, H. Boehm, B. Li, V. Vogel, J. P. Spatz, F. M. Watt and W. T. S. Huck, Nat. Mater., 2012, 11, 642–649 CrossRef CAS PubMed.
- J. D. Mih, A. S. Sharif, F. Liu, A. Marinkovic, M. M. Symer and D. J. Tschumperlin, PLoS One, 2011, 6, e19929 CrossRef CAS PubMed.
- B. Díaz-Bello, A. X. Monroy-Romero, D. Pérez-Calixto, D. Zamarrón-Hernández, N. Serna-Marquez, G. Vázquez-Victorio and M. Hautefeuille, ACS Biomater. Sci. Eng., 2019, 5, 4219–4227 CrossRef.
- R. Lei, E. A. Akins, K. C. Y. Wong, N. A. Repina, K. J. Wolf, G. E. Dempsey, D. V. Schaffer, A. Stahl and S. Kumar, ACS Biomater. Sci. Eng., 2021, 7, 2453–2465 CrossRef CAS PubMed.
- C.-M. Lo, H.-B. Wang, M. Dembo and Y.-L. Wang, Biophys. J., 2000, 79, 144–152 CrossRef CAS PubMed.
- S. Kidoaki and T. Matsuda, J. Biotechnol., 2008, 133, 225–230 CrossRef CAS.
- A. Ueki and S. Kidoaki, Biomaterials, 2015, 41, 45–52 CrossRef CAS PubMed.
- H. Ebata and S. Kidoaki, Biomaterials, 2021, 274, 120860 CrossRef CAS PubMed.
- J. C. Kohn, A. Chen, S. Cheng, D. R. Kowal, M. R. King and C. A. Reinhart-King, J. Biomech., 2016, 49, 1447–1453 CrossRef PubMed.
- J. A. VanderBurgh, H. Hotchkiss, A. Potharazu, P. V. Taufalele and C. A. Reinhart-King, Integr. Biol., 2018, 10, 734–746 CrossRef CAS PubMed.
- M. C. Lampi, M. Guvendiren, J. A. Burdick and C. A. Reinhart-King, ACS Biomater. Sci. Eng., 2017, 3, 3007–3016 CrossRef CAS.
- A. R. Cameron, J. E. Frith and J. J. Cooper-White, Biomaterials, 2011, 32, 5979–5993 CrossRef CAS PubMed.
- A. R. Cameron, J. E. Frith, G. A. Gomez, A. S. Yap and J. J. Cooper-White, Biomaterials, 2014, 35, 1857–1868 CrossRef CAS.
- M. Murrell, R. Kamm and P. Matsudaira, Biophys. J., 2011, 101, 297–306 CrossRef CAS PubMed.
- J. Y. Zheng, S. P. Han, Y.-J. Chiu, A. K. Yip, N. Boichat, S. W. Zhu, J. Zhong and P. Matsudaira, Biophys. J., 2017, 113, 1585–1598 CrossRef CAS PubMed.
- O. Chaudhuri, L. Gu, M. Darnell, D. Klumpers, S. A. Bencherif, J. C. Weaver, N. Huebsch and D. J. Mooney, Nat. Commun., 2015, 6, 7365 CrossRef PubMed.
- O. Chaudhuri, L. Gu, D. Klumpers, M. Darnell, S. A. Bencherif, J. C. Weaver, N. Huebsch, H.-P. Lee, E. Lippens, G. N. Duda and D. J. Mooney, Nat. Mater., 2016, 15, 326–334 CrossRef CAS PubMed.
- Z. Gong, S. E. Szczesny, S. R. Caliari, E. E. Charrier, O. Chaudhuri, X. Cao, Y. Lin, R. L. Mauck, P. A. Janmey, J. A. Burdick and V. B. Shenoy, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E2686–E2695 CAS.
- K. Adebowale, Z. Gong, J. C. Hou, K. M. Wisdom, D. Garbett, H. P. Lee, S. Nam, T. Meyer, D. J. Odde, V. B. Shenoy and O. Chaudhuri, Nat. Mater., 2021, 20, 1290–1299 CrossRef CAS PubMed.
- J.-H. Seo, S. Kakinoki, Y. Inoue, T. Yamaoka, K. Ishihara and N. Yui, J. Am. Chem. Soc., 2013, 135, 5513–5516 CrossRef CAS PubMed.
