Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Conformational transition in SPR experiments: impact of spacer length, immobilization mode and aptamer density on signal sign and amplitude†
Marina
Pons
a,
Marine
Perenon
a,
Hugues
Bonnet
a,
Emilie
Gillon
b,
Celio
Vallée
a,
Liliane
Coche-Guérente
 a,
Eric
Defrancq
a,
Eric
Defrancq
 a,
Nicolas
Spinelli
a,
Nicolas
Spinelli
 a,
Angeline
Van der Heyden
*a and
Jérôme
Dejeu
a,
Angeline
Van der Heyden
*a and
Jérôme
Dejeu
 *ac
*ac
aUniv. Grenoble Alpes, CNRS, DCM UMR-5250, F-38000 Grenoble, France. E-mail: Jerome.dejeu@univ-grenoble-alpes.fr; angeline.van-der-heyden@univ-grenoble-alpes.fr
bUniv. Grenoble Alpes, CERMAV-CNRS, 601 rue de la chimie, F-38610 Gières, France
cFEMTO-ST Institute, CNRS UMR-6174, Université de Bourgogne Franche-Comté, F-25000 Besançon, France
First published on 3rd August 2022
Abstract
Surface plasmon resonance (SPR) is an optical, real-time and label-free technique which represents a standard to study biomolecular interactions. While SPR signals are usually positive upon recognition, a few cases of negative signals have been reported because of significant conformational transition of the receptor upon the recognition of the target. In this study, we reported on the observation of negative or null SPR signals for an aptamer recognition with its low molecular weight target. The introduction of a spacer group for the aptamer immobilization led to a null SPR signal despite the device sensitivity and effective target recognition (a KD around 200 nM as demonstrated using a quartz crystal microbalance with dissipation monitoring and isothermal titration calorimetry). We demonstrated that this unconventional signal could be attributed to two opposite contributions: a positive one is afforded by the aptamer recognition and folding whereas a negative one results from the refractive index increment (RII) deviation upon the formation of the complex (ligand/analyte). We also demonstrated that the RII deviation is sensitive to the modification of the sequence flexibility and therefore depends on the anchoring procedure and the spacer length between the anchoring function and the site of recognition.
Introduction
SPR is an optical technique that measures changes in the refractive index (RI) in close proximity to a metal surface. As the RI depends on the composition and molecular weight of molecules, this technique allows the detection of biomolecular recognition events between analytes and a ligand immobilized on the surface in real time and without analyte labelling. While the detection of small molecules is still challenging, thanks to the high sensitivity and optimal microfluidic system, the latest generation of devices enables the monitoring of interactions involving such molecules. Indeed, the number of fragments or small molecules screened and characterized using SPR has dramatically increased during the last decade.1 Usually, upon analyte interaction with the immobilized ligand, positive signals are measured with an intensity concordant with the values defined by Wilson's formula.2 When null SPR signals are recorded, a non-interaction is usually considered. Moreover, in some specific cases, the SPR responses could be enhanced3–8 or on the contrary could be negative3,9–13 depending on the interaction mechanism. Such unexpected signals are usually observed when the ligand immobilized on the surface undergoes a significant conformational transition upon analyte recognition, i.e. folding3–7,13 or stretching.9–12 Indeed, in addition to its composition, the structural conformation of a protein has been recently identified as a parameter that could influence its refractive index increment (RII).14 As a consequence, for recognition systems involving the significant conformational transition of one partner, the additivity of the RII (the ligand/analyte RII equal to the sum of the target and ligand RII) no longer applies: a deviation of the ligand/analyte complex RII is observed in solution.15 This RII non-additivity has also been demonstrated on the surface.16 To date, prediction of the RII deviation is not possible considering only size variations as initially proposed by Gestwicki et al.3 Indeed, negative SPR signals and thus negative RII deviations were reported for calcium binding protein stretching or folding5 Moreover, aptamer folding can also induce positive16 or negative13 RII variations. Indeed, we have recently demonstrated that the recognition of a low molecular weight target (L-tyrosinamide, L-Tym) by a 49-mer aptamer attached on the surface through biotin/streptavidin binding8,13,16 led to a positive RII deviation and so an enhancement of the SPR signals was observed. On the contrary, L-Tym recognition by a shortened aptamer sequence (23-mer) anchored on the surface via thiol–gold interaction gave negative RII deviation and negative sensorgrams were recorded upon interaction.13 For this latter aptamer, a 6 thymine spacer was included between the thiol anchoring function and the recognition sequence. In order to further identify the parameters influencing the RII deviation among the length of the aptamer sequence, the spacer length and the anchoring procedure, we report herein a study of L-Tym recognition by the 23-mer aptamer sequence immobilized on a streptavidin platform under various conditions. We demonstrated that the SPR signal was affected by the immobilization mode (a biotinylated aptamer adsorbed on a 2D streptavidin monolayer versus a thiolated aptamer directly adsorbed on a gold surface) and by the presence of a spacer between the anchoring function and the recognition sequence of the aptamer. The signal could be negative or even null despite the occurrence of target recognition. This variation was explained by the modification of the RII deviation which is sensitive to parameters affecting the flexibility of the recognition sequence.Materials and methods
Solutions
