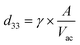Open Access Article
Open Access ArticlePiezoelectric nanocomposite bioink and ultrasound stimulation modulate early skeletal myogenesis†
Claudia
Paci
 *ab,
Federica
Iberite
*ab,
Federica
Iberite
 ab,
Lorenzo
Arrico
ab,
Lorenzo
Arrico
 ab,
Lorenzo
Vannozzi
ab,
Lorenzo
Vannozzi
 ab,
Paola
Parlanti
ab,
Paola
Parlanti
 c,
Mauro
Gemmi
c,
Mauro
Gemmi
 c and
Leonardo
Ricotti
c and
Leonardo
Ricotti
 ab
ab
aThe BioRobotics Institute, Scuola Superiore Sant'Anna, Piazza Martiri della Libertà 33, 56127 Pisa, Italy. E-mail: claudia.paci@santannapisa.it; Tel: +39 050 883074
bDepartment of Excellence in Robotics & AI, Scuola Superiore Sant'Anna, Piazza Martiri della Libertà 33, 56127 Pisa, Italy
cIstituto Italiano di Tecnologia, Center for Materials Interfaces, Electron Crystallography, Viale Rinaldo Piaggio 34, 56025 Pontedera, Italy
First published on 1st August 2022
Abstract
Despite the significant progress in bioprinting for skeletal muscle tissue engineering, new stimuli-responsive bioinks to boost the myogenesis process are highly desirable. In this work, we developed a printable alginate/Pluronic-based bioink including piezoelectric barium titanate nanoparticles (nominal diameter: ∼60 nm) for the 3D bioprinting of muscle cell-laden hydrogels. The aim was to investigate the effects of the combination of piezoelectric nanoparticles with ultrasound stimulation on early myogenic differentiation of the printed structures. After the characterization of nanoparticles and bioinks, viability tests were carried out to investigate three nanoparticle concentrations (100, 250, and 500 μg mL−1) within the printed structures. An excellent cytocompatibility was confirmed for nanoparticle concentrations up to 250 μg mL−1. TEM imaging demonstrated the internalization of BTNPs in intracellular vesicles. The combination of piezoelectric nanoparticles and ultrasound stimulation upregulated the expression of MYOD1, MYOG, and MYH2 and enhanced cell aggregation, which is a crucial step for myoblast fusion, and the presence of MYOG in the nuclei. These results suggest that the direct piezoelectric effect induced by ultrasound on the internalized piezoelectric nanoparticles boosts myogenesis in its early phases.
1. Introduction
Skeletal muscle is one of the most abundant tissues in the human body, constituting 40–45% of the adult human total mass. When subjected to minor injuries, skeletal muscle tissue has the innate capability to self-regenerate without therapeutic intervention. Despite that, when severe traumatic injuries occur with volumetric muscle loss (VML) over 20%, the remaining muscle tissue is unable to fully restore its function and native mass, leading to the formation of fibrous scar tissue.1 Strategies to improve myogenesis on differentiating myoblasts are desirable to foster healing or regeneration of the native skeletal muscle tissue.2,3 Such strategies may also be beneficial to produce physiologically relevant on-chip models of the skeletal muscle for drug testing and personalized medicine.4In recent years, significant progress has been made concerning skeletal muscle tissue engineering (SMTE) to address the reconstruction of skeletal muscle in vitro and in vivo. In this regard, different approaches have been reported. Biophysical stimuli (physical, biochemical, biological, electrical, mechanical, magnetic, or their combinations) during the in vitro cell culture represent a promising route to obtain more efficient development of the skeletal muscle tissue.5 Another crucial aspect is mimicking the ultra-organized structure of the native skeletal muscle tissue.6
In the context of biophysical stimulations performed during in vitro cell culture, the use of piezoelectric nanoparticles in combination with ultrasound (US) stimulation, a wireless source of mechanical energy, is currently regarded as an intriguing and promising strategy.7 Indeed, piezoelectric nanoparticles can act as nanoscale transducers enabling the conversion of a mechanically induced deformation into an electrical signal at the intracellular or extracellular level. Notably, electrical inputs are known to promote the development and maturation of the skeletal muscle.8
The paradigm of US-stimulated piezoelectric nanoparticles as an indirect electrical cell stimulation mediator has already shown beneficial effects in promoting the differentiation of neural, osteogenic, and muscular precursors on 2D cell culture models. In one of the first attempts in this vein, Ciofani et al. demonstrated a remarkable increment of developed neurites in US-stimulated neuronal-like PC12 cells incubated with boron nitride nanotubes (BNNTs).9 Similar results were obtained by Marino et al., who carried out neural stimulation of neuroblastoma-derived SH-SY5Y cells by combining tetragonal barium titanate nanoparticles (BTNPs) and US.10 US-activated piezoelectric nylon-11 nanoparticles have also been tested on dental pulp stem cells to promote osteogenic differentiation.11 Genchi et al. tested the culture of SH-SY5Y cells on polyvinylidene fluoride-trifluoroethylene films doped with BTNPs and their stimulation with US, and they found significantly (p < 0.05) augmented neurite length with respect to the non-stimulated control.12 The only attempt to apply the paradigm of US-stimulated piezoelectric nanoparticles on skeletal muscle differentiation was reported by Ricotti et al. They investigated the effects of US and piezoelectric BNNTs on myoblasts, demonstrating that the expression of myogenesis genes and the development of myotubes were considerably enhanced by combining these elements.13
In order to properly mimic the skeletal muscle structure, numerous fabrication approaches can be used to create tissue-like topographies on 2D substrates.3,6,14–17 However, 3D fabrication techniques are necessary to more closely reproduce the complexity of the tissue and its functions. In this regard, 3D tissue-engineered skeletal muscle constructs can also be used as in vitro disease models to study the causes of skeletal muscle diseases, such as VML and Duchenne muscular dystrophy, and for drug testing.18 3D bioprinting is a promising approach in this context, allowing the combination of biomaterials, cells and growth factors by precisely depositing them in a pre-designed structure.19–21 The composition of bioinks plays a fundamental role in 3D bioprinting.22 Hence, several biomaterials or their combinations have been investigated for SMTE applications.23
As mentioned above, biophysical stimuli can help to obtain more efficient development of the skeletal muscle tissue. Bioinks can be designed to respond to external biophysical stimuli by playing on the material chemistry24 or by including responsive fillers. In the second case, nano-sized elements can guarantee responsiveness to outer inputs by directly interacting with cells at the nanoscale to promote their differentiation.25,26 Although it has been claimed that the next-generation bioinks will necessarily be ground on hydrogels embedding multifunctional nanoparticles,27 to the best of our knowledge, no research group has explored the myogenic bioeffects induced by the inclusion of piezoelectric nanoparticles in skeletal muscle cell-laden constructs fabricated through 3D bioprinting, and remotely triggered by US.
In this work, BTNPs were chosen as the piezoelectric nano-sized element to be embedded in a 3D printed alginate/Pluronic (ALG/PLU)-based bioink. Among natural and synthetic polymers, alginate has been widely used to promote cell adhesion, proliferation, and differentiation due to its high biocompatibility and its ability to form gels that can support cell encapsulation and survival during the 3D printing process.22,28 Several examples of alginate-based bioinks used for 3D bioprinting of skeletal muscle constructs are reported in the literature.29–34 However, due to its poor mechanical properties, alginate is typically co-printed with other synthetic polymers such as Pluronic® F-127 to improve them, when skeletal muscle applications are targeted.29 After material characterization and cytocompatibility assessment, we stimulated printed piezoelectric and bare (non-doped) structures with a dose-controlled US source to investigate the bioeffects on myoblast differentiation.
2. Materials and methods
2.1 Preparation and sterilization of the bioink
The ALG/PLU-based bioink was prepared as previously described.29 Briefly, a 2% w/w alginate solution was prepared by slowly adding sodium alginate (W201502, Sigma-Aldrich) to a diluted solution of Dulbecco's Modified Eagle's Medium (DMEM, D5796, Biosigma) in deionized water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 v/v). Then, Pluronic® F-127 (P2443, Sigma-Aldrich) was dissolved in the alginate solution at a concentration of 20% w/w while maintaining in an ice bath under mild stirring until the polymer was completely dissolved. Finally, the solution was sterilized by autoclaving (1 atm, 121 °C, 20 min) and maintained at 4 °C before use.
5 v/v). Then, Pluronic® F-127 (P2443, Sigma-Aldrich) was dissolved in the alginate solution at a concentration of 20% w/w while maintaining in an ice bath under mild stirring until the polymer was completely dissolved. Finally, the solution was sterilized by autoclaving (1 atm, 121 °C, 20 min) and maintained at 4 °C before use.
2.2 Bioink characterization through FT-IR and DSC
Autoclaved and non-autoclaved ALG/PLU-based bioinks were characterized using Fourier transform infrared spectroscopy (FT-IR). The FT-IR spectra were also recorded for sodium alginate and Pluronic® F-127 as reference materials.Before FT-IR analysis, samples were freeze-dried using a FreeZone Plus 2.5 Liter freeze dry system (Labconco, Kansas City, MO, USA) to remove water. Then, FT-IR spectra were recorded in attenuated total reflection (ATR) mode, using an IRAffinity-1 FT-IR spectrophotometer equipped with an ATR MIRacle-10 accessory (Shimadzu Scientific Instruments, Japan). The spectra were collected in the range 4000–500 cm−1 with 4 cm−1 and 128 scan resolution. The background spectrum of air was recorded immediately before collecting the FT-IR spectrum of each sample.
Autoclaved and non-autoclaved ALG/PLU-based bioinks were also characterized through differential scanning calorimetry (DSC). As for the FT-IR analysis, samples were freeze-dried before starting DSC analysis; then, 8 mg of samples was sealed in hermetic stainless-steel crucibles. The DSC analysis was recorded using a Mettler Toledo STARe DSC 1 system (Mettler Toledo, Columbus, OH, USA). Firstly, samples were maintained at 4 °C for 2 min. Then, the temperature was increased to 125 °C at 10 °C min−1 and maintained at this temperature for 1 min. The temperature was then decreased to 4 °C at 20 °C min−1 and maintained at this temperature for 1 min. A second heating step was performed from 4 °C to 125 °C at 2 °C min−1.
2.3 Nanoparticle preparation and characterization through TEM and DLS
BTNPs (nominal diameter: ∼60 nm) were graciously supplied by PlasmaChem GmbH (Berlin, Germany).For transmission electron microscopy (TEM) characterization, two drops of BTNP water suspension (100 μg mL−1) were deposited onto 300-mesh carbon-coated copper grids (TedPella). The TEM analysis was carried out using a Libra 120 Plus microscope (Carl Zeiss, Oberkochen, Germany) operating at an accelerating voltage of 120 keV, equipped with an in-column omega filter for energy filtered imaging, and with a bottom mounted 12 bit 2k × 2k CCD camera (TRS).
Propylene glycol alginate (PGA, YP58644, Biosynth Carbosynth) was added at three different concentrations (100, 250, and 500 μg mL−1) separately to deionized water or DMEM, then sterilized by filtration through a 0.22 μm polyethersulfone filter (16532-K, Sartorius). BTNPs were sterilized by autoclaving (1 atm, 121 °C, 20 min), then diluted at three different concentrations (100, 250, and 500 μg mL−1) in filtered deionized water or DMEM, and mixed with the PGA solution at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 BTNPs/PGA.