- J.-H. Seo and N. Yui, Biomaterials, 2013, 34, 55–63 CrossRef CAS PubMed.
- J.-H. Seo, M. Hirata, S. Kakinoki, T. Yamaoka and N. Yui, RSC Adv., 2016, 6, 35668–35676 RSC.
- L. Yu, Y. Hou, W. Xie, J. L. C. Camacho, C. Cheng, A. Holle, J. Young, B. Trappmann, W. Zhao, M. F. Melzig, E. A. Cavalcanti-Adam, C. Zhao, J. P. Spatz, Q. Wei and R. Haag, Adv. Mater., 2020, 32, 2002566 CrossRef CAS PubMed.
- A. C. Chang, K. Uto, K. Homma and J. Nakanishi, Biomaterials, 2021, 274, 120861 CrossRef CAS PubMed.
- A. C. Chang, K. Uto, S. A. Abdellatef and J. Nakanishi, Langmuir, 2022, 38, 5307–5314 CrossRef CAS PubMed.
- M. D. Rosenberg, in Cellular Control Mechanisms and Cancer, ed. P. Emmelot and O. Mohbock, Elsevier, Amsterdam, 1964, pp. 146–164 Search PubMed.
- I. Giaever and C. R. Keese, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 219–222 CrossRef CAS PubMed.
- C. R. Keese and I. Giaever, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 5622–5626 CrossRef CAS.
- D. Kong, K. D. Q. Nguyen, W. Megone, L. Peng and J. E. Gautrot, Faraday Discuss., 2017, 204, 367–381 RSC.
- K. Minami, T. Mori, W. Nakanishi, N. Shigi, J. Nakanishi, J. P. Hill, M. Komiyama and K. Ariga, ACS Appl. Mater. Interfaces, 2017, 9, 30553–30560 CrossRef CAS PubMed.
- D. Kong, L. Peng, S. Di Cio, P. Novak and J. E. Gautrot, ACS Nano, 2018, 12, 9206–9213 CrossRef CAS.
- L. Peng and J. E. Gautrot, Mater. Today Bio, 2021, 12, 100159 CrossRef CAS PubMed.
- X. Jia, K. Minami, K. Uto, A. C. Chang, J. P. Hill, J. Nakanishi and K. Ariga, Adv. Mater., 2020, 32, 1905942 CrossRef CAS.
- C. Jensen and Y. Teng, Front. Mol. Biosci., 2020, 7, 33 CrossRef CAS PubMed.
- M. P. Lutolf, J. L. Lauer-Fields, H. G. Schmoekel, A. T. Metters, F. E. Weber, G. B. Fields and J. A. Hubbell, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5413–5418 CrossRef CAS.
- Q. Xing, Z. Qian, W. Jia, A. Ghosh, M. Tahtinen and F. Zhao, ACS Biomater. Sci. Eng., 2017, 3, 1462–1476 CrossRef CAS PubMed.
- T. Panciera, A. Citron, D. Di Biagio, G. Battilana, A. Gandin, S. Giulitti, M. Forcato, S. Bicciato, V. Panzetta, S. Fusco, L. Azzolin, A. Totaro, A. P. Dei Tos, M. Fassan, V. Vindigni, F. Bassetto, A. Rosato, G. Brusatin, M. Cordenonsi and S. Piccolo, Nat. Mater., 2020, 19, 797–806 CrossRef CAS PubMed.
- S. Tang, B. M. Richardson and K. S. Anseth, Prog. Mater. Sci., 2021, 120, 100738 CrossRef CAS.
- B. J. Adzima, H. A. Aguirre, C. J. Kloxin, T. F. Scott and C. N. Bowman, Macromolecules, 2008, 41, 9112–9117 CrossRef CAS PubMed.
- F. Liu, F. Li, G. Deng, Y. Chen, B. Zhang, J. Zhang and C.-Y. Liu, Macromolecules, 2012, 45, 1636–1645 CrossRef CAS.
- V. Yesilyurt, A. M. Ayoob, E. A. Appel, J. T. Borenstein, R. Langer and D. G. Anderson, Adv. Mater., 2017, 29, 1605947 CrossRef PubMed.
- S. Tang, H. Ma, H.-C. Tu, H.-R. Wang, P.-C. Lin and K. S. Anseth, Adv. Sci., 2018, 5, 1800638 CrossRef PubMed.
- D. J. Andrew and A. J. Ewald, Dev. Biol., 2010, 341, 34–55 CrossRef CAS PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS PubMed.