All aqueous solutions were prepared with ultrapure water (Purelab UHQ Elga). (11-Mercaptoundecyl)hexa(ethylene glycol) biotinamide (HS-(CH2)11-EG6-biotin) was purchased from Prochimia (Poland). All the other chemical products (NaCl, MgCl2, Tween 20, Trizma base, (11-mercaptoundecyl)tetra(ethylene glycol) (HS-(CH2)11-EG4-OH)) were purchased from Sigma-Aldrich. Running buffer (20 mM Trizma base, 50 mM NaCl, 5 mM MgCl2, Tween 20, 0.5% v/v, pH = 7.5) in ultrapure water was used as the buffer in each experiment. This composition was previously optimized.17Oligonucleotide synthesis
The syntheses of the L-Tym aptamer (5′-TGT GGT GTG TGA GTG CGG TGC CC (Y)n-Biot-3′, where Y represents the nucleotide that is used as spacer (T or A), n the number of nucleotides and Biot the biotin moiety with triethylene glycol (TEG), similar to the one selected by Vianini et al.18) and a random sequence (5′-ATG ACC CTA CCT GCT GAT GCG TA Biot-3′) were performed on a 3′-BiotinTEG-CPG resin as previously reported.16 The length of the spacer was modulated by the number of thymines, either 0, 3, 6, 9 or 16. The number of adenines was 6. The aptamers are noted as Apt23-TX-Biot or Apt23-A6-Biot, with X being the number of thymine groups.Sensor chip surface generation
Upon removal from storage at 4 °C, the bare gold sensor chip was rinsed with ultrapure water, dried through a nitrogen gas stream and exposed for 10 min to UV–ozone cleaning (model 42-220, Jelight Company Inc.). The cleaned gold surface was then dipped overnight in the mixture solution of thiols (90% of 1 mM HS-(CH2)11-EG4-OH and 10% of 1 mM HS-(CH2)11-EG6-biotin) in ethanol. Next, the surface was carefully cleaned with ethanol and dried over gaseous nitrogen. It has been demonstrated that 10% of biotinylated thiol is necessary to obtain a streptavidin layer with maximum surface coverage.19 The functionalized gold sensor chip was mounted on the sample holder for immediate usage by QCM-D or SPR.Quartz crystal microbalance with dissipation monitoring (QCM-D) measurements
QCM-D measurements were performed using Q-Sense E1 or E4 instruments (Biolin Scientific) equipped with one or four flow modules, respectively. In addition to measurement of the bound mass, which is provided by changes in the resonance frequency, f, of the sensor crystal, the QCM-D technique also provided structural information for biomolecular films via the changes in the energy dissipation, D, of the sensor crystal. f and D were measured at the fundamental resonance frequency (4.95 MHz) as well as at the third, fifth, seventh, ninth, eleventh, and thirteenth overtones (i = 3, 5, 7, 9, 11 and 13). Normalized frequency shifts Δf = Δfi/i and dissipation shifts ΔD = ΔDi were presented.Experiments were conducted in a continuous flow of running buffer without Tween 20 with a flow rate of 50 μL min−1 using a peristaltic pump (ISM935C, Ismatec, Switzerland). The temperature of the QCM-D platform and all solutions was stabilized at 25 °C to ensure stable operation. All buffers were previously degassed in order to avoid air bubble formation in the fluidic system. For each aptamer, at least two independent experiments were performed on new surfaces. At least six concentrations of L-Tym solution were injected on each surface. The frequency variation of the 7th overtone for each concentration was taken 10 minutes after injection of the L-Tym and averaged over 2 minutes.
Surface plasmon resonance measurement
All SPR measurements were performed at 25 °C in a four flow-cells Biacore T200 instrument (Cytiva). The experimental conditions were similar to those previous reported.13,16Isothermal titration microcalorimetry
ITC experiments were performed with an ITC200 titration calorimeter (Malvern). The experiments were carried out at 25 °C. All compounds were dissolved in running buffer without Tween 20. The aptamers were placed in the microcalorimeter cell (200 μL) at a concentration of 25 μM. A total of 20 injections of 0.5 μL for the first and 2 μL for the other injections of L-tyrosinamide at a concentration of 250 μM were performed with an interval of 120 s between each injection while stirring at 750 rpm. The experimental data were fitted to a theoretical titration curve using the Microcal PEAQ-ITC analysis software, with ΔH (enthalpy change) and Ka (equilibrium association constant) as the adjustable parameters. The equilibrium dissociation constant (KD) and the entropy contributions (TΔS) were derived from the previous ones. Two independent titrations were performed for each aptamer tested.Results and discussion
Previous studies reported on the recognition of L-Tym by a 49-mer aptamer. This aptamer was further sequentially truncated at both ends to identify a minimal binding sequence (MBS) able to recognize L-Tym. The resulting MBS was a 23-mer corresponding to the Apt49 truncated at it 5′ end. Moreover, it has been demonstrated that the recognition is an induced-fit process, i.e. the binding of the target first occurs with the unstructured aptamer and thus triggers its folding somehow directed from the 5′-end to the 3′end.21 An increase in the affinity for L-Tym was obtained for the MBS compared with the Apt49.17,21 SPR analysis of the recognition of L-Tym by these immobilized aptamers allowed demonstration of the non-additivity of the refractive index increment (RII) upon the L-Tym/aptamer recognition as reported by Bornhop et al. for interaction in the solution involving large structural or hydration modification of one of the partners.15 The structural modification of the aptamer upon recognition induced a RII deviation for L-Tym/Apt49-T0-Biot recognition that enhanced the SPR signal while negative signals were observed for the L-Tym/Apt23-T6-SH one.13,16 Since the affinity of this aptamer is closely linked to the efficiency of its conformational transition, parameters affecting affinity could also impact the RII deviation of the aptamer.Influence of the aptamer length – Apt49-T0-Biot vs. Apt23-T0-Biot