1 BTNPs/PGA.
Dynamic light scattering (DLS) analysis was performed using a Zetasizer Nano ZS90 (Malvern Instruments Ltd, Malvern, UK). Before the analysis, samples (prepared in both deionized water and DMEM) were sonicated using an ultrasonic homogenizer Sonopuls HD 4050 (Bandelin, Berlin, Germany) for 5 min at 25% of the maximum power, then transferred in a disposable cuvette (634–1067, VWR™) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution. The measurement parameters were set as follows: temperature = 25 °C; medium viscosity = 0.8872 and 0.94 mPa s for deionized water and DMEM, respectively; medium refractive index = 1.33 and 1.345 for deionized water and DMEM, respectively;35 nanoparticle absorption index = 0.200;36 and nanoparticle refractive index = 2.4.37 The DLS measurements were conducted every 10 min for a total of 1 h. Each sample type was analyzed in triplicate, using independent samples.
10 dilution. The measurement parameters were set as follows: temperature = 25 °C; medium viscosity = 0.8872 and 0.94 mPa s for deionized water and DMEM, respectively; medium refractive index = 1.33 and 1.345 for deionized water and DMEM, respectively;35 nanoparticle absorption index = 0.200;36 and nanoparticle refractive index = 2.4.37 The DLS measurements were conducted every 10 min for a total of 1 h. Each sample type was analyzed in triplicate, using independent samples.
2.4 Assessment of piezoelectric properties
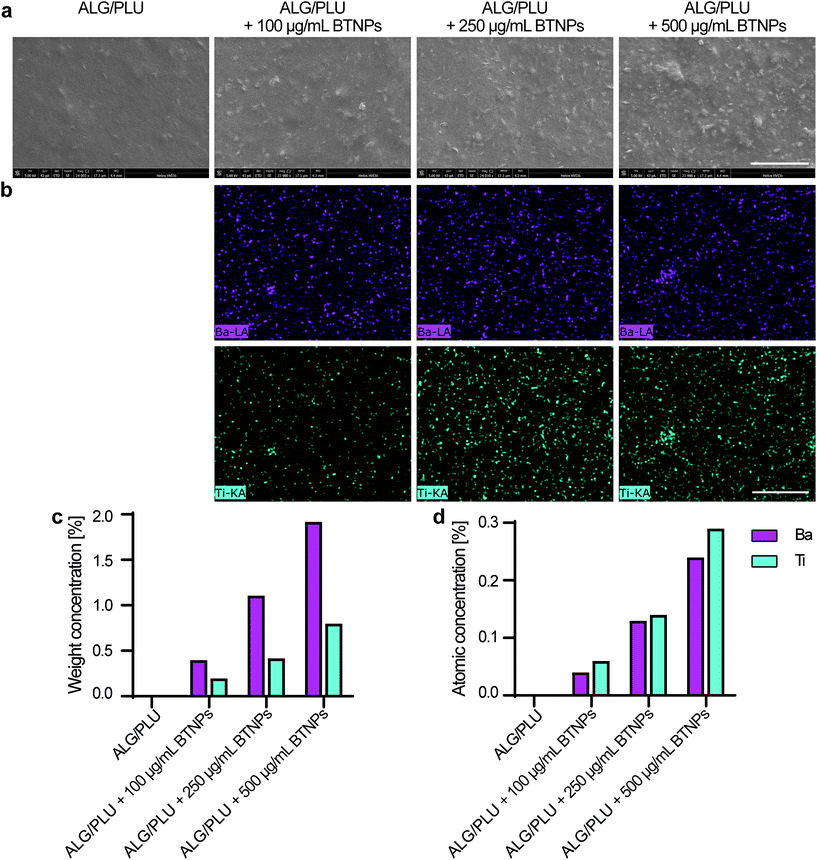
The piezoelectric properties of BTNPs were investigated through piezoelectric force microscopy (PFM). PFM measurements were performed using an Icon Bruker AFM system (Dimension Icon, Bruker Co., USA) working in peak force PFM mode (scan frequency = 0.2 Hz; scanning area = 0.25 × 0.25 μm2). The peak force modality was employed to investigate the indirect piezoelectric response when the material was subjected to an electrical stimulus. A silicon probe with a Pt–Ir coating (SCM-PIT-V2, Bruker, Billerica, MA, USA) with a measured spring constant of 2.5 N m−1, resonant frequency of 60 kHz, and deflection sensitivity of 118.2 nm V−1 was used. The amplitude and phase of the piezoelectric signals were acquired in the vertical direction via lock-in detection by applying to the tip an alternating current voltage (Vac) of 2 V at 60 kHz. Before the measurements, BTNPs were autoclaved (1 atm, 121 °C, 20 min). Then, to keep them stable during the scan, they were mixed with a diluted solution (5 mg mL−1) of polylactic acid (PLLA) in dichloromethane (concentration of BTNPs: 100 μg mL−1). The dispersion in the PLLA solution allowed fabricating a composite ultra-thin film by spin coating (speed = 4000 rpm for 30 s) over a silicon wafer previously covered by gold through sputtering to immobilize the nanoparticle and facilitate the analysis. The nanomaterial surface was exposed for the analysis as the PLLA film thickness was smaller than 50 nm. Five independent samples were analyzed and the average value of the piezoresponse amplitude (d33 piezoelectric coefficient) in the PFM maps was calculated as follows:where γ is a correction factor (6) and A is the amplitude signal (pm). A reference sample made of polyvinyl fluoride (PVDF) in the form of thin film (Goodfellow, thickness: 28 μm, d33: ∼−20 pC N−1) was also analyzed to properly calibrate the PFM amplitude signal.
2.5 Preparation and 3D printing of piezoelectric bioinks
For further experiments, the ALG/PLU-based bioink was prepared as previously described (see section 2.1). Then, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 BTNP/PGA solution was prepared by mixing the autoclaved BTNPs (10 mg mL−1) and a filtered solution of PGA in deionized water. The solution was then sonicated through an ultrasonic homogenizer, as previously described, underneath a laminar hood to ensure a sterile environment. After sonication, appropriate aliquots of the nanoparticle solution were mixed with the ALG/PLU-based bioink by gently pipetting to obtain the following final BTNP concentrations in the bioink: 100, 250, and 500 μg mL−1 (0.01, 0.025, and 0.05% w/v, respectively).
1 BTNP/PGA solution was prepared by mixing the autoclaved BTNPs (10 mg mL−1) and a filtered solution of PGA in deionized water. The solution was then sonicated through an ultrasonic homogenizer, as previously described, underneath a laminar hood to ensure a sterile environment. After sonication, appropriate aliquots of the nanoparticle solution were mixed with the ALG/PLU-based bioink by gently pipetting to obtain the following final BTNP concentrations in the bioink: 100, 250, and 500 μg mL−1 (0.01, 0.025, and 0.05% w/v, respectively).
Murine myoblasts (C2C12 cells, CRL-1772™, ATCC, Rockefeller, New York, NY, USA) were cultured and expanded in vented flasks until 80% confluence using DMEM supplemented with 10% fetal bovine serum (FBS, F4135, Sigma-Aldrich) and 1% penicillin/streptomycin (P/S P0781, Sigma-Aldrich), hereinafter referred to as “growth medium”. Cells were detached by trypsinization and centrifuged at 500g for 5 min; then, the pellet was homogeneously suspended in the ALG/PLU-based bioink without BTNPs or with BTNPs at the abovementioned concentrations.
3D printing was performed with a 3D-Bioplotter® Manufacturer Series (EnvisionTEC GmbH, Gladbeck, Germany) placed underneath a laminar hood. The control of the 3D-Bioplotter® consisted of two modules: (i) the PerFactory software for standard triangulation language (STL) data import and slicing, and (ii) the VisualMachines software for material parameters and machine control. The bioink was loaded into a 30 cc polypropylene syringe (7012134, Nordson EFD) and heated at 37 °C before printing. Then, the bioink was printed through a 250-μm cone-shaped polypropylene nozzle (7018370, Nordson EFD) under a pressure of 0.4 bar and a deposition speed of 10 mm s−1, depositing one single layer of parallelly aligned filaments in a 24-well plate (055429, Corning Costar). Immediately after printing, the printed structures were crosslinked by adding 1 mL of a filtered 25 mM CaCl2 (C1016, Sigma-Aldrich) solution in deionized water and leaving them for 10 min at room temperature. Then, the crosslinking solution was replaced with 1 mL of growth medium.
2.6 SEM, TEM, and EDX analyses
An FEI Helios 600i Dual Beam SEM-FIB microscope (FEI Company, Hillsboro, OR, USA) equipped with a Bruker Quantax 200 EDX detector (Bruker Nano GmbH, Berlin Germany) was used to obtain the scanning electron microscopy (SEM) images and energy dispersive X-ray (EDX) spectra of the printed structures. SEM imaging was carried out under high vacuum conditions by setting a beam voltage of 5 kV and a current of 43 pA. For EDX spectra acquisition, a beam voltage of 10 kV and a current of 86 pA were set. The working distance was set at ∼4 mm. Before the analyses, the printed structures were air-dried for 24 h and mounted onto standard stubs. Then, they were gold-sputtered using a Quorum 150R ES sputter coater (Quorum Technologies Ltd, Laughton, ES, UK) for 30 s and applying a current of 30 mA. SEM images and EDX spectra were acquired on printed structures in the absence of cells without BTNPs and doped with 100, 250, and 500 μg mL−1 BTNPs to analyze the scaffold morphology and nanoparticle distribution. Furthermore, to analyze the hydrogel porosity and the internal microstructure, a protocol similar to that of Cvek et al. was used.38 Briefly, the printed structures without BTNPs were freeze-dried overnight at −80 °C under low vacuum (0.002 mbar). To analyze the internal structure of the samples, the printed structures were cut crosswise into slices with a sharp scalpel and placed onto standard stubs, with the cross-section of the printed structures facing up. Then, the samples were gold-sputtered and imaged via SEM operating at an accelerating voltage of 15 kV.TEM imaging and EDX analysis were also performed at the end of the differentiation experiment with LIPUS stimulation (see section 2.9) in order to investigate the position of the BTNPs with respect to the cells. Doped printed structures were fixed at room temperature for 2 h with 2% v/v paraformaldehyde + 2% v/v glutaraldehyde diluted in a 25 mM CaCl2 solution, then rinsed and fixed overnight at 4 °C with 2% v/v glutaraldehyde in a 25 mM CaCl2 solution. On the following day, samples were rinsed, post-fixed for 1 h with 2% v/v osmium tetroxide in a 25 mM CaCl2 solution, rinsed and treated for 1 h with a home-made staining solution.39 Samples were dehydrated and embedded in epoxy resin (Epon 812, EMS, Hatfield, PA, USA), which was cured for 48 h at 60 °C. Finally, samples were cut into 90 nm thin slices using a UC7 ultramicrotome (Leica Microsystem, Wetzlar, Germany) equipped with a 45° diamond knife (DiATOME, Nidau, Switzerland), and sections were collected on 300 mesh copper grids (EMS, Hatfield, PA, USA). The TEM analysis was carried out using a Libra 120 Plus microscope (Carl Zeiss, Oberkochen, Germany) operating at an accelerating voltage of 120 keV, equipped with an in-column omega filter for energy-filtered imaging, a bottom mounted 12 bit 2k × 2k CCD camera (TRS), and a Bruker XFlash 6T-60 SDD detector for EDX spectroscopy.
2.7 Rheological analyses and shear stress estimation
An Anton Paar Physica MCR 302 rheometer (Anton Paar GmbH, Ostfildern, Germany) equipped with an H-PTD 200 temperature control device was used with a plate–plate geometry (diameter = 25 mm, measure gap height = 1 mm) to perform rheological measurements. The measurements were performed to evaluate bioink printability, to estimate the shear stress to which cells were exposed during the 3D bioprinting process, and the yield stress of the bioink. The bare ALG/PLU-based bioink and the doped ones (with 100, 250, and 500 μg mL−1 BTNPs) were loaded with 2 × 106 cells per mL and then analyzed in terms of strain sweep, temperature sweep, and shear rate sweep tests. For each test, the samples were prepared and deposited on a preheated/cooled Peltier plate for 5 min before starting the test. Then, the superior plate was lowered until contacting the sample surface and the testing sequence was applied to the sample. Each test was conducted on three independent samples for each sample type.Strain sweep tests ranging from 0.01 to 1000% were conducted measuring 10 points per decade at 37 °C to determine the shear elastic modulus G′ and the shear loss modulus G′′. Temperature sweep tests were performed following a heating ramp of 2 °C min−1 from 4 °C to 40 °C at a shear rate of 100 s−1. The dynamic behavior of viscosity and shear stress were also measured as a function of the shear rate ranging from 0.01 s−1 to 1000 s−1 at 37 °C, measuring 10 points per decade.
