 Open Access Article
Open Access ArticleMix-and-read, one-minute SARS-CoV-2 diagnostic assay: development of PIFE-based aptasensor†
J. Michelle Lee‡
ab,
Chae Rin Kim‡ab,
Sion Kimbc,
Junhong Min d,
Min-Ho Lee*d and
SangWook Lee
d,
Min-Ho Lee*d and
SangWook Lee *be
*be
aDepartment of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139, USA
bPCL, Inc., Rm 701, Star Valley, 99, Digital-ro-9-gil, Ge-umcheon-gu, Seoul, 08510, Republic of Korea. E-mail: swlee@pclchip.com
cCollege of LSA, University of Michigan, Ann Arbor, MI 48104, USA
dSchool of Integrative Engineering, Chung-Ang University, 84 Heukseok-ro, Dongjak-gu, Seoul, 06974, Republic of Korea. E-mail: mhlee7@cau.ac.kr
ee Bio-health Product Research Center, Inje University, Gimhae-si, 50834, Korea
First published on 15th September 2021
Abstract
We developed a one-minute, one-step SARS-CoV-2 antigen assay based on protein-induced fluorescence enhancement of a DNA aptamer. The system showed significant selectivity and sensitivity towards both nucleocapsid protein and SARS-CoV-2 virus lysate, but with marked improvements in speed and manufacturability. We hence propose this platform as a mix-and-read testing strategy for SARS-CoV-2 that can be applied to POC diagnostics in clinical settings, especially in low- and middle-income countries.
Real-time reverse transcription-polymerase chain reaction (RT-PCR) has been the gold standard for SARS-CoV-2 diagnosis, but the urgency of the ongoing COVID-19 pandemic has called for more accessible, less labour-intensive, and faster diagnostic tools, shifting the focus to the development of point-of-care (POC) detection assays.1–7 In particular, the needs for POC tests are more acute in low- and middle-income countries (LMICs) with fragile health systems, yet LMICs rely exclusively on global supply chains for diagnostic kits. To address inequity in pandemic response, there is a need for a POC diagnostics technology that is accessible, generalizable, and adaptable by all countries.8
Various rapid antigen-detection POC diagnostics tests for COVID-19 have been developed using antigen–antibody reactions. They are able to directly detect spike (S) and nucleocapsid (N) proteins of SARS-CoV-2 on-site and allow instantaneous reporting without having to send in patient samples to centralized facilities, enabling faster implementation of strategies to control the viral spread.2,5
Most antigen tests for SARS-CoV-2 detection use a conventional immunochromatographic lateral flow assay (LFA) format with immobilized coated SARS-CoV-2 anti-bodies serving as detecting agents and colloidal gold or other visible dyes as indicators.9–11 However, the colorimetric LFA detection methods often come with lower sensitivity (5 ng μL−1)12 with high false-negative results, proving them clinically insufficient.13,14 LFA format can be coupled with alternative detecting methods such as surface-enhanced Raman Scattering (SERS) to lead to enhanced performance, but detecting SERS requires the use of advanced, specialized and costly equipment.15,16 To overcome these limitations, development of simple yet accurate assay formats is in urgent need. Distance-dependent, liquid-platform fluorescence enhancement technology such as FRET (Fluorescence Resonance Energy Transfer) or PIFE (protein-induced fluorescence enhancement) offers a promising alternative.
FRET read-out relies on the distance-dependent transfer of energy from a donor molecule to an acceptor molecule.17 Using this mechanism, a rapid, simple, homogenous assay for the detection of SARS-CoV-2 N proteins (NP) and S proteins (SP) was developed utilizing two anti-antigen antibodies conjugated with donor and acceptor dye, offering a one-step “mix-and-read” format that does not require washing steps nor a complex detecting machine.18 This significantly simplifies the assay procedure with enhanced sensitivity (2.5 pg μL−1 for NP and 87.5 pg μL−1 for SP), as compared to standard, solid-phase LFA.
However, FRET-based assays require conjugation of fluorescent dyes to two different antibodies, posing a significant deadlock.17–19 Fluorescent tagging of proteins is a labour-intensive process that suffers from low yield and inconsistency, and antibody production involves sacrificing animals, which not only raises ethical issues but also batch-to-batch variability.20,21 Hence FRET-based assays face difficulties in standardization of manufacturing, which confer limitation in repeatability and uniformity of the assay. Moreover, given that FRET is a two-component process that involves the interaction between two fluorophores, there is a complication with crosstalk.22
PIFE is a promising alternative. The excited state of Cy3 can exist either as a trans or cis isomer, with only the trans form having a significant fluorescence quantum yield.23 In close proximity to a protein, Cy3 is preferentially locked into its trans conformational state, resulting in fluorescence enhancement (Fig. 1b).24 PIFE has a higher spatial resolution and specificity than FRET since it detects direct binding of the protein to the DNA, showing a sharp response only within a 0–30 Å range, as compared to 10–100 Å in FRET.17,25
 | ||
| Fig. 1 Protein-induced fluorescence enhancement-based assay for detection of SARS-CoV-2 (a) scheme of our rapid one-step assay (b) principle of PIFE. | ||
As a result, PIFE allows Cy3-labelled aptamer to monitor protein binding in liquid phase with no immobilization or washing steps.20 Compared to antibody-based assays, aptamer-based detection methods offer higher specificity and sensitivity due to the aptamer's high affinity to the target proteins. Aptamers are also cheaper and easier to manufacture and label in larger scale.21,26 Therefore, a Cy3-labelled aptamer can single-handedly act as a biosensor, dramatically simplifying the design of the assay. In our study, we utilized an aptamer that was previously shown to have a high binding affinity (KD = 0.70 nM) toward N protein as our single-component biosensor for N protein detection.27
The specificity and efficiency of binding between SARS-CoV-2 N proteins and N protein aptamers was confirmed with aptamer-equivalent direct ELISA (Enzyme-Linked Immunosorbent Assay) utilizing Cy5-labelled aptamers. Enhancement of fluorescent signal appeared upon adding each four aptamers to N protein-coated wells and washing (Fig. S1, ESI†). No increase of fluorescent signals was observed when the same concentrations of aptamers were added in the BSA-coated wells, confirming the specificity of the aptamers toward NP. Similarly, no increase of fluorescent signals were observed when a scramble aptamer was incubated with NP. Possibility of non-specific protein–nucleic acid interaction between can therefore be excluded. We noted that PCL-Apto3 and PCL-Apto4 showed the highest binding affinity followed by PCL-Apto1 and PCL-Apto2 (Fig. S1a and b, ESI†). PCL-Apto3 was selected for use in further experiments.
We first seeked to optimize the position of Cy3 within the aptamer by testing four different aptamers with Cy3 labelled at four different positions within PCL-Apto3. 20, 40 nM NP and 1 nM of each aptamer were mixed in a 100 μL solution for the induction of PIFE behavior of Cy3. We determined that 5′-labelled Cy3-PCL-Apto3 to be the best labelling position due to its linearity of signal increase and the ease of labelling (Fig. S2, ESI†).
We then explored the kinetics of PIFE by mixing NP with 5′-Cy3-labeled PCL-Apto3. Upon adding 60 nM of 5 μL NP into the 100 μL aptamer solution in the well, the fluorescent signals were measured for 10 minutes. Immediate signal enhancement was observed, and by t = 2 min, maximum fluorescence enhancement is reached (Fig. 2). We also tested dynamics of signal enhancement of two other different concentration of NP (20 and 40 nM). Similar kinetic behaviours were observed, where there is an immediate, concentration-dependent increase in the fluorescence intensity (Fig. S4, ESI†).
The selectivity of this assay was evaluated by investigating changes of fluorescent signal of the buffer solution, BSA, and spike protein (SP) of SARS-CoV-2. Despite adding three solutions in each well to the same final volume and concentration, no fluorescence enhancements were observed for ten minutes (Fig. 2). Curiously, the addition of SP and BSA resulted in a slight decrease of fluorescent signal. This may be caused by the high concentrations of unbound proteins blocking or absorbing fluorescence of the Cy3 dye, but the possibility of non-specific binding can be excluded given the specificity displayed in our aptamer-equivalent ELISA (Fig. S1, ESI†). Overall, our result shows that the assay is highly selective for NP, demonstrating a proof-of-concept of PIFE that originates from specific, tight binding of NP with the aptamer (KD = 0.70 nM) that places Cy3 close to NP.27
For the evaluation of the system's performance over a wide concentration range, we measured signal intensities of aptamer upon the addition of varied concentrations of NP (0 nM, 1 nM, 5 nM, 10 nM, 20 nM, 40 nM) after two minutes when fluorescent signals were saturated. As the concentration of NP increases the fluorescence intensities increase, demonstrating an excellent linear relationship (R2 = 0.93) with an equation of (F − F0)/F0 = 0.017 × CNP (nM) + 0.093 (Fig. 3).
 | ||
| Fig. 3 Normalized fluorescence signal plotted against different concentrations of NP. LOD is 50 fmol of N protein. | ||
Encouraged by the assay's rapid, sensitive detection toward N protein, the assay's sensitivity and selectivity for SARS-CoV-2 viral lysate was examined as a target to optimize the assay to be used in the clinical field. In the same way, 5 μL of prepared virus samples were added into 100 μL of aptamer solution in the wells. As shown in Fig. 4, upon the addition of virus samples, there is an immediate, concentration-dependent increase in the fluorescence intensity for four-fold and forty-fold dilutions of SARS-CoV-2 virus lysate. In contrast, Influenza A and B resulted in negligible signal enhancement. The selectivity of our assay toward SARS-CoV-2 is statistically significant (Fig. S6, ESI,†P = 0.0416, n = 14). It is noteworthy to remark that there was no sample preparation or washing steps, in that all virus lysates had not been purified, which means they contained other viral components.
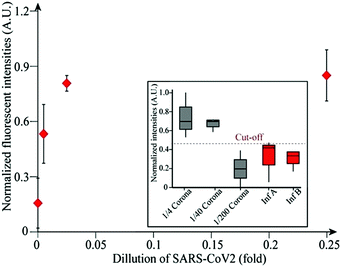 | ||
| Fig. 4 Detection of SARS-CoV-2 virus lysate. LOD is 5.8 × 104 infectious doses [TCID50 mL−1]. Cut-off line was drawn above the highest negative control signal. | ||
To accurately capture the limit of detection of the assay, a pool of N protein samples was mixed with 1 nM of the aptamer at various volumes and concentrations as low as 1 nM of N protein samples. The lowest concentration producing a readily detectable signal was 1 nM of N protein at 50 μL total volume or 50 fmol. The detection limit of the assay was then calculated to be 2.5 ng of N protein or 0.05 ng μL−1. For the detection of SARS-CoV-2, the LOD was estimated to be approximately 5.8 × 104 infectious doses [TCID50 mL−1] (Fig. 4).
This PIFE-based detection system enabled us to reduce the assay time to less than 120 seconds at hundred-fold improved performance (LOD = 0.05 ng μL−1) for the detection of N Proteins as compared to LFA (LOD = 5 ng μL−1).12 The limit of detection of our assay for NP is comparable to FRET-based detection (2.5 pg μL−1), and LOD for SARS-CoV-2 virus (5.8 × 104 infectious doses [TCID50 mL−1]) is also comparable to Ellume's antibody-based antigen detection kit (a 15 minute POCT kit with an LOD at 6.3 × 103 infectious doses [TCID50 mL−1]), approved for commercial use under FDA's Emergency Use Authorization (EUA).7 However, our assay bypasses the need for protein labelling, is much simpler in design (single-component aptasensor) and faster (two minutes), which offers a significant advantage.
There has recently been a push towards POCT, with growing evidence that a frequent POC tests are sufficient, without requiring the analytical sensitivity seen in nucleic acid amplification tests (NAAT).28 There is also a constant, urgent need to expand accessibility of POCT to target a wider global audience. This calls for a simpler design that is scalable and adaptable by many countries, lowering the technological barrier required for manufacturing other assays (e.g., complex conjugation steps). Our work addresses these challenges with a fluorescence-based detection model that detects N protein in liquid-phase, utilizing the principles of PIFE.
The key advantage of liquid-phase assay over solid-phase assay is the speed of interaction between reagents, given that diffusion is much faster in liquid-phase. While traditional fluorescence-based detection methods tend to display poor signal to noise (S/N) levels, PIFE is an established aptamer-based sensing platform that has demonstrated its promise to be highly sensitive and specific, owing to its distance sensitivity.20,29–32
Our assay was specific toward N proteins as well as SARS-CoV-2 virus without any added sample preparation steps. When tested against unpurified influenza A and B viruses, the assay showed no significant signal (Fig. 4). Regardless of the number of viral fragments and proteins present in unpurified virus lysates, our assay responded only to SARS-CoV-2 virus with high signal-to-noise (S/N) ratio. This is a remarkable feat given that liquid-phase assays tend to have high background signal, as biological samples fluoresce between 190 and 650 nm.30
The PIFE-based assay dramatically reduced the complexity of the assay. FRET-based assay requires protein labelling – a significant bottleneck that limits the yield, scale, and consistency of the assay. Lateral flow immunoassay similarly requires conjugation of antibodies to a tag, which is a costly process. In contrast, the only major modification required in our platform is the conjugation of a Cy3 dye to the DNA aptamer, which is a highly standardized process that can be scaled up relatively easily. This is in addition to the simplicity of the assay procedure itself—performing the assay only requires a standard buffer, Cy3-conjugated DNA aptamers, and a simple, low-cost fluorescence reader, and the signal saturates just within two minutes. Therefore, this PIFE-based assay can be developed into a one-step, ultrafast, “mix-and-read” diagnostics kit, where the virus sample is simply mixed with a liquid buffer containing the aptamer for on-site, immediate detection of SARS-CoV-2 viruses.
Our work shows a proof-of-concept that shows how PIFE can be applied as a simple, rapid diagnostics tool for SARS-CoV-2 antigen detection. Despite the remarkable simplicity and speed of the assay, no significant compromise was made on the performance of the assay. In future studies, we aim to further test the system under clinical trials, optimize performances, and ultimately develop this platform as a widely accessible, point-of-care, commercial testing method, especially for use in LMICs.
Another key feature of the PIFE-based platform lies in its generalizability—as long as there is a sequence of an aptamer that specifically binds to a protein, PIFE-based detection system can theoretically be applied for the detection of any proteins. Leveraging this generalizability, we expect the sensitivity of the assay to be further enhanced by multiplexing the detection of antigens, simultaneously utilizing different aptamers that detect different antigens unique to SARS-CoV-2. Moreover, we expect that this PIFE-based aptasensor platform has a potential to serve as a model for a rapid, accessible diagnostics tool for other diseases.
In conclusion, we combined the principles of PIFE with the highly scalable and modifiable properties of DNA aptamers and demonstrated the ability of our aptasensor platform to act as a precise detection system for the N protein of SARS-CoV-2. As a fast, quantitative solution that is relatively easy to manufacture and prepare for on-site use, the proof-of-concept we demonstrated is concurrent with a need to develop a highly accessible diagnostics platform that can be manufactured and widely adopted worldwide to combat the COVID-19 pandemic.
Conflicts of interest
There are no conflicts to declare.Notes and references
- B. D. Kevadiya, J. Machhi, J. Herskovitz, M. D. Oleynikov, W. R. Blomberg, N. Bajwa, D. Soni, S. Das, M. Hasan, M. Patel, A. M. Senan, S. Gorantla, J. McMillan, B. Edagwa, R. Eisenberg, C. B. Gurumurty, S. P. Reid, C. Punyadeera, L. Chang and H. E. Gendelman, Nat. Mater., 2021, 20, 593–605 CrossRef CAS.
- B. Udugama, P. Kadhiresan, H. N. Kozlowski, A. Malekjahani, M. Osborne, V. Y. Li, H. Chen, S. Mubareka, J. B. Gubbay and W. C. Chan, ACS Nano, 2020, 14, 3822–3835 CrossRef CAS.
- Q. Song, X. Sun, Z. Dai, Y. Gao, X. Gong, B. Zhou, J. Wu and W. Wen, Lab Chip, 2021, 21, 1634 RSC.
- H. Y. Mu, Y. L. Lu, T. H. Hsiao and J. H. Huang, Biomicrofluidics, 2020, 14, 061504 CrossRef CAS PubMed.
- D. B. Larremore, B. Wilder, E. Lester, S. Shehata, J. Burke, J. A. Hay, M. Tambe, M. J. Mina and P. Paker, Sci. Adv., 2021, 7, eabd5393 CrossRef CAS PubMed.
- T. Parupudi, N. Panchagnula, S. Muthukumar and S. Prasad, Biotechniques, 2020, 70, 59–68 Search PubMed.
- A. Ghaffari, R. Meurant and A. Ardakani, Diagnostics, 2021, 11, 115 CrossRef CAS PubMed.
- O. Vadenberg, D. Martiny, O. Rochas, A. V. Belkum and Z. Kozlakidis, Nat. Rev. Microbiol., 2021, 19, 171 CrossRef PubMed.
- D. Wang, S. He, X. Wang, Y. Yan, J. Liu, S. Wu, S. Liu, Y. Lei, M. Chen, L. Li, J. Zhang, L. Zhang, X. Hu, X. Zheng, J. Bai, Y. Zhang, Y. Zhang, M. Song and Y. Tang., Nat. Biomed. Eng., 2020, 4, 1150–1158 CrossRef CAS PubMed.
- B. D. Grant, C. E. Anderson, J. R. Williford, L. F. Alonzo, V. A. Glukhova, D. S. Boyle, B. H. Weigl and K. P. Nichols, Anal. Chem., 2020, 92, 11305–11309 CrossRef CAS.
- M. Derakhshan, A. Amani and R. Faridi-Majidi, ACS Appl. Mater. Interfaces, 2021, 13, 14816–14843 CrossRef CAS PubMed.
- A. N. Baker, S. J. Richards, C. S. Guy, T. R. Congdon, M. Hasan, A. J. Zwetsloot, A. Gallo, J. R. Lewandowski, P. J. Stansfeld, A. Straube, M. Walker, S. Chessa, G. Pergolizzi, S. Dedola, R. A. Field and M. I. Gibson, ACS Cent. Sci., 2020, 6, 2046–2052 CrossRef CAS.
- G. C. Mak, P. K. Cheng, S. S. Lau, K. K. Wong, C. S. Lau, E. T. Lam, R. C. Chan and D. N. Tsang, J. Clin. Virol., 2020, 129, 104500 CrossRef CAS PubMed.
- A. Scohy, A. Anantharajah, M. Bodéus, B. Kabamba-Mukadi, A. Verroken and H. Rodriguez-Villalobos, J. Clin. Virol., 2020, 129, 104455 CrossRef CAS.
- H. Chen, S. G. Park, N. Choi, H. J. Kwon, T. Kang, M. K. Lee and J. Choo, ACS Sens., 2021, 6, 2378–2385 CrossRef CAS PubMed.
- S. Yadav, M. A. Sadique, P. Ranjan, N. Kumar, A. Singhal, A. K. Srivastava and R. Khan, ACS Appl. Bio Mater., 2021, 4, 2974–2995 CrossRef CAS.
- L. Wu, C. Huang, B. Emery, A. Sedgwick, S. Bull, X. P. He, H. Tian, J. Yoon, J. Sessler and T. James, Chem. Soc. Rev., 2020, 49, 5110–5139 RSC.
- J. Rusanen, L. Kareinen, L. Szirovicza, H. Uğurlu, L. Levanov, A. Jääskeläinen, M. Ahava, S. Kurkela, K. Saksela, K. Hedman, O. Vapalahti and J. Hepojoki, mBio, 2021, 12, e00902-21 CrossRef.
- B. T. Bajar, E. S. Wang, S. Zhang, M. Z. Lin and J. Chu, Sensors, 2016, 16, 1488 CrossRef PubMed.
- H. Hwang and S. Myong, Chem. Soc. Rev., 2014, 43, 1221–1229 RSC.
- V. Thiviyanathan and D. G. Gorenstein, Proteomics: Clin. Appl., 2016, 6, 565 Search PubMed.
- D. Piston and G. J. Kremers, Trends Biochem. Sci., 2007, 32, 407–414 CrossRef CAS PubMed.
- M. Levitus and S. Ranjit, Q. Rev. Biophys., 2011, 44, 123–151 CrossRef CAS PubMed.
- E. M. S. Stennett, M. A. Ciuba, S. Lin and M. Levitus, J. Phys. Chem. Lett., 2015, 6, 1819–1823 CrossRef CAS PubMed.
- S. Valuchova, J. Fulnecek, A. P. Petrov, K. Tripsianes and K. Riha, Sci. Rep., 2016, 6, 39653 CrossRef CAS PubMed.
- Z. Chen, Q. Wu, J. Chen, X. Ni and J. Dai, Virol. Sin., 2020, 35, 351–354 CrossRef CAS PubMed.
- L. Zhang, X. Fang, X. Liu, H. Ou, H. Zhang, J. Wang, Q. Li, H. Cheng, W. Zhang and Z. Luo, Chem. Commun., 2020, 56, 10235–10238 RSC.
- M. J. Mina, R. Parker and D. B. Larremore, N. Engl. J. Med., 2020, 383, e120 CrossRef CAS.
- S. Umrao, V. Jain, B. Chakraborty and R. Roy, Sens. Actuators, B, 2018, 267, 294–301 CrossRef CAS.
- D. Song, T. Graham and J. Lopar, Nat. Commun., 2016, 7, 2 Search PubMed.
- H. Hwang, H. Kim and S. Myong, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 7414–7418 CrossRef CAS.
- S. Myong, S. Cui, P. V. Cornish, A. Kirchhofer, M. U. Gack, J. U. Jung, K.-P. Hopfner and T. Ha, Science, 2009, 323, 1070–1074 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, information of aptamers and figures. See DOI: 10.1039/d1cc04066a |
| ‡ Both authors are co-first authors. |
| This journal is © The Royal Society of Chemistry 2021 |