- R. Singhvi, A. Kumar, G. P. Lopez, G. N. Stephanopoulos, D. I. C. Wang, G. M. Whitesides and D. E. Ingber, Science, 1994, 264, 696–698 CrossRef CAS PubMed.
- K. K. Parker, A. L. Brock, C. Brangwynne, R. J. Mannix, N. Wang, E. Ostuni, N. A. Geisse, J. C. Adams, G. M. Whitesides and D. E. Ingber, FASEB J., 2002, 16, 1195–1204 CrossRef CAS PubMed.
- M. Thery, V. Racine, A. Pepin, M. Piel, Y. Chen, J. B. Sibarita and M. Bornens, Nat. Cell Biol., 2005, 7, 947–953 CrossRef CAS PubMed.
- M. Thery, A. Jimenez-Dalmaroni, V. Racine, M. Bornens and F. Julicher, Nature, 2007, 447, 493–U496 CrossRef CAS PubMed.
- R. McBeath, D. M. Pirone, M. N. Celeste, K. Bhadriraju and C. S. Chen, Dev. Cell, 2004, 6, 483–495 CrossRef CAS.
- K. A. Kilian, B. Bugarija, B. T. Lahn and M. Mrksich, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4872–4877 CrossRef CAS PubMed.
- S. Dupont, L. Morsut, M. Aragona, E. Enzo, S. Giulitti, M. Cordenonsi, F. Zanconato, J. Le Digabel, M. Forcato, S. Bicciato, N. Elvassore and S. Piccolo, Nature, 2011, 474, 179–183 CrossRef CAS PubMed.
- K. L. Ellefsen, J. R. Holt, A. C. Chang, J. L. Nourse, J. Arulmoli, A. H. Mekhdjian, H. Abuwarda, F. Tombola, L. A. Flanagan, A. R. Dunn, I. Parker and M. M. Pathak, Commun. Biol., 2019, 2, 298 CrossRef.
- T. C. von Erlach, S. Bertazzo, M. A. Wozniak, C.-M. Horejs, S. A. Maynard, S. Attwood, B. K. Robinson, H. Autefage, C. Kallepitis, A. del Río Hernández, C. S. Chen, S. Goldoni and M. M. Stevens, Nat. Mater., 2018, 17, 237–242 CrossRef CAS.
- C. M. Nelson, R. P. Jean, J. L. Tan, W. F. Liu, N. J. Sniadecki, A. A. Spector and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11594–11599 CrossRef CAS PubMed.
- E. W. Gomez, Q. K. Chen, N. Gjorevski and C. M. Nelson, J. Cell. Biochem., 2010, 110, 44–51 CAS.
- L. Q. Wan, K. Ronaldson, M. Park, G. Taylor, Y. Zhang, J. M. Gimble and G. Vunjak-Novakovic, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12295–12300 CrossRef CAS PubMed.
- A. S. Chin, K. E. Worley, P. Ray, G. Kaur, J. Fan and L. Q. Wan, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 12188–12193 CrossRef CAS PubMed.
- C. M. Nelson, M. M. Vanduijn, J. L. Inman, D. A. Fletcher and M. J. Bissell, Science, 2006, 314, 298–300 CrossRef CAS PubMed.
- O. Y. Dudaryeva, A. Bucciarelli, G. Bovone, F. Huwyler, S. Jaydev, N. Broguiere, M. al-Bayati, M. Lütolf and M. W. Tibbitt, Adv. Funct. Mater., 2021, 31, 2104098 CrossRef CAS.
- J. Lee, T. G. Molley, C. H. Seward, A. A. Abdeen, H. Zhang, X. Wang, H. Gandhi, J.-L. Yang, K. Gaus and K. A. Kilian, Commun. Biol., 2020, 3, 341 CrossRef CAS.
- N. Gjorevski, M. Nikolaev, T. E. Brown, O. Mitrofanova, N. Brandenberg, F. W. DelRio, F. M. Yavitt, P. Liberali, K. S. Anseth and M. P. Lutolf, Science, 2022, 375, eaaw9021 CrossRef CAS.
- R. G. Harrison, J. Exp. Zool., 1914, 17, 521–544 CrossRef.
- P. Weiss, J. Exp. Zool., 1934, 68, 393–448 CrossRef.