To estimate the shear stress, the bioink viscosity was modeled in the region in which the shear rate ranged from 0.1 to 1000 s−1 according to the following power law:
η = K![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) n−1 n−1 | (1) |
![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) is the shear rate of the fluid, and K and n are the consistency index and the flow behavior index, respectively. Through linear interpolation of the shear rate sweep test curve, K and n were determined. Based on these constants, obtained from rheological analyses, and considering the printing parameters such as the inner diameter of the nozzle (d = 250 μm) and flow rate (Q = 2 mm3 s−1), the shear stress τ was estimated as follows:40
is the shear rate of the fluid, and K and n are the consistency index and the flow behavior index, respectively. Through linear interpolation of the shear rate sweep test curve, K and n were determined. Based on these constants, obtained from rheological analyses, and considering the printing parameters such as the inner diameter of the nozzle (d = 250 μm) and flow rate (Q = 2 mm3 s−1), the shear stress τ was estimated as follows:40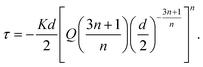 | (2) |
Furthermore, to estimate the yield stress τ0, the dependence of the shear stress on the shear rate was plotted using the Herschel–Bulkley model shown in eqn (3), which is recommended for the investigation of the yield stress for hydrogel structures.41 The yield stress was calculated using a least squares method:
τ = τ0 + K![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) n. n. | (3) |
2.8 Viability and metabolic activity assays
Cell viability in cell-laden structures was analyzed to evaluate the effects of the BTNP presence. The bare ALG/PLU-based bioink and the doped ones with 100, 250, and 500 μg mL−1 BTNPs were loaded with 2 × 106 cells per mL and printed by depositing one single layer (7 × 7 mm2), using the printing and crosslinking parameters reported in section 2.5. Then, the printed structures were placed in an incubator (37 °C, 5% CO2) for up to 5 days.Cell viability and metabolic activity were evaluated using the LIVE/DEAD™ viability/cytotoxicity kit (L3224, Invitrogen) and the PrestoBlue™ cell viability reagent kit (A13262, Invitrogen) after 1, 3, and 5 days of culture.
For LIVE/DEAD™, at the time points, for each sample, the growth medium was removed and replaced with 1 mL of DMEM containing 0.5 μL mL−1 calcein AM and 2 μL mL−1 EthD-1. Samples were then incubated for 30 min at 37 °C in the dark. Representative fluorescence images were taken for each experimental condition with a Leica DMi8 microscope (Leica Microsystems, Wetzlar, Germany) using a Leica lamp and equipped with FITC and TRITC filters. Three independent samples were analyzed for each time point and each sample type.
For PrestoBlue™, at the time points, the growth medium was removed for each sample, the assay solution was mixed with DMEM at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 volume ratio, then 1 mL of this mixture was added to each structure. Samples were incubated for 90 min at 37 °C in the dark. After incubation, fluorescence intensity was measured using a VICTOR X multilabel plate reader (PerkinElmer, Waltham, MA, USA) at the excitation/emission of 560 nm/590 nm. Three independent samples were analyzed for each time point and each sample type, and fluorescence reading was performed in triplicate for each sample.
10 volume ratio, then 1 mL of this mixture was added to each structure. Samples were incubated for 90 min at 37 °C in the dark. After incubation, fluorescence intensity was measured using a VICTOR X multilabel plate reader (PerkinElmer, Waltham, MA, USA) at the excitation/emission of 560 nm/590 nm. Three independent samples were analyzed for each time point and each sample type, and fluorescence reading was performed in triplicate for each sample.
2.9 Differentiation experiment with US stimulation
Based on the results of the viability tests (see section 3.3), only the non-doped ALG/PLU-based bioink and the doped bioink with 250 μg mL−1 BTNPs were considered for the differentiation experiment, which is depicted in Fig. 1.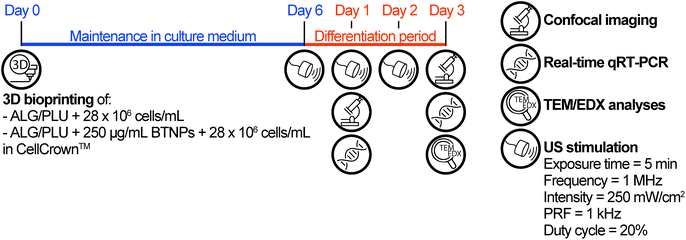 | ||
| Fig. 1 Timeline of the differentiation experiment. ALG/PLU: alginate/Pluronic-based bioink. BTNPs: barium titanate nanoparticles. US: ultrasound. PRF: pulse repetition frequency. | ||
In this experiment, the two bioinks were loaded with 28 × 106 cells per mL and printed by depositing one single layer (5.5 × 5.5 mm2), using the printing and crosslinking parameters reported in section 2.5. For this experiment, the printing surface for the structure was a 0.029 mm-thick polystyrene film (ST311029, Goodfellow) mounted on a CellCrown™ insert (Z742381-12EA, Sigma), to enable subsequent US stimulation. Before use, the polystyrene films and CellCrown™ inserts were washed under sterile conditions with ethanol 70%, rinsed with sterile water, and exposed to ultraviolet (UV) light for 30 min. After structure printing and crosslinking, the printed structures were maintained in growth medium for 6 days. A preliminary differentiation test was carried out to identify the most suitable medium to be used in the differentiation experiment with US stimulation. The details of this preliminary differentiation test are reported in the ESI, section S1.† Briefly, two different differentiation media, namely differentiation medium 1 (DM1) and differentiation medium 2 (DM2) were tested. DM1 consisted of DMEM supplemented with 1% FBS, 1% P/S, and 1% insulin-transferrin-selenium (ITS, I3146, Sigma-Aldrich). DM2 consisted of DMEM supplemented with 10% horse serum (HS, ECS0091L, Euroclone), 1% P/S, and 50 ng mL−1 insulin-like growth factor (IGF-1, 250-19, PeproTech).42–44 For the differentiation experiment with US stimulation, DM1 was used and changed every day for 3 days. For this experiment, non-doped and doped printed structures were divided into two groups: one group received no additional stimuli, while the other was stimulated every day by US, as described below.
US stimulation of the printed structures was performed using a system for LIPUS, described in the work of Fontana et al.,45 which allowed precisely controlling the dose delivered to cells. A key element of this system was the biological retaining system, designed to be water-proof and transparent to the US waves. This feature enabled the control of the US dose and avoided undesired reflections, attenuations, and formation of standing waves. For stimulating the samples, the CellCrown™ inserts hosting the cell-laden printed structures were moved from the 24-well plate, used to keep the printed structures in the incubator, to the dedicated chamber of the retaining system. Then, each chamber was filled with 3.5 mL of growth medium (to avoid air interfaces, which may hamper US wave transmission) and samples were subjected to the stimulation (Movie S1†). LIPUS stimulations were performed every day during the differentiation period setting the parameters as follows: exposure time = 5 min; frequency = 1 MHz; spatial average pulse average intensity = 250 mW cm−2; pulse repetition frequency (PRF) = 1 kHz; and duty cycle = 20%. Finally, at the end of the stimulation, CellCrown™ inserts were moved back into the 24-well plate and placed in the incubator to continue the culture.
At day 1 of differentiation (before the second LIPUS stimulation) and at the end-point (day 3 of differentiation), immunofluorescence imaging and real-time qRT-PCR were performed. For immunofluorescence staining, the printed structures were gently washed with 1 mL of 0.9% w/v NaCl solution (746398, Sigma-Aldrich). Then, they were fixed in 500 μL of 4% v/v paraformaldehyde (28908, Thermo Scientific) diluted in a 25 mM CaCl2 and 0.9% w/v NaCl solution (15 min at room temperature). Samples were washed three times with 1 mL of a 25 mM CaCl2 solution and then incubated twice in 500 μL of 0.1% v/v Triton X-100 (T8787, Sigma-Aldrich) in 25 mM CaCl2 to permeabilize the cells (10 min at room temperature). After incubation with 500 μL of blocking solution (25 mM CaCl2 solution with 0.1% v/v Triton X-100 and 10% v/v FBS) for 2 h, samples were washed four times with 1 mL of 25 mM CaCl2 solution with 0.1% v/v Triton X-100 and incubated with 250 μL of primary antibody solution (mouse anti-myogenin antibody, clone F5D, ab1835, Abcam) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) overnight at 4 °C. Then samples were washed four times with 1 mL of 25 mM CaCl2 solution with 0.1% v/v Triton X-100 and incubated with 250 μL of secondary antibody solution (goat anti-mouse Alexa Fluor 488, A-11029, Invitrogen) (1
500) overnight at 4 °C. Then samples were washed four times with 1 mL of 25 mM CaCl2 solution with 0.1% v/v Triton X-100 and incubated with 250 μL of secondary antibody solution (goat anti-mouse Alexa Fluor 488, A-11029, Invitrogen) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) overnight at 4 °C protected from light. After washing four times with 1 mL of 25 mM CaCl2 solution with 0.1% v/v Triton X-100 and then four times with 1 mL of 25 mM CaCl2 solution, samples were stained. The staining solution was prepared by diluting tetramethylrhodamine (TRITC)-conjugated phalloidin (P1951, Sigma-Aldrich) (1
500) overnight at 4 °C protected from light. After washing four times with 1 mL of 25 mM CaCl2 solution with 0.1% v/v Triton X-100 and then four times with 1 mL of 25 mM CaCl2 solution, samples were stained. The staining solution was prepared by diluting tetramethylrhodamine (TRITC)-conjugated phalloidin (P1951, Sigma-Aldrich) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) and Hoechst 33342 (H1399, Invitrogen) (1
400) and Hoechst 33342 (H1399, Invitrogen) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) in 25 mM CaCl2 solution. All samples were incubated in the staining solution for 80 min at room temperature. Finally, immunofluorescence images were acquired using a Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany). Representative images were selected for each experimental group.
1000) in 25 mM CaCl2 solution. All samples were incubated in the staining solution for 80 min at room temperature. Finally, immunofluorescence images were acquired using a Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany). Representative images were selected for each experimental group.
For real-time qRT-PCR, printed structures were gently washed with 1 mL of 0.9% w/v NaCl solution, then dissolved in 500 μL of sodium citrate buffer composed of 50 mM sodium citrate (S4641, Sigma-Aldrich) and 100 mM sodium chloride, with pH 7.4 in deionized water, as described by Mozetic et al.29 Then, cells were collected by centrifugation (500g, 5 min), the supernatant was removed, and total RNA was isolated using 500 μL TRIzol™ reagent (15596018, Thermo Scientific) and extracted using RNeasy Micro kit (74004, Qiagen) according to the manufacturer's instructions. The extracted RNA was quantified with a Nanodrop™ 2000 (Thermo Scientific, Waltham, MA, USA) and reverse-transcribed using SuperScript™ IV VILO™ Master Mix (11756050, Thermo Scientific), according to the manufacturer's protocol. Real-time qRT-PCR was subsequently performed with PowerUP™ SYBR™ Green Master Mix (A25742, Thermo Fisher Scientific) in a Rotor-Gene Q (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), myogenic differentiation 1 (MYOD1), myogenin (MYOG), cyclin D3 (CCND3), cyclin-dependent kinase inhibitor 1A (CDKN1A), myogenic factor 5 (MYF5), actin alpha 1, skeletal muscle (ACTA1), cysteine and glycine-rich protein 3 (CSRP3), myosin heavy chain 2 (MYH2), myosin heavy chain 4 (MYH4), and actinin alpha 2 (ACTN2) genes were analyzed with real-time qRT-PCR. All the primers used are listed in Table S1.†GAPDH was used as a housekeeping gene and relative gene expression quantification was performed following the 2−ΔCt method. At least three independent samples were analyzed for each experimental group.