To explore the influence of the aptamer length on the RII deviation, an SPR study of L-Tym recognition of the MBS was performed using the same immobilization procedure as used for the Apt49-T0-Biot.8,16,22 Monolayers of the anti-L-Tym aptamer and its corresponding scramble sequence (used as a negative control for the reference surface) were formed through streptavidin–biotin interaction. First, the gold surface was functionalized ex situ by co-adsorption of two pegylated alkanethiols, one exhibiting a biotin terminal group while the other is a hydroxyl terminated alkanethiol. The pegylated function was introduced to shield the surface from nonspecific adsorption while a 20% molar ratio of biotinylated thiol was selected to allow the formation of a saturated streptavidin (SA) layer.23 After immobilization of the SA on the thiol layer, the biotinylated aptamers were adsorbed on the SA layer. This architecture has already demonstrated its excellent stability and the appropriate orientation of the biomolecule toward the solution23 for the study of different kinds of biomolecular recognition system.24The recognition was first evaluated on a SA surface saturated with Apt23-T0-Biot. Injection of the Apt23-T0-Biot on the SA platform led to a saturation signal of 480 RU. According to Jung's equation (see the details of the calculation in the ESI, eqn (SI-1) and (SI-2) and Fig. SI-1†), this signal corresponds to a surface density of around 4 pmol cm−2, similar to the one obtained for Apt49-T0-Biot on the SA layer.22 Indeed, the surface density in small biotinylated ligands is governed by the SA binding sites directed to the solution.23
Simultaneous injection of L-Tym was then performed on the aptamer layer and on the scramble one. After a double-reference-subtraction procedure,20 the SPR signals for the aptamer layer were found to be negative (Fig. 1A) while the maximal SPR expected was around 11 RU according to the Wilson equation (see Eqn (SI-3)† and Table 1). The latter expected value was calculated with the hypothesis that all the aptamer recognized the target. It should be noted that if all the aptamer was not accessible for recognition it would induce a weaker maximal signal but not a negative one. Commonly, the untreated SPR signals on the active flow cell first increase due to the RII of the injected solution, known as the bulk effect (also observed on the reference surface) and then a positive curvature is observed due to the target recognition (that should not be observed on the reference surface in the absence of non-specific recognition). After the double subtraction procedure, the bulk effect is cancelled and the SPR signals increase as they are only related to the recognition. In the present case, square profiles of the sensorgrams with a response increasing with the concentration of L-Tym were recorded on the scramble surface (Fig. SI-2A†). These signals confirmed the absence of non-specific interaction between L-Tym and the scramble layer and are attributed to the difference in refractive index increment between the RB and the injection solution as previously detailed.13 However, the raw signal recorded upon L-Tym injection on the aptamer surface led to unexpected profiles on the sensorgrams. Indeed, the raw signals decreased for L-Tym concentration from 10 nM to 25 μM and it should also be noted that after an initial increase in the signal due to the bulk effect, the curvature of the signal was negative (Fig. SI-2B and D†). For L-Tym concentrations above 25 μM, raw SPR signals increased (Fig. SI-2C and D†).
| Ligand | Apt23-T0-Biot | Apt23-T3-Biot | Apt23-T6-Biot | Apt23-A6-Biot | Apt23-T9-Biot | Apt23-T12-Biot |
|---|---|---|---|---|---|---|
a The RII values was determined using SPR in solution as previously described13,25 (see eqn (SI-3) and details in the ESI – Fig. SI-1†). The RII value was measured using the aptamers without the biotin function for Apt23-T6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 and Apt23-A6 (ESI†).
b Calculated for a saturated aptamer layer (RUL = 500 RU). These values also correspond to the maximal SPR signals expected from Dejeu's equation considering no folding of the aptamer and no RII deviation, i.e. with ρ and x equal to 1 and 0 respectively in eqn (1).
c Calculated on a saturated aptamer layer (RUL = 500 RU) considering aptamer folding and no RII deviation, i.e. with ρ and x equal to 0.73 (ref. 13) and 0 respectively in eqn (1). 13 and Apt23-A6 (ESI†).
b Calculated for a saturated aptamer layer (RUL = 500 RU). These values also correspond to the maximal SPR signals expected from Dejeu's equation considering no folding of the aptamer and no RII deviation, i.e. with ρ and x equal to 1 and 0 respectively in eqn (1).
c Calculated on a saturated aptamer layer (RUL = 500 RU) considering aptamer folding and no RII deviation, i.e. with ρ and x equal to 0.73 (ref. 13) and 0 respectively in eqn (1).
|
||||||
| MWL (g mol−1) | 7728 | 8641 | 9553 | 9607 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 466 466 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 378 378 |
| (dn/dc)′ (cm3 g−1)a | 0.252 | 0.190 | 0.238 | 0.245 | 0.193 | 0.196 |
| B = MWA (dn/dc)′A/MWL (dn/dc)′L | 0.0307 | 0.0274 | 0.0184 | 0.0247 | 0.0226 | 0.0208 |
| R max (RU),b Wilson's equation | 10.7 | 12.7 | 9.1 | 8.9 | 10.3 | 9.4 |
| R max (RU),c Dejeu's equation considering only folding of the aptamer | 12.8 | 14.8 | 11.2 | 10.9 | 12.3 | 11.4 |
For low L-Tym concentrations, the effect of RII variation of the solution was negligible, and the signal depended only on the recognition process: target recognition and structural conformation variation. The unconventional signals observed on the raw active surface could be mainly attributed to the structural conformation variation of the aptamer upon recognition, whose contribution to the SPR signal is predominant for concentrations up to 25 μM (Fig. SI-2B†). For concentrations above 25 μM, the SPR signals increased due to the non-negligible contribution of the refractive index of the L-Tym solution at these high concentrations (Fig. SI-2C and D†). Similar behavior has been reported by SPR for L-Tym recognition by Apt23-T6-SH immobilized on a gold surface with the raw signal on the active flow cell being negative.16 With Apt23-T0-Biot, the raw signals remained positive or null due to a lower contribution of the structural conformation variation, and after the double subtraction procedure, the SPR signals became negative.
Injection of D-Tym led to a square profile on both flow cells and to a null signal after the double referencing procedure (Fig. SI-3†). The latter result confirmed that the recognition is enantiomeric.
