Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of measurement mode and sample processing for FTIR microspectroscopy in skin cancer research
Bijay Ratna
Shakya
 *a,
Hanna-Riikka
Teppo
*a,
Hanna-Riikka
Teppo
 bcd and
Lassi
Rieppo
bcd and
Lassi
Rieppo
 a
a
aResearch Unit of Medical Imaging, Physics and Technology, Faculty of Medicine, University of Oulu, Aapistie 5 A, 90220, Oulu, Finland. E-mail: bijay.shakya@oulu.fi
bCancer Research and Translational Medicine Research Unit, University of Oulu, Aapistie 5 A, 90220, Oulu, Finland
cDepartment of Pathology, Oulu University Hospital, Oulu, Finland
dMedical Research Center Oulu, Oulu University Hospital and University of Oulu, Finland
First published on 31st January 2022
Abstract
The use of Fourier Transform Infrared (FTIR) microspectroscopy to study cancerous cells and tissues has gained popularity due to its ability to provide spatially resolved information at the molecular level. Transmission and transflection are the commonly used measurement modes for FTIR microspectroscopy, and the tissue samples measured in these modes are often paraffinized or deparaffinized. Previous studies have shown that variability in the spectra acquired using different measurement modes and sample processing methods affect the result of the analysis. However, there is no protocol that standardizes the mode of measurement and sample processing method to achieve the best classification result. This study compares the spectra of primary (IPC-298) and metastatic (SK-MEL-30) melanoma cell lines acquired in both transmission and transflection modes using paraffinized and deparaffinized samples to determine the optimal combination for accurate classification. Significant differences were observed in the spectra of the same cell line measured in different modes and with or without deparaffinization. The PLS-DA model built for the classification of two cell lines showed high accuracy in each case, suggesting that both modes and sample processing alternatives are suitable for differentiating cultured cell samples using supervised multivariate analysis. The biochemical information contained in the cells capable of discriminating two melanoma cell lines is present regardless of mode or sample type used. However, the paraffinized samples measured in transflection mode provided the best classification.
Introduction
Fourier transform infrared (FTIR) microspectroscopy has become popular in histopathological and cytopathological research due to its capability to assess biochemical composition and molecular structure of unstained cells and tissues. Several groups have used FTIR microspectroscopy to diagnose skin cancers1 and other cancers, such as breast, cervical, colorectal, lung, brain, bladder, head and neck cancers.2,3 These studies have demonstrated the ability of FTIR microspectroscopy to differentiate between diseased and non-diseased conditions. Biochemical changes occurring in the cells and tissues are seen as alterations in the prominent bands of FTIR spectra.4 These variations in spectra may be used as markers to discriminate between different diseased and non-diseased conditions.4 In addition, machine learning algorithms are nowadays commonly used to classify spectra into various disease classes.5–7FTIR microspectroscopy measurements are most often conducted either in transmission or transflection mode.1,8 In transmission mode, the infrared beam is passed through the sample and the transmitted energy is measured as a function of frequency to generate the infrared absorption spectrum. Thin tissue sections are placed on substrates transparent to infrared light, such as CaF2, BaF2 or ZnSe, for the transmission mode measurements. These substrates are expensive and brittle, making them unfavourable for integration with traditional automated tissue sample preparation methods.9 Due to these drawbacks, transflection mode has become popular in FTIR microspectroscopic studies. Low-emissivity slides with infrared reflective surfaces are used for transflection mode measurements. These slides are cheaper than those used for transmission mode measurements and have similar dimensions as standard light microscope slides, making them easy to handle during sample preparation. A transflectance spectrum is generated by measuring the beam passing through the sample and reflecting back from the slide surface through the sample. As a result, the beam passes through the sample twice, leading to double absorbance. The drawback of transflection mode is the electric field standing wave (EFSW) effect which occurs due to interference between the incident and reflected beams.10 The EFSW can distort the spectra so that it affects, e.g., the classification of spectra.11 The problem can be particularly persistent in the measurement of cultured cell samples as there is less biological material in the sample compared to tissue sections.8 In an effort to solve problems related with substrates of both modes, studies have also been conducted using the traditional glass slides.9,12 According to these studies, glass slides are suitable for studying a narrow transmission zone at 2500–3800 cm−1 where stretching vibrations of N–H, O–H, and C–H bonds are observed. This higher wavenumber region has diagnostic value, and a robust classifier with high accuracy can be constructed using this region for discriminating malignant and non-malignant cells. Furthermore, recent studies using glass coverslips as substrates has revealed that additional spectral information from the fingerprint region between 1800 cm−1 and 1350 cm−1 can now be obtained.13,14
Tissue samples used in FTIR microspectroscopy are most often fixed with formalin and embedded in paraffin, i.e., formalin-fixed paraffin embedded (FFPE) samples.1 Paraffin serves as a support medium during sectioning and aids in preserving the sample for future analysis. Thin sections are cut with a microtome and placed on a suitable substrate, depending upon the mode used. Prior to the spectroscopic measurement, the sections are often treated with organic solvents like xylene or hexane to remove the paraffin. This allows the samples to rehydrate and reduces the contribution of paraffin in acquired spectra.15 However, the paraffin remnants can often still be observed in sample spectra even after dewaxing, making the analysis of the affected bands unreliable.16 The dewaxing procedure also alters the chemical composition of the hydrophobic constituents of samples, such as lipids. FTIR spectroscopic measurements can also be conducted without the dewaxing procedure. However, the acquired spectra will have high paraffin contribution masking some of the molecular vibrations associated with the samples and making the spectral interpretation more difficult.17 Fortunately, these paraffin peaks are relatively narrow and can either be truncated or digitally removed from the spectra without chemical dewaxing procedures.17 Additionally, leaving paraffin on the samples reduces the scattering effect caused by variations in the refractive index across the sample.18
Despite the emergence of FTIR spectroscopy in cell and tissue research,19 there are no standard protocols, which could improve reproducibility of the results between different laboratories, for the measurements. It is essential to understand the influence of different measurement modes and sample processing methods on the acquired spectra for reliable interpretation and accurate diagnosis of the diseased conditions. The challenges for selecting the suitable measurement mode (transmission or transflection) and the sample processing procedure (paraffinized or deparaffinized) for the measurement of samples should therefore be addressed. This study aims to evaluate the impact of different measurement modes and the presence of paraffin using cultured cell samples as a representative model to optimize the protocol for tissue samples. Here, we compare the spectra of two different melanoma cell lines acquired from paraffinized and deparaffinized samples measured in both transmission and transflection modes.
Materials and methods
Cell lines
Two melanoma cell lines, IPC-298 (primary) and SK-MEL-30 (metastatic), were used in the study. IPC-298 is derived from a primary melanoma tumour of a 64-year-old female, and SK-MEL-30 is derived from a subcutaneous metastasis of a 67-year-old male with melanoma.The cell lines were purchased from Leibniz-Institut, DSMZ, Braunschweig, Germany and were cultured in RPMI-1640 with 10% fetal bovine serum and 100 IU ml−1 penicillin and streptomycin. A more detailed description regarding the cell lines can be found in our previous study.6
Sample preparation
Both cell lines were formalin-fixed and paraffin-embedded. The detailed procedure of paraffin infiltration and embedding process is described in our previous article.6 Briefly, the procedure was conducted using a Tissue Tek processor (Tissue-Tek VIP 5 Jr, Sakura, The Netherlands), where the cells were sequentially immersed in formalin, water and absolute alcohol for 30 minutes in each container, followed by immersion in 5 containers of absolute alcohol for 60 minutes in each. Next, the cells were immersed in two containers of xylene for 60 and 90 minutes, respectively, followed by two containers of molten paraffin for 60 minutes in each. Finally, the cells were manually transferred to an embedding mould filled with molten paraffin, which was subsequently allowed to solidify for 120 minutes. For the study, four different sets of samples were prepared, each set consisting of two 5 μm-thick sections of each cell line: (1) the sections were transferred to 2 mm-thick CaF2 windows for transmission mode measurements, (2) the sections were transferred to 2 mm-thick CaF2 windows and deparaffinized with xylene before the transmission mode measurements, (3) the sections were transferred to low-E microscope slides for transflection mode measurements, and (4) the sections were transferred to low-E microscope slides and deparaffinized with xylene prior to the transflection mode measurements. The samples were deparaffinized by immersing them for 3 minutes in three different xylene-filled containers. To begin rehydration, the samples were immersed in three different containers filled with absolute alcohol for 3 minutes, followed by 2 minutes in two containers filled with 96% ethanol. To complete the rehydration procedure, the samples were washed with water.Data acquisition
Nicolet iN10 MX (Thermo Scientific, Wisconsin, MA, USA) FTIR microscope equipped with nitrogen-cooled mercury cadmium telluride (MCT) detector was used for the data acquisition. Measurements were conducted in both transmission and transflection modes. The sections placed on CaF2 windows were measured in transmission mode, while sections placed on low-E slides were measured in transflection mode. From each sample, spectra of individual cells (i.e., cells not connected with other cells) were acquired in a spectral range of 900–4000 cm−1 with a spectral resolution of 8 cm−1 using an aperture size of 20 × 20 μm2. It took approximately 4 minutes to complete one measurement with 256 scans (including background collection). A background spectrum was collected from a clean area of the CaF2 window for transmission mode and a clean surface of the low-E slide for transflection mode. A total of 2080 cell spectra were collected from four sets of samples: (1) 260 (primary), 260 (metastatic), (2) 260 (primary), 260 (metastatic), (3) 260 (primary), 260 (metastatic), and (4) 260 (primary), 260 (metastatic).Data pre-processing
All spectral pre-processing was carried out using MATLAB (R2017b, MathWorks, Natick, MA, USA). A simple offset correction for baseline was first carried out on all spectra to eliminate any negative values. Baseline distortions due to scattering of infrared radiation were removed using the resonant Mie scattering correction (RMieS) algorithm.20 The spectral regions affected by paraffin, i.e., 1350–1490 cm−1 and 2700–3000 cm−1, were down-weighted during the RMieS correction for all sample sets. Otherwise, the correction options available for the RMieS algorithm were all set to default values. The spectra were then truncated to 1000–1800 cm−1. Furthermore, the CH3 region (1350–1490 cm−1) was removed from the spectra as it is influenced by paraffin and the spectra were vector normalized before univariate analysis. For multivariate analysis, second derivative spectra were first calculated using the Savitzky–Golay algorithm (polynomial order = 2 and smoothing points = 7) followed by truncation and removal of CH3 region as described earlier. The second derivative spectra were then vector normalized.Data analysis
Mean spectra were used to visualize the spectral differences between the same cell line measured using different modes, sample preparation procedures and the differences between the two cell lines. For univariate analysis, integrated areas of the prominent peaks and their possible shifts were measured. An independent t-test was used to determine whether the differences were statistically significant. The limit of statistical significance was set at p < 0.05. In multivariate analysis, principal component analysis (PCA) was performed to study the differences in the spectra due to measurement modes and paraffin. Furthermore, Partial Least Squares Discriminant Analysis (PLS-DA) models with nine latent variables were built to differentiate the two cell lines. PLS-DA models were built using 60% of the spectra from each cell line for training the model and the remaining 40% as the test set. The model was run 25 times, with the spectra in both sets being assigned at random in each run.Results
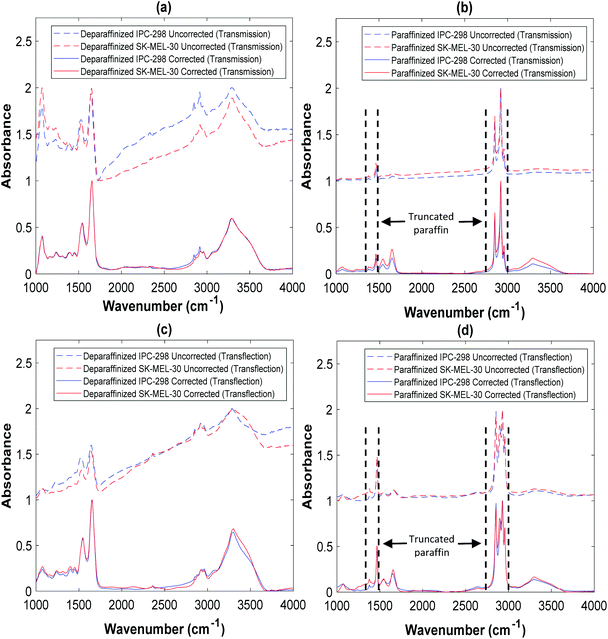
The melanoma FTIR spectra were investigated in the fingerprint region, i.e., the region of 1000–1800 cm−1. The vibrations due to paraffin were evident in the 1350–1490 cm−1 and 2700–3000 cm−1 regions in paraffinized samples (Fig. 1B and D). Consequently, the affected region was truncated from all spectra from further analysis to make the results comparable.Comparison of transmission and transflection spectra (of same cell line)
The average spectrum of each cell line measured in transmission and transflection modes before and after deparaffinization are presented in Fig. 2. Four prominent spectral bands, i.e., symmetric phosphate (1000–1138 cm−1), antisymmetric phosphate (1200–1273 cm−1), amide II (1497–1593 cm−1), and amide I (1597–1720 cm−1) were well characterized in both modes. Compared with the transmission mode spectra, the absorbances of the prominent bands were lower in transflection mode spectra for the deparaffinized samples from both cell lines. In addition, amide II peak shifted to a higher wavenumber (from 1543 cm−1 to 1547 cm−1) in transflection mode for IPC-298 cell line while amide I peak shifted to a lower wavenumber (from 1655 cm−1 to 1651 cm−1) for SK-MEL-30 cell line. For the paraffinized samples, the absorbances of all bands (except for symmetric phosphate) were lower in transflection mode compared with transmission mode. The shifts in amide I (from 1651 cm−1 to 1655 cm−1) and amide II peaks (from 1543 cm−1 to 1547 cm−1) of the IPC-298 cell line were also observed in transflection mode for paraffinized samples. The peak positions of the prominent spectral bands and their integrated areas for deparaffinized and paraffinized samples measured using both modes are shown in Table 1.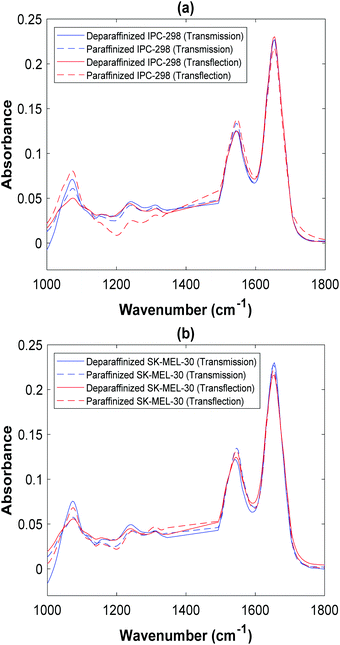 | ||
| Fig. 2 Mean spectrum of two cell lines processed using two different methods and measured using two different modes. (a) IPC-298 (b) SK-MEL-30. | ||
| Primary melanoma cells (IPC-298) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Peak assignment | Band positions | Integrated area (mean) | p-value | |||||||
| DP_Tm | DP_Tf | P_Tm | P_Tf | DP_Tm | DP_Tf | P_Tm | P_Tf | DP_Tm vs. DP_Tf & P_Tm vs. P_Tf | DP_Tm vs. P_Tm & DP_Tf vs. P_Tf | |
| Phosphate (symmetric) | 1072 | 1072 | 1072 | 1072 | 3.92 | 1.88 | 2.62 | 3.73 | <0.0001* | <0.0001* |
| Phosphate (anti-symmetric) | 1238 | 1238 | 1238 | 1238 | 0.41 | 0.39 | 0.41 | 0.27 | <0.0001* | 0.6, <0.0001 |
| Amide II | 1543 | 1547 | 1543 | 1547 | 3.34 | 3.20 | 3.51 | 3.41 | <0.0001* | <0.0001* |
| Amide I | 1655 | 1655 | 1651 | 1655 | 10.06 | 9.96 | 10.03 | 8.67 | <0.0001* | 0.19, <0.0001 |
| Metastatic melanoma cells (SK-MEL-30) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DP_Tm | DP_Tf | P_Tm | P_Tf | DP_Tm | DP_Tf | P_Tm | P_Tf | DP_Tm vs. DP_Tf & P_Tm vs. P_Tf | DP_Tm vs. P_Tm & DP_Tf vs. P_Tf | |
| DP_Tm: deparaffinized sample measured in transmission mode, DP_Tf: deparaffinized sample measured in transflection mode, P_Tm: paraffinized sample measured in transmission mode; P_Tf: paraffinized sample measured in transflection mode; *value for both comparisons. | ||||||||||
| Phosphate (symmetric) | 1072 | 1072 | 1072 | 1072 | 4.37 | 2.02 | 2.42 | 3.21 | <0.0001* | <0.0001* |
| Phosphate (anti-symmetric) | 1238 | 1238 | 1238 | 1238 | 0.47 | 0.35 | 0.39 | 0.36 | <0.0001* | <0.0001, 0.08 |
| Amide II | 1543 | 1543 | 1547 | 1547 | 3.25 | 2.95 | 3.60 | 3.34 | <0.0001* | <0.0001* |
| Amide I | 1655 | 1651 | 1651 | 1651 | 10.17 | 9.50 | 10.07 | 9.80 | <0.0001* | <0.0001* |
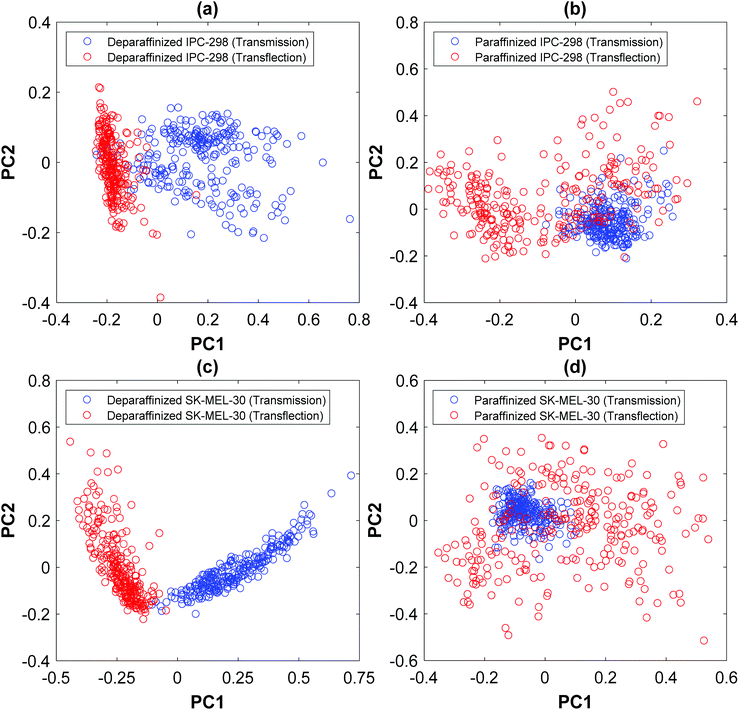
PCA was used to visualize the spectral differences occurring between the two modes. The PCA was applied to normalized 2nd derivative spectra (excluding the paraffin region) of each cell line separately. The score plot was created using the first two PCs, which accounted for 78% (PC1: 66.64%, PC2: 11.21%), 72% (PC1: 43.36%, PC2: 27.73%) and 84% (PC1: 64.64%, PC2: 18.91%), 75% (PC1: 45.48%, PC2: 28.92%) of variance for deparaffinized IPC-298, paraffinized IPC-298 and deparaffinized SK-MEL-30, paraffinized SK-MEL-30, respectively. In Fig. 3, each point in the plot represents the transformed variable values of each spectrum known as scores. The score plot showed separate clusters with some overlap for both cell lines, suggesting significant spectral variations due to measurement modes. Loadings of the first two PCs (plot not shown) revealed that the difference in the score of the spectra acquired from two modes were primarily due to variation in carbohydrate, nucleic acid, and amide regions. As the samples were identical, the changes in this region are not related to differences in biochemical composition in the cells but the variations are either the result of EFSW or Mie scattering due to incomplete RMieS correction. Another possible explanation for the variation is the presence of paraffin during the measurement.
Comparison of deparaffinized and paraffinized cell spectra (of same cell line)
The average spectra of both cell lines with and without dewaxing are shown in Fig. 2. The integrated areas of the prominent bands showed an increase in the absorbance of the symmetric phosphate, antisymmetric phosphate, and amide I and decrease in the absorbance of amide II for deparaffinized samples from both cell lines compared with paraffinized samples spectra acquired in transmission mode. In addition, there was a shift of amide I peak to lower wavenumber (from 1655 cm−1 to 1651 cm−1) in both cell lines. The independent t-test showed statistically significant differences in the same prominent bands between the two sample processing methods (except antisymmetric phosphate and amide I in the IPC-298 cell line). In the samples measured in transflection mode, an increase in absorbance was observed for all prominent bands (except antisymmetric phosphate and amide I in IPC-298 cell line) for paraffinized samples compared with deparaffinized ones in both cell lines. No peak shifts were observed in the transflection mode of either cell line. The peak positions of the prominent spectral bands and their integrated area are shown in Table 1.PCA performed for spectra acquired from the same cell lines but with different sample processing procedures showed two defined clusters in both measurement modes indicating a significant spectral difference between samples. As mentioned earlier, this variation must be due to presence of paraffin during the measurement because the samples measured were identical. The PCA score plot comparing two different sample processing processes is shown in Fig. 4. The plot was computed using first two PCs, which accounted for 72% (PC1: 59.24%, PC2: 12.73%), 67% (PC1: 40.27%, PC2: 25.94%) and 87% (PC1: 78.66%, PC2: 7.74%), 68% (PC1: 41.05%, PC2: 26.36%) of variance for IPC-298 and SK-MEL-30 samples measured in transmission and transflection mode, respectively.
Differentiation between two malignant melanoma cell lines
The differentiation between the two cell lines in each set of the samples was conducted using multivariate analysis to identify the most appropriate mode and processing method for accurate classification. Fig. 5 illustrates the average normalized second derivative spectra of two cell lines in each set of samples used for the analysis. | ||
| Fig. 5 Mean normalized second derivative spectra of two cell lines acquired from each set of samples. (a) In transmission mode (b) in transflection mode. | ||
PLS-DA models were developed to differentiate the two cell lines. The built PLS-DA models showed high recognition rates in discriminating the two cell lines from all sample sets. The mean (± SD) percentages of the classification accuracy for each set are shown in Table 2. In addition, the regression vectors plotted for each set of samples with significant wavenumbers are shown in Fig. 6. Carbohydrate, nucleic acid and protein regions show large regression coefficients indicating that the discrimination of the two melanoma cell lines is based on the overall spectral fingerprint rather than on individual features.
| Transmission mode | ||
|---|---|---|
| Cell type | Predicted as | |
| IPC-298 (deparaffinized) | SK-MEL-30 (deparaffinized) | |
| IPC-298 (deparaffinized) | 94.52 ± 2.44 | 5.47 ± 2.44 |
| SK-MEL-30 (deparaffinized) | 4.46 ± 2.56 | 95.54 ± 2.56 |
| IPC-298 (paraffinized) | SK-MEL-30 (paraffinized) | |
| IPC-298 (paraffinized) | 90.38 ± 2.89 | 9.61 ± 2.89 |
| SK-MEL-30 (paraffinized) | 22.83 ± 4.23 | 77.16 ± 4.23 |
| Transflection mode | ||
|---|---|---|
| Cell type | Predicted as | |
| IPC-298 (deparaffinized) | SK-MEL-30 (deparaffinized) | |
| IPC-298 (deparaffinized) | 98.80 ± 1.07 | 1.19 ± 1.07 |
| SK-MEL-30 (deparaffinized) | 1.50 ± 1.09 | 98.50 ± 1.09 |
| IPC-298 (paraffinized) | SK-MEL-30 (paraffinized) | |
| IPC-298 (paraffinized) | 98.68 ± 0.98 | 1.31 ± 0.98 |
| SK-MEL-30 (paraffinized) | 0.36 ± 0.48 | 99.64 ± 0.48 |
Discussion
The present study uses cultured cell samples as a representative model in an attempt to optimize the measurement geometry and sample processing for FTIR microspectroscopic measurements of skin biopsy samples. For this purpose, two melanoma cell lines were studied to create a controlled set up for comparison. The study compared two different FTIR microspectroscopy measurement modes and two sample processing alternatives for differentiation between two melanoma cell lines. In the first phase, the univariate analysis was used to evaluate the differences in the prominent bands of the cell spectra measured in different modes, then in the spectra acquired using differently processed samples, and finally between the two melanoma cell lines. Significant differences were observed in all prominent spectral bands of the same cell line measured in different modes and between two differently processed samples (except for anti-symmetric phosphate and amide I band of IPC-298 samples measured in transmission mode and anti-symmetric phosphate of SK-MEL-30 in transflection mode). Univariate analysis conducted to distinguish between two melanoma cell lines showed significant differences in the integrated absorbance of all prominent bands acquired from all sample sets. Good classification results were also achieved through multivariate analysis. The developed PLS-DA models differentiated the two melanoma cell lines with high accuracy in each case, suggesting that both modes and sample processing alternatives are suitable for cultured cell samples. However, the samples measured in transflection mode with paraffin provided the best classification in this study.The spectra obtained in transmission and transflection were visually similar. However, the univariate analysis of the prominent spectral bands revealed significant differences between the two sampling geometries. The differences are likely attributed to the principle underlying spectral acquisition, and the use of separate substrates for each mode with different chemical and optical properties. In particular, the presence of EFSW in transflection mode measurement can cause non-linear changes in the spectral characteristics of acquired spectra with respect to sample thickness that are dependent on the wavelength of the radiation. Furthermore, it causes variations in the baseline comparable to Mie scattering.8,10 In addition, EFSW is not confined to the sample surface but can also occur within the sample layer, resulting in interference in acquired spectra leading to alteration of peak absorbance.21 Another possible explanation for the variation in the spectra of two geometries is the use of a different substrate for sample measurement, which also contributes to the occurrence of artefacts. The refractive index of CaF2 and low-e slides are 1.4 and 2.0, respectively. This EFSW effect has shown to increase when the refractive indices of the utilized substrate increase.21 The variation in the spectra due to the use of different substrates is present for both incoherent and coherent light sources, implying that the final acquired spectrum is reliant not only on the choice of substrate and measurement geometry but also on the infrared light source characteristics.22 EFSW has been shown to produce interference in transmission mode as well, resulting in periodic variations in the acquired spectra as well as interference fringes that are likewise dependent on sample thickness. This leads to the spectral variation that causes changes in absorbance of the spectral bands.23 PCA conducted on transmission and transflection spectra of the same cell line formed two separate clusters suggesting apparent change in biochemical information due to different principle of spectra acquisition. This is significant considering that all spectra were acquired from the same cultured samples. FFPE tissue samples are most commonly used in FTIR microspectroscopy. In most studies, paraffin is removed before the measurements with organic solvents. This procedure has been shown to alter the lipids in the sample. To overcome the issue, studies have also been conducted on paraffinized samples.24 In addition, digital dewaxing methods for paraffinized samples have been developed to address the inefficiency of chemical dewaxing.17 Nevertheless, this adds an extra step to the pre-processing process, and its effectiveness in removing paraffin content has yet to be established. A study by Hughes et al.25 suggests that unbound lipids in the tissue are already removed in the early sample processing stages during the FFPE procedure. Only the solvent-resistant lipids remain, which are bound to tissue proteins during the FFPE procedure. As a result, the remaining lipid bands after deparaffinization of the samples can still be used for diagnostic purposes, provided that the paraffin is removed to the point where the methylene vibrations are at stable baseline.
Our study compared the paraffinized and deparaffinized samples of each cell line to determine the spectral difference caused by sample processing. After the removal of the paraffin bands at 1350–1490 cm−1, the spectra from both samples showed visually similar spectral profiles. However, significant differences in the absorbances of the prominent spectral bands in each mode were observed between the two samples. The differences could be due to the changes in optical parameters of the system caused by the presence of paraffin. The differences between the two samples are also highlighted in the PCA score plots, as two separate clusters are formed in both modes for each cell line. Paraffin has been reported to reduce the scattering and EFSW problem caused by the variation of the refractive index and sample heterogeneity throughout the sample surface. In this study, the spectra of dewaxed samples suffered from high baseline distortion occurring due to resonant Mie scattering. The effect was much lower but still present in paraffinized samples (Fig. 1). Tight clusters for paraffinized samples in transmission mode may be the reason of this reduced scattering. Paraffin has been reported to reduce the scattering problem caused by the variation of the refractive index throughout the sample surface.26 Contradicting with this, the clusters of paraffinized samples in transflection mode are more diffused compared with deparaffinized samples. This could be due to the presence of EFSW in the acquired spectra. However, no visual differentiation due to EFSW was observed in acquired spectra from either cell line, suggesting that the EFSW effect was minor in this study. The increase in sample homogeneity along the surface due to the paraffin layer may be the reason for this reduced influence.
The PLS-DA models developed to discriminate two melanoma cell lines displayed good classification with high sensitivity and specificity in each mode with both sample types. This suggests that when supervised multivariate methods are used for classification, both modes and sample processing methods perform well, and physical phenomena such as scattering and EFSW have only minor effect on classification accuracy. The biochemical content of the cells that leads to the separation of two cell lines remains in the acquired spectra regardless of the processing method or mode used. Even though more than 90% sensitivity and specificity were achieved in each case, it should be noted that the sensitivity of the paraffinized samples measured on transmission mode was only 77.16%, which is relatively low when compared to other cases. We speculate that the decrease in this rate is due to a weaker signal in transmission mode compared with transflection mode that exhibits double absorbance due to infrared beams passing twice through the section during measurement, which is beneficial for samples with low amount of biological material, such as cultured cells. The depth of penetration of the infrared beam on the measured sample has also been demonstrated to have a significant effect on the classification result. According to a study by Song et al.,27 measuring the spectra from the shallow layer at the surface of the sample yields higher accuracy in discriminating diseased samples from healthy ones when compared with the spectra measured from the thicker region of the sample with higher penetration depth. The overlap of paraffin signals may have also caused some alteration to the spectral data, as they can still have an effect on remaining spectral regions despite the fact that the most prominent paraffin band were truncated during data analysis. The best classification was achieved for paraffinized samples measured in transflection mode. Similar results have been shown in the study by Kochan et al.28 in which histiocytic sarcoma samples from the canine liver were used. The study compared two measurement modes to differentiate between cancerous and healthy regions with both deparaffinized and paraffinized samples and indicated that the choice of the substrate does not affect the diagnostic capability of multivariate analysis of FTIR spectra. They further proposed the use of Mirr-IR substrates with paraffin for better classification of the cancerous region from a healthy region in liver sarcoma. Additionally, a study by Fullwood et al.29 suggest that dewaxed tissue placed on low-E slides does not retain paraffin (remaining paraffin contributions) and the spectra acquired during measurement of such samples are not influenced by paraffin contributions.
Another key finding of this study is that the pre-processing methods used on the spectra do not produce comparable results between the different measurement modes and sample processing methods. This implies that our pre-processing protocol, which was similar to protocols used in other current studies, did not account for all factors associated with different measurement modes and sample types. This may not be a problem if only one type of dataset is used, as evidenced by the fact that the classification works well on all data sets in this study. However, if different datasets are used for a study, the impact of different measurement modes and sample types should be considered.
Although it is beyond the scope of this manuscript, the other vibrational spectroscopic method, i.e., Raman spectroscopy, has also been extensively researched as a diagnostic tool in various in vitro and in vivo studies of skin cancer.30–33 These studies have demonstrated the high sensitivity of Raman spectroscopy in the early diagnosis of melanoma, the ability to distinguish melanoma from melanocytes, predict the susceptibility of melanoma cells to anticancer drugs and differentiate it from other pigmented lesions and nonmelanoma skin cancers. In addition, Raman spectroscopy has already been commercialized for in vivo skin analysis for determining the molecular concentrations profiles (https://www.riverd.com/). For FTIR microspectroscopy to be used as a new alternative tool for automated classification, a crucial step would be to use the same sample type and mode across all research units in order to acquire comparable spectra, allowing for the development of a comprehensive library of infrared spectra of respective tissues and cell types that can be used to build better machine learning models capable of accurate classification.
Conclusion
This study investigated the differences in acquired FTIR spectra of two melanoma cell lines caused by two measurement modes and sample processing methods and compared each case in differentiation between the two melanoma cell lines. The univariate analysis showed significant differences between the spectra acquired in different modes and due to the presence of paraffin. PLS-DA models developed to differentiate between the two melanoma cell lines showed high accuracy in each case, highlighting the diagnostic potential and flexibility of FTIR microspectroscopy. The result suggests that the biochemical information relevant for classification is retained in each mode with both sample types. Although all options seem to allow for differentiation between the two cell lines, the use of the transflection mode with paraffinized samples may be beneficial given the economic advantage of low-E slides, greater ease of handling, and no further alteration of samples with the treatment of organic solvents. Furthermore, with appropriate sample thickness, spectral features present in the cells can be significant, reducing the impact of EFSW resulting from transflection measurement. The combination also presented the best result in the study. Despite the high contribution of paraffin in the acquired spectra, the spectral region not affected by paraffin can be identified and used for accurate classification. Currently, every research unit follows its own designed methods for spectroscopic analysis. This study contributes to the standardization of sample processing and measurement geometry. However, for FTIR spectroscopy to be used as a routine laboratory protocol, standardization of other procedures such as sample collection, handling, preparation methods, and storage needs to be addressed as well to achieve reproducibility among the research units and laboratories.Author contributions
Bijay Ratna Shakya: conceptualization, methodology, software, formal analysis, investigation, validation, data curation, writing – original draft, writing – review and editing. Hanna-Riikka Teppo: conceptualization, methodology, resources, writing – review & editing, supervision. Lassi Rieppo: conceptualization, methodology, software, validation, resources, data curation, writing – review & editing, supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Erja Tomperi for her technical knowledge in sample preparation. This research was supported by Finnish Cultural Foundation (grant number: 00211005), Finland.Notes and references
- B. R. Shakya, P. Shrestha, H. Teppo and L. Rieppo, Appl. Spectrosc. Rev., 2020, 56(5), 1–33 Search PubMed.
- A. Kumari, J. Kaur and S. Bhattacharyya, Am. J. Biomed. Sci., 2018, 10(3), 139–148 CrossRef CAS.
- M. Paraskevaidi, B. J. Matthew, B. J. Holly, B. J. Hugh, C. P. V. Thulya, C. Loren, C. StJohn, G. Peter, G. Callum, K. G. Sergei, K. Kamila, K. Maria, L. M. G. Kássio, M. H. L. Pierre, P. Evangelos, P. Savithri, A. A. John, S. Alexandra, S. Marfran, S. S. Josep, T. Gunjan, W. Michael and W. Bayden, Appl. Spectrosc. Rev., 2021, 56, 804–868 CrossRef CAS.
- Z. Hammody, R. K. Sahu, S. Mordechai, E. Cagnano and S. Argov, Thescientificworldjournal, 2005, 5, 173–182 CrossRef CAS PubMed.
- N. Wald, N. Bordry, P. G. Foukas, D. E. Speiser and E. Goormaghtigh, Biochim. Biophys. Acta, Mol. Basis Dis., 2016, 1862, 202–212 CrossRef CAS PubMed.
- B. R. Shakya, H. Teppo and L. Rieppo, Spectrochim. Acta, Part A, 2020, 254, 119665 CrossRef PubMed.
- M. J. Pilling, A. Henderson and P. Gardner, Anal. Chem., 2017, 89, 7348–7355 CrossRef CAS PubMed.
- J. Filik, M. D. Frogley, J. K. Pijanka, K. Wehbe and G. Cinque, Analyst, 2012, 137, 853–861 RSC.
- P. Bassan, J. Mellor, J. Shapiro, K. J. Williams, M. P. Lisanti and P. Gardner, Anal. Chem., 2014, 86, 1648–1653 CrossRef CAS PubMed.
- P. Bassan, J. Lee, A. Sachdeva, J. Pissardini, K. M. Dorling, J. S. Fletcher, A. Henderson and P. Gardner, Analyst, 2013, 138, 144–157 RSC.
- M. J. Pilling, P. Bassan and P. Gardner, Analyst, 2015, 140, 2383–2392 RSC.
- M. J. Pilling, A. Henderson, J. H. Shanks, M. D. Brown, N. W. Clarke and P. Gardner, Analyst, 2017, 142, 1258–1268 RSC.
- A. V. Rutter, J. Crees, H. Wright, M. Raseta, D. G. van Pittius, P. Roach and J. Sulé-Suso, Appl. Spectrosc., 2020, 74, 178–186 CrossRef CAS PubMed.
- L. M. Dowling, P. Roach, A. V. Rutter, I. Yousef, S. Pillai, D. Latham, D. G. van Pittius and J. Sulé-Suso, Appl. Spectrosc., 2021, 75, 343–350 CrossRef CAS PubMed.
- E. Ó. Faoláin, M. B. Hunter, J. M. Byrne, P. Kelehan, H. A. Lambkin, H. J. Byrne and F. M. Lyng, J. Histochem. Cytochem., 2005, 53, 121–129 CrossRef PubMed.
- C. W. Meuse and P. E. Barker, Appl. Immunohistochem. Mol. Morphol., 2009, 17, 547–552 CrossRef CAS PubMed.
- E. Ly, O. Piot, R. Wolthuis, A. Durlach, P. Bernard and M. Manfait, Analyst, 2008, 133, 197–205 RSC.
- E. Goormaghtigh, Biomed. Spectrosc. Imaging, 2016, 5, 325–346 Search PubMed.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood, K. A. Heys, C. Hughes, P. Lasch, P. L. Martin-Hirsch, B. Obinaju, G. D. Sockalingum, J. Sulé-Suso, R. J. Strong, M. J. Walsh, B. R. Wood, P. Gardner and F. L. Martin, Nat. Protocols, 2014, 9, 1771–1791 CAS.
- P. Bassan, A. Kohler, H. Martens, J. Lee, H. J. Byrne, P. Dumas, E. Gazi, M. Brown, N. Clarke and P. Gardner, Analyst, 2010, 135, 268–277 RSC.
- T. G. Mayerhöfer and J. Popp, Spectrochim. Acta, Part A, 2018, 191, 283–289 CrossRef PubMed.
- B. M. DeVetter, S. Kenkel, S. Mittal, R. Bhargava and T. P. Wrobel, Vib. Spectrosc., 2017, 91, 119–127 CrossRef CAS.
- T. G. Mayerhöfer, H. Mutschke and J. Popp, ChemPhysChem, 2017, 18, 2916–2923 CrossRef PubMed.
- A. Tfayli, O. Piot, A. Durlach, P. Bernard and M. Manfait, Biochim. Biophys. Acta, 2005, 1724, 262–269 CrossRef CAS PubMed.
- C. Hughes, L. Gaunt, M. Brown, N. W. Clarke and P. Gardner, Anal. Methods, 2014, 6, 1028–1035 RSC.
- B. Bird, K. Bedrossian, N. Laver, M. Miljković, M. J. Romeo and M. Diem, Analyst, 2009, 134, 1067–1076 RSC.
- C. L. Song and S. G. Kazarian, Analyst, 2019, 144, 2954–2964 RSC.
- K. Kochan, P. Heraud, M. Kiupel, V. Yuzbasiyan-Gurkan, D. McNaughton, M. Baranska and B. R. Wood, Analyst, 2015, 140, 2402–2411 RSC.
- L. M. Fullwood, D. Griffiths, K. Ashton, T. Dawson, R. W. Lea, C. Davis, F. Bonnier, H. J. Byrne and M. J. Baker, Analyst, 2014, 139, 446–454 RSC.
- I. P. Santos, R. van Doorn, P. J. Caspers, T. C. Bakker Schut, E. M. Barroso, T. E. C. Nijsten, V. Noordhoek Hegt, S. Koljenovic and G. J. Puppels, Br. J. Cancer, 2018, 119, 1339–1346 CrossRef PubMed.
- E. Brauchle, S. Noor, E. Holtorf, C. Garbe, K. Schenke-Layland and C. Busch, Clin. Exp. Dermatol., 2014, 39, 636–645 CrossRef CAS PubMed.
- Y. Zhang, A. J. Moy, X. Feng, H. Nguyen, K. R. Sebastian, J. S. Reichenberg, C. O. Wilke, M. K. Markey and J. W. Tunnell, Molecules, 2020, 25, 2852 CrossRef CAS PubMed.
- X. Feng, A. J. Moy, H. T. Nguyen, Y. Zhang, J. Zhang, M. C. Fox, K. R. Sebastian, J. S. Reichenberg, M. K. Markey and J. W. Tunnell, J. Biomed. Opt., 2018, 23, 057002 Search PubMed.
| This journal is © The Royal Society of Chemistry 2022 |