2.10 Statistical analyses
Experimental data were subjected to a D'Agostino–Pearson normality test. Then, normally distributed data were analyzed by one-way analysis of variance (ANOVA), followed by Tukey's post hoc testing for multiple comparisons. Instead, non-normally distributed data were analyzed by Kruskal–Wallis, followed by Dunn's post hoc testing for multiple comparisons. Results are expressed as mean values ± standard deviations. Differences between experimental groups were considered statistically significant at p < 0.05.3. Results and discussion
3.1 Nanoparticle characterization
TEM imaging allowed measuring the BTNP dimensions (Fig. 2a). The nanoparticles had a diameter of 61.6 ± 16.5 nm. The piezoelectric properties of the BTNPs were also investigated by PFM analysis. Fig. 2b shows a topographic image of a single BTNP entrapped in the PLLA ultra-thin layer. This layer allowed keeping the nanoparticles stable during the analysis while keeping their surface exposed to the PFM tip. By applying a voltage (2 V), the amplitude response of the piezoelectric material (Fig. 2c) was considerably different from that of the background. This analysis allowed quantifying the piezoelectric coefficient d33, which resulted 88.1 ± 23.7 pm V−1. Such a value matches the typical ones reported in the literature for BTNPs.46BTNPs were chosen as piezoelectric nanocomponents to be included in the ALG/PLU-based bioink due to their strong piezoelectric properties and biocompatibility.47 However, BTNPs tend to aggregate;48 thus, a coating to stabilize them in an aqueous solution is needed to guarantee their homogeneous dispersion in the bioink. Therefore, BTNPs were coated by polymer wrapping of PGA, which was chosen being FDA-approved for various biomedical applications.49
Fig. 2d and e summarize the results obtained on the nanoparticles in terms of hydrodynamic diameter and polydispersity index (PDI), both in deionized water and DMEM. The hydrodynamic diameter values measured with the DLS analysis were higher than the diameters measured through TEM imaging. In terms of hydrodynamic diameter, no differences could be appreciated among different BTNP concentrations in deionized water (∼150 nm for all samples), while in DMEM slight differences (p < 0.05) were observed only between the BTNP concentration of 100 and 250 μg mL−1 (296.2 ± 5.8 nm, 249.2 ± 8.5 nm, and 273.3 ± 10.7 nm for 100, 250, and 500 μg mL−1 BTNPs, respectively). The hydrodynamic diameter measured in DMEM resulted higher in comparison with the one in deionized water. This can be explained by the fact that DMEM has a higher ionic strength and it is composed of a greater quantity of salts and other molecules (i.e., amino acids, vitamins, glucose) that can interact with BTNPs and PGA, resulting in a higher hydrodynamic diameter than the deionized water case, as also reported in other state-of-the-art examples.50–54 The hydrodynamic diameter remained almost constant over time for each experimental condition, as well as the PDI, thus demonstrating the suitability of the PGA coating to guarantee a homogeneous dispersion of BTNPs in a nanocomposite bioink during the whole printing procedure.
No insights are reported in the state of the art so far regarding the use of PGA for BTNP coating. In the literature, PGA has been proposed for stabilizing graphene oxide nanoflakes,55 demonstrating high biocompatibility and the ability to keep these nanomaterials well dispersed in aqueous media. However, the dimensions of these flakes (lateral dimension: ∼10 μm, thickness: ∼1.5 nm) were not comparable with the ones of BTNPs used in this study. Other surfactants have been reported to promote BTNP dispersion and stabilization in aqueous suspensions, such as ammonium polyacrylate,56–59 polymethacrylic acid,56,60 block copolymers of poly(vinyl alcohol) and poly(acrylic acid),61 polyaspartic acid,62 glycol chitosan,63 or poly-L-lysine.50 These surfactants proved to be effective in dispersing BTNPs. However, none of them was FDA-approved for medical use. Our results show the opportunity to use this certified polymer as a reliable alternative for this purpose.
3.2 Bare bioink and doped bioink characterization
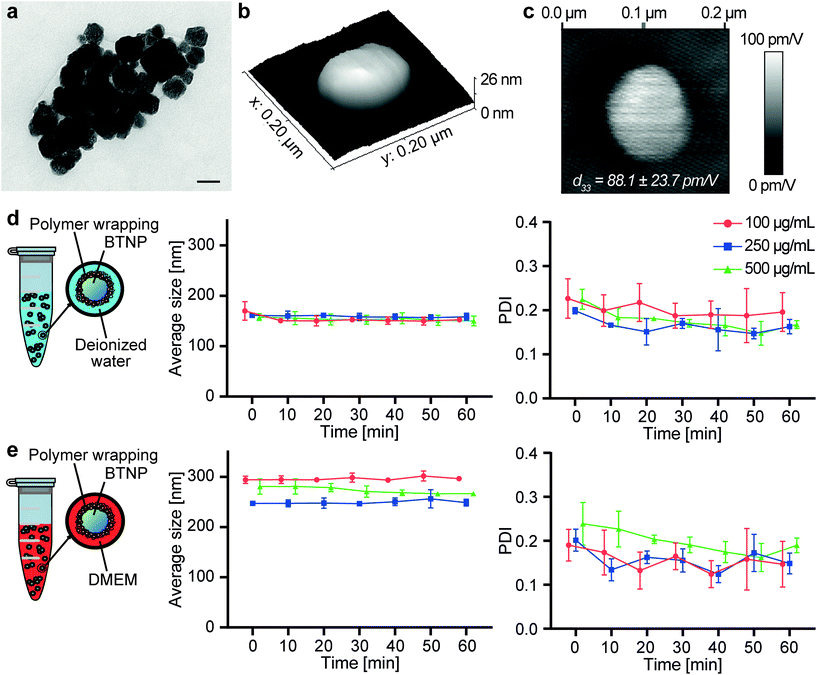
The FT-IR spectra of sodium alginate, Pluronic® F-127, autoclaved ALG/PLU-based bioink, and non-autoclaved ALG/PLU-based bioink are shown in Fig. 3a. The autoclaved and non-autoclaved bioink spectra showed a combination of the spectral bands of both Pluronic® F-127 and sodium alginate. In particular, the bands at 2881, 1465, 1342, and 1097 cm−1 are associated with Pluronic® F-127: the band at 2881 cm−1 represents the stretching vibration of C–H bonds, those at 1465 and 1342 cm−1 correspond to the bending vibrations of C–H, while the spectral band at 1097 cm−1 is related to the stretching vibration of C–O. The band at 1595 cm−1 is related to sodium alginate, corresponding to the asymmetric stretching vibration of the carboxyl group. Comparing the spectra of the autoclaved and non-autoclaved ALG/PLU-based bioinks, no differences could be appreciated, thus suggesting that the sterilization process did not cause any significant change in the chemical nature of the material. As expected, a noticeable similarity of the bioinks spectra with the one of Pluronic® F-127 was observed, being Pluronic® F-127 their most abundant component.Fig. 3b shows the DSC results for autoclaved and non-autoclaved bioinks. Both of them presented an exothermic peak at around 28 °C and an endothermic peak at around 57 °C. The endothermic peak was associated with Pluronic® F-127 melting process occurring during the heating scan, while the exothermic peak was related to the Pluronic® F-127 crystallization process occurring during the cooling scan.64 Sodium alginate transitions are generally found at higher temperatures, which are out of the temperature range analyzed in this study.65 Except for these two peaks in the DSC curves of the autoclaved and non-autoclaved bioinks, no other significant transitions were observed. No differences were appreciated between autoclaved and non-autoclaved bioink curves, indicating that the sterilization process did not alter the thermal behavior of the bioinks.
Pictures of the printed structures obtained through the procedure described in section 2.5 are shown in Fig. S1.†
The internal microstructure of the bioink was investigated by SEM imaging. Fig. 3c shows the SEM images of the internal section of the printed structures at different magnifications (400×, 1000×, and 2750×). The samples exhibited porosity, particularly appreciable at 1000× and 2750× magnifications. The pores were interconnected and homogeneously distributed throughout the volume. The nanoparticle presence did not alter the biomaterial porosity (results not shown). Similar internal microstructure and porosity were observed by Cabrini et al., who analyzed freeze-dried hydrogels composed of 20% w/v Pluronic® F-127.66 An architecture of such high porosity and interconnected pores is desirable to promote cell migration, proliferation, and diffusion of oxygen and nutrients inside the printed structures.67
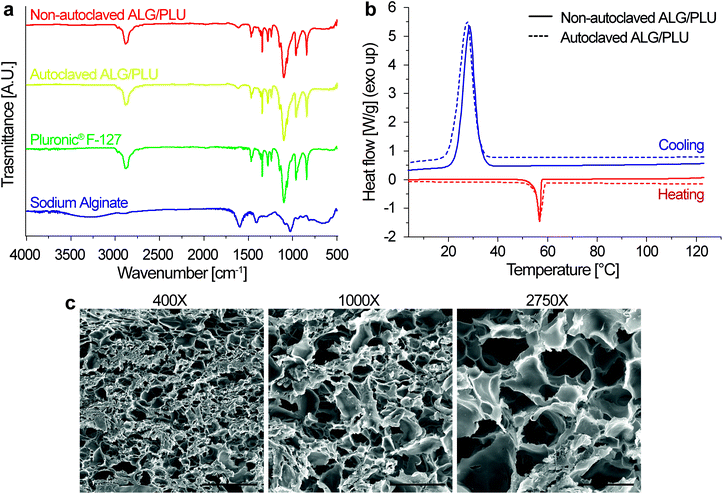
Fig. 4a shows 24000× magnification SEM images of a small surface portion of the printed structures without BTNPs and also doped with 100, 250, and 500 μg mL−1 BTNPs, respectively (without cells embedded). Sample surfaces qualitatively appeared more irregular by increasing the BTNP concentration. The EDX analysis performed on the same surface portion showed the co-localized presence of barium (Ba) and titanate (Ti) only in the doped bioinks, thus confirming the presence of the nanoparticles and their homogeneous distribution in the ALG/PLU matrix (Fig. 4b). Also, the EDX analysis quantifying weight concentration % and atomic concentration % of Ba and Ti confirmed an increased content of the two elements in the bioinks with higher nanoparticle concentrations (Fig. 4c and d).