The negative sensorgrams recorded for L-Tym injection were thus attributed to the conformational transition of the aptamers. Indeed, the conformation transition of Apt23-T0-Biot was confirmed by circular dichroism (CD) (Fig. SI-4A†). As expected, the introduction of a biotin residue at the 3′ aptamer extremity did not hamper the folding of the MBS upon L-Tym in solution.21 To confirm that the structural modification still occurred after immobilization of Apt23-T0-Biot on this SA platform, the same experiment was performed using QCM-D as a non-optical transducing technique. The aptamer and the scramble were immobilized on gold according to the same procedure. No significant QCM-D signal variations were observed for 1 mM L-Tym solution on the scramble surface (Fig. SI-5A†), confirming the absence of nonspecific recognition of L-Tym on the reference surface, as observed by SPR. The QCM-D responses of the aptasensor for L-Tym concentrations ranging from 10 nM to 500 μM gave signals of increasing magnitude (Fig. 1B). These signals were weak (about 1.8 Hz at 10 μM L-Tym concentration) but exploitable. As for the Apt49-T0-Biot, instead of negative shifts in frequency corresponding to binding of the analyte, positive shifts in frequency and negative shifts in dissipation (blue and red curves in Fig. 1B, respectively) were measured. These results confirmed that L-Tym binding induces a folding of the aptamer that promotes a decrease in the Apt23-T0-Biot layer thickness, corresponding to a release of water and thus a loss of mass from the sensing layer.16,22 This behavior was also reported in the literature in the case of the binding of calcium ions to a calmodulin-functionalized surface.26
The SPR experiments allowed determination of the equilibrium dissociation constant KD 1.2 ± 0.5 μM (Fig. 1C), while an apparent KD of 3.3 ± 0.9 μM was determined using QCM-D8,22 (Fig. SI-5B†). Indeed, the KD value obtained using QCM-D should be considered as an apparent KD as it was indirectly obtained from the quantification of the expelled water upon L-Tym binding.22 To identify the impact of the immobilization procedure on the recognition event, evaluation of the interaction on solution by isothermal titration microcalorimetry (ITC) measurements was performed under the same experimental conditions, i.e. buffer composition and temperature (Fig. SI-6A and Table SI-1†). A KD value of 0.42 ± 0.02 μM was obtained This value is close to those determined previously in solution for this MBS: 0.3 μM, 0.8 μM and 0.4 μM by ITC, electrochemical titration and fluorescence polarization, respectively, at 20 °C in a slightly different buffer (10 mM Tris, 50 mM NaCl and 10 mM MgCl2).21 The lower affinity observed for the immobilized aptamer could be linked to steric hindrance that hampers the optimized structural transition of the recognition sequence. Steric hindrance could originate from the proximity of the other aptamer on the SA layer or from the proximity of the MBS to the SA layer.8 Indeed, previous studies have demonstrated that the recognition efficiency of immobilized aptamers could be improved by variation of the immobilized molecule density depending on the recognition partners8,27,28 or by the introduction of a spacer between the anchoring function and the aptamer sequence in order to move it away from the surface.29–32 The impact of the two latter parameters on the affinity and on the RII deviation was thus studied.
Influence of the aptamer density
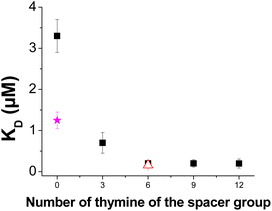
To improve the affinity between L-Tym and the immobilized Apt23-T0-Biot, the aptamer surface density was decreased by adjusting the immobilization time. QCM-D was not sensitive enough to obtain exploitable signals when the aptamer surface density was decreased. However, using SPR, negative sensorgrams were also obtained for lower Apt23-T0-Biot surface densities (Fig. SI-7†). The surface density of Apt23-T0-Biot has a weak influence on the KD (Fig. 1D) as well as on the kinetic constants of the interaction (Fig. SI-8†). Indeed, regardless of the Apt23-T0-Biot surface density, the KD was near 1.2 ± 0.5 μM; the kinetic dissociation and association constants were almost constant and close to 0.02 s−1 and 1.8 × 104 M−1 s−1, respectively. These kinetic constants were concordant with the values obtained in solution using the kinetic rotating droplet electrochemical method or kinITC: koff around 0.02 s−1 and kon 2.2–5.5 × 104 M−1 s−1.21 Interestingly, Apt49-T0-Biot immobilized on SA exhibited a better affinity for L-Tym at lower surface densities. Indeed, an increase in the kon from around 400 M−1 s−1 to 1500 M−1 s−1 upon a decrease in the aptamer density was observed while the koff remained constant around 0.04 s−1.8 In contrast to what was observed for Apt49-T0-Biot, a decrease in the surface density did not improve the affinity of this shorter sequence. Apt23-T0-Biot grafted on the streptavidin platform seemed to be spaced enough to rule out interligand steric hindrance, even at the highest aptamer surface density.Influence of the linker length
Another option to improve the affinity of an immobilized aptamer is to take away its recognition sequence from the surface. Indeed, recognition of L-Tym by the thiolated MBS (Apt23-T6-SH) immobilized on gold required the addition of a T6 spacer.21 Moreover, a lower equilibrium dissociation constant (0.2 μM)13,21 was determined for Apt23-T6-SH compared with the present immobilization of Apt23-T0-Biot on the SA layer (1.2 μM).The proximity between the aptamer and the streptavidin platform could reduce the flexibility of the aptamer sequence and hamper its structural modification upon target recognition. Indeed, a previous experiment in solution has demonstrated that the affinity of the 3′ biotinylated MBS decreases by a factor of 5 when linked to avidin.21 On the surface, this effect should be exacerbated. As a consequence, we studied the impact of a poly(dT) spacer length by varying the thymine number from three to twelve (T3, T6, T9 and T12; Fig. 2). The chemical nature of the spacer was also evaluated by replacing the thymine spacer with an adenosine one (T6 replaced by A6). Indeed, the influence of poly(dT) spacers vs. poly(dA) was reported for aptamer immobilization on glass.30 It should also be noted that poly(dA) groups can act as an anchoring function on the gold surface.33,34
First, ITC measurements demonstrated that the introduction of the spacer has a weak impact on the recognition in solution (Fig. SI-6 and Table SI-1†). The KD value (Fig. 2, blue circle) varied slightly from 0.42 μM to 0.13 μM upon increasing the linker length (between T0 and T6) and changed to 0.26 μM upon replacing T6 with A6. For all spacers, a high negative enthalpic contribution is associated with a major contribution of the entropy term that mainly originates from the significant conformational and dehydration changes. The CD spectra also confirmed that a conformation transition upon L-Tym recognition still occurred in solution in the presence of the spacers (Fig. SI-4†).