- C. Leclech and C. Villard, Front. Bioeng. Biotechnol., 2020, 8, 551505 CrossRef PubMed.
- M. M. Machado-Paula, M. A. F. Corat, M. Lancellotti, G. Mi, F. R. Marciano, M. L. Vega, A. A. Hidalgo, T. J. Webster and A. O. Lobo, Mater. Sci. Eng., C, 2020, 111, 110706 CrossRef CAS PubMed.
- T. J. Sill and H. A. von Recum, Biomaterials, 2008, 29, 1989–2006 CrossRef CAS PubMed.
- C. N. LaFratta, J. T. Fourkas, T. Baldacchini and R. A. Farrer, Angew. Chem., Int. Ed., 2007, 46, 6238–6258 CrossRef CAS.
- D.-H. Kim, P. P. Provenzano, C. L. Smith and A. Levchenko, J. Cell Biol., 2012, 197, 351–360 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS.
- R. J. McMurray, N. Gadegaard, P. M. Tsimbouri, K. V. Burgess, L. E. McNamara, R. Tare, K. Murawski, E. Kingham, R. O. C. Oreffo and M. J. Dalby, Nat. Mater., 2011, 10, 637–644 CrossRef CAS.
- P. M. Tsimbouri, R. J. McMurray, K. V. Burgess, E. V. Alakpa, P. M. Reynolds, K. Murawski, E. Kingham, R. O. C. Oreffo, N. Gadegaard and M. J. Dalby, ACS Nano, 2012, 6, 10239–10249 CrossRef CAS PubMed.
- J. Song, X. Jia, K. Minami, J. P. Hill, J. Nakanishi, L. K. Shrestha and K. Ariga, ACS Appl. Nano Mater., 2020, 3, 6497–6506 CrossRef CAS.
- B. Zhang, Y. Xiao, A. Hsieh, N. Thavandiran and M. Radisic, Nanotechnology, 2011, 22, 494003 CrossRef PubMed.
- M. Tallawi, R. Rai, R. Boccaccini and K. E. Aifantis, Tissue Eng., Part B, 2015, 21, 157–165 CrossRef CAS PubMed.
- P.-Y. Wang, H.-T. Yu and W.-B. Tsai, Biotechnol. Bioeng., 2010, 106, 285–294 CrossRef CAS PubMed.
- Kshitiz, J. Afzal, S.-Y. Kim and D.-H. Kim, Cell Adhes. Migr., 2015, 9, 300–307 CrossRef CAS.
- D.-H. Kim, K. Han, K. Gupta, K. W. Kwon, K.-Y. Suh and A. Levchenko, Biomaterials, 2009, 30, 5433–5444 CrossRef CAS PubMed.
- M. K. Driscoll, X. Sun, C. Guven, J. T. Fourkas and W. Losert, ACS Nano, 2014, 8, 3546–3555 CrossRef CAS PubMed.
- R. S. Fischer, X. Sun, M. A. Baird, M. J. Hourwitz, B. R. Seo, A. M. Pasapera, S. B. Mehta, W. Losert, C. Fischbach, J. T. Fourkas and C. M. Waterman, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2021135118 CrossRef CAS PubMed.
- A. B. C. Buskermolen, T. Ristori, D. Mostert, M. C. van Turnhout, S. S. Shishvan, S. Loerakker, N. A. Kurniawan, V. S. Deshpande and C. V. C. Bouten, Cell Rep. Phys. Sci., 2020, 1, 100055 CrossRef.
- A. Ray, O. Lee, Z. Win, R. M. Edwards, P. W. Alford, D.-H. Kim and P. P. Provenzano, Nat. Commun., 2017, 8, 14923 CrossRef CAS PubMed.
- C. Leclech and A. I. Barakat, Cytoskeleton, 2021, 78, 284–292 CrossRef PubMed.
- M. Arnold, E. A. Cavalcanti-Adam, R. Glass, J. Blummel, W. Eck, M. Kantlehner, H. Kessler and J. P. Spatz, ChemPhysChem, 2004, 5, 383–388 CrossRef CAS PubMed.
- J. P. Spatz and B. Geiger, Methods Cell Biol., 2007, 83, 89–111 CAS.
- D. Aydin, I. Louban, N. Perschmann, J. Blummel, T. Lohmuller, E. A. Cavalcanti-Adam, T. L. Haas, H. Walczak, H. Kessler, R. Fiammengo and J. P. Spatz, Langmuir, 2010, 26, 15472–15480 CrossRef CAS PubMed.