The rheological curves obtained for the different bioinks are reported in Fig. S2.† In particular, Fig. S2a and S2b† report the moduli (i.e., G′ and G′′) upon strain %: in each sample condition, the storage modulus G′ exhibited an initial plateau phase followed by a decreasing phase for higher strain % values, while the loss modulus G′′ revealed a positive peak before the decreasing phase. In the linear viscoelastic region (strain < 1%), G′ values resulted in 10.9 ± 3.4, 13.1 ± 1.3, 12.5 ± 1.2, and 11.1 ± 3.7 kPa (p > 0.05) for the ALG/PLU-based bioink and the ALG/PLU-based bioinks doped with 100, 250, and 500 μg mL−1 BTNPs, respectively, while G′′ values were 0.9 ± 0.2, 1.2 ± 0.2, 1.3 ± 0.2, and 1.2 ± 0.3 kPa (p > 0.05). The G′ and G′′ values were close to the ones reported by Abrami et al., who characterized a hydrogel made of 2% w/w sodium alginate and 18% w/w Pluronic® F-127 in deionized water.68 Temperature sweep results (Fig. S2c†) showed a sigmoidal trend of the viscosity, highlighting that bioinks underwent a gelation process from 15 °C on up, with an exponential increase of viscosity at 20–25 °C. This allowed us to select 37 °C as the printing temperature as it maintained the bioink in its gel state while ensuring good shape retention during the 3D printing process and avoiding filament collapse or spreading after extrusion. Additionally, cells encapsulated in gel state bioinks have no tendency to sediment, thus yielding to homogeneous cell distribution in the printed material. Setting the printing temperature to 37 °C also allowed us to avoid an abrupt change in temperature to the cells embedded in the bioinks. At 37 °C, measured viscosity values were 2.6 ± 0.5, 3.2 ± 0.4, 2.6 ± 0.1, and 2.4 ± 0.5 Pa s (p > 0.05) for the ALG/PLU-based bioink and the ALG/PLU-based bioinks doped with 100, 250, and 500 μg mL−1 BTNPs, respectively. Viscosity decreased linearly as the shear rate increased (Fig. S2d†) for all the analyzed samples, thus all bioinks exhibited a shear-thinning behavior typical of non-Newtonian fluids, such as hydrogels.69 Shear-thinning properties lead to reduced shear stress during printing, thus guaranteeing both the facile deposition of the bioink and cell survival.70
The rheological properties of the bare ALG/PLU-based bioink and the doped ones with 100, 250, and 500 μg mL−1 BTNPs were similar to the ones of Pluronic® F-127,71 which represented the most abundant component.
Overall, the rheological properties were similar for all bioinks, showing that the BTNP presence did not significantly influence their rheological behavior. Indeed, the BTNP concentrations used in this work were significantly lower than 1% w/v, which is considered the threshold above which nanoparticles induce drastic changes in the mechanical properties of hydrogels.72
The shear rate sweep test curves (Fig. S2d†) were fitted to eqn (1) using linear regression. The fitted values of K and n are listed in Table 1. The shear stress to which cells were exposed during the 3D bioprinting process was estimated by applying eqn (2), considering the diameter of the nozzle used for 3D bioprinting and the flow rate applied. Table 1 shows that τ values were similar for all bioinks (p > 0.05). For all of them, cells were exposed to τ values much smaller than 5 kPa, which is considered the safety threshold to guarantee cell viability during the printing process.73
| K [Pa sn] | n | τ [Pa] | τ 0 [Pa] | |
|---|---|---|---|---|
| Results are expressed in terms of mean values ± standard deviations. ALG/PLU: alginate/Pluronic-based bioink. BTNPs: barium titanate nanoparticles. | ||||
| ALG/PLU | 228.81 ± 35.17 | 0.10 ± 0.06 | 626.46 ± 301.22 | 245.10 ± 51.76 |
| ALG/PLU + 100 μg mL−1 BTNPs | 201.62 ± 31.59 | 0.08 ± 0.03 | 436.08 ± 109.59 | 201.80 ± 22.63 |
| ALG/PLU + 250 μg mL−1 BTNPs | 196.36 ± 20.67 | 0.06 ± 0.05 | 381.83 ± 136.51 | 177.37 ± 29.27 |
| ALG/PLU + 500 μg mL−1 BTNPs | 170.38 ± 4.04 | 0.07 ± 0.12 | 338.40 ± 41.90 | 167.77 ± 12.29 |
To quantify the yield stress of the ALG/PLU-based bioink and the ALG/PLU-based bioinks doped with 100, 250, and 500 μg mL−1 BTNPs, the experimental data of the shear stress on the shear rate curves (Fig. S2e†) were fitted to the Herschel–Bulkley model – eqn (3). The calculated yield stress values are listed in Table 1: no significant differences were found between the bioinks. The obtained yield stress values were larger than the ones reported for other bioinks used for skeletal muscle tissue engineering74,75 and they were consistent with the values obtained with Pluronic® F-127-based hydrogels with similar composition.76 As recently reported,77 yield stress higher than 100 Pa allows for material retention and shape fitting even before crosslinking, thus preventing layer spreading after extrusion and increasing print fidelity and homogeneous distribution of cells within the bioink.78,79
3.3 Viability and metabolic activity of cells embedded in the bioinks
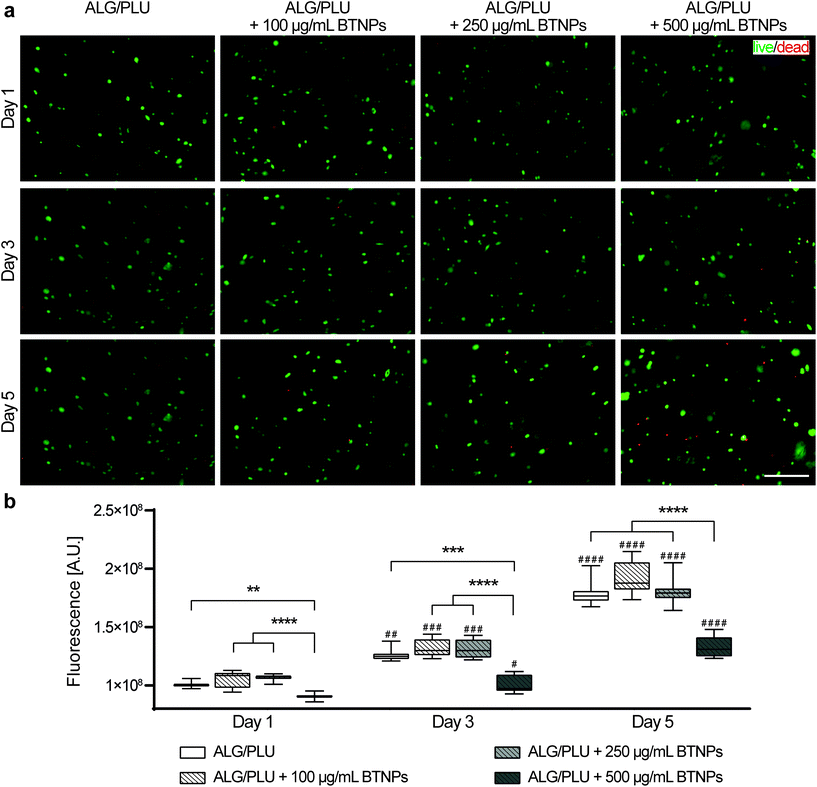
Cell viability and metabolic activity results are shown in Fig. 5. The fluorescence images highlighted live (green) and dead/necrotic (red) cells embedded in the bioinks (Fig. 5a). An excellent cytocompatibility of BTNPs was demonstrated up to a concentration of 250 μg mL−1 throughout the whole analyzed period. Almost no dead cells were observed in the printed structures, similar to the non-doped control. A slightly higher number of dead cells were observed in the bioink doped with 500 μg mL−1 BTNPs.Qualitative viability results were confirmed by the measurements of cell metabolic activity (Fig. 5b). No statistically significant differences were found between the metabolic activity of cells embedded in the bare bioink and the ones doped with BTNPs, with concentrations up to 250 μg mL−1. A statistically significant decrease in the cell metabolic activity was observed instead for bioinks doped with 500 μg mL−1 BTNPs on day 1, 3, and 5. Overall, cells remained metabolically active over 5 days, which is demonstrated by a significant increase in metabolic activity from day 1 to day 5 for all sample types. However, cells in the bioink doped with 500 μg mL−1 BTNPs showed a less marked increase in the metabolic activity compared to the other experimental conditions.
Based on these results, further differentiation experiments were performed with the cell-laden bioink without BTNPs and the cell-laden bioink doped with 250 μg mL−1 to maximize the effects induced by the synergetic interaction between US and nanoparticles. This was the highest concentration of BTNPs that was found to be non-toxic to cells.
3.4 Ultrasound stimulation and cell differentiation in the piezoelectric bioinks
A preliminary differentiation test was carried out to identify the most suitable medium to be used in the differentiation experiment and to preliminarily evaluate the effects of BTNPs embedded into the printed structures. The results of this test are reported in ESI, section S1.† Here, gene expression analyses of myogenic markers pointed out the greater myogenic potential of the medium based on DMEM, 1% FBS, 1% P/S, and 1% ITS. Notably, under the same condition of printed structures cultured in this medium, higher expression levels of MYOD1, MYOG, ACTA1, and CSRP3, which are key genes in the early myogenesis phases,80 were measured for printed structures embedded with BTNPs, highlightling a possible promyogenic role of the nanoparticles.Then, we performed a differentiation experiment stimulating the printed structures with LIPUS.
This phase was crucial to evaluate the effects of the piezoelectric bioink combined with the US treatment (thought to induce the generation of local electrical charges)7 on the bioprinted cells. Four different experimental groups were investigated on independent samples: (i) non-doped and non-LIPUS treated, (ii) non-doped and LIPUS treated, (iii) doped and non-LIPUS treated, and (iv) doped and LIPUS treated. The results obtained are reported in Fig. 6 and 7.
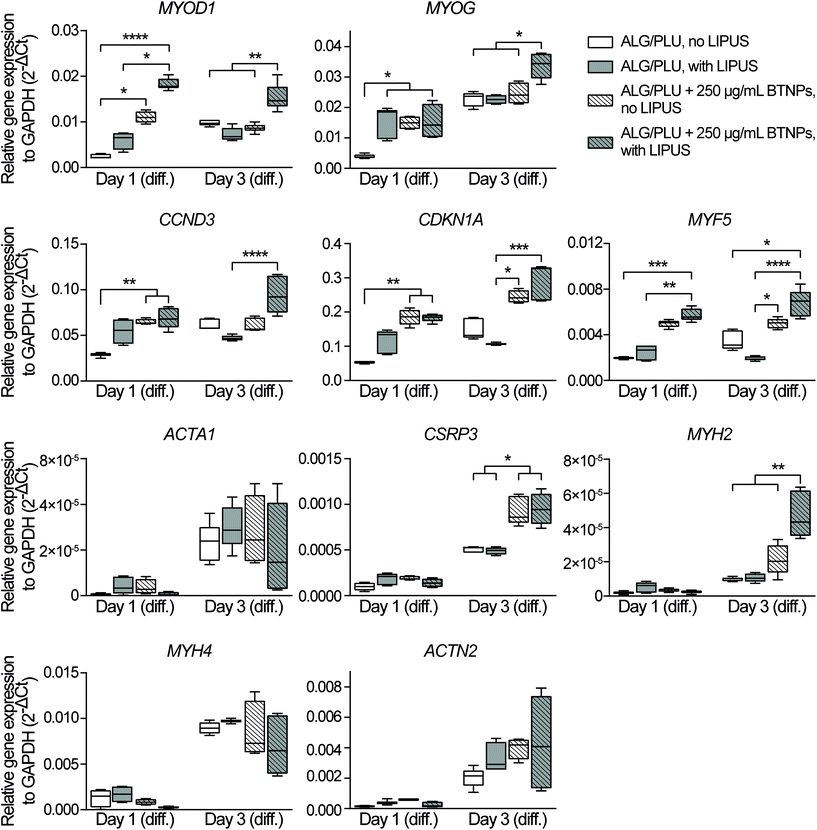
Myogenic differentiation consists of several processes, including the initial cell cycle arrest and subsequent activation of various genes for the acquisition of skeletal muscle-specific functions. In the early phase of differentiation, also named specification, the myogenic regulatory factors have an essential role.81 More specifically, MYOD1 is the master gene of skeletal myogenesis and it is in charge of committing cells to a myogenic program; MYOG follows the up-regulation of MYOD1 and activates the expression of late differentiation genes; while MYF5, together with MYOD1, regulates myoblast commitment.81–83 Cell cycle arrest is a key step to start myogenesis, and MYOD1 itself activates the expression of CCND384 and CDKN1A,85 whose upregulation is needed to induce and maintain a terminal cell cycle withdrawal.
