Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
SERS multiplexing of methylxanthine drug isomers via host–guest size matching and machine learning†
Weng-I Katherine
Chio
 abc,
Jia
Liu
ab,
Tabitha
Jones
abc,
Jia
Liu
ab,
Tabitha
Jones
 ab,
Jayakumar
Perumal
c,
U. S.
Dinish
ab,
Jayakumar
Perumal
c,
U. S.
Dinish
 c,
Ivan P.
Parkin
c,
Ivan P.
Parkin
 b,
Malini
Olivo
c and
Tung-Chun
Lee
b,
Malini
Olivo
c and
Tung-Chun
Lee
 *ab
*ab
aInstitute for Materials Discovery, University College London (UCL), London WC1H 0AJ, UK. E-mail: tungchunlee@ucl.ac.uk
bDepartment of Chemistry, University College London (UCL), London WC1H 0AJ, UK
cTranslational Biophotonics Laboratory, Institute of Bioengineering and Bioimaging (IBB), Agency for Science Technology and Research (A*STAR), Singapore 138667, Singapore
First published on 13th August 2021
Abstract
Multiplexed detection and quantification of structurally similar drug molecules, methylxanthine MeX, incl. theobromine TBR, theophylline TPH and caffeine CAF, have been demonstrated via solution-based surface-enhanced Raman spectroscopy (SERS), achieving highly reproducible SERS signals with detection limits down to ∼50 nM for TBR and TPH, and ∼1 μM for CAF. Our SERS substrates are formed by aqueous self-assembly of gold nanoparticles (Au NPs) and supramolecular host molecules, cucurbit[n]urils (CBn, n = 7, 8). We demonstrate that the binding constants can be significantly increased using a host–guest size matching approach, which enables effective enrichment of analyte molecules in close proximity to the plasmonic hotspots. The dynamic range and the robustness of the sensing scheme can be extended using machine learning algorithms, which shows promise for potential applications in therapeutic drug monitoring, food processing, forensics and veterinary science.
Introduction
Theobromine (TBR, 3,7-dimethylxanthine), theophylline (TPH, 1,3-dimethylxanthine) and caffeine (CAF, 1,3,7-trimethylxanthine), which are structurally similar family members of purine alkaloids, are naturally present in foods and beverages such as chocolate, tea and coffee. Interestingly, TBR and TPH are two of the three major metabolites of CAF which coexist in blood plasma and urine.1,2 Methylxanthines (MeX) act as central nervous system stimulants for sustaining alertness by blocking adenosine receptors and inhibiting phosphodiesterases,3,4 while showing antitumoral and anti-inflammatory properties.5 For instance, TBR and TPH are active ingredients of bronchodilator drugs taken to widen the airways in the lungs for asthma and other respiratory tract problems,6 whereas CAF is widely used in the formulations of prescription and over-the-counter medications. Although MeX are generally safe for human consumption except in the cases of severe overdose,7 they are potentially toxic to small animals such as cats and dogs.8Development of a high-performance MeX sensor with multiplexing ability is thus crucial for effective diagnosis of caffeine intoxication (a.k.a caffeinism)2 and other diseases, therapeutic drug monitoring, quality control of consumer products in the food and pharmaceutical industries, as well as for forensics and veterinary science. Conventional methods for MeX detection are based on high-performance liquid chromatography, near infrared spectroscopy, immunoassay, voltammetry and fluorescence,6,9–17 but not all of these methods allow multiplexed detection of MeX in real-time with minimal sample preparation and high performance.
Surface-enhanced Raman spectroscopy (SERS) is an analytical technique capable of quantitatively discriminating multiple structurally similar analyte molecules located in close proximity to the plasmonic nanostructures (e.g. Au NPs and Ag NPs) via their vibrational fingerprints, with additional advantages such as rapid response, high sensitivity, selectivity and reproducibility. Surprisingly, most of the previous SERS studies on MeX detection were based on Ag NPs18–27 for their relatively strong signals despite Au NPs having higher chemical stability, reproducibility and biocompatibility.28 This is probably due to the poor sensitivity and reproducibility of the SERS signals resulted from bare Au NPs or uncontrolled aggregation triggered by NaCl.1,29 The interparticle spacing between Au NPs can be precisely controlled via cucurbit[n]uril (CBn) mediated aggregation, thus localising the analyte molecules at the centre of or in close proximity to the plasmonic hotspots via formation of host–guest complexes.30–36 Previous work on MeX detection mainly focuses on CAF, while TBR and TPH, in particular their multiplexed SERS sensing, remain largely unexplored.
Herein, the host–guest complexations between CBn (n = 7, 8) and structurally similar drug molecules, MeX (TBR, TPH and CAF), were investigated in solution as to quantify their key binding parameters. While precedent examples of Au NP: CB SERS systems mostly focus on using CB7 as the supramolecular host, the use of the larger homologue CB8 as the host which can effectively encapsulate larger biomolecules, i.e. MeX in our case, is relatively rare. In particular, we showed a significant increase in binding constants by matching the molecular size of the host–guest pair (Fig. 1 and 2). Quantitative SERS detection of MeX was demonstrated with highly reproducible signals via formation of precise plasmonic hotspots within the Au NP: CB nanoaggregates. Thanks to the strong surface enrichment effect of CBs, the detection limit of CAF is down to ∼1 μM while those of its demethylated analogues, TBR and TPH, have reached ∼50 nM, which is at least an order of magnitude better than other similar SERS techniques in the literature (see Table S1 for details, ESI†)1,20–22,26,27 and covers the lower limit of MeX in human urine samples.2 Notably, multiplexed detection of TBR and TPH at sub-μM concentrations was also demonstrated with CB8 using our SERS sensing platform. Finally, we showed that the dynamic range and the robustness of our sensing scheme can be further enhanced by machine learning algorithms, partial least squared regression (PLSR) and artificial neural networks (ANNs), which effectively modelled the signal non-linearity at higher analyte concentrations.