- R. Oria, T. Wiegand, J. Escribano, A. Elosegui-Artola, J. J. Uriarte, C. Moreno-Pulido, I. Platzman, P. Delcanale, L. Albertazzi, D. Navajas, X. Trepat, J. M. García-Aznar, E. A. Cavalcanti-Adam and P. Roca-Cusachs, Nature, 2017, 552, 219 CrossRef CAS PubMed.
- S. R. Coyer, A. Singh, D. W. Dumbauld, D. A. Calderwood, S. W. Craig, E. Delamarche and A. J. Garcia, J. Cell Sci., 2012, 125, 5110–5123 CAS.
- R. Changede, H. Cai, S. J. Wind and M. P. Sheetz, Nat. Mater., 2019, 18, 1366–1375 CrossRef CAS PubMed.
- M. Thery, V. Racine, M. Piel, A. Pepin, A. Dimitrov, Y. Chen, J. B. Sibarita and M. Bornens, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19771–19776 CrossRef CAS PubMed.
- F. Senger, A. Pitaval, H. Ennomani, L. Kurzawa, L. Blanchoin and M. Thery, J. Cell Sci., 2019, 132, jcs236604 CrossRef CAS PubMed.
- H.-Y. Lou, W. Zhao, Y. Zeng and B. Cui, Acc. Chem. Res., 2018, 51, 1046–1053 CrossRef CAS PubMed.
- S. G. Higgins, M. Becce, A. Belessiotis-Richards, H. Seong, J. E. Sero and M. M. Stevens, Adv. Mater., 2020, 32, 1903862 CrossRef CAS.
- Y. Chen, J. Wang, X. Li, N. Hu, N. H. Voelcker, X. Xie and R. Elnathan, Adv. Mater., 2020, 32, 2001668 CrossRef CAS PubMed.
- H.-Y. Lou, W. Zhao, X. Li, L. Duan, A. Powers, M. Akamatsu, F. Santoro, A. F. McGuire, Y. Cui, D. G. Drubin and B. Cui, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 23143–23151 CrossRef CAS.
- W. Zhao, L. Hanson, H.-Y. Lou, M. Akamatsu, P. D. Chowdary, F. Santoro, J. R. Marks, A. Grassart, D. G. Drubin, Y. Cui and B. Cui, Nat. Nanotechnol., 2017, 12, 750–756 CrossRef CAS PubMed.
- S. Gopal, C. Chiappini, J. Penders, V. Leonardo, H. Seong, S. Rothery, Y. Korchev, A. Shevchuk and M. M. Stevens, Adv. Mater., 2019, 31, 1806788 CrossRef PubMed.
- H. Liang, H. Mu, F. Jean-Francois, B. Lakshman, S. Sarkar-Banerjee, Y. Zhuang, Y. Zeng, W. Gao, A. M. Zaske, D. V. Nissley, A. A. Gorfe, W. Zhao and Y. Zhou, Life Sci. Alliance, 2019, 2, e201900343 CrossRef.
- K. Uto, J. H. Tsui, C. A. DeForest and D.-H. Kim, Prog. Polym. Sci., 2017, 65, 53–82 CrossRef CAS.
- A. M. Rosales and K. S. Anseth, Nat. Rev. Mater., 2016, 1, 15012 CrossRef CAS PubMed.
- C. Bao, L. Zhu, Q. Lin and H. Tian, Adv. Mater., 2015, 27, 1647–1662 CrossRef CAS PubMed.
- H. Zhao, E. S. Sterner, E. B. Coughlin and P. Theato, Macromolecules, 2012, 45, 1723–1736 CrossRef CAS.
- A. Specht and M. Goeldner, Angew. Chem., Int. Ed., 2004, 43, 2008–2012 CrossRef CAS PubMed.
- H. M. D. Bandara and S. C. Burdette, Chem. Soc. Rev., 2012, 41, 1809–1825 RSC.
- L. Kortekaas and W. R. Browne, Chem. Soc. Rev., 2019, 48, 3406–3424 RSC.
- S. R. Trenor, A. R. Shultz, B. J. Love and T. E. Long, Chem. Rev., 2004, 104, 3059–3078 CrossRef CAS PubMed.
- I. Novak, L. Klasinc and S. P. McGlynn, Chem. Phys. Lett., 2019, 728, 50–52 CrossRef CAS.