The gene expression analyses performed on day 1 of differentiation showed that the presence of BTNPs (even without LIPUS) implies an initial boost in the differentiation induction compared to the normal differentiation condition, shown by the upregulation of MYOD1 (p < 0.05), MYOG (p < 0.05), CCND3 (p < 0.01), and CDKN1A (p < 0.01) (Fig. 6). Interestingly, the doped and LIPUS treated printed structures exhibited considerably higher gene expression levels of MYOD1 (p < 0.0001), MYOG (p < 0.05), CCND3 (p < 0.01), CDKN1A (p < 0.01), and MYF5 (p < 0.001) compared to the non-doped and non-stimulated printed structures. The activation of these genes is a clear signal of the cell cycle withdrawal and the onset of the myogenesis, pathways that are boosted at day 1 of differentiation by the combination of BTNPs and LIPUS stimulation. On day 1 of differentiation, LIPUS stimulation on its own showed no effects: no significant differences were detected in the gene expression levels. For markers featuring later differentiation phases (ACTA1, CSRP3, MYH2, MYH4, and ACTN2) no differences were detected among the four experimental groups at this time point.
On day 3 of differentiation, the piezoelectric nanoparticles in combination with LIPUS stimulation led to a considerable upregulation of MYOD1 (p < 0.01), MYOG (p < 0.05), and MYH2 (p < 0.01) with respect to all the other samples, including the BTNPs-doped ones. Actually, the correlation between MYOD1, MYOG, and MYH2 is widely known, since MYOD1 is a myogenic factor expressed in the early stages of myogenesis, while MYOG mediates the differentiation process and it is associated with the expression of MYH2, which is a later muscle differentiation marker.80,86 These results confirmed the pro-differentiative effect that the combination of piezoelectric nanoparticles and LIPUS stimulation has on the early myogenesis of 3D bioprinted cells. ACTA1, MYH4, and ACTN2 gene expression levels increased when compared with the corresponding values on day 1 of differentiation; however, for these genes, no significant differences were found among the experimental groups, probably because these markers feature later differentiation stages, not analyzed in these experiments. LIPUS stimulation on its own showed no significant differences in terms of gene expression levels, with respect to the non-stimulated control.
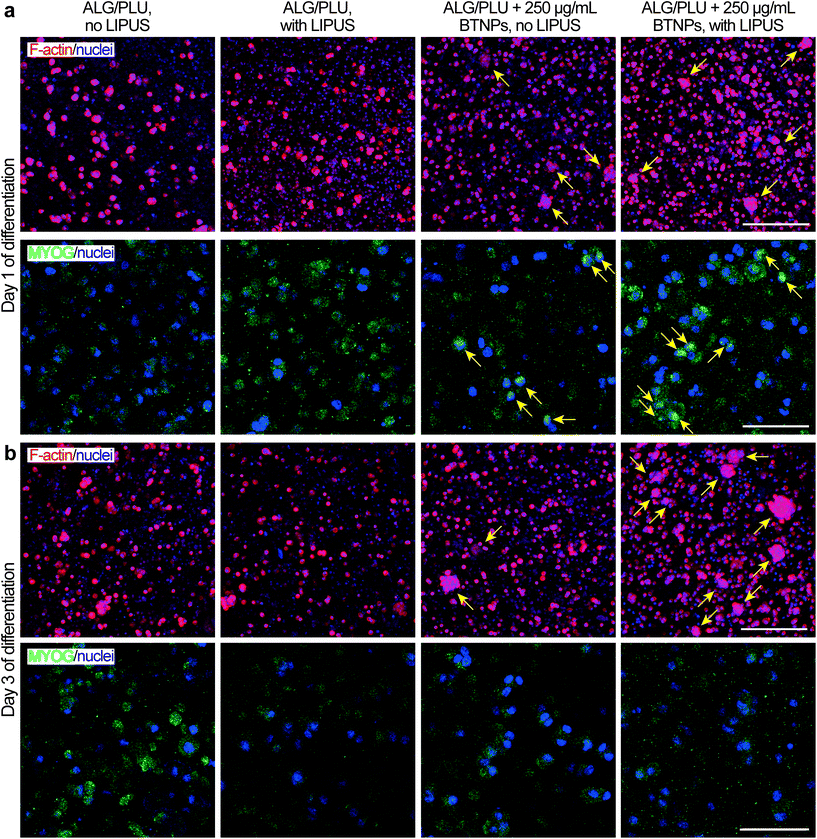
On day 1 of differentiation, immunofluorescence images (showing F-actin and nuclei in Fig. 7a) confirmed that cells were well distributed in the printed structures. Notably, the presence of BTNPs led to a marked formation of cell aggregates, which were even more evident in the LIPUS-stimulated sample. The formation of cell aggregates went with a higher presence of MYOG in the nuclei, highlighted by the colocalization of the green and blue signals, representing MYOG and nuclei, respectively (Fig. 7a). This expression pattern is in agreement with the role of MYOG in promoting myocyte fusion.87 Ferri et al. observed that MYOG is expressed in all undifferentiated cells, mainly in the cytoplasm. Then, after differentiation induction, MYOG is progressively concentrated in the nucleus to activate the expression of late differentiation genes for myotube formation and the contractile apparatus protein synthesis.88 Therefore, the higher presence of MYOG in the nuclei in the presence of BTNPs suggests a boost in differentiation induction due to the BTNPs.
On day 3 of differentiation, cell aggregates increased in size and they were still more evident in the presence of BTNPs, and even more in the doped and LIPUS treated printed structures (Fig. 7b). Immunostaining for MYOG was still found to be positive, matching with previous findings for early phases of C2C12 differentiation,89 but no differences were observed between the different experimental conditions.
The results obtained in this study are in agreement with previous evidence shown by the authors,13 in which the differentiation process of C2C12 was accelerated once treated with piezoelectric BNNTs (internalized by the cells) and US waves (wavelength: 40 kHz).
To ascertain the role of US-stimulated BTNPs in the differentiation process, the voltage generated by the interaction between the US waves and the BTNPs inside the printed structures was estimated through an electroelastic model developed by Marino et al.10 Briefly, the model provided the maximum voltage φR generated by the US stimulation on the surface of a single BTNP:
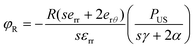 | (4) |
According to eqn (4), when stimulating BTNPs with an intensity of 250 mW cm−2, the maximum voltage generated on the surface of a single BTNP is ∼0.09 mV. However, the local voltage may increase if BTNPs are accumulated in clusters, thus increasing the overall diameter.
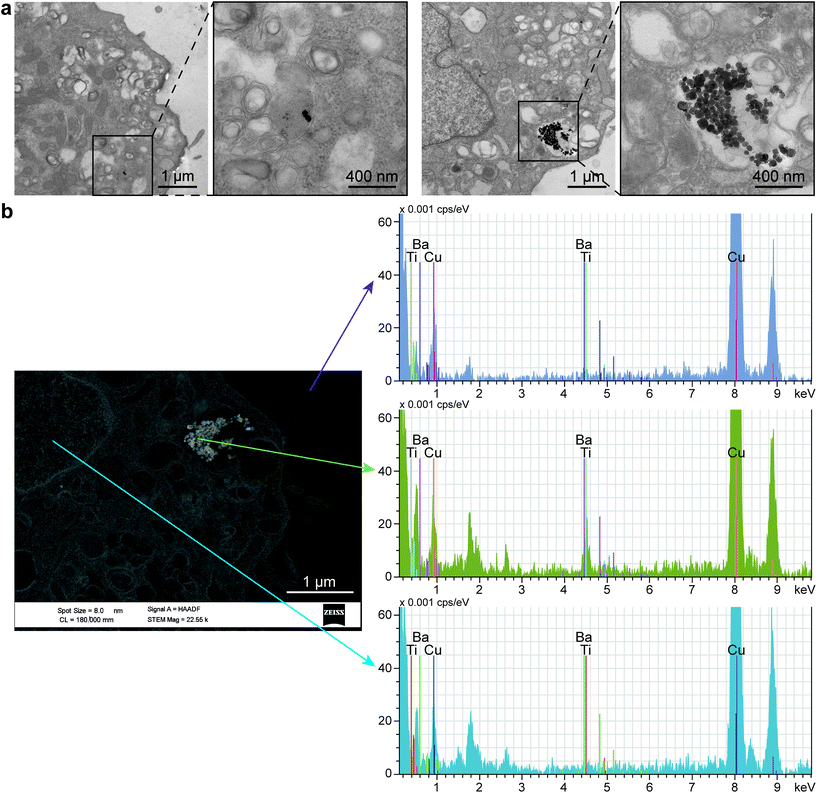
Interestingly, TEM images revealed the internalization of BTNPs within the cell membrane and entrapped in intracellular vesicles in groups from a few to a hundred of BTNPs (Fig. 8a). An EDX analysis performed on the formed clusters confirmed the presence of BTNPs (Fig. 8b). Probably, the relatively small size of the BTNPs (diameter: 60 nm) contributes to their endocytosis, as also demonstrated by Zhao et al. (diameter: 66 nm),90 different from what was found by Marino et al. in which BTNPs (diameter: 300 nm) were accumulated onto the cell membrane.10
The presence of a cluster implies an increase in the estimated voltage, supposing the superposition property. In our case, such a voltage ranges from 0.18 mV to 0.91 mV, for clusters having a diameter from 120 nm to 600 nm, respectively, as visible in Fig. 8a. As known in the state of the art, the localized voltage generated by the US-mediated BTNP stimulation induces the enhancement of the Ca2+ ions influx, according to the prospect that a Ca2+ channel opening increases exponentially with membrane depolarization.11,90 Indeed, it has been shown that even 2 mV has a chance to open them up to a relevant fraction (∼30%).91 Furthermore, it is worth mentioning that such probability is a continuous function of the membrane potential, with no threshold.92 Thus, it is likely that even a relatively small intracellular potential (hundreds of μV or even tens of μV), like the one estimated in our case, may contribute to the activation of the myoblast voltage gate channels, which have an important role in skeletal muscle differentiation.93,94
LIPUS has been recently employed to evaluate possible beneficial effects on various cell cultures, including muscle cells.95,96 In particular, Salgarella et al. found optimal frequency at 1 MHz after developing an in-house stimulation system that minimized reflections and attenuations, allowing precise control of ultrasound delivery. Despite that, the analysis was performed only on the morphological shape of differentiated myotubes cultured over a 2D substrate. In our study, the LIPUS stimulation has been applied to a printed structure for the first time, and results showed that LIPUS promoted the expression of MYOD1, MYOG, and MYH2 in doped bioinks, thus enabling the modulation of myogenic markers.
Overall, our results show the opportunity of using piezoelectric nanoparticles and LIPUS stimulation as a pro-myogenic paradigm for accelerating the differentiation/maturation of printed skeletal muscle structures. This paradigm was demonstrated in our case by using an alginate/Pluronic-based bioink, which was proposed in the literature as a material keeping good shape retention at physiological conditions and guaranteeing high cell viability and expression of myogenic genes with higher performance of 3D constructs compared to cells cultured in two-dimensional conditions.29 However, the same paradigm may be easily applied in the future to a variety of bioinks that proved their high potential as matrices for 3D skeletal muscle constructs, e.g., hyaluronic acid, gelatin, and fibrinogen,74,97 gelatin, fibrinogen, hyaluronic acid, and glycerol,98–100 or decellularized extracellular matrices (dECM).101
Our findings may have important future implications in drug testing and treatment of VML diseases or muscle defect injuries. Indeed, US-stimulated piezoelectric materials combined with 3D bioprinting may allow to obtain mature 3D printed skeletal muscle constructs more rapidly, by boosting the myogenesis of myoblasts embedded in a 3D bioprinted construct. This paradigm also has the promising perspective of a possible wireless stimulation in vivo, after implantation, exploiting the minimal invasiveness of ultrasound. This opportunity can make this technology attractive for future preclinical and clinical translation of this technology.
4. Conclusion
In this study, we demonstrated the possibility of developing a printable piezoelectric bioink based on an alginate/Pluronic hydrogel and doped with barium titanate nanoparticles (BTNPs, diameter: ∼60 nm, d33 piezoelectric coefficient: ∼88 pm V−1) and stimulating it through ultrasound waves to boost myoblast differentiation. A non-covalent wrapping of propylene glycol alginate allowed obtaining a stable aqueous dispersion of the nanoparticles, which guaranteed their homogeneous dispersion in the printed nanocomposite bioink. Rheological analyses evidenced that both the bare and doped bioinks allowed a safe bioprinting procedure, with estimated shear stresses well below the safety threshold of 5 kPa. Single-layer structures embedding C2C12 cells were bioprinted, showing that cell metabolism and viability were not negatively affected by the nanoparticle presence in the bioink up to a concentration of 250 μg mL−1. When subjected to ultrasound stimulation for three consecutive days during differentiation, cells embedded in the doped printed structures showed more marked skeletal muscle differentiation than the controls, with the formation of cell aggregates and the over-expression of MYOD1, MYOG, and MYH2. Gene expression analyses suggest that this behaviour may be due to the activation of the cell cycle arrest.These results highlight that piezoelectric nanoparticle-mediated ultrasound stimulation can be exploited to boost myoblast differentiation in 3D bioprinted constructs. Thus, in future experiments, we will consider exploring the combination of piezoelectric nanoparticles and LIPUS stimulation to different bioinks, also shaping them into a more complex architecture, based on multiple layers. The results of these investigations will allow deeper elucidation of the mechanisms of action of the combinations of the two stimuli, in view of further in vitro application and in vivo translations.
Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
The authors would like to acknowledge Carlo Filippeschi for technical assistance during SEM imaging and EDX analysis, Pasqualantonio Pingue for his support during PFM data acquisition, Chiara Ippolito for technical support during confocal microscope imaging, Francesco Fontana for his support during LIPUS stimulations, and Carsten Jost and Yirij Fedutik (PlasmaChem GmbH) for providing piezoelectric nanoparticles.This work received both financial and technical support by INAIL, in the framework of the project MIO-PRO (Engineered patient-specific muscles for the restoration of myoelectric channels and prosthesis control), grant number: PR19-CR-P1.
References
- B. J. Kwee and D. J. Mooney, Curr. Opin. Biotechnol., 2017, 47, 16–22 CrossRef CAS PubMed.
- J. M. Grasman, M. J. Zayas, R. L. Page and G. D. Pins, Acta Biomater., 2015, 25, 2–15 CrossRef CAS PubMed.
- J. Liu, D. Saul, K. O. Böker, J. Ernst, W. Lehman and A. F. Schilling, Biomed Res. Int., 2018, 2018, 1984879 Search PubMed.
- J. Wang, A. Khodabukus, L. Rao, K. Vandusen, N. Abutaleb and N. Bursac, Biomaterials, 2019, 221, 119416 CrossRef CAS PubMed.
- C. Mueller, M. Trujillo-Miranda, M. Maier, D. E. Heath, A. J. O'Connor and S. Salehi, Adv. Mater. Interfaces, 2021, 8, 2001167 CrossRef.
- S. Ostrovidov, V. Hosseini, S. Ahadian, T. Fujie, S. P. Parthiban, M. Ramalingam, H. Bae, H. Kaji and A. Khademhosseini, Tissue Eng., Part B, 2014, 20, 403–436 CrossRef PubMed.
- A. Cafarelli, A. Marino, L. Vannozzi, J. Puigmartí-Luis, S. Pané, G. Ciofani and L. Ricotti, ACS Nano, 2021, 15, 11066–11086 CrossRef CAS PubMed.
- S. Ahadian, S. Ostrovidov, V. Hosseini, H. Kaji, M. Ramalingam, H. Bae and A. Khademhosseini, Organogenesis, 2013, 9, 87–92 CrossRef PubMed.
- G. Ciofani, S. Danti, D. D'Alessandro, L. Ricotti, S. Moscato, G. Bertoni, A. Falqui, S. Berrettini, M. Petrini, V. Mattoli and A. Menciassi, ACS Nano, 2010, 4, 6267–6277 CrossRef CAS PubMed.
- A. Marino, S. Arai, Y. Hou, E. Sinibaldi, M. Pellegrino and Y. Chang, ACS Nano, 2015, 9, 7678–7689 CrossRef CAS PubMed.
- B. Ma, F. Liu, Z. Li, J. Duan, Y. Kong, M. Hao, S. Ge, H. Jiang and H. Liu, J. Mater. Chem. B, 2019, 7, 1847–1854 RSC.
- G. G. Genchi, L. Ceseracciu, A. Marino, M. Labardi, S. Marras, F. Pignatelli, L. Bruschini, V. Mattoli and G. Ciofani, Adv. Healthcare Mater., 2016, 5, 1808–1820 CrossRef CAS PubMed.
- L. Ricotti, T. Fujie, H. Vazão, G. Ciofani, R. Marotta, R. Brescia, C. Filippeschi, I. Corradini, M. Matteoli, V. Mattoli, L. Ferreira and A. Menciassi, PLoS One, 2013, 8, e71707 CrossRef CAS PubMed.
- L. Ricotti, A. Polini, G. G. Genchi, G. Ciofani, D. Iandolo, H. Vazão, V. Mattoli, L. Ferreira, A. Menciassi and D. Pisignano, Biomed. Mater., 2012, 7, 035010 CrossRef PubMed.
- L. Altomare, N. Gadegaard, L. Visai, M. C. Tanzi and S. Farè, Acta Biomater., 2010, 6, 1948–1957 CrossRef CAS PubMed.
- A. Hasebe, Y. Suematsu, S. Takeoka, T. Mazzocchi, L. Vannozzi, L. Ricotti and T. Fujie, ACS Biomater. Sci. Eng., 2019, 5, 5734–5743 CrossRef CAS PubMed.
- F. Greco, T. Fujie, L. Ricotti, S. Taccola, B. Mazzolai and V. Mattoli, ACS Appl. Mater. Interfaces, 2013, 5, 573–584 CrossRef CAS PubMed.
- A. Khodabukus, Front. Physiol., 2021, 12, 619710 CrossRef PubMed.
- S. Derakhshanfar, R. Mbeleck, K. Xu, X. Zhang, W. Zhong and M. Xing, Bioact. Mater., 2018, 3, 144–156 CrossRef PubMed.
- T. H. Qazi, D. J. Mooney, M. Pumberger, S. Geißler and G. N. Duda, Biomaterials, 2015, 53, 502–521 CrossRef CAS PubMed.
- M. Askari, M. A. Naniz, M. Kouhi, A. Saberi, A. Zolfagharian and M. Bodaghi, Biomater. Sci., 2021, 9, 535–573 RSC.
- S. Ostrovidov, S. Salehi, M. Costantini, K. Suthiwanich, M. Ebrahimi, R. B. Sadeghian, T. Fujie, X. Shi, S. Cannata, C. Gargioli, A. Tamayol, M. R. Dokmeci, G. Orive, W. Swieszkowski and A. Khademhosseini, Small, 2019, 15, 1–14 CrossRef PubMed.
- P. Zhuang, J. An, C. K. Chua and L. P. Tan, Mater. Des., 2020, 193, 108794 CrossRef CAS.
- R. T. Shafranek, S. C. Millik, P. T. Smith, C. U. Lee, A. J. Boydston and A. Nelson, Prog. Polym. Sci., 2019, 93, 36–67 CrossRef CAS.
- T. Gonzalez-Fernandez, P. Sikorski and J. K. Leach, Acta Biomater., 2019, 96, 20–34 CrossRef CAS PubMed.
- P. Lavrador, M. R. Esteves, V. M. Gaspar and J. F. Mano, Adv. Funct. Mater., 2021, 31, 2005941 CrossRef CAS.
- S. M. Bakht, A. Pardo, M. Gómez-Florit, R. L. Reis, R. M. A. Domingues and M. E. Gomes, J. Mater. Chem. B, 2021, 9, 5025–5038 RSC.
- A. Popov, S. Malferrari and D. M. Kalaskar, J. 3D Print. Med., 2017, 1, 191–211 CrossRef CAS.
- P. Mozetic, S. M. Giannitelli, M. Gori, M. Trombetta and A. Rainer, J. Biomed. Mater. Res., Part A, 2017, 105, 2582–2588 CrossRef CAS PubMed.
- M. Costantini, S. Testa, P. Mozetic, A. Barbetta, C. Fuoco, E. Fornetti, F. Tamiro, S. Bernardini, J. Jaroszewicz, W. Święszkowski, M. Trombetta, L. Castagnoli, D. Seliktar, P. Garstecki, G. Cesareni, S. Cannata, A. Rainer and C. Gargioli, Biomaterials, 2017, 131, 98–110 CrossRef CAS PubMed.
- M. Yeo, H. Lee and G. H. Kim, Biofabrication, 2016, 8, 1–12 CrossRef PubMed.
- F. L. Ronzoni, F. Aliberti, F. Scocozza, L. Benedetti, F. Auricchio, M. Sampaolesi, G. Cusella, I. N. Redwan, G. Ceccarelli and M. Conti, J. Tissue Eng. Regener. Med., 2022, 16, 484–495 CrossRef CAS PubMed.
- R. Seyedmahmoud, B. Çelebi-Saltik, N. Barros, R. Nasiri, E. Banton, A. Shamloo, N. Ashammakhi, R. M. Dokmeci and S. Ahadian, Micromachines, 2019, 10, 679 CrossRef PubMed.
- T. Distler, A. A. Solisito, D. Schneidereit, O. Friedrich, R. Detsch and A. R. Boccaccini, Biofabrication, 2020, 12, 045005 CrossRef CAS PubMed.
- E. Fröhlich, G. Bonstingl, A. Höfler, C. Meindl, G. Leitinger, T. R. Pieber and E. Roblegg, Toxicol. In Vitro, 2013, 27, 409–417 CrossRef PubMed.
- A. Rached, M. A. Wederni, K. Khirouni, S. Alaya, R. J. Martín-Palma and J. Dhahri, Mater. Chem. Phys., 2021, 267, 124600 CrossRef CAS.
- K. Buse, S. Riehemann, S. Loheide, H. Hesse, F. Mersch and E. Krätzig, Phys. Status Solidi, 1993, 135, K87–K89 CrossRef CAS.
- M. Cvek, A. Zahoranova, M. Mrlik, P. Sramkova, A. Minarik and M. Sedlacik, Colloids Surf., B, 2020, 190, 110912 CrossRef CAS PubMed.
- A. Moscardini, S. Di Pietro, G. Signore, P. Parlanti, M. Santi, M. Gemmi and V. Cappello, Sci. Rep., 2020, 10, 1–11 CrossRef PubMed.
- H. Kim, J. Jang, J. Park, K. P. Lee, S. Lee, D. M. Lee, K. H. Kim, H. K. Kim and D. W. Cho, Biofabrication, 2019, 11, 35017 CrossRef CAS PubMed.
- M. M. Lübtow, M. Mrlik, L. Hahn, A. Altmann, M. Beudert, T. Lühmann and R. Luxenhofer, J. Funct. Biomater., 2019, 10, 36 CrossRef PubMed.
- L. Grant, R. Raman, C. Cvetkovic, M. C. Ferrall-Fairbanks, G. J. Pagan-Diaz, P. Hadley, E. Ko, M. O. Platt and R. Bashir, Tissue Eng., Part A, 2019, 25, 1023–1036 CrossRef CAS PubMed.
- R. Raman, L. Grant, Y. Seo, C. Cvetkovic, M. Gapinske, A. Palasz, H. Dabbous, H. Kong, P. P. Pinera and R. Bashir, Adv. Healthcare Mater., 2017, 6, 1–9 Search PubMed.
- C. Cvetkovic, R. Raman, V. Chan, B. J. Williams, M. Tolish, P. Bajaj, M. S. Sakar, H. H. Asada, M. T. A. Saif and R. Bashir, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 10125–10130 CrossRef CAS PubMed.
- F. Fontana, F. Iberite, A. Cafarelli, A. Aliperta, G. Baldi, E. Gabusi, P. Dolzani, S. Cristino, G. Lisignoli, T. Pratellesi, E. Dumont and L. Ricotti, Ultrasonics, 2021, 106495 CrossRef CAS PubMed.
- M. Acosta, N. Novak, V. Rojas, S. Patel, R. Vaish, J. Koruza and G. A. Rossetti, Appl. Phys. Rev., 2017, 4, 041305 Search PubMed.
- F. R. Baxter, C. R. Bowen, I. G. Turner and A. C. E. Dent, Ann. Biomed. Eng., 2010, 38, 2079–2092 CrossRef CAS PubMed.
- G. Ciofani and A. Menciassi, Piezoelectric nanomaterials for biomedical applications, Springer, Berlin, 2012 Search PubMed.
- M. Okamoto and B. John, Prog. Polym. Sci., 2013, 38, 1487–1503 CrossRef CAS.
- G. Ciofani, S. Danti, S. Moscato, L. Albertazzi, D. D'Alessandro, D. Dinucci, F. Chiellini, M. Petrini and A. Menciassi, Colloids Surf., B, 2010, 76, 535–543 CrossRef CAS PubMed.
- A. C. Sabuncu, J. Grubbs, S. Qian, T. M. Abdel-Fattah, M. W. Stacey and A. Beskok, Colloids Surf., B, 2012, 95, 96–102 CrossRef CAS PubMed.
- A. Barreto, L. G. Luis, A. V. Girão, T. Trindade, A. M. Soares and M. Oliveira, J. Nanopart. Res., 2015, 17, 1–13 CrossRef CAS.
- L. S. Franqui, M. A. De Farias, R. V. Portugal, C. A. R. Costa, R. R. Domingues, A. G. Souza Filho, V. R. Coluci, A. F. P. Leme and D. S. T. Martinez, Mater. Sci. Eng., C, 2019, 100, 363–377 CrossRef CAS PubMed.
- K. Strojan, A. Leonardi, V. B. Bregar, I. Križaj, J. Svete and M. Pavlin, PLoS One, 2017, 12, e0169552 CrossRef PubMed.
- L. Vannozzi, E. Catalano, M. Telkhozhayeva, E. Teblum, A. Yarmolenko, E. S. Avraham, R. Konar, G. D. Nessim and L. Ricotti, Nanomaterials, 2021, 11, 2105 CrossRef CAS PubMed.
- J. H. Jean and H. R. Wang, J. Am. Ceram. Soc., 2000, 83, 277–280 CrossRef CAS.
- C. Gómez-Yáñez, H. Balmori-Ramírez and F. Martínez, Ceram. Int., 2000, 26, 609–616 CrossRef.
- J. Jean and H. Wang, J. Mater. Res., 1998, 13, 2245–2250 CrossRef CAS.
- Z. G. Shen, J. F. Chen, H. K. Zou and J. Yun, J. Colloid Interface Sci., 2004, 275, 158–164 CrossRef CAS PubMed.
- J. H. Jean and H. R. Wang, J. Am. Ceram. Soc., 1998, 81, 1589–1599 CrossRef CAS.
- Y. Hu, S. Gong and D. Zhou, Mater. Sci. Eng., B, 2003, 99, 520–522 CrossRef.
- X. Wang, B. I. Lee and L. Mann, Colloids Surf., A, 2002, 202, 71–80 CrossRef CAS.
- G. Ciofani, S. Danti, D. D'Alessandro, S. Moscato, M. Petrini and A. Menciassi, Nanoscale Res. Lett., 2010, 5, 1093–1101 CrossRef CAS PubMed.
- A. A. Barba, M. D'Amore, M. Grassi, S. Chirico, G. Lamberti and G. Titomanlio, J. Appl. Polym. Sci., 2009, 114, 688–695 CrossRef CAS.
- J. P. Soares, J. E. Santos, G. O. Chierice and E. T. G. Cavalheiro, Ecletica Quim., 2004, 29, 57–64 CrossRef CAS.
- F. M. Cabrini, M. Champeau and M. G. de Oliveira, J. Appl. Polym. Sci., 2020, 137, 49056 CrossRef CAS.
- J. Zeltinger, J. K. Sherwood, D. A. Graham, R. Müeller and L. G. Griffith, Tissue Eng., 2001, 7, 557–572 CrossRef CAS PubMed.
- M. Abrami, I. D'Agostino, G. Milcovich, S. Fiorentino, R. Farra, F. Asaro, R. Lapasin, G. Grassi and M. Grassi, Soft Matter, 2014, 10, 729–737 RSC.
- S. C. Lee, G. Gillispie, P. Prim and S. J. Lee, Chem. Rev., 2020, 120, 10834–10886 CrossRef CAS PubMed.
- N. Paxton, W. Smolan, T. Böck, F. Melchels, J. Groll and T. Jungst, Biofabrication, 2017, 9, 044107 CrossRef PubMed.
- V. Lenaerts, C. Triqueneaux, M. Quartern, F. Rieg-Falson and P. Couvreur, Int. J. Pharm., 1987, 39, 121–127 CrossRef CAS.
- R. Michel and R. Auzély-Velty, Biomacromolecules, 2020, 21, 2949–2965 CrossRef CAS PubMed.
- A. Blaeser, D. F. Duarte Campos, U. Puster, W. Richtering, M. M. Stevens and H. Fischer, Adv. Healthcare Mater., 2016, 5, 326–333 CrossRef CAS PubMed.
- R. Mestre, T. Patiño, X. Barceló, S. Anand, A. Pérez-Jiménez and S. Sánchez, Adv. Mater. Technol., 2019, 4, 1–13 Search PubMed.
- W. J. Kim, C. H. Jang and G. H. Kim, Chem. Eng. J., 2021, 419, 129570 CrossRef CAS.
- E. Gioffredi, M. Boffito, S. Calzone, S. M. Giannitelli, A. Rainer, M. Trombetta, P. Mozetic and V. Chiono, Procedia CIRP, 2016, 49, 125–132 CrossRef.
- J. M. Townsend, E. C. Beck, S. H. Gehrke, C. J. Berkland and M. S. Detamore, Prog. Polym. Sci., 2019, 91, 126–140 CrossRef CAS PubMed.
- D. Chimene, C. W. Peak, J. L. Gentry, J. K. Carrow, L. M. Cross, E. Mondragon, G. B. Cardoso, R. Kaunas and A. K. Gaharwar, ACS Appl. Mater. Interfaces, 2018, 10, 9957–9968 CrossRef CAS PubMed.
- H. Tetsuka and S. R. Shin, J. Mater. Chem. B, 2020, 8, 2930–2950 RSC.
- J. Chal and O. Pourquié, Development, 2017, 144, 2104–2122 CrossRef CAS PubMed.
- X. Shen, J. M. Collier, M. Hlaing, L. Zhang, E. H. Delshad, J. Bristow and H. S. Bernstein, Dev. Dyn., 2003, 226, 128–138 CrossRef CAS PubMed.
- I. Delgado, X. Huang, S. Jones, L. Zhang, R. Hatcher, B. Gao and P. Zhang, Genomics, 2003, 82, 109–121 CrossRef CAS PubMed.
- D. A. Bergstrom and S. J. Tapscott, Mol. Cell. Biol., 2001, 21, 2404–2412 CrossRef CAS PubMed.
- G. De Luca, R. Ferretti, M. Bruschi, E. Mezzaroma and M. Caruso, Stem Cells, 2013, 31, 2478–2491 CrossRef CAS PubMed.
- O. Halevy, B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach and A. B. Lassar, Science, 1995, 267, 1018–1021 CrossRef CAS PubMed.
- G. H. Yang, W. Kim, J. Kim and G. H. Kim, Theranostics, 2020, 11, 48 CrossRef PubMed.
- M. Ganassi, S. Badodi, H. P. Ortuste Quiroga, P. S. Zammit, Y. Hinits and S. M. Hughes, Nat. Commun., 2018, 9, 1–17 CrossRef CAS PubMed.
- P. Ferri, E. Barbieri, S. Burattini, M. Guescini, A. D'Emilio, L. Biagiotti, P. Del Grande, A. De Luca, V. Stocchi and E. Falcieri, J. Cell. Biochem., 2009, 108, 1302–1317 CrossRef CAS PubMed.
- Y. Chen, V. Stegaev, V. P. Kouri, T. Sillat, P. L. Chazot, H. Stark, J. G. Wen and Y. T. Konttinen, Mol. Med. Rep., 2015, 11, 2624–2630 CrossRef CAS PubMed.
- D. Zhao, P. Feng, J. Liu, M. Dong, X. Shen, Y. Chen and Q. Shen, Adv. Mater., 2020, 32, 2003800 CrossRef CAS PubMed.
- M. T. Nelson, J. B. Patlak, J. F. Worley and N. B. Standen, Am. J. Physiol.: Cell Physiol., 1990, 259, C3–C18 CrossRef CAS PubMed.
- B. Hille, Biophys. J., 1978, 22, 283–294 CrossRef CAS PubMed.
- E. E. Spangenburg, D. K. Bowles and F. W. Booth, Endocrinology, 2004, 145, 2054–2063 CrossRef CAS PubMed.
- A. M. Afzali, T. Ruck, A. M. Herrmann, J. Iking, C. Sommer, C. Kleinschnitz, C. Preuβe, W. Stenzel, T. Budde, H. Wiendl, S. Bittner and S. G. Meuth, Am. J. Physiol.: Cell Physiol., 2016, 311, C583–C595 CrossRef PubMed.
- Y. S. Chan, K. Y. Hsu, C. H. Kuo, S. Da Lee, S. C. Chen, W. J. Chen and S. W. N. Ueng, Ultrasound Med. Biol., 2010, 36, 743–751 CrossRef PubMed.
- A. R. Salgarella, A. Cafarelli, L. Ricotti, L. Capineri, P. Dario and A. Menciassi, Ultrasound Med. Biol., 2017, 43, 1452–1465 CrossRef PubMed.
- T. K. Merceron, M. Burt, Y. J. Seol, H. W. Kang, S. J. Lee, J. J. Yoo and A. Atala, Biofabrication, 2015, 7, 35003 CrossRef PubMed.
- H. W. Kang, S. J. Lee, I. K. Ko, C. Kengla, J. J. Yoo and A. Atala, Nat. Biotechnol., 2016, 34, 312–319 CrossRef CAS PubMed.
- J. H. Kim, Y. J. Seol, I. K. Ko, H. W. Kang, Y. K. Lee, J. J. Yoo, A. Atala and S. J. Lee, Sci. Rep., 2018, 8, 1–15 Search PubMed.
- J. H. Kim, I. Kim, Y. J. Seol, I. K. Ko, J. J. Yoo, A. Atala and S. J. Lee, Nat. Commun., 2020, 11, 1–12 CrossRef PubMed.
- Y. J. Choi, T. G. Kim, J. Jeong, H. G. Yi, J. W. Park, W. Hwang and D. W. Cho, Adv. Healthcare Mater., 2016, 5, 2636–2645 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1bm01853a |
| This journal is © The Royal Society of Chemistry 2022 |