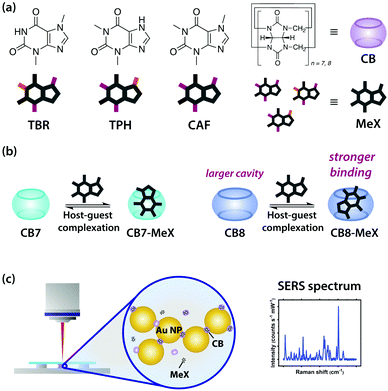 | ||
Fig. 1 Schematic illustrations of (a) MeX (TBR, TPH and CAF) and CB, (b) the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB7-MeX and CB8-MeX host–guest complexes, (c) the SERS measurement of the Au NP-CB-MeX sample solution. 1 CB7-MeX and CB8-MeX host–guest complexes, (c) the SERS measurement of the Au NP-CB-MeX sample solution. | ||
Materials and methods
Materials
40 nm citrate-stabilised gold nanoparticles (Au NPs) were purchased from nanoComposix. Paraformaldehyde, HCl, theobromine (TBR), theophylline (TPH) and caffeine (CAF) were purchased from Sigma-Aldrich. Methanol and ethanol were purchased from VWR. Glycoluril was purchased from Acros Organics. Cucurbit[n]urils (CBn, n = 7–8) were synthesised and isolated according to literature.37 All chemicals were used as received without further purification. 18.2 MΩ Milli-Q water was used in all experiments.Nuclear magnetic resonance (NMR) spectroscopy
1 mM of CB7-TBR, CB7-TPH and CB7-CAF solutions were prepared in D2O with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. Similarly, 1 mM of CB8-TBR, CB8-TPH and CB8-CAF solutions were prepared in 10 mM of DCl with 1
1 molar ratio. Similarly, 1 mM of CB8-TBR, CB8-TPH and CB8-CAF solutions were prepared in 10 mM of DCl with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. 1H NMR spectra were measured using a Bruker Avance III 400 spectrometer. Chemical shifts (in ppm) were referenced to D2O with δ = 4.80 ppm for 1H NMR.
1 molar ratio. 1H NMR spectra were measured using a Bruker Avance III 400 spectrometer. Chemical shifts (in ppm) were referenced to D2O with δ = 4.80 ppm for 1H NMR.
UV-Visible spectroscopy
UV-Vis measurements were performed on Agilent Cary 500 UV-Vis-NIR spectrophotometer in a cuvette with 10 mm optical path length. For CB7-MeX binding studies, a small volume of 2 mM CB7 solution was added to 4 μM MeX solution sequentially up to 5 or more equivalents. For CB8-MeX binding studies, a small volume of 1 mM CB8 solution (pre-dissolved in 10 mM HCl) was added to 4 μM MeX solution (pre-dissolved in 10 mM HCl) up to 2 or more equivalents. The concentrations of CB7 and CB8 were increased gradually while those of MeX were kept approximately constant. Prior to fitting, proportionated absorbance of CB7 and CB8 were subtracted accordingly to eliminate the effect that might be caused by their UV-Vis absorption around 190 nm. Additionally, spectra for samples containing CB8 were aligned by setting the absorbance at 500 nm equal to zero to minimise the effect of turbidity at high equivalents of CB8.Simulations
Density functional theory (DFT) calculations were performed using Gaussian and Spartan 18 Parallel Suite. Force-field calculations were performed using Chem3D. Geometry optimisation was first performed using MMFF94, followed by full optimisation at the required level of theory, in our case wB97XD/6-31G* followed by CPCM/wB97X-D/6-31G*. Restricted (closed-shell) models were used in all quantum mechanical calculations. The binding energies of the [CB-MeX-H]+ inclusion complexes were calculated from the energy difference between the complex and the total energies of CB and [MeX-H]+ which were optimised and calculated at the same level of theory.Raman and surface-enhanced Raman spectroscopy (SERS)
Raman and SERS spectra were acquired using a Reinshaw Raman InVia Microscope with a 633 nm He–Ne laser (9.3 mW). The laser was focused onto the sample via a 50 × objective lens (N.A. = 0.75). The grating used was 1800 lines mm−1 which gave a spectral resolution of 1 cm−1. All spectra were calibrated with respect to Si and acquired at room temperature. Stock solutions were prepared by mixing CB and MeX. For CB7 studies, 1 mL of the TBR stock solution with varying concentration of 0–0.2 mM was added to 1 mL of a 0.2 mM CB7 solution in a 2 mL Eppendorf tube. 20 μL of the stock solution was then added to a 180 μL Au NP solution in a 0.5 mL tube to give a final TBR concentration of 0–10 μM. For CB8 studies, 1 mL of the TBR stock solution with varying concentration of 0–0.1 mM was added to 1 mL of a 0.1 mM CB8 solution (prepared in 10 mM HCl) in a 2 mL Eppendorf tube. 20 μL of the stock solution was then added to a 180 μL Au NP solution in a 0.5 mL tube to give a final TBR concentration of 0–5 μM. Similar procedures were used to prepare sample solutions of TPH and CAF. The sample solution was vortexed for 30 s before dropping 15 μL onto a custom-made sample holder for SERS measurements. Three accumulations of 30 s scan were acquired on each measurement and five measurements were taken across different regions of interest per sample. The spectra were averaged and baseline corrected using an asymmetric least squares plugin in Origin.Results and discussion
Supramolecular chemistry of CB and MeX
The conjugated acids of theobromine (TBR) and theophylline (TPH), [TBR-H]+ and [TPH-H]+, have pKa of 10.0 and 8.8 respectively,4 implying that they exist in their protonated forms in solution under our experimental condition. Caffeine (CAF) also exists in its protonated form in our studies as its conjugated acid has a pKa of 10.4.4 The binding of CB7 to CAF was first suggested by Issacs and co-workers in 2009 but without reporting the binding constant,38 meanwhile the host–guest complexations of CB with TBR and TPH were studied for the first time.When CB7 and TBR were mixed in D2O with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, characteristic upfield shifts of the TBR proton signals (Hb and Hc) were observed in the 1H NMR spectra, verifying the formation of host–guest complexes between CB7 and TBR (Fig. S1b, ESI†). In particular, the NMR signal of Hb was significantly shifted upfield and broadened after host–guest complexation (Δδb = −0.099 ppm), indicating that Hb is located deep inside the CB7 cavity and that the binding kinetics fall into the intermediate exchange regime on the NMR time scale at 298 K, respectively. This observation also implies that the Ha signal may be too broad to be observed in the NMR spectra. In a separate measurement, TBR was mixed with the larger CB homologue, CB8, in 10 mM DCl with 1
1 stoichiometry, characteristic upfield shifts of the TBR proton signals (Hb and Hc) were observed in the 1H NMR spectra, verifying the formation of host–guest complexes between CB7 and TBR (Fig. S1b, ESI†). In particular, the NMR signal of Hb was significantly shifted upfield and broadened after host–guest complexation (Δδb = −0.099 ppm), indicating that Hb is located deep inside the CB7 cavity and that the binding kinetics fall into the intermediate exchange regime on the NMR time scale at 298 K, respectively. This observation also implies that the Ha signal may be too broad to be observed in the NMR spectra. In a separate measurement, TBR was mixed with the larger CB homologue, CB8, in 10 mM DCl with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. Characteristic upfield shifts of the TBR proton signals (Ha, Hb and Hc) were observed in the NMR spectra, verifying the formation of 1
1 stoichiometry. Characteristic upfield shifts of the TBR proton signals (Ha, Hb and Hc) were observed in the NMR spectra, verifying the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB8-TBR host–guest complexes (Fig. S2b, ESI†). Δδ for Ha (Δδa = −0.014 ppm) is greater than that for Hc/Hb (Δδc = −0.010 ppm and Δδb = −0.011 ppm) which indicates Ha is deeper inside the CB8 cavity, whereas Hc/Hb are closer to the carbonyl portals. Compared to the CB7 case, the smaller upfield shift can be attributed to the larger cavity of CB8 which exhibits a weaker shielding effect. Meanwhile all proton signals appear to be in the fast exchange regime, which is consistent to the wider portal and thus weaker constrictive binding of CB8.
1 CB8-TBR host–guest complexes (Fig. S2b, ESI†). Δδ for Ha (Δδa = −0.014 ppm) is greater than that for Hc/Hb (Δδc = −0.010 ppm and Δδb = −0.011 ppm) which indicates Ha is deeper inside the CB8 cavity, whereas Hc/Hb are closer to the carbonyl portals. Compared to the CB7 case, the smaller upfield shift can be attributed to the larger cavity of CB8 which exhibits a weaker shielding effect. Meanwhile all proton signals appear to be in the fast exchange regime, which is consistent to the wider portal and thus weaker constrictive binding of CB8.
Although TPH and TBR are isomers, strong broadening of proton signals after host–guest complexation between CB and TPH was not observed in the 1H NMR spectra (Fig. S3b, ESI†). When CB7 and TPH were mixed in D2O with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry, characteristic shifts of the TPH proton signals (Ha, Hb and Hc) were observed, verifying the host–guest complexation between CB7 and TPH. In particular, Hc/Hb (Δδc = −0.003 ppm and Δδb = −0.002 ppm) were upfield shifted whereas Ha was downfield shifted (Δδa = 0.021 ppm) after host–guest complexation, implying Hc/Hb should be within the CB7 cavity while Ha should be around the portal region. Notably, the host–guest complexation of CB8-TPH is very similar to that of CB7-TPH (Fig. S4b, ESI†).
1 stoichiometry, characteristic shifts of the TPH proton signals (Ha, Hb and Hc) were observed, verifying the host–guest complexation between CB7 and TPH. In particular, Hc/Hb (Δδc = −0.003 ppm and Δδb = −0.002 ppm) were upfield shifted whereas Ha was downfield shifted (Δδa = 0.021 ppm) after host–guest complexation, implying Hc/Hb should be within the CB7 cavity while Ha should be around the portal region. Notably, the host–guest complexation of CB8-TPH is very similar to that of CB7-TPH (Fig. S4b, ESI†).
On the other hand, the relatively bulky MeX, CAF, shows smaller upfield shifts of the proton signals when mixing with CB7 in D2O with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S5b, ESI†), suggesting weaker binding of CAF to CB7. Interestingly, splitting of the NMR signal of Hd was observed after host–guest complexation, resulting in two different values for the change in chemical shift (Δδ) for Hd (Δδd = −0.002 ppm and −0.010 ppm). Δδ for Hd is greater than that for Hc/Hb/Ha (Δδc = −0.002 ppm, Δδb = −0.002 ppm and Δδa = −0.003 ppm) which indicates Hd is deep inside the CB7 cavity, whereas Hc/Hb/Ha are much closer to the carbonyl portal. Similar results were observed in the 1H NMR spectra when CAF was mixed with CB8 with 1
1 stoichiometry (Fig. S5b, ESI†), suggesting weaker binding of CAF to CB7. Interestingly, splitting of the NMR signal of Hd was observed after host–guest complexation, resulting in two different values for the change in chemical shift (Δδ) for Hd (Δδd = −0.002 ppm and −0.010 ppm). Δδ for Hd is greater than that for Hc/Hb/Ha (Δδc = −0.002 ppm, Δδb = −0.002 ppm and Δδa = −0.003 ppm) which indicates Hd is deep inside the CB7 cavity, whereas Hc/Hb/Ha are much closer to the carbonyl portal. Similar results were observed in the 1H NMR spectra when CAF was mixed with CB8 with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry (Fig. S6b, ESI†).
1 stoichiometry (Fig. S6b, ESI†).
Binding constants of the CB7-TBR and CB8-TBR complexes were obtained from UV-Vis titrations by fitting 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
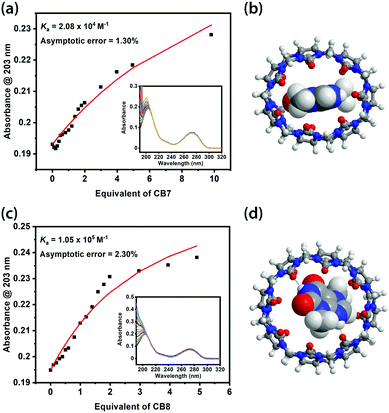
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding models (Fig. 2a and c). Notably, the binding constant of CB8-TBR (1.05 × 105 M−1) was found to be five times larger than that of CB7-TBR (2.08 × 104 M−1). Similar results were observed for TPH while the binding between CAF and CB8 is on par with that of CB7 (Fig. S3–S8, ESI†). We note that it remains challenging to minimise the error in the fitting, despite multiple attempts in UV-Vis titration, due to the interfering UV absorption of CBs at ∼190 nm and, in the case of CB8, the effect of turbidity at high CB equivalents. Meanwhile the change in absorbance for the peak of MeX at ∼275 nm is too small to be precisely measured. Nevertheless, the fitted binding constants are considered sufficient for qualitative comparison in our context. Accurate binding constant data might be obtained using isothermal titration calorimetry (ITC). However, ITC measurements could not be performed due to unavailability of the instrument but could be a potential approach for further study. In particular, the generally larger binding constants of CB8-MeX complexes can be rationalised by host–guest size matching effects as described in the Rebek's 55% rule, where a packing coefficient (PC) of 55% gives the best binding affinity for host–guest complexes in solution, with a lower or higher PC resulting in a lower binding affinity.39 For instance, the packing coefficients of CB7-MeX are 0.70–0.78 while those of CB8-MeX are 0.46–0.51 (see Table S2 for details, ESI†). Indeed upon binding with TBR, the molecular shape of CB7 becomes highly distorted and thus conformationally destabilised, whereas CB8 retains a round conformation resembling that of the ground state of its empty form (Fig. 2b and d).
1 binding models (Fig. 2a and c). Notably, the binding constant of CB8-TBR (1.05 × 105 M−1) was found to be five times larger than that of CB7-TBR (2.08 × 104 M−1). Similar results were observed for TPH while the binding between CAF and CB8 is on par with that of CB7 (Fig. S3–S8, ESI†). We note that it remains challenging to minimise the error in the fitting, despite multiple attempts in UV-Vis titration, due to the interfering UV absorption of CBs at ∼190 nm and, in the case of CB8, the effect of turbidity at high CB equivalents. Meanwhile the change in absorbance for the peak of MeX at ∼275 nm is too small to be precisely measured. Nevertheless, the fitted binding constants are considered sufficient for qualitative comparison in our context. Accurate binding constant data might be obtained using isothermal titration calorimetry (ITC). However, ITC measurements could not be performed due to unavailability of the instrument but could be a potential approach for further study. In particular, the generally larger binding constants of CB8-MeX complexes can be rationalised by host–guest size matching effects as described in the Rebek's 55% rule, where a packing coefficient (PC) of 55% gives the best binding affinity for host–guest complexes in solution, with a lower or higher PC resulting in a lower binding affinity.39 For instance, the packing coefficients of CB7-MeX are 0.70–0.78 while those of CB8-MeX are 0.46–0.51 (see Table S2 for details, ESI†). Indeed upon binding with TBR, the molecular shape of CB7 becomes highly distorted and thus conformationally destabilised, whereas CB8 retains a round conformation resembling that of the ground state of its empty form (Fig. 2b and d).
Energy-minimised molecular models of the [CB7-MeX-H]+ and [CB8-MeX-H]+ complexes calculated based on density functional theory (DFT) at CPCM/wB97X-D/6-31G* level of theory support the host–guest binding geometries derived from NMR (Fig. S1a–S6a, ESI†). The dispersion-corrected DFT functional wB97X-D was chosen to accurately estimate the van der Waals interactions, which were expected to contribute significantly to the stability of these complexes. Implicit water model CPCM was selected to effectively account for the solvation energy of the protonated guests and complexes with a formal charge of +1. Moreover, the binding energies of the [CB-MeX-H]+ complexes (see Table S3 and S4 for details, ESI†) are consistent to similar host–guest complexes reported in literature.34,40 TBR, TPH and CAF are fully or almost fully encapsulated within the cavity of CB7 and CB8, leaving both of the CB portals available for binding to the surface of Au NPs and thus localising them at the centre of or in close proximity to the plasmonic hotspots, which is critical to the subsequent SERS studies.
Raman spectroscopy of MeX and SERS of CB
The Raman spectrum of TBR powder is characterised by two major peaks at 622 cm−1 and 1334 cm−1, which are attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C deformation and imidazole ring stretching vibration respectively (Fig. S9a, ESI†).41,42 The Raman spectrum of TPH powder is characterised by a main peak at 557 cm−1, which corresponds to pyrimidine ring deformation + C–N–C deformation + CH3 rocking (Fig. S9b, ESI†). The other peaks at 668 cm−1 and 1316 cm−1 are assigned to O
C–C deformation and imidazole ring stretching vibration respectively (Fig. S9a, ESI†).41,42 The Raman spectrum of TPH powder is characterised by a main peak at 557 cm−1, which corresponds to pyrimidine ring deformation + C–N–C deformation + CH3 rocking (Fig. S9b, ESI†). The other peaks at 668 cm−1 and 1316 cm−1 are assigned to O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N deformation + pyrimidine imidazole ring deformation and imidazole ring stretching vibration, respectively.41 Similarly, the Raman spectrum of CAF powder is characterised by two major peaks at 558 cm−1 and 1330 cm−1, corresponding to pyrimidine ring deformation + C–N–C deformation + CH3 rocking and imidazole ring stretching vibration respectively (Fig. S9c, ESI†).26,41
C–N deformation + pyrimidine imidazole ring deformation and imidazole ring stretching vibration, respectively.41 Similarly, the Raman spectrum of CAF powder is characterised by two major peaks at 558 cm−1 and 1330 cm−1, corresponding to pyrimidine ring deformation + C–N–C deformation + CH3 rocking and imidazole ring stretching vibration respectively (Fig. S9c, ESI†).26,41
The two characteristic Raman peaks of CB7 at 444 cm−1 and 833 cm−1, which correspond to ring scissor and ring deformation modes, were clearly observed in the SERS spectra of CB7 at 447 cm−1 and 833 cm−1 (Fig. S9d, ESI†). The slight shifts in peak position and peak broadening could be due to the solution effect and the molecular interaction between Au NPs and CB7. Notably, the ring scissor mode of CB8 is at a slightly lower wavenumber of 444 cm−1 whereas its ring deformation mode is at a slightly higher wavenumber of 834 cm−1 than that of CB7, which is consistent with the previous report.43
SERS sensing of CB-MeX host–guest complexes
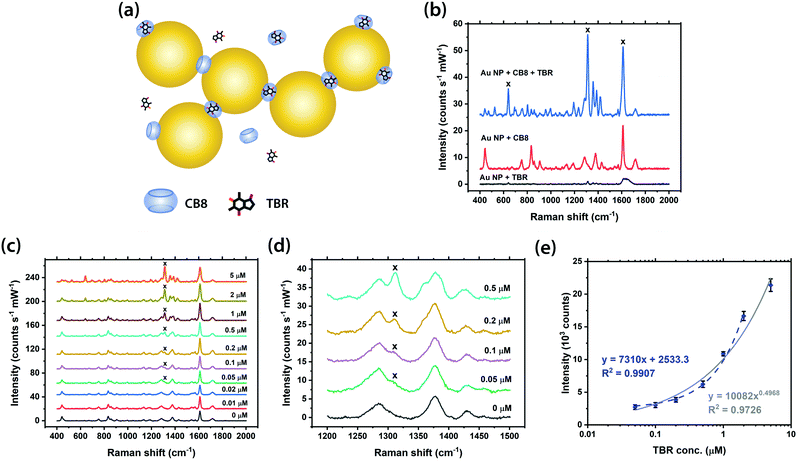
The SERS detection of MeX was first performed with CB7 and TBR by adding a pre-mixed CB7-TBR solution into a 40 nm citrate-capped Au NP solution. The concentration of CB7 in the final mixture was kept constant at 10 μM for all cases in order to ensure the formation of reproducible nanoaggregates, i.e. SERS substrates, since the aggregation kinetics is delicately determined by the ratio of Au NP: CB.34 As CB7 defines precise nanojunctions between Au NPs while TBR is fully or almost fully encapsulated within the CB7 cavity, TBR is localised at the plasmonic hotspots within the Au NP: CB7 nanoaggregates (Fig. S10a, ESI†). It should be noted that no aggregation of Au NPs can be triggered, and no SERS signals can be observed in the absence of CB7 (Fig. S10b, ESI†), thus illustrating the importance of CB on TBR sensing. In the Au NP: CB7 system, the characteristic Raman peak of TBR at 1312 cm−1, which is attributed to imidazole ring stretching vibration, can be clearly observed in the SERS spectra down to 0.5 μM (Fig. S10c and d, ESI†). A strong linear correlation (R2 > 0.99) between the SERS intensity and concentration of TBR (up to 2 μM) was found, while the full range was fitted well by power law (R2 ∼0.94) (Fig. S10e, ESI†).Notably, the sensitivity of our SERS system for TBR detection can be significantly enhanced by using the larger CB homologue, i.e. CB8. The SERS detection of TBR was performed by adding a pre-mixed CB8-TBR solution into the Au NP solution, at a constant CB8 concentration of 5 μM, to form precise plasmonic nanojunctions as in the case of CB7 (Fig. 3a). It is noted that the pre-mixed CB8-TBR solution contained 10 mM HCl to facilitate the dissolution of CB8. As expected, no aggregation or SERS signals can be observed in the absence of CB8 (Fig. 3b). The detection limit of TBR was found to be 10-fold lower (i.e. down to ∼50 nM) in the Au NP: CB8 system (Fig. 3c and d), which is the lowest among all similar SERS platforms in the literature,1,20 with a strong linear correlation (R2 ∼0.99) between the SERS intensity and concentration of TBR up to 2 μM (Fig. 3e). The full range was also fitted by power law with a good correlation (R2 ∼0.97).
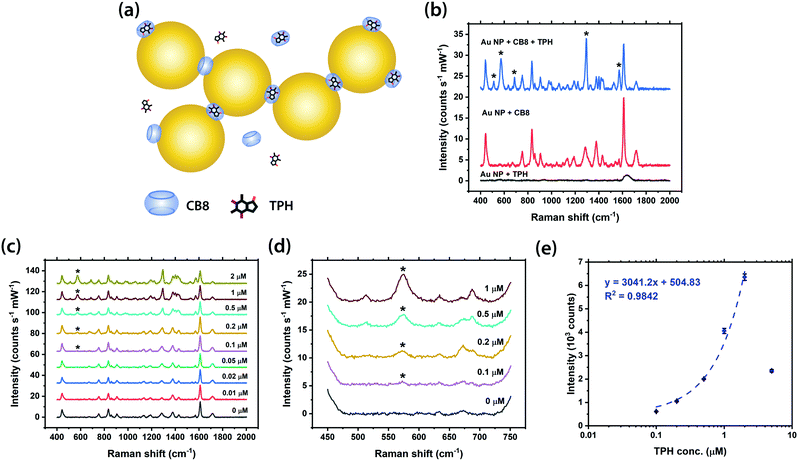
The SERS detection of the isomer of TBR, i.e. TPH, was subsequently investigated in our studies, at a constant concentration of CB7 or CB8. Similarly, TPH is fully or almost fully encapsulated within the cavity of CB7 and CB8 as in the case of TBR while no aggregation of Au NPs can be triggered in the absence of CB (Fig. 4a, b and Fig. S11a, b, ESI†). The characteristic Raman peak of TPH at 573 cm−1, which corresponds to pyrimidine ring deformation + C–N–C deformation + CH3 rocking, can be clearly observed in the SERS spectra down to 50 nM in the presence of CB7 (Fig. S11c and d, ESI†), with a strong linear correlation (R2 ∼0.99) between the SERS intensity and concentration of TPH up to 2 μM and a good correlation (R2 ∼0.98) for the full range fitted by power law (Fig. S11e, ESI†). Interestingly, the detection limit of TPH in the Au NP: CB7 SERS system is 10-fold better than that of its isomer, TBR, despite being less Raman-active (Fig. S9a and b, ESI†). This could be due to the difference in the binding geometries of the two complexes. Moreover, the [CB8-TPH-H]+ complex has a similar binding geometry to that of the [CB7-TPH-H]+ complex, with a detection limit of ∼0.1 μM (Fig. 4c and d) and a very strong linear correlation (R2 ∼0.98) between the SERS intensity and concentration of TPH from 0.1 to 2 μM (Fig. 4e). It should be noted that the 5 μM data point deviates from the trendline, probably due to the LSPR peak shifting away from the 633 nm excitation or the difference in pH.
The SERS detection of CAF was also investigated in our studies as control experiments (Fig. S12 and S13, ESI†). Similar to the case of other MeX, no aggregation of Au NPs can be triggered in the absence of CB (Fig. S12b and S13b, ESI†). The SERS signals of CAF are relatively weaker than those of its demethylated analogues, TBR and TPH, probably due to the nature of its bulkier size. The characteristic Raman peak of CAF at 1326 cm−1, which is attributed to imidazole ring stretching vibration, can be observed down to 5 μM in the Au NP: CB7 system (Fig. S12c and d, ESI†) and 1.25 μM in the Au NP: CB8 nanoaggregates respectively (Fig. S13c and d, ESI†). A good correlation (R2 ∼0.98) between the SERS intensity and concentration of CAF in the Au NP: CB8 system was found (Fig. S13e, ESI†).
Multiplexed SERS sensing of drug isomers
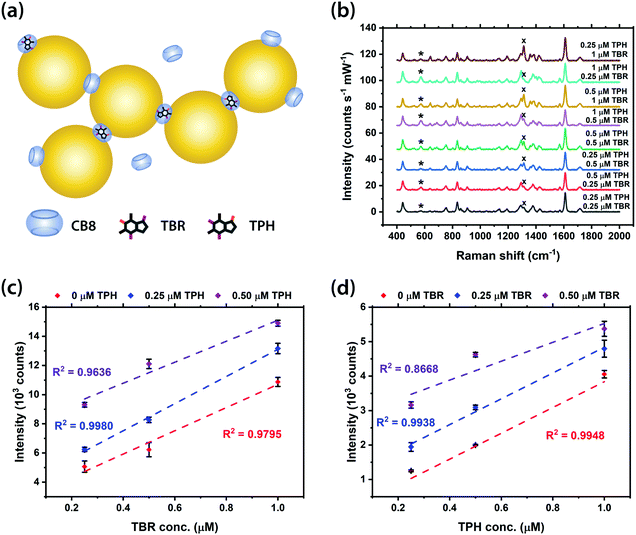
Furthermore, the potential multiplexed detection of structurally similar molecules using our SERS system was demonstrated with the drug isomers, TBR and TPH, at various concentrations within the Au NP: CB8 nanoaggregates (Fig. 5a). The main peaks of TPH and TBR, at 573 cm−1 and 1312 cm−1 respectively, can be clearly observed in all SERS spectra (Fig. 5b). Good linear correlations (R2 ∼0.87–>0.99) between the SERS intensity and concentration of TBR and TPH were found for all mixtures with a combined concentration of MeX between 0.25 and 1 μM (Fig. 5c). Meanwhile, the small error bars (∼1–18% error) in Fig. 3–5 and Fig. S10, S11 (ESI†) indicate the high reproducibility of the SERS signals in our sensing scheme. Notably, the multiplexed detection of MeX at sub-μM levels can be performed using Au NP-based SERS system, in contrast to precedent examples using Ag NPs,1,20 with improved detection limit. Therefore, it is possible to distinguish isomers using our SERS sensors by identifying their characteristic Raman peaks, as opposed to other techniques which do not allow molecular fingerprinting.Multiplexed quantification using machine learning techniques
Machine learning techniques were employed in further analysing the Au NP: CB8 dataset, with an aim of tackling the nonlinear correlations at high analyte concentrations and therefore extending the detection range. It should be noted that the employed full dataset consists of all SERS spectra of TBR and TPH measured using the Au NP: CB8 system, including those of single analyte samples (TBR as shown in Fig. 3 and TPH as shown in Fig. 4) and binary mixtures (as shown in Fig. 5). Two different algorithms, partial least squared regression (PLSR) and artificial neural networks (ANNs), were used to predict the metabolite concentrations from unseen spectra (Fig. S17A and B, ESI†). The models were trained and tested via the bootstrapping random resampling procedure (Fig. S17D, ESI†). A total of 1000 bootstrapping iterations were performed and the mean R2 and root mean square error of prediction (RMSEP) values were calculated to assess the models.At higher analyte concentrations (above 1 μM), the magnitude of the SERS peaks increases less significantly, and in some cases even decreases, due to disruption in the Au NP aggregation. To form plasmonic nanojunctions, both portals of a CB molecule need to bind to Au NPs. When host–guest complexes are formed between CB8 and TBR or TPH, one of the portals of the CB8 molecule could become partially hindered, weakening its ability to aggregate the Au NPs. At high analyte concentrations, there may not be sufficient free CB8 to mediate the fast aggregation kinetics of the Au NPs, so the SERS signals could become significantly weaker. This means that the magnitudes of the characteristic peaks are no longer linearly proportional to the analyte concentrations.
For the Au NP: CB8 dataset, the relationships between the intensity of the characteristic peaks and the analyte concentrations are linear up to 1 μM and start to become nonlinear at concentrations above 1 μM (Fig. S14, ESI†). To assess the ability of the machine learning algorithms to model linear and nonlinear relationships, the dataset was split into two versions: one which contained solutions with individual analyte concentrations up to 1 μM and another which included all of the solutions analysed (concentrations up to 5 μM). PLSR and ANN models were trained and tested using both versions of the dataset.
After optimisation, the number of components used in the PLSR models were 6 and 8 for the 0–1 μM and 0–5 μM datasets respectively and the final ANN architectures were 1167-16-128-2 and 1167-32-32-128-16-2 (Fig. S17A and B, ESI†). More components/layers are required to model the nonlinear dataset as the relationship between the spectra and the analyte concentrations is more complex. The results for the different datasets and models are shown in Table 1 and Fig. S15–S18 (ESI†). The partial least squares regression model performed well (R2 = 0.939) for the linear dataset (0–1 μM). However, it struggled to correctly predict the analyte concentrations for the nonlinear dataset (0–5 μM). This is to be expected as the PLSR algorithm is based on linear regression. In contrast, the ANNs achieved better results with R2 values of 0.953 and 0.948 for both the 0–1 μM and 0–5 μM datasets respectively. The X-loadings for each component of a PLSR model built with 6 components are included in Fig. S17J (ESI†). Our results are comparable to previous work using similar techniques for SERS quantification of neurotransmitters.44
| Detection Range | Theobromine | Theophylline | |||
|---|---|---|---|---|---|
| R 2 | RMSEP (M) | R 2 | RMSEP (M) | ||
| 0–1 μM | PLSR | 0.952 | 8.22 × 10−8 | 0.926 | 1.02 × 10−7 |
| ANN | 0.968 | 6.74 × 10−8 | 0.939 | 9.24 × 10−8 | |
| 0–5 μM | PLSR | 0.840 | 3.96 × 10−7 | 0.875 | 3.45 × 10−7 |
| ANN | 0.962 | 1.93 × 10−7 | 0.934 | 2.52 × 10−7 | |
To further investigate how the algorithms interact with the spectra, PLSR and ANN models were built using spectra in which the characteristic TBR and TPH peaks had been removed (Fig. S20, ESI†). The performance of these models was only slightly worse (less than a 0.035 reduction in R2) than the models trained on the original spectra which contained the characteristic peaks (Fig. S21 and S22, ESI†). This finding highlights the ability of the machine learning algorithms to extract subtle information from datasets and justifies using the entire spectra to make robust predictions.
For the ANN trained on the larger dataset, the root mean square errors of prediction (RMSEPs), which can also be interpreted as the detection limits,1 were 1.93 × 10−7 M and 2.52 × 10−7 M for TBR and TPH, which are consistent with those determined directly by SERS titration experiments (Fig. 3d and 4d). Therefore, using the ANN model has extended the quantification range for multiplexed TBR and TPH from 1 μM to 5 μM without compromising the prediction accuracy. It has also enabled the simultaneous quantification of both analytes. These results are significant because they demonstrate that the Au NP: CB8 system can achieve comparable detection limits to analytical techniques such as high-performance liquid chromatography (0.07 mg L−1 (0.39 μM) for TBR and 0.08 mg L−1 (0.44 μM) for TPH using HPLC-UV)45,46 whilst also being portable, quicker to run and more cost-effective.47,48
Conclusions
A novel multiplexed SERS sensing platform that utilises various host–guest complexes of CBn (n = 7, 8) to detect and quantify structurally similar MeX drug molecules, TBR, TPH and CAF, has been developed. The key binding parameters of the six different 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB-MeX complexes have been quantified using NMR, UV-Vis and supported by DFT molecular models. The potential SERS detection of TBR, TPH and CAF has been demonstrated by using CB7 or CB8 to mediate the formation of precise plasmonic hotspots within the Au NP: CB nanoaggregates. We also showed that the binding constants can be significantly enhanced using a host–guest size matching approach. The detection limits of TBR and TPH using our system have reached ∼50 nM with highly reproducible SERS signals, as opposed to the relatively weaker signals of CAF due to its larger size. The capability of our SERS system to simultaneously quantify multiple structurally similar molecules has also been successfully demonstrated with the drug isomers, TBR and TPH, in proof-of-concept experiments, where the dynamic range of the sensor can be extended using machine learning algorithms. Hence, our SERS sensor holds great potential for a wide range of applications including therapeutic drug monitoring, food processing, forensics and veterinary science.
1 CB-MeX complexes have been quantified using NMR, UV-Vis and supported by DFT molecular models. The potential SERS detection of TBR, TPH and CAF has been demonstrated by using CB7 or CB8 to mediate the formation of precise plasmonic hotspots within the Au NP: CB nanoaggregates. We also showed that the binding constants can be significantly enhanced using a host–guest size matching approach. The detection limits of TBR and TPH using our system have reached ∼50 nM with highly reproducible SERS signals, as opposed to the relatively weaker signals of CAF due to its larger size. The capability of our SERS system to simultaneously quantify multiple structurally similar molecules has also been successfully demonstrated with the drug isomers, TBR and TPH, in proof-of-concept experiments, where the dynamic range of the sensor can be extended using machine learning algorithms. Hence, our SERS sensor holds great potential for a wide range of applications including therapeutic drug monitoring, food processing, forensics and veterinary science.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
TCL is grateful to the support from the Research Project Grant (RPG-2016-393) funded by the Leverhulme Trust, the Royal Society Research Grant 2016 R1 (RG150551) and the UCL BEAMS Future Leader Award funded through the Institutional Sponsorship award by the EPSRC (EP/P511262/1). WIKC, IPP, MO and TCL are grateful to the Studentship funded by the A*STAR-UCL Research Attachment Programme through the EPSRC M3S CDT (EP/L015862/1). TJ and TCL acknowledge Camtech Innovations for contributing to TJ's studentship. WIKC, JK, DUS and MO thank IBB TBL for use of facilities and resources. JP, DUS and MO would like to acknowledge the support from A*STAR BMRC Central Research Fund (UIBR). We acknowledge the use of the UCL Thomas high performance computing facility (Thomas@UCL), and the associated support services, for the completion of this work.References
- O. Alharbi, Y. Xu and R. Goodacre, Anal. Bioanal. Chem., 2015, 407, 8253 CrossRef CAS.
- M. E. Rybak, M. R. Sternberg, C.-I. Pao, N. Ahluwalia and C. M. Pfeiffer, J. Nutr., 2015, 145, 766 CrossRef CAS PubMed.
- E. Martinez-Pinilla, A. Onatibia-Astibia and R. Franco, Front. Pharmacol., 2015, 6, 30 Search PubMed.
- P. Sanphui and A. Nangia, J. Chem. Sci., 2014, 126, 1249 CrossRef CAS.
- N. Sugimoto, S. Miwa, Y. Hitomi, H. Nakamura, H. Tsuchiya and A. Yachie, Nutr. Cancer, 2014, 66, 419 CrossRef CAS.
- M. S. Bispo, M. C. C. Veloso, H. L. C. Pinheiro, R. F. S. De Oliveira, J. O. N. Reis and J. B. De Andrade, J. Chromatogr. Sci., 2002, 40, 45 CAS.
- P. Holmgren, L. Norden-Pettersson and J. Ahlner, Forensic Sci. Int., 2004, 139, 71 CrossRef CAS.
- C. Cortinovis and F. Caloni, Front. Vet. Sci., 2016, 3, 26 Search PubMed.
- C. W. Huck, W. Guggenbichler and G. K. Bonn, Anal. Chim. Acta, 2005, 538, 195 CrossRef CAS.
- M. R. Brunetto, L. Gutierrez, Y. Delgado, M. Gallignani, A. Zambrano, A. Gomez, G. Ramos and C. Romero, Food Chem., 2007, 100, 459 CrossRef CAS.
- J. J. Carvalho, M. G. Weller, U. Panne and R. J. Schneider, Anal. Bioanal. Chem., 2010, 396, 2617 CrossRef CAS PubMed.
- N. Chiem and D. J. Harrison, Anal. Chem., 1997, 69, 373 CrossRef CAS.
- V. K. Gupta, A. K. Jain and S. K. Shoora, Electrochim. Acta, 2013, 93, 248 CrossRef CAS.
- Y. Peng, W. Zhang, J. Chang, Y. Huang, L. Chen, H. Deng, Z. Huang and Y. Wen, Food Anal. Methods, 2017, 10, 3375 CrossRef.
- F. Bates and M. del Valle, Microchim. Acta, 2015, 182, 933 CrossRef CAS.
- W. Xu, T.-H. Kim, D. Zhai, J. C. Er, L. Zhang, A. A. Kale, B. K. Agrawalla, Y.-K. Cho and Y.-T. Chang, Sci. Rep., 2013, 3, 2255 CrossRef PubMed.
- A. K. Mahapatra, P. Sahoo, S. Goswami, S. Chantrapromma and H.-K. Fun, Tetrahedron Lett., 2009, 50, 89 CrossRef CAS.
- P. Liu, R. Liu, G. Guan, C. Jiang, S. Wang and Z. Zhang, Analyst, 2011, 136, 4152 RSC.
- H. Zheng, D. Ni, Z. Yu, P. Liang and H. Chen, Sens. Actuators, B, 2016, 231, 423 CrossRef CAS.
- M. Wu, H. Li, D. Lv and F. Lu, Anal. Bioanal. Chem., 2017, 409, 5569 CrossRef CAS PubMed.
- J. Kang, H. Gu, L. Zhong, Y. Hu and F. Liu, Spectrochim. Acta, Part A, 2011, 78, 757 CrossRef.
- I. Pavel, A. Szeghalm, D. Moigno, S. Cinta and W. Kiefer, Biopolymers, 2003, 72, 25 CrossRef CAS PubMed.
- R. Hu, R. Tang, J. Xu and F. Lu, Anal. Chim. Acta, 2018, 1034, 176 CrossRef CAS PubMed.
- Y. Kurokawa, Y. Imai and Y. Tamai, Analyst, 1997, 122, 941 RSC.
- W. W. Y. Lee, V. A. D. Silverson, C. P. McCoy, R. F. Donnelly and S. E. J. Bell, Anal. Chem., 2014, 86, 8106 CrossRef CAS PubMed.
- X. Chen, H. Gu, G. Shen, X. Dong and J. Kang, J. Mol. Struct., 2010, 975, 63 CrossRef CAS.
- R. Sato-Berru, R. Redon, A. Vazquez-Olmos and J. M. Saniger, J. Raman Spectrosc., 2009, 40, 376 CrossRef CAS.
- H.-H. Jeong, E. Choi, E. Ellis and T.-C. Lee, J. Mater. Chem. B, 2019, 7, 3480 RSC.
- M. Velicka, S. Adomaviciute, E. Zacharovas and V. Sablinskas, Proc. SPIE, 2020, 11257, 112570S Search PubMed.
- T.-C. Lee and O. A. Scherman, Chem. Commun., 2010, 46, 2438 RSC.
- T.-C. Lee and O. A. Scherman, Chem. – Eur. J., 2012, 18, 1628 CrossRef CAS PubMed.
- R. W. Taylor, T.-C. Lee, O. A. Scherman, R. Esteban, J. Aizpurua, F. M. Huang, J. J. Baumberg and S. Mahajan, ACS Nano, 2011, 5, 3878 CrossRef CAS PubMed.
- W.-I. K. Chio, W. J. Peveler, K. I. Assaf, S. Moorthy, W. M. Nau, I. P. Parkin, M. Olivo and T.-C. Lee, J. Phys. Chem. C, 2019, 123, 15769 CrossRef CAS PubMed.
- W.-I. K. Chio, S. Moorthy, J. Perumal, U. S. Dinish, I. P. Parkin, M. Olivo and T.-C. Lee, J. Mater. Chem. C, 2020, 8, 7051 RSC.
- W.-I. K. Chio, G. Davison, T. Jones, J. Liu, I. P. Parkin and T.-C. Lee, J. Visualized Exp., 2020, 164, e61682 Search PubMed.
- J. Wu and L. Issacs, Chem. – Eur. J., 2009, 15, 11675 CrossRef CAS PubMed.
- J. Lagona, P. Mukhopadhyay, S. Chakrabarti and L. Issacs, Angew. Chem., Int. Ed., 2005, 44, 4844 CrossRef CAS PubMed.
- W. A. Ameku, W. R. de Araujo, C. J. Rangel, R. A. Ando and T. R. L. C. Paixao, ACS Appl. Nano Mater., 2019, 2, 5460 CrossRef CAS.
- S. Mecozzi and J. Rebek, Chem. – Eur. J., 1998, 4, 1016 CrossRef CAS.
- T.-C. Lee, E. Kalenius, A. I. Lazar, K. I. Assaf, N. Kuhnert, C. H. Grun, J. Janis, O. A. Scherman and W. M. Nau, Nat. Chem., 2013, 5, 376 CrossRef CAS PubMed.
- H. G. M. Edwards, T. Munshi and M. Anstis, Spectrochim. Acta, Part A, 2005, 61, 1453 CrossRef CAS.
- S. Gunasekaran, G. Sankari and S. Ponnusamy, Spectrochim. Acta, Part A, 2005, 61, 117 CrossRef CAS.
- S. Mahajan, T.-C. Lee, F. Biedermann, J. T. Hugall, J. J. Baumberg and O. A. Scherman, Phys. Chem. Chem. Phys., 2010, 12, 10429 RSC.
- S. Kasera, L. O. Herrmann, J. D. Barrio, J. J. Baumberg and O. A. Scherman, Sci. Rep., 2014, 4, 6785 CrossRef CAS PubMed.
- B. Srdjenovic, V. Djordjevic-Milic, N. Grujic, R. Injac and Z. Lepojevic, J. Chromatogr. Sci., 2008, 46, 144 CAS.
- F. Buiarelli, F. Bernardini, G. Simonetti, P. Di Filippo, D. Pomata, C. Riccardi and R. Risoluti, J. AOAC Int., 2019, 102, 865 CrossRef CAS PubMed.
- M. W. Dong, Modern HPLC for Practicing Scientists, John Wiley & Sons, 2006 Search PubMed.
- J. A. Dietrich, A. E. McKee and J. D. Keasling, Annu. Rev. Biochem., 2010, 79, 563 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Limit of detection, NMR, computational details, UV-Vis, Raman, SERS and machine learning. See DOI: 10.1039/d1tc02004h |
| This journal is © The Royal Society of Chemistry 2021 |