The immobilization of the MBS with the different spacers was first performed on the saturated SA layer. For all the spacers, the QCM-D responses depended on the L-Tym concentrations (Fig. SI-9†) and confirmed the recognition of L-Tym by the MBS. Folding of the aptamer was still operating, as depicted by the increase in the resonance frequency and the decrease in the dissipation (Fig. SI-9†). The QCM-D signal recorded with the different L-Tym concentrations was weak, whatever the length and the composition of the spacer, but allowed an estimation of the apparent KD for each spacer used (Fig. SI-9 to SI-11 and Table SI-2†).
As shown in Fig. 2 (black square), a decrease in the KD,app of up to 0.2 μM was observed when a spacer was introduced between the MBS and the anchoring function. Replacement of the T6 spacer with an A6 spacer had no influence on the affinity as a similar KD,app value was determined. This observation suggested that poly(dT) and poly(dA) were not part of the recognition sequence but that they impacted the recognition by taking away the MBS from the SA layer. Interestingly, the KD,app obtained using the T6 spacer was similar to the one measured in solution using ITC. This result suggested that the T6 spacer almost removed the influence of steric hindrance resulting from the close proximity of the MBS to the SA layer. The addition of a spacer moved the MBS away from the SA platform, resulting in an improvement in the apparent measured affinity. These results supported the idea that without a spacer, the MBS was too close to the streptavidin layer, which impeded its folding for target recognition. Indeed, in the domain of aptasensors, spacers are usually added to separate the recognition sequence of aptamers from the surface in order to improve either the response of the transducer and/or the affinity.29–32,35–37 As an example, Martin et al. studied the influence of various spacer lengths and compositions on the recognition capacity of aptamers against immunoglobulin E, thrombin and riboflavin. They demonstrated that poly(dT) linkers are quite efficient spacers for the different aptamer/target pairs in comparison with poly(dA), poly(dC) and poly(dG) linkers. They also identified that a minimal spacer length may be required for reliable binding data to be obtained.30 Poly(dT) spacers were also preferred to dodecyl ones for the recognition of two aptamers against thrombin.31 Other types of spacers are also reported in the literature and reveal that depending on the surface, the structure and length of the spacer need to be investigated for each aptamer/target pair.36 It should be noted that spacer lengths are also reported to modulate the affinity of other recognition systems.38–40 Concerning the present MBS/L-Tym recognition system, even in solution, the presence of the bulky anchoring function alters the recognition of L-Tym by hampering the conformational transition.21 This steric hindrance is thus exacerbated on the surface and a spacer is required to minimize it. Given the results presented in Fig. 2, the T6 spacer seems to be optimum in our case as no further decrease in KD,app is observed for the addition of a T9 or T12 spacer.
SPR experiments were performed on the same saturated sensing layers. The sensorgrams in Fig. 3A–D show the effect of the thymine spacer length for an aptamer immobilization level close to 500 RU and L-Tym concentrations ranging from 10 nM to 25 μM.
 | ||
| Fig. 3 Sensorgrams recorded during the interaction of L-Tym (concentrations ranging from 10 nM to 25 μM) on the active flow cell after the double-reference-subtraction procedure for: (A) Apt23-T3-Biot, (B) Apt23-T6-Biot, (C) Apt23-T9-Biot and (D) Apt23-T12-Biot. T = 25 °C. Flow rate: 30 μL min−1. Sensorgrams for Apt23-A6-Biot are presented in Fig. SI-12.† | ||
No variation in the SPR signal was observed during L-Tym injection on the saturated Apt23-Tx-Biot layer (Fig. 3) and on Apt23-A6-Biot layer (Fig.-SI-12†). The absence of a signal could not be attributed to the absence of recognition since an affinity of up to 250 nM was measured for the Apt23-Tx-Biot by QCM-D using the same immobilization procedure. Moreover, the sensitivity of the SPR device was adequate to detect the calculated maximal SPR signal according to the Wilson formula (from 8.9 to 12.7 RU, eqn (SI-2)† and Table 1)2 related to this interaction. This intensity increased from 10.9 to 14.8 if the aptamer folding was considered. However, while variations of signals were observed on both reference and active flow cells (Fig. SI-13†), the SPR responses after the double-reference-subtraction procedure were almost null whatever the L-Tym concentration (Fig. 3). A possible explanation could be that the addition of a spacer between the MBS and the biotin function could generate steric hindrance between aptamer strands on the SA layer. Experiments were thus performed with lower surface densities of aptamer and no signal variations were observed whatever the Apt23-Tx-Biot density used (data not shown).