- T. Furuta, S. S. H. Wang, J. L. Dantzker, T. M. Dore, W. J. Bybee, E. M. Callaway, W. Denk and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 1193–1200 CrossRef CAS.
- B. D. Fairbanks, M. P. Schwartz, C. N. Bowman and K. S. Anseth, Biomaterials, 2009, 30, 6702–6707 CrossRef CAS.
- T. F. Scott, A. D. Schneider, W. D. Cook and C. N. Bowman, Science, 2005, 308, 1615–1617 CrossRef CAS PubMed.
- J. Zhao, H. Su, G. E. Vansuch, Z. Liu, K. Salaita and R. B. Dyer, ACS Nano, 2019, 13, 515–525 CrossRef CAS PubMed.
- Y. Nihongaki, Y. Furuhata, T. Otabe, S. Hasegawa, K. Yoshimoto and M. Sato, Nat. Methods, 2017, 14, 963–966 CrossRef CAS.
- D. Niopek, P. Wehler, J. Roensch, R. Eils and B. Di Ventura, Nat. Commun., 2016, 7, 10624 CrossRef CAS PubMed.
- H. Yumerefendi, A. M. Lerner, S. P. Zimmerman, K. Hahn, J. E. Bear, B. D. Strahl and B. Kuhlman, Nat. Chem. Biol., 2016, 12, 399–401 CrossRef CAS PubMed.
- Y. Shin, J. Berry, N. Pannucci, M. P. Haataja, J. E. Toettcher and C. P. Brangwynne, Cell, 2017, 168, 159–171 CrossRef CAS.
- D. Bracha, M. T. Walls, M.-T. Wei, L. Zhu, M. Kurian, J. L. Avalos, J. E. Toettcher and C. P. Brangwynne, Cell, 2018, 175, 1467–1480 CrossRef CAS.
- T.-J. Oh, H. Fan, S. S. Skeeters and K. Zhang, Adv. Biol., 2021, 5, 2000180 CrossRef PubMed.
- J. I. Spiltoir and C. L. Tucker, Curr. Opin. Struct. Biol., 2019, 57, 1–8 CrossRef CAS PubMed.
- Y. Li, B. Xue and Y. Cao, ACS Macro Lett., 2020, 9, 512–524 CrossRef CAS PubMed.
- P. Klán, T. Šolomek, C. G. Bochet, A. Blanc, R. Givens, M. Rubina, V. Popik, A. Kostikov and J. Wirz, Chem. Rev., 2013, 113, 119–191 CrossRef PubMed.
- S. Yamamoto, H. Ikegami, K. Yamaguchi and J. Nakanishi, ChemPhotoChem, 2018, 2, 786–790 CrossRef CAS.
- A. A. Beharry, O. Sadovski and G. A. Woolley, J. Am. Chem. Soc., 2011, 133, 19684–19687 CrossRef CAS PubMed.
- C. A. DeForest and K. S. Anseth, Nat. Chem., 2011, 3, 925–931 CrossRef CAS.
- A. D. Rape, M. Zibinsky, N. Murthy and S. Kumar, Nat. Commun., 2015, 6, 8129 CrossRef.
- M. T. Frey and Y.-L. Wang, Soft Matter, 2009, 5, 1918–1924 RSC.
- C. Yang, M. W. Tibbitt, L. Basta and K. S. Anseth, Nat. Mater., 2014, 13, 645 CrossRef CAS PubMed.
- C. J. Walker, C. Crocini, D. Ramirez, A. R. Killaars, J. C. Grim, B. A. Aguado, K. Clark, M. A. Allen, R. D. Dowell, L. A. Leinwand and K. S. Anseth, Nat. Biomed. Eng., 2021, 5, 1485–1499 CrossRef CAS PubMed.
- J. Winkler, A. Abisoye-Ogunniyan, K. J. Metcalf and Z. Werb, Nat. Commun., 2020, 11, 5120 CrossRef CAS PubMed.
- M. Guvendiren and J. A. Burdick, Nat. Commun., 2012, 3, 792 CrossRef.
- M. G. Ondeck, A. Kumar, J. K. Placone, C. M. Plunkett, B. F. Matte, K. C. Wong, L. Fattet, J. Yang and A. J. Engler, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 3502–3507 CrossRef CAS.
- A. M. Rosales, S. L. Vega, F. W. DelRio, J. A. Burdick and K. S. Anseth, Angew. Chem., Int. Ed., 2017, 56, 12132–12136 CrossRef CAS PubMed.
- I. N. Lee, O. Dobre, D. Richards, C. Ballestrem, J. M. Curran, J. A. Hunt, S. M. Richardson, J. Swift and L. S. Wong, ACS Appl. Mater. Interfaces, 2018, 10, 7765–7776 CrossRef CAS PubMed.
- A. M. Rosales, C. B. Rodell, M. H. Chen, M. G. Morrow, K. S. Anseth and J. A. Burdick, Bioconjugate Chem., 2018, 29, 905–913 CrossRef CAS PubMed.
- K. Homma, A. C. Chang, S. Yamamoto, R. Tamate, T. Ueki and J. Nakanishi, Acta Biomater., 2021, 132, 103–113 CrossRef CAS.
- R. Tamate, T. Ueki, Y. Kitazawa, M. Kuzunuki, M. Watanabe, A. M. Akimoto and R. Yoshida, Chem. Mater., 2016, 28, 6401–6408 CrossRef CAS.
- K. A. Gunay, T. L. Ceccato, J. S. Silver, K. L. Bannister, O. J. Bednarski, L. A. Leinwand and K. S. Anseth, Angew. Chem., Int. Ed., 2019, 58, 9912–9916 CrossRef.
- L. Liu, J. A. Shadish, C. K. Arakawa, K. Shi, J. Davis and C. A. DeForest, Adv. Biosyst., 2018, 2, 1800240 CrossRef PubMed.
- M. Hörner, K. Raute, B. Hummel, J. Madl, G. Creusen, O. S. Thomas, E. H. Christen, N. Hotz, R. J. Gübeli, R. Engesser, B. Rebmann, J. Lauer, B. Rolauffs, J. Timmer, W. W. A. Schamel, J. Pruszak, W. Römer, M. D. Zurbriggen, C. Friedrich, A. Walther, S. Minguet, R. Sawarkar and W. Weber, Adv. Mater., 2019, 31, 1806727 CrossRef PubMed.
- Y. Zheng, M. K. Liong Han, Q. Jiang, B. Li, J. Feng and A. del Campo, Mater. Horiz., 2020, 7, 111–116 RSC.
- O. Friedrich, A. L. Merten, D. Schneidereit, Y. Guo, S. Schurmann and B. Martinac, Front. Bioeng. Biotechnol., 2019, 7, 55 CrossRef PubMed.
- Y. S. Zhang, G. Haghiashtiani, T. Hübscher, D. J. Kelly, J. M. Lee, M. Lutolf, M. C. McAlpine, W. Y. Yeong, M. Zenobi-Wong and J. Malda, Nat. Rev. Methods Primers, 2021, 1, 75 CrossRef CAS.
- J. Nakanishi, Chem. Rec., 2017, 17, 611–621 CrossRef CAS PubMed.
- J. Nakanishi, Chem. – Asian J., 2014, 9, 406–417 CrossRef CAS PubMed.
- Y. Kikuchi, J. Nakanishi, H. Nakayama, T. Shimizu, Y. Yoshino, K. Yamaguchi, Y. Yoshida and Y. Horiike, Chem. Lett., 2008, 37, 1062–1063 CrossRef CAS.
- J. Nakanishi, Y. Kikuchi, T. Takarada, H. Nakayama, K. Yamaguchi and M. Maeda, J. Am. Chem. Soc., 2004, 126, 16314–16315 CrossRef CAS PubMed.
- J. Nakanishi, Y. Kikuchi, S. Inoue, K. Yamaguchi, T. Takarada and M. Maeda, J. Am. Chem. Soc., 2007, 129, 6694–6695 CrossRef CAS.
- J. Nakanishi, Y. Kikuchi, T. Takarada, H. Nakayama, K. Yamaguchi and M. Maeda, Anal. Chim. Acta, 2006, 578, 100–104 CrossRef CAS.
- M. Kamimura, O. Scheideler, Y. Shimizu, S. Yamamoto, K. Yamaguchi and J. Nakanishi, Phys. Chem. Chem. Phys., 2015, 17, 14159–14167 RSC.
- Y. Shimizu, M. Kamimura, S. Yamamoto, S. A. Abdellatef, K. Yamaguchi and J. Nakanishi, Anal. Sci., 2016, 32, 1183–1188 CrossRef CAS PubMed.
