 Open Access Article
Open Access ArticleTowards practical and sustainable SERS: a review of recent developments in the construction of multifunctional enhancing substrates
Chunchun
Li
,
Yiming
Huang
,
Xinyuan
Li
,
Yingrui
Zhang
,
Qinglu
Chen
,
Ziwei
Ye†
,
Zarah
Alqarni
 ,
Steven E. J.
Bell
,
Steven E. J.
Bell
 and
Yikai
Xu
and
Yikai
Xu
 *
*
School of Chemistry and Chemical Engineering, Queen's University Belfast, University Road, Belfast BT7 1NN, UK. E-mail: yxu18@qub.ac.uk
First published on 19th July 2021
Abstract
Surface-enhanced Raman spectroscopy (SERS) is a powerful analytical technique, which allows quantitative detection of chemical species with molecular specificity and single-molecule sensitivity. These useful properties can be further combined with portable Raman spectrometers which allow SERS to be potentially employed at the point-of-care. As a result, SERS has found a wide range of potential applications in both real-life chemical analysis and fundamental mechanistic studies. Despite these advantages, true applications of SERS have been limited due to its high cost, which arises mainly from the fact that SERS relies on expensive single-use Ag/Au enhancing substrates suitable only for the analysis of pure samples. A viable approach to address this issue is to develop multifunctional SERS substrates, which in addition to providing Raman signal enhancement, is armed with other practical functionalities that simplifies the analysis and/or allows the substrate to be regenerated for repeated use. This review gives an account of the recent progress in the fabrication of multi-functional SERS substrates, namely flexible, separation-enhancement-in-one, calibration-enhancement-in-one and regeneration-enhancement-in-one substrates. Specific focus is placed on summarizing and discussing the most widely used strategies to incorporate each type of functionality and their respective advantages and drawbacks. Finally, we present our perspectives on the future challenges and potential opportunities in the development of smart multifunctional SERS sensors for achieving sustainable and wide-spread application of SERS.
1. Introduction
Raman scattering consists of inelastic light scattered from molecular systems, which has energy unique to the structure and scattering intensity proportional to the number density of the probed molecules.1,2 Moreover, since Raman scattering can be directly generated from gas, solid and liquid samples, this makes Raman spectroscopy a versatile and non-destructive technique for both qualitative and quantitative chemical analysis.3 Despite the advantages listed above, wide-spread application of Raman spectroscopy in real-life detection has been limited since normal Raman scattering is intrinsically weak and only occurs for one in every 106–108 scattered photons.1Surface-enhanced Raman scattering is a phenomenon associated with significant amplification of the Raman signals of analytes located near the surface of signal-enhancing materials. The enhancement of Raman signals observed in SERS has been attributed mainly to electromagnetic and chemical enhancements, with the former being widely acknowledged as the dominant factor.4 More specifically, electromagnetic enhancement arises from the amplification of Raman scattering by electromagnetic fields localized on the surface of the enhancing material (also known as localized surface-plasmon resonance, LSPR) owing to the excitation of surface-electron oscillations.5 Chemical enhancement stems from charge-transfer between the analyte molecule and enhancing substrate which leads to a change in the polarizability of the molecule, resulting in the enhancement of its Raman scattering.6 The combination of both enhancing pathways gives SERS significantly improved sensitivity that can reach single-molecule levels while retaining the main advantages of the parent Raman spectroscopy.7,8
Clearly, the most crucial element of SERS is the enhancing substrate and resultingly the tale of SERS has largely been the development of signal-enhancing materials, in particular plasmonic substrates which provide intense electromagnetic enhancement.9–11 In general, the inherent plasmonic properties of Ag and Au make them the most efficient, and thus the most widely applied enhancing materials for SERS.5,12,13 Equally important is the geometry of the substrate material since strong SPR can only be localized and sustained on the surface of nanostructures while the SPR generated on bulk plasmonic metals is evanescent.5,11 Historically, the first class of substrate that satisfied the conditions above were Ag and Au electrodes, which were chemically roughened to carry plasmonic nanostructures, reported nearly 50 years ago.14–17 Studies based on roughened metals led to the establishment of SERS and the fundamentals of the electromagnetic theory.18–21 However, the inconsistency of this crude approach also meant that early SERS enhancement obtained with roughened electrodes were generally weak and plagued by irreproducibility. As a result, one of the main themes in SERS research has been the fabrication of enhancing substrates with improved signal enhancement ability and reproducibility. Now, after several decades of research and benefiting from the immense development in nanotechnology, SERS substrates have evolved from crude roughened electrodes to nanomaterials with tailored morphologies.22–24 More specifically, as shown in Fig. 1, examples of these nanomaterials include lithographically etched nanostructures,25–29 chemically synthesized colloidal nanoparticles30–34 and their multi-dimensional assemblies.35–39 These substrates typically carry well-defined plasmonic hot spots, which enable routine quantitative SERS analysis of a decent range of analytes by SERS specialists in ideal laboratory environments with high reproducibility.40,41 The development in substrate fabrication has also led to the emergence of a small but growing selection of commercial SERS substrates,42–44 which coupled with cheap and convenient portable Raman spectrometers, has helped introduce SERS to the wider scientific community as a promising analytical technique with diverse potential applications, which include environmental monitoring,45,46 bio-analysis,47,48 forensic analysis,49,50 food quality control,40,51 material characterizations,52,53etc.54
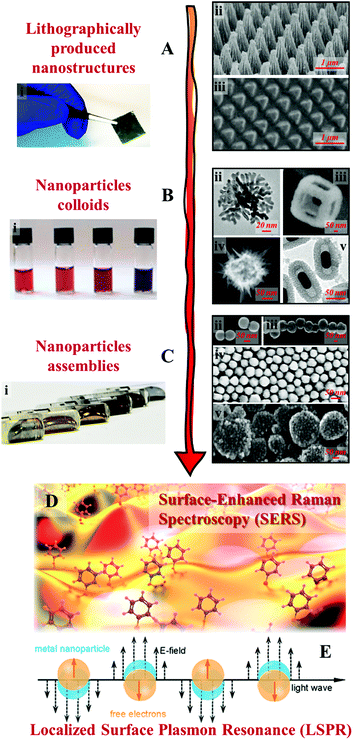 | ||
| Fig. 1 Examples of SERS substrates: (A) solid films carrying lithographically etched nanostructures. Adapted with permission from ref. 26, copyright 2020 Elsevier Ltd and ref. 29, copyright 2016 Tsinghua University Press and Springer-Verlag Berlin Heidelberg; (B) chemically synthesized plasmonic nanoparticle colloids. (ii–v) were adapted with permission from ref. 30, copyright 2010 American Chemical Society; ref. 31, copyright 2015 American Chemical Society; ref. 32, copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim; ref. 33, copyright 2016 The Royal Society of Chemistry; (C) multi-dimensional assemblies of plasmonic nanoparticles. (i–v) were adapted with permission from ref. 35, copyright 2016 American Chemical Society; ref. 36, copyright 2010 American Chemical Society; ref. 37, copyright 2015 American Chemical Society; ref. 38, copyright 2020 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim; ref. 39, copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim; (D) artistic illustration of SERS effect taking place on a nanostructured surface; (E) schematic illustrations of the localized surface plasmon effect on the surface of metal nanoparticles excited by light. Panel (D and E) were reproduced with permission from ref. 40, copyright 2017 American Chemical Society. | ||
Based on these developments, the next grand challenge is to further develop SERS to simplify its application process and enable its routine use by non-specialists. The main roadblock between SERS and actual applications is the difficulty in analysing complex real-life samples, especially in a cost-efficient manner. This challenge stems from the fact that real-life samples typically contain a wide range of chemical species and micro/macro contaminants in addition to the target analyte molecules. These contaminants are often present in much higher concentrations than the analyte and can interfere with the analysis, for example by chemically competing with the analyte for the enhancing surface or physically blocking the probe laser, leading to significantly reduced sensitivity and reproducibility. One method to address this issue is to couple SERS with established purification/separation techniques, such as chromatography and centrifugation.55,56 This has been demonstrated to be effective for applications, which range from pharmacokinetic studies to food-quality analysis, and can be automated to increase sampling efficiency.57–61 However, this approach relies on sophisticated equipment for sample pre-treatment which restrains the analysis to a laboratory setting and having to be operated by trained professionals. Therefore, while combined SERS techniques have seen some success, they are currently limited by their high running cost, as well as their inefficacy in point-of-care analysis. An alternative approach to simplify SERS detection and realize the potential of SERS as a rapid, sensitive and cost-efficient analytical technique for point-of-care analysis is through the construction of smart and sustainable enhancing substrates, which possess practical functionalities in addition to providing Raman signal enhancement.62,63 More specifically, as illustrated in Fig. 2, the multifunctional substrates can be categorized into four main types, namely, flexible substrates, separation-enhancement-in-one substrates, calibration-enhancement-in-one substrates and regeneration-enhancement-in-one substrates. As an important area of research, there are excellent reviews which give comprehensive accounts of the progress that has been made in the fabrication of particular types of multifunctional SERS substrates.64–67 Therefore, this review is not intended to be comprehensive but rather targets to bring together the state-of-the-art research trends in constructing the four main types of multifunctional SERS substrates listed above with the hopes to inspire novel substrate designs which move SERS closer towards real-life practice. More specifically, we discuss research performed within the past five years and highlight the key strategies used for constructing each type of multifunctional SERS substrates, as well as their advantages, effectiveness and practicality. The drawbacks of the current generation of multifunctional SERS substrates are also outlined.
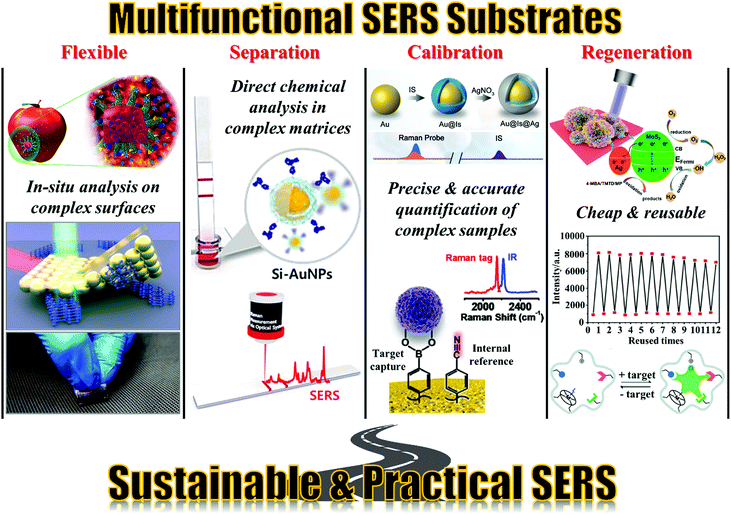 | ||
| Fig. 2 Schematic overviews of the four types of multifunctional SERS substrates discussed in this review. The images in the “Flexible” panel were reproduced with permission from ref. 26, copyright 2020 Elsevier Ltd, ref. 68, copyright 2016 American Chemical Society and ref. 69, copyright 2018 Elsevier Ltd. The images in the “Separation” panel were reproduced with permission from ref. 70, copyright 2020 Elsevier Ltd. The images in the “Calibration” panel were reproduced with permission from ref. 71, copyright 2020 The Royal Society of Chemistry and ref. 72, copyright 2021 American Chemical Society. The images in the “Regeneration” panel were reproduced with permission from ref. 73, copyright 2020 American Chemical Society, ref. 74 copyright 2016 The Royal Society of Chemistry and ref. 75, copyright 2020 American Chemical Society. | ||
2. Flexible SERS substrates for the analysis of chemical residues on complex surfaces
Chemical analysis of surface-residues is of significance in a variety of areas including health and safety, homeland security, forensics, etc. With conventional rigid or colloidal SERS substrates, the analyte must first be extracted from the surface with solvents to form sample solutions which can then be analyzed. This tedious process is not only unsuitable for in-field analysis, but also risks introducing contamination and dilutes the analyte. Although it has been demonstrated that colloidal dispersions or pastes of enhancing nanoparticles can be physically deposited onto complex sample surfaces for in situ analysis,75,76 this approach is unsuitable for analyzing precious samples, such as artworks, or samples where the spatial distribution of the analytes holds important information, such as forensic fingerprint samples.A more ideal approach for analyzing chemical compounds on complex surfaces with SERS is to use flexible enhancing substrates, which are typically composed of exposed enhancing nanoparticles sitting on the surface of flexible and robust polymeric substrate materials. Despite being a relatively young research area, flexible SERS substrates have already been demonstrated useful for the non-invasive detection of chemical compounds, which range from small molecules to biological materials, on a variety of sample surfaces including fruits, textiles and biological tissues.66,77,78 The majority of recent flexible substrates designed for point-of-care analysis fall into two sub-categories, sticky “SERS tapes” and adsorptive “SERS swabs”. SERS tapes are typically flexible and adhesive plastic films, which carry plasmonic nanoparticles on their surfaces. Importantly, the adhesive feature allows SERS tapes to be pressed and peeled from the sample surface to contact and extract analyte molecules for in situ/ex situ analysis. SERS swabs are typically flexible and adsorptive materials, such as cellulose and cotton, which are infused with plasmonic nanoparticles. During analysis, the SERS swab can be used to dab the sample surface to collect chemical compounds for further ex situ analysis. The following section introduces the main strategies for fabricating SERS tapes and swabs, and discusses their performance in analyzing surface residues.
2.1 Deposition of preformed plasmonic nanoparticles
Arguably the most straightforward method to construct flexible SERS substrates is to deposit preformed plasmonic nanoparticle colloids on flexible films.79–83 For example, Huang, Chen, Guo et al. demonstrated that flexible and adhesive “SERS tape” can be conveniently generated by drying droplets of colloidal Au nanoparticles on commercial adhesive tapes, as shown in Fig. 3A.84 More specifically, the SERS tapes were fabricated by drying 25 nm diameter citrate-reduced Au colloids onto a variety of commercial tapes. By using 4-mercaptopyridine as the model analyte, it was determined that SERS tapes fabricated from transparent polypropylene tape containing acrylate adhesive generated the strongest signal enhancement and lowest fluorescent noise, which allowed 4-mercaptopyrine deposited from 10 μL droplets to be detected down to 10−9 M. Despite the simplicity of this fabrication method, the product SERS tape showed decent signal uniformity, with the relative standard deviation (RSD) of the signal intensity measured on ten random spots being less than 15%, as shown in Fig. 3B. Moreover, the authors proposed that the sticky properties of the SERS tape would allow rapid and convenient extraction of analyte molecules from complex surfaces which was successfully demonstrated through the detection of pesticides on the surface of fruits and vegetables, as shown in Fig. 3C. By simply pressing and peeling the SERS tapes on the surface of cucumber peels, the detection limit for parathion-methyl, thiram and chloropyrifos pesticides were determined to be 2.60 ng cm−2, 0.24 ng cm−2 and 3.51 ng cm−2, respectively, which were significantly lower than the maximum residue limits allowed by China and European Union. Importantly, this work pioneered the concept of using sticky SERS tapes for rapid analyte extraction and detection on complex surfaces which has led to an outburst of research based on this approach.81,85,86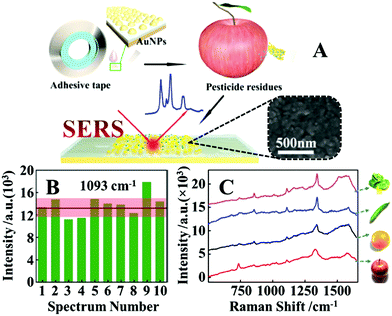 | ||
| Fig. 3 (A) Schematic illustrations of the fabrication of SERS tapes and the extraction of target analytes from fruit peels using SERS tapes. Inset shows the scanning electron microscopy (SEM) image of the Au nanoparticles deposited on the surface of a SERS tape sample; (B) bar chart showing the SERS signal intensity of the 1093 cm−1 vibration band from 4-mercaptopyridine acquired from 10 random sites on a piece of SERS tape; (C) SERS spectra of parathion-methyl extracted from the surfaces of different fruits and vegetables using SERS tape. Adapted with permission from ref. 84, copyright 2016 American Chemical Society. | ||
A major advantage that comes with the colloidal deposition approach is that it can be generally used to create flexible SERS substrates from colloidal nanoparticles with tailored morphologies, which for example include Ag/Au nanorods,87,88 Au bipyramids,89 Ag/Au nanoprisms,79,90 Au@Ag nanospheres,91etc., and therefore improved plasmonic properties. Moreover, the plasmonic nanomaterials can be combined with other materials, such as semiconductors or carbonaceous materials to create SERS tapes carrying hybrid nanomaterials with enhanced functionalities.92,93 For example, Liu, Wang, Ju et al. demonstrated a “three-in-one” SERS tape for rapid sampling, photo-controlled release, and SERS detection of wound infectious pathogens.94 As shown in Fig. 4A the plasmonic component of the SERS tape was constructed by sandwiching a densely packed layer of Au nanostars modified with 4-mercaptobenzoic acid as SERS internal standards between two graphene layers. The outer graphene layers not only improved the biocompatibility and SERS stability of the Au nanostars, but more crucially, acted as a substrate for functionalizing o-nitrobenzyl derivative molecules, which enabled photo-controlled capturing/release of pathogens for SERS analysis (Fig. 4B). As illustrated in Fig. 4C, in a typical analysis, the Au@graphene SERS tape was applied directly to the wound surface to capture infectious pathogens via electrostatic attraction, after which the SERS tape was peeled from the sample and transferred onto the surface of a solid culture medium and irradiated for two minutes with UV light. This led to the photocleavage of the charged o-nitrobenzyl moiety which resulted in the release of the pathogens into the culture medium, where the pathogens were cultivated to a concentration that could be detected by SERS. Impressively, as shown in Fig. 4D, the authors demonstrated that this method allowed simultaneous detection of P. aeruginosa and S. aureus on the burnt wound of mice to be achieved in a significantly shortened analytical period compared to conventional techniques.
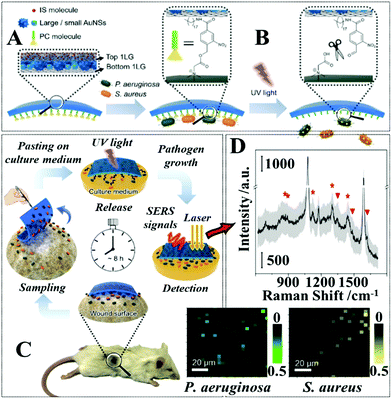 | ||
| Fig. 4 (A) Schematic illustrations of the functional components within the SERS adhesive tape for pathogen capture and release; (B) schematic illustrations of the UV-induced cleavage of the o-nitrobenzyl moiety for the release of captured pathogens; (C) schematic illustrations of the pathogen sampling process using SERS adhesive tape; (D) typical SERS spectrum of the SERS adhesive tape after sampling from 1 × 108 CFU per mL of S. aureus contaminated agar after 4 h of pathogen growth. The light gray region represents the standard deviation of the signal intensity obtained from 60 independent measurements. The red asterisks indicate the characteristic SERS peaks of S. aureus. Adapted with permission from ref. 94, copyright 2019 American Chemical Society. | ||
Similarly, since colloidal deposition can be generally performed on any substrate surface, the substrate materials can also be upgraded to provide enhanced or incorporate new functionalities.95–97 For example, Fan et al. demonstrated a flexible SERS sensor for rapid and in-field detection of the surface residues of a commonly used explosive, 2,2′,4,4′,6,6′-hexanitrostilbene (HNS).98 More specifically, the authors showed that commercially available sticky and flexible gel pads composed mainly of polyurethane could be used to extract HNS from complex sample surfaces through the formation of Meisenheimer-alike complexes between HNS and the amine moieties on the surface of the gel substrate which are visually identifiable as orange taints. Depositing aggregated citrate-reduced Ag nanoparticles directly onto the surface of the orange taints generated intense SERS signals of the analyte–substrate complexes which allowed indirect detection of HNS to be achieved. Importantly, the method was determined to be one of the most sensitive methods for detecting HNS in literature and was successfully utilized for the detection of HNS residues within fingerprints and on the surface of bags with the lowest detectable amounts being 5 and 200 ng, respectively.
While the deposition of colloidal nanoparticles on flexible substrates is a simple and versatile approach to construct flexible SERS substrates from different combinations of plasmonic nanoparticles and substrate materials, it offers little control over the ordering of the deposited nanoparticles and the long-range uniformity of the product enhancing materials due to the “coffee ring effect”, particularly for hydrophobic plastic substrates, which limits the reproducibility and signal uniformity of the product substrate. To combat this, Zhang et al. developed a two-step synthesis to produce SERS tape with a uniform layer of canted Ag nanorods on the surface.99 More specifically, a uniform forest of tilted Ag rods ca. 100 nm in diameter were grown in situ on the surface of Si(100) substrates through glancing angle physical vapor deposition. This nanorod forest was transferred onto the surface of commercial tapes by pressing the sticky side of the adhesive tape onto the nanoparticle layer. The product SERS tape were applied for the detection of benzotriazole, which acted as corrosion inhibitors, on the surface of aluminium films. By immersing the SERS tape in model solutions, the limit of detection of benzotriazole was found to be 1.2 × 10−6 g L−1. Moreover, using this test system, the RSD of the signal intensity acquired from ten random spots on the SERS tape was determined to be within 10%.
Alternatively, Sun et al. demonstrated that SERS tape carrying a highly uniform layer of Au@Ag nanospheres can be obtained with the aid of interfacial nanoparticle self-assembly.100 More specifically, Au@Ag colloidal particles ca. 40 nm in diameter were assembled into monolayer arrays at water–oil interfaces which could be transferred onto the surface of silicon wafers and finally onto the surface of adhesive tape, as shown in Fig. 5A. Since the interfacial self-assembly generated densely packed and uniform nanoparticle arrays which were retained throughout the fabrication process, this allowed the production of SERS tapes with excellent plasmonic activity, uniformity and reproducibility. As shown in Fig. 5B, the limit of detection for thiram on the surface of apples, tomatoes and cucumbers were determined to be 5 ng cm−2, which exceeded the permissive maximum residue limits set by China, European Union and USA. Impressively, by using 4-mercaptobenzoic acid as the probe molecules, the RSD of the signal intensity collected on 48 randomly selected spots from 5 batches of the product SERS tape were measured to be as low as 11.57%, as shown in Fig. 5C.
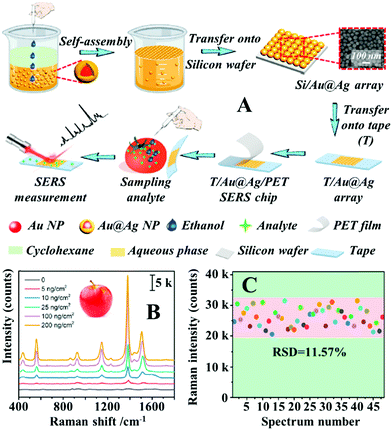 | ||
| Fig. 5 (A) Schematic illustrations of the self-assembly and substrate transfer process for the fabrication of SERS tapes carrying a 2D layer of densely packed Au@Ag nanospheres. Inset shows a SEM image of the 2D nanoparticle array on the surface of the SERS tape; (B) SERS spectra of different amounts of thiram residue extracted from apple peels obtained using SERS tape; (C) bar chart showing the SERS signal intensity of the 1581 cm−1 vibration band from 4-mercaptobenzoic acid acquired from 48 random sites on 5 different batches of SERS tape. Reproduced with permission from ref. 100, copyright 2021 Elsevier Ltd. | ||
Li, Fodjo et al. demonstrated a scalable screening printing technique to produce SERS swabs composed of uniformly deposited citrate-reduced Ag nanoparticles and graphene oxide on cellulose paper, which exhibited excellent SERS signal uniformity.101 Using 4-aminothiophenol as the probe analyte, the RSD of the SERS signals acquired from 20 random sites on the swabs were 3.2%. Using the screen-printed SERS swabs the detection limit of several pesticides including thiram, thiabendazole and metyl parathion on the surface of fruits were measured to be 0.25 ng cm−2, 28 ng cm−2, and 7.4 ng cm−2, respectively, which were comparable to the limit of detection obtained using gas chromatography coupled with mass spectroscopy.
2.2 In situ formation of plasmonic nanoparticles
Another widely researched approach to fabricate flexible substrates for SERS analysis on complex surfaces is to form plasmonic nanoparticles on flexible substrates in situ. The most straightforward way to generate plasmonic nanostructures on substrate materials in situ is through physical vapour deposition (PVD).102,103 Similar to the deposition of preformed colloids, PVD allows a wide selection of substrate materials and convenient control over the total amount of enhancing material introduced onto the substrate surface but in addition, offers greatly improved deposition uniformity compared to depositing preformed colloids. However, this additional advantage comes at the price of having less control over the morphology of the deposited particles. As a result, the product plasmonic materials are typically films of caterpillar like nanostructures, which are formed from several fused spherical particles, with inter-caterpillar hot spots that provide only average plasmonic enhancement. A common approach to regulate the morphology of the nanostructures produced from PVD to enhance their plasmonic performance is to use solid substrates which carry tailored surface morphologies to template the deposition process.26,104–107 For example, Li et al. reported the synthesis of flexible and transparent polyethylene terephthalate (PET) SERS stamps with crossing nanoporous Au structures on their surfaces for detection of thiram and 4-aminothiophenol on fruit surfaces, as illustrated in Fig. 6A.108 To create crossing nanopatterns on the surface of PET stamps, the surface of the PET were first covered with a 500 nm thick layer of SiO2. After this, a layer of inter-crossing carbon nanotubes (CNT) was deposited on the surface of the SiO2. Chemical etching of the CNT covered SiO2 surface led to the exposed parts of the SiO2 being etched away, which resulted in the formation of inter-crossing nano-passageways on the SiO2 surface. Finally, the CNT layer was removed via sonication and the PET films carrying the etched SiO2 surface layer were coated with Au nanoparticles through PVD. Impressively, by using crystal violet as the model analyte the authors showed that the introduction of micropatterns improved the SERS signals obtained from Au PET films by several orders of magnitudes. Moreover, the RSD of the SERS signals of 4-aminothiophenol measured from a total of 81 spots spread over a 2.5 × 2.5 mm2 area was as low as 9.6%, which showed the excellent uniformity of the product films. As shown in Fig. 6B and C, the Au PET films could be pressed directly onto the surface of a tomato sprayed with crystal violet to detect the dye down to 2.9 ng cm−2.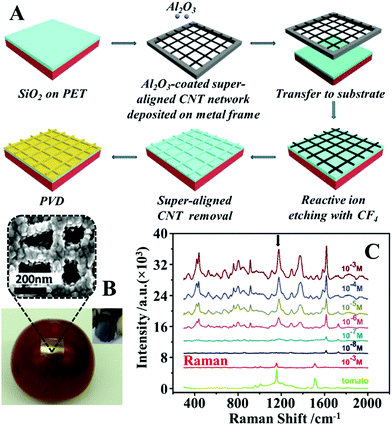 | ||
| Fig. 6 (A) Schematic illustrations of the fabrication of PET SERS stamps with super-aligned CNTs as the template; (B) photograph of a PET SERS stamp pressed on the surface of a tomato during SERS analysis. Inset shows the photograph of a bent piece of PET SERS stamp demonstrating the flexibility of the substrate. SEM image of the plasmonic nanostructure on the surface of the PET SERS stamp; (C) SERS spectra of different amounts of crystal violet residue on the surface of tomatoes obtained with PET SERS stamps. The two spectra at the bottom correspond to the Raman spectra of the bare tomato and the tomato sprayed with 10−3 M of crystal violet solution. Adapted with permission from ref. 108, copyright 2018. The Royal Society of Chemistry. | ||
Although templated PVD allows highly plasmonically active nanostructures to be formed on the surface of flexible substrates in situ with excellent reproducibility and uniformity, the process is generally sophisticated and expensive. An alternative method to generate more plasmonically active nanomaterials on the surface of flexible substrates is to grow the nanoparticles in situ. This method typically involves in situ chemical reduction of noble metal precursors physically dispersed in a flexible and adsorptive matrix, such as paper or textile.109–112 For example, Ge et al. reported a large-scale synthesis of flexible cotton SERS wipes for trace analysis of surface residues.113 The cotton SERS wipes were produced by dipping cotton sheets in AgNO3 solution and then in a solution of ascorbic acid reducing agent, which led to the formation of Ag nanoparticles on the surface of the cotton fibers. The authors showed that the average size and density of the product Ag nanoparticles could be controlled by simply altering the concentration of the metal precursor solution. The optimal concentration led to the formation of a dense layer of Ag nanoparticles with diameters ranging between 40 to 80 nm which allowed two common SERS analytes, p-aminothiophenol and rhodamine 6G to be detected down to 10−12 M and 10−9 M, respectively. However, due to the large size distribution of the Ag nanoparticles, the RSD of the SERS signals of p-aminothiophenol measured from 10 random spots on the substrate was only acceptable, at ca. 20%.
In the quest for improved signal uniformity and plasmonic activity, a number of approaches which combine several of the methods discussed above have been developed.68,114–117 For example, Jiang, Wei et al. developed a flexible polydimethylsiloxane (PDMS) substrate carrying a surface-layer of plasmonic tentacles for multiphase SERS analysis which included the detection of pesticide residues on food surfaces.118 As shown in Fig. 7A, the PDMS enhancing substrates were fabricated by a multistep procedure. More specifically, flexible PDMS films produced by spin-coating were treated with oxygen plasma and then 3-aminopropyltriethoxysilane solution to introduce positively charged surface-groups. Immersing the positively charged PDMS films in a colloidal solution of negatively charged citrate-reduced Au nanoparticles led to the assembly of a uniform monolayer of Au nanoparticles on the surface of the substrate. These Au nanoparticles acted as seeds which allowed a forest of Au nanowires to be formed through in situ chemical reduction in a growth solution containing 3-mercaptobenzoic acid growth directing agent, HAuCl4 metal precursor and L-ascorbic acid reducing agent. Finally, Ag nanoparticles were coated onto the surface of the Au nanowires through PVD to further improve the plasmonic activity of the product films. The enhancing properties of the PDMS films were tested using 4-nitrothiophenol as the model analyte, and the limit of detection for 4-nitrophenol adsorbed from 50 μL of sample solution was determined to be ca. 10−11 M. Moreover, the RSD of the SERS signals of 4-nitrophenol collected from 676 spots within a 500 × 500 μm2 area was 5.38%, which showed that the PDMS films were highly plasmonically active and uniform. As shown in Fig. 7B and C, the PDMS films were pressed directly onto the surface of tomatoes sprayed with methyl parathion solution to quantitatively detect the pesticide from 1 to 10−6 mg mL−1.
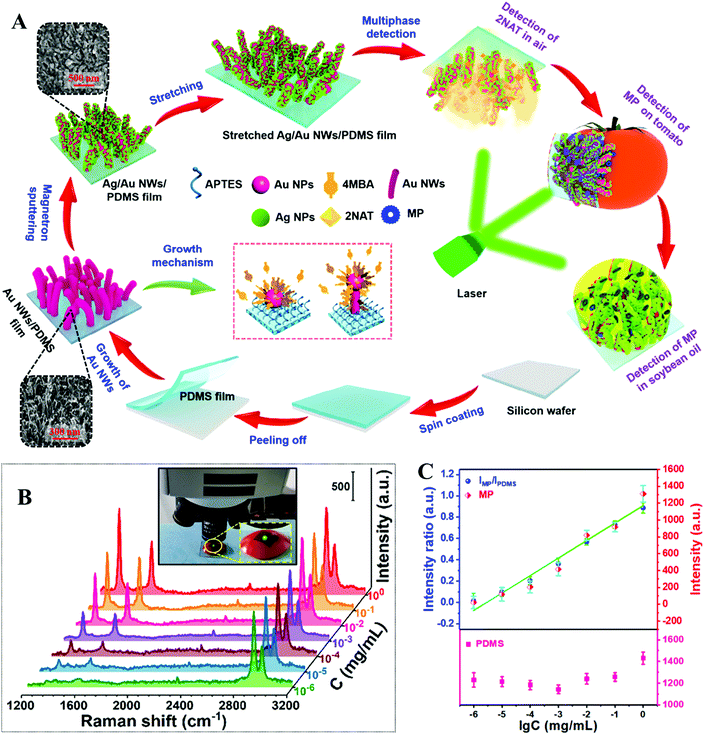 | ||
| Fig. 7 (A) Schematic illustrations of the fabrication of flexible PDMS films carrying a surface-layer of plasmonic tentacles. Inset shows SEM images of the tentacle structures on the surface of the films at different stages of the fabrication process; (B) SERS spectra of different amounts of methyl parathion (MP) residue on the surface of tomatoes detected with the PDMS films. The inset shows the PDMS film pressed on the surface of a tomato sample during SERS analysis; (C) SERS signal intensity of methyl parathion (represented using the 1345 cm−1 vibration band), and the relative intensity ratio between methyl parathion and PDMS (represented using the 2905 cm−1 vibration band) plotted against the log concentration of methyl parathion, respectively. Adapted with permission from ref. 118, copyright 2020 Elsevier Ltd. | ||
2.3 One-pot formation of flexible and plasmonic films
Although methods which combine colloidal deposition and in situ nanoparticle formation allow highly active and uniform layers of plasmonic materials to be formed on flexible substrates, their widespread application is limited by the high complexity of the fabrication procedures. An emerging strategy which allows flexible SERS substrates with uniform and active enhancing layers to be conveniently produced is the self-assembly of nanoparticles at water–oil interfaces.119,120 For two immiscible fluids, their interaction at the interface is purely repulsive. Therefore, mixing an aqueous colloid with an immiscible oil creates a high-energy interface where solid nanoparticles are driven to minimize the interfacial area and, in turn, reduce interfacial energy.120 It is worth noting that quite often plasmonic colloidal nanoparticles are protected by charged capping ligands, which provide interparticle electrostatic repulsion that prevents the formation of tightly packed plasmonic arrays.26 However, this issue can be overcome through the addition of small amounts of chemical compounds to screen or remove the electrostatic charge on the surface of the nanoparticles.26,120 Importantly, the nature of this process means that the self-assembly approach can be generally applied to produce densely packed 2-dimensional arrays from various types of plasmonic nanoparticles despite their morphology and surface chemistry which can then be conveniently transferred onto the surface of flexible substrates.100,114,121 Moreover, the fluid phases can act as the media to introduce polymeric materials which allows the soft liquid-based nanoparticle arrays to be transformed into flexible enhancing substrates for SERS through in situ film formation.122–124 For example, Chin et al. showed that Ag nanocubes can be induced to self-assembled at the water–cyclohexane interface with ethanol to form a densely packed monolayer film.125 Subsequently, part of the cyclohexane phase was replaced with equivalent amounts of PDMS/cyclohexane solution and the entire solution was cured at 65 °C for 2 hours to allow evaporation of the cyclohexane. This led to the formation of a flexible PDMS film, which could be directly handled with a tweezer, with a densely packed monolayer of nanocubes anchored on the surface. By immersing the Ag nanocube@PDMS films in a solution of 4-aminothiophenol, the limit of detection for the model analyte was determined to be 10−7 M and the RSD calculated from the SERS signals collected from 25 randomly selected points on a film was 11.69%. To demonstrate their feasibility as a flexible substrate for SERS analysis of surface-residues, the Ag nanocube@PDMS films were applied for the detection of crystal violet spiked on the surface of fish, which allowed the dye to be detected down to ca. 10−6 M.Similarly, we have shown that densely packed 2-dimensional arrays of plasmonic nanoparticles can be formed at the interface between water and dichloromethane and further converted into flexible polystyrene-based films through evaporation induced polymer deposition.126 However, different from conventional methods, which introduce the polymer solution post-nanoparticle self-assembly, we showed that the polystyrene/DCM solution can be used directly as the oil phase for the formation of densely packed 2-dimensional nanoparticle arrays, as shown in Fig. 8A. Moreover, since dichloromethane is highly volatile even at room-temperature, this allowed large sheets of flexible polystyrene films with a densely packed monolayer of plasmonic nanoparticles anchored on the surface (termed surface-exposed nanoparticle sheets, SENSs) to be produced at room-temperature within minutes. By using thiophenol as the model test analyte, the RSD measured from a total of 20 randomly selected points on two different pieces of SENSs were measured to be as low as 8%, which showed the high reproducibility and uniformity of the product films.127 Since the enhancing hot spots in the SENSs are not only exposed and physically accessible but also firmly anchored onto a robust and flexible plastic substrate, this allowed various solid crystals of nonadsorbing analyte molecules to be pressed physically into the enhancing hot spots in SENSs to generate intense SERS signals for solvent-free analysis (Fig. 8B).69 Using deposited crystals of crystal violet, adenine, trinitrotoluene (TNT) and 3,4-methylenedioxymethamphetamine (MDMA, also known as “ecstasy”) as the model analytes, the limits of detection were determined to be ca. 0.002 ng, 4 ng, 0.129 ng and 0.102 ng, respectively. Moreover, the exposed hot spots in SENSs also allowed direct SERS analysis of layered samples, such as crossing ink lines. Determining the order of crossing ink lines is a significant and long-standing problem in forensic analysis, in particular the identification of altered documents. As shown in Fig. 8C, when normal Raman measurements were performed on dried crossing blue and green ink lines drawn on polystyrene cardboard, the signals from different ink lines did not give sufficient discrimination and were dominated by fluorescence and the Raman signals of the polystyrene cardboard. Conversely, pressing a piece of SENS onto the surface of the same sample enabled intense and distinct SERS signals to be generated from the two ink lines. Moreover, the SERS signals collected at crossing points were identical to the SERS signals of the ink in the uppermost line, which allowed the order of ink lines to be rapidly and confidently determined without damaging the sample. It is worth noting that before this, the analysis of surface-residue with flexible SERS substrates relied on solvents as well as strong chemical interactions between the analyte and enhancing metal to promote analyte adsorption into the enhancing hot spots, which had limited the analysis to a small subset of adsorbing analyte molecules distributed on solvent-inert surfaces. Therefore, the ability to carry out solvent-free SERS with SENSs significantly expands the applicability of flexible SERS substrates for chemical analysis of surface residues.
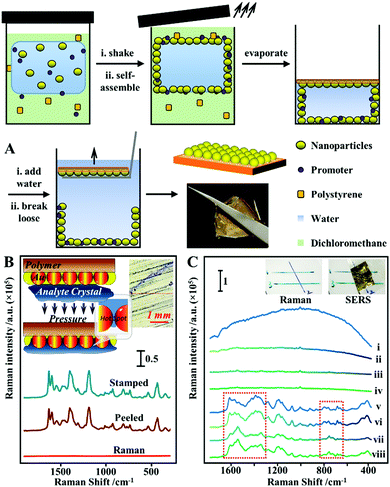 | ||
| Fig. 8 (A) Schematic illustrations of the one-pot fabrication process of surface-exposed nanoparticle sheets (SENSs). The photograph shows a typical piece of Au SENS; (B) spectra set showing the Raman spectrum obtained directly from a sample of crystal violet crystals, the SERS spectrum obtained in situ by pressing a piece of SENS onto the crystal violet sample and the SERS spectrum obtained ex situ from the SENS after it was peeled off the crystal violet sample. Inset shows the schematic illustration of analyte crystals being physically pressured into the plasmonic hot spots in SENSs, and an optical image of the crystal violet sample from the Raman microscope; (C) spectra set showing the application of SENSs for determining the order of crossing ink lines. Spectra (i–iv) correspond to the Raman spectrum of blue ink, blue ink on top of green ink, green ink, green ink on top of blue ink, respectively. Spectra v–viii correspond to the SERS spectra of blue ink, blue ink on top of green ink, green ink, green ink on top of blue ink, respectively, obtained by pressing a piece of SENS onto the same sample. Inset show photographs of the sample. Panel (A) adapted with permission from ref. 126, copyright 2017 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. Panel (B and C) adapted with permission from ref. 69, copyright 2018 Elsevier Ltd. | ||
In summary, this section introduced the three main strategies for constructing flexible SERS substrates for SERS analysis of surface residues. Additional examples from recent literature for each of the aforementioned strategies are summarized in Table 1. In general, the deposition of preformed plasmonic nanoparticles allows the morphology of the plasmonic components to be conveniently adjusted which paves the way for producing flexible SERS substrates with excellent plasmonic properties. Conversely, in situ formation of plasmonic nanomaterials generates plasmonic films with excellent uniformity but are typically less plasmonically active. While the plasmonic activity of the films formed through in situ particle growth can be increased by templating the surface of the substrate material or by growing larger enhancing particles, this normally comes at the price of higher production cost and/or lower substrate uniformity. In this regard, interfacial self-assembly stands out particularly since it produces uniform enhancing surfaces and allows the full range of colloidal nanomaterials to be utilized.
| Substrate | Synthesis | Uniformity (RSD) | Sampling | Sample & LOD |
|---|---|---|---|---|
| Abbreviations: limit of detection (LOD); polydimethylsiloxane (PDMS); nanoparticle (NP); nanowire (NW). | ||||
| Ag nano-triangles decorated cotton fabric79 | Colloid deposition | 12.9–17% (10 points on one substrate) | Swab wet sample for ex situ analysis | Unspecified volume of 10−5 M carbaryl on apples |
| Ag colloidal aggregates decorated cotton swab80 | Colloid deposition | 7.43% (21 points on one substrate) | Swab dry sample for ex situ analysis | 1 ng cm−2 thiabendazole or thiram on bitter gourds |
| Au@Ag nanorods PDMS paper substrate81 | Self-assembly + colloid deposition | 12.6% (400 points in 1 mm2) | Swab wet sample for ex situ analysis | Unspecified volume of 0.5 ppm thiram on orange |
| Lotus seedpod like Ag heterogeneous arrays107 | In situ growth | 10.3–13.8% (Raman mapping over 4900 μm2, number of sampling points unspecified) | Press dry sample for in situ analysis | 10 μL of 10−10 M thiram on apple surface |
| Ag NPs decorated polyurethane sponge109 | In situ growth | 4.49% (20 points on one substrate) | Swab dry sample for ex situ analysis | 0.13 ng (ClO4−), 0.13 ng (ClO3−) and 0.11 ng (NO3−) on aluminum foil |
| Ag@Au NPs polyimide film110 | In situ growth | 8.7% (30 points on one substrate) | Press on and peel from wet sample for ex situ analysis | 10 ng cm−2 thiram apple |
| Ag & Au NPs decorated PDMS nanotentacle arrays68 | Colloid deposition + in situ growth | 5.8% (30 points on five substrates) | Press on and peel from wet sample for ex situ analysis | Thiram (ng cm−2): 1.6 on apple; 10 on cucumber; 10 on grape. |
| Methyl parathion (ng cm−2): 25 on apple; 60 on cucumber; 40 on grape. | ||||
| Malachite green (ng cm−2): 0.4 on apple; 8 on cucumber; 1.6 on grape. | ||||
| Ag NPs@Ag NWs embedded in PDMS115 | Colloid deposition + in situ growth | 4.22% (10 points on one substrate) | Press on and peel from wet sample for ex situ analysis | 0.19 ng cm−2 thiram on leaf; 0.58 ng cm−2 malachite green on live fish |
| Au nanobush PDMS film117 | Colloid deposition + in situ growth | 5.56% (15 points on one substrate) | Press or swab wet sample for in situ or ex situ analysis | 20 μL analyte solution on cherry. |
| Thiabendazole: 0.64 ng mL−1in situ; 3.7 ng mL−1ex situ. | ||||
| Carbaryl: 0.77 ng mL−1in situ; 5.8 ng mL−1ex situ | ||||
| Au NPs decorated micro-hemisphere array PDMS film121 | Self-assembly + colloid deposition | 4.89% (50 points on one substrate) | Press on and peel from wet sample for ex situ analysis | Sample soaked in 2 mL analyte solution and then dried. |
| 10−10 M malachite green on fish; 10−7 M carbendazol or parathion on tomatoes | ||||
| Au NPs polyvinylchloride film123 | One-pot self-assembly | 10.24% (Raman mapping over 1600 μm2, number of sampling points unspecified) | Press on wet sample for in situ analysis | 10 ng cm−2 thiram on apple |
3. Separation-enhancement-in-one SERS substrates for direct analysis of samples in complex matrices
In real-life samples the target analyte is often dispersed within a matrix, most commonly a solvent, with complex chemical and/or physical environments, which interferes with SERS analysis. For example, chemical compounds, such as protein macromolecules in bio-samples, can interfere with SERS analysis by chemically competing with analyte molecules for the enhancing surfaces,128 while the high ionic concentration of sea water samples can interfere with SERS analysis by disrupting the electric double layers that stabilizes colloidal SERS substrates.129 Alternatively, particulates, such as sediments in food samples or sand in river water samples, can interfere with SERS analysis by physically blocking the probe laser and/or the scattered Raman light.130 As discussed above, conventional approaches to analyse samples dispersed in complex matrices typically involves multiple sample clean-up steps using chromatography and/or centrifugation, which are expensive and non-applicable in field. Within this context, a variety of enhancing substrates, which have incorporated mechanisms that allow the Raman signals of analyte molecules to be selectively enhanced, have been developed to enable direct and sensitive analysis of complex real-life samples. These enhancing substrates are typically hybrid materials which include noble metal nanoparticles that provide SERS enhancement and additional functional materials that separate the analyte molecule from the complex sample matrix.45,131–134 Depending on the components within the sample mixture the main strategies used for analyte separation are based on polarity differences, intermolecular interactions, size exclusion, wettability, magnetism or a combination of the several of the above, which are summarized using typical examples in this section.3.1 Magnetism and wettability-based separation
Quite often in SERS analysis, particularly in environmental analysis, the target analyte is dispersed in an aqueous solution with both organic and inorganic macro-particulates which physically interfere with SERS analysis. To analyse these samples without performing tedious sample pre-treatment, magnetic SERS substrates can be dispersed into and then conveniently extracted from sample solutions using magnetic force. For example, a number of research groups have designed hybrid colloidal nanomaterials composed of magnetic nanoparticles and plasmonic noble metal nanoparticles to enable SERS detection of a variety of important analytes dispersed in a range of physically challenging samples, such as honey and hair dye.135–137 To further improve the practicality of the magnetic SERS substrates, we have recently shown a facile one-pot method to generate powders of micro-polymer particles embedded with magnetic nanoparticles and externally covered in a layer of enhancing noble metal nanoparticles, termed nano-micro-particles (NMPs).138 As shown in Fig. 9A, in a typical synthesis, aqueous colloid containing poly(vinylpolypyrrolidone) (PVP) capped Au/Ag nanoparticles was shaken with polystyrene/dichloromethane solution containing micromolar concentrations of promoter ions and magnetic nanoparticles. This led to the formation of oil-in-water Pickering emulsions with a densely packed layer of plasmonic nanoparticles on the surface. Further evaporation of the dichloromethane led to the formation of a solid polystyrene micro-particle core with the magnetic nanoparticles embedded inside and a layer of plasmonically active nanoparticles anchored on the surface. Importantly, the product NMPs can be dried to form free-flowing powders for easy-storage and direct handling. In use, a single magnetic NMP particle can be placed into the sample solution to capture and enrich analyte molecules and then extracted using magnetic force. Using this method, crystal violet could be quantitatively detected down to 5 × 10−7 M in clear river water (Fig. 9B) and 10−6 M in muddy river water, which were several orders of magnitudes better than the limits of detection achieved by conventional aggregated colloids.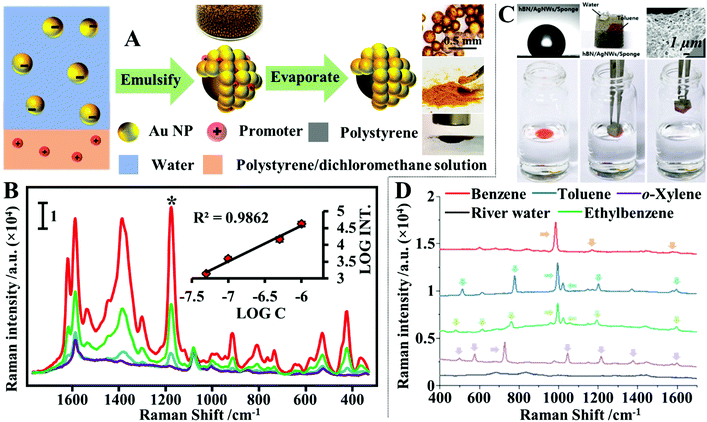 | ||
| Fig. 9 (A) Schematic illustrations of the fabrication of magnetic nano-micro-particles (NMPs). Photograph of Au Pickering emulsions and the product magnetic Au NMPs; (B) SERS spectra of river water samples spiked with varying concentrations of crystal violet, each measured using a single magnetic Au NMP. Inset shows the liner relationship between the SERS intensity of the starred crystal violet vibration band and the concentration of crystal violet; (C) photograph showing the contact angle of a water droplet sitting on the surface of a hydrophobic sponge. Photograph showing the hydrophobic sponge selectively adsorbing a droplet of toluene spiked with Sudan III but not adsorbing a water droplet. SEM image of a fibre coated with Ag nanowires and hexagonal boron nitride nanoparticles in a hydrophobic sponge sample. Photograph set showing the separation of stained toluene from water using a hydrophobic sponge; (D) spectra set demonstrating the simultaneous separation and detection of organic oils in river water using hydrophobic sponges. Panel (A and B) adapted with permission from ref. 138, copyright 2019 Elsevier Ltd. Panel (C and D) adapted with permission from ref. 143, copyright 2019 The Royal Society of Chemistry. | ||
Another type of challenging sample commonly encountered within environmental analysis is inhomogeneous solutions of water–oil mixtures. These samples can be probed directly with superhydrophobic SERS substrates, which separate the water and oil phases so that they can be analysed independently.139–142 More specifically, water and oil mixtures can be effectively separated based on wettability differences using adsorptive materials, such as paper, sponge or textile, which have been rendered superhydrophobic so that they selectively adsorb the oil but not the water to achieve sample separation. For example, Jung, Lee, Kim et al. designed a plasmonic and hydrophobic sponge as a reusable SERS substrate for simultaneous separation and detection of organic pollutants in water.143 The plasmonic and hydrophobic sponges were fabricated by simply dip coating melamine sponges in colloidal solutions of Ag nanowires and hexagonal boron nitride nanoparticles, sequentially, which led to the adsorption of plasmonic Ag nanowires and hydrophobic hexagonal boron nitride nanoparticles on the surface of the sponge fibres. As shown in Fig. 9C and D, this enabled the plasmonic and hydrophobic sponges to selectively attract and adsorb organic solvents mixed within water, which allowed a variety of organic solvents including benzene, toluene, ethylbenzene and xylene to be identified at hundreds-of-ppm levels in river and sea water samples within several seconds using a portable Raman spectrometer.
3.2 Polarity-based separation
A simple and reproducible method that can be used in-field with no specialist equipment to extract and enrich hydrophobic dipolar molecules from complex aqueous samples is liquid–liquid extraction.144–146 For example, Liu, Tan et al. showed that simply shaking fruit juice spiked with thiabendazole with chloroform allowed rapid extraction of the dipolar pesticide from the juice sample into chloroform.147 As shown in Fig. 10A, the separated thiabendazole chloroform solution could be used directly as the oil phased for nanoparticle self-assembly by mixing and shaking it with a colloidal solution of Au nanorods (see Section 2.3 for detail mechanism). This led to the formation of a densely packed and highly plasmonically active 2-dimensional array of Au nanorods at the water–oil interface which allowed thiabenzadole in fresh juice to be quantitatively detected from 104 to 1 ppm. Importantly, the authors demonstrated that the whole process could be completed within 10 minutes using a portable Raman spectrometer which showed the high practicality of the approach. Similarly, Tian, Yang, Li et al. showed that n-butanol can be used to extract creatinine, which is a key biomarker for kidney disease, from artificial urine.148 The separated creatinine m-butanol solution can be mixed with an aqueous solution of aggregated Au nanoparticles which allowed the SERS signals of creatinine in artificial urine samples to be quantitatively detected between 10 mg L−1 to 260 mg L−1 with excellent reproducibility. Importantly, this approach was faster and more sensitive than the analytical technique currently employed by hospitals, which demonstrated the practicality of this novel SERS platform.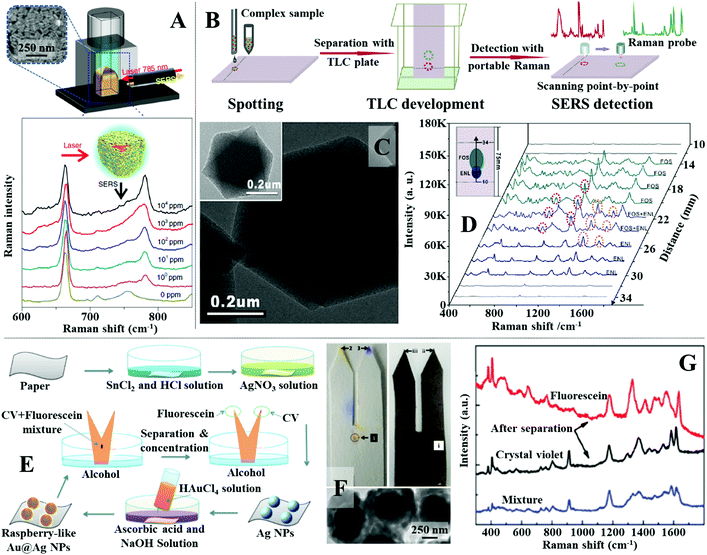 | ||
| Fig. 10 (A) Schematic illustrations of the experimental set-up used for SERS analysis with plasmonic interfacial arrays. Spectra set showing the SERS signals of thiabendazole dissolved at different concentrations in the oil phase obtained using the interfacial nanoparticle arrays as the enhancing material; (B) schematic illustrations of the separation and detection of complex samples using TLC-SERS substrate; (C) transmission electron microscopy (TEM) images of MIL-101 MOF nanoparticles with Au nanoparticles embedded within; (D) SERS spectra set and schematic illustrations showing the separation and detection of FOS and ENL in invisible and overlapping sample points using TLC-SERS; (E) schematic illustrations of the fabrication of fork paper substrates and its sampling process; (F) photograph of a pure paper and a paper substrate coated with Au@Ag nanoparticles used for chromatographic separation of fluorescence and crystal violet. TEM image of the Au@Ag nanoparticles; (G) SERS spectra obtained from the tips and middle of the paper strip after chromatographic separation. Panel (A) adapted with permission from ref. 147, copyright 2018 Springer Nature Limited. Panel (B–D) adapted with permission from ref. 156, copyright 2018 Elsevier Ltd. Panel (E–G) adapted with permission from ref. 163, copyright 2021 The Royal Society of Chemistry. | ||
Although convenient, liquid–liquid extraction requires the polarity of the target analyte to be highly distinct from the contaminant molecules. For more chemically challenging samples, simultaneous separation and analysis of each of the individual chemical components can be achieved using thin layer chromatography (TLC)-based SERS approaches. The most straightforward method to couple TLC with SERS is to deposit colloidal nanoparticles directly onto the separated sample spots on the TLC plate. This method has been demonstrated useful for quantitative SERS detection of a variety of analytes, including aflatoxins, pesticides and herbal extracts, dispersed within a wide range of complex real-life samples, including paint, cooking oils and mouldy food.149–154 However, colloidal deposition is a dynamic process, which leads to time-dependent SERS responses, and the product deposits are generally non-uniform, which results in irreproducible measurements.155,156 Moreover, successful deposition of the enhancing colloid onto the sample requires knowledge of the positions of the separated sample spots on the TLC board. This makes analysing overlapping or invisible sample spots, which is a commonly occurring phenomenon in TLC separation, extremely challenging using simple colloidal deposition. To tackle this issue and further improve the practicality and sensitivity of TLC-SERS methods, multifunctional SERS substrates, which combine TLC and Raman enhancement in-one have been developed.61,157,158 For example, Lu, Li et al. developed a TLC-SERS-in-one substrate which contained a mixture of SiO2 microparticles and MIL-101 metal–organic framework (MOF) microparticles incorporated with Au nanoparticles, as shown in Fig. 10B and C.156 In use, the SiO2 microparticles acted as the solid phase for TLC while the Au-modified MOF microparticles provided Raman enhancement. Using rhodamine 6G as the model analyte, the authors showed through contrast experiments that the TLC-SERS-in-one substrate allowed faster sampling and stronger SERS signals to be obtained compared to the conventional approach of depositing colloidal nanoparticles onto commercial TLC plates. The improved functionality of the multifunctional substrate was attributed to the difference between the physical arrangement of the Au enhancing nanoparticles dispersed within MOFs and deposited directly from colloids. To demonstrate the advantage of the TLC-SERS-in-one substrate over the conventional method of depositing colloids on TLC plates, direct SERS analysis of chemical compounds in botanical dietary supplements were performed. It was shown that performing Raman scanning across the surface of the TLC-SERS-in-one substrate allowed fosinopril sodium and enalaprilat, which generated overlapping and invisible sample spots, to be successfully identified (Fig. 10D).
In addition to TLC plates, paper has also been demonstrated as an effective substrate for separating complex samples for SERS analysis.159–162 For example, Man, Leng et al. designed a fork-shaped paper substrate functionalized with Au@Ag raspberry nanoparticles for separation and SERS detection of mixtures of charged dyes.163 The fabrication and application of the substrate is illustrated in Fig. 10E. The fork paper substrates were functionalized with raspberry-like Au@Ag nanoparticles, as shown in Fig. 10E and F, through multiple steps of in situ chemical growth. Importantly, the tips of the fork paper substrates were functionalized with positively-charged poly(allylamine hydrochloride) and negatively charged poly(styrenesulfonate), respectively, which allowed mixtures of oppositely charged dyes to be separated and concentrated onto different fork tips for SERS analysis, as shown in Fig. 10F and G. The practicality of the fork paper substrate was demonstrated through the analysis of malachite green in fishpond water with fluorescein as the contaminant. Impressively, it was shown that nearly all the malachite green could be separated from the sample mixture and concentrated onto the tip of the substrate, which allowed its detection down to 1 pM concentration.
3.3 Intermolecular interactions-based separation
The separation of chemical compounds within complex sample matrices can also be achieved by modifying the surface of the enhancing nanomaterials with a thin layer of functional materials.131–134 More specifically, these functional materials enable the separation of the analyte molecules from the bulk sample and localize them near the enhancing surface through intermolecular interactions, such as electrostatic, hydrophobic, hydrogen bonding, π–π, van der Waals or a combination of several of the above. Arguably, the most widely employed class of functional material for surface modification of SERS enhancing substrates is the self-assembled monolayer (SAM) of small organic molecules.164–166 For example, Ling et al. designed a “SERS taster” for profiling wine flavours by harnessing vibrational information of the intermolecular interactions between flavour molecules and SAMs of organic chemical receptor molecules functionalized on the surface of plasmonically active Ag nanocube films, as illustrated in Fig. 11A.167Fig. 11B shows the four different types of chemical receptors used for the analysis. Apart from the bare Ag nanoparticle receptor which provided SERS information by interacting directly with the flavour molecule, the other receptors were small molecules, which identified flavour molecules through intermolecular interactions that generated spectral changes. Importantly, each receptor was carefully designed so that they interacted with a different functional moiety of the same flavour molecule through a different type of intermolecular interaction. Therefore, combining the spectral changes generated by the same flavour molecule on different receptors allowed a complete chemical profile, termed “SERS superprofile”, of the analyte to be obtained, as shown in Fig. 11A and C. Finally, the authors showed that the SERS superprofiles can be analysed using principle-component analysis (PCA), which allowed quantitative multiplex profiling of five different wine flavour molecules to be achieved at parts-per-million levels with ≥95% accuracy.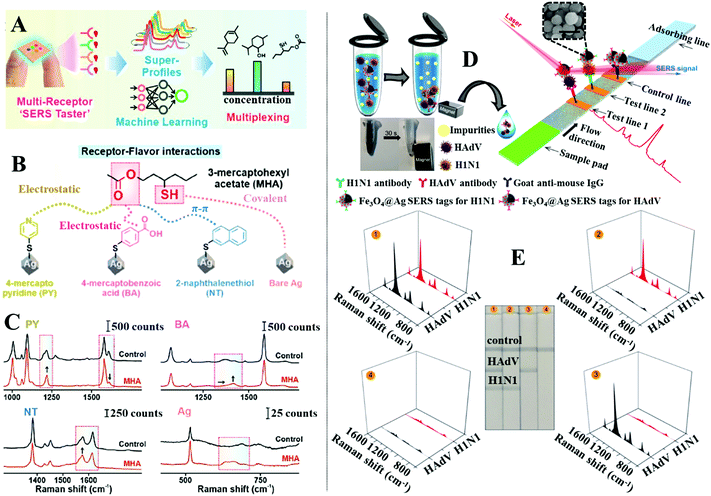 | ||
| Fig. 11 (A) Schematic illustrations of the working principles of the “SERS taster”; (B) schematic illustrations of the different types of intermolecular interactions between different receptors and the same flavour molecule; (C) spectra set showing the spectral changes induced by intermolecular interactions between different receptors and the flavour molecule 3-mercaptohexyl acetate; (D) schematic illustrations of the working principles of the SERS lateral flow assay platform. Photograph demonstrating the magnetic properties of the SERS active magnetic nanoparticles. SEM image of a typical batch of the SERS active magnetic nanoparticles. The scale bar in the SEM image corresponds to 250 nm; (E) photographs of the lateral flow assay SERS platform treated with samples, which contained (1) 105 pfu per mL of HAdV and 105 pfu per mL of H1N1, (2) 0 pfu per mL of HAdV and 105 pfu per mL of H1N1, (3) 0 pfu per mL of HAdV and 0 pfu per mL of H1N1, (4) 105 pfu per mL of HAdV and 0 pfu per mL of H1N1, respectively. Spectra sets showing the corresponding SERS signals obtained from each paper strip. Panels (A–C) adapted with permission from ref. 167, copyright 2021 American Chemical Society. Panel (D and E) adapted with permission from ref. 175, copyright 2019 American Chemical Society. | ||
The surface of SERS enhancing nanomaterials can also be functionalized with a self-assembled monolayer of biomolecules, in particular aptamers and antibodies, which selectively capture target analytes mixed within complex samples.168–171 To introduce the sample and facilitate target recognition the functionalized enhancing particles are typically used in combination with carefully designed paper substrates to form separation-enhancement-in-one lateral flow assay platforms.134,172–174 For example, Wang and Wang et al. designed a lateral flow assay platform which utilized SERS active magnetic nanoparticles for rapid simultaneous detection of two respiratory viruses, influenza A (H1N1) and human adenovirus (HAdV), in real biological samples, as illustrated in Fig. 11D.175 The enhancing nanoparticles were prepared through a multistep process to incorporate magnetic, plasmonic and target recognition functionalities. More specifically, a solvothermal reaction was used to prepare magnetic Fe3O4 nanoparticles ca. 120 nm in diameter. The surface of the Fe3O4 nanoparticles were modified with positively charged polyethyleneimine (PEI) to attract negatively charged Au nanoparticles ca. 3 nm in diameter. This led to the formation of Fe3O4@Au core–shell satellites, which were treated with 5,5-dithiobis-(2-nitrobenzoic acid) (DTNB) to modify the surface of the Au nanoparticles with DTNB SERS tags. After this a continuous Ag shell was formed on the outside of the Fe3O4@Au satellite particles through in situ chemical reduction to produce Fe3O4@Au@Ag nanoparticles with an encapsulated SAM of DTNB. Finally, the surface of the Fe3O4@Au@Ag nanoparticles was modified with a mixed layer of DTNB and capture antibodies to produce magnetic SERS tags that bound selectively to matching viruses within bio-samples. In use, the magnetic SERS tags were dispersed directly into biofluids to capture H1N1 and HAdV viruses. After the magnetic SERS tags complexed with viruses, they were magnetically separated from the bulk sample to form the final sample solution. Applying the final sample solution to the sample pad on one end of the paper substrate allowed the magnetic SERS tags complexed with viruses to be carried to different test lines on the paper substrate by lateral flow. Since the test lines were functionalized with antibodies which bound specifically to a matching virus or antibody, this allowed the bare magnetic SERS tags and SERS tags complexed with H1N1 or HAdV to be selectively captured on different test lines for SERS analysis. The concentration of each of the viruses could be indirectly determined by measuring the SERS signal intensity of the DTNB tags on the corresponding test lines, as shown in Fig. 11E. This allowed H1N1 and HAdV to be quantitatively detected down to 50 and 10 pfu per mL, respectively, which were more than 2000× more sensitive than standard lateral flow assays based on simple colloidal Au nanoparticles.
In addition to molecular monolayers the enhancing surface can also be functionalized with thin layers of molecularly imprinted polymers (MIPs) to facilitate target capture within complex sample matrices.132,176,177 MIPs are often described as the synthetic analogues to antibody–antigen systems, since they function through a similar “lock and key” mechanism to selectively capture the template molecule used during MIP production, as illustrated in Fig. 12A.178 For example, Liu, Yang et al. designed a MIP-based magnetic SERS substrate for highly selective quantitative SERS analysis of paclobutrazol pesticide residues in complex environments.75 As shown in Fig. 12B, the SERS substrates were fabricated by assembling a uniform layer of Au@Ag nanoparticles with 12 nm interparticle spacing on the surface of Fe3O4@SiO2 microparticles, which were then encapsulated within a layer of MIP to form the final (Fe3O4@SiO2–Au@Ag)@MIP (FSAA@MIP) microparticles. As shown in the inset of Fig. 12B, transmission microscopy analysis of the FSAA@MIP microparticles revealed that the MIP was uniformly coated as a ∼15 nm thick shell on the surface of the FSAA particles. This was significantly thinner compared to other MIP-based SERS substrates previously reported in literature which allowed the SERS activity of the noble metal nanoparticles to be fully retained. In use, the FSAA@MIP microparticles could be incubated with soil samples spiked with paclobutrazol and then extracted conveniently with magnetic separation, which allowed the pesticide to be quantitatively detected down to 1.5 ng. As shown in Fig. 12C, since the MIPs bound selectively to paclobutrazol, no interference was observed within the SERS signals even when the soil samples were also spiked with other organic pollutants including thiram, malachite green and methylene blue, which typically adsorbed strongly to noble metal nanomaterials. To demonstrate the versatility of the FSAA@MIP as a practical SERS substrate, the authors demonstrated that they could be sprayed directly onto the surface of a mango or incubated with river water and then collected with magnetic force to detected paclobutrazol.
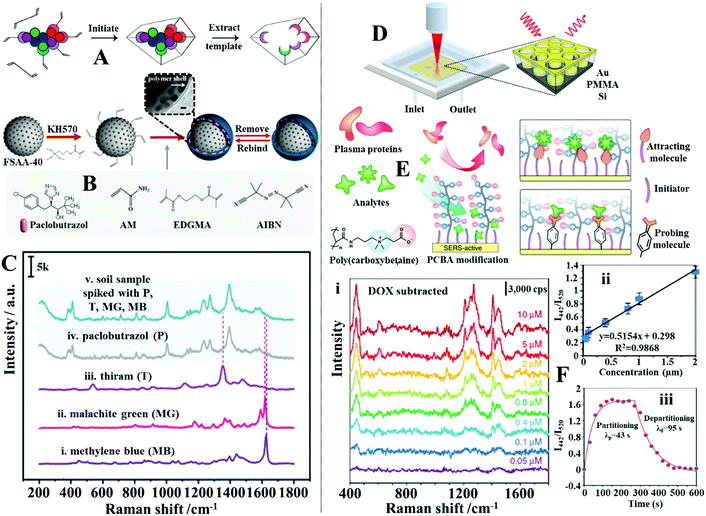 | ||
| Fig. 12 (A) Schematic illustrations of the working principles of molecular imprinted polymers (MIPs); (B) schematic illustration of the preparation of Fe3O4@SiO2–Au@Ag (FSAA) MIPs. Inset shows a TEM image of the product FSAA MIPs; (C) SERS spectra set demonstrating the selectivity of the FSAA MIP sensors. Spectra (i–iv) correspond to the SERS signals of the target analyte paclobutrazol and three contaminants, namely, thiram, malachite green and methylene blue. Spectrum v corresponds to the SERS signals obtained from a soil sample spiked with all four chemicals; (D) schematic illustrations of the SERS optofluidic system; (E) schematic illustrations of the different types of mixed self-assembled monolayers functionalized on the surface of the enhancing material to achieve anti-fouling and target recognition; (F) panel (i) show SERS spectra of Doxorubicin in plasma ultrafiltrate control at concentrations ranging from 0.05 to 10 mM. The background signals from plasma have been subtracted. Panel (ii) shows the linear region of the Doxorubicin detection curve generated by plotting the average intensity ratio of the doxorubicin peak at 442 cm−1 and plasma peak at 520 cm−1 as a function of doxorubicin concentration. The error bars stand for standard deviations obtained from three replicates. Panel (iii) shows partitioning and departitioning kinetics of 20 mM Doxorubicin in plasma on the anti-fouling SERS sensor. Panel (A) reproduced with permission from ref. 178, copyright 2017 American Chemical Society. Panel (B and C) adapted with permission from ref. 75, copyright 2020 American Chemical Society. Panel (D–F) adapted with permission from ref. 182, copyright Springer Nature Limited. | ||
Alternatively, polymeric macromolecules, such as oligo(ethylene glycol) or zwitterionic polymers can also be used to repel contaminants from the enhancing surface to enable direct SERS analysis of bio-samples.179–181 For example, Jiang, Yu et al. designed an optofluidic system functionalized with antifouling mixed SAMs which allowed quantitative SERS analysis of a variety of clinically relevant molecules in plasma to be achieved by flowing the drug containing plasma samples through the SERS enhancing channels, as shown in Fig. 12D.182 More specifically, the enhancing surface was first modified with a SAM of mercaptoundecyl bromoisobutyrate “initiator”, which were used to graft chains of antifouling zwitterionic poly(carboxybetaine acrylamide) to form a mixed monolayer of initiators and zwitterionic polymers, as shown in Fig. 12E. Using this set-up, it was demonstrated that a strongly adsorbing anticancer drug, doxorubicin, could be quantitatively detected between 0.05 and 2 μM in plasma, which covered most of its clinically relevant concentration region (Fig. 12F(i and ii)). Moreover, it was shown that the sensor could be recovered in 95 seconds by fluxing the doxorubicin-complexed enhancing surface with pure plasma to facilitate analyte desorption (Fig. 12F(iii)). To demonstrate the versatility of this approach, the authors showed that the initiator within the mixed SAM could be replaced with “attracting molecules” (such as 3-mercaptopropionic acid and 1-propane-thiol) or “probing molecules” (such as 4-mercaptobenzoic acid and 4-mercaptophenylboronic acid) to enable SERS detection of weakly adsorbing drug molecules (such as amitriptyline hydrochloride and carbamazepine) or indirect SERS detection of bio-analytes with small Raman cross-sections (such as fructose) in plasma, respectively.
3.4 Size-exclusion-based separation
Another feasible approach to separate small analyte molecules from micro/macro-contaminants is through size exclusion. For example, to remove the interference of macro-particulates in liquid samples, plasmonic nanomaterials can be introduced on the surface of commercial filtering materials, such as filter paper, to construct multifunctional SERS substrates that allow sample filtration and SERS enhancement.130,183,184 For more challenging samples, such as bio-fluids which contain polymeric protein molecules that compete with the analyte for enhancing surface, the SERS substrate can be functionalized with molecular sieves to preclude the micro-sized contaminants from the enhancing surface (Fig. 13A).185–187 For example, Yang et al. showed that colloidal Au nanorods@metal organic frameworks (MOFs) could be used as the SERS substrate for direct detection of small molecules in whole blood.188 In their work, a simple wet-synthesis was used to produce Au nanorods@zeolitic imidazolate frameworks-8 (Au@ZIF-8), as shown in Fig. 13B. Since the ZIF-8 shell were measured to be ca. 143 nm thick, it was found that they prohibited the formation of SERS hot spots between Au nanorods. Regardlessly, drying a droplet of the Au@ZIF-8 nanoparticles on a slippery polydimethylsiloxane (PDMS) surface allowed sufficient enrichment of the enhancing particles, which enabled a test analyte, 4-nitrobenzene thiol, to be detected down to 0.1 nM. To test the ZIF-8 functionality as a molecular sieve, the Au@ZIF-8 nanoparticles were dried with 10 μL of an analyte solution containing a mixture of 4-nitrobenzene thiol, 1-naphthalene thiol and rhodamine 6G. Since the aperture size of the ZIF-8 were ca. 3.4 Å, which were significantly smaller than the hydrodynamic diameter of rhodamine 6G (10 Å), only the SERS signals of 4-nitrobenzene thiol and 1-naphthalene thiol were observed. To demonstrate the practicality of the Au@ZIF-8 nanoparticles, they were dried with whole blood spiked with 10 nM of 4-nitrobenzene thiol. As shown in Fig. 13B–D, the SERS signals acquired from Au@ZIF-8 nanoparticles only contained the vibrational bands of 4-nitrobenzene thiol while strong interference signals were observed in contrast experiments performed with plain Au nanorods. Alternatively, the enhancing nanoparticles can also be encased in nano-porous polymers to achieve size-selective SERS detection. For example, Kim, Kim et al. designed a microfluidic synthesis to produce colloidal dispersions of charged microgels containing Au nanoparticle agglomerates for direct SERS analysis of toxic chemicals in eggs.189 The microgels were not only nano-porous but could also be functionalized with either acrylic acid (−) or acrylamide (+) functional groups to render them charged, which allowed the microgels to attract oppositely charged target molecules while rejecting similarly charged contaminants and any macromolecules with hydrodynamic diameters larger than the mesh size, as shown in Fig. 13E. This allowed fipronil sulfone, which is a toxic metabolite, to be directly detected in fresh eggs with SERS using the charged microgels without any pre-treatment of the egg. As shown in Fig. 13F, the limit of detection of fipronil sulfone in eggs was determined to be 0.05 ppm which was comparable to the maximum residual level of 0.02 ppm designated by the United States Environmental Protection Agency.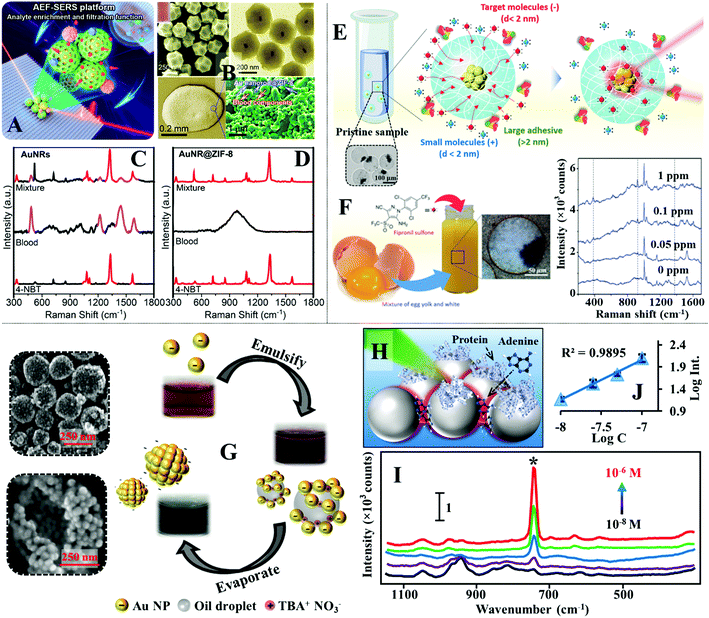 | ||
| Fig. 13 (A) Schematic illustrations of SERS detection performed by drying Au nanorods@MOF (AuNR@ZIF-8) enhancing particles on a slippery surface; (B) SEM and TEM images of the AuNR@ZIF-8 particles. Inset shows a photograph of the AuNR@ZIF-8 colloid. Optical and SEM images of an aggregate composed of dried AuNR@ZIF-8 particles and complex blood components on the slippery surface; (C and D) SERS spectra of whole blood and whole blood spiked with 10 nM of 4-nitrobenzene thiol obtained with AuNRs and AuNR@ZIF-8 particles as the SERS substrates; (E) schematic illustrations of the working principles of SERS detection using charged microgels incorporated with Au nanoparticle agglomerates. Inset shows an optical microscopy image of the charged microgels containing Au agglomerates; (F) schematic and photograph showing positively charged microgel suspended in a raw egg sample spiked with fipronil sulfone. SERS spectra of varying concentrations of fipronil sulfone in eggs measured using charged microgels; (G) schematic illustrations of the formation process of stable plasmonic aggregates. The photographs show the evolution of the colloid at different stages of the fabrication process. SEM images of the stable plasmonic aggregates; (H) schematic illustration of proteins being excluded from the interparticle hot spots due to steric congestion; (I) SERS spectra of varying concentrations of adenine in artificial serum obtained with stable plasmonic aggregates; (J) the corresponding calibration curve for adenine concentration versus the SERS signal intensity of the vibration band at 731 cm−1. Panel (A–D) adapted with permission from ref. 188, copyright 2020 American Chemical Society. Panel (E and F) adapted with permission from ref. 189, copyright 2018 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. Panel (G–I) adapted with permission from ref. 39, copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
For many commonly used plasmonic nanoparticles, such as nanospheres and nanocubes, the maximum plasmonic enhancement is achieved at interparticle nano-junctions, which are only several nanometres wide.190 Therefore, it is also possible to construct SERS enhancing substrates which contain plasmonic nano-junctions that act simultaneously as SERS “hot spots” and molecular sieves.191,192 For example, we have recently demonstrated an emulsion-templated method to produce stable quasi-spherical Au/Ag colloidal aggregates, as shown in Fig. 13G.39 Since the plasmonic nanoparticles within each aggregate were packed tightly to form nano-junctions which precluded the adsorption of protein macro-molecules, this allowed the colloidal aggregates to be used directly as the SERS substrate for quantitative detection of adenine and phenytoin in artificial serum. Moreover, since the protein molecules were limited to adsorb only onto the less-enhancing outer surface areas of the nanoparticles, this in turn enriched the adenine molecules within the plasmonic hot spots, as illustrated in Fig. 13H, which led to a 3× SERS signal increase compared to contrast experiments performed without protein. As shown in Fig. 13I and J, the limit of detection of adenine in artificial serum was determined to be 10−8 M, and the linear quantification range was determined to be between 10−8–10−7 M.
Additional examples of each of the aforementioned strategies from recent literature are provided in Table 2. In summary, this section introduced the four main strategies to produce separation-enhancement-in-one SERS substrates for direct and rapid SERS detection of analytes dispersed in complex sample matrices. In general, the choice of separation mechanism is greatly dependant on the composition of the sample i.e., the physical and chemical properties of the analyte molecules and contaminants. Magnetic and wettability separation enables the extraction of analyte molecules from physically complex sample matrices but are not effective for separating analytes from contaminants which compete for the enhancing surface. In this regard, competitively adsorbing macro-contaminants, such as protein can be physically excluded from the enhancing surface with size-exclusion provided by nano-porous functional materials, which has been demonstrated useful for SERS analysis of bio-samples, such as serum. If the identity of the target and/or contaminant is known, direct SERS detection can also be achieved via target-specific intermolecular interactions, which capture the analyte molecules, or via antifouling layers, which selectively reject the contaminants. Alternatively, complex samples can be treated using liquid–liquid extraction, which is a method that allows certain analytes to be conveniently extracted from the sample matrice. This method also has the potential to allow analyte enrichment by developing SERS methodologies based on single-drop microextraction.193–196 For a more thorough analysis, the samples can be treated with TLC-based SERS substrates, which separates the mixture into individual components based on polarity differences allowing SERS analysis of each of the chemical species within the sample.
| Substrate | Separation strategy | Sample & challenge | Target analyte | Performance in practice | Performance with model solution |
|---|---|---|---|---|---|
| Abbreviations: limit of detection (LOD); nanoparticle (NP); 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (4,4′-DDT); graphene oxide (GO); 4-anilino-N-phenethyl-piperidine (4-ANPP); poly(ethylene glycol) thiolate (PEG-S); metal organic framework (MOF); molecular imprinted polymer (MIP). | |||||
| Fe3O4@GO@Ag136 | Magnetic enrichment & separation | Honey: viscous food matrix containing 77% sugars and several other minor components | Chloramphenicol | Demonstrated 1.0 × 10−8 M | LOD: 1.0 × 10−10 M |
| Au/Ag nanostar decorated superhydrophobic–superoleophilic polystyrene membrane142 | Wettability | Artificial water–oil mixtures: insoluble oil presents challenges to the analysis; the oil and dissolved dyes interferes with SERS | Rhodamine 6G | LOD: 10−14 M | Unspecified |
| Au superparticle144 | Liquid–liquid extraction | Farmland, river and fishpond water: unknown contaminants | 4,4′-DDT, chlordane, tetradifon, α-endosulfan | Demonstrated 5 nM (4,4′-DDT, chlordane, tetradifon), 6 nM (α-endosulfan) | LOD: 0.46 nM (4,4′-DDT), 0.59 nM (chlordane), 0.97 nM (tetradifon), 0.87 nM (α-endosulfan) |
| Silicon nanowire array decorated with Ag nanodendrite157 | TLC | Milk: competitively adsorbing proteins | Melamine | LOD: 2.5 ppm | Unspecified |
| Au sputtered chromatography paper159 | Paper chromatography | Pond water: unknown contaminants; rice paste: viscous sample with unknown contaminants | Malachite green, crystal violet, methylene blue (pond water); Cd2+, Cu2+, Ni2+ (rice paste) | 10−10 M (malachite green and crystal violet), 10−9 M (methylene blue), 10−6 M (Cd2+, Cu2+, Ni2+) | Unspecified |
| Polyadenine–DNA & alkyne reporter modified Au NP164 | Coordination | Artificial water sample: unspecified amounts of dye contaminants | Hg+, Ag+ | LOD: 0.77 nM (Hg+), 0.86 nM (Ag+) | LOD: 0.36 nM (Hg+), 0.06 nM (Ag+) |
| Superhydrophobic Au NPs/nickel foam165 | Hydrophobic interactions | Tap water: unknown contaminants | Pyrene, 1-naphthol | Demonstrated 10−5 M (pyrene, 1-naphthol) | LOD: 10−8 M (pyrene), 10−6 M (1-naphthol) |
| Raman reporter labelled Au NPs-based lateral flow assay strip174 | Bio-recognition | Bacterial pathogen solution: unknown bio-contaminants | Y. pestis, F. tularensis, B. anthracis | LOD: 43.4 CFU per mL (Y. pestis), 45.8 CFU per mL (F. tularensis), 357 CFU per mL (B. anthracis) | Not applicable |
| Magnetic-based MIP decorated with Au NPs176 | Hydrogen bonding | Milk: competitively adsorbing proteins; tab water: unknown contaminants | 2,4-Dichlorophenoxyacetic acid | Demonstrated 1 ng mL−1 in milk and tab water | 0.00147 ng mL−1 |
| PEG-S functionalized Au triangular nanoprisms patch180 | Antifouling | Diluted human plasma: bio-contaminants, mainly protein | Fentanyl, 4-ANPP, cocaine, heroin, cannabinoids | LOD: 3.0 pg mL−1 (fentanyl); demonstrated 1 nM (4-ANPP, cocaine, heroin, cannabinoids) | Unspecified |
| Metal NPs embedded hydrogel micropellet186 | Size exclusion | Milk: competitively adsorbing proteins; whole blood: bio-contaminants, mainly protein | Melamine, glucose | LOD: 10 nM (melamine in milk); 0.01 mM (glucose in whole blood) | LOD: 0.01 mM (glucose) |
| Polydopamine@Au nanowaxberry191 | Size exclusion | Soil: particulate matter | Thiram | LOD: 0.31 μg g−1 | LOD: 0.5 nM |
4. Calibration-enhancement-in-one SERS substrates for precise quantitative analysis in complex sample matrices
As discussed in Section 3, the surface accessibility and plasmonic properties of enhancing substrates are often compromised by the contaminants within complex real-life samples. This often leads to irreproducible SERS signals, and in turn presents a significant challenge for quantitative analysis. Although it is possible to remove the interference from complex matrices with sample pre-treatment, this significantly raises the cost of analysis. Therefore, a variety of SERS substrates with self-calibration functions have been developed, some of which have been combined with the functional materials discussed in Section 3 to build separation-calibration-enhancement-in-one SERS substrates to allow direct quantitative SERS analysis of complex samples with improved precision and accuracy. While the enhancing substrates incorporated with calibration functionality come in various forms, the strategies for incorporating internal standards generally fall into two categories, surface modification with self-assembled monolayers or utilization of intrinsic reference materials which are discussed in detail below.4.1 Calibration with adsorbed self-assembled monolayers
The most widely used strategy to introduce self-calibration functionality in SERS substrates is to modify the enhancing surface with a self-assembled monolayer of Raman tag molecules.197–200 Typical examples of Raman tag molecules include 4-mercaptobenzoic acid (MBA) and 5,5-dithio-bis-(2-nitrobenzoic acid) (DTNB, also known as “Ellman's Reagent”) which adsorb strongly to Ag/Au surfaces to generate intense and distinct Raman vibration bands that could be compared against the SERS signals of analytes to improve the accuracy of quantitative detection.94,201 However, a problem with these conventional Raman tag molecules is that their signature peaks often lie within the Raman fingerprint region and therefore overlaps with the SERS signals of the analyte, which complicates data analysis. To tackle this issue, several research groups have introduced new Raman tag molecules which retain the advantages of traditional tag molecules but generate distinct vibration bands within the Raman silent region.71,72 For example, Li et al. demonstrated that Prussian blue (PB) could be used as an internal standard to improve the quantitative performance of Au colloids in the SERS analysis of dyes and bio-molecules dispersed in real-life samples.201 As shown in Fig. 14A, the Prussian blue internal standard was chemically grown onto the enhancing surface of the colloidal Au nanoparticles by treating citrate-stabilized Au colloid with an aqueous solution of K3[Fe(CN)6] followed by an aqueous mixture containing K4[Fe(CN)6] and FeCl3, which led to the formation of a 2–3 nm Prussian blue shell around the Au nanoparticles. Importantly, the SERS signals of the Au@PB nanoparticles consisted of a single intense Raman vibration band at 2155 cm−1 arising from the Prussian blue shell, as shown in Fig. 14B. Moreover, the Prussian blue shell and its SERS signals remained highly stable within a wide pH (3.2–7.5) and temperature (25–100 °C) range, which make it an excellent candidate as an internal standard. Using the Au@PB colloids as the enhancing substrate allowed crystal violet and dopamine in model aqueous solutions to be quantitatively detected from 2.0 to 100 nmol L−1 and 500 to 8000 nmol L−1, respectively. More importantly, a significant improvement in the linearity and precision of the SERS quantification curve could be observed when the calibration curves were plotted using the relative intensity of the characteristic peaks between the analyte and Prussian blue internal standard rather than just the SERS signal intensity of the analyte. Lin, Feng et al. demonstrated that 4-mercaptobenzonitrile could be used as an internal standard to significantly improve the SERS quantitative performance of Au nanoparticles.202 Similar to Prussian blue, the 4-mercaptobenzonitrile contained C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N functional groups, which when functionalized on the enhancing surface of the Au nanoparticles generated an intense and distinct vibration band at 2224 cm−1 in the Raman silent region of the spectra. As shown in Fig. 14C spectra set i, this led to a significant improvement of the precision of the indirect SERS quantification of a tumour marker, carcinoembryonic antigen in clinical serum samples (Fig. 14C spectra set ii and iii).
N functional groups, which when functionalized on the enhancing surface of the Au nanoparticles generated an intense and distinct vibration band at 2224 cm−1 in the Raman silent region of the spectra. As shown in Fig. 14C spectra set i, this led to a significant improvement of the precision of the indirect SERS quantification of a tumour marker, carcinoembryonic antigen in clinical serum samples (Fig. 14C spectra set ii and iii).
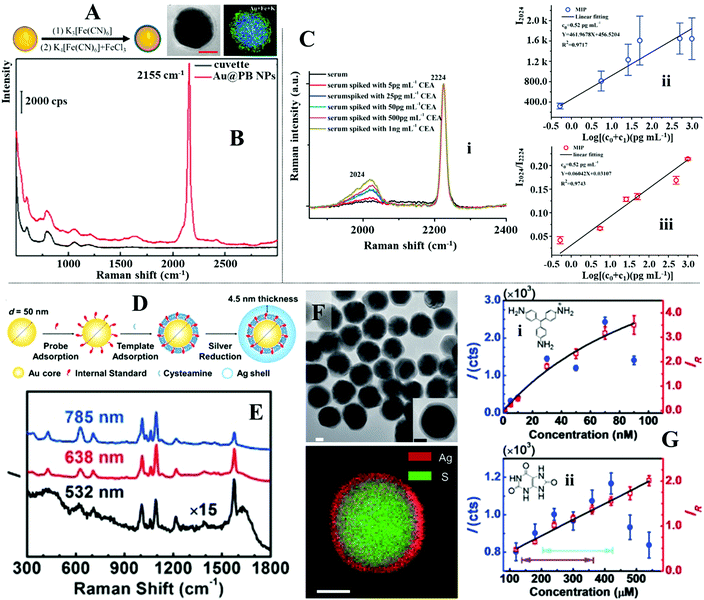 | ||
| Fig. 14 (A) Schematic illustrations of the synthetic process of Au@Prussian Blue (PB) nanoparticles. TEM and energy-dispersive X-ray (EDX) mapping of a Au@PB nanoparticle; (B) SERS spectra of the Au@PB particles; (C) spectra set (i) shows SERS spectra of carcinoembryonic antigen (CEA) in serum obtained with the enhancing substrate functionalized with 4-mercaptobenzonitrile internal standards. Plots (ii–iii) compares the calibration curves obtained by plotting the signal intensity of CEA and the intensity ratio between CEA and the internal standard as a function of CEA concentration; (D) schematic illustrations of the fabrication process of Au@Ag particles with an embedded layer of internal standard molecules; (E) SERS spectra of the Au@Ag particles with an embedded layer of 4-mercaptopyridine internal standards acquired at different excitation wavelengths; (F) TEM and EDX mapping of the Au@Ag nanoparticles; (G) plots comparing the calibration curves obtained by plotting the signal intensity of the analyte or the SERS intensity ratio between the analyte and the internal standard as a function of analyte concentration. Panel (A and B) adapted with permission from ref. 201, copyright 2019 American Chemical Society. Panel (C) adapted with permission from ref. 202, copyright 2019 Elsevier Ltd. Panel (D–G) adapted with permission from ref. 203, copyright 2015 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
A common issue that arises from functionalizing the enhancing surface with Raman tag molecules is that they compete with adsorbing analyte molecules for the enhancing surface. This not only leads to weaker SERS signals, and in turn poorer limit of detection of the analyte molecules but also jeopardizes the calibration functionality of the internal standard. This has limited the applications of SERS substrates with externally functionalized internal standards to the in-direct detection of weakly adsorbing analyte molecules, which do not compete with the internal standard for enhancing surface. To combat this issue, Ren et al. designed colloidal core–shell nanoparticles with embedded internal standards.203 As illustrated in Fig. 14D, the core–shell nanoparticles were synthesized by functionalizing the surface of plasmonic Au nanoparticles with a mixed monolayer of 4-mercaptopyridine, which acted as the internal standard, and cysteamine, which acted as the molecular linker for the chemical growth of an Ag shell surrounding the thiol-modified Au cores. As shown in Fig. 14E, the core–shell structure with embedded internal standards were verified with transmission microscopy and energy-dispersive X-ray spectroscopy which showed that the mixed-thiol layer on the surface of the Au cores retained its integrity and was encapsulated within a 10 nm thick Ag shell. More importantly, strong SERS signals of the 4-mercaptopyridine internal standard could be obtained from the product particles with 532, 638 and 785 nm excitation wavelengths, as shown in Fig. 14F. An important advantage that comes with embedding the Raman tags within the core–shell particles is that this leaves the enhancing surface accessible to analyte molecules. This paved the way for the adsorption, and in turn SERS detection of analyte molecules, such as 1,4-phenylene diisocyanide, pararosaniline (also known as “basic red 9”) and uric acid, which have low affinity towards Ag/Au surfaces. Moreover, normalizing the SERS signal intensity of the analytes against the embedded internal standards in the core–shell nanoparticles allowed the huge signal fluctuations induced by colloidal aggregation to be removed. This led to significant improvements in the accuracy and precision of the quantitative SERS measurements without compromising the limit of detection, as shown in Fig. 14G. This work pioneered the synthesis of core–shell plasmonic nanomaterials with embedded internal standards which have now been used both directly as the enhancing material for quantitative SERS and as the functional component for building more sophisticated SERS sensors.71,204–207
4.2 Calibration with intrinsically present reference materials
In some cases, the SERS/Raman signals of materials, which are intrinsically present within the SERS sensor can be used as a convenient and stable reference for signal calibration.208–211 For example, Liu et al. showed that the SERS signals of acetone which were present in the environment of interfacial Au nanoparticle arrays could be used as a reliable reference to calibrate the SERS signals of analyte molecules.212 As shown in Fig. 15A, the acetone molecules were part of the solution in which the plasmonic nanoparticle arrays resided, and as a result, were physically present within the enhancing hot spots between adjacent Au nanoparticles. Correspondingly, the presence of acetone within the plasmonic hot spots were reflected by the presence of a clear vibration band at 1430 cm−1 within the SERS spectrum obtained from the Au nanoparticle arrays. As shown in Fig. 15C, calibrating the SERS signal of the analyte molecule, thiram, against the 1430 cm−1 peak from acetone allowed the RSD of the SERS signals collected on 40 different spots of the enhancing substrate to be improved significantly from 14.5% to 5.9%. The improvement in the precision of the quantitative measurements was also clearly reflected in the calibration curves of thiram established with and without acetone calibration (Fig. 15B). Similarly, Jiang et al. showed that the polydimethylsiloxane (PDMS) polymer in Au nanowire/PDMS films can be used as the internal standard to improve the precision of SERS quantification.213 As shown in Fig. 15D, the PDMS generated a stable and distinct characteristic peak at 2905 cm−1, which when used as an internal standard, enabled SERS quantification of 2-naphthalenethiol (2NAT) with notably improved precision (Fig. 15E). | ||
| Fig. 15 (A) Schematic illustrations of acetone internal standards residing along with the analyte molecules in the interparticle hot spots within interfacial nanoparticle arrays; (B) plots comparing the calibration curves obtained by plotting the signal intensity of thiram or the intensity ratio between thiram and the acetone internal standard as a function of thiram concentration; (C) plots comparing the relative standard deviation of the intensity of the 1386 cm−1 vibration band from thiram on 40 different points of the SERS substrate before and after calibration with acetone internal standard; (D) SERS spectra of varying concentrations of 2-naphthalenethiol (2-NAT) obtained on PDMS-based SERS substrate; (E) plots comparing the calibration curves obtained by plotting the signal intensity of 2-NAT or the intensity ratio between 2-NAT and the PDMS internal standard as a function of 2-NAT concentration. Panel (A–C) adapted with permission from ref. 212, copyright 2018 American Chemical Society. Panel (D and E) reproduced with permission from ref. 213, copyright 2020 American Chemical Society. | ||
Table 3 lists additional examples of calibration-enhancement-in-one SERS substrates from recent literature which fall into the two categories discussed above. Another worth-noting strategy that has recently been proposed for calibrating SERS signals is the use of the plasmon-enhanced electronic Raman scattering signals from the enhancing metal as the internal standard. This approach has been successfully utilized to improve the accuracy of quantitative SERS analysis performed in living biological systems.214,215 In summary, utilizing the SERS/Raman signals of intrinsically present reference materials as an internal standard is a convenient approach to construct calibration-enhancement-in-one SERS substrates. Moreover, the approach does not rely on surface modifications of the enhancing materials which leaves the enhancing substrate accessible for analyte molecules. However, this approach is not generally applicable since it relies on the existence of a suitable reference material within the SERS system. The generally applicable strategy is to rationally functionalize the enhancing substrate with Raman tag molecules which generate intense and distinct Raman vibration bands. However, this process is not straightforward since the internal standards must be firmly bound onto the enhancing surface but not hinder the interaction between the enhancing surface and analyte molecules. This challenge has been recognized and largely resolved with the development of core–shell nanoparticles with an embedded inner layer of internal standard molecules. It should be noted that both strategies discussed above are focused on calibrating fluctuations in the plasmonic properties of the SERS substrates but overlook the potential change in SERS signals brought about by the change in the surface chemistry of the enhancing material during analysis. In fact, the change in the surface properties of the enhancing materials is of particular significance in real-life analysis, since a major challenge faced by SERS sensors deployed in complex real-life samples is surface passivation by adsorbing contaminants and/or oxidation which might not drastically change the plasmonic properties of the enhancing substrate but would significantly inhibit the adsorption of analyte molecules.216,217 This can be partially resolved by adding structurally tailored internal standards, which possess similar affinities to the enhancing surface as the target analyte molecule, to co-adsorb with the analyte molecules.218,219 However, this approach must be further developed so that the structurally tailored internal standards can be integrated into the SERS substrate to increase the practicality of the approach.
| Substrate | Internal type | Internal material & characteristic vibration | Sample | Performance without internal | Performance with internal |
|---|---|---|---|---|---|
| Abbreviations: limit of detection (LOD); nanoparticle (NP); self-assembled monolayer (SAM); internal standard (IS); 4,4′-dipyridyl (4,40-DP), 5,5′-dithiobis (2-nitrobenzoic acid) (DTNB); graphene oxide (GO). | |||||
| Plasmonic Au film72 | External SAM | 4-Mercapto benzonitrile (2223 cm−1) | Bacteria in culture media and human blood | Unspecified | Dynamic detection range between 101 to 107 CFU per mL; 6.70% RSD on 900 points |
| Silicon nanohybrid SERS chip198 | External SAM | 4-Amino thiophenol (1079 cm−1) | Pb2+ and Hg2+ in water | Poor linear quantitation. Pb2+: R2 = 0.975; Hg2+R2 = 0.753 | Same quantitation range. Pb2+: R2 = 0.993; Hg2+R2 = 0.991 |
| Au@IS@Ag core–satellite nano-assembly71 | Internal SAM | 4-Mercapto benzonitrile (2221 cm−1) | Prostate-specific antigen in clinical blood samples | Unspecified | Linear quantitation between 7.69 to 173.1 ng mL−1; R2 = 0.99; RSD 6.514% |
| Au fractal SERS probe204 | Internal SAM | 4-Mercapto benzoic acid (1078 cm−1) | DNA in buffer solution | Unsuccessful quantitation; 1.75 fM LOD | Linear quantitation between 6.77 to 50 fM; R2 = 0.947 |
| GO coated Fe3O4@TiO2@AuAg NP208 | Intrinsic material | TiO2 (149 cm−1) | Methylene blue, saccharin sodium, carmine acid in water | Adequate linear quantitation. Methylene blue: R2 = 0.977; saccharin sodium: R2 = 0.986; carmine acid: R2 = 0.974 | Same quantitation range. Methylene blue: R2 = 0.996; saccharin sodium: R2 = 0.998; carmine acid: R2 = 0.994 |
| Biogenetic Au@Ag nano island210 | Intrinsic material | Biomolecules on bacteria: phospholipid (713 cm−1), tyrosine (853 cm−1), phenylalanine (1006 cm−1) | Rhodamine 6G, malachite green, uric acid in water | Unsuccessful quantitation | Rhodamine 6G: linear quantitation between 0.01 to 1 μM with phospholipid internal (R2 = 0.9528), 1 to 100 μM with phenylalanine internal (R2 = 0.9904); malachite green: linear quantitation between 1 to 100 nM with phospholipid internal (R2 = 0.9819); uric acid: linear quantitation between 100 to 500 μM with tyrosine internal (R2 = 0.9877) |
| Alkyne-modified graphene-isolated Au NP211 | Intrinsic material | Acetonitrile (2263 cm−1) | Alkaline phosphatase in buffer solution | Unsuccessful quantitation | Linear quantitation between 0.002 U L−1 to 1 U L−1; R2 = 0.994 |
5. Reusable regeneration-enhancement-in-one SERS substrates
As a surface technique SERS relies on the strong interactions between molecules and the enhancing surface to confine analytes within the localized surface plasmon resonance, as well as to facilitate charge transfer, to achieve signal enhancement. This allows the Raman signals of extremely small amounts of analyte molecules adsorbed on the enhancing surface to be detected but also makes SERS particularly sensitive to surface contamination.216,217,220 As a result, SERS substrates have typically been used for single measurements to ensure optimum sensitivity and quantification accuracy which has significantly driven up the price of SERS analysis. This is not helped by the fact that the most widely used enhancing materials are expensive Ag/Au noble metals, which require additional fine processing to generate well-defined nanostructures with tailored surface properties to enable optimal enhancement. In an effort to lower the cost of SERS, a variety of strategies to create reusable SERS substrates have been proposed by the SERS community and are summarized in the following section.5.1 Simple substrate recycling procedures
Up to now, a variety of post-analysis treatments, both chemical and physical, have been developed to regenerate SERS substrates. Amongst these methods, the most straightforward is to wash the enhancing surface with common solvents, such as water and ethanol, after SERS analysis.115,221,222 To increase the effectiveness of this approach and shorten the treatment time, the washing process is sometimes accompanied by sonication.223–225 This approach has typically been demonstrated with model dye molecules, such as rhodamine 6G and malachite green,222,224,225 which are adsorbed on the surface of the enhancing substrate through weak electrostatic attraction but has also been demonstrated with less effectiveness for more strongly adsorbing analytes, such as thiram.115,221 Strongly adsorbed analytes can be fully removed from the enhancing surface via thermal treatment.226–228 However, since this process typically takes place at temperatures >250 °C and lasts for tens of minutes, the surface of the enhancing noble metal nanoparticles must be functionalized with additional heat-resistant inorganic surface-layers, such as metal oxides and graphene, to preserve the morphology of the enhancing particles during thermal treatment. Alternatively, strongly adsorbed chemical species can be removed from the surface of enhancing substrates using oxidative plasma cleaning, which can be completed within seconds and is generally applicable to solid enhancing substrates.229–231 However, this requires the use of expensive laboratory-based specialist equipment which is undesirable for the routine applications of SERS in-field.A cheap, mild and rapid method, which has been demonstrated to be highly effective even for the removal of strongly adsorbed analytes, such as thiols and pyridines, on both Ag and Au enhancing surfaces, is treatment with sodium borohydride solution.122,123,232,233 This method was pioneered by Zhang et al. who showed that the cleaning ability of borohydride solution originated from the hydride ions derived from sodium borohydride having a strong affinity to Au nanoparticles.234 This allowed a variety of adsorbing analytes which included, rhodamine, poly(N-vinylpyrrolidone) (PVP), adenine, I−, Br−, p-methylbenzenethiol, 2-napthalenethiol, p-benzenedithiol and homocysteine to be completely displaced from the surface of colloidal Au nanoparticles. Naturally, the rate of analyte desorption was positively correlated to the concentration of NaBH4 solution, it was found that complete desorption of the thiol analytes occurred within 10 minutes when the concentration of NaBH4 was above 25 mM. After displacing the analytes, the hydride anions can be desorbed from the enhancing surface through mild acid treatment to reactivate the SERS substrate.
5.2 Photocatalytic regeneration
Although post-analysis cleaning procedures allow effective removal of even strongly adsorbed analytes from the enhancing surfaces, these procedures typically require careful handling by specialists, which significantly limit their routine applications. Even treatment with NaBH4 solutions is not straightforward, since the NaBH4 solution must be freshly made to generate hydride ions. To increase the practicality of reusable substrates, multifunctional SERS substrates with integrated self-cleaning functionalities have been designed. Amongst these self-cleaning substrates, arguably the most widely researched are hybrid substrates which contain plasmonic noble metals that provide SERS enhancement and photocatalytically active materials that enable catalytic degradation of the adsorbed analyte molecules.64 For example, Jiang et al. designed multifunctional Fe3O4@TiO2@Ag microparticles as recyclable SERS substrates for direct detection of antigens in human serum.235 The multifunctional Fe3O4@TiO2@Ag microparticles were prepared by a multi-step reaction, which allowed the thickness of the intermediate TiO2 layer and the density of the outer layer of plasmonic Ag nanoparticles to be adjusted. The Ag nanoparticles not only provided SERS enhancement, which allowed antigens containing a mixture of ferritin and carcinoembryonic to be quantitatively detected between 10−4 to 10−10 g mL−1 in human serum, but also acted as electron-sinks, which significantly enhanced the photocatalytic efficiency of the TiO2 catalyst. The catalytic efficiency of the microparticles could be further improved by concentrating the microparticles through external magnetic force. Using human serum containing 10 μg mL−1 of antigens as test samples, the authors showed that the adsorbed antigens could be degraded down to an undetectable level with 100 minutes of UV irradiation. This allowed the microparticles to be used repeatedly for 9 cycles of SERS analysis without any observable decrease in their photocatalytic and SERS efficiency, as shown in Fig. 16A. To further validate their practicality as reusable SERS substrates, the same batch of microparticles were used as the enhancing substrate to analyse 5 clinical serum samples from different patients. As shown in Fig. 16B, the results of the SERS measurements were highly consistent with the results obtained from conventional chemiluminescent immunoassay kits used in hospitals. In addition to TiO2, a variety of other photocatalytically active materials, such as MoS2, ZnO and WO3−x, have been used to construct SERS substrates, which can be regenerated with UV irradiation.73,236–238 However, the prolonged regeneration process along with the requirement for special irradiation sources make these substrates less ideal in practice. This has led to the development of visible-light-induced self-cleaning SERS substrates, which are potentially more sustainable and often offer faster regeneration speed compared to their UV-excited counterparts.239–242 For example, Jiang et al. designed SERS active reduced graphene oxide (RGO)@MoS2@Ag nanopowders which can be regenerated with solar-light irradiation, as shown in Fig. 16C.243 The nanocomposite powders were fabricated by in situ hydrothermal growth of MoS2 nanoflowers onto the surface of GO sheets. This was followed by in situ growth of AgNO3 nanoparticles on the surface of the MoS2 nanoflowers via photo-reduction. The product RGO@MoS2@Ag showed excellent SERS efficiency which allowed methylene blue to be detected down to 10−10 M with just 20 μL of analyte solution. Moreover, the nanocomposites exhibited excellent photocatalytic activity. As shown in Fig. 16D, the methylene blue in 20 μL of 10−2 M methylene blue solution could be photocatalytically degraded down to a concentration undetectable by SERS with 45 minutes of artificial sunlight irradiation. To demonstrate their practicality, the RGO@MoS2@Ag powders were applied as enhancing substrates for the detection of chemical additives in milk. As shown in Fig. 16E, the limit of detection for melamine, vanillin (VA) and bisphenol A (BPA) in milk were determined to be 10−8 M, 10−8 M and 10−7 M, respectively. To demonstrate the reusability of the RGO@MoS2@Ag enhancing substrates, the enhancing powders were treated with 20 μL of milk containing a mixture of the three analytes and then photo-cleansed for eight repeated cycles. As shown in Fig. 16F, the SERS signals obtained for each cycle were nearly identical with only ca. 9.8% intensity deviation. | ||
| Fig. 16 (A) Plot showing the SERS activity of the Fe3O4@TiO2@Ag microparticles after 9 cycles of photo-regeneration; (B) plot comparing SERS results obtained from Fe3O4@TiO2@Ag microparticles and chemiluminescent results obtained from commercial immunoassay kits; (C) schematic illustrations of the photodegradation process on RGO@MoS2@Ag nanocomposites. Inset shows the SEM image of a typical RGO@MoS2@Ag particle; (D) SERS spectra showing the photodegradation of methylene blue catalysed by RGO@MoS2@Ag nanocomposites; (E) plots showing the change in SERS signal intensity of melamine, vanillin and bisphenol A in milk as a function of analyte concentration; (F) SERS spectra of a milk sample spiked with a mixture of melamine, vanillin and bisphenol A obtained using the same batch of RGO@MoS2@Ag nanocomposites after 8 cycles of photo-regeneration. Panel (A and B) reproduced with permission from ref. 235, copyright 2020 Elsevier Ltd. Panel (C–F) adapted with permission from ref. 243, copyright 2021 Elsevier Ltd. | ||
It is worth noting that visible-light induced substrate regeneration can also be achieved with just noble metal nanomaterials using plasmon-induced photocatalytic reactions.244,245 For example, Postnikov, Lyutakov et al. designed bimetallic Au/Pt films, which enabled the detection of dibenzothiophene in biodiesel and could be regenerated with solar-light irradiation.245 As shown in Fig. 17A, the bimetallic films were constructed by spin coating SU-8 polymer onto glass substrates, which allowed periodical patterns to be formed via laser lithography. After this the substrates were made plasmonically active by evaporating a 25 nm thick layer of Au on its surface. Finally, the Au layer was covered with a 2.5 nm thick layer of Pt, which promoted the adsorption of the target analytes. Using the bimetallic films as the enhancing substrate allowed dibenzothiophene to be quantitatively detected between 10−4 M to 10−12 M in biodiesel. The adsorbed dibenzothiophene molecules could be effectively removed down to concentrations, which were undetectable by SERS, using plasmon-induced reactions in cyclohexane. As shown in Fig. 17B, the degradation could be completed within 15 minutes of irradiation using 250 W of artificial sunlight. Moreover, the SERS signal intensity of dibenzothiophene only decreased by 4.8% after five cycles of detection-regeneration, as shown in Fig. 17C.
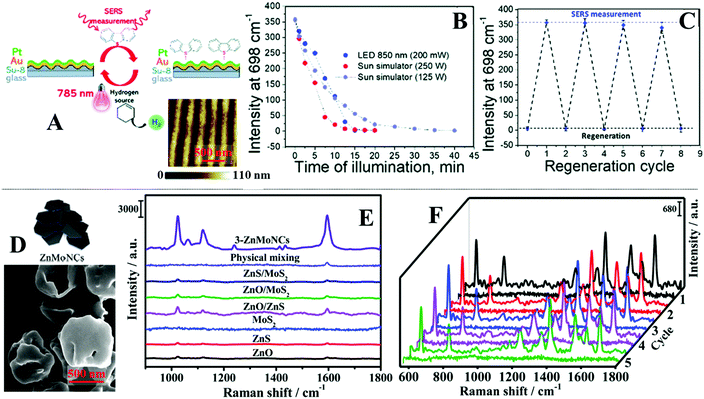 | ||
| Fig. 17 (A) Schematic illustrations of the bimetallic Au/Pt films. Atomic force microscopy image showing the surface morphology of the bimetallic Au/Pt films; (B) plot showing the photo-degradation of dibenzothiophene monitored by SERS; (C) plot showing the SERS activity of the bimetallic Au/Pt films after 4 cycles of photo-regeneration; (D) schematic representation and TEM image of ZnO/ZnS/MoS2 nanocomposites; (E) SERS spectra of 4-mercaptopyridine adsorbed on ZnO/ZnS/MoS2 nanocomposites compared to other types of materials; (F) SERS spectra of rhodamine 6G adsorbed on the same batch of ZnO/ZnS/MoS2 nanocomposite over 5 cycles of photo-regeneration. Panel (A–C) adapted with permission from ref. 245, copyright 2019 The Royal Society of Chemistry. Panel (D–F) adapted with permission from ref. 251, copyright 2019 The Royal Society of Chemistry. | ||
The recent development of SERS substrates has also led to the emergence of a variety of novel noble metal-free enhancing materials.246 Amongst these materials, enhancing substrates based on semiconductors, such as ZnO, TiO2, and MoS2, stand out particularly since they are not only potentially cheap and stable alternatives to Ag and Au but are also photocatalytically active which allows for the construction of reusable substrates.247–250 For example, in a proof-of-concept demonstration, Liu, Gao et al. designed photocatalytically active ternary ZnO/ZnS/MoS2 nanocomposites, as shown in Fig. 17D, which could be used as reusable substrates for SERS.251 The nanocomposites were synthesized through in situ chemical growth using phosphomolybdic acid hydrate (PMo12)@zeolitic imidazolate frameworks-8 (ZIF-8) as the precursor. Importantly, forming the nanocomposites via this approach allowed nanoscale contact points between the ZnO, ZnS and MoS2 in the nanocomposites to be generated with higher efficiency compared to physically mixing the individual components which is significant for achieving strong SERS and photocatalytic activity (Fig. 17E). By immersing the nanocomposite powders in 4 mL of different analyte solutions, the limit of detection for a model analyte, rhodamine 6G, under resonant conditions were determined to be 10−9 M. Moreover, the enhancing surface of the nanocomposites could be regenerated with 30 min of UV irradiation, which allowed 5 repeated cycles of SERS analysis to be performed without any observable loss in the signal intensity of the analyte, as shown in Fig. 17F.
5.3 Superhydrophobic self-cleaning
Although photocatalytically active SERS substrates have integrated regeneration mechanisms which make them more convenient to operate by non-expert users, the extensive regeneration period and the requirement of additional light sources limit their applications in practice. Conversely, superhydrophobic self-cleaning enhancing substrates can typically be regenerated easily and rapidly by rinsing with water.252–255 Inspired by nature, superhydrophobic enhancing substrates are typically composed of enhancing noble metal nanoparticles dispersed on the surface of micro-structures to mimic the surface structure of self-cleaning biomaterials, such as lotus leaves. In a typical display, Li et al. designed non-pinning superhydrophobic films with plasmonic hierarchical microscale and nanoscale surface structures produced by laser lithography as reusable SERS substrates (Fig. 18A).256 Using rhodamine 6G as the model analyte, the authors demonstrated that the enhancing surface could be regenerated by simply immersing the substrate in ethanol, as shown in Fig. 18B. Moreover, as shown in Fig. 18A and D, the large contact angle of the water droplets on non-pinning superhydrophobic surfaces enables droplets of aqueous analyte solutions to dry into a concentrated spot instead of a dispersed coffee-ring structure on the enhancing surface, which allows significant enrichment of the analyte, thus presenting a unique advantage for SERS. As shown in Fig. 18E, this led to a significant increase of SERS signal intensity by 28.3× when 20 μL of rhodamine 6G was dried onto a non-pinning superhydrophobic plasmonic surface instead of a hydrophilic plasmonic surface. As a result, the limit of detection of rhodamine 6G deposits on the non-pinning superhydrophobic enhancing substrates reached 10−14 M, as shown in Fig. 18C. Although evaporation of the analyte droplet on non-pinning superhydrophobic enhancing substrates allows analyte enrichment, it also requires extensive periods of drying. For example, Yin et al. showed that complete evaporation of just 3 μL of aqueous analyte solution under ambient conditions took ca. 15 minutes.257 Therefore, in an effort to shorten the analysis period of superhydrophobic enhancing substrates, the authors designed superhydrophobic Cu(OH)2 nanoneedle arrays decorated with Ag nanoparticles which allowed direct analysis of analytes dissolved in solution droplets without the need for solvent evaporation. As shown in Fig. 18F, arrays of Cu(OH)2 needles were deposited electrochemically on the surface of copper foils to form microscopic structures. The surface of the Cu(OH)2 needles were decorated with 20 nm diameter Ag nanoparticles through electrostatic attraction to form plasmonic hot spots. Finally, the surface of the Cu(OH)2 needles and Ag nanoparticles were functionalized with a self-assembled monolayer of 1H,1H,2H,2H-per-fluorodecyltriethoxysilane, which rendered the surface superhydrophobic. In use, a 3 μL droplet of analyte solution could be deposited on the surface of the enhancing substrate and analyzed with SERS directly, which allowed melamine in milk and thiram in apple juice to be detected down to 0.1 ppm (Fig. 18G). Moreover, the substrate could be regenerated by simply blowing away the analyte solution droplet with air. As shown in Fig. 18H, this allowed crystal violet and rhodamine 6G to be detected sequentially for 5 cycles without any obvious decrease in the SERS activity of the enhancing substrate.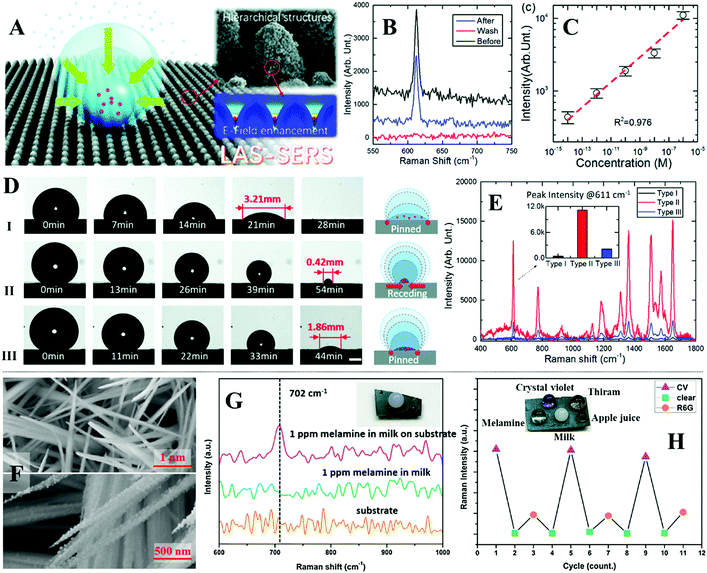 | ||
| Fig. 18 (A) Schematic illustrations of an aqueous sample droplet drying into a concentrated spot on the surface of a non-pinning superhydrophobic film. SEM image showing the hierarchical micro and nanoscale structure on the film surface. Finite element simulations showing the distribution of the electromagnetic field enhancement on the film surface; (B) spectra set showing the SERS signals of rhodamine 6G obtained before and after washing; (C) plot showing the signal intensity of rhodamine 6G as a function of concentration; (D) optical images and schematic illustrations of a water droplet drying on a hydrophilic surface (i), a pinning hydrophobic surface (ii) and a non-pinning hydrophobic surface (iii); (E) SERS spectra obtained by drying an aqueous droplet of rhodamine 6G onto (i–iii) surfaces; (F) SEM images of the Cu(OH)2 nanoneedle array before and after being coated with Ag nanoparticles; (G) SERS and Raman spectra of milk spiked with 1 ppm melamine and the background Raman signals of the enhancing substrate; (H) plot showing the Raman intensity of crystal violet and rhodamine 6G after 5 cycles of substrate regeneration. The inset photograph shows varying types of sample droplets sitting on the surface of the Ag decorated Cu(OH)2 films. Panel (A–E) adapted with permission from ref. 256, copyright 2017 The Royal Society of Chemistry. Panel (F–H) adapted with permission from ref. 257, copyright 2018 Elsevier Ltd. | ||
5.4 Reversible intermolecular interactions
An emerging strategy for constructing reusable SERS substrates is through reversible intermolecular interactions. This is generally achieved by functionalizing the enhancing surface with a layer of functional material, which interact selectively with the target analyte through reversible interactions that can be reflected in the SERS signals. For example, Alvarez-Puebla, Romo-Herrera et al. designed reusable SERS sensors for the real-time monitoring of CO based on its reversible interactions with iron porphyrins.258 The SERS sensor was composed of xerogel foam embedded with concave Au nanocubes, which have been modified with Fe(II) metalized 5-[(triisopropylsilyl)thio]-10,20-diphenylporphyrin (Fe-TDPP), confined in a capillary glass tube. The Fe-TDPP ligands formed complexes with CO through polar pocket effects which led to the appearance of several new vibrational bands in the SERS spectra that allowed CO to be detected quantitatively (Fig. 19A). Since CO is a weakly adsorbing analyte with an inherently weak Raman cross-section, this meant that the changes observed in the SERS signals were contributed solely by the complexation of CO with Fe-TDPP. This allowed the quantitative detection range of the CO sensor to be adjusted to 10–40 ppm to match the minimum hazardous concentration of CO in air, by simply varying the amount of Fe-TDPP ligand modified on the enhancing surface. Moreover, since Fe-TDPP also complexed with O2, this meant that the sensor could be regenerated by simply flowing through dry air for 32 seconds, which allowed 5 cycles of CO detection without any notable decrease in the activity of the sensor (Fig. 19B).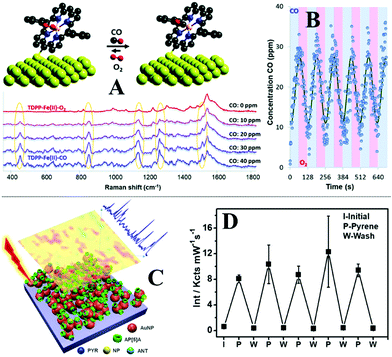 | ||
| Fig. 19 (A) Schematic illustrations of the reversibility of the CO sensor. SERS spectra of the sensor as a function of the concentration of CO in the flowing air; (B) plot showing the SERS reversibility of the sensor upon exposure to several cycles of CO and air; (C) schematic illustrations of the reusable supramolecular plasmonic films; (D) plot showing the SERS activity of the supramolecular sensor after 5 cycles of regeneration. Panel (A and B) reproduced with permission from ref. 258, copyright 2021 The Royal Society of Chemistry. Panel (C and D) reproduced with permission from ref. 263, copyright 2017 American Chemical Society. | ||
The surface of the enhancing material can also be functionalized with macromolecules to capture target analytes through reversible host–guest interactions.259–261 More specifically, the macromolecules capture and place the analyte molecules within the enhancing surface plasmon through weak intermolecular interactions, such as cation–dipole interactions or hydrophobic force.262 Therefore, this approach is most useful for SERS analysis of analyte molecules with a decent Raman cross-section but low affinity to the enhancing surface. For example, Pérez-Juste, Pastoriza-Santos et al. designed reusable supramolecular plasmonic films composed of multiple layers of plasmonic Au nanospheres, which have been modified with ammonium pillar[5]arene (AP[5]A) macrocycles for the detection of polyaromatic hydrocarbons, as illustrated in Fig. 19C.263 The plasmonic films were fabricated based on electrostatic layer-by-layer deposition of citrate-stabilized Au nanoparticles and AP[5]A macrocycles, which led to the formation of randomly dispersed plasmonically active Au nanoparticle aggregates functionalized with AP[5]A molecules. The AP[5]A macrocycles captured polyaromatic hydrocarbons through favourable host–guest intermolecular interactions, which enabled quantitative SERS detection of pyrene, nitropyrene and anthracene in aqueous solutions down to 5 nM, 0.05 μM and 50 nM, respectively. Non-quantitative SERS detection of the three analytes at sub-nanomolar concentrations, as well as multiplex detection, were also demonstrated. As shown in Fig. 19D, the enhancing surface could be regenerated by soaking the substrate in N,N-dimethylformamide for 4 hours which led to the release of the analyte from the host AP[5]A macrocycles. This allowed 5 cycles of SERS detection of pyrene in 0.1 μM solutions, without any significant decrease in the SERS activity of the plasmonic films.
Another popular type of functional material for achieving reusability is molecular imprinted polymers (MIPs).179,264–266 As introduced in Section 3.3, MIPs not only enable selective binding of analytes within complex sample matrices but can also be regenerated through solvent treatment, which make them excellent candidates for constructing reusable SERS substrates for practical applications. For example, Yu et al. designed reusable Fe3O4@Ag@MIPs nanocomposites for SERS detection of sibutramine illegal additives in oral substances.267 The Fe3O4@Ag@MIP nanocomposites captured sibutramine analyte molecules through favourable intermolecular interactions between the analyte and the carboxylate functional moieties of the MIP, which enriched the analyte molecules near the surface of the plasmonic Ag layer. This allowed sibutramine to be quantitatively detected from 10−5 to 10−9 M using a portable Raman spectrometer by dispersing the Fe3O4@Ag@MIP nanocomposites in 5 mL of model sibutramine solution. After SERS analysis, the Fe3O4@Ag@MIP nanocomposites could be conveniently extracted magnetically from the sample solution and redispersed into organic solvents for regeneration. This allowed the nanocomposites to be reused for 10 detection cycles with just a slight decrease in SERS activity. To demonstrate their practicality, the Fe3O4@Ag@MIP nanocomposites were applied for the SERS analysis of three commercial slimming supplements, which revealed the presence of sibutramine. More impressively, the amount of sibutramine within each of the positive samples predicted by SERS were highly consistent with the results obtained using conventional high-performance liquid chromatography coupled with mass spectroscopy.
In some cases, the analyte molecules can react with the functional materials on the enhancing surface leading to reversible structural changes of the functional materials, which is reflected in the SERS spectra and can therefore be utilized for reversible SERS analysis.268–271 For example, Ji et al. designed a reusable SERS sensor for the quantitative detection of F− in real-life samples based on a pH-reversible reaction between F− and 4-mercaptoboric acid.272 As illustrated in Fig. 20A, the SERS substrates were composed of a densely packed layer of plasmonic Ag nanoparticles chemical grown on the surface of a silicon wafer. Importantly, the surface of the enhancing Ag nanoparticles was functionalized with a self-assembled monolayer of 4-mercaptoboric acid Raman tag, which reacted specifically with F− through the boronic acid moieties leading to structural, and in turn, spectral changes of the Raman tags that could be used to indirectly quantify the amount of F− within the sample. Analyzing the area ratio between the SERS characteristic peak of fluoride-bond and unreacted 4-mercaptobenzoic acid Raman tags allowed F− in model aqueous samples to be quantitatively detected between 10−4–10−7 M, as shown in Fig. 20B. The limit of detection of F− was determined to be 10−8 M, which exceeded the maximum guideline values of F− in drinking water set by the World Health Organization by >3 orders of magnitudes. The binding of F− ions could be reversed by adjusting the pH of the chemical environment since OH− and F− ions competed for the boron center. This enabled the enhancing substrate to be regenerated by washing with sodium hydroxide solution, which allowed it to be used for 6 cycles without any observable degradation in SERS activity, as shown in Fig. 20C. To demonstrate the practicality of the F− sensor, the enhancing substrate were used for the detection of F− in samples of natural water and toothpastes. Impressively, the recovery rate of F− in these samples ranged between 91.43–111.4%, and the relative standard deviations were less than 5.0%. Li et al. designed a SERS sensor for the quantitative detection of a significant biomarker, tyrosinase, in serum based on an electro-chemically reversible reaction of p-thiol catechol Raman tags catalyzed by the tyrosinase analyte.273 As shown in Fig. 20D, the SERS substrates were fabricated by chemically modifying citrate-reduced Au nanoparticles onto the surface of indium tin oxide electrodes. To enable indirect detection of tyrosinase, the surface of the plasmonic Au nanoparticles was functionalized with p-thiol catechol, which converted to p-thiol benzoquinone upon interacting with tyrosinase. This induced significant spectral changes to the SERS signals of the Raman tags which in turn allowed tyrosinase to be quantitatively detected between 0.1 to 100 U mL−1. Based on the principle of signal-to-noise ratio being ≥3, the theoretical limit of detection of tyrosinase was calculated to be 0.07 U mL−1. Since the oxidation of the p-thiol catechol Raman tags were selectively induced by tyrosinase, the tyrosinase SERS sensor exhibited high selectivity to tyrosinase even in complex biological samples containing other chemical species such as inorganic salts, small molecules and macro-biomolecules, as shown in Fig. 20E. Importantly, the p-thiol benzoquinone Raman tags could be conveniently reduced to p-thiol catechol electrochemically by applying −0.3 V potential for 120 s, which allowed the tyrosinase SERS sensor to be reused for 10 cycles while retaining excellent reproducibility, as shown in Fig. 20F. To demonstrate their practicality, the tyrosinase SERS sensors were applied for the quantitative detection of tyrosinase in real serum samples. The results were highly consistent with the data obtained from commercial ELISA kits, which showed the accuracy of the SERS sensor.
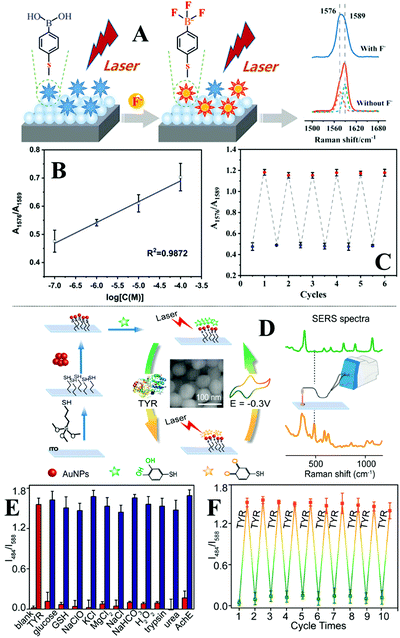 | ||
| Fig. 20 (A) Schematic illustration of the working principles of the pH-reversible F− sensor; (B) plot showing the average area ratio between the SERS peak at 1576 cm−1 and 1589 cm−1 as a function of the log concentration of F−; (C) plot showing the SERS activity of the sensor after 6 cycles of regeneration; (D) schematic illustrations of the fabrication of the electro-reversible tyrosinase sensor. Inset shows a SEM image of the enhancing Au nanoparticles on the surface of the sensor; (E) bar chart showing the ratio value between the SERS intensity of the vibration bands at 484 cm−1 and 588 cm−1 as a function of sample type. The red histograms correspond to the signals obtained from blank reference samples and pure samples of the chemicals labelled below. The blue histograms correspond to samples containing a mixture of tyrosinase and the chemicals labelled below. The error bars represent the standard deviation calculated from 3 independent measurements; (F) plot showing the SERS activity of the sensor after 10 cycles of electro-regeneration. Panel (A–C) reproduced with permission from ref. 272, copyright 2019 American Chemical Society. Panel (D–F) reproduced with permission from ref. 273, copyright 2019 American Chemical Society. | ||
Table 4 lists additional examples of reusable SERS substrates from recent literature, which fall into the categories discussed above. Examples of other less researched substrate regeneration strategies include the replacement of the enhancing nanoparticles and voltage-induced desorption of analyte molecules through reverse bias or electro-oxidation.274–278 In summary, different strategies for substrate regeneration come with different advantages and disadvantages. More specifically, treatment with sodium borohydride solution offers a convenient and effective solution for substrate regeneration. However, the practicality and generality of this approach is limited by the fact that the washing process also endangers any functional molecules which have been deliberately introduced on the enhancing surface. Amongst the different types of multifunctional SERS substrates with incorporated self-cleaning functionality, photocatalytic self-cleaning substrates are the most widely researched. However, the current generation of photocatalytic SERS substrates typically require extensive periods of UV exposure, which is in itself unsustainable and impractical. Conversely, it has been demonstrated that the SERS activity of superhydrophobic substrates can be regenerated within minutes or even seconds by simply rinsing with water or blowing with air. However, the nature of this regeneration process and the extreme sensitivity of surface-hydrophobicity to adsorbed surface species means that self-cleaning superhydrophobic SERS substrates are inherently limited to the analysis of weakly adsorbing analytes dispersed in samples without strongly adsorbing contaminants. Within this context, reusable substrates based on reversible intermolecular interactions seem the readiest for real-life applications. This is because the intermolecular interactions are typically target selective, which provides separation functionality that enables direct SERS detection in complex real-life samples. Moreover, the molecule layer which provides reversibility can be rationally engineered so that the spectral changes observed during SERS analysis arise from the structural changes of the functional molecule layer rather than the analyte molecule itself, which enables indirect SERS analysis of chemical species with low Raman cross-sections and/or weak affinity to the enhancing surface.
| Substrate | Sample & target analyte | Regeneration strategy | Regeneration procedure | Reusability |
|---|---|---|---|---|
| Abbreviations: nanoparticle (NP); reduced graphene oxide (rGO); metal organic framework (MOF); nicotinamide adenine dinucleotide (NAD) + hydrogen (H) (NADH). | ||||
| Nanoporous Ag sheet224 | 1 μM lindane or rhodamine B water solution | Solvent treatment | Immerse in ethanol for 5 min – sonicate in water for 8 min | Analyte fully removed; SERS activity fully maintained for 22 cycles |
| Graphene-veiled Au(Ag) nanoarray228 | 10−5 M rhodamine 6G, 10−2 M amoxicillin, unspecified amount of rhodamine B and methylene blue water solutions | Annealing | Heated at 300 °C for 30 min | Analyte fully removed; SERS activity maintained for 15 cycles with rhodamine 6G; fluctuation in SERS activity due to change in particle morphology; 2 regeneration cycles demonstrated for amoxicillin, rhodamine B and methylene blue |
| Au nanorod array on Si wafer230 | 10 mg L−1 thiabendazole methanol solution | Plasma | Treatment for 1 min | SERS activity fully maintained for 7 cycles; RSD between regeneration cycles was 4.87% |
| Au NPs decorated cicada wing233 | 10−5 M p-amino thiophenol, crystal violet, methyl orange, nile blue A, rhodamine 6G water solution | Treatment with NaBH4 water solution | Treatment with unspecified concentration of NaBH4 solution for 5 min – wash with water 3 times – dry in vacuum at 30 °C | Analyte fully removed; SERS activity fully maintained for 4 cycles |
| Ag/ZnO/Au 3D hybrid film236 | 10−6 M rhodamine 6G water solution, 200 ng μL−1 λ-DNA ethanol solution | UV light photocatalytic degradation | UV irradiation (8 W, 254 nm) for 40 (100) min while immersed in 4 mL of water for rhodamine 6G (λ-DNA) | Analyte fully removed; slight drop off in SERS activity after each regeneration cycle, demonstrated 8 and 4 regeneration cycles for rhodamine 6G and λ-DNA, respectively |
| Chestnut-like Ag/WO3−x NPs237 | Unspecified concentration of malachite green, 1 μM thiram water–ethanol solution | Visible light photocatalytic degradation | Rinse with deionized water for 5 s – irradiation with visible light (1000 W xenon lamp, 400–800 nm) for 50 min – rinse with deionized water for 5 s – blow dry with nitrogen gas | Near-full removal of analyte; slight drop off in SERS activity after each regeneration cycle, maintained more than 70% SERS activity after 5 cycles using malachite green as analyte; maintained more than 83% SERS activity using thiram as analyte |
| MoS2/Fe2O3 heterojunction nanocomposite248 | 10−4 M bisphenol A water solution | UV light photocatalytic degradation | Incubation of sample with substrate for 10 min – UV (details unspecified) irradiation for 50 min – magnetic separation | Near-full removal of analyte; slight drop off in SERS activity after each regeneration cycle after 5 cycles |
| TiO2-decorated chiral carbonaceous nanotube film249 | 10−4 M methylene blue | Visible light photocatalytic degradation | Drop 10 μL water on substrate – irradiation with visible light (300 W xenon lamp, AM1.5 air mass filter) for 20 min – blow dry with nitrogen gas | Analyte fully removed; SERS activity fully maintained for 4 cycles |
| rGO–TiO2–Fe3O4 nanohybrid250 | 1 × 10−3 M 4-mercaptobenzoic acid, 4-mercaptopyridine, 6-mercaptopurine monohydrate, 1,2-di(4-pyridyl) ethylene, p-aminobenzoic acid | UV light photocatalytic degradation | Disperse used substrate in ultrapure water – UV irradiation (details unspecified) for 2 h with stirring – magnetic separation – rinse with anhydrous ethanol | Analyte fully removed; SERS activity fully maintained for 8 cycles demonstrated with 4-mercaptobenzoic acid; detection and regeneration also demonstrated by shuffling between different analytes |
| Ag flower and MOF leaf decorated carbon cloth253 | 1.00 × 10−6 M rhodamine 6G | Superhydrophobicity | Sonicated in 30 mL of deionized water for 10 min | Analyte fully removed; SERS activity decreased by 17.3% after 6 cycles |
| Si wafer decorated with hierarchical Ag nanostructures on indium oxide nano-branches254 | 2.1 × 10−5 M rhodamine 6G water solution | Superhydrophobicity | Sonicated in isopropyl-alcohol bath for 10 min | SERS activity fully maintained for 10 cycles |
| MIP@Ag266 | 1 × 10−5 M bisphenol A in acetonitrile solution | Intermolecular interaction | Treatment with methanol–acetic acid solution (details unspecified) | 29% drop off in SERS activity after 4 cycles of regeneration |
| Ag decorated nanotip270 | Unspecified concentration of NADH water solution | Intermolecular interaction | Treatment with 1.0 mM FeCl3 solution for 20–30 min | Raman tag fully reversed; slight drop off in signal intensity after 4 regeneration cycles |
6. Conclusions and perspectives
In conclusion, this review introduced the main strategies for constructing multifunctional enhancing substrates designed for practical SERS analysis. Depending on their functionality, the multifunctional SERS substrates were categorized into flexible substrates for in situ analysis of surface residues, separation-enhancement-in-one substrates for direct chemical analysis of complex samples, calibration-enhancement-in-one substrates for direct quantitative analysis of complex samples and regeneration-enhancement-in-one SERS substrates for repeated use. In general, some success has been achieved in incorporating the above functionalities into enhancing substrates which allows reproducible and quantitative SERS analysis of certain complex real-life samples to be performed. Moving forward, the next step is to develop multifunctional substrates which combine several of the functionalities discussed above. In this regard, there have already been some successful demonstrations of substrates with combined functionalities.94,199,202,208,213 In addition, other important metrics, such as suitability for automated mass production and stability during storage must also be taken into consideration. While the importance of these properties has been recognized to some degree by the SERS community,162,279–281 more rigorous standards must be established in order to facilitate the routine application of SERS in practice. For example, a SERS substrate should not be defined “cheap” simply by the apparent cost of the raw materials within the substrate but as a whole, which includes the cost of production, application, storage and disposal. To reach practical standards, the stability of the substrates should be assessed at least by months rather than days. These challenges can likely be addressed by smart substrate engineering combined with the discovery of novel enhancing materials, such as semiconductors and plasmonic polymers,247,282 which offer similar magnitudes of signal enhancement to Ag/Au but at a reduced cost. Finally, standardized procedures and methodologies must be developed to determine substrate performance, as well as for the application of SERS in practice,283,284 since these are crucial preconditions for promoting SERS to non-specialist users.Author contributions
C. L. contributed to conceptualization, data curation and drafting of the manuscript. Y. H. contributed drafting of the manuscript. X. L. contributed to conceptualization and data curation. Y. Z. contributed to drafting and reviewing of the manuscript. Q. C. contributed to data curation. Z. Y. contributed to conceptualization, data curation and reviewing of the manuscript. Z. A. contributed to reviewing of the manuscript. S. B. contributed to supervision and funding acquisition. Y. X. contributed to conceptualization, data curation, visualization, supervision, funding acquisition and drafting, reviewing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Z. Y. and C. L. acknowledges the University Special Research Scholarship (Q. U. B.) for financial support. Y. X. acknowledges the Leverhulme Trust (Early Career Fellowship) for funding support. Y. Z. acknowledges the Chinese Scholarship Council (202008370188) for funding support. Z. A. acknowledges the Jeddah University Saudi Arabia for funding support. The authors thank Ms Yuhua Deng for her assistance with English.Notes and references
- D. A. Long, The Raman Effect, John Wiley & Sons, Ltd, Chichester, UK, 2002 Search PubMed.
- E. J. Heller, R. Sundberg and D. Tannor, J. Phys. Chem., 1982, 86, 1822–1833 CrossRef CAS.
- R. S. Das and Y. K. Agrawal, Vib. Spectrosc., 2011, 57, 163–176 CrossRef CAS.
- J. R. Lombardi and R. L. Birke, Acc. Chem. Res., 2009, 42, 734–742 CrossRef CAS PubMed.
- S. Y. Ding, E. M. You, Z. Q. Tian and M. Moskovits, Chem. Soc. Rev., 2017, 46, 4042–4076 RSC.
- N. Valley, N. Greeneltch, R. P. Van Duyne and G. C. Schatz, J. Phys. Chem. Lett., 2013, 4, 2599–2604 CrossRef CAS.
- N. P. Pieczonka and R. F. Aroca, Chem. Soc. Rev., 2008, 37, 946–954 RSC.
- E. C. Le Ru and P. G. Etchegoin, Annu. Rev. Phys. Chem., 2012, 63, 65–87 CrossRef CAS PubMed.
- R. Panneerselvam, G. K. Liu, Y. H. Wang, J. Y. Liu, S. Y. Ding, J. F. Li, D. Y. Wu and Z. Q. Tian, Chem. Commun., 2017, 54, 10–25 RSC.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguie, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. Garcia de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Kall, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J. F. Li, X. Y. Ling, S. A. Maier, T. Mayerhofer, M. Moskovits, K. Murakoshi, J. M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlucker, L. L. Tay, K. G. Thomas, Z. Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzan, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- S.-Y. Ding, J. Yi, J.-F. Li, B. Ren, D.-Y. Wu, R. Panneerselvam and Z.-Q. Tian, Nat. Rev. Mater., 2016, 1, 16021 CrossRef CAS.
- B. Sharma, R. R. Frontiera, A.-I. Henry, E. Ringe and R. P. Van Duyne, Mater. Today, 2012, 15, 16–25 CrossRef CAS.
- M. Moskovits, Rev. Mod. Phys., 1985, 57, 783–826 CrossRef CAS.
- M. P. Fleischmann, P. J. Hendra and A. J. Mcquillan, Chem. Phys. Lett., 1974, 26, 163–166 CrossRef CAS.
- D. L. Jeanmaire and R. P. Van Duyne, J. Electroanal. Chem. Interfacial Electrochem., 1977, 84, 1–20 CrossRef CAS.
- M. G. Albrecht and J. A. Creighton, J. Am. Chem. Soc., 1977, 99, 5215–5217 CrossRef CAS.
- T. E. Furtak, Solid State Commun., 1978, 28, 903–906 CrossRef CAS.
- M. Moskovits, Notes Rec. Roy. Soc., 2012, 66, 195–203 CrossRef.
- M. Moskovits, J. Chem. Phys., 1978, 69, 4159–4161 CrossRef CAS.
- P. F. Liao, J. G. Bergman, D. S. Chemla, A. Wokaun, J. Melngailis, A. M. Hawryluk and N. P. Economou, Chem. Phys. Lett., 1981, 82, 355–359 CrossRef CAS.
- T. E. Furtak and J. Reyes, Surf. Sci., 1980, 93, 351–382 CrossRef CAS.
- M. F. Cardinal, E. Vander Ende, R. A. Hackler, M. O. McAnally, P. C. Stair, G. C. Schatz and R. P. Van Duyne, Chem. Soc. Rev., 2017, 46, 3886–3903 RSC.
- H. K. Lee, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, C. L. Lay, H. Y. F. Sim, Y. C. Kao, Q. An and X. Y. Ling, Chem. Soc. Rev., 2019, 48, 731–756 RSC.
- G. Demirel, H. Usta, M. Yilmaz, M. Celik, H. A. Alidagi and F. Buyukserin, J. Mater. Chem. C, 2018, 6, 5314–5335 RSC.
- I. Izquierdo-Lorenzo, S. Jradi and P.-M. Adam, RSC Adv., 2014, 4, 4128–4133 RSC.
- R. Gao, X. Song, C. Zhan, C. Weng, S. Cheng, K. Guo, N. Ma, H. Chang, Z. Guo, L.-B. Luo and L. Yu, Sens. Actuators, B, 2020, 314, 128081 CrossRef CAS.
- N. Gisbert Quilis, M. Lequeux, P. Venugopalan, I. Khan, W. Knoll, S. Boujday, M. Lamy de la Chapelle and J. Dostalek, Nanoscale, 2018, 10, 10268–10276 RSC.
- L. Qin, Y. Huang, F. Xia, L. Wang, J. Ning, H. Chen, X. Wang, W. Zhang, Y. Peng, Q. Liu and Z. Zhang, Nano Lett., 2020, 20, 4916–4923 CrossRef CAS PubMed.
- K. Hasna, A. Antony, J. Puigdollers, K. R. Kumar and M. K. Jayaraj, Nano Res., 2016, 9, 3075–3083 CrossRef CAS.
- M. Yang, R. Alvarez-Puebla, H. S. Kim, P. Aldeanueva-Potel, L. M. Liz-Marzan and N. A. Kotov, Nano Lett., 2010, 10, 4013–4019 CrossRef CAS PubMed.
- W. S. Chew, S. Pedireddy, Y. H. Lee, W. W. Tjiu, Y. Liu, Z. Yang and X. Y. Ling, Chem. Mater., 2015, 27, 7827–7834 CrossRef CAS.
- A. J. Blanch, M. Doblinger and J. Rodriguez-Fernandez, Small, 2015, 11, 4550–4559 CrossRef CAS PubMed.
- Z. Wu, Q. Zeng and H. Wang, J. Mater. Chem. C, 2016, 4, 2614–2620 RSC.
- P. Yang, J. Zheng, Y. Xu, Q. Zhang and L. Jiang, Adv. Mater., 2016, 28, 10508–10517 CrossRef CAS PubMed.
- Y. Xu, M. P. Konrad, W. W. Lee, Z. Ye and S. E. Bell, Nano Lett., 2016, 16, 5255–5260 CrossRef CAS PubMed.
- W. Li, P. H. C. Camargo, L. Au, Q. Zhang, M. Rycenga and Y. Xia, Angew. Chem., Int. Ed., 2010, 49, 164–168 CrossRef CAS PubMed.
- F. Zou, H. Zhou, T. V. Tan, J. Kim, K. Koh and J. Lee, ACS Appl. Mater. Interfaces, 2015, 7, 12168–12175 CrossRef CAS PubMed.
- C. Li, Y. Xu, X. Li, Z. Ye, C. Yao, Q. Chen, Y. Zhang and S. E. J. Bell, Adv. Mater. Interfaces, 2020, 7, 2000391 CrossRef CAS.
- Z. Ye, C. Li, Q. Chen, Y. Xu and S. E. J. A. C. Bell, Angew. Chem., Int. Ed., 2019, 131, 19230–19235 CrossRef.
- R. Shi, X. Liu and Y. Ying, J. Agric. Food Chem., 2018, 66, 6525–6543 CrossRef CAS PubMed.
- M. Fan, G. F. S. Andrade and A. G. Brolo, Anal. Chim. Acta, 2020, 1097, 1–29 CrossRef CAS PubMed.
- M. Liszewska, B. Bartosewicz, B. Budner, B. Nasiłowska, M. Szala, J. L. Weyher, I. Dzięcielewski, Z. Mierczyk and B. J. Jankiewicz, Vib. Spectrosc., 2019, 100, 79–85 CrossRef CAS.
- Y. Liu, Y. Zhang, M. Tardivel, M. Lequeux, X. Chen, W. Liu, J. Huang, H. Tian, Q. Liu, G. Huang, R. Gillibert, M. L. de la Chapelle and W. Fu, Plasmonics, 2019, 15, 743–752 CrossRef.
- S. Almaviva, S. Botti, L. Cantarini, R. Fantoni, S. Lecci, A. Palucci, A. Puiu and A. Rufoloni, J. Raman Spectrosc., 2014, 45, 41–46 CrossRef CAS.
- D. Song, R. Yang, F. Long and A. Zhu, J. Environ. Sci., 2019, 80, 14–34 CrossRef PubMed.
- H. Wei, S. M. Hossein Abtahi and P. J. Vikesland, Environ. Sci.: Nano, 2015, 2, 120–135 RSC.
- C. Zong, M. Xu, L. J. Xu, T. Wei, X. Ma, X. S. Zheng, R. Hu and B. Ren, Chem. Rev., 2018, 118, 4946–4980 CrossRef CAS PubMed.
- H. Chen, A. Das, L. Bi, N. Choi, J. I. Moon, Y. Wu, S. Park and J. Choo, Nanoscale, 2020, 12, 21560–21570 RSC.
- C. Muehlethaler, M. Leona and J. R. Lombardi, Anal. Chem., 2016, 88, 152–169 CrossRef CAS PubMed.
- J. Wu, L. Zhang, F. Huang, X. Ji, H. Dai and W. Wu, J. Hazard. Mater., 2020, 387, 121714 CrossRef CAS PubMed.
- R. Pilot, J. Raman Spectrosc., 2018, 49, 954–981 CrossRef CAS.
- J. R. Anema, J. F. Li, Z. L. Yang, B. Ren and Z. Q. Tian, Annu. Rev. Anal. Chem., 2011, 4, 129–150 CrossRef CAS PubMed.
- S. Dick, M. P. Konrad, W. W. Lee, H. McCabe, J. N. McCracken, T. M. Rahman, A. Stewart, Y. Xu and S. E. Bell, Adv. Mater., 2016, 28, 5705–5711 CrossRef CAS PubMed.
- I. J. Jahn, A. Muhlig and D. Cialla-May, Anal. Bioanal. Chem., 2020, 412, 5999–6007 CrossRef CAS PubMed.
- Y. Zhang, S. Zhao, J. Zheng and L. He, TrAC, Trends Anal. Chem., 2017, 90, 1–13 CrossRef.
- G. Ochoa-Vazquez, B. Kharisov, A. Arizmendi-Morquecho, A. Cario, C. Aymonier, S. Marre and I. Lopez, IEEE Trans. Nanobioscience, 2019, 18, 558–566 Search PubMed.
- A. Subaihi, D. K. Trivedi, K. A. Hollywood, J. Bluett, Y. Xu, H. Muhamadali, D. I. Ellis and R. Goodacre, Anal. Chem., 2017, 89, 6702–6709 CrossRef CAS PubMed.
- G. C. Phan-Quang, E. H. Z. Wee, F. Yang, H. K. Lee, I. Y. Phang, X. Feng, R. A. Alvarez-Puebla and X. Y. Ling, Angew. Chem., Int. Ed., 2017, 56, 5565–5569 CrossRef CAS PubMed.
- G. Trachta, B. Schwarze, B. Sägmüller, G. Brehm and S. Schneider, J. Mol. Struct., 2004, 693, 175–185 CrossRef CAS.
- Y. Wang, Q. Ruan, Z. C. Lei, S. C. Lin, Z. Zhu, L. Zhou and C. Yang, Anal. Chem., 2018, 90, 5224–5231 CrossRef CAS PubMed.
- O. Durucan, K. Wu, M. Viehrig, T. Rindzevicius and A. Boisen, ACS Sens., 2018, 3, 2492–2498 CrossRef CAS PubMed.
- L. Yang, P. Li and J. Liu, RSC Adv., 2014, 4, 49635–49646 RSC.
- J.-A. Huang, Y.-L. Zhang, H. Ding and H.-B. Sun, Adv. Opt. Mater., 2015, 3, 618–633 CrossRef CAS.
- J. Prakash, Int. Rev. Phys. Chem., 2019, 38, 201–242 Search PubMed.
- Y. Yuan, N. Panwar, S. H. K. Yap, Q. Wu, S. Zeng, J. Xu, S. C. Tjin, J. Song, J. Qu and K.-T. Yong, Coord. Chem. Rev., 2017, 337, 1–33 CrossRef CAS.
- Z. Li, X. Huang and G. Lu, J. Mater. Chem. C, 2020, 8, 3956–3969 RSC.
- D. Li, D. Yao, C. Li, Y. Luo, A. Liang, G. Wen and Z. Jiang, TrAC, Trends Anal. Chem., 2020, 127, 115885 CrossRef CAS.
- P. Wang, L. Wu, Z. Lu, Q. Li, W. Yin, F. Ding and H. Han, Anal. Chem., 2017, 89, 2424–2431 CrossRef CAS PubMed.
- Y. Xu, Z. Ye, C. Li, H. McCabe, J. Kelly and S. E. J. Bell, Appl. Mater. Today, 2018, 13, 352–358 CrossRef.
- J. Jeon, S. H. Lee, Y. Joung, K. Kim, N. Choi and J. Choo, Sens. Actuators, B, 2020, 321, 128521 CrossRef CAS.
- Q. Wu, G. Chen, S. Qiu, S. Feng and D. Lin, Nanoscale, 2021, 13, 7574–7582 RSC.
- X. Gao, Y. Yin, H. Wu, Z. Hao, J. Li, S. Wang and Y. Liu, Anal. Chem., 2021, 93, 1569–1577 CrossRef CAS PubMed.
- Y. Chen, H. Liu, Y. Tian, Y. Du, Y. Ma, S. Zeng, C. Gu, T. Jiang and J. Zhou, ACS Appl. Mater. Interfaces, 2020, 12, 14386–14399 CrossRef CAS PubMed.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- Y. Kou, T. Wu, H. Zheng, N. R. Kadasala, S. Yang, C. Guo, L. Chen, Y. Liu and J. Yang, ACS Sustainable Chem. Eng., 2020, 8, 14549–14556 CrossRef CAS.
- J. F. Li, Y. F. Huang, Y. Ding, Z. L. Yang, S. B. Li, X. S. Zhou, F. R. Fan, W. Zhang, Z. Y. Zhou, D. Y. Wu, B. Ren, Z. L. Wang and Z. Q. Tian, Nature, 2010, 464, 392–395 CrossRef CAS PubMed.
- D. Zhang, H. Pu, L. Huang and D.-W. Sun, Trends Food Sci. Technol., 2021, 109, 690–701 CrossRef CAS.
- K. Xu, R. Zhou, K. Takei and M. Hong, Adv. Sci., 2019, 6, 1900925 CrossRef PubMed.
- D. Cheng, M. He, J. Ran, G. Cai, J. Wu and X. Wang, Sens. Actuators, B, 2018, 270, 508–517 CrossRef CAS.
- L. Kong, M. Huang, J. Chen and M. Lin, New J. Chem., 2020, 44, 12779–12784 RSC.
- S. Lin, W. Hasi, S. Han, X. Lin and L. Wang, Anal. Methods, 2020, 12, 2571–2579 RSC.
- K. Wang, M. Huang, J. Chen, L. Lin, L. Kong, X. Liu, H. Wang and M. Lin, J. Raman Spectrosc., 2018, 49, 493–498 CrossRef CAS.
- J. Chen, M. Huang, L. Kong and M. Lin, Carbohydr. Polym., 2019, 205, 596–600 CrossRef CAS PubMed.
- J. Chen, Y. Huang, P. Kannan, L. Zhang, Z. Lin, J. Zhang, T. Chen and L. Guo, Anal. Chem., 2016, 88, 2149–2155 CrossRef CAS PubMed.
- J. Jiang, S. Zou, Y. Li, F. Zhao, J. Chen, S. Wang, H. Wu, J. Xu, M. Chu, J. Liao and Z. Zhang, Microchim. Acta, 2019, 186, 603 CrossRef PubMed.
- H. Zhao, W. Hasi, L. Bao, Y. Liu, S. Han and D. Lin, J. Raman Spectrosc., 2018, 49, 1469–1477 CrossRef CAS.
- N. Yang, T. T. You, Y. K. Gao, C. M. Zhang and P. G. Yin, J. Agric. Food Chem., 2018, 66, 6889–6896 CrossRef CAS PubMed.
- J. Jiang, F. Zhao, S. Shi, Y. Du, J. Chen, S. Wang, J. Xu, C. Li and J. Liao, ACS Omega, 2019, 4, 12319–12324 CrossRef CAS PubMed.
- H. Wu, Y. Luo, C. Hou, D. Huo, Y. Zhou, S. Zou, J. Zhao and Y. Lei, Sens. Actuators, B, 2019, 285, 123–128 CrossRef CAS.
- T. Liyanage, A. Rael, S. Shaffer, S. Zaidi, J. V. Goodpaster and R. Sardar, Analyst, 2018, 143, 2012–2022 RSC.
- S. Lin, X. Lin, S. Han, Y. Liu, W. Hasi and L. Wang, Anal. Chim. Acta, 2020, 1108, 167–176 CrossRef CAS PubMed.
- W. A. Tegegne, W.-N. Su, M.-C. Tsai, A. B. Beyene and B.-J. Hwang, Appl. Mater. Today, 2020, 21, 100871 CrossRef.
- G. Xiao, Y. Li, W. Shi, L. Shen, Q. Chen and L. Huang, Appl. Surf. Sci., 2017, 404, 334–341 CrossRef CAS.
- J. Guo, Z. Zhong, Y. Li, Y. Liu, R. Wang and H. Ju, ACS Appl. Mater. Interfaces, 2019, 11, 36399–36408 CrossRef CAS PubMed.
- D. Wang, F. Wang and H. Yang, Sens. Actuators, B, 2018, 274, 676–681 CrossRef CAS.
- C. Wang, K. W. Wong, Q. Wang, Y. Zhou, C. Tang, M. Fan, J. Mei and W. M. Lau, Talanta, 2019, 191, 241–247 CrossRef CAS PubMed.
- J. Sun, L. Gong, Y. Lu, D. Wang, Z. Gong and M. Fan, Analyst, 2018, 143, 2689–2695 RSC.
- W. Fan, S. Yang, Y. Zhang, B. Huang, Z. Gong, D. Wang and M. Fan, ACS Sens., 2020, 5, 3599–3606 CrossRef CAS PubMed.
- L. Ma, J. Wang, C. Ren, P. Ju, Y. Huang, F. Zhang, F. Zhao, Z. Zhang and D. Zhang, Sens. Actuators, B, 2020, 321, 128617 CrossRef CAS.
- K. Wang, D. W. Sun, H. Pu and Q. Wei, Talanta, 2021, 223, 121782 CrossRef CAS PubMed.
- Y. Ma, Y. Wang, Y. Luo, H. Duan, D. Li, H. Xu and E. K. Fodjo, Anal. Methods, 2018, 10, 4655–4664 RSC.
- M. Wang, G. Shi, Y. Zhu, Y. Wang and W. Ma, Nanomaterials, 2018, 8, 289 CrossRef PubMed.
- X. Bian, J. Xu, J. Yang, K.-L. Chiu and S. Jiang, Surf. Interfaces, 2021, 23, 100995 CrossRef CAS.
- G. C. Shi, M. L. Wang, Y. Y. Zhu, L. Shen, W. L. Ma, Y. H. Wang and R. F. Li, Sci. Rep., 2018, 8, 6916 CrossRef PubMed.
- V. Suresh, L. Ding, A. B. Chew and F. L. Yap, ACS Appl. Nano Mater., 2018, 1, 886–893 CrossRef CAS.
- Y. Kalachyova, M. Erzina, P. Postnikov, V. Svorcik and O. Lyutakov, Appl. Surf. Sci., 2018, 458, 95–99 CrossRef CAS.
- Y. Wang, M. Zhang, L. Feng, B. Dong, T. Xu, D. Li, L. Jiang and L. Chi, Small, 2019, 15, e1804527 CrossRef PubMed.
- Y. Wang, Y. Jin, X. Xiao, T. Zhang, H. Yang, Y. Zhao, J. Wang, K. Jiang, S. Fan and Q. Li, Nanoscale, 2018, 10, 15195–15204 RSC.
- J. Liu, T. Si and Z. Zhang, Talanta, 2019, 204, 189–197 CrossRef CAS PubMed.
- X. Liu, J. Ma, P. Jiang, J. Shen, R. Wang, Y. Wang and G. Tu, ACS Appl. Mater. Interfaces, 2020, 12, 45332–45341 CrossRef CAS PubMed.
- A. Nenashkina, S. Koltsov, O. Y. Orlova, A. A. Nikitina, D. A. Kirilenko, D. V. Andreeva, M. Blanco-Formoso, N. Pazos-Perez, R. Alvarez-Puebla and E. V. Skorb, Nanoscale, 2021, 13, 7375–7380 RSC.
- V. T. N. Linh, J. Moon, C. Mun, V. Devaraj, J.-W. Oh, S.-G. Park, D.-H. Kim, J. Choo, Y.-I. Lee and H. S. Jung, Sens. Actuators, B, 2019, 291, 369–377 CrossRef CAS.
- Y. Chen, F. Ge, S. Guang and Z. Cai, Appl. Surf. Sci., 2018, 436, 111–116 CrossRef CAS.
- W. Zhao, Y. Zhang, J. Yang, J. Li, Y. Feng, M. Quan, Z. Yang and S. Xiao, Nanoscale, 2020, 12, 18056–18066 RSC.
- W. Wei, Y. Du, L. Zhang, Y. Yang and Y. Gao, J. Mater. Chem. C, 2018, 6, 8793–8803 RSC.
- X. He, S. Yang, T. Xu, Y. Song and X. Zhang, Biosens. Bioelectron., 2020, 152, 112013 CrossRef CAS PubMed.
- Y. Ma, Y. Chen, Y. Tian, C. Gu and T. Jiang, J. Agric. Food Chem., 2021, 69, 1975–1983 CrossRef CAS PubMed.
- Y. Ma, Y. Du, Y. Chen, C. Gu, T. Jiang, G. Wei and J. Zhou, Chem. Eng. J., 2020, 381, 122710 CrossRef CAS.
- L. Song, Y. Huang, Z. Nie and T. Chen, Nanoscale, 2020, 12, 7433–7460 RSC.
- Z. Ye, C. Li, Q. Chen, Y. Xu and S. E. J. Bell, Nanoscale, 2021, 13, 5937–5953 RSC.
- L. Zhang, X. Li, W. Liu, R. Hao, H. Jia, Y. Dai, M. Usman Amin, H. You, T. Li and J. Fang, J. Mater. Chem. C, 2019, 7, 15259–15268 RSC.
- L. B. Zhong, Q. Liu, P. Wu, Q. F. Niu, H. Zhang and Y. M. Zheng, Environ. Sci. Technol., 2018, 52, 5812–5820 CrossRef CAS PubMed.
- P. Wu, L. B. Zhong, Q. Liu, X. Zhou and Y. M. Zheng, Nanoscale, 2019, 11, 12829–12836 RSC.
- Y. Chen, K. J. Si, D. Sikdar, Y. Tang, M. Premaratne and W. Cheng, Adv. Opt. Mater., 2015, 3, 919–924 CrossRef CAS.
- L. Li and W. S. Chin, ACS Appl. Mater. Interfaces, 2020, 12, 37538–37548 CrossRef CAS PubMed.
- Y. Xu, M. P. Konrad, J. L. Trotter, C. P. McCoy and S. E. Bell, Small, 2017, 13, 1602163 CrossRef PubMed.
- C. Li, Q. Chen, H. McCabe, J. Kelly, Z. Ye, Y. Xu and S. E. J. Bell, Data Brief, 2019, 23, 103746 CrossRef PubMed.
- M. Wang, C. Fu, X. Liu, Z. Lin, N. Yang and S. Yu, Nanoscale, 2015, 7, 15191–15196 RSC.
- Q. Zhang, D. Li, X. Cao, H. Gu and W. Deng, Anal. Chem., 2019, 91, 11192–11199 CrossRef CAS PubMed.
- H. S. Siebe, Q. Chen, X. Li, Y. Xu, W. R. Browne and S. E. J. Bell, Analyst, 2021, 146, 1281–1288 RSC.
- S. Wang, B. Sun, J. Feng, F. An, N. Li, H. Wang and M. Tian, Anal. Methods, 2020, 12, 5657–5670 RSC.
- X. Guo, J. Li, M. Arabi, X. Wang, Y. Wang and L. Chen, ACS Sens., 2020, 5, 601–619 CrossRef CAS PubMed.
- T. Yaseen, H. Pu and D.-W. Sun, Trends Food Sci. Technol., 2018, 72, 162–174 CrossRef CAS.
- K. Kim, L. Kashefi-Kheyrabadi, Y. Joung, K. Kim, H. Dang, S. G. Chavan, M.-H. Lee and J. Choo, Sens. Actuators, B, 2021, 329, 129214 CrossRef CAS.
- H. Zhang, H. Lai, X. Wu, G. Li and Y. Hu, Anal. Chem., 2020, 92, 4607–4613 CrossRef CAS PubMed.
- S. Yu, Z. Liu, H. Li, J. Zhang, X. X. Yuan, X. Jia and Y. Wu, Analyst, 2018, 143, 883–890 RSC.
- Z. Liu, Y. Wang, R. Deng, L. Yang, S. Yu, S. Xu and W. Xu, ACS Appl. Mater. Interfaces, 2016, 8, 14160–14168 CrossRef CAS PubMed.
- Z. Ye, C. Li, N. Skillen, Y. Xu, H. McCabe, J. Kelly, P. Robertson and S. E. J. Bell, Appl. Mater. Today, 2019, 15, 398–404 CrossRef.
- C. Xu, W. Lu, M. Li, Y. Cao, H. Pang, C. Gong and S. Cheng, Adv. Mater. Interfaces, 2019, 6, 1900836 CrossRef CAS.
- C. Liu, F. Lei, J. Li, Y. Wei, Z. Li, C. Zhang, B. Man and J. Yu, Appl. Surf. Sci., 2021, 542, 148735 CrossRef CAS.
- C. Liu, M. Yang, J. Yu, F. Lei, Y. Wei, Q. Peng, C. Li, Z. Li, C. Zhang and B. Man, Sens. Actuators, B, 2020, 307, 127663 CrossRef.
- C. Xu, D. Cao, W. Lu, J. Sun and S. Cheng, New J. Chem., 2018, 42, 11660–11664 RSC.
- H. S. Jung, E. H. Koh, C. Mun, J. Min, W. Sohng, H. Chung, J.-Y. Yang, S. Lee, H. J. Kim, S.-G. Park, M.-Y. Lee and D.-H. Kim, J. Mater. Chem. C, 2019, 7, 13059–13069 RSC.
- R. Li, M. Chen, H. Yang, N. Hao, Q. Liu, M. Peng, L. Wang, Y. Hu and X. Chen, Anal. Chem., 2021, 93, 4657–4665 CrossRef CAS PubMed.
- N. E. Markina, A. V. Markin, K. Weber, J. Popp and D. Cialla-May, Anal. Chim. Acta, 2020, 1109, 61–68 CrossRef CAS PubMed.
- L. Morelli, L. Serioli, F. A. Centorbi, C. B. Jendresen, M. Matteucci, O. Ilchenko, D. Demarchi, A. T. Nielsen, K. Zor and A. Boisen, Lab Chip, 2018, 18, 869–877 RSC.
- L. Tian, M. Su, F. Yu, Y. Xu, X. Li, L. Li, H. Liu and W. Tan, Nat. Commun., 2018, 9, 3642 CrossRef PubMed.
- W. Zhu, B. Y. Wen, L. J. Jie, X. D. Tian, Z. L. Yang, P. M. Radjenovic, S. Y. Luo, Z. Q. Tian and J. F. Li, Biosens. Bioelectron., 2020, 154, 112067 CrossRef CAS PubMed.
- L. L. Qu, Q. Jia, C. Liu, W. Wang, L. Duan, G. Yang, C. Q. Han and H. Li, J. Chromatogr. A, 2018, 1579, 115–120 CrossRef CAS PubMed.
- Y. Kang, T. Wu, W. Chen, L. Li and Y. Du, Food Chem., 2019, 270, 494–501 CrossRef CAS PubMed.
- B. Campanella, J. Botti, T. Cavaleri, F. Cicogna, S. Legnaioli, S. Pagnotta, F. Poggialini, T. Poli, D. Scalarone and V. Palleschi, Microchem. J., 2020, 152, 104292 CrossRef CAS.
- Q. Zhu, M. Chen, L. Han, Y. Yuan and F. Lu, Anal. Methods, 2017, 9, 1595–1602 RSC.
- Y. Li, C. Zhao, C. Lu, S. Zhou, G. Tian, L. He, Y. Bao, M. L. Fauconnier, H. Xiao and J. Zheng, Food Chem., 2021, 338, 128115 CrossRef CAS PubMed.
- J. Chen, Y. W. Huang and Y. Zhao, J. Mater. Chem. B, 2015, 3, 1898–1906 RSC.
- Y. Zhao, A. Tan, K. Squire, K. Sivashanmugan and A. X. Wang, Sens. Actuators, B, 2019, 299, 126902 CrossRef CAS PubMed.
- B. B. Zhang, Y. Shi, H. Chen, Q. X. Zhu, F. Lu and Y. W. Li, Anal. Chim. Acta, 2018, 997, 35–43 CrossRef CAS PubMed.
- B. S. Lee, D. Z. Lin, C. H. Huang and T. J. Yen, J. Raman Spectrosc., 2018, 49, 1920–1927 CrossRef CAS.
- B. S. Lee, P. C. Lin, D. Z. Lin and T. J. Yen, Sci. Rep., 2018, 8, 516 CrossRef PubMed.
- Y. Song, Z. Ma, H. Fang, Q. Zhang, Q. Zhou, Z. Chen, H. Yang and F. Wang, ACS Sens., 2020, 5, 1455–1464 CrossRef CAS PubMed.
- J. D. Weatherston, R. K. O. Seguban, D. Hunt and H. J. Wu, ACS Sens., 2018, 3, 852–857 CrossRef CAS PubMed.
- A. G. Berger, S. M. Restaino and I. M. White, Anal. Chim. Acta, 2017, 949, 59–66 CrossRef CAS PubMed.
- S. M. Restaino and I. M. White, Anal. Chim. Acta, 2019, 1060, 17–29 CrossRef CAS PubMed.
- Y. Xu, X. Gao, C. Yang, B. Man and J. Leng, J. Mater. Chem. C, 2021, 9, 2763–2774 RSC.
- Y. Zeng, J. Ren, A. Shen and J. Hu, ACS Appl. Mater. Interfaces, 2016, 8, 27772–27778 CrossRef CAS PubMed.
- H. Zhao, J. Jin, W. Tian, R. Li, Z. Yu, W. Song, Q. Cong, B. Zhao and Y. Ozaki, J. Mater. Chem. A, 2015, 3, 4330–4337 RSC.
- H. Yang, Y. Xiang, X. Guo, Y. Wu, Y. Wen and H. Yang, Sens. Actuators, B, 2018, 271, 118–121 CrossRef CAS.
- Y. X. Leong, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, I. Y. Phang and X. Y. Ling, Nano Lett., 2021, 21, 2642–2649 CrossRef CAS PubMed.
- H. He, D. W. Sun, H. Pu and L. Huang, Food Chem., 2020, 324, 126832 CrossRef CAS PubMed.
- Z. Zhou, R. Xiao, S. Cheng, S. Wang, L. Shi, C. Wang, K. Qi and S. Wang, Anal. Chim. Acta, 2021, 1160, 338421 CrossRef CAS PubMed.
- S. Pal, S. Harmsen, A. Oseledchyk, H. T. Hsu and M. F. Kircher, Adv. Funct. Mater., 2017, 27, 1606632 CrossRef PubMed.
- Q. Wang, Y. Hu, N. Jiang, J. Wang, M. Yu and X. Zhuang, Bioconjugate Chem., 2020, 31, 813–820 CrossRef CAS PubMed.
- D. Zhang, L. Huang, B. Liu, H. Ni, L. Sun, E. Su, H. Chen, Z. Gu and X. Zhao, Biosens. Bioelectron., 2018, 106, 204–211 CrossRef CAS PubMed.
- E. Sheng, Y. Lu, Y. Xiao, Z. Li, H. Wang and Z. Dai, Biosens. Bioelectron., 2021, 181, 113149 CrossRef CAS PubMed.
- R. Wang, K. Kim, N. Choi, X. Wang, J. Lee, J. H. Jeon, G.-E. Rhie and J. Choo, Sens. Actuators, B, 2018, 270, 72–79 CrossRef CAS.
- C. Wang, C. Wang, X. Wang, K. Wang, Y. Zhu, Z. Rong, W. Wang, R. Xiao and S. Wang, ACS Appl. Mater. Interfaces, 2019, 11, 19495–19505 CrossRef CAS PubMed.
- Y. Xu, M. M. Hassan, S. Ali, H. Li and Q. Chen, Mikrochim. Acta, 2020, 187, 454 CrossRef CAS PubMed.
- Y. Zhu, L. Wu, H. Yan, Z. Lu, W. Yin and H. Han, Anal. Chim. Acta, 2020, 1101, 111–119 CrossRef CAS PubMed.
- H. R. Culver and N. A. Peppas, Chem. Mater., 2017, 29, 5753–5761 CrossRef CAS PubMed.
- K. Sivashanmugan, P. C. Liu, K. W. Tsai, Y. N. Chou, C. H. Lin, Y. Chang and T. C. Wen, Nanoscale, 2017, 9, 2865–2874 RSC.
- A. N. Masterson, S. Hati, G. Ren, T. Liyanage, N. E. Manicke, J. V. Goodpaster and R. Sardar, Anal. Chem., 2021, 93, 2578–2588 CrossRef CAS PubMed.
- S. S. Panikar, N. Banu, J. Haramati, G. Y. Gutierrez-Silerio, B. E. Bastidas-Ramirez, M. C. Tellez-Banuelos, T. A. Camacho-Villegas, S. D. Toro-Arreola and E. De la Rosa, Anal. Chim. Acta, 2020, 1138, 110–122 CrossRef CAS PubMed.
- F. Sun, H. C. Hung, A. Sinclair, P. Zhang, T. Bai, D. D. Galvan, P. Jain, B. Li, S. Jiang and Q. Yu, Nat. Commun., 2016, 7, 13437 CrossRef CAS PubMed.
- S. Fateixa, M. Raposo, H. I. S. Nogueira and T. Trindade, Talanta, 2018, 182, 558–566 CrossRef CAS PubMed.
- S. D. Dryden, S. Anastasova, G. Satta, A. J. Thompson, D. R. Leff and A. Darzi, Sci. Rep., 2021, 11, 8802 CrossRef CAS PubMed.
- Z. Yang, T. Liu, W. Wang and L. Zhang, Chem. Commun., 2020, 56, 3065–3068 RSC.
- D. Sun, F. Cao, H. Wang, S. Guan, A. Su, W. Xu and S. Xu, Sens. Actuators, B, 2021, 327, 128943 CrossRef CAS.
- M. Chen, W. Luo, Z. Zhang, R. Wang, Y. Zhu, H. Yang and X. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 42156–42166 CrossRef CAS PubMed.
- Q. Ding, J. Wang, X. Chen, H. Liu, Q. Li, Y. Wang and S. Yang, Nano Lett., 2020, 20, 7304–7312 CrossRef CAS PubMed.
- D. J. Kim, S. G. Park, D. H. Kim and S. H. Kim, Small, 2018, 14, 1802520 CrossRef PubMed.
- N. J. Halas, S. Lal, W. S. Chang, S. Link and P. Nordlander, Chem. Rev., 2011, 111, 3913–3961 CrossRef CAS PubMed.
- D. Chen, X. Zhu, J. Huang, G. Wang, Y. Zhao, F. Chen, J. Wei, Z. Song and Y. Zhao, Anal. Chem., 2018, 90, 9048–9054 CrossRef CAS PubMed.
- E. Dumont, C. De Bleye, J. Cailletaud, P. Y. Sacre, P. B. Van Lerberghe, B. Rogister, G. A. Rance, J. W. Aylott, P. Hubert and E. Ziemons, Talanta, 2018, 186, 8–16 CrossRef CAS PubMed.
- L. Cai, J. Dong, Y. Wang and X. Chen, Electrophoresis, 2019, 40, 2041–2049 CrossRef CAS PubMed.
- S. A. Majeed, Analyst, 2020, 145, 6744–6752 RSC.
- E. B. Santos, C. Valsecchi, J. L. S. Goncalves, L. F. Ávila and J. W. Menezes, Sensors, 2019, 19, 4394 CrossRef CAS PubMed.
- L. Xue, L. Chen, J. Dong, L. Cai, Y. Wang and X. Chen, Talanta, 2020, 208, 120360 CrossRef CAS PubMed.
- R. Mei, Y. Wang, Q. Yu, Y. Yin, R. Zhao and L. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 2059–2066 CrossRef CAS PubMed.
- Y. Shi, N. Chen, Y. Su, H. Wang and Y. He, Nanoscale, 2018, 10, 4010–4018 RSC.
- K. Yuan, Q. Mei, X. Guo, Y. Xu, D. Yang, B. J. Sanchez, B. Sheng, C. Liu, Z. Hu, G. Yu, H. Ma, H. Gao, C. Haisch, R. Niessner, Z. Jiang, Z. Jiang and H. Zhou, Chem. Sci., 2018, 9, 8781–8795 RSC.
- X. Yan, P. Li, B. Zhou, X. Tang, X. Li, S. Weng, L. Yang and J. Liu, Anal. Chem., 2017, 89, 4875–4881 CrossRef CAS PubMed.
- M. Li, J. Y. Wang, Q. Q. Chen, L. H. Lin, P. Radjenovic, H. Zhang, S. Y. Luo, Z. Q. Tian and J. F. Li, Anal. Chem., 2019, 91, 15025–15031 CrossRef CAS PubMed.
- X. Lin, Y. Wang, L. Wang, Y. Lu, J. Li, D. Lu, T. Zhou, Z. Huang, J. Huang, H. Huang, S. Qiu, R. Chen, D. Lin and S. Feng, Biosens. Bioelectron., 2019, 143, 111599 CrossRef CAS PubMed.
- W. Shen, X. Lin, C. Jiang, C. Li, H. Lin, J. Huang, S. Wang, G. Liu, X. Yan, Q. Zhong and B. Ren, Angew. Chem., Int. Ed., 2015, 54, 7308–7312 CrossRef CAS PubMed.
- X. Wang, B. Liu, M. Xiao, Y. Zou, W. Lai, H. Pei, M. F. Alam, W. Zhang, Y. Wan and L. Li, Biosens. Bioelectron., 2020, 156, 112130 CrossRef CAS PubMed.
- X. Dai, L. Lu, X. Zhang, Z.-L. Song, W. Song, Q. Chao, Q. Li, W. Wang, J. Chen, G.-C. Fan and X. Luo, Sens. Actuators, B, 2021, 334, 129605 CrossRef CAS.
- E. Hahm, M. G. Cha, E. J. Kang, X. H. Pham, S. H. Lee, H. M. Kim, D. E. Kim, Y. S. Lee, D. H. Jeong and B. H. Jun, ACS Appl. Mater. Interfaces, 2018, 10, 40748–40755 CrossRef CAS PubMed.
- S. Lin, X. Lin, Y. Liu, H. Zhao, W. Hasi and L. Wang, Anal. Methods, 2018, 10, 4201–4208 RSC.
- Y. Tian, H. Liu, Y. Chen, C. Gu, G. Wei and T. Jiang, ACS Sustainable Chem. Eng., 2020, 8, 16990–16999 CrossRef CAS.
- M. Chen, W. Luo, Q. Liu, N. Hao, Y. Zhu, M. Liu, L. Wang, H. Yang and X. Chen, Anal. Chem., 2018, 90, 13647–13654 CrossRef CAS PubMed.
- J. Liu, Z. Hong, W. Yang, C. Liu, Z. Lu, L. Wu, M. F. Foda, Z. Yang, H. Han and Y. Zhao, ACS Appl. Bio Mater., 2020, 4, 2009–2019 CrossRef.
- X. Zhao, S. Zhao, Z.-L. Song, X. Zhang, S. Zhang, W. Song and Z. Chen, Sens. Actuators, B, 2021, 331, 129373 CrossRef CAS.
- F. Yu, M. Su, L. Tian, H. Wang and H. Liu, Anal. Chem., 2018, 90, 5232–5238 CrossRef CAS PubMed.
- Y. Ma, H. Liu, Y. Chen, Y. Du, C. Gu, Z. Zhao, H. Si, G. Wei, T. Jiang and J. Zhou, ACS Appl. Nano Mater., 2020, 3, 4610–4622 CrossRef CAS.
- W. Nam, Y. Zhao, J. Song, S. Ali Safiabadi Tali, S. Kang, W. Zhu, H. J. Lezec, A. Agrawal, P. J. Vikesland and W. Zhou, J. Phys. Chem. Lett., 2020, 11, 9543–9551 CrossRef CAS PubMed.
- W. Nam, X. Ren, I. Kim, J. Strobl, M. Agah and W. Zhou, Anal. Chem., 2021, 93, 4601–4610 CrossRef CAS PubMed.
- A. Matikainen, T. Nuutinen, T. Itkonen, S. Heinilehto, J. Puustinen, J. Hiltunen, J. Lappalainen, P. Karioja and P. Vahimaa, Sci. Rep., 2016, 6, 37192 CrossRef CAS PubMed.
- K. L. Norrod and K. L. Rowlen, Anal. Chem., 1998, 70, 4218–4221 CrossRef CAS PubMed.
- A. Mechlińska, L. Wolska, J. Namieśnik and L. Wolska, TrAC, Trends Anal. Chem., 2010, 29, 820–831 CrossRef.
- N. Itoh and S. E. Bell, Analyst, 2017, 142, 994–998 RSC.
- X. M. Lin, Y. Cui, Y. H. Xu, B. Ren and Z. Q. Tian, Anal. Bioanal. Chem., 2009, 394, 1729–1745 CrossRef CAS PubMed.
- S. Chen, B. Liu, X. Zhang, Y. Mo, F. Chen, H. Shi, W. Zhang, C. Hu and J. Chen, Electrochim. Acta, 2018, 274, 242–249 CrossRef CAS.
- J. Wu, Y. Du, C. Wang, S. Bai, T. Zhang, T. Chen and A. Hu, Appl. Surf. Sci., 2019, 494, 583–590 CrossRef CAS.
- T. K. Naqvi, A. Bajpai, M. S. S. Bharati, M. M. Kulkarni, A. M. Siddiqui, V. R. Soma and P. K. Dwivedi, J. Hazard. Mater., 2021, 407, 124353 CrossRef CAS PubMed.
- H. Chi, C. Wang, Z. Wang, H. Zhu, V. S. D. Mesias, X. Dai, Q. Chen, W. Liu and J. Huang, Analyst, 2020, 145, 5158–5165 RSC.
- C. Byram, S. S. B. Moram and V. R. Soma, J. Raman Spectrosc., 2019, 50, 1103–1113 CrossRef CAS.
- L. Ma, H. Wu, Y. Huang, S. Zou, J. Li and Z. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 27162–27168 CrossRef CAS PubMed.
- H. Chong, G. Gao and G. Li, New J. Chem., 2020, 44, 704–708 RSC.
- X. Zhang, S. Si, X. Zhang, W. Wu, X. Xiao and C. Jiang, ACS Appl. Mater. Interfaces, 2017, 9, 40726–40733 CrossRef CAS PubMed.
- V. Shvalya, G. Filipič, D. Vengust, J. Zavašnik, M. Modic, I. Abdulhalim and U. Cvelbar, Appl. Surf. Sci., 2020, 517, 146205 CrossRef CAS.
- G. Fu, D. W. Sun, H. Pu and Q. Wei, Talanta, 2019, 195, 841–849 CrossRef CAS PubMed.
- R. B. Chevalier and J. R. Dwyer, J. Raman Spectrosc., 2020, 52, 608–615 CrossRef.
- Y. Yang, Y. Wang, X. Zhang, G. Qi, S. Xu and W. Xu, J. Opt., 2015, 17, 075003 CrossRef.
- M. Y. Lv, H. Y. Teng, Z. Y. Chen, Y. M. Zhao, X. Zhang, L. Liu, Z. Wu, L. M. Liu and H. J. Xu, Sens. Actuators, B, 2015, 209, 820–827 CrossRef CAS.
- S. M. Ansar, F. S. Ameer, W. Hu, S. Zou, C. U. Pittman Jr and D. Zhang, Nano Lett., 2013, 13, 1226–1229 CrossRef CAS PubMed.
- Y. Du, H. Liu, Y. Chen, Y. Tian, X. Zhang, C. Gu, T. Jiang and J. Zhou, Appl. Surf. Sci., 2020, 528, 146953 CrossRef CAS.
- A. K. Pal, S. Pagal, K. Prashanth, G. K. Chandra, S. Umapathy and D. Bharathi Mohan, Sens. Actuators, B, 2019, 279, 157–169 CrossRef CAS.
- J. Huang, D. Ma, F. Chen, D. Chen, M. Bai, K. Xu and Y. Zhao, ACS Appl. Mater. Interfaces, 2017, 9, 7436–7446 CrossRef CAS PubMed.
- X. He, H. Wang, Z. Li, D. Chen, J. Liu and Q. Zhang, Nanoscale, 2015, 7, 8619–8626 RSC.
- H. Qiu, J. Guo, M. Wang, S. Ji, M. Cao, M. A. Padhiar and A. S. Bhatti, Colloids Surf., A, 2019, 560, 9–19 CrossRef CAS.
- L. L. Qu, Z. Q. Geng, W. Wang, K. C. Yang, W. P. Wang, C. Q. Han, G. H. Yang, R. Vajtai, D. W. Li and P. M. Ajayan, J. Hazard. Mater., 2019, 379, 120823 CrossRef CAS PubMed.
- J. Dong, J. Huang, A. Wang, G. V. Biesold-McGee, X. Zhang, S. Gao, S. Wang, Y. Lai and Z. Lin, Nano Energy, 2020, 71, 104579 CrossRef CAS.
- Q. Liu, P. Chen, Y. Ye, Y. Wan, B. Wang and Z. Luo, Adv. Mater. Interfaces, 2019, 6, 1900659 CrossRef.
- Y. Chen, H. Liu, X. Li, S. Tang, C. Gu, G. Wei, T. Jiang and X. Zhou, Sens. Actuators, B, 2021, 339, 129856 CrossRef CAS.
- J. Huang, C. Tang, G. Chen, Z. He, T. Wang, X. He, T. Yi, Y. Liu, L. Zhang and K. Du, ACS Appl. Mater. Interfaces, 2021, 13, 7735–7744 CrossRef CAS PubMed.
- O. Guselnikova, R. Samant, P. Postnikov, A. Trelin, V. Svorcik and O. Lyutakov, J. Mater. Chem. C, 2019, 7, 14181–14187 RSC.
- X. Tan, J. Melkersson, S. Wu, L. Wang and J. Zhang, ChemPhysChem, 2016, 17, 2630–2639 CrossRef CAS PubMed.
- B. Yang, S. Jin, S. Guo, Y. Park, L. Chen, B. Zhao and Y. M. Jung, ACS Omega, 2019, 4, 20101–20108 CrossRef CAS PubMed.
- M. Hu, Y. Quan, S. Yang, R. Su, H. Liu, M. Gao, L. Chen and J. Yang, Microsyst. Nanoeng., 2020, 6, 1–10 CrossRef.
- B. Qiu, M. Xing, Q. Yi and J. Zhang, Angew. Chem., Int. Ed., 2015, 54, 10643–10647 CrossRef CAS PubMed.
- X. Jiang, Q. Sang, M. Yang, J. Du, W. Wang, L. Yang, X. Han and B. Zhao, Phys. Chem. Chem. Phys., 2019, 21, 12850–12858 RSC.
- D. Yin, M.-L. Wang, Y.-Z. Wang, X. Hu, B. Liu, H. Liu, L. Ma and G.-G. Gao, J. Mater. Chem. C, 2019, 7, 9856–9864 RSC.
- Y. Wang, J. Ma, J. Zhu, N. Ye, X. Zhang and H. Huang, Water Res., 2016, 92, 104–112 CrossRef PubMed.
- Q. Wang, Z. Xu, Y. Zhao, H. Zhangsun, T. Bu, C. Zhang, X. Wang and L. Wang, Sens. Actuators, B, 2021, 329, 129080 CrossRef CAS.
- K. Min, K. S. Choi, W. J. Jeon, D. K. Lee, S. Oh, J. Lee, J.-Y. Choi and H. K. Yu, RSC Adv., 2018, 8, 12927–12932 RSC.
- D. Das, S. Senapati and K. K. Nanda, ACS Sustainable Chem. Eng., 2019, 7, 14089–14101 CrossRef CAS.
- A. Wang, L. Jiang, X. Li, Q. Xie, B. Li, Z. Wang, K. Du and Y. Lu, J. Mater. Chem. B, 2017, 5, 777–784 RSC.
- Y. Gao, N. Yang, T. You, C. Zhang and P. Yin, Sens. Actuators, B, 2018, 267, 129–135 CrossRef CAS.
- I. B. Becerril-Castro, F. Munoz-Munoz, A. B. Castro-Cesena, A. L. Gonzalez, R. A. Alvarez-Puebla and J. M. Romo-Herrera, Nanoscale, 2021, 13, 1738–1744 RSC.
- C. Wang, L. Tian, W. Zhu, S. Wang, N. Gao, K. Zhou, X. Yin, W. Zhang, L. Zhao and G. Li, Chem. Sci., 2018, 9, 889–895 RSC.
- G. Y. Chen, Y. B. Sun, P. C. Shi, T. Liu, Z. H. Li, S. H. Luo, X. C. Wang, X. Y. Cao, B. Ren, G. K. Liu, L. L. Yang and Z. Q. Tian, Light: Sci. Appl., 2021, 10, 85 CrossRef CAS PubMed.
- S. Kasera, L. O. Herrmann, J. del Barrio, J. J. Baumberg and O. A. Scherman, Sci. Rep., 2014, 4, 6785 CrossRef CAS PubMed.
- L. L. Tan, M. Wei, L. Shang and Y. W. Yang, Adv. Funct. Mater., 2020, 31, 2007277 CrossRef.
- V. Montes-Garcia, B. Gomez-Gonzalez, D. Martinez-Solis, J. M. Taboada, N. Jimenez-Otero, J. de Una-Alvarez, F. Obelleiro, L. Garcia-Rio, J. Perez-Juste and I. Pastoriza-Santos, ACS Appl. Mater. Interfaces, 2017, 9, 26372–26382 CrossRef CAS PubMed.
- E. Ekmen, M. Bilici, E. Turan, U. Tamer and A. Zengin, Sens. Actuators, B, 2020, 325, 128787 CrossRef CAS.
- X. Ren and X. Li, Anal. Methods, 2020, 12, 2858–2864 RSC.
- Z. Wang, R. Yan, S. Liao, Y. Miao, B. Zhang, F. Wang and H. Yang, Appl. Surf. Sci., 2018, 457, 323–331 CrossRef CAS.
- Z. Liu, Y. Gao, L. Jin, H. Jin, N. Xu, X. Yu and S. Yu, ACS Sustainable Chem. Eng., 2019, 7, 8168–8175 CrossRef CAS.
- D. Sun, G. Qi, F. Cao, W. Xu, Q. Chen and S. Xu, Talanta, 2017, 171, 159–165 CrossRef CAS PubMed.
- B. Han, Y.-L. Zhang, L. Zhu, X.-H. Chen, Z.-C. Ma, X.-L. Zhang, J.-N. Wang, W. Wang, Y.-Q. Liu, Q.-D. Chen and H.-B. Sun, Sens. Actuators, B, 2018, 270, 500–507 CrossRef CAS.
- J. Chen, J. Wang, Y. Geng, J. Yue, W. Shi, C. Liang, W. Xu and S. Xu, ACS Sens., 2021, 6, 1663–1670 CrossRef CAS PubMed.
- L. Jiang, L. Wang, D. S. Zhan, W. R. Jiang, E. K. Fodjo, M. E. Hafez, Y. M. Zhang, H. Zhao, R. C. Qian and D. W. Li, Biosens. Bioelectron., 2021, 175, 112918 CrossRef CAS PubMed.
- X. Yue, Y. Su, X. Wang, L. Li, W. Ji and Y. Ozaki, ACS Sens., 2019, 4, 2336–2342 CrossRef CAS PubMed.
- L. Wang, Z. F. Gan, D. Guo, H. L. Xia, F. T. Patrice, M. E. Hafez and D. W. Li, Anal. Chem., 2019, 91, 6507–6513 CAS.
- Y. Kang, T. Wu, X. Han, H. Gu and X. Zhang, Mikrochim. Acta, 2018, 185, 504 CrossRef PubMed.
- M. Viehrig, S. T. Rajendran, K. Sanger, M. S. Schmidt, T. S. Alstrom, T. Rindzevicius, K. Zor and A. Boisen, Anal. Chem., 2020, 92, 4317–4325 CrossRef CAS PubMed.
- S. Cong, Z. Wang, W. Gong, Z. Chen, W. Lu, J. R. Lombardi and Z. Zhao, Nat. Commun., 2019, 10, 678 CrossRef CAS PubMed.
- K. Wu, T. Li, M. S. Schmidt, T. Rindzevicius, A. Boisen and S. Ndoni, Adv. Funct. Mater., 2018, 28, 1704818 CrossRef.
- X. Peng, D. Li, Y. Li, H. Xing and W. Deng, J. Mater. Chem. B, 2021, 9, 1123–1130 RSC.
- J. F. Betz, W. W. Yu, Y. Cheng, I. M. White and G. W. Rubloff, Phys. Chem. Chem. Phys., 2014, 16, 2224–2239 RSC.
- P. Joshi and V. Santhanam, RSC Adv., 2016, 6, 68545–68552 RSC.
- C. Zhang, P. Yi, L. Peng, X. Lai, J. Chen, M. Huang and J. Ni, Sci. Rep., 2017, 7, 39814 CrossRef CAS PubMed.
- M. Yilmaz, E. Babur, M. Ozdemir, R. L. Gieseking, Y. Dede, U. Tamer, G. C. Schatz, A. Facchetti, H. Usta and G. Demirel, Nat. Mater., 2017, 16, 918–924 CrossRef CAS PubMed.
- S. E. J. Bell, G. Charron, E. Cortes, J. Kneipp, M. L. de la Chapelle, J. Langer, M. Prochazka, V. Tran and S. Schlucker, Angew. Chem., Int. Ed., 2020, 59, 5454–5462 CrossRef CAS PubMed.
- S. Fornasaro, F. Alsamad, M. Baia, L. A. E. Batista de Carvalho, C. Beleites, H. J. Byrne, A. Chiado, M. Chis, M. Chisanga, A. Daniel, J. Dybas, G. Eppe, G. Falgayrac, K. Faulds, H. Gebavi, F. Giorgis, R. Goodacre, D. Graham, P. La Manna, S. Laing, L. Litti, F. M. Lyng, K. Malek, C. Malherbe, M. P. M. Marques, M. Meneghetti, E. Mitri, V. Mohacek-Grosev, C. Morasso, H. Muhamadali, P. Musto, C. Novara, M. Pannico, G. Penel, O. Piot, T. Rindzevicius, E. A. Rusu, M. S. Schmidt, V. Sergo, G. D. Sockalingum, V. Untereiner, R. Vanna, E. Wiercigroch and A. Bonifacio, Anal. Chem., 2020, 92, 4053–4064 CrossRef CAS PubMed.
Footnote |
| † Present address: Key Laboratory for Advanced Materials and Feringa Nobel Prize Scientist Joint Research Center, Institute of Fine Chemicals, School of Chemistry and Molecular Engineering, East China University of Science & Technology, 130 Meilong Road, Shanghai 200237, China. |
| This journal is © The Royal Society of Chemistry 2021 |