Influence of RII deviation
As recently demonstrated, the non-additivity of the RII could be observed for recognition events involving a structural rearrangement of one of the partners as suggested by Bornhop15 for interactions in solution or by Dejeu for interactions on the surface.16 So the negative SPR signal for Apt23-T0-Biot as well as the null ones for Apt23-TX-Biot could be explained by the occurrence of two opposite effects: (1) an increase in the signal due to the target recognition and the aptamer folding that is counterbalanced by (2) a decrease in the signal triggered by a negative RII deviation from the sum of the RII of the individual entities for the L-Tym/aptamer complex formation.16 Indeed, the folding of the aptamer should also contribute to an enhancement of the signal of around 2 RU for each aptamer layer at saturation (Table 1, and Fig. SI-14 and SI-15†).16To obtain a concordance between the experimental maximal SPR response and its expected value, Dejeu's relationship was used to calculate the RII correction factor x of the L-Tym/aptamer complex RII (see eqn (SI-4) to (SI-6)†):
 | (1) |
The correction factor x was calculated from the Dejeu relationship for all the aptamer layers at each surface density (Fig. SI-14 and SI-15†) and is presented in Fig. 4 as a function of the spacer length. Negative sensorgrams obtained for the Apt23-T0-Biot could be attributed to a negative RII deviation of around −4.2% (Fig. 4), whatever the surface density is (Fig. SI-14†). This result is in agreement with the similar affinity measured for each surface density, suggesting that the conformation transition of the aptamer was not hampered by lateral steric hindrance on the SA layer.
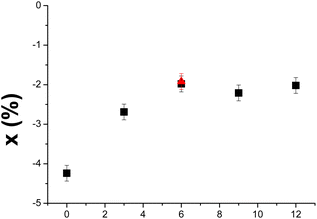 | ||
| Fig. 4 Influence of the spacer length on the deviation factor x of the aptamer RII defined by Dejeu's equation.16 The red triangle and the full black squares correspond to spacers composed of adenosine and thymine respectively. | ||
The RII of Apt23-T0-Biot was modified from 0.252 cm3 g−1 to 0.241 cm3 g−1 upon interaction with L-Tym. When a spacer was incorporated, the null SPR signal could be explained by an x variation between −2.0% and −2.7% depending on the length of the poly(dT) spacer. The RII of the aptamer shifted from 0.190 cm3 g−1 to 0.185 cm3 g−1 for a T3 spacer and from 0.196 cm3 g−1 to 0.192 cm3 g−1 for a T12 spacer when it was complexed with the target. A value of −2.0% must be considered for Apt23-T6-Biot. It seems that this parameter reached an asymptotic value of near −2%, which is correlated with an optimal KDapp of near 0.2 μM. Replacement of the T6 spacer with an A6 spacer also leads to null sensorgrams (Fig. SI-12†). Thus, Apt23-A6-biot undergoes a RII deviation similar to Apt23-T6-Biot upon interaction with L-Tym. It is noteworthy that a negative RII deviation upon interaction between Apt23-T6 and L-Tym was also previously highlighted in solution.13 To further demonstrate the RII deviation, determination of aptamer RII alone (Table 1) and complexed with L-Tym was performed in homogeneous media using SPR (Fig. SI-1†). To perform this experiment, the aptamer was first injected alone at different concentrations on a gold surface coated with HS-(CH2)11-EG4-OH to avoid nonspecific adsorption on the surface (Fig. SI-1† (full point)). Then similar injections of aptamer, at 100 μM, were performed in the presence of L-Tym at 1 mM to ensure total complexation of the aptamer strands (Fig. SI-1† (open circle)). A decrease of the SPR signal for Apt23-T3-Biot and Apt23-A6, in solution in the presence of L-Tym 1 mM suggested that a negative RII deviation also occurred in solution (Fig. 5), as already observed for Apt23-T3-SH.13
 | ||
| Fig. 5 SPR response recorded for the injection of Apt23 bearing various spacers on a gold surface protected by a pegylated self-assembled monolayer. (Plain) 100 μM of aptamer; (hatched) 100 μM of aptamer with 1 mM of L-Tym in running buffer. T = 25 °C. Apt23-T3 (Red), Apt23-T6 (Blue),13 Apt23-A6 (green). | ||
Interestingly, the RII deviation on the surface is also dependent on the immobilization mode. Indeed, for the Apt23-T6 sequence, a deviation of −2.0% is here calculated when a streptavidin/biotin coupling is used while a deviation of −2.8% was calculated for this sequence immobilized via a direct thiol coupling on a gold surface.13 Indeed, a subtle change in the flexibility of the MBS, in solution or on the surface, generates modification of its RII upon L-Tym recognition. These results are in agreement with the work of Khago that demonstrated that the RII of proteins also depends on their conformation.14 They have also observed a positive correlation between the solvent-exposed surface area of hydroxyl groups and the deviation from predicted dn/dc values for proteins suggesting that protein hydration also plays a role in the RII value.14
Conclusion
In this study, we have demonstrated that unconventional SPR signals, negative or null, could be observed for the recognition of a small analyte by the same aptamer sequence depending on its immobilization mode. Indeed, the aptamer undergoing a conformation transition upon interaction generates non-additivity of the RII that impacts the SPR signal. The positive contributions to the SPR signal by the target capture and the reduction of the aptamer layer thickness were compensated for by a negative RII deviation induced by the conformational transition of the aptamer. This latter contribution is dominated by a negative deviation of the aptamer RII during the recognition which is a function of the length of the spacer group used between the anchoring function and the aptamer sequence. We demonstrated that parameters affecting the flexibility of the recognition sequence, such as the aptamer length and the immobilization procedure by means of the anchoring function (thiol or biotin) and the length of the spacer (poly(dT) or poly(dA)), modify the RII deviation and consequently the SPR signal. Indeed, the introduction of a poly(dT) or a poly(dA) spacer led to a null SPR signal despite a recognition confirmed by QCM-D and the expected measurable SPR signals. The present model was validated with the MBS/L-Tym recognition system and should now be enlarged to other biomolecular recognition systems undergoing significant conformational or hydration modifications upon interaction. Nevertheless, these results offer new insight into the impact of the non-linearity of RII induced by significant conformational transitions of biomolecular molecules for their detection and quantification using optical biosensors. This work also points out the necessity of well-designed and controlled experiments in order to get a deep understanding of how several parameters, such as surface proximity, spacer addition, significant conformational and hydration modifications, impact transducer responses and the determination of affinities.Author contributions
M. P., H. B., C. V., E. G.: Investigation, L. C.-G., N. S.: Investigation, writing – review & editing; E. D.: Funding acquisition, writing – review & editing; A. V. d. H.: Writing – original draft, writing – review & editing, J. D.: Investigation, formal analysis, supervision, writing – original draft, writing – review & editing. The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was partially supported by the French National Agency (ANR) under ECSTASE, contract ANR-10-blan-1517 (Rational design of a sensitive and enantiospecific electrocatalytically-amplified aptasensor for amphetamine derivatives drugs), under ARCANE and CBH-EUR-GS (ANR-17-EURE-0003) and the Université Grenoble Alpes. The authors wish to acknowledge the support from the ICMG (UAR2607) Chemistry Nanobio Platform, Grenoble. Prof. P. Labbé is acknowledged for fruitful discussions and E. Laigre for the drawing of the graphical abstract. The authors thank Dr A. Imberty for the ITC facility.Notes and references
- R. B. M. Schasfoort, Handbook of Surface Plasmon Resonance, 2nd edn, 2017, pp. 001–524 Search PubMed.