- M. Kamimura, M. Sugawara, S. Yamamoto, K. Yamaguchi and J. Nakanishi, Biomater. Sci., 2016, 4, 933–937 RSC.
- S. Yamamoto, K. Okada, N. Sasaki, A. C. Chang, K. Yamaguchi and J. Nakanishi, Langmuir, 2019, 35, 7459–7468 CrossRef CAS PubMed.
- S. A. Abdellatef and J. Nakanishi, Biomaterials, 2018, 169, 72–84 CrossRef CAS PubMed.
- A. Ravikrishnan, E. W. Fowler, A. J. Stuffer and X. Jia, ACS Biomater. Sci. Eng., 2021, 7, 4305–4317 CrossRef CAS.
- Y. Ohmuro-Matsuyama and Y. Tatsu, Angew. Chem., Int. Ed., 2008, 47, 7527–7529 CrossRef CAS.
- S. Petersen, J. M. Alonso, A. Specht, P. Duodu, M. Goeldner and A. del Campo, Angew. Chem., Int. Ed., 2008, 47, 3192–3195 CrossRef CAS PubMed.
- M. J. Salierno, L. García-Fernandez, N. Carabelos, K. Kiefer, A. J. García and A. D. Campo, Biomaterials, 2016, 82, 113–123 CrossRef CAS.
- R. G. Wylie, S. Ahsan, Y. Aizawa, K. L. Maxwell, C. M. Morshead and M. S. Shoichet, Nat. Mater., 2011, 10, 799–806 CrossRef CAS PubMed.
- I. Batalov, K. R. Stevens and C. A. DeForest, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2014194118 CrossRef CAS.
- E. Dolgin, Nature, 2018, 555, 300–302 CrossRef CAS.
- C. Matthaeus and J. W. Taraska, Front. Cell Dev. Biol., 2021, 8, 614472 CrossRef PubMed.
- Y. Shimizu, H. Boehm, K. Yamaguchi, J. P. Spatz and J. Nakanishi, PLoS One, 2014, 9, e91875 CrossRef PubMed.
- S. De Martino, F. Mauro and P. A. Netti, La Rivista del Nuovo Cimento, 2020, 43, 599–629 CrossRef CAS.
- S. De Martino, W. Zhang, L. Klausen, H.-Y. Lou, X. Li, F. S. Alfonso, S. Cavalli, P. A. Netti, F. Santoro and B. Cui, Nano Lett., 2020, 20, 577–584 CrossRef CAS PubMed.
- S. De Martino, S. Cavalli and P. A. Netti, Adv. Healthcare Mater., 2020, 9, 2000470 CrossRef CAS.
- L. F. Kadem, K. G. Suana, M. Holz, W. Wang, H. Westerhaus, R. Herges and C. Selhuber-Unkel, Angew. Chem., Int. Ed., 2017, 56, 225–229 CrossRef CAS PubMed.
- Z. Liu, Y. Liu, Y. Chang, H. R. Seyf, A. Henry, A. L. Mattheyses, K. Yehl, Y. Zhang, Z. Huang and K. Salaita, Nat. Methods, 2016, 13, 143–146 CrossRef CAS PubMed.
- A. N. Ramey-Ward, H. Su and K. Salaita, ACS Appl. Mater. Interfaces, 2020, 12, 35903–35917 CrossRef CAS PubMed.
- Y. Zheng, M. K. L. Han, R. Zhao, J. Blass, J. Zhang, D. W. Zhou, J.-R. Colard-Itté, D. Dattler, A. Çolak, M. Hoth, A. J. García, B. Qu, R. Bennewitz, N. Giuseppone and A. del Campo, Nat. Commun., 2021, 12, 3580 CrossRef CAS PubMed.
- L. Yu, Y. Hou, W. Xie, J. L. Cuellar-Camacho, Q. Wei and R. Haag, Adv. Mater., 2020, 32, e2006986 CrossRef.
- J. Ricken, R. Medda and S. V. Wegner, Adv. Biosyst., 2019, 3, 1800302 CrossRef.
- D. Xu, J. Ricken and S. V. Wegner, Chem. – Eur. J., 2020, 26, 9859–9863 CrossRef CAS PubMed.
- J. Zhang, S. H. D. Wong, X. Wu, H. Lei, M. Qin, P. Shi, W. Wang, L. Bian and Y. Cao, Adv. Mater., 2021, 33, 2105765 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |