Limited-resource preparable chitosan magnetic particles for extracting amplification-ready nucleic acid from complex biofluids†
Sayantan
Tripathy
a,
Ashish Kumar
Chalana
a,
Arunansu
Talukdar
b,
P. V.
Rajesh
c,
Abhijit
Saha
 c,
Goutam
Pramanik
*c and
Souradyuti
Ghosh
c,
Goutam
Pramanik
*c and
Souradyuti
Ghosh
 *ad
*ad
aDepartment of Chemistry, Bennett University, Greater Noida, Uttar Pradesh 201310, India. E-mail: souradyuti.ghosh@bennett.edu.in; Tel: +91 9959849754
bDepartment of Medicine, Medical College and Hospital, Kolkata, West Bengal, India
cUGC-DAE CSR, Kolkata Centre, Sector III, LB-8, Bidhan Nagar, Kolkata – 700 106, India
dDepartment of Biotechnology, Bennett University, Greater Noida, Uttar Pradesh 201310, India
First published on 16th November 2021
Abstract
Extraction and concentration of pure nucleic acid from complex biofluids are the prerequisite for nucleic acid amplification test (NAAT) applications in pathogen detection, biowarfare prevention, and genetic diseases. However, conventional spin-column mediated nucleic acid extraction is constricted by the requirement for costly power-intensive centralized lab infrastructure, making it unsuitable for limited-resource settings. Significant progress in lab-on-a-chip devices or cartridges (e.g., Cepheid GeneXpert®) that integrate nucleic acid extraction and amplification has been made, but these approaches either require additional equipment or are costly. Similarly, their complexities make them difficult to fabricate in low-resource settings by the end-user themselves. The application of magnetic particles such as silica-coated iron oxide beads for nucleic acid extraction is relatively instrument-free, rapid, user-friendly, and amenable to automation. But, they rely on hazardous chaotropic salt chemistry and ethanol desalting that could limit their efficacy for downstream NAATs. Recent advances in several types of novel material (e.g., polyamine) coated magnetic bead-based chaotropic salt-free extraction methods offer a possible solution to this problem. However, these materials also involve multistep synthesis impermissible in limited-resource settings. To offer a possible instrument-free magnetic particle-based nucleic acid extraction doable at limited-resource settings, we investigated the nucleic acid capture ability of two chitosan-coated magnetic particles that are preparable by minimally trained personnel using only a water bath and a magnetic stirrer within 6–8 h. We quantitatively probed the efficiency of the passive (without any electrical shaking or vortex-aided) DNA magnetocapture (i.e., binding to chitosan magnetic particles, physical separation from its sample of origin, and release from the particles) using UV260. To explore their suitability towards clinically relevant sensitive downstream NAATs, 100–1000 copies (i.e., in the order of zeptomole) of Escherichia coli (E. coli) or human genomic DNA from aqueous solution, crude cell lysate, and fetal bovine serum were extracted by them and then successfully detected using quantitative real-time loop-mediated isothermal amplification (LAMP) or real-time polymerase chain reaction (PCR). Alongside, their suitability with gel-based LAMP, colorimetric LAMP, and in situ (on beads) LAMP was also probed. The required optimization of the amplification methods has been discussed. Overall, the turnaround time for the magnetocapture combined with NAAT was 1.5–2 h and is thus expected to aid in rapid clinical decision making. With the ease of preparation, reproducibility, and compatibility with downstream NAATs, we anticipate that these magnetic particles would facilitate the expansion and decentralization of nucleic acid-based diagnosis for limited-resource settings.
Introduction
According to the World Health Organization estimate,1 the frequency of occurrence of infectious diseases is predicted to increase significantly in near future from the destruction of natural wildlife habitats,2 globalization,3 unplanned urbanization,4 and climate change.5 In the last 20 years, this has already seen manifestation in the occurrence of several lethal viruses of particularly zoonotic origin such as severe acute respiratory syndrome (SARS), H1N1 influenza, Middle East respiratory syndrome (MERS), Ebola, NIPAH, and the ongoing SARS-CoV-2.2 The resulting outbreaks, epidemics, and pandemics have caused significant loss of human life and disruption of the economy. These diseases have so far appeared and are predicted to appear in short notices without giving significant lead timing,1 often overwhelming the local health agencies to contain such outbreaks. As it was seen in the early phase of the SARS-CoV-2 pandemic, it would risk incapacitating the detection and therapeutic capability of a region.6 In addition, such outbreaks are more likely to affect populations at remote and resource constrained settings due to their limited access to centralized labs, necessity of sample transport to faraway detection centers, and resulting loss of time for treatment initiation.7 Despite the vulnerability, there is inadequate published research, innovation, or open access resources to help a potential end-user at a limited resource setting for improvising or fabricating devices capable of sensitive molecular diagnosis.Nucleic acid amplification tests (NAATs) are the primary analytical tools in pathogen identification, environmental monitoring, food contamination, genetic disease detection, and biowarfare prevention.8 In the event of a disease outbreak, NAATs such as real-time polymerase chain reaction (real-time PCR) are always the first type of diagnostic intervention. Isothermal NAATs (iNAATs) have further helped expand the scope of nucleic acid detection based molecular diagnosis due to their independence of thermal cyclers.9 Thanks to the invention of a diversity of readouts in iNAATs such as colorimetry, electrochemistry, turbidometry, etc. in addition to the traditional real-time fluorescence,9 the democratization of molecular diagnosis has become further achievable. However, despite the innovations in the nucleic acid amplification and readout steps in NAATs or iNAATs, a critical step is the extraction of polymerase inhibitor-free nucleic acid from complex samples. The prevailing method for nucleic acid extraction is solid-phase spin-columns suited for centralized high-speed centrifuges.10 Commonly based on silica or cellulose, these spin columns capture the nucleic acid from complex biofluids or cellular lysates using chaotropic salt.11,12 It then removes the excess salt and remnants of complex biofluid components utilizing successive alcohol washes. Finally, low to moderate salt conditions are used to desorb the nucleic acid from a solid phase support in the columns for downstream application. However, the use of hazardous chaotropic salt and ethanol, both of which are known polymerase inhibitors, necessitates multiple wash steps, which in turn increases extraction time. Due to their restrictive ability to handle 0.5–1 mL liquid, the spin columns are also unsuitable for high dilution biofluid samples such as urine. In addition, silica column-based extraction is synonymous with the use of high-speed centrifuges, which confines their application to well-equipped centralized labs. In the event of a sudden outbreak overwhelming the supply chain, these columns are difficult to be fabricated by the end-user themselves. Therefore, despite their wide and almost universal adoption, silica or cellulose-based spin columns are unsuitable for nucleic acid extraction in NAAT-based molecular diagnosis in limited-resource settings.
To address these shortcomings, several research groups have come up with innovative non-silica-based solutions that actuate nucleic acid extraction without compromising its compatibility with downstream NAATs. These included several paper-based microfluidic interventions from the Whitesides group,13 origami-based microdevices for simultaneous viable cell detection and isothermal NAAT,14 and a nylon-cross-linked chitosan device capable of capturing tens of copies nucleic from very large sample volumes15 among others. Despite the ingenuity of these devices, they would often require additional equipment to control the liquid flow for generating an analytically sensitive nucleic acid extraction. An alternative to these microdevices is magnetic particle-based extraction systems. In this, a coated magnetic particle specifically binds to DNA/RNA and in the process physically separates the nucleic acid from the rest of the solution. After successive washing away of undesirable components such as polymerase inhibitors and cell debris, the amplification-ready nucleic acid is released from the beads. The magnetic particle-based extraction is advantageous due to its independence or near independence from electrical equipment, relatively low cost, and operability by moderately skilled personnel. Furthermore, open-source initiatives such as bio-on-magnetic-beads (BOMB) introduced low-cost and easily adoptable methods for preparing silica and carboxyl coated ferrite (Fe3O4) magnetic particles.16 Despite these remarkable advancements, silica- or carboxyl-based magnetic beads still suffer from the same shortcomings as their spin column counterparts, i.e., usage of chaotropic salts and ethanol for removing polymerase inhibitors. However, recent advancements have been made in alternative materials for nucleic acid extractions such as polyamines or chitosan-coated magnetic particles.17,18 These magnetic particles accomplished the capture of nucleic acid using pH modulation while avoiding the use of chaotropic salt, alcohol, or extreme pH. At the same time, the captured nucleic acid remains suitable for downstream NAATs. Some of the recent chitosan magnetic particle-based extraction systems have also documented in situ (on-bead/on-particle) amplification, significantly reducing steps and time of the ultrasensitive nucleic acid detection.18 A comprehensive summary of chitosan-based magnetic particles and other microdevices is documented in Table S1.† It comprises the preparation, capture efficiency, and application of extracted nucleic acid using downstream NAAT/iNAAT. As shown in Table S1†, their preparations would involve multiple processing steps, sophisticated instruments,18 prior hydrolysis (or purchase) of the chitosan ingredient into chitosan oligosachharide,19 or long (12–24 h) preparation time.20,21 Some devices also require additional equipment like syringe pumps for operation. Additionally, most of these magnetic particle-based nucleic acid capture systems are yet to be commercialized. Despite their proven utility in ultrasensitive nucleic acid capture/detection, they therefore may not reach an end-user at limited-resource settings in the event of a sudden outbreak. Especially in cases where such extraction devices may require deployment at short notice, they could be challenging to be prepared by the end-user themselves due to the lack of sophisticated instruments or necessary time to improvise. Additionally, the associated cost of assembling such devices may also turn out to be a barrier.
Manual handling of nucleic acid extraction or related reagent preparation is generally undesirable and inefficient. However, ease of preparation of critical molecular diagnosis components such as the nucleic acid extraction-capable magnetic particles, even if manual, would be considered lifesaving in times of exigency. The primary motivation behind this work was thus to explore methods that can synthesize chitosan-coated magnetic particles in resource-limited settings within a few hours preferably by minimally trained personnel. The preparation time of such magnetic particles should be less than 8 h (within a day), so that it can be repeatedly prepared in hundreds of milligram scales within short notice during an urgent situation (e.g., a pandemic). Despite the ease of synthesis, these particles should not compromise the quality of the extracted nucleic acid for downstream procedures. Therefore, they should not only be able to extract nucleic acid from complex biofluids, but the extracted nucleic acid should also be NAAT/iNAAT compatible. In this work, we evaluated two such methods with synthesis and characterization and then quantified their ability for electricity-free passive magnetocapture (i.e., benchtop incubation without instrument-enabled mixing) of the nucleic acid of two different genome sizes (E. coli and human). The detectability of magnetocapture-isolated nucleic acid with downstream NAATs was probed through the extraction of the low copy of DNA from aqueous solution, crude cell lysate, and complex biofluid (serum) coupled with follow-up gel-based loop-mediated isothermal amplification (LAMP), in situ (on-bead or on-particle) LAMP, real-time LAMP with homemade master mix, commercial colorimetric LAMP mix, and real-time PCR.
Results and discussion
Preparation and characterization of chitosan magnetic particles
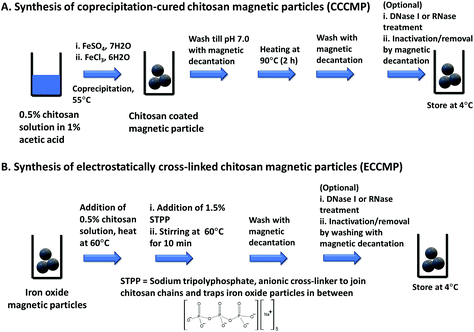
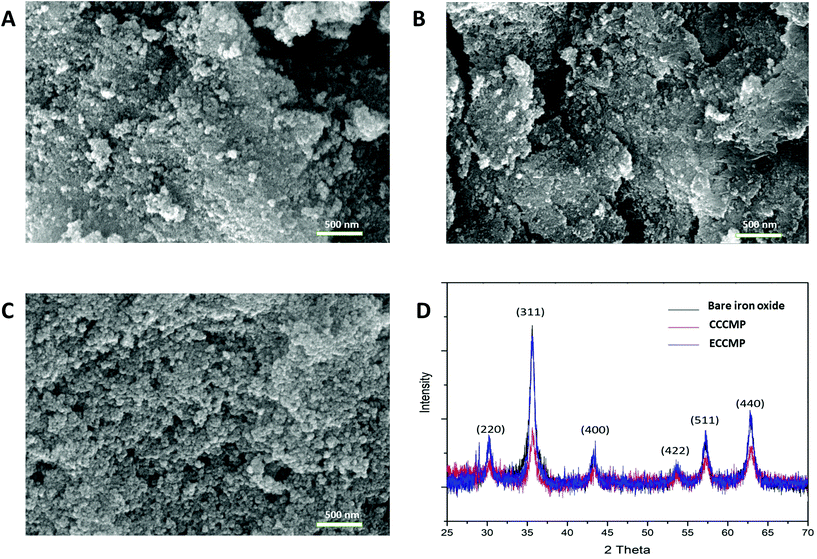
The primary motivation behind this work was to utilize magnetic particles that could extract amplification-ready nucleic acid in limited-resource settings. To ensure that such magnetic particles become employable in a sudden health emergency (e.g., an outbreak), we opted for synthesis protocols that enabled the preparation of magnetic particles in at least 0.1 g scale with bare minimum equipment, within a short time (6–8 h), and by minimally trained personnel. An attractive choice of coating for such magnetic particles is chitosan due to its proven pH-modulated nucleic acid binding ability, low cost, and availability. Exploring the literature for available methods that would fulfill the above criteria, we shortlisted and synthesized two types of chitosan-coated magnetic nanoparticles, namely coprecipitation-cured chitosan magnetic nanoparticles (CCCMP)22 and electrostatically cross-linked chitosan magnetic particles (ECCMP).23 Interestingly, we could not find any report utilizing these magnetic particles for amplification-ready ultrasensitive nucleic acid extraction from the crude lysate or complex biofluid (see Table S1†). For the synthesis of CCCMP, chitosan was dissolved in acetic acid solution and then subjected to alkaline coprecipitation with Fe(II)/Fe(III) salts followed by curing (90 °C). This would cause chitosan coating on ferrite (Fe3O4) nanoparticles.24 For ECCMP preparation, ionic cross-linker sodium tripolyphosphate was utilized to trap pre-synthesized ferrite iron oxide nanoparticles within a chitosan polymer. To mimic conditions as in resource constrained settings and a short-notice preparation, we even abstained from any nitrogen purging of the solutions prior to the reactions. With molecular diagnosis as the objective, the synthesized chitosan magnetic particles could be subjected to an optional DNase I or RNase treatment followed by heat inactivation and washing using magnetic decantation. Both synthesis protocols could produce chitosan-coated magnetic particles using nothing but a magnetic stirrer and a water bath (or a hot plate-magnetic stirrer) and were completed within 5–6 h (Scheme 1, Fig. S1†).The characterization of CCCMP, ECCMP, and coprecipitation synthesized bare iron oxide magnetic particles was carried out using scanning electron microscopy (SEM), EDX, FT-IR, dynamic light scattering (DLS), zeta potential, and XRD. EDX spectrum analysis showed the presence of carbon and nitrogen in CCCMP and ECCMP (consistent with chitosan coating) but the absence of the same in bare iron oxide (Fig. S2†). Intense peaks at 538 cm−1 (in bare iron oxide) and 545 cm−1 bands (in CCCMP and ECCMP) characteristic of the Fe–O bending frequency were found in FT-IR investigations, confirming the presence of iron oxide in all three substrates (Fig. S3†). Broad peaks due to possible moisture absorbance were detected at 3260, 3267, and 3175 cm−1 in bare iron oxide, CCCMP, and ECCMP, respectively. In addition, peaks from C–O stretching in primary alcohols (1028 cm−1 in CCCMP and 1066 cm−1 in ECCMP), N–H bending in chitosan amine (1585 cm−1 in CCCMP and 1531 cm−1 in ECCMP), and CH3 symmetrical deformation (1370 cm−1 in CCCMP and 1378 cm−1 in ECCMP) substantiated the chitosan coating on the iron oxide. The SEM images of the MPs showed agglomerated spherical particles with some unusual filament-like structure present in some regions of CCCMP (Fig. 1A–C and Fig. S4†). Magnification indicated the presence of spherical-shaped nanoparticles consistent with coprecipitation synthesized iron oxide nanoparticles (Fig. 1A–C). The crystalline structure of Fe3O4 nanoparticles was characterized by XRD as shown in Fig. 1D. All three Fe3O4 nanoparticles showed diffraction peaks at 30.32° (220), 35.60° (311), 43.24° (400), 53.62° (422), 57.34° (511), and 62.83° (440) which are associated with Fe3O4. The observed peaks of Fe3O4 were consistent with the database in ICDD (International Centre for Diffraction Data) PDF Card – 01-071-6337 and revealed that the pure Fe3O4 phase was a spinal structure. The average diameter of the Fe3O4 nanoparticles calculated by using Debye-Scherrer's equation to the (311) peak at 2θ = 35.60° was about 15.5 nm. XRD results showed that the chitosan coating does not alter the crystallinity of the Fe3O4 core (Fig. 1D). Overall, the FE-SEM, EDX, and FT-IR characterization of magnetic particles indicated the successful coating of chitosan on iron oxide magnetic particles while the XRD established their crystalline structure.
The hydrodynamic diameter of ferrite (Fe3O4) nanoparticles (NPs) in aqueous solution was determined by DLS in pH 5.2 buffer as the DNA binding was to be carried out under this buffer condition. The hydrodynamic diameter of uncoated bare iron oxide NPs was 162 ± 24.18 nm, which is significantly larger than the determined value by Debye-Scherrer's equation from XRD results (Table 1, Fig. S5A†). The increase in particle size might be attributed to the aggregation of bare iron oxide NPs due to van der Waals forces and magnetic dipole–dipole interactions generated from residual magnetic moments.25 The increased hydrodynamic radius of both CCCMP and ECCMP compared to bare iron oxide, once again, indicated successful coating of chitosan coating on bare iron oxide NPs (Table 1, Fig. S5†).26,27
| Particles | Hydrodynamic radius (Rh) (nm) | Zeta potential (ζ) (mV) at pH 5.2 |
|---|---|---|
| CCCMP | 253.5 ± 38.08 | 18.9 ± 1.63 |
| ECCMP | 275.0 ± 106.4 | 7.35 ± 1.36 |
| Bare iron oxide | 162 ± 24.18 | −12.3 ± 1.63 |
The zeta potential of bare iron oxide, CCCMP, and ECCMP was measured at pH 5.2 to understand the role of surface charge in DNA binding (Table 1). The bare iron oxide nanoparticle zeta potential is negative at pH 5, which is consistent with the previously reported literature.28 The positive zeta potential of both CCCMP and ECCMP at pH 5.2 confirms the presence of cationic protonated ammonium groups of chitosan on the particle surface. However, CCCMP showed higher zeta potential compared to ECCMP, probably due to the partial neutralization of positively charged primary amino groups by the negatively charged tripolyphosphate (pKa 1,0, 2.2, 2.3, 5.7, 8.5)29 crosslinker in ECCMP.
DNA binding study
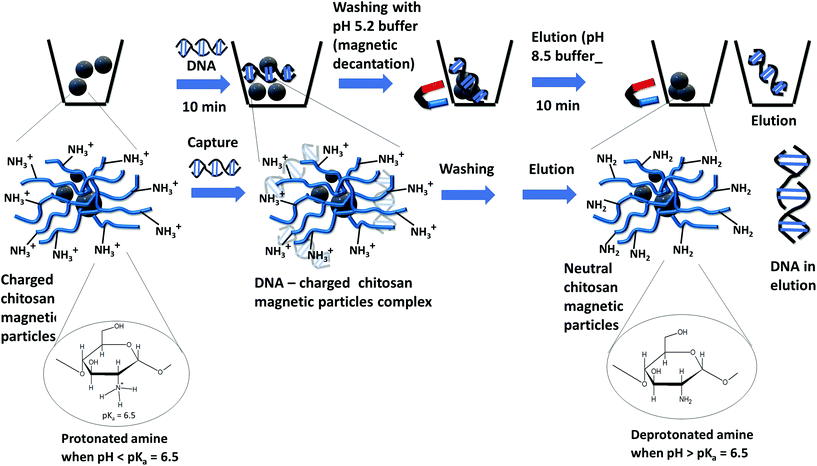
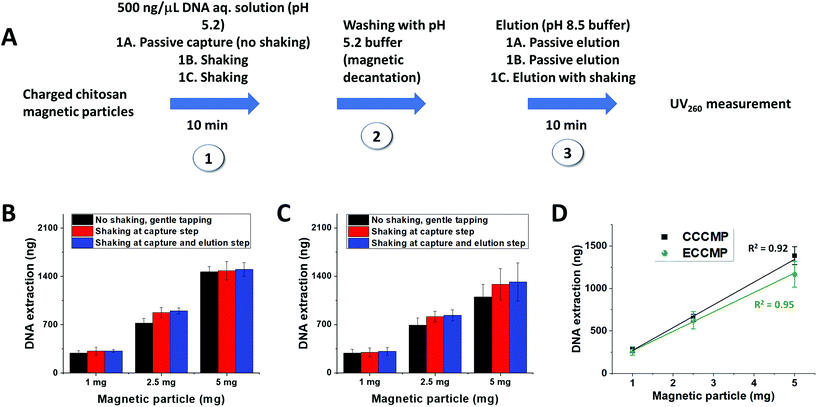
Next, we assessed the CCCMP and ECCMP for their ability to extract nucleic acid from the solution. Due to the presence of the amino group with a pKa ∼6.4 in aqueous solutions, chitosan owes its nucleic acid binding capability on solution pH variation. Quantitatively, it relies on the Henderson–Hasselbach equation (pH = pKa + log10([A−]/[HA]) that correlates solution pH with the ratio of deprotonated (NH2) to protonated (NH3+) form. As a result, a chitosan-based material would acquire a backbone positive charge below pH 6–6.5 and a predominantly charge-neutral state above pH 7–7.5. It would therefore bind negatively charged nucleic acid below pH 6–6.5 from the solution and release the same at solution pH 8.5. Prior reports on chitosan-based microdevices and magnetic particles as well as gene delivery studies have utilized this charge switching property for nucleic acid capture and transport.30 The 25–30 min magnetocapture assay illustratively described in Scheme 2 was therefore composed of nucleic acid adsorption (referred to as “capture”) by the positively charged chitosan backbone under the influence of a pH 5.2 buffer (10 min), magnetic decantation washing with the same buffer (5–10 min), and desorption of nucleic acid from chitosan-magnetic particles (referred to as “elution”) using a pH 8.5 buffer (10 min). We assessed the nucleic acid extraction ability of CCCMP and ECCMP under three different combinations of capture and elution (Fig. 2A). Under the first condition, both the capture and elution steps were carried out under passive mixing conditions employing only gentle tapping (i.e., without any mechanical shaking) (condition 1A, steps 1 and 3, Fig. 2A). In the second, mechanical shaking (using a vortex) was employed at the capture step while the passive elution was utilized (condition 1B, steps 1 and 3, Fig. 2A). In the third, both capture and elution steps utilized mechanical shaking (condition 1C, steps 1 and 3, Fig. 2A). When vortex-aided shaking was employed at either capture or capture as well as elution steps, slightly higher DNA binding was observed. Interestingly, the increase in the amount of magnetocaptured DNA by shaking was not significantly higher than compared to those found for passive mixing. This could be due to a lesser degree of chitosan coating as evident in the zeta potential measurement (Table 1). This experiment also demonstrated that the DNA extraction efficiency during passive mixing would not be significantly affected despite the absence of mechanical shaking. Utilizing the passive DNA binding and elution assay, 1–5 mg wet CCCMP or ECCMP also demonstrated a linear genomic DNA extraction (capture and elution) ability from aqueous solution with a mean extraction efficiency of 277.0 ± 21.2 and 233.3 ± 30.5 ng mg−1, respectively (Fig. 2C). In contrast, the DNA capture ability of bare iron oxide magnetic particles was 11.2 ± 7.2 ng mg−1 (measured for 5 mg bare iron oxide). The absorbance for 1 and 2.5 mg bare iron oxide was below the limit of detection of nanodrops (data not shown). The superior DNA capture by chitosan magnetic particles, compared to a poor DNA binding ability of bare iron oxide, thus justified the importance of chitosan coating on iron oxide for nucleic acid capture. It also validated the passive DNA binding capacity of both the CCCMP and ECCMP materials besides demonstrating that the DNA extraction efficiency would not be significantly affected despite the absence of mechanical shaking.The superior DNA extraction efficiency of CCCMP compared to ECCMP could be attributed to the higher zeta potential of CCCMP at pH 5 (Table 1). The higher zeta potential of CCCMP indicated a greater number of positively charged amino groups exposed at the surface of the CCCMP than that of ECCMP. The higher number of exposed positively charged amino groups increased the ionic interaction with the negatively charged phosphate backbone of DNA and subsequently improves the extraction efficiency of the CCCMP. On the other hand, the CCCMP and ECCMP average hydrodynamic particle sizes did not significantly differ from each other (Table 1). Although the DNA binding capacity is reported to have a direct correlation with particle size (and in turn with the surface to volume ratio),31 the similar particle sizes of CCCMP and ECCMP, in this case, might not have a direct association with their DNA binding ability.
Detection of magnetocaptured DNA using gel-based LAMP, colorimetric LAMP, and real-time LAMP
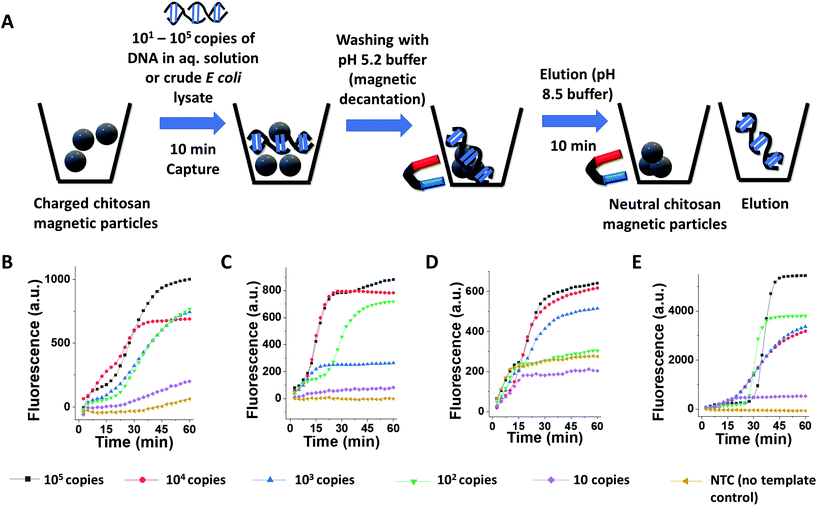
Direct detection instruments using UV260 absorbance or direct fluorescence (e.g., in qubit instrument) can neither detect a clinically relevant ultralow quantity (hundreds to thousands of copies32) of target nucleic acid nor identify specific sequence elements in it. NAATs or iNAATs, in contrast, would specifically identify the presence or absence of the clinically relevant amount of biomarker nucleic acid sequence while also acting as a billion-fold (or higher) signal amplifier element. However, their performance as specific and sensitive bioanalytical tools is contingent on the purity of extracted nucleic acid and may get attenuated because of the presence of impurities such as PCR inhibitors.33 An effective nucleic acid extraction assay is thus expected to rescue amplification-ready DNA/RNA from complex biofluids through the physical separation of nucleic acid from polymerase inhibitors. The second key goal of this study was therefore to assess the compatibility of the CCCMP and ECCMP extracted nucleic acid (including those isolated from complex biofluids) for downstream NAAT/iNAAT applications. To probe this qualitatively, magnetocapture followed by loop-mediated isothermal amplification (LAMP) was performed on E. coli genomic DNA present in an aqueous solution and crude cell lysate. The reason for selecting LAMP over other isothermal amplifications was its high sensitivity and compatibility with several readout methods which included but were not limited to turbidimetry, electrochemistry, colorimetry, surface plasmon resonance, lateral flow assay, and real-time fluorescence.34 To mimic conditions similar to those found in limited-resource settings, we opted to utilize our own homemade LAMP master mix over the commercially available ones as the latter is not as commonly available as their real-time PCR counterparts. While utilizing an earlier reported set of LAMP primers (E. coli malB gene,35 see Table S2† for primer sequence), non-specific LAMP amplification was encountered during agarose gel electrophoresis (Fig. S6A†). We cross-checked our findings with that of an earlier publication by the Whitesides group that used the same primer set and recorded non-specific amplification for longer assays or as a function of betaine concentration (see Fig. S1 panel b and c in their paper).36 From this, we inferred that the non-specific amplification could be due to the absence of betaine in our homemade amplification mix causing primer-dimer formation (Fig. S6A†). The non-specific LAMP was mitigated after the removal of loop primers from the 10× primer mix and characteristic ladder-like amplification patterns in gel electrophoresis were noted only in those containing the template (E. coli genomic DNA, Fig. S6B†). Without any further optimization in magnetocapture or LAMP protocol, 109 copies of genomic DNA in an aqueous solution were subjected to magnetocapture followed by LAMP (Fig. S7A†). Similarly, 109E. coli cells were heat lysed and then subjected to magnetocapture followed by LAMP (Fig. S7A†). Elutions from CCCMP and ECCMP magnetocapture on both aqueous genomic DNA solution and crude lysate demonstrated successful LAMP amplifications in agarose gel electrophoresis (Fig. S7B and S7D†). We then investigated whether LAMP could also be performed on the DNA-bound magnetic particles prior to the elution step (i.e., an “in situ” or “on-particle” LAMP). If successful, it would reduce the assay timing by at least 10 min. To investigate this, we subjected the DNA-bound magnetic particles (following magnetic decantation wash but before pH 8.5 elution) themselves to LAMP. In agarose gel, ladder-like patterns characteristic of LAMP reactions were visualized for these experiments (Fig. S7C and S7E†). Overall, these assays qualitatively established the compatibility of magnetocapture extraction using both CCCMP and ECCMP with LAMP experiments, showing that DNA from aqueous solution as well as crude lysate can be extracted. An interesting observation was that LAMP can even be performed “in situ” on the DNA-bound magnetic particles themselves without elution. This could probably be attributed to the alkaline pH of the LAMP buffer (pH 8.8) causing some degree of elution, amplification on the surface of the magnetic particle, or the combination of both. Although highly promising in terms of reducing assay time, this aspect was not pursued in this work any further and would be investigated in a follow-up study.While gel electrophoresis-mediated visualization of LAMP can qualitatively confirm an amplification experiment, this method of readout is neither sensitive enough to detect low copy target nucleic acid nor a quantitative technique, both of which are standard requirements in molecular diagnosis assays.37,38 We, therefore, opted for real-time LAMP using our homemade mastermix to establish the limit of detection (LoD or analytical sensitivity) of magnetocapture followed by LAMP assays. While experimenting to establish proof-of-concept assays using 106 copies of target E. coli genomic DNA, our homemade real-time LAMP mastermix was successfully able to distinguish target from no template control (NTC) experiments (Fig. S8A†). However, the NTC itself still was producing large fluorescence despite the LAMP reactions at this point being devoid of loop primers (Fig. S8A†). We attributed the large fluorescence in NTC LAMP to a possible minor primer-dimer formation otherwise undetectable in agarose gel electrophoresis. We deduced that a higher temperature LAMP initiation would further mitigate non-specific interactions like primer-dimer formation. Therefore, the standard real-time LAMP protocol (66 °C 1 min for 60 cycles with fluorescence monitoring at each cycle) was modified to a touchdown-like LAMP protocol (Fig. S8B†). In the latter, the temperature at each cycle underwent a stepwise decrease from 69 to 66 °C with fluorescence monitoring at the final step. Although it reduced the fluorescence in both positive control as well as NTC, the latter was attenuated over 6-fold. Next, we subjected 101–105 copies of E. coli DNA (as pure genomic DNA or in the form of crude lysate) to magnetocapture with the elution being analyzed using follow-up real-time LAMP (Fig. 3A). For CCCMP magnetocapture with either genomic DNA in aqueous solution or as a crude lysate, the LoD was 102 copies (established using a characteristic “S” shaped amplification profile at Fig. 3B and C with corresponding melt curve analysis in Fig. S9A and S9B†). On the other hand, the LoD for ECCMP magnetocapture followed by real-time LAMP was recorded as 103 copies for aqueous genomic DNA and 102 copies for crude lysate (amplification profile at Fig. 3D and E with melt curve analysis in Fig. S9C and S9D†). Encouragingly, the LAMP assays could reproducibly distinguish “yes/no” samples within 45 min, making the total turnaround time for magnetocapture and amplification 1.5–2 h. The instrument-free nature of the magnetocapture combined with an overall low turnaround time is anticipated to facilitate swift “yes/no” clinical decision-making in limited-resource settings. One limitation of our LAMP assays was that the cycle threshold (Ct) varied considerably to obtain a mean Ct with a low (less than 10%) standard deviation, preventing any potential pathogen load estimation. This could be due to our use of homemade LAMP mastermix, use of touchdown-like LAMP, minor but detectable primer-dimer formation mediated amplification (as seen in melt-curve analysis in Fig. S9†), or a combination of all these factors. It could be likely that the use of a commercial LAMP mastermix may lead to the generation of mean Ct with a low standard deviation suitable for pathogen load determination. Altogether, these assays established that the DNA extraction using both magnetic particles from either aqueous solution or crude lysate was compatible with real-time LAMP with clinically relevant analytical sensitivity.
Next, we investigated whether the colorimetric LAMP protocol can be integrated with magnetocapture assays. The polymerase activity during the LAMP reaction would generate protons, causing a pH drop, and trigger the color change in a pH indicator dye present in the solution.39 The proprietary WarmStart Colorimetric LAMP 2X Mastermix employs this principle to provide a fast and clear visual (pink to yellow) readout of positive LAMP reactions.39 The pH change (with resulting color change) also relies on low concentration (25–400 μM) of Tris providing very weak buffering ability. This assay has recently been utilized in the rapid visual detection of SARS-CoV-2.40 We anticipated that the integration of colorimetric LAMP with magnetocapture assays would provide a highly utilitarian and rapid NAAT molecular diagnosis platform suitable for limited-resource settings. Therefore, CCCMP magnetocapture of 2 × 101–106 copies of E. coli genomic DNA from aqueous solution followed by colorimetric LAMP on the elution was investigated. Within 10 min of starting the LAMP reaction, perceivable color changes to various degrees were observed (Fig. S10† tubes 1–6). Additionally, the elution from the negative control “mock” magnetocapture (on buffer alone but no DNA) had a visual difference from those containing DNA (Fig. S10† tube 7). Similarly, addition of elution buffer alone (without magnetocapture) retained the pink color of the commercial mastermix (tube 8, Fig. S10†). Although this apparently suggested a successful assay with LoD in the order of 10 copies, we experimented further with the conditions. Interestingly, when we added the “neutralized” (i.e., by addition of elution buffer) magnetic particles from the “mock” magnetocapture to the colorimetric amplification mix, it also changed color from pink to yellow within 10 min of starting the amplification (Fig. S11† tube 1). This suggested that the color change could be due to release of remnant protons bound to chitosan matrix amines and might not be from the LAMP reaction. To test further, we carried out CCCMP magnetocapture on 106 copies of genomic DNA, eluted the same, resuspended the CCCMP particles to water (pH 7) post-elution. Then we subjected the particles as well as elution to colorimetric LAMP. Both the reactions become yellow within 10 min (tubes 2 for particles and 3 for elution, Fig. S11†). It again hinted that the protons bound that even so-called “neutralized” particles could be responsible for the color change. Similarly, “in situ” (“on-bead”) LAMP on magnetic particles from another “mock” (no genomic DNA) CCCMP magnetocapture but without incubation with “neutralizing” elution buffer also demonstrated color change (Tube 4, Fig. S11†). The experiments thus confirmed that the protons bound to chitosan itself could cause pH reduction, resulting in a color change in the indicator dye present in the colorimetric master mix. Furthermore, the studies indicated that incubation with 10 mM Tris-HCl pH 8.5 would probably be incompletely neutralizing chitosan. In the process, it was causing color change (i.e., a false positive readout) in colorimetric LAMP even in the absence of eluted DNA (as in tube 7, Fig. S10†). A false positive readout might also originate from the inadvertent transfer of a trace amount of chitosan particles into the LAMP reaction tube as the partially neutralized particles would release proton due to the alkaline pH and heating during the assays. Overall, these reactions indicated that colorimetric pH change-based LAMP could be incompatible with 10 mM Tris-HCl pH 8.5 as the elution buffer at least in the magnetocapture assays of this work. We tried changing the concentration of neutralizing elution buffer to 25 mM Tris-HCl pH 8.5 without changing the incubation time of elution. This elution buffer, however, only recorded color change for 106 copies. Possibly, tris buffer at 25 mM might have been probably too concentrated (therefore having higher buffering capacity) for its pH to be altered by LAMP proton release (tube 1–6, Fig. S12†). The experiments hinted at possible novel correlations about interactions between chitosan and the neutralizing “elution” buffer including a possible role of neutralization kinetics. It suggested the need for further optimization in terms of elution buffer composition as well as neutralization time for the magnetocapture for integration with colorimetric LAMP. No further experiments with colorimetric LAMP in this work were conducted and would be pursued as part of future studies.
Detection of magnetocaptured DNA using real-time PCR
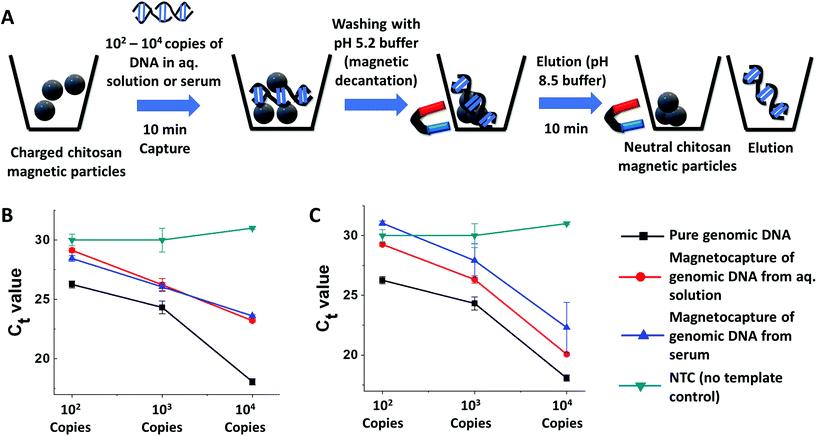
Real-time PCR assays are still the mainstay of clinical NAAT molecular diagnosis and are used in several automated instruments such as GeneXPert®.41 However, the target nucleic acid in clinical samples is often present in complex biofluids like urine, cough, swab, and serum-containing various PCR inhibitors that inhibit downstream real-time PCR application. Traditional spin-column-based procedures to extract nucleic acid from these biofluids rely on centralized high-speed centrifuges. We hypothesized that charged CCCMP or ECCMP should be able to rescue nucleic acid from such complex biofluids in a centrifuge-free manner and make it suitable for downstream real-time PCR analysis. Unlike a homemade mastermix used in the real-time LAMP, we employed commercial real-time PCR mastermix due to their commonplace availability. We opted for serum as the complex biofluid of choice due to the presence of PCR inhibitory as well as fluorescence quenching immunoglobulin (1 mg mL−1) and hemoglobin (20 mg mL−1) in it.42,43 Additionally, the serum is a primary ingredient in viral transport media, commonly being used in SARS-CoV-2 detection.44 The inhibitory role of serum was immediately apparent when real-time PCR assays on 102–104 copies of purified genomic DNA spiked into 50% fetal bovine serum failed to generate an amplification curve (Fig. S13†). An assay succeeding in the extraction of amplification-compatible nucleic acid from serum would therefore be useful in real-life clinical assays involving SARS-CoV-2 detection.Before utilizing the CCCMP and ECCMP towards extracting DNA spiked in serum, their amenability with real-time PCR was investigated with their ability to capture 102–104 copies of genomic DNA from an aqueous solution (Fig. 4A). When compared for cycle threshold (Ct) values, there was a consistent increase between the same copies of pure genomic DNA before and after magnetocapture from aqueous solution (Fig. 4B and C, melt curve analysis in Fig. S14A–E†). The increase in the Ct value could be attributed to the reduction in analytical sensitivity from the loss of some DNA molecules during the wash steps or from the failure to get all bound DNA desorbed during elution. In the latter, these DNA molecules probably failed to get released in the “neutralizing” elution buffer and might still have remained electrostatically bound to partially neutralized chitosan (as shown in the colorimetric study), decreasing analytical sensitivity. When challenged with the “rescue” of 102–104 copies of DNA from serum, both CCCMP and ECCMP magnetocapture were able to restore real-time PCR amplification albeit with a slight further increase of Ct value (Fig. 4B and C, melt curve analysis in Fig. S12D and E†). The increase of the Ct value at this instance could be from the minute presence of PCR inhibitors that could not be completely removed in the magnetocapture process as well as loss of genomic DNA. The magnetocapture with follow-up real-time PCR together had an LoD in the order of 102 copies for CCCMP and 103 copies for ECCMP for both aqueous or serum samples. They were therefore sufficiently sensitive to detect a clinically relevant low copy number of target nucleic acid in molecular diagnosis assays.32 Interestingly, the LoD from the magnetocapture in combination with real-time PCR was similar to that for magnetocapture with follow-up real-time LAMP reactions, validating the reproducibility of the methods. Furthermore, the better LoD of CCCMP was in accordance with its zeta potential as well as higher DNA extraction efficiency. These experiments demonstrated that the magnetic particles could be utilized for identifying (through capture and amplification) nucleic acid from complex biofluid, and for physically separating it from PCR inhibitors present therein.
Comparison with other published methods and limitations
We compared our method with published chitosan-dependent or independent nucleic acid extraction devices and magnetic particles in the context of preparation time, equipment required for preparation, application for nucleic acid extraction from complex biofluid, post-extraction detection using NAAT/iNAAT, the limit of detection (LoD), and the prospect of in situ amplification (Table S1†). According to the table, our method is the only one that could prepare chitosan-coated magnetic particles within 6 h using inexpensive equipment like the magnetic stirrer and water bath. Despite the ease of synthesis, our method was only among the few that could capture nucleic acid from crude lysate and complex biofluid. To the best of our knowledge, our work is the first to report the amenability of extracted nucleic acid with both downstream NAAT and iNAAT along with a highly sensitive LoD for each. We also reported the prospect of in situ (“on particle”) amplification involving magnetic particle, a unique feature only shared by two other published magnetic particle reports.18,21 The cost of the preparation (based on the raw material prices and excluding electrical power usage or downstream NAAT applications) of 1 g of wet CCCMP and ECCMP was $0.50 and $0.45, respectively. Therefore, the cost of a single nucleic acid extraction assay using 2.5 mg–5 mg of these magnetic particles would be a few cents, significantly lower than that of commercial magnetic particle.16 Despite its inexpensiveness or ease of synthesis, it is important to underscore that our assay did not compromise the necessary analytical sensitivity of downstream NAAT assays. Additionally, our methods, like other chitosan-based devices and unlike silica or cellulose-based magnetic particles, performed nucleic acid extraction sans the use of polymerase inhibitor chemicals such as chaotropic salt and ethanol. Altogether, the comparison showed that our method investigated and expanded the possibility of the clinically relevant quantity of low-cost nucleic acid extraction from complex biofluid at a possible limited resource setting.We found that both the CCCMP and ECCMP systems lost their nucleic acid capture ability after 2 weeks under 4 °C storage conditions. It could be due to the weak electrostatic nature of the interaction between chitosan and iron oxide. We did not check whether they are stable at lower storage temperatures (e.g., −20 °C). However, the inexpensiveness of the precursor chemicals, minimal equipment necessity for synthesis, and amenability to preparation in a short time would possibly outweigh this factor. The extraction system in its present form is incompatible with colorimetric LAMP and possibly other pH-based readout systems. Likely, changing the elution buffer concentration, pH, or adoption of a different colorimetric pH indicator dye would resolve this issue. However, these factors were not explored in this work. In our experiments, the real-time LAMP experiments in combination with melt curve analysis reproducibly detected the presence of analyte nucleic acid to as low as clinically relevant 100–1000 copies. Presumably due to the use of homemade LAMP mix, however, the standard deviation associated with the mean Ct values was above 10%. Therefore, the magnetocapture followed by real-time LAMP experiments at present can only act as a “Yes or No” system without the ability of quantification (e.g., finding bacterial or viral load).
Future studies
Although the beneficial role of the chitosan surface charge (as a function of pH) in the DNA binding is qualitatively well-known, the molecular level interaction behind this remains poorly understood. We could not find a published report quantitatively correlating the amount of chitosan coating on nanoparticles, net surface charge, and DNA binding capacity. Our experiments reported a decreasing DNA extraction capacity as a function of zeta potential and that in turn controlled the sensitivity of downstream NAAT/iNAAT assays. Previous studies involving DLS measurement and molecular tweezers have indicated a possible role of cooperative effect on chitosan-DNA binding.45,46 On the other hand, a permanent positive charge on chitosan magnetic particles was found to benefit DNA binding as well as in situ NAAT.18 Our colorimetric LAMP experiment also hinted at a role of chitosan deprotonation kinetics that may in principle affect DNA extraction efficiency. Future studies would therefore comprise of in-depth investigations for correlating chitosan coating on nanoparticles with DNA binding capacity, optimizations to improve the stability of magnetic particles beyond 2 weeks, the kinetics of chitosan-DNA electrostatic complexation, experimenting with buffer conditions for colorimetric LAMP readout, and obtaining Ct values for real-time LAMP. We would also be exploring the ability of magnetic particles to extract RNA, associated reverse transcription, and TaqMan probe-based detection. We also noted but chose not to explore the possibility of “in situ” (on-bead/on-particle) amplification for LAMP experiments which would minimize the extraction timing by at least 10 min. The experimental optimization for “in situ” optimization would also be taken up in future studies.Conclusions
In this work, we evaluated the application of two chitosan-coated magnetic particles for the extraction of amplification-ready nucleic acid from complex biofluids for application in limited-resource settings. The magnetic particles are preparable within 6–8 h by minimally trained personnel using only a water bath and magnetic stirrer using commonly available inexpensive chemicals. The resulting CCCMP and ECCMP particles showed a surface charge-dependent DNA extraction capacity in the order of 233–277 ng μL−1. The particles were able to capture and physically separate genomic DNA from aqueous solutions, crude cell lysate, and fetal bovine serum without any mechanized shaking, mixing, or stirring. The extraction assay was completed within 30 min, purifying nucleic acid that can be detected using NAATs/iNAATs such as real-time LAMP and real-time PCR. Within a combined 1.5–2 h turnaround time, the magnetocapture method in combination with real-time LAMP was able to extract and detect 102–103 copies (i.e., in the order of zeptomole) of genomic DNA reproducibly from both aqueous solution and crude cell lysate. In combination with real-time PCR assays, the magnetocapture methods were able to extract and detect 102–103 copies of spiked human genomic DNA in 50% fetal bovine serum with few-cycle threshold values (Ct values) higher than that of pure genomic DNA. Assays involving the CCCMP consistently demonstrated slightly higher analytical sensitivity than ECCMP and could be due to the marginally higher DNA binding capacity of the former. To the best of our knowledge, this work is the first to report the application of coprecipitation synthesized or electrostatically cross-linked chitosan-coated iron oxide magnetic particles to demonstrate their application in nucleic acid magnetocapture from cell lysate and complex biofluid. It is also probably the first report about the compatibility of the low copies of extracted nucleic acid with both real-time LAMP and real-time PCR. The compatibility of the magnetocapture methods with real-time LAMP and real-time PCR highlighted the potential application of the method with rapid pathogen detection, biowarfare prevention, cell-free nucleic acid detection, genetic disease identification, mutation screening, and digital PCR. Overall, the study eliminates the necessity for sophisticated instruments for magnetic particle synthesis and then demonstrates the utility of high-efficiency magnetocapture of the clinically relevant low copy of nucleic acids. Altogether, we anticipate that this study would help expand NAAT applications in limited-resource settings and democratize nucleic acid-based diagnosis.Experimental
Detailed experimental methods and materials, additional material characterization and spectra, LAMP primer optimization, gel analysis, colorimetric LAMP data, LAMP melt curve analysis, and real-time PCR melt curve analysis have been included in the ESI.†Author contributions
S.T. and S.G. designed the study and have conducted the nucleic acid extraction and NAAT experiments. A.T. provided guidance about clinical perspectives of molecular diagnosis and requirements in limited resource settings. A.K.C. helped analyze the FE-SEM and EDX results. A.S., P.V.R. and G.P. helped conduct the other material characterization studies (XRD, DLS, and zeta potential) of the magnetic particles. G.P. helped analyze the material characterization data and interpret their role in DNA binding and downstream analysis. S.T., S.G., and G.P. wrote the paper. All authors edited and approved the final manuscript.Conflicts of interest
Dr. Souradyuti Ghosh and Mr. Sayantan Tripathy have filed an Indian patent application based on this work.Acknowledgements
The authors are thankful to Mission on Nano Science and Technology, Department of Science and Technology, Government of India for Nanomission grant (ref no DST/NM/NB/2018 /189), and Bennett University scholarship for Mr. Sayantan Tripathy. The authors are grateful to Prof. Rajinder S. Chauhan and Prof. Debajyoti Mukhopadhyay for their constant scholarly support, and Prof. Mrittika Sengupta for her inputs on microbiology. The authors are thankful to the editor and reviewers for helping improve the manuscript through scholarly critique.References
- WHO EMRO, The next flu pandemic: a matter of ‘when’, not ‘if’ | News | Epidemic and pandemic diseases, http://www.emro.who.int/pandemic-epidemic-diseases/news/the-next-flu-pandemic-a-matter-of-when-not-if.html, (accessed May 11, 2021).
- G. T. Keusch, M. Pappaioanou, M. C. Gonzalez, K. A. Scott and P. Tsai, National Research Council (US) Committee on Achieving Sustainable Global Capacity for Surveillance and Response to Emerging Diseases of Zoonotic Origin, National Academies Press, USA, 2009 Search PubMed.
- Institute of Medicine (US) Forum on Microbial Threats, S. Knobler, A. Mahmoud, S. Lemon and L. Pray, The Impact of Globalization on Infectious Disease Emergence and Control: Exploring the Consequences and Opportunities: Workshop Summary, National Academies Press, USA, 2006 Search PubMed.
- C. J. Neiderud, Infect. Ecol. Epidemiol., 2015, 5, 27060 Search PubMed.
- J. A. Patz and M. B. Hahn, in One Health: The Human-Animal-Environment Interfaces in Emerging Infectious Diseases: Food Safety and Security, and International and National Plans for Implementation of One Health Activities, ed. J. S. Mackenzie, M. Jeggo, P. Daszak and J. A. Richt, Springer, Berlin, Heidelberg, 2013, pp. 141–171 Search PubMed.
- V. Tangcharoensathien, M. T. Bassett, Q. Meng and A. Mills, Br. Med. J., 2021, 372, n83 Search PubMed.
- COVID-19 and Rural Communities, https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/other-at-risk-populations/rural-communities.html (accessed May 11, 2021).
- CDC, Nucleic Acid Amplification Tests (NAATs) – CDC, https://www.cdc.gov/coronavirus/2019-ncov/lab/naats.html, (accessed May 11, 2021).
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- S. C. Tan and B. C. Yiap, J. Biomed. Biotechnol., 2009, 2009, e574398 Search PubMed.
- S. A. Thatcher, Clin. Chem., 2015, 61, 89–99 CrossRef CAS PubMed.
- R. Boom, C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen and J. van der Noordaa, J. Clin. Microbiol., 1990, 28, 495–503 CrossRef CAS PubMed.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Angew. Chem., Int. Ed., 2007, 46, 1318–1320 CrossRef CAS PubMed.
- P. T. Trieu and N. Y. Lee, Anal. Chem., 2019, 91, 11013–11022 CrossRef CAS PubMed.
- T. S. Schlappi, S. E. McCalla, N. G. Schoepp and R. F. Ismagilov, Anal. Chem., 2016, 88, 7647–7653 CrossRef CAS PubMed.
- P. Oberacker, P. Stepper, D. M. Bond, S. Höhn, J. Focken, V. Meyer, L. Schelle, V. J. Sugrue, G.-J. Jeunen, T. Moser, S. R. Hore, F. von Meyenn, K. Hipp, T. A. Hore and T. P. Jurkowski, PLoS Biol., 2019, 17, e3000107 CrossRef PubMed.
- A. Arakaki, K. Shibata, T. Mogi, M. Hosokawa, K. Hatakeyama, H. Gomyo, T. Taguchi, H. Wake, T. Tanaami, T. Matsunaga and T. Tanaka, Polym. J., 2012, 44, 672–677 CrossRef CAS.
- K. R. Pandit, I. A. Nanayakkara, W. Cao, S. R. Raghavan and I. M. White, Anal. Chem., 2015, 87, 11022–11029 CrossRef CAS PubMed.
- W. Cao, C. J. Easley, J. P. Ferrance and J. P. Landers, Anal. Chem., 2006, 78, 7222–7228 CrossRef CAS PubMed.
- B. G. Maciel, R. J. da Silva, A. E. Chávez-Guajardo, J. C. Medina-Llamas, J. J. Alcaraz-Espinoza and C. P. de Melo, Carbohydr. Polym., 2018, 197, 100–108 CrossRef CAS PubMed.
- I. A. Nanayakkara, W. Cao and I. M. White, Anal. Chem., 2017, 89, 3773–3779 CrossRef CAS PubMed.
- T. Ahn, J. H. Kim, H.-M. Yang, J. W. Lee and J. D. Kim, J. Phys. Chem. C, 2012, 116, 6069–6076 CrossRef CAS.
- V. Zamora-Mora, M. Fernández-Gutiérrez, J. San Román, G. Goya, R. Hernández and C. Mijangos, Carbohydr. Polym., 2014, 102, 691–698 CrossRef CAS PubMed.
- T. Iwasaki, N. Mizutani, S. Watano, T. Yanagida and T. Kawai, J. Exp. Nanosci., 2010, 5, 251–262 CrossRef CAS.
- A. Zhu, L. Yuan and T. Liao, Int. J. Pharm., 2008, 350, 361–368 CrossRef CAS PubMed.
- A. Díaz-Hernández, J. Gracida, B. E. García-Almendárez and C. Regalado, J. Nanomater., 2018, 2018, 9468574 Search PubMed.
- G. H. Podrepšek, Ž. Knez and M. Leitgeb, Nanomaterials, 2020, 10, E1913 CrossRef PubMed.
- M. J. Hasan, F. A. Petrie, A. E. Johnson, J. Peltan, M. Gannon, R. T. Busch, S. O. Leontsev, E. S. Vasquez and E. E. Urena-Benavides, Cellulose, 2021, 28, 4807–4823 CrossRef CAS.
- D. E. C. Corbridge, Phosphorus: An Outline of its Chemistry, Biochemistry and Uses, Elsevier, 1995, vol. 20, pp. 169–305 Search PubMed.
- D. Zhao, S. Yu, B. Sun, S. Gao, S. Guo and K. Zhao, Polymers, 2018, 10, 462–463 CrossRef PubMed.
- S. Petralia, E. L. Sciuto and S. Conoci, Analyst, 2016, 142, 140–146 RSC.
- S. O. Kelley, ACS Sens., 2017, 2, 193–197 CrossRef CAS PubMed.
- J. Hedman and P. Rådström, Methods Mol. Biol., 2013, 943, 17–48 CrossRef CAS PubMed.
- Y.-P. Wong, S. Othman, Y.-L. Lau, S. Radu and H.-Y. Chee, J. Appl. Microbiol., 2018, 124, 626–643 CrossRef CAS PubMed.
- J. Hill, S. Beriwal, I. Chandra, V. K. Paul, A. Kapil, T. Singh, R. M. Wadowsky, V. Singh, A. Goyal, T. Jahnukainen, J. R. Johnson, P. I. Tarr and A. Vats, J. Clin. Microbiol., 2008, 46, 2800–2804 CrossRef CAS PubMed.
- J. T. Connelly, J. P. Rolland and G. M. Whitesides, Anal. Chem., 2015, 87, 7595–7601 CrossRef CAS PubMed.
- B. R. Bista, C. Ishwad, R. M. Wadowsky, P. Manna, P. S. Randhawa, G. Gupta, M. Adhikari, R. Tyagi, G. Gasper and A. Vats, J. Clin. Microbiol., 2007, 45, 1581–1587 CrossRef CAS PubMed.
- F. Maruyama, T. Kenzaka, N. Yamaguchi, K. Tani and M. Nasu, Appl. Environ. Microbiol., 2003, 69, 5023–5028 CrossRef CAS PubMed.
- N. A. Tanner, Y. Zhang and T. C. Evans, BioTechniques, 2015, 58, 59–68 CrossRef CAS PubMed.
- B. A. Rabe and C. Cepko, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24450–24458 CrossRef CAS PubMed.
- C. A. Gaydos, Expert Rev. Mol. Diagn., 2014, 14, 135–137 CrossRef CAS PubMed.
- M. Sidstedt, J. Hedman, E. L. Romsos, L. Waitara, L. Wadsö, C. R. Steffen, P. M. Vallone and P. Rådström, Anal. Bioanal. Chem., 2018, 410, 2569–2583 CrossRef CAS PubMed.
- Fetal Bovine Serum USA origin, sterile-filtered, suitable for cell culture, suitable for hybridoma | Sigma-Aldrich, http://www.sigmaaldrich.com/, (accessed June 20, 2021).
- CDC Standard Operating Procedure for Creating Viral Transport Media, https://www.cdc.gov/csels/dls/locs/2020/new_sop_for_creating_vtm.html.
- Y. Wang, X. Zhang and G. Yang, RSC Adv., 2015, 5, 29594–29600 RSC.
- F. Ma, Y. Wang and G. Yang, Polymers, 2019, 11, 646 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01150b |
| This journal is © The Royal Society of Chemistry 2022 |