- T. M. Davis and W. D. Wilson, Anal. Biochem., 2000, 284, 348–353 CrossRef CAS PubMed.
- J. E. Gestwicki, H. V. Hsieh and J. B. Pitner, Anal. Chem., 2001, 73, 5732–5737 CrossRef CAS PubMed.
- D. Dell'Orco, S. Sulmann, S. Linse and K. W. Koch, Anal. Chem., 2012, 84, 2982–2989 CrossRef PubMed.
- S. Sulmann, D. Dell'Orco, V. Marino, P. Behnen and K. W. Koch, Chem. – Eur. J., 2014, 20, 6756–6762 CrossRef CAS PubMed.
- D. Dell'Orco and K. W. Koch, ACS Chem. Biol., 2016, 11, 2390–2397 CrossRef PubMed.
- D. Dell'Orco, M. Muller and K. W. Koch, Chem. Commun., 2010, 46, 7316–7318 RSC.
- H. MacDonald, H. Bonnet, A. Van der Heyden, E. Defrancq, N. Spinelli, L. Coche-Guérente and J. Dejeu, J. Phys. Chem. C, 2019, 123, 13561–13568 CrossRef CAS.
- M. Geitmann, K. Retra, G. E. de Kloe, E. Homan, A. B. Smit, I. J. P. de Esch and U. H. Danielson, Biochemistry, 2010, 49, 8143–8154 CrossRef CAS PubMed.
- S. Mesch, D. Moser, D. S. Strasser, A. Kelm, B. Cutting, G. Rossato, A. Vedani, H. Koliwer-Brandl, M. Wittwer, S. Rabbani, O. Schwardt, S. Kelm and B. Ernst, J. Med. Chem., 2010, 53, 1597–1615 CrossRef CAS PubMed.
- D. Stokmaier, O. Khorev, B. Cutting, R. Born, D. Ricklin, T. O. G. Ernst, F. Böni, K. Schwingruber, M. Gentner, M. Wittwer, M. Spreafico, A. Vedani, S. Rabbani, O. Schwardt and B. Ernst, Bioorg. Med. Chem., 2009, 17, 7254–7264 CrossRef CAS PubMed.
- C. Crauste, N. Willand, B. Villemagne, M. Flipo, E. Willery, X. Carette, M. M. Dimala, A.-S. Drucbert, P.-M. Danze, B. Deprez and A. R. Baulard, Anal. Biochem., 2014, 452, 54–66 CrossRef CAS PubMed.
- H. Bonnet, L. Coche-Guérente, E. Defrancq, N. Spinelli, A. Van der Heyden and J. Dejeu, Anal. Chem., 2021, 93, 4134–4140 CrossRef CAS PubMed.
- D. Khago, J. C. Bierma, K. W. Roskamp, N. Kozlyuk and R. W. Martin, J. Phys.: Condens. Matter, 2018, 30, 435101 CrossRef PubMed.
- D. J. Bornhop, M. N. Kammer, A. Kussrow, R. A. Flowers and J. Meiler, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E1595–E1604 CAS.
- J. Dejeu, H. Bonnet, N. Spinelli, E. Defrancq, L. Coche-Guérente, A. Van der Heyden and P. Labbé, J. Phys. Chem. C, 2018, 122, 21521–21530 CrossRef CAS.
- Z. Zhu, T. Schmidt, M. Mahrous, V. Guieu, S. Perrier, C. Ravelet and E. Peyrin, Anal. Chim. Acta, 2011, 707, 191–196 CrossRef CAS PubMed.
- E. Vianini, M. Palumbo and G. Barbara, Bioorg. Med. Chem., 2001, 9, 2543–2548 CrossRef CAS PubMed.
- L. Sandrin, D. Thakar, C. Goyer, P. Labbe, D. Boturyn and L. Coche-Guerente, J. Mater. Chem. B, 2015, 3, 5577–5587 RSC.
- R. L. Rich and D. G. Myszka, Curr. Opin. Biotechnol., 2000, 11, 54–61 CrossRef CAS PubMed.
- L. Challier, R. Miranda-Castro, B. Barbe, C. Fave, B. Limoges, E. Peyrin, C. Ravelet, E. Fiore, P. Labbé, L. Coche-Guérente, E. Ennifar, G. Bec, P. Dumas, F. Mavré and V. Noël, Anal. Chem., 2016, 88, 11963–11971 CrossRef CAS PubMed.
- A. Osypova, D. Thakar, J. Dejeu, H. Bonnet, A. Van der Heyden, G. V. Dubacheva, R. P. Richter, E. Defrancq, N. Spinelli, L. Coche-Guérente and P. Labbé, Anal. Chem., 2015, 87, 7566–7574 CrossRef CAS PubMed.
- G. V. Dubacheva, C. Araya-Callis, A. Geert Volbeda, M. Fairhead, J. Codée, M. Howarth and R. P. Richter, J. Am. Chem. Soc., 2017, 139, 4157–4167 CrossRef CAS PubMed.
- T. M. A. El-Aziz, C. Ravelet, J. Molgo, E. Fiore, S. Pale, M. Amar, S. Al-Khoury, J. Dejeu, M. Fadl, M. Ronjat, G. S. Taiwe, D. Servent, E. Peyrin and M. De Waard, Sci. Rep., 2017, 7, 7202 CrossRef PubMed.
- T. Tumolo, L. Angnes and M. S. Baptista, Anal. Biochem., 2004, 333, 273–279 CrossRef CAS PubMed.
- X. Wang, J. S. Ellis, E.-L. Lyle, P. Sundaram and M. Thompson, Mol. Biosyst., 2006, 2, 184–192 RSC.
- L. Simon, Z. Bognár and R. E. Gyurcsányi, Electroanalysis, 2020, 32, 851–858 CrossRef CAS.
- F. V. Oberhaus, D. Frense and D. Beckmann, Biosensors, 2020, 10, 45 CrossRef CAS PubMed.
- K. A. Edwards and A. J. Baeumner, Anal. Bioanal. Chem., 2010, 398, 2635–2644 CrossRef CAS PubMed.
- J. A. Martin, Y. Chushak, J. L. Chávez, J. A. Hagen and N. Kelley-Loughnane, J. Nucleic Acids, 2016, 2016, 11 Search PubMed.
- Y.-H. Lao, K. Peck and L.-C. Chen, Anal. Chem., 2009, 81, 1747–1754 CrossRef CAS PubMed.
- S. Balamurugan, A. Obubuafo, R. L. McCarley, S. A. Soper and D. A. Spivak, Anal. Chem., 2008, 80, 9630–9634 CrossRef CAS PubMed.
- A. Opdahl, D. Y. Petrovykh, H. Kimura-Suda, M. J. Tarlov and L. J. Whitman, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 9–14 CrossRef PubMed.
- D. Zhu, P. Song, J. W. Shen, S. Su, J. Chao, A. Aldalbahi, Z. Zhou, S. P. Song, C. H. Fan, X. L. Zuo, Y. Tian, L. H. Wang and H. Pei, Anal. Chem., 2016, 88, 4949–4954 CrossRef CAS PubMed.
- C. Flores, N. W. Woodbury and E. Katilius, Nucleic Acids Res., 2007, 35, 7626–7635 CrossRef PubMed.
- M. Witt, J.-G. Walter and F. Stahl, Microarrays, 2015, 4, 115–132 CrossRef CAS PubMed.
- R. Thevendran and M. Citartan, Talanta, 2022, 238, 122971 CrossRef CAS PubMed.
- A. Imran, B. S. Moyer, A. J. Wolfe, M. S. Cosgrove, D. E. Makarov and L. Movileanu, J. Phys. Chem. Lett., 2022, 13, 4021–4028 CrossRef CAS PubMed.
- H. L. Birchenough, M. J. Swann, E. Zindy, A. J. Day and T. A. Jowitt, Nanoscale Adv., 2020, 2, 1625–1633 RSC.
- I. R. Olmsted, A. Kussrow and D. J. Bornhop, Anal. Chem., 2012, 84, 10817–10822 CrossRef CAS PubMed.
Footnote |
† Electronic supplementary information (ESI) available: Determination of the refractive index increment (RII) of the aptamer in solution by SPR alone and in the presence of L-Tym, expression of the Jung equation and Wilson formula, raw SPR signals recorded during L-Tym injection on Apt23-T0-Biot and reference flow cells, SPR signals recorded during D-Tym injection on the Apt23-T0-Biot surface, circular dichroism (CD) of a different aptamer (Apt23-Tx-Biot) alone and in the presence of L-Tym, the apparent thermodynamic equilibrium constant for L-Tym/Apt23-T0-Biot interaction determined by QCM-D, ITC experiments for L-Tym recognition by Apt23 bearing various spacers (Apt23-T0, Apt23-T3, Apt23-T6, Apt23-A6), the SPR signal recorded on active flow cells exhibiting different Apt23-T0-Biot surface densities, kinetic constants for Apt23-T0-Biot upon interaction with L-Tym, the QCM-D signal recorded upon L-Tym injection on biotinylated Apt23 layers with different spacers, variation of the resonance frequency as a function of L-Tym concentrations for the 7th overtone and fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding curve with a linear and logarithm X axis, the SPR signal recorded on Apt23-A6-Biot and on the reference flow cell upon L-Tym injection, the raw SPR signal recorded on Apt23-T6-Biot and on the reference flow cell upon L-Tym injection, refractive index increment correction in the model developed by Dejeu et al., comparison of the experimental and expected maximal SPR signal for Apt23-T0-Biot upon interaction with L-Tym, comparison of the experimental and expected maximal SPR signal for Apt23-T6-Biot upon interaction with L-Tym. See DOI: https://doi.org/10.1039/d2an00824f 1 binding curve with a linear and logarithm X axis, the SPR signal recorded on Apt23-A6-Biot and on the reference flow cell upon L-Tym injection, the raw SPR signal recorded on Apt23-T6-Biot and on the reference flow cell upon L-Tym injection, refractive index increment correction in the model developed by Dejeu et al., comparison of the experimental and expected maximal SPR signal for Apt23-T0-Biot upon interaction with L-Tym, comparison of the experimental and expected maximal SPR signal for Apt23-T6-Biot upon interaction with L-Tym. See DOI: https://doi.org/10.1039/d2an00824f |
| This journal is © The Royal Society of Chemistry 